Abstract
Hepatocellular carcinoma (HCC) is a common and deadly cancer worldwide. Many factors contribute to mortality and place an individual at high risk of developing HCC, including viral infection, alcohol intake, metabolic-associated disease, autoimmunity and genetic liver disorders. Although there are many therapeutics available, much about this disease remains to be understood. This is most evident when investigating the tumour microenvironment (TME). Both innate and adaptive immune cells have been associated with carcinogenesis within the TME of HCC patients. The ability to interrogate the TME more thoroughly with spatial technologies continues to improve, both at the experimental and analytical stages. This review provides insight into technologies available to investigate the TME, and how such technologies are beneficial for improving our understanding of HCC carcinogenesis.
Keywords: hepatocellular carcinoma, histopathology, immunohistochemistry, transcriptomics, tumour microenvironment
Background: The disease burden
In 2020, hepatocellular carcinoma (HCC) was the sixth most common cancer worldwide and the third most common cause of cancer death. 1 The highest incidence rates globally are seen in Asia and Africa with Mongolia demonstrating the highest incidence worldwide. 1 HCC is rapidly growing in prevalence, with a 350% increase between 1982 and 2014 in Australia. 2 Similar increases have been observed in India, the Americas and most of Europe, although decreased rates have been seen across many Asian countries and Italy. 3 Primary liver cancer incidence has been predicted to increase in 30 countries by 2030, including the Americas, most of Europe and Oceania, with Asian and some European countries predicted to decrease. 4 This is despite an increase in therapeutics available to treat HCC. 5 Over 75% of HCC cases develop on a background of chronic liver disease (mainly cirrhosis). 1 The causes of chronic liver disease include viral hepatitis, alcohol, metabolic-associated fatty liver disease (MAFLD), autoimmune and genetic liver disorders. 6 These risk factors vary across geographical regions. Hepatitis B virus is the primary aetiological factor in most parts of Asia and Africa, whereas the hepatitis C virus is the leading cause in Japan, Italy and Egypt. 1 Since the advent of successful antiviral therapies for hepatitis B and C virus infections, in the USA, MAFLD is now the most common background liver disease predisposing an individual to HCC. 7
Although survival varies according to disease stage and treatment, the 5-year survival for patients with HCC is less than 20%. 8 Although curative therapies can be delivered in early-stage disease (hepatic resection or liver transplant), this is only applicable to a small number of patients. 6 The inability of these strategies to decrease the prevalence of HCC cases indicates that a greater understanding of the disease is still required. Differences in the immune landscape have been observed in patients with hepatitis B-associated HCC, 9 MAFLD-associated HCC 10 and patients with microvascular invasion. 11 Employing high-dimensional techniques to interrogate the systemic and local cancer and immune landscape of HCC will provide greater comprehension of disease pathology specific to each aetiology. By phenotyping cells and determining their potential role, more targeted therapeutics may be developed capable of inhibiting or promoting selected immune cells.
Tumour microenvironment of HCC
The development of HCC is related to chronic hepatic inflammation, liver tissue injury, hepatocyte regeneration and the subsequent formation of fibrosis. This leads to the malignant transformation of hepatocytes and a tumour microenvironment (TME) that promotes HCC development and progression. 6
The TME in HCC, as in other cancers, is thought overall to be immunosuppressive, allowing the tumour to escape immune killing and facilitate its growth and spread. The TME inflammatory response may include anti-inflammatory cytokines such as interleukin (IL)-4, IL-5, IL-8, IL-10 and transforming growth factor (TGF)-β. Chemokines, such as CXCL12, CL20 and CCL22, are also thought to be mechanisms involved in tumour immune-evasion. 12
Many immune cells of both innate (including neutrophils,13–18 γδT cells,19,20 mucosal-associated invariant T (MAIT) cells, 21 monocytes/macrophages,22–26 natural killer (NK) cells27–30 and innate lymphoid cells (ILC)31,32) and adaptive (including T cells9,33–37 and B cells38–42) immune responses are associated with the prognostic outcome of HCC, as either detrimental or protective. Major effector functions within tumour-infiltrating lymphocytes (TILs) are thought to be blunted by the expression of immune-checkpoint molecules such as PD-1 and immunosuppressive PD-1 ligands, PD-L1 (B7-H1) and PD-L2 (B7-DC). 12 Other cell types, such as tumour-associated macrophages,23–26 tumour-associated neutrophils13–18 and myeloid-derived suppressor cells,22,43 are also thought to suppress tumour killing. In the past, these studies have used traditional and well-established tissue-staining and imaging techniques such as immunohistochemistry (IHC) or immunofluorescence (IF). However, accurately identifying and understanding these cells in detail and how they are spatially organised can be studied using relatively new methods that allow the description of novel cell subsets and how they interact with themselves, other cells and the tumour itself.
Interrogating the immune landscape in HCC
Tissue dissociates
To circumvent the limited number of markers available for tissue staining and the species barriers associated with primary antibodies, a common technique to gain further insight into the TME is to stain tumour dissociates by flow cytometry. Although spatial information is lost, it previously allowed for greater dimensionality into the immune cell makeup within tissue regions of interest. Zhang et al. 41 separated tumour and non-tumour regions from HCC patients, and then used flow cytometry to interrogate B-cell subsets. This analysis revealed non-tumour regions have a higher density of naïve B cells and plasma cells compared to tumour regions. A similar approach was utilised to investigate ILC by Heinrich et al., 31 where tissue dissociates of non-tumour, margin and tumour were collected. This study found ILC2 levels to be greatest within tumour, whereas ILC3 were at the lowest compared to non-tumour and margin. This technique is a useful alternative to identify and characterise a large number of cell subsets within tissue. Table 1 highlights the multiple subsets that have been associated with prognostic outcomes, and the technologies used to identify these correlations.
Table 1.
Immune cells within the TME and their association with prognostic outcome of HCC.
| Subset | Technology | High-density prognostic outcome | Reference(s) |
|---|---|---|---|
| Tumour-infiltrating lymphocytes | IHC | Better recurrence-free survival | Atanasov et al. 25 |
| CD3+ T cells | IHC, IF | Better overall survival | Garnelo et al. 44 |
| CD8+ CD103+ tissue resident T cells | mIHC | Better overall survival | Lim et al. 9 |
| PD-L1+ CD8+ TIL | IHC | Better overall survival | Sideras et al. 45 |
| Gal-9+ CD8+ TIL | IHC | Better overall survival | Sideras et al. 45 |
| TIM-3(hi) T cells | IF | Worsened overall survival | Li et al. 46 |
| CD4+ Foxp3+ regulatory T cells | IHC, mIHC | Worsened overall survival | Lim et al., 9 Gao et al. 47 |
| γδT cells | IHC | Better overall and recurrence-free survival | Cai et al. 20 |
| MAIT cells | IF | Worsened overall survival | Duan et al. 21 |
| CD20+ B cells | IHC, IF, mIHC | Better overall survival | Brunner et al., 40 Zhang et al., 41 Garnelo, Tan, 44 Shi, Gao 48 |
| Granzyme B+ cells | IHC | Better overall survival | Gao et al. 47 |
| Tumour-associated macrophages | IHC | Better recurrence-free survival | Atanasov et al. 25 |
| TIM-3+ CD14+ monocytes | IF | Worsened overall survival | Yan et al. 49 |
| CD66b+ Neutrophils | IHC | Worsened overall and recurrence-free survival | Gao et al., 13 Zhou et al., 14 Zhou et al. 50 |
| neutrophil extracellular traps | IHC | Worsened overall survival | Guan et al. 17 |
| ILC2 | IF | Worsened overall survival | Xu, et al. 32 |
| CD27+ cells | IHC, IF | Better overall survival | Garnelo et al. 44 |
| CD38+ cells | IHC, IF | Better overall survival | Garnelo et al. 44 |
| Siglec-10(hi) cells | IF | Worsened overall survival | Xiao et al. 51 |
mIHC, multiplex IHC.
