Abstract
The necessity to understand the influence of global ocean change on biota has exposed wide-ranging gaps in our knowledge of the fundamental principles that underpin marine life. Concurrently, physiological research has stagnated, in part driven by the advent and rapid evolution of molecular biological techniques, such that they now influence all lines of enquiry in biological oceanography. This dominance has led to an implicit assumption that physiology is outmoded, and advocacy that ecological and biogeochemical models can be directly informed by omics. However, the main modeling currencies are biological rates and biogeochemical fluxes. Here, we ask: how do we translate the wealth of information on physiological potential from omics-based studies to quantifiable physiological rates and, ultimately, to biogeochemical fluxes? Based on the trajectory of the state-of-the-art in biomedical sciences, along with case-studies from ocean sciences, we conclude that it is unlikely that omics can provide such rates in the coming decade. Thus, while physiological rates will continue to be central to providing projections of global change biology, we must revisit the metrics we rely upon. We advocate for the co-design of a new generation of rate measurements that better link the benefits of omics and physiology.
Keywords: physiology, omics, co-design, marine biogeochemistry, ocean change
INTRODUCTION
A major challenge for ocean scientists is to address key questions on future ecosystem services. For example, how will global climate change alter low latitude primary productivity and hence food security? A powerful tool to address these global-scale questions is Earth system models, such as those within the Coupled Model Intercomparison Project (CMIP6) (Kwiatkowski et al., 2020). The CMIP currencies include the critically important rates at which metabolism occurs in living organisms (i.e. physiological rates) and the biogeochemical fluxes of bioactive elements. It is unlikely that these currencies will change in the coming decade, for example when CMIP7 is developed. Currently, the accuracy of the model projections is hindered by two issues: (i) computational limitations to developing more complex parameterisations for processes such as nitrogen (N) fixation (Kwiatkowski et al., 2020) and (ii) our inability to untangle how marine life responds to complex ocean change (Weisse and Montagnes, 2021). For the latter, we need to decipher the fundamental physiological rules that govern biological responses to ocean change. These include the metabolic co-dependencies in response to multiple stressors, and strategies to buffer responses to rapid change, such as phenotypic plasticity and microevolution.
The physiological metrics used to quantify biological rates that are the cornerstones of Earth system models, such as primary productivity, have not fundamentally changed in decades. In contrast, omics techniques have evolved rapidly this century and have the potential to supersede physiological metrics as the main approach to study the fundamental principles driving marine life. With this dominance has come an implicit assumption by some that measuring physiological rates directly is obsolete, as they can be inferred from omics (Hellweger, 2020; McCain et al., 2021). However, omics provides a surfeit of data, at a level of detail that is often difficult to relate to the information provided by physiological rate measurements and the current needs of Earth system models. This growing mismatch between the currencies of global-scale models (rates and fluxes) and the aspirations of omics (coupling cellular potential via omics to Earth system model projections) must be addressed urgently.
Here, we ask: How do we translate the wealth of information on physiological potential from omics-based studies to quantifiable physiological rates and, ultimately, to biogeochemical processes and their representation in Earth system models? We employ three approaches to address this question. First, we use ocean N2 fixation as an illustrative example of research that has evolved through joint advances in physiology and omics (Fig. 1). Second, we examine the recent trajectory of biomedical research to forecast how ocean sciences might evolve in the next decade. Third, we broaden our view by examining insights that can be gained for understanding the ocean phosphorus (P) and iron (Fe) cycles by better linking omics and physiology. We conclude with advocacy for the co-design of better physiological tools.
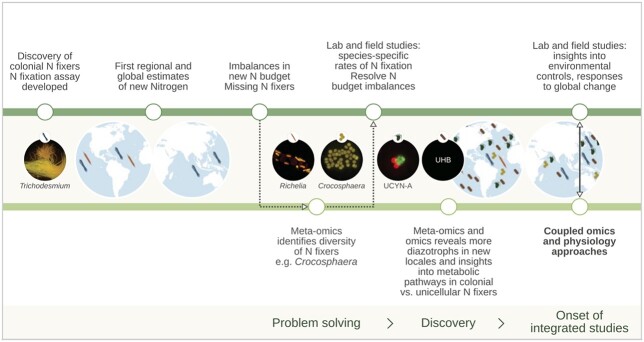
Fig. 1.
The contributions of physiology and omics to understanding the role of diazotrophy in the ocean N cycle (based on Zehr and Capone, 2020). Key events in the physiology timeline (top green line) include the estimation of N fluxes through nitrogenase activity (Dugdale and Dugdale, 1962), initial estimates of global marine N2 fixation rates (Capone et al., 2005) and the combining of lab and field measurements to understand individual diazotrophs and community contributions and constraints. Pivotal events in the omics timeline (lower green line) include problem solving (Zehr and Montoya, 2007) and discovery of diazotroph diversity including in unicellular cyanobacteria group A (UNCYN-A) and diverse uncultured heterotrophic bacteria (Martínez-Pérez et al., 2016). Recent examples of more integrated physiological and omics co-designed studies (Walworth et al., 2016; Qu et al., 2022) offer an important way forward.
LESSONS FROM MARINE DIAZOTROPHY
Here, we use the history of N2 fixation (diazotrophy) research to reveal the benefits and limitations of physiological rate measurements, and how these measurements are complemented by more recent omics approaches (Fig. 1).
The contribution of diazotrophy to the supply of new N is an important facet of the ocean N cycle (Fogg, 1942; Dugdale et al., 1961). Physiological studies played an important early role by quantifying rates of diazotrophy (e.g. Dilworth, 1966). These measurements provided the integrated rates necessary to estimate global biogeochemical fluxes of N (Karl et al., 2002) and to identify the environmental drivers of N2 fixation (see Carpenter and Capone, 2008), including how climate change may alter future diazotrophy (Garcia et al., 2015; Hutchins et al., 2013) leading to improved model projections (see Fig. 3 in Hutchins et al., 2013). Still, imbalances in these N fluxes have uncovered unidentified N sources, and the subsequent application of genetic tools has identified additional diazotrophic taxa that contribute to ocean N2 fixation (Zehr and Capone, 2020).
