CONSPECTUS:
Tailored biomaterials with tunable functional properties are crucial for a variety of task-specific applications ranging from healthcare to sustainable, novel bio-nano-devices. To generate polymeric materials with predictive functional outcomes, exploiting designs from nature while morphing them towards non-natural systems offers an important strategy. Silks are Nature’s building blocks, that are produced by arthropods for a variety of uses that are essential for their survival. Due to the genetic control of encoded protein sequence, mechanical properties, biocompatibility, and biodegradability, silk proteins have been selected as prototype models to emulate for the tunable designs of biomaterial systems.
The bottom up strategy of material design opens important opportunities to create predictive functional outcomes, following the exquisite polymeric templates inspired by silks. Recombinant DNA technology provides a systematic approach to recapitulate, vary and evaluate the core structure peptide motifs in silks, and then biosynthesize silk-based polymers by design. Post biosynthesis processing allows for another dimension of material design by controlled or assisted assembly. Multiscale modeling, from the theoretical prospective, provides strategies to explore interactions at different length scales, leading to selective material properties. Synergy among experimental and modeling approaches can provide new and more rapid insights into the most appropriate structure-function relationships to pursue while also furthering our understanding in terms of the range of silk-based systems that can be generated. This approach utilizes nature as a blueprint for initial polymer designs with useful functions (e.g., silk fibers), but also employs modeling-guided experiments to expand the initial polymer designs into new domains of functional materials that do not exist in nature. The overall path to these new functional outcomes is greatly accelerated via the integration of modeling with experiment.
In this Account, we summarize recent advances in understanding and functionalization of silk-based protein systems, with a focus on the integration of simulation and experiment for biopolymer design. Spider silk was selected as an exemplary protein to address the fundamental challenges in polymer designs, including specific insights into the role of molecular weight, hydrophobic/hydrophilic partitioning, and shear stress for silk fiber formation. To expand current silk designs toward bio-interfaces and stimuli responsive materials, peptide modules from other natural proteins were added to silk designs to introduce new functions, exploiting the modular nature of silk proteins and fibrous proteins in general. The integrated approaches explored suggest that protein folding, silk volume fraction and protein amino acid sequence changes (e.g., mutations) are critical factors for functional biomaterial designs.
In summary, the integrated modeling-experimental approach described in this Account suggests a more rationally directed and more rapid method for the design of polymeric materials. It is expected that this combined use of experimental and computational approaches has a broad applicability not only for silk-based systems, but also for other polymer and composite materials.
Graphical Abstract
1. INTRODUCTION
The design and synthesis of new polymeric materials is a key for future needs in science and technology. To generate polymeric materials with predictive functional outcomes, exploiting designs from nature while morphing them towards non-natural systems offers an important strategy. Silks are exquisite polymeric material systems that have been optimized in nature through evolution, resulting in a balance of chain length, sequence chemistry and aqueous processing. Despite the relatively simple amino acid building blocks, materials generated from silk proteins exhibit remarkable mechanical properties.1-4 Silk based biomaterials are also useful in a wide range of applications.2,5-8 Therefore, silks have been selected as a prototype polymer model to emulate for the future material designs.9-12
Silks are highly modular protein polymers, with large internal repetitive sequences flanked by short non-repetitive N- and C-termini domains.13,14 Within the internal repetitive sequences, highly conserved poly-(Gly-Ala) and poly-Ala motifs form hydrophobic β-sheet crystalline domains, and the glycine-rich motifs form hydrophilic non-crystalline domains (e.g., random coil, helices, β-turn).3,5 These motifs self-assemble into hierarchical architecture, where stiff orderly crosslinked β-sheet nanocrystals are confined within an elastic semi-amorphous protein matrix, making silk fibers one of the toughest and most versatile materials known.15,16
The use of recombinant DNA technology to emulate and expand the modular silk templates provide new opportunities to build materials from the molecular level. In addition to altering sequence, a second level of control of material assembly is achieved via the use of post biosynthesis processing. Significant progress has been achieved over the past decade to address the fundamental challenges in biopolymer design and processing by using recombinant silk mimetic peptides, to provide specific insight into the roles of molecular weight,11,17 domain sizes and distributions,18-20 hydrophobic/hydrophilic partitioning,11 on protein self-assembly and the resulting mechanical properties.14,21,22 Importantly, recombinant DNA technology has also been used to construct silk biomaterials de novo through the addition of key modules from other proteins or functional groups for chemical and physical processing to introduce new functions, exploiting the modular nature of silk proteins. For example, silk-elastin-like proteins (SELPs) were developed as new polymers for targeted drug delivery23 and chemically modified into soft stimuli responsive hydrogel actuators.24 Silk-silica fusion proteins were designed as biomaterials for bone regeneration.25 However, conventional recombinant protein methods are limited by generally tedious and low-yielding cloning and biosynthesis methods because of the repetitive nature and length of the silk sequences. In addition, the serial design process, where new genetic/protein constructs are prepared and characterized in sequential fashion, also leads to a slower, trial-and-error process to reach specific functional goals for the materials. Therefore, there is an unmet need for developing new predictive tools, such as modeling, to accelerate the material development process.