Importance of structural and spatial information
When imaging cells in tissue, the structural landscape is maintained. This is essential for the identification of specific areas or structures of interest in the TME such as tumour, stroma, tertiary lymphoid structures (TLSs) or vessels. These structures are of importance and their presence or absence have been associated with clinical outcomes. For example, blood vessels are sometimes seen encapsulating tumour clusters. These have been called vascular endothelial tumour clusters (VETCs) and are sinusoid-like vessels that form a cobweb pattern surrounding tumour clusters. HCC patients with VETCs have higher metastasis and reoccurrence rates compared to HCC patients absent of VETCs. 52 Furthermore, sorafenib was shown to prolong survival in VETC(+) HCC patients, but not in VETC(–) patients. 53 Despite this, VETC(+) HCC patients have more severe clinical features compared to VETC(–) patients, including higher alpha-foetoprotein levels, larger tumour size and poorer differentiation. 54 Similar results have been found with CD34 staining, defining micro-vessel density, whereby high levels are associated with worse prognosis. 55 These studies highlight association between vessel formation and carcinogenesis.
TLSs are another important structure associated with many cancers, including HCC. The prognostic significance of TLS in HCC, however, remains unclear. One report suggested that TLSs located in non-tumour regions were associated with worse prognosis. 56 However, another study found TLSs in adjacent tissue of HCC had no correlation and instead intra-tumoural TLSs were associated with lower incidence of reoccurrence (<2 years after surgery). Interestingly in the same study, no correlation was found with TLSs and late reoccurrence. 57 These conflicting results may be explained by Li et al., 58 who found germinal centre formation within TLS of the peri-tumour improved prognostic outcome, which improved further if both intra-tumoural and peri-tumoural TLS were formed. It is only with techniques that allow for in situ analysis resulting in spatial data so that such structures and their significance can be elucidated.
Areas outside of the TME can also be informative. HCC patients with high numbers of S100+ dendritic cells (DCs) within peri-tumour or non-tumour regions have worse prognostic outcomes, but intra-tumoural S100+DCs have no correlation with prognosis. 39 High numbers of CD20+B cell at the interface and low levels within the stroma were associated with better overall survival in HCC.40,48 These findings are interesting to consider together, as DC have been reported to induce B-cell production of anti-inflammatory IL-10, 39 which may contribute to an immunosuppressed TME.
High numbers of intra- or peri-tumoural γδT cells were also associated with better overall survival; however, only high levels of peri-tumoural (but not intra-tumoural) γδT cells were associated with a lower probability of recurrence in HCC, suggesting they provide a beneficial anti-tumour response. 20 In contrast, TIM-3-expressing CD14+ monocytes, 49 CD4+, and CD8+T cells 46 are found at greater numbers within the tumour of HCC when compared to adjacent tissues, with high levels of TIM-3+ cells associated with worse overall survival, suggesting TIM-3+ cells contribute to carcinogenesis.
Similar results were reported for ILC2, whereby a higher proportion of ILC2 observed within the tumour or non-tumour regions were associated with decreased overall survival. 32 As ILC2 numbers were generally greater within the tumour region when compared to non-tumour regions and normal tissue, it was hence suggested ILC2 are immunoregulatory within the tumour and promote carcinogenesis.
Within the peri-tumour of HCC, there were higher densities of IL-21+ tissue-resident TFH-like cells compared to non- and intra-tumour areas, which also positively correlated with CD68+ and CD138+ cell density. 59 Furthermore, the same study also found HCC patients with high densities of IL-21+ TFH-like cells within the non- and peri-tumour regions had decreased overall and disease-free survival. Although there was no correlation with intra-tumoural levels, the authors suggest these IL-21+ TFH cells are pro-tumour via M2 macrophage polarisation.
Some of the differences between tumour regions and peri- or non-tumoural regions may not have prognostic values but be informative about the pathogenesis of HCC. IL-17+ cells were primarily found in peri-tumoural stroma (compared to intra-tumoural tissue) and positively correlated with monocyte/macrophage densities, 23 which could impact tumour control mechanisms. Tumour regions of HCC patients have greater CXCR3 (CD183) expression compared to non-tumour adjacent regions, 60 suggesting that increased Th1 or IFN-γ responses could be dominant. This is also supported by the fact that CXCL9/10 were more highly expressed in HCC tumours than normal tissue (whilst CXCL2/12/14 were lower). 61 However, it is worth noting that these increased levels were not associated with outcomes. IgA+ cells were found within the tumour of HCC patients, but not outside of it. 62 In this study, IgA+ cells induced CD8+ T-cell exhaustion in a murine HCC model, suggesting IgA+ cells contribute to an immunoregulatory environment. Even the microbiota of HCC patients differs between tumour and non-tumour regions, 63 and there is increasing evidence pointing to a key role for microbiota in shaping local and systemic immune responses. 64 Therefore, the ability to identify and compare specific tissue regions is paramount in understanding the molecular pathogenesis of HCC.
Cell subsets and their interactions
The ability to differentiate between cell subsets is an important aspect to consider when interrogating the immune landscape, as there can be contrasting roles for cells that have slight phenotypical differences during carcinogenesis. This is particularly important for myeloid cells due to their large heterogeneity. When looking at total CD68+ tumour-associated macrophages by IHC, high numbers are associated with decreased overall survival. However, when looked at in greater phenotypical detail allowing subsets to be correctly differentiated, high numbers of CD86+ macrophages but low numbers of CD206+ macrophages were associated with improved overall survival. 24 In a similar instance, tumour-infiltrating TIM-3+ CD8+ PD-1(hi) T-cell levels were associated with worsened overall and disease-free survival, whereas high proportions of TIM-3(–) CD8+ PD-1(hi) T cells were associated with better overall survival. 36 The authors suggest increased PD-1 and TIM-3 expression results in CD8+ T-cell dysfunction and worse survival for HCC. HCC-infiltrating CD103+ CD69+ HBV-specific CD8+ T cells are associated with patient relapse-free survival in HBV-associated HCC, suggesting they are involved in anti-tumour responses. 65 This same study was able to identify five distinct antigen-specific resident memory T-cell subsets within liver.