Nitrogen fixation provides clear examples of both the limitations and benefits of non-targeted omics-based discoveries (Fig. 1). Nitrogenase (nif) genes can be used to detect N2 fixation potential, and their expression is used as an index of N2 fixation activity (Zehr et al., 1996; Zehr and Montoya, 2007). Omics has revealed diverse N2 fixers including the unicellular cyanobacteria Crocosphaera and UCYN-A, and endosymbiotic and heterotrophic diazotrophs (Mehta et al., 2003; Church et al., 2005; Martínez-Pérez et al., 2016). But, nif gene abundance does not directly equate to either diazotroph abundance- and biomass-based biogeographic “currencies” (Meiler et al., 2022) or to N2 fixation rates (Turk-Kubo et al., 2013). Transcriptomics and proteomics targeting nif genes provide more relevant information about nitrogenase activity than genomics. However, taxon-specific dynamics can complicate the estimates of community N2 fixation rates (Church et al., 2005), and measurements of nif expression are not well correlated with 15N-based rates of N2 fixation (Turk et al., 2011).
Thus, despite the insights gained from omics, critical gaps remain in our understanding of the phylogenies, distribution and physiology of marine N2 fixers, and accurate global estimates of N2 fixation remain elusive (Zehr and Capone, 2020). Measuring N2 fixation remains critical to estimate the biogeochemical processing and ecological fates of new N. Yet, N2 fixation is not included in the CMIP6 models, which currently project declining productivity in low latitude oceans in coming decades (Kwiatkowski et al., 2020). Therefore, both rates and omics will be needed increasingly to reveal and quantify currently unknown (but biogeochemically important) pathways for the turnover of N (Fig. 1) to improve global models.
Resolving these unknowns will require combined measurements of nif gene expression with rate measurements based on nitrogenase enzyme activity (e.g. Turk et al., 2011). Broader application of flow-through high-throughput rate measurements can improve the spatial and temporal coverage of N2 fixation (Cassar et al., 2018). Rates, when coupled with omics approaches to N2 fixation research (Tang et al., 2020), will continue to expand our understanding of diazotroph diversity and could help focus N2 fixation rate measurements on these emerging diazotrophic groups (Zehr and Capone, 2020). Mechanistic controls on diazotrophy can be revealed through variations in nif gene expression (Church et al., 2005), supporting prior conclusions that local environmental conditions influence N2 fixation rates (Capone, 1993; Carpenter and Capone, 2008). Such environmental controls could be further explored using targeted proteomics analyses (e.g. Saito et al., 2011).
The history of N2 fixation research clearly demonstrates the rapid progress that can be made by bringing together omics and physiology. Walworth et al. (2016) in a lab-based investigation of long-term (i.e. multi-year) adaptation by Trichodesmium to high carbon dioxide levels demonstrated that genetic assimilation may be driving adaptation, but also identified the metabolic pathways that were initially altered and then maintained during this adaptation. In another lab-based diazotroph study, Qu et al. (2022) provide a mechanistic understanding of how the diazotrophs Crocosphaera and Trichodesmium acclimate or adapt to ocean global change using phenotypic metrics in conjunction with whole-genome sequencing and variant analysis.
THE STATUS OF OMICS
Both marine and biomedical sciences study the genome, transcriptome, proteome and metabolome, with most research on the first three. In the field of meta-omics (where “meta-” refers to a population of organisms rather than a single organism), marine metagenomics has set the pace and is directly influencing research into the human microbiome (Poceviciute and Ismagilov, 2019). Here, we focus on genomics through to proteomics at the cellular level where, in contrast to meta-omics, biomedical research has led the way (Okada and Kuroda, 2019). Genomics is the most distal to physiological activity. It demonstrates the breadth of possible gene functions but only catalogues the functional potential of an organism (Sunagawa et al., 2015). Transcriptomics is a popular approach to explore how organisms respond to environmental change by characterizing shifts in mRNA abundance (Evans, 2015). Feder and Walser (2005) offered a pointed description of the major issues facing the use of transcriptomics in finding the genes that matter for environmental adaptation. Their critique concentrated on three major issues: (i) genes with large impacts on fitness are rare and therefore unlikely to be identified with transcriptomics, (ii) the relationship between gene expression and fitness is unreliable and (iii) fitness is primarily determined by proteins, and mRNA abundance is a poor proxy for protein abundance. Proteomics, on the other hand, provides taxonomically specific information on metabolic enzymes and is therefore more proximal to physiological activity. Proteomics has advanced methodologically, with more accurate standardized quantitative analyses (Collins et al., 2017; Pino et al., 2020) and protein identifications that allow metabolic profiling (Nunn et al., 2013; Mikan et al., 2020).
Numerous efforts have been made to identify correlations between omics layers. However, evidence from both marine and biomedical science reveals that making these linkages is not straightforward. For example, in marine sciences, it is well recognized that the amplitude and timing of the mRNA pool does not align with protein expression. This misalignment was illustrated in Waldbauer et al. (2012) while tracking diel changes in the transcriptome and proteome within a single cyanobacteria species (Fig. 2). Subsequent research on the diatom Phaeodactylum tricornutum used multiple omics layers to explore the regulation of N limitation and again reported mismatches between transcript, protein and metabolite abundance (Remmers et al., 2018). In the further advanced biomedical field, it remains difficult to obtain mechanistic and functional insights by simply integrating multiomics data (Okada and Kuroda, 2019). As far back as the late eighties, Kurland and Ehrenberg (1987) discussed the challenges of linking cellular design and molecular design (such as via enzyme expression) in the context of physiology. More recently, Lalanne et al. (2018) uncovered post-transcriptional controls that ensure the maintenance of the protein stoichiometries required for specific biological pathways. This compensatory mechanism rectifies divergences in regulation driven by changes of internal promoters and terminators. Hence, even in advanced biomedical research, there are confounding issues, driven by post-transcriptional and post-translational modifications to enzymes, in deriving metabolic rates from omics.