The benefit of studying tissues in situ using a technique that allows the detection of multiple markers into a single panel not only allows identification of more defined cell subsets but also provides insight into cell location within the tissue structure and cellular interactions. HCC patients who respond to combined cabozantinib and nivolumab treatment have a closer proximity between proliferative macrophages and lymphocytes (including B cells, CD4+ and CD8+ T cells). In contrast, non-responders have a closer interaction between Arg-1(hi) macrophage and lymphocytes within tumours. 66 The cellular networks formed within tumours may also influence prognostic outcomes, whereby high levels of Kupffer cell neighbourhoods were associated with worsened overall survival, whereas high levels of infiltrating macrophages provide improved overall survival. 67
Limitations of spatial analysis
One major limit to studying tissue sections is the fact they are a snapshot in time. From a tissue section alone, it is not possible to study the entire course of the disease from its inception. However, by providing a more thorough phenotypic interrogation of cell types, a clearer picture for their role can be ascertained. When combined with systemic investigations, such as analysing circulating immune cells, a more complete picture can be drawn. Interrogating activation markers can provide a wealth of insight into the function of immune cells.
Available technologies for spatially resolving HCC TME
Immunostaining techniques can provide unprecedented insight into the spatial organisation of cells within tissues. It can reveal cellular interactions that are lost when analysing dissociated tissues, as well as providing an in-depth view of the immune landscape. This is particularly important considering the success of immunotherapies that can inhibit pro-tumour immune responses whilst enhancing anti-tumour immune responses. Although immunostaining has been around for decades, the ability to interrogate many markers in a single panel in human tissue has only been made possible recently. Each technique has advantages and disadvantages, and are best utilised as complementary technologies, not alternative.
Multiplexed IHC
Opal™ multiplex immunohistochemistry (mIHC) staining (Akoya Biosciences, Marlborough, MA, USA) allows for individual IHC antibody stains to be multiplexed onto the same tissue regardless of antibody host species resulting in simultaneous detection of up to eight markers. This platform utilises tyramide signal amplification (TSA), which uses horseradish peroxidase bound to primary antibody to catalyse covalent bonds of chosen fluorescent labels to be embedded into the tissue. 68 This allows optimised staining for each antibody, as each marker can be stained under different optimisation conditions. TSA can provide 2–3 log sensitivity enhancement. Embedded fluorescent labels retain their fluorescence for much longer than conventional fluorescent antibody conjugates, so tissues can be imaged years after initial staining. Opal™ mIHC was used to investigate TILs in HCC patients and found infiltrating T and B cells localised closely to each other and their densities positively correlating with better overall survival. 44
mIHC does take longer to achieve than classic IHC or IF protocols as markers are stained individually. A major limitation of this technique, however, is the fact that antibody complexes need to be embedded by heat treatment for sequential staining, and during this process, the tissues can easily be damaged.
Another limitation of using multiple markers in a single panel is the bleeding/spill-over of fluorochromes, particularly when dealing with autofluorescent tissues such as the liver. Opal™ mIHC uses fluorescent signalling and detection but is capable of isolating specific tissue autofluorescence for its removal and detecting and measuring fluorescence over the whole spectrum using a spectral camera combined with fluorescence microscopy. This system requires a licensed software program to perform the spectral-unmixing. The advent of spectral microscopy allows the differentiation of fluorochromes with similar emission spectra. As such, a larger number of markers can be incorporated into a single panel. It can do this by recording the signal intensity across a wider spectrum, allowing finer differentiation between fluorochrome spectra. Spectral microscopy can also provide clearer differentiation from background staining. By recording the signal intensity of unstained cells/tissue, the autofluorescence will be known and can be differentiated from specific antibody staining. mIHC staining utilising TSA can amplify signals and provide high resolution images.
Cyclic mIHC
It is also possible to use mIHC in a cyclical manner, whereby tissue samples are stained, imaged and then bleached before repeating with a different marker. Although cyclic mIHC has similar problems to mIHC (particularly tissue damage after each bleach), it can minimise spectral overlap by imaging as little as two fluorochromes at a time. For example, PhenoCycler (formerly CODEX) (Akoya Biosciences) imaging involves staining a section with antibodies conjugated to unique oligonucleotides, allowing countless markers to be stained at once. Two unique fluorescent deoxynucleoside triphosphates (dNTP) are added to bind to two specified antibodies, which can then be fluorescently imaged. The tissue is bleached to remove the fluorophores, so a second round of fluorescent dNTP can be used and imaged, until all markers have been imaged and stacked on top of each other for analysis. 69 Phillips et al. 70 developed a 56-marker PhenoCycler panel for interrogating the TME within cutaneous T-cell lymphoma patients. 70 Although this process is time-consuming (particularly when dealing with large panels of antibodies) and expensive (limited suppliers of oligo-conjugated antibodies), it provides a great amount of detail and resolution.
Spatial transcriptomics and mass spectrometry
Spatial transcriptomics is a technique that allows interrogation of the transcriptome on a section of tissue that retains the spatial information. This involves a glass slide with oligo(dT) primers adhered to the surface. Each primer is barcoded to correlate with different locations on the slide. The tissue section is then applied to the slide, such that the primers are taken up by tissue transcripts. The tissue may then undergo sequencing (including the spatial barcoding), such that the transcriptome can be interrogated whilst maintaining spatial data. It has a resolution of ~100 µm. 71 This was recently used to interrogate the TME in HCC patients, whereby a six-gene signature was identified that could be used for prognostic prediction in HCC. 72 By looking at transcripts, it is possible to identify and differentiate between cell subsets that are indistinguishable using antibodies. In liver, Kupffer cells are indistinguishable from macrophages. However, the gene VSIG4 was recently identified to identify Kupffer cells in humans. 73 Although transcriptome levels/signatures are not always indicative of protein expression, the ability to interrogate the proteome and transcriptome whilst maintaining the spatial arrangement of tissue is an extremely powerful complementary technique when combined with proteomics.
Matrix-assisted laser desorption/ionisation imaging mass spectrometry (MALDI IMS) uses mass spectrometry whilst maintaining tissue spatial arrangement, allowing the identification and localisation of peptides and proteins without the need of antibodies.74,75 Many studies have utilised this technology to identify differences between tumour and non-tumour regions in HCC,76–78 including a higher level of ubiquitin within tumour regions. 79 The same approach found histone H4 to have different forms in HCC patients with microvascular invasion compared to patients without. 80 Although MALDI IMS provides large unrestricted data, validation is still required to confirm protein identification and differences between groups.