Fig. 2.
Examples of the potential for mismatches in transcriptomics versus proteomics in a pico-prokaryote over the diurnal light:dark cycle. (A) The diel cycling and amplitudes of transcripts and proteins in Prochlorococcus for Ribonucleotide reductase (nrdJ), the large sub-unit of Rubisco (rbcL) and Geranylgeranyl diphosphate reductase (chlP). (B) Histogram of lag-times for proteins and their transcripts for a 312 gene dataset. Antiphase refers to genes that are offset by ~12 h (i.e. 50%) of the diel cell cycle. Redrawn from Waldbauer et al. (2012).
In the marine context, omics has clearly demonstrated large-scale patterns in microbial diversity across oceanic provinces and provided insights into which metabolic pathways are active (Fig. 1). But, omics-based approaches provide static “snap-shots” of physiological potential, and we need to improve our quantitative, process-level understanding of the roles of marine microbes in biogeochemical cycles. Indeed, it is physiological activity—as modified by biological species differences, environmental drivers and the interactions between the two—that ultimately drives biogeochemical cycles.
LINKING PHYSIOLOGY AND OMICS: THE NEED FOR CO-DESIGN
We propose that physiological rates can bridge biogeochemistry and omics. Physiological rates quantify the integrated activity of proteins that drive marine biogeochemical cycles in units that modelers can use (Fig. 3). Research into the ocean’s N cycle reveals the potential of using the joint expertise of the physiology and omics communities (i.e. co-design) to guide future research (Fig. 1). We can extend this complementary approach to use omics datasets to develop new targeted physiological metrics that improve the parameterization of biogeochemical processes. Here, we explore the feasibility of co-design using case studies of the ocean P and Fe cycles that illustrate how physiological metrics may act as a “currency converter” to link omics datasets and biogeochemical models.
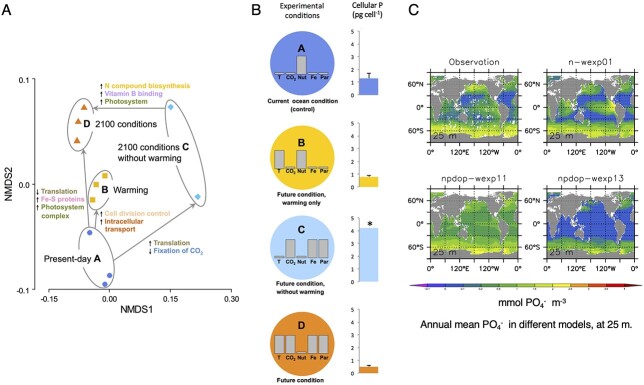
Fig. 3.
An example illustrating the utility of physiological metrics as a “currency converter” to link omics and biogeochemical modeling. (A) Higher (downward arrows) and lower expression (upwards arrows) of proteins in 4 treatments within a climate change manipulation experiment measured with proteomics (Boyd et al., 2015). Warming results in higher expression of P-containing proteins associated with translation. (B) Corresponding changes to the cellular P quotas (no change in cell size was observed) of the study subject, a lab culture of a subantarctic diatom, across the treatments A–D. This physiological metric reveals the causal link between lower expression of translation proteins and decreased P quotas (as previously described by Toseland et al., 2013). (C) A subset of global model projections of upper ocean phosphate (PO4−) stocks across biogeochemical models of different complexity (Kriest et al., 2010). The approaches employed in panels A and C can be linked using the cellular P quotas obtained from panel B.
In the case of P, a lab study used proteomics and physiological metrics to explore the cumulative effect of five climate-change stressors on a subpolar diatom (Boyd et al., 2015). A central finding was that the effect of decreased nutrient supply in a future ocean was offset by warming. Proteomics revealed that a decreased need for P was driven by lower expression of P-containing proteins associated with translation (Fig. 3). Physiological metrics corroborated this finding, with lower cellular P quotas under warming. Hence, P quotas acted as a currency converter between protein synthesis and the biogeochemical cycle of P. They showed serendipitously a link between protein synthesis and P quotas. In the future, we must actively seek conceptual linkages, rather than uncovering them by chance. Better links from omics via physiology to biogeochemistry would benefit from input from the modeling and biogeochemical research communities.
Physiology was established earlier than omics or biogeochemistry, which begs the question: are we currently measuring the best physiological metrics to mesh omics with biogeochemistry? Two examples that begin to straddle the gaps between omics and physiology come from Saito et al. (2011) and Wu et al. (2019). The former revealed diel changes in the proteome, including Fe-metalloproteins involved in N2 fixation and photosynthesis of Crocosphaera watsonii resulting in more efficient use of Fe, which is essential for N2 fixation. In the latter case, protein expression and physiological metrics were coupled to examine the influence of Fe and manganese on Phaeocystis antarctica.
Although our current choice of physiological metrics needs urgent scrutiny, there is compelling evidence of the utility of long-established assays, such as those used to determine the macromolecular P content of cells from Liefer et al. (2019), for more innovative phytoplankton cellular P models (Inomura et al., 2020). But can we be inventive and use omics to interpret P physiology in a more holistic manner (Fig. 4)? Physiology can provide valuable insights, even when considering only a few components of the cellular P cycle. Imagine the progress if we developed better metrics jointly with omics (Feng et al., 2014; Lin et al., 2016). Therefore, the way ahead may be to use molecular biology to “reverse engineer” the most pertinent physiological metrics (Fig. 4). For example, a useful point of departure would be to select processes in which protein abundance correlates with quantifiable metabolic activity. It is likely that a range of approaches—from metabolic theory, ocean observations of key metabolites along with improved maps of cellular functioning from omics (e.g. iPath)—can help jointly refine the targeting of key biochemicals most suitable for reverse-engineering of better metrics. In some cases, for targeted research questions, the range of approaches presented as illustrative examples in Fig. 4 may need to be expanded upon or modified. Such co-design, in our opinion, will further facilitate the transition from lab- to field-based omics and will lead in the coming decades to incorporation of omics into biogeochemical models.
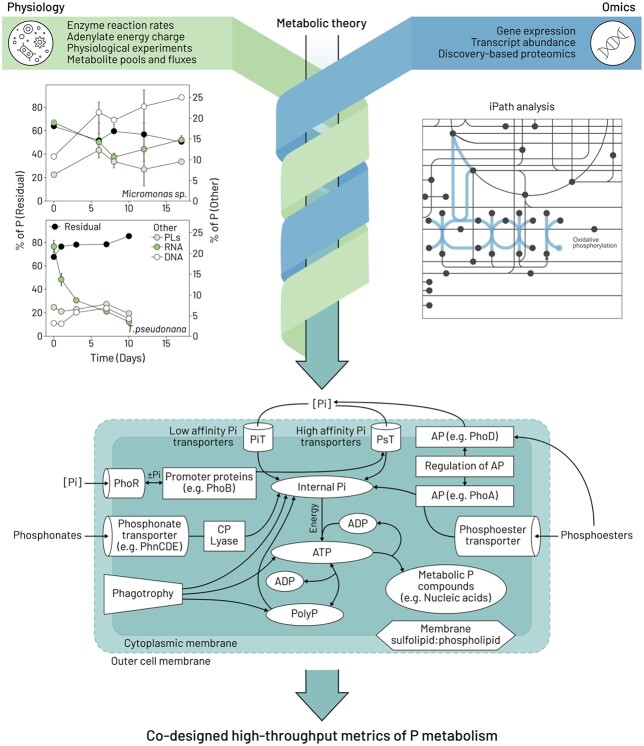
Fig. 4.
The potential of reverse-engineering physiological metrics to provide better linkages with molecular tools using the example of phosphorus (P). Findings from a physiological study (top left; redrawn from Liefer et al., 2019. PLs = phospholipids) using a cluster of long-established metrics (residual P pools/intracellular storage of inorganic P) to compare the P allocation strategies of a diatom (Thalassiosira pseudonana) and a prasinophyte (Micromonas sp.). Other physiological approaches (top left green box) include measuring enzyme reaction rates, clever experimental design (Bell, 2019), quantifying adenylate energy charge (Karl, 1980) and tracking metabolite pools and fluxes (Moran et al., 2022). The top right figure is a Kyoto Encyclopedia of Genes and Genomes (KEGG) map from Interactive Pathways Explorer v3 (iPath) (https://pathways.embl.de; Letunic et al., 2008; Darzi et al., 2018). iPath is a web-based tool for the visualization and analysis of cellular pathways from omics (e.g. see Nunn et al., 2013 for Fe replete versus Fe deplete proteomes). In this example, the search terms “phosphate” and “oxidative phosphorylation” returned compound C00009 (https://www.genome.jp/dbget-bin/www_bget?C00009) and map00190 (https://www.genome.jp/dbget-bin/www_bget?map00190), respectively. The results are shown in the iPath panel as the blue dot (phosphate; C00009) and the blue line (map00190) connecting compounds (nodes) with enzymes (lines). Combining approaches—denoted by the green and blue intersecting arrows—will improve underpinning biochemical theory and identify candidate enzymes and pathways for the co-design of new physiological assays, as depicted in the bottom cartoon summarizing putative phosphorus uptake, metabolism and storage in phytoplankton (redrawn and simplified from Fig. 4 in Lin et al., 2016). The present cartoon is for a cyanobacterium, which in addition to the plasma membrane, also contains a semipermeable outer membrane (shown as a dotted line). Abbreviations: ADP = adenosine diphosphate; ATP = adenosine triphosphate; AP = alkaline phosphatase; Pi = inorganic phosphorus; PolyP = polyphosphate; gene names as described in Lin et al., 2016).
The transition to field studies will face additional challenges that centre on how marine biota integrate environmental history (i.e. cellular status imposed by conditions encountered prior to sampling; Fig. 2) (Prairie et al., 2012; Deutschmann et al., 2021). This requires a multi-stranded approach. Placing the sampling locale in a wider environmental context will be necessary (Fig. 5A). For example, profiling robotic floats with multiple sensors are providing synoptic snapshots of spatial variability in ocean properties along with the prior seasonal dynamics of key resources such as nutrients (Claustre et al., 2021). It will also be essential to determine how such prior oceanic conditions set cellular status, for example the degree of Fe stress (Fig. 5B). An open question is whether the relationship between environmental forcing and cellular status is instantaneous or lagged (Fig. 2). Will such co-designed metrics reconcile a biological product with a chemical residual since different physiological metrics display a range of response times (Boyd et al., 2005; Baker et al., 2018), as do different omics layers (Waldbauer et al., 2012)? One promising approach to probe environmental history and cellular status is physiological titration, for example by manipulating Fe availability to contextualize cellular Fe status (Fig. 5B).
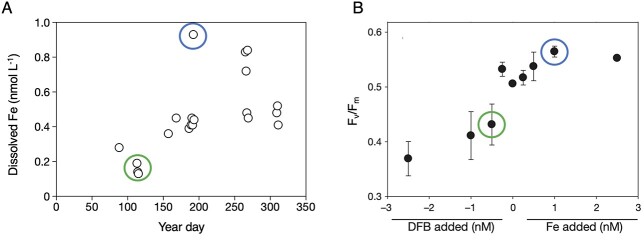
Fig. 5.