Techniques using mass cytometry
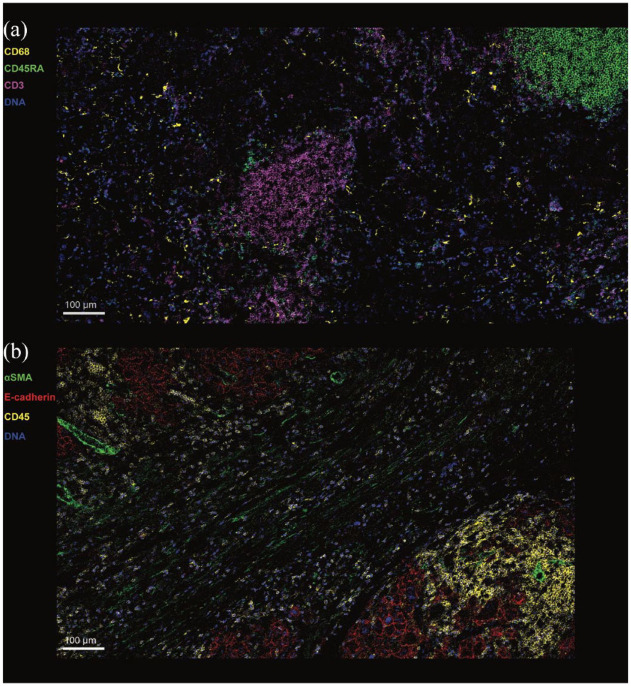
Imaging mass cytometry (IMC) (e.g. Hyperion™ (Fluidigm, South San Francisco, CA, USA) and MIBI™ (Ionpath Inc., Menlo Park, CA, USA)) uses antibodies conjugated to heavy metals (as opposed to fluorochromes). After staining, sections will go into a chamber attached to a mass cytometer, where the tissue is ablated with a laser, nebulising a small region of tissue (typically ~1–3 µm). Nebulised particulates are then passed through a mass spectrometer that will identify and differentiate individual heavy metals that correlate with markers of interest. As it does not use fluorochromes, it avoids the issues associated with spill-over/bleeding effect and is not affected by autofluorescence issues associated with tissues such as liver. It can accommodate >40 markers in a single panel, providing unprecedented insight into a range of cell subsets. Figure 1 shows an example of IMC staining on human liver from HCC patients. Traum et al. 81 similarly used the high dimensionality of IMC to compare between cells within HCC tumour and clinical data, including serum alanine aminotransferase (ALT). This showed a positive correlation between serum ALT and activated/memory immune cells, including CD45RO+ T cells, B cells and macrophages.
Figure 1.
Imaging mass cytometry of Hepatocellular carcinoma liver.
Complex immune microenvironment in HCC. Imaging mass cytometry was used to visualise the immune landscape of HCC. (a) Reveals a cluster of CD3+ T cells (purple) in the centre of the image and another cluster of B cells (green) in the top right corner of the image. By contrast, CD68+ macrophages (yellow) are seen dispersed throughout the tumour. (b) Combination of immune, stromal and adhesion molecule markers reveals two tumour nodules (top left corner and bottom right corner) with a fibrous band between the two. Within the tumour nodules, hepatoma cells are expressing E-cadherin (red). CD45+ immune cells (yellow) are seen predominantly not only within the tumour nodules but also in the fibrous band. αSMA expression (green) is seen on the vascular structures within the tumour nodules and in the stroma of the fibrous band.
Although extremely powerful, there are some limitations. IMC is less sensitive and has a lower resolution compared to mIHC and IF staining. After imaging, it is not possible to re-visit the same section, for further staining/imaging or RNA/DNA extraction. Once ablated, the tissue is lost. Finally, only selected imaging mass cytometers can be used to analyse stained sections, so is less accessible than conventional staining. Furthermore, the cost and analysis time involved with IMC is much greater.
Together, these technologies can very nicely complement each other (Table 2). IMC can provide an overview of the whole immune landscape, as it can incorporate a larger range of markers. mIHC staining is useful for detecting lowly expressed markers and producing higher resolution images. Spatial transcriptomics can be useful for identifying transcripts of proteins that cannot be detected with conventional proteomics. Incorporating such technologies allows the combination of high dimensionality with specificity and resolution.
Table 2.
Comparison of imaging techniques.
| IHC | IF | Multiplex IHC | Imaging mass cytometry | Spatial protein and transcriptomics | |
|---|---|---|---|---|---|
| Example technologies | –/– | –/– | Opal™ | Hyperion™/MIBI™ | Visium/NanoString |
| Number of markers | 1–2 | 3–4 | Up to eight protein markers | >40 protein markers | Protein and gene expression |
| Sample type | FFPE or Fresh frozen | FFPE or Fresh frozen | FFPE or Fresh frozen | FFPE or Fresh frozen | Fresh frozen (Visium) FFPE or Fresh frozen (NanoString) |
| Antibody availability/format | Purified antibody | Purified or fluorochrome-conjugated antibody | Any purified antibody suitable for IHC | Metal-tagged antibodies (commercially available or self-conjugation) | Slide layered with DNA probes |
| Staining | Time: <1 h Cost: low Difficulty: low |
Time: <1 h Cost: low Difficulty: low |
Time: up to 5 days Cost: low Difficulty: low |
Time: up to 2 days Cost: mid-high (dependent on region size for ablation) Difficulty: mid-high (requires specific instrument) |
Time: up to 2 days Cost: high Difficulty: high (various instruments required) |
| Imaging/data collection | Conventional microscope | Immunofluorescent microscope | Mantra™ imaging: manual Vectra® Polaris™ imaging: automated slide scanning |
Hyperion™ ablation: 1 mm2 tissue approx. 2.5 h MIBI™: 1 mm2 tissue approx. 5 h |
Microscope (protein) RNA sequencing |
| Resolution | Dependent on microscope | Dependent on microscope | 200 nm (dependent on microscope) |
1000 nm | 100 µm |
Data analysis
When it comes to spatial analysis of HCC tissue, similar approaches may be used as with other tissues. Individual cells need to first be identified and the signal intensity of each marker to be calculated. This can be achieved by cell segmentation tools such as CellProfiler 82 and ilastik, 83 which have been successfully utilised to classify cells of the heterogeneous TME.66,67,81 Once cells are identified, their densities and interactions with other cell subsets can be quantified. Cellular neighbourhood interactions have been interrogated with freely available tools, including histoCAT 84 and Spectre. 85 mIHC (Opal™) requires the use of inForm® (Akoya Biosciences) software for spectral unmixing and image extraction prior to use in other analysis platforms. inForm® can also be used for cellular phenotyping, quantification and limited spatial analysis. In addition, the purchasable HALO® image analysis platform (Indica Labs, Albuquerque, NM, USA) provides a range of tools for downstream analysis of all antibody-staining techniques including IHC, IF, mIHC and IMC for cell segmentation and spatial analysis, including that of liver. 86 This is an ever-adapting field, with many new approaches being developed for the various technologies being made available.87,88
Current therapies in advanced HCC
The treatment landscape of HCC is rapidly evolving. A greater understanding of hepatocarcinogenesis and the TME of HCC has led to the development of several molecularly targeted agents for advanced HCC. For much of the last decade, monotherapy with multi-kinase inhibitors have been recommended in both first-line (sorafenib and lenvatinib) and second-line (regorafenib, cabozantinib and apatinib) treatment settings based on positive phase III clinical trials showing modest, but significant, improvements in overall survival of 2–3 months.89–93
More recently, the introduction of immune checkpoint inhibitors has been a major development in the treatment of many advanced cancers, including HCC. 94 Despite achieving objective response rates of 20%, initial phase III trials of checkpoint inhibitor monotherapy, both first-line (PD-1 inhibitor nivolumab) and second-line (PD-1 inhibitor pembrolizumab) treatments have failed to demonstrate significantly superior outcomes compared to standard of care (sorafenib and placebo, respectively).95,96 Recently, combination therapies with a checkpoint inhibitor (either PD-1 or PD-L1 inhibitor) paired with an anti-vascular endothelial growth factor (VEGF) monoclonal antibody became the first regimens in over a decade to demonstrate both improved overall and progression-free survival compared to sorafenib as a first-line treatment.97,98 This combination now forms the new standard of care for advanced HCC worldwide. Other novel combinations, such as cabozantinib plus atezolizumab (PD-L1 inhibitor) or the use of two checkpoint inhibitors together (nivolumab plus CTLA-4 inhibitor ipilimumab), have also shown promise in first- and second-line treatment settings, respectively.99,100 In particular, the combination of durvalumab (anti-PD-L1) and tremelimumab (anti-CTLA-4) has demonstrated a significant improvement in overall survival compared to sorafenib alone. 101
Clearly, we are entering into a new era of advanced HCC therapies involving checkpoint inhibitors as a backbone in combination with VEGF inhibitors, multi-kinase inhibitors and/or other checkpoint inhibitors.