Utility of environmental context to define the present physiological status of cells in relation to prior oceanic conditions. (A) Dissolved Fe time series for the upper ocean in the subtropical Atlantic (BATS site) that reveals conspicuous aerosol Fe inputs (>0.5 nmol L−1) along with the influence of eddy activity (<0.3 nmol L−1) on dissolved Fe concentrations (Sedwick et al., 2020). (B) Photosynthetic efficiency of PSII (Fv/Fm) measured in deckboard incubation experiments “titrated” with dissolved Fe concentrations by either lowering bioavailable Fe using the fungal siderophore desferrioxamine B (DFB) or increasing it with chelated inorganic Fe addition (from Wilhelm et al., 2013). The circles denote putative linkages between chemical stocks and biological responses (blue = high Fe; green = low Fe).
CONCLUSIONS AND FUTURE DIRECTIONS
We conclude with recent field-leading examples from ocean sciences that seek to derive metabolic rates from omics, explored through the lens of biomedical sciences. Saito et al. (2020) conducted metaproteomic analysis on subsurface biota in the Tropical North Pacific to pinpoint commonly occurring enzymes. They reported that nitrite oxidoreductase associated with the bacterium Nitrospina was abundant in this stratum and explored whether they could estimate rates of nitrite oxidation using wide-ranging methods, including biochemistry (specific activity), physiology (Michaelis–Menten kinetics) and omics. Despite employing this innovative suite of approaches, derived rates ranged >200-fold, pointing to the need to develop targeted physiological assays (c.f. Fig. 4). There are also promising initial developments from the emergence of phenomenological models based on simple geochemical/taxonomic principles that yield phytoplankton growth rates assuming steady-state growth (McCain et al., 2021).
The latest developments in biomedical and model-system omics suggest that obtaining rates from omics is still under development. First, holistic investigations of well-characterized model organisms have tracked every metabolite and protein to generate enzyme-directed functional rates in the bacterium Escherichia coli (Taniguchi et al., 2010) and the yeast Saccharomyces cerevisiae (Ho et al., 2018), but this approach is restricted to the organisms for which the function of every gene and protein is known. Second, expression-fitness landscapes (linking enzyme expression with growth rate) have revealed that enzyme expression can have a “ripple” effect across layers of biological organization ranging from mechanistic, regulatory to systemic (Lalanne et al., 2021), which adds further complexity to deriving growth rates from enzymatic fluxes. Third, sophisticated microbiome studies (from cheese to the human gut) (Poceviciute and Ismagilov, 2019), which are more akin to oceanic microbial systems, reveal that there are still a high number of metabolic functions that remain uncharacterized (Price et al., 2018). Fourth, progress in tackling cell regulatory mechanisms using multiomic modeling has been made but requires complex computing using deep neural networks such as GEMS (Genome-scale metabolic models) (Okada and Kuroda, 2019).
These four categories of advanced well-resourced research point to challenges yet to be surmounted in obtaining physiological rates from omics for biomedical sciences. But, they also provide cautionary lessons for ocean sciences. In our opinion, it may be more straight-forward to co-design targeted physiological metrics that better link omics with marine biogeochemistry. We advocate for better communication across these research communities that could be readily facilitated through co-design workshops and other forums to ascertain the best ways to reverse-engineer a new generation of physiological metrics, in tandem with the development of high-throughput technologies to promote “co-measurement” (Fig. 4), that better exploit the power of molecular biology to answer the most pressing questions in ocean sciences.
Contributor Information
Robert F Strzepek, Australian Antarctic Program Partnership (AAPP), Institute for Marine and Antarctic Studies, University of Tasmania, 20 Castray Esplanade, Hobart, TAS 7004, Australia.
Brook L Nunn, Department of Genome Sciences, University of Washington, Foege Building S113 3720 15th Ave NE, Seattle, WA 98195, USA.
Lennart T Bach, Institute for Marine and Antarctic Studies, University of Tasmania, Hobart, TAS 7004, Australia.
John A Berges, Department of Biological Sciences and School of Freshwater Sciences, University of Wisconsin-Milwaukee, 3209 N. Maryland Avenue, Milwaukee, WI 53211, USA.
Erica B Young, Department of Biological Sciences and School of Freshwater Sciences, University of Wisconsin-Milwaukee, 3209 N. Maryland Avenue, Milwaukee, WI 53211, USA.
Philip W Boyd, Australian Antarctic Program Partnership (AAPP), Institute for Marine and Antarctic Studies, University of Tasmania, 20 Castray Esplanade, Hobart, TAS 7004, Australia; Institute for Marine and Antarctic Studies, University of Tasmania, Hobart, TAS 7004, Australia.
Dedication
We dedicate this article to the late Paul J. Harrison, a phytoplankton physiologist who influenced and inspired generations of scientists, including several of the authors.
Funding
Australian Antarctic Program Partnership (ASCI000002 to R.F.S. and P.W.B.), National Science Foundation (IOS-2041497 to B.L.N.), the Australian Research Council received through a Future Fellowship (FT200100846 to L.T.B.), by a Wisconsin Sea Grant award (R/HCE-33) and a RACAS grant from the University of Wisconsin-Milwaukee (to E.B.Y. and J.A.B.), and by a University of Tasmania Visiting Scholarship (to J.A.B.).