Predictors of response to immunotherapies
Although the recent successes of immunotherapy to treat advanced HCC are unprecedented, an objective response is only seen in a minority of patients (30% with any response and 5% with complete response). 97 To date, it has been difficult to predict which patients will respond and which will not respond due in part to the heterogeneous nature of HCC. HCC arising from non-viral aetiologies of liver disease (in particular non-alcoholic steatohepatitis (NASH)) might be less responsive to checkpoint inhibitors compared to viral aetiologies (hepatitis B or C). 10 Early pre-clinical data showed that tumour immune surveillance in NASH-induced HCC is impaired because CD8+ T cells inducted NASH-HCC rather than invigorating the immune response. 10 These findings were supported by a meta-analysis of three large randomised controlled phase III trials in patients with advanced HCC where neither nivolumab, pembrolizumab nor combined atezolizumab and bevacizumab (anti-VEGFA) immunotherapy improved survival in patients with non-viral HCC. 10 The effectiveness of checkpoint inhibitors was similarly improved with other virally induced cancers, including EBV-associated non-Hodgkin lymphoma 102 and HPV-induced head and neck squamous cell carcinoma. 103 Conversely, Asian patients and those with high alpha-foetoprotein levels treated with checkpoint inhibitors have slightly better overall survival.95,96 Furthermore, it is likely that different combination therapies will emerge that result in improved response rates in the future, and these combinations may also be adopted into earlier (intermediate) stages of HCC as an adjunct to locoregional therapies. 104 Therefore, clinicians will increasingly need guidance on which combination therapy shows the best efficacy for each (sub)group of HCC patients.
The development of predictive biomarkers to inform patient treatment is currently an area of active research. As mentioned above, the TME plays a vital role in the development and progression of HCC. To date, no biomarkers of the TME (or otherwise) have demonstrated robust predictive value adequate for routine clinical use. Intra-tumoural PD-L1 expression was not significantly associated with improved objective response in clinical trials of checkpoint inhibitors during HCC.105,106 Similarly, non-significant trends have been observed between PD-1 expression levels and objective response or overall survival from therapy. 107 In terms of TILs, CD3+, CD4+ and CD8+ T-cell infiltration have only shown weak correlations with patient survival.107,108 Although intra-tumoural CD8+ T cells can be significantly increased by immunotherapy, they may become dysfunctional in the TME and be ineffective for tumour surveillance. 10 Thus, current biomarkers of single-cell types within the TME are not clinically helpful, hampering personalised medicine in advanced HCC. Indeed, the TME is complex, comprising many different immune, stromal and vascular cell types that interact. 109 Hence, there is an urgent need for more sophisticated techniques to study the TME and its interactions more broadly to find better biomarkers to guide patient selection. Using technologies such as IMC to spatially and quantitatively analyse tumour tissue can aid clinicians in identifying patients with cellular markers and neighbourhoods predictive of treatment response, whilst providing invaluable information about tumour heterogeneity and its impact on treatment outcomes.
Concluding remarks
Despite recent advances in HCC therapeutics, overall response rates are low and there remains a lack of validated biomarkers to reliably predict treatment response. The aforementioned techniques allow for a more sophisticated and comprehensive analysis of the TME beyond that of cellular quantification. Spatial resolution-based phenotyping helps to resolve tumour heterogeneity and allows for a topological analysis of the HCC TME through the identification of cellular neighbourhoods and cell-to-cell interactions. Not only will this enhance our current understanding of the tumour biology of HCC, but it may also lead to an improved classification of HCC, identification of immune and gene signatures predictive of treatment response and the discovery of novel targets for future therapies.
Acknowledgments
FM-W is supported by the International Society for the Advancement of Cytometry (ISAC) Marylou Ingram Scholars Program. We gratefully acknowledge subsidised access to Sydney Cytometry and thank the support staff in this core facility for their assistance.
Footnotes
ORCID iD: Felix Marsh-Wakefield  https://orcid.org/0000-0002-6839-7628
https://orcid.org/0000-0002-6839-7628
Contributor Information
Felix Marsh-Wakefield, Liver Injury & Cancer Program, Centenary Institute, Royal Prince Alfred Hospital, Missenden Road, Sydney, NSW, 2050, Australia; Human Immunology Laboratory, The University of Sydney, Sydney, NSW, Australia.
Angela L Ferguson, Liver Injury & Cancer Program, Centenary Institute, Sydney, NSW, Australia; Human Immunology Laboratory, The University of Sydney, Sydney, NSW, Australia.
Ken Liu, Liver Injury & Cancer Program, Centenary Institute, Sydney, NSW, Australia; A.W. Morrow Gastroenterology and Liver Centre, Royal Prince Alfred Hospital, The University of Sydney, Sydney, NSW, Australia.
Cositha Santhakumar, Liver Injury & Cancer Program, Centenary Institute, Sydney, NSW, Australia; A.W. Morrow Gastroenterology and Liver Centre, Royal Prince Alfred Hospital, The University of Sydney, Sydney, NSW, Australia.
Geoffrey McCaughan, Liver Injury & Cancer Program, Centenary Institute, Sydney, NSW, Australia; A.W. Morrow Gastroenterology and Liver Centre, Royal Prince Alfred Hospital, The University of Sydney, Sydney, NSW, Australia.
Umaimainthan Palendira, Human Immunology Laboratory, The University of Sydney, Sydney, NSW, Australia.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contributions: Felix Marsh-Wakefield: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Angela L. Ferguson: Investigation, Methodology, Writing – original draft, Writing – review & editing.
Ken Liu: Investigation, Writing – original draft, Writing – review & editing
Cositha Santhakumar: Methodology, Writing – review & editing.
Geoffrey McCaughan: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Umaimainthan Palendira: Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Competing Interests: The authors declare that there is no conflict of interest.
Availability of data and materials: Not applicable.