References
- Baker, K. G., Radford, D. T., Evenhuis, C., Kuzhiumparam, U., Ralph, P. J. and Doblin, M. A. (2018) Thermal niche evolution of functional traits in a tropical marine phototroph. J. Phycol., 54, 799–810. 10.1111/jpy.12759. [DOI] [PubMed] [Google Scholar]
- Bell, T. (2019) Next-generation experiments linking community structure and ecosystem functioning. Environ. Microbiol. Rep., 11, 20–22. 10.1111/1758-2229.12711. [DOI] [PubMed] [Google Scholar]
- Boyd, P., Dillingham, P., McGraw, C., Armstrong, E. A., Cornwall, C. E., Feng, Y.-Y., Hurd, C. L., Gault-Ringold, M.et al. (2015) Physiological responses of a Southern Ocean diatom to complex future ocean conditions. Nat. Clim. Chang., 6, 207–213. 10.1038/nclimate2811. [DOI] [Google Scholar]
- Boyd, P. W., Strzepek, R., Takeda, S., Jackson, G., Wong, C. S., McKay, R. M., Law, C., Kiyosawa, H.et al. (2005) The evolution and termination of an iron-induced mesoscale bloom in the northeast subarctic Pacific. Limnol. Oceanogr., 50, 1872–1886. [Google Scholar]
- Capone, D. G., Burns, J. A., Montoya, J. P., Subramaniam, A., Mahaffey, C., Gunderson, T., et al. (2005) Nitrogen fixation by Trichodesmium spp.: an important source of new nitrogen to the tropical and subtropical North Atlantic Ocean. Global Biogeochem. Cycles, 19, GB2024. 10.1029/2004GB002331. [DOI] [Google Scholar]
- Capone, D. G. (1993) Determination of nitrogenase activity in aquatic samples using the acetylene reduction procedure. In Kemp, P. F., Sherr, B. F., Sherr, E. B. and Cole, J. J. (eds.), Handbook of Methods in Aquatic Microbial Ecology, CRC Press, Boca Raton, pp. 621–631. [Google Scholar]
- Carpenter, E. J. and Capone, D. G. (2008) Nitrogen fixation in the marine environment. In: Capone, D. G., Bronk, D. A., Mulholland, M. R., and Carpenter, E. J. (eds) Nitrogen in the Marine Environment (2nd edn), Academic Press, p. 141-198, 10.1016/B978-0-12-372522-6.00004-9. [DOI] [Google Scholar]
- Cassar, N., Tang, W., Gabathuler, H. and Huang, K. (2018) Method for high frequency underway N2 fixation measurements: flow-through incubation acetylene reduction assays by cavity ring down laser absorption spectroscopy (FARACAS). Anal. Chem., 90, 2839–2851. 10.1021/acs.analchem.7b04977. [DOI] [PubMed] [Google Scholar]
- Church, M. J., Short, C. M., Jenkins, B. D., Karl, D. M. and Zehr, J. P. (2005) Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol., 71, 5362–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claustre, H., Legendre, L., Boyd, P. W. and Levy, M. (2021) The oceans’ biological carbon pumps: framework for a research observational community approach. Front. Mar. Sci., 8, 780052. 10.3389/fmars.2021.780052. [DOI] [Google Scholar]
- Collins, B. C., Hunter, C. L., Liu, Y., Schilling, B., Rosenberger, G., Bader, S. L., Chan, D. W., Gibson, B. W.et al. (2017) Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun., 8, e1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzi, Y., Letunic, I., Bork, P. and Yamada, T. (2018) iPath3.0: interactive pathways explorer v3. Nucleic Acids Res., 46, W510–W513. 10.1093/nar/gky299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschmann, I. M., Krabberød, A. K., Benites, L. F., Latorre, F., Delage, E., Marrasé, C. V.et al. (2021) Disentangling temporal associations in marine microbial networks. bioRxiv 2021.07.13.452187. 10.1101/2021.07.13.452187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth, M. J. (1966) Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianum. Biochim. Biophys. Acta, 127, 285–294. [DOI] [PubMed] [Google Scholar]
- Dugdale, R. C., Goering, J. J. and Ryther, J. H. (1961) Nitrogen fixation in the Sargasso Sea. Deep-Sea Res., 7, 298–300. [Google Scholar]
- Dugdale, V. A. and Dugdale, R. C. (1962) Nitrogen metabolism in lakes II. Role of nitrogen fixation in Sanctuary Lake, Pennsylvania. Limnol. Oceanogr., 7, 170–177. 10.4319/lo.1962.7.2.0170. [DOI] [Google Scholar]
- Evans, T. G. (2015) Considerations for the use of transcriptomics in identifying the 'genes that matter' for environmental adaptation. J. Exp. Biol., 218, 1925–1935. 10.1242/jeb.114306. [DOI] [PubMed] [Google Scholar]
- Feder, M. E. and Walser, J.-C. (2005) The biological limitations of transcriptomics in elucidating stress and stress responses. J. Evol. Biol., 18, 901–910. 10.1111/j.1420-9101.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- Feng, T.-Y., Yang, Z.-K., Zheng, J.-W., Xie, Y., Li, D.-W., Murugan, S. B., Yang, W. D., Liu, J. S.et al. (2014) Examination of metabolic responses to phosphorus limitation via proteomic analyses in the marine diatom Phaeodactylum tricornutum. Sci. Rep., 5, 10373. 10.1038/srep10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg, G. E. (1942) Studies on nitrogen fixation by blue-green algae: I. Nitrogen fixation by Anabaena cylindrica Lemm. J. Exp. Biol., 19, 78–87. 10.1242/jeb.19.1.78. [DOI] [Google Scholar]
- Garcia, N., Fu, F., Sedwick, P. and Hutchins, D. A. (2015) Iron deficiency increases growth and nitrogen-fixation rates of phosphorus-deficient marine cyanobacteria. ISME J., 9, 238–245. 10.1038/ismej.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellweger, F. (2020) Combining molecular observations and microbial ecosystem modeling: a practical guide. Annu. Rev. Mar. Sci., 12, 267–289. 10.1146/annurev-marine-010419-010829. [DOI] [PubMed] [Google Scholar]
- Ho, B., Baryshnikova, A. and Brown, G. W. (2018) Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Systems, 6, 192–205.e3. [DOI] [PubMed] [Google Scholar]