References
- 1. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71: 209–249. [DOI] [PubMed] [Google Scholar]
- 2. Wallace MC, Preen DB, Short MW, et al. Hepatocellular carcinoma in Australia 1982-2014: increasing incidence and improving survival. Liver Int 2019; 39: 522–530. [DOI] [PubMed] [Google Scholar]
- 3. McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of hepatocellular carcinoma. Hepatology 2021; 73(Suppl 1): 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Valery PC, Laversanne M, Clark PJ, et al. Projections of primary liver cancer to 2030 in 30 countries worldwide. Hepatology 2018; 67: 600–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gallage S, García-Beccaria M, Szydlowska M, et al. The therapeutic landscape of hepatocellular carcinoma. Med 2021; 2: 505–552. [DOI] [PubMed] [Google Scholar]
- 6. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021; 7: 6. [DOI] [PubMed] [Google Scholar]
- 7. Estes C, Razavi H, Loomba R, et al. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018; 67: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brar G, Greten TF, Graubard BI, et al. Hepatocellular carcinoma survival by etiology: A SEER-Medicare database analysis. Hepatol Commun 2020; 4: 1541–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim CJ, Lee YH, Pan L, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut 2019; 68: 916–927. [DOI] [PubMed] [Google Scholar]
- 10. Pfister D, Nunez NG, Pinyol R, et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 2021; 592: 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Du M, Cai YM, Yin YL, et al. Evaluating tumor-infiltrating lymphocytes in hepatocellular carcinoma using hematoxylin and eosin-stained tumor sections. World J Clin Cases 2022; 10: 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oura K, Morishita A, Tani J, et al. Tumor immune microenvironment and immunosuppressive therapy in hepatocellular carcinoma: A Review. Int J Mol Sci 2021; 22: 5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao Q, Zhao YJ, Wang XY, et al. CXCR6 upregulation contributes to a proinflammatory tumor microenvironment that drives metastasis and poor patient outcomes in hepatocellular carcinoma. Cancer Res 2012; 72: 3546–3556. [DOI] [PubMed] [Google Scholar]
- 14. Zhou SL, Zhou ZJ, Hu ZQ, et al. Tumor-associated neutrophils recruit macrophages and T-Regulatory cells to promote progression of hepatocellular carcinoma and resistance to Sorafenib. Gastroenterology 2016; 150: 1646–1658.e17. [DOI] [PubMed] [Google Scholar]
- 15. Zhou SL, Yin D, Hu ZQ, et al. A positive feedback loop between cancer stem-like cells and tumor-associated neutrophils controls hepatocellular carcinoma progression. Hepatology 2019; 70: 1214–1230. [DOI] [PubMed] [Google Scholar]
- 16. Choi WM, Kim JY, Choi J, et al. Kinetics of the neutrophil-lymphocyte ratio during PD-1 inhibition as a prognostic factor in advanced hepatocellular carcinoma. Liver Int 2021; 41: 2189–2199. [DOI] [PubMed] [Google Scholar]
- 17. Guan X, Lu Y, Zhu H, et al. The crosstalk between cancer cells and neutrophils enhances hepatocellular carcinoma metastasis via neutrophil extracellular traps-associated cathepsin G component: A potential therapeutic target. J Hepatocell Carcinoma 2021; 8: 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rich NE, Parvathaneni A, Sen A, et al. High Neutrophil-lymphocyte ratio and delta Neutrophil-lymphocyte ratio are associated with increased mortality in patients with hepatocellular cancer. Dig Dis Sci 2022; 67: 2666–2676. [DOI] [PubMed] [Google Scholar]
- 19. Yi Y, He HW, Wang JX, et al. The functional impairment of HCC-infiltrating γδ T cells, partially mediated by regulatory T cells in a TGFβ- and IL-10-dependent manner. J Hepatol 2013; 58: 977–983. [DOI] [PubMed] [Google Scholar]
- 20. Cai XY, Wang JX, Yi Y, et al. Low counts of γδ T cells in peritumoral liver tissue are related to more frequent recurrence in patients with hepatocellular carcinoma after curative resection. Asian Pac J Cancer Prev 2014; 15: 775–780. [DOI] [PubMed] [Google Scholar]
- 21. Duan M, Goswami S, Shi JY, et al. Activated and exhausted MAIT cells foster disease progression and indicate poor outcome in hepatocellular carcinoma. Clin Cancer Res 2019; 25: 3304–3316. [DOI] [PubMed] [Google Scholar]
- 22. Hoechst B, Ormandy LA, Ballmaier M, et al. A new population of myeloid-derived suppressor cells in hepatocellular carcinoma patients induces CD4(+)CD25(+)Foxp3(+) T cells. Gastroenterology 2008; 135: 234–243. [DOI] [PubMed] [Google Scholar]
- 23. Kuang DM, Peng C, Zhao Q, et al. Activated monocytes in peritumoral stroma of hepatocellular carcinoma promote expansion of memory T helper 17 cells. Hepatology 2010; 51: 154–164. [DOI] [PubMed] [Google Scholar]
- 24. Dong P, Ma L, Liu L, et al. CD86+/CD206+, diametrically polarized tumor-associated macrophages, predict hepatocellular carcinoma patient prognosis. Int J Mol Sci 2016; 17: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Atanasov G, Dino K, Schierle K, et al. Immunologic cellular characteristics of the tumour microenvironment of hepatocellular carcinoma drive patient outcomes. World J Surg Oncol 2019; 17: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng HHM, Lee RY, Goh S, et al. Immunohistochemical scoring of CD38 in the tumor microenvironment predicts responsiveness to anti-PD-1/PD-L1 immunotherapy in hepatocellular carcinoma. J Immunother Cancer 2020; 8: e000987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoechst B, Voigtlaender T, Ormandy L, et al. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology 2009; 50: 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang P, Lu X, Tao K, et al. Siglec-10 is associated with survival and natural killer cell dysfunction in hepatocellular carcinoma. J Surg Res 2015; 194: 107–113. [DOI] [PubMed] [Google Scholar]
- 29. Tao L, Wang S, Yang L, et al. Reduced siglec-7 expression on NK cells predicts NK cell dysfunction in primary hepatocellular carcinoma. Clin Exp Immunol 2020; 201: 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zecca A, Barili V, Rizzo D, et al. Intratumor regulatory noncytotoxic NK cells in patients with hepatocellular carcinoma. Cells 2021; 10: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heinrich B, Gertz EM, Schäffer AA, et al. The tumour microenvironment shapes innate lymphoid cells in patients with hepatocellular carcinoma. Gut 2022; 71: 1161–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu X, Ye L, Zhang Q, et al. Group-2 innate lymphoid cells promote HCC progression through CXCL2-Neutrophil-Induced immunosuppression. Hepatology 2021; 74: 2526–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu J, Xu D, Liu Z, et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 2007; 132: 2328–2339. [DOI] [PubMed] [Google Scholar]
- 34. Zhang JP, Yan J, Xu J, et al. Increased intratumoral IL-17-producing cells correlate with poor survival in hepatocellular carcinoma patients. J Hepatol 2009; 50: 980–989. [DOI] [PubMed] [Google Scholar]
- 35. Zheng C, Zheng L, Yoo JK, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017; 169: 1342–1356.e16. [DOI] [PubMed] [Google Scholar]
- 36. Ma J, Zheng B, Goswami S, et al. PD1(Hi) CD8(+) T cells correlate with exhausted signature and poor clinical outcome in hepatocellular carcinoma. J Immunother Cancer 2019; 7: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fessas P, Spina P, Boldorini RL, et al. Phenotypic characteristics of the tumour microenvironment in primary and secondary hepatocellular carcinoma. Cancers 2021; 13: 2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu RX, Wei Y, Zeng QH, et al. Chemokine (C-X-C motif) receptor 3-positive B cells link interleukin-17 inflammation to protumorigenic macrophage polarization in human hepatocellular carcinoma. Hepatology 2015; 62: 1779–1790. [DOI] [PubMed] [Google Scholar]
- 39. Ouyang FZ, Wu RQ, Wei Y, et al. Dendritic cell-elicited B-cell activation fosters immune privilege via IL-10 signals in hepatocellular carcinoma. Nat Commun 2016; 7: 13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brunner SM, Itzel T, Rubner C, et al. Tumor-infiltrating B cells producing antitumor active immunoglobulins in resected HCC prolong patient survival. Oncotarget 2017; 8: 71002–71011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Z, Ma L, Goswami S, et al. Landscape of infiltrating B cells and their clinical significance in human hepatocellular carcinoma. OncoImmunology 2019; 8: e1571388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang S, Liu Z, Wu D, et al. Single-cell RNA-Seq analysis reveals microenvironmental infiltration of plasma cells and hepatocytic prognostic markers in HCC with Cirrhosis. Front Oncol 2020; 10: 596318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chiu DKC, Xu IMJ, Lai RKH, et al. Hypoxia induces myeloid-derived suppressor cell recruitment to hepatocellular carcinoma through chemokine (C-C motif) ligand 26. Hepatology 2016; 64: 797–813. [DOI] [PubMed] [Google Scholar]
- 44. Garnelo M, Tan A, Her Z, et al. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut 2017; 66: 342–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sideras K, Biermann K, Verheij J, et al. PD-L1, galectin-9 and CD8(+) tumor-infiltrating lymphocytes are associated with survival in hepatocellular carcinoma. OncoImmunology 2017; 6: e1273309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H, Wu K, Tao K, et al. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology 2012; 56: 1342–1351. [DOI] [PubMed] [Google Scholar]
- 47. Gao Q, Qiu SJ, Fan J, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol 2007; 25: 2586–2593. [DOI] [PubMed] [Google Scholar]
- 48. Shi JY, Gao Q, Wang ZC, et al. Margin-infiltrating CD20(+) B cells display an atypical memory phenotype and correlate with favorable prognosis in hepatocellular carcinoma. Clin Cancer Res 2013; 19: 5994–6005. [DOI] [PubMed] [Google Scholar]
- 49. Yan W, Liu X, Ma H, et al. Tim-3 fosters HCC development by enhancing TGF-β-mediated alternative activation of macrophages. Gut 2015; 64: 1593–1604. [DOI] [PubMed] [Google Scholar]
- 50. Zhou SL, Dai Z, Zhou ZJ, et al. Overexpression of CXCL5 mediates neutrophil infiltration and indicates poor prognosis for hepatocellular carcinoma. Hepatology 2012; 56: 2242–2254. [DOI] [PubMed] [Google Scholar]
- 51. Xiao N, Zhu X, Li K, et al. Blocking siglec-10(hi) tumor-associated macrophages improves anti-tumor immunity and enhances immunotherapy for hepatocellular carcinoma. Exp Hematol Oncol 2021; 10: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fang JH, Zhou HC, Zhang C, et al. A novel vascular pattern promotes metastasis of hepatocellular carcinoma in an epithelial-mesenchymal transition-independent manner. Hepatology 2015; 62: 452–465. [DOI] [PubMed] [Google Scholar]
- 53. Fang JH, Xu L, Shang LR, et al. Vessels that Encapsulate tumor clusters (VETC) pattern is a predictor of sorafenib benefit in patients with hepatocellular carcinoma. Hepatology 2019; 70: 824–839. [DOI] [PubMed] [Google Scholar]
- 54. Renne SL, Woo HY, Allegra S, et al. Vessels encapsulating tumor clusters (VETC) is a powerful predictor of aggressive hepatocellular carcinoma. Hepatology 2020; 71: 183–195. [DOI] [PubMed] [Google Scholar]
- 55. Tanigawa N, Lu C, Mitsui T, et al. Quantitation of sinusoid-like vessels in hepatocellular carcinoma: its clinical and prognostic significance. Hepatology 1997; 26: 1216–1223. [DOI] [PubMed] [Google Scholar]
- 56. Finkin S, Yuan D, Stein I, et al. Ectopic lymphoid structures function as microniches for tumor progenitor cells in hepatocellular carcinoma. Nat Immunol 2015; 16: 1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Calderaro J, Petitprez F, Becht E, et al. Intra-tumoral tertiary lymphoid structures are associated with a low risk of early recurrence of hepatocellular carcinoma. J Hepatol 2019; 70: 58–65. [DOI] [PubMed] [Google Scholar]
- 58. Li H, Liu H, Fu H, et al. Peritumoral tertiary lymphoid structures correlate with protective immunity and improved prognosis in patients with hepatocellular carcinoma. Front Immunol 2021; 12: 648812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen MM, Xiao X, Lao XM, et al. Polarization of tissue-resident TFH-Like cells in human hepatoma bridges innate monocyte inflammation and m2b macrophage polarization. Cancer Discov 2016; 6: 1182–1195. [DOI] [PubMed] [Google Scholar]
- 60. Ding Q, Xia Y, Ding S, et al. An alternatively spliced variant of CXCR3 mediates the metastasis of CD133+ liver cancer cells induced by CXCL9. Oncotarget 2016; 7: 14405–14414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin T, Zhang E, Mai PP, et al. CXCL2/10/12/14 are prognostic biomarkers and correlated with immune infiltration in hepatocellular carcinoma. Biosci Rep 2021; 41: BSR20204312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shalapour S, Lin XJ, Bastian IN, et al. Erratum: inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 2017; 552: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Komiyama S, Yamada T, Takemura N, et al. Profiling of tumour-associated microbiota in human hepatocellular carcinoma. Sci Rep 2021; 11: 10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yoo JY, Groer M, Dutra SVO, et al. Gut Microbiota and immune system interactions. Microorganisms 2020; 8: 1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cheng Y, Gunasegaran B, Singh HD, et al. Non-terminally exhausted tumor-resident memory HBV-specific T cell responses correlate with relapse-free survival in hepatocellular carcinoma. Immunity 2021; 54: 1825–1840.e7. [DOI] [PubMed] [Google Scholar]
- 66. Ho WJ, Zhu Q, Durham J, et al. Neoadjuvant cabozantinib and nivolumab convert locally advanced hepatocellular carcinoma into resectable disease with enhanced antitumor immunity. Nature Cancer 2021; 2: 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sheng J, Zhang J, Wang L, et al. Topological analysis of hepatocellular carcinoma tumour microenvironment based on imaging mass cytometry reveals cellular neighbourhood regulated reversely by macrophages with different ontogeny. Gut 2022; 71: 1176–1191. [DOI] [PubMed] [Google Scholar]
- 68. Faget L, Hnasko TS. Tyramide signal amplification for immunofluorescent enhancement. Methods Mol Biol 2015; 1318: 161–172. [DOI] [PubMed] [Google Scholar]
- 69. Goltsev Y, Samusik N, Kennedy-Darling J, et al. Deep profiling of mouse splenic architecture with CODEX Multiplexed Imaging. Cell 2018; 174: 968–981.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Phillips D, Schürch CM, Khodadoust MS, et al. Highly multiplexed phenotyping of immunoregulatory proteins in the tumor microenvironment by CODEX Tissue Imaging. Front Immunol 2021; 12: 687673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ståhl PL, Salmén F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016; 353: 78–82. [DOI] [PubMed] [Google Scholar]