- Hutchins, D., Fu, F. X., Webb, E., Walworth, N. and Tagliabue, A. (2013) Taxon-specific response of marine nitrogen fixers to elevated carbon dioxide concentrations. Nat. Geosci., 6, 790–795. 10.1038/ngeo1858. [DOI] [Google Scholar]
- Inomura, K., Omta, A. W., Talmy, D., Bragg, J., Deutsch, C. and Follows, M. J. (2020) A mechanistic model of macromolecular allocation, elemental stoichiometry, and growth rate in phytoplankton. Front. Microbiol., 11, 86. 10.3389/fmicb.2020.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl, D., Michaels, A., Bergman, B., Capone, D., Carpenter, E., Letelier, R., Lipschultz, F., Paerl, H.et al. (2002) Dinitrogen fixation in the world’s oceans. In: Boyer E.W., Howarth R.W. (eds) The Nitrogen Cycle at Regional to Global Scales. Springer, Dordrecht. 10.1007/978-94-017-3405-9_2 [DOI] [Google Scholar]
- Karl, D. M. (1980) Cellular nucleotide measurements and applications in microbial ecology. Microbiol. Rev., 44, 739–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriest, I., Khatiwala, S. and Oschlies, A. (2010) Towards an assessment of simple global marine biogeochemical models of different complexity. Prog. Oceanogr., 86, 337–360. 10.1016/j.pocean.2010.05.002. [DOI] [Google Scholar]
- Kurland, C. G. and Ehrenberg, M. (1987) Growth-optimizing accuracy of gene expression. Annu. Rev. Biophys. Biophys. Chem., 16, 291–317. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski, L., Torres, O., Bopp, L., Aumont, O., Chamberlain, M., Christian, J. R., Dunne, J. P., Gehlen, M.et al. (2020) Twenty-first century ocean warming, acidification, deoxygenation, and upper-ocean nutrient and primary production decline from CMIP6 model projections. Biogeosciences, 17, 3439–3470. 10.5194/bg-17-3439-2020. [DOI] [Google Scholar]
- Lalanne, J. B., Parker, D. J. and Li, G. W. (2021) Spurious regulatory connections dictate the expression-fitness landscape of translation factors. Mol. Syst. Biol., 17, e10302. 10.15252/msb.202110302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne, J. B., Taggart, J. C., Guo, M. S., Herzel, L., Schieler, A. and Li, G. W. (2018) Evolutionary convergence of pathway-specific enzyme expression stoichiometry. Cell, 173, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., Yamada, T., Kanehisa, M. and Bork, P. (2008) iPath: interactive exploration of biochemical pathways and networks. Trends Biochem. Sci., 33, 101–103. [DOI] [PubMed] [Google Scholar]
- Liefer, J. D., Garg, A., Fyfe, M. H., Irwin, A. J., Benner, I., Brown, C. M., Follows, M. J., Omta, A. W.et al. (2019) The macromolecular basis of phytoplankton C:N:P under nitrogen starvation. Front. Microbiol., 10, 763. 10.3389/fmicb.2019.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S., Litaker, R. W. and Sunda, W. G. (2016) Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J. Phycol., 52, 10–36. 10.1111/jpy.12365. [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez, C., Mohr, W., Löscher, C. R., Dekaezemacker, J., Littmann, S., Yilmaz, P., Lehnen, N., Fuchs, B. M.et al. (2016) The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat. Microbiol., 1, 16163. 10.1038/nmicrobiol.2016.163. [DOI] [PubMed] [Google Scholar]
- McCain, J. S. P., Tagliabue, A., Susko, E., Achterberg, E. P., Allen, A. E. and Bertrand, E. M. (2021) Cellular costs underpin micronutrient limitation in phytoplankton. Sci. Adv., 7, eabg6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, M. P., Butterfield, D. A. and Baross, J. A. (2003) Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl. Environ. Microbiol., 69, 960–970. 10.1128/AEM.69.2.960-970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler, S., Britten, G. L., Dutkiewicz, S., Gradoville, M. R., Moisander, P. H., Jahn, O. and Follows, M. J. (2022) Constraining uncertainties of diazotroph biogeography from nifH gene abundance. Limnol. Oceanogr., 67, 816–829. 10.1002/lno.12036. [DOI] [Google Scholar]
- Mikan, M. P., Harvey, H. R., Timmins-Schiffman, E., Riffle, M., May, D. H., Salter, I., Noble, W. S. and Nunn, B. L. (2020) Metaproteomics reveal that rapid perturbations in organic matter prioritize functional restructuring over taxonomy in western Arctic Ocean microbiomes. ISME J., 14, 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, M. A., Kujawinski, E. B., Schroer, W. F., Amin, S. A., Bates, N. R., Bertrand, E. R., Braakman, R., Brown, C. T.et al. (2022) Microbial metabolites in the marine carbon cycle. Nat. Microbiol., 7, 508–523. 10.1038/s41564-022-01090-3. [DOI] [PubMed] [Google Scholar]
- Nunn, B. L., Faux, J. F., Hippmann, A. A., Maldonado, M. T., Harvey, H. R., Goodlett, D. R., Boyd, P. W. and Strzepek, R. F. (2013) Diatom proteomics reveals unique acclimation strategies to mitigate Fe limitation. PLoS One, 8, e75653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, M. and Kuroda, S. (2019) Editorial overview: regulatory mechanism from multi-omic data. Curr. Opin. Syst. Biol., 15, iv–vi. 10.1016/j.coisb.2019.06.002. [DOI] [Google Scholar]
- Pino, L. K., Searle, B. C., Yang, H.-Y., Hoofnagle, A. N., Noble, W. S. and MacCoss, M. J. (2020) Matrix-matched calibration curves for assessing analytical figures of merit in quantitative proteomics. J. Proteome Res., 19, 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poceviciute, R. and Ismagilov, R. F. (2019) Human-gut-microbiome on a chip. Nat. Biomed. Eng., 3, 500–501. [DOI] [PubMed] [Google Scholar]
- Prairie, J. C., Sutherland, K. R., Nickols, K. J. and Kaltenberg, A. M. (2012) Biophysical interactions in the plankton: a cross-scale review. Limnol. Oceanogr. Fluids Environ., 2, 121–145. 10.1215/21573689-1964713. [DOI] [Google Scholar]