- 72. Zhao N, Zhang Y, Cheng R, et al. Spatial maps of hepatocellular carcinoma transcriptomes highlight an unexplored landscape of heterogeneity and a novel gene signature for survival. Cancer Cell Int 2022; 22: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Guilliams M, Bonnardel J, Haest B, et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 2022; 185: 379–396.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Karas M, Bachmann D, Bahr U, et al. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int J Mass Spectrom Ion Process 1987; 78: 53–68. [Google Scholar]
- 75. Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem 1997; 69: 4751–4760. [DOI] [PubMed] [Google Scholar]
- 76. Han EC, Lee YS, Liao WS, et al. Direct tissue analysis by MALDI-TOF mass spectrometry in human hepatocellular carcinoma. Clin Chim Acta 2011; 412: 230–239. [DOI] [PubMed] [Google Scholar]
- 77. Powers TW, Neely BA, Shao Y, et al. MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 2014; 9: e106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Marquardt C, Tolstik T, Bielecki C, et al. MALDI imaging-based classification of hepatocellular carcinoma and non-malignant lesions in fibrotic liver tissue. Z Gastroenterol 2015; 53: 33–39. [DOI] [PubMed] [Google Scholar]
- 79. Le Faouder J, Laouirem S, Chapelle M, et al. Imaging mass spectrometry provides fingerprints for distinguishing hepatocellular carcinoma from cirrhosis. J Proteome Res 2011; 10: 3755–3765. [DOI] [PubMed] [Google Scholar]
- 80. Poté N, Alexandrov T, Le Faouder J, et al. Imaging mass spectrometry reveals modified forms of histone H4 as new biomarkers of microvascular invasion in hepatocellular carcinomas. Hepatology 2013; 58: 983–994. [DOI] [PubMed] [Google Scholar]
- 81. Traum D, Wang YJ, Schwarz KB, et al. Highly multiplexed 2-dimensional imaging mass cytometry analysis of HBV-infected liver. JCI Insight 2021; 6: e146883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006; 7: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Berg S, Kutra D, Kroeger T, et al. Ilastik: interactive machine learning for (bio)image analysis. Nat Methods 2019; 16: 1226–1232. [DOI] [PubMed] [Google Scholar]
- 84. Schapiro D, Jackson HW, Raghuraman S, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods 2017; 14: 873–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ashhurst TM, Marsh-Wakefield F, Putri GH, et al. Integration, exploration, and analysis of high-dimensional single-cell cytometry data using Spectre. Cytometry Part A J Int Soc Anal Cytol 2022; 101: 237–253. [DOI] [PubMed] [Google Scholar]
- 86. Horai Y, Mizukawa M, Nishina H, et al. Quantification of histopathological findings using a novel image analysis platform. J Toxicol Pathol 2019; 32: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Parra ER. Methods to determine and analyze the cellular spatial distribution extracted from multiplex immunofluorescence data to understand the tumor microenvironment. Front Mol Biosci 2021; 8: 668340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wu Y, Cheng Y, Wang X, et al. Spatial omics: navigating to the golden era of cancer research. Clin Transl Med 2022; 12: e696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. New Engl J Med 2008; 359: 378–390. [DOI] [PubMed] [Google Scholar]
- 90. Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017; 389: 56–66. [DOI] [PubMed] [Google Scholar]
- 91. Abou-Alfa GK, Meyer T, Cheng AL, et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. New Engl J Med 2018; 379: 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018; 391: 1163–1173. [DOI] [PubMed] [Google Scholar]
- 93. Qin S, Li Q, Gu S, et al. Apatinib as second-line or later therapy in patients with advanced hepatocellular carcinoma (AHELP): a multicentre, double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Gastroenterol Hepatol 2021; 6: 559–568. [DOI] [PubMed] [Google Scholar]
- 94. Xu W, Liu K, Chen M, et al. Immunotherapy for hepatocellular carcinoma: recent advances and future perspectives. Ther Adv Med Oncol 2019; 11: 1758835919862692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sangro B, Park J, Finn R, et al. LBA-3 checkmate 459: Long-term (minimum follow-up 33.6 months) survival outcomes with nivolumab versus sorafenib as first-line treatment in patients with advanced hepatocellular carcinoma. Ann Oncol 2020; 31: S241–S2. [Google Scholar]
- 96. Finn RS, Ryoo BY, Merle P, et al. Pembrolizumab as second-line therapy in patients with advanced hepatocellular carcinoma in KEYNOTE-240: a randomized, double-blind, Phase III trial. J Clin Oncol 2020; 38: 193–202. [DOI] [PubMed] [Google Scholar]
- 97. Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. New Engl J Med 2020; 382: 1894–1905. [DOI] [PubMed] [Google Scholar]
- 98. Ren Z, Xu J, Bai Y, et al. Sintilimab plus a bevacizumab biosimilar (IBI305) versus sorafenib in unresectable hepatocellular carcinoma (ORIENT-32): a randomised, open-label, phase 2–3 study. Lancet Oncol 2021; 22: 977–990. [DOI] [PubMed] [Google Scholar]
- 99. Yau T, Kang YK, Kim TY, et al. Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib: the checkmate 040 randomized clinical trial. JAMA Oncol 2020; 6: e204564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kelley RK, Yau T, Cheng AL, et al. VP10-2021: cabozantinib (C) plus atezolizumab (A) versus sorafenib (S) as first-line systemic treatment for advanced hepatocellular carcinoma (aHCC): results from the randomized phase III COSMIC-312 trial. Ann Oncol 2022; 33: 114–116. [Google Scholar]
- 101. Abou-Alfa GK, Chan SL, Kudo M, et al. Phase 3 randomized, open-label, multicenter study of tremelimumab (T) and durvalumab (D) as first-line therapy in patients (pts) with unresectable hepatocellular carcinoma (uHCC): HIMALAYA. J Clin Oncol 2022; 40: 379–379. [Google Scholar]
- 102. Kim SJ, Hyeon J, Cho I, et al. Comparison of efficacy of pembrolizumab between Epstein-barr Virus–Positive and –negative relapsed or refractory Non-Hodgkin lymphomas. Cancer Res Treat 2019; 51: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ghanizada M, Jakobsen KK, Grønhøj C, et al. The effects of checkpoint inhibition on head and neck squamous cell carcinoma: a systematic review. Oral Oncol 2019; 90: 67–73. [DOI] [PubMed] [Google Scholar]
- 104. Prince D, Liu K, Xu W, et al. Management of patients with intermediate stage hepatocellular carcinoma. Ther Adv Med Oncol 2020; 12: 1758835920970840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (checkmate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017; 389: 2492–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhu AX, Finn RS, Edeline J, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol 2018; 19: 940–952. [DOI] [PubMed] [Google Scholar]
- 107. Sangro B, Melero I, Wadhawan S, et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol 2020; 73: 1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sia D, Jiao Y, Martinez-Quetglas I, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 2017; 153: 812–826. [DOI] [PubMed] [Google Scholar]
- 109. Santhakumar C, Gane EJ, Liu K, et al. Current perspectives on the tumor microenvironment in hepatocellular carcinoma. Hepatol Int 2020; 14: 947–957. [DOI] [PubMed] [Google Scholar]