- Price, M. N., Wetmore, K. M., Waters, R. J., Callaghan, M., Ray, J., Liu, H., Kuehl, J. V., Melnyk, R. A.et al. (2018) Mutant phenotypes for thousands of bacterial genes of unknown function. Nature, 557, 503–509. 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- Qu, P.-P., Fu, F.-X., Wang, X.-W., Kling, J. D., Elghazzawy, M., Huh, M., Zhou, Q. Q., Wang, C.et al. (2022) Two co-dominant nitrogen-fixing cyanobacteria demonstrate distinct acclimation and adaptation responses to cope with ocean warming. Environ. Microbiol. Rep., 14, 203–217. 10.1111/1758-2229.13041. [DOI] [PubMed] [Google Scholar]
- Remmers, I. M., D’Adamo, S., Martens, D. E., deVos, R. C. H., Mumm, R., America, A. H. P., Cordewener, J. H. G., Bakkerb, L. V.et al. (2018) Orchestration of transcriptome, proteome and metabolome in the diatom Phaeodactylum tricornutum during nitrogen limitation. Algal Res., 35, 33–49. 10.1016/j.algal.2018.08.012. [DOI] [Google Scholar]
- Saito, M. A., Bertrand, E. M., Dutkiewicz, S., Bulygin, V. V., Moran, D. M., Monteiro, F. M., Follows, M. J., Valois, F. W.et al. (2011) Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc. Natl. Acad. Sci., 108, 2184–2189. 10.1073/pnas.1006943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, M. A., McIlvin, M. R., Moran, D. M., Santoro, A. E., Dupont, C. L., Rafter, P. A., Saunders, J. K., Kaul, D.et al. (2020) Abundant nitrite-oxidizing metalloenzymes in the mesopelagic zone of the tropical Pacific Ocean. Nat. Geosci., 13, 355–362. 10.1038/s41561-020-0565-6. [DOI] [Google Scholar]
- Sedwick, P. N., Bowie, A. R., Church, T. M., Cullen, J. T., Johnson, R. J., Lohan, M. C., Marsay, C. M., McGillicuddy, D. J.Jr.et al. (2020) Dissolved iron in the Bermuda region of the subtropical North Atlantic Ocean: seasonal dynamics, mesoscale variability, and physicochemical speciation. Mar. Chem., 219, 103748. 10.1016/j.marchem.2019.103748. [DOI] [Google Scholar]
- Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., Djahanschiri, B., Zeller, G.et al. (2015) Structure and function of the global ocean microbiome. Science, 348, 1261359. [DOI] [PubMed] [Google Scholar]
- Tang, W., Cerdán-García, E., Berthelot, H., Polyviou, D., Wang, S., Baylay, A., Whitby, H., Planquette, H.et al. (2020) New insights into the distributions of nitrogen fixation and diazotrophs revealed by high-resolution sensing and sampling methods. ISME J., 14, 2514–2526. 10.1038/s41396-020-0703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, Y., Choi, P. J., Li, G. -W., Chen, H., Babu, M., Hearn, J., Emili, A. and Xie, X. S. (2010) Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science, 329, 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toseland, A., Daines, S. J., Clark, J. R., Kirkham, A., Strauss, J., Uhlig, C., Lenton, T. M., Valentin, K.et al. (2013) The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Chang., 3, 979–984. [Google Scholar]
- Turk, K. A., Rees, A. P., Zehr, J. P., Pereira, N., Swift, P., Shelley, R., Lohan, M., Woodward, E. M. S.et al. (2011) Nitrogen fixation and nitrogenase (nifH) expression in tropical waters of the eastern North Atlantic. ISME J., 5, 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk-Kubo, K. A., Achilles, K. M., Serros, T. R. C., Ochiai, M., Montoya, J. P. and Zehr, J. P. (2013) Nitrogenase (nifH) gene expression in diazotrophic cyanobacteria in the Tropical North Atlantic in response to nutrient amendments. Front. Microbiol., 3, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldbauer, J. R., Rodrigue, S., Coleman, M. L. and Chisholm, S. W. (2012) Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS One, 7, e43432. 10.1371/journal.pone.0043432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth, N. G., Lee, M. D., Fu, F. X., Hutchins, D. A. and Webb, E. A. (2016) Molecular and physiological evidence of genetic assimilation to high CO2 in the marine nitrogen fixer Trichodesmium. Proc. Natl. Acad. Sci. U. S. A., 113, E7367–E7374. 10.1073/pnas.1605202113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisse, T. and Montagnes, D. J. S. (2021) Ecology of planktonic ciliates in a changing world: concepts, methods, and challenges. J. Eukaryot. Microbiol., 2021, e12879. 10.1111/jeu.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, S. W., King, A. L., Twining, B. S., LeCleir, G. R., DeBruyn, J. M., Strzepek, R. F., Breene, C. L., Pickmere, S.et al. (2013) Elemental quotas and physiology of a southwestern Pacific Ocean plankton community as a function of iron availability. Aquat. Microb. Ecol., 68, 185–194. [Google Scholar]
- Wu, M., McCain, J. S. P., Rowland, E., Middag, R., Sandgren, M., Allen, A. E. and Bertrand, E. M. (2019) Manganese and iron deficiency in Southern Ocean Phaeocystis antarctica populations revealed through taxon-specific protein indicators. Nat. Commun., 10, 3582. 10.1038/s41467-019-11426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr, J. P., Braun, S., Chen, Y. B. and Mellon, M. (1996) Nitrogen fixation in the marine environment: relating genetic potential to nitrogenase activity. J. Exp. Mar. Biol. Ecol., 203, 61–73. [Google Scholar]
- Zehr, J. P. and Capone, D. G. (2020) Changing perspectives in marine nitrogen fixation. Science, 368, eaay9514. 10.1126/science.aay9514. [DOI] [PubMed] [Google Scholar]
- Zehr, J. P. and Montoya, J. P. (2007) Measuring N2 fixation in the field. In Bothe, H., Ferguson, S. J., and Newton W. E. (eds) Biology of the Nitrogen Cycle. Elsevier, Netherlands, p. 193-205. 10.1016/B978-044452857-5.50014-X [DOI] [Google Scholar]