Bottom-up multiscale material simulations provide information into the mechanisms behind changes in material properties starting with the interactions of atoms to the assembly of clusters of atoms based on primary sequence, chain folding and interactions, and processing condition. Integrating computational modeling at early stages of material design suggests a time- and cost-efficient solution to generate function from the molecular building blocks. A synergistic approach, which starts from a specific functional goal, combines inputs from both simulations and experiments, provides new insights into structure-function relationships11,13,14 and guides functional material designs.24,26 Such an approach is also amenable to combinatorial strategies and offers potential to serve as a roadmap for innovations in polymer, polymer alloy and polymer composite systems.
This Account summarizes recent advances in the understanding and functionalization of silk-based systems, with an emphasis on the power of integrated simulation and experimental approaches for biopolymer design. This review is based on previous work on silk-based systems that focused on the structure and biomedical applications of silks,2-8 and highlight more recent studies of novel silk designs achieved through a combined genetic engineering and computational modeling approach. Challenges and opportunities for this approach are also discussed in each section.
2. INTEGRATING EXPERIMENTAL APPROACHES AND MULTI-SCALE MODELING FOR BIOPOLYMER DESIGN
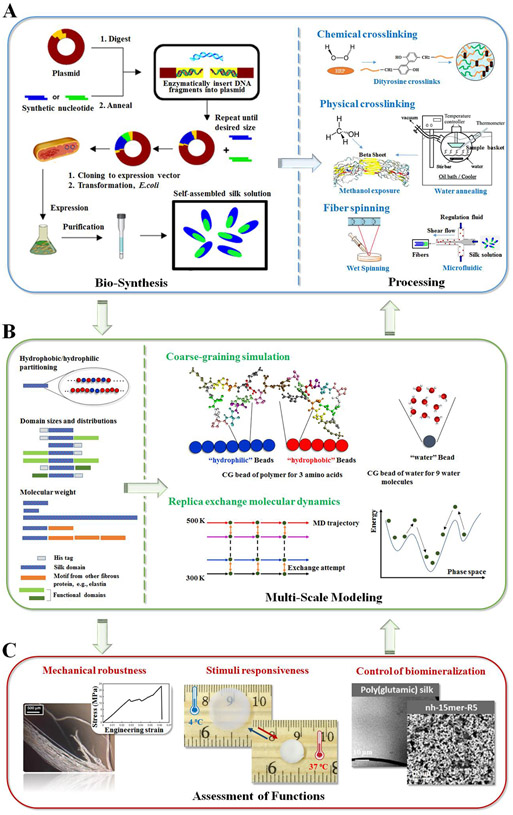
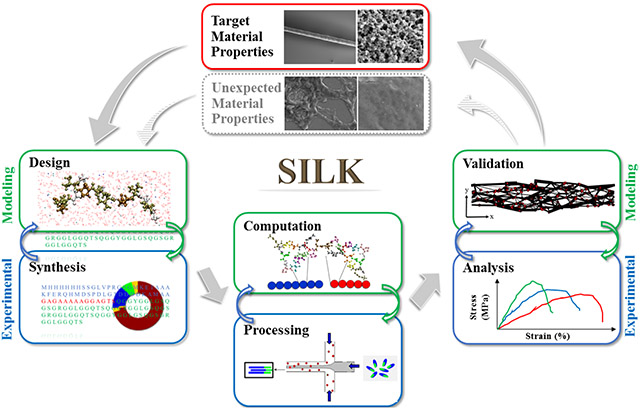
Developing fundamental tools and insight into biomaterial designs for predictive functional outcomes should be key to propelling polymer discovery forward. The integration of bio-synthesis, processing, multiscale modeling, and experimental validation provides a path towards the de novo design of silk-based materials with tailored properties (Figure 1). Each aspect of this process informs further understanding of material properties and also feeds into the other steps, towards a more optimized or predictable material outcome.
Figure 1.
Synergetic integration of genetic engineering, polymer processing, multi-scale modeling, and functional assessment towards the de novo design of new functional biomaterials. (A) General bio-synthesis scheme of genetically engineered proteins using recombinant DNA technology, and common polymer processing methods of protein-based biomaterials using chemical/physical crosslinking approaches and shear flow spinning process. (B) Multi-scale modeling, illustrated by coarse-grained simulation and replica exchange molecular dynamics modeling, bridges the gap between molecular design, processing conditions and target macroscopic physical properties. (C) Assessment of functions provide a feedback loop to revise the simulation models during the iteration. Adapted with permission from ref. 11, 24, 26, and 27. Copyright 2015 Nature Publishing Group, 2011 American Chemical Society, 2016 WILEY-VCH, and 2016 American Chemical Society.
2.1. BIO-SYNTHESIS OF SILKS
As summarized in Figure 1A, the main steps to generate recombinant silks by design are gene design, cloning, expression, and protein purification. A variety of cloning strategies have been used towards this goal, including step by step directional ligation and recursive directional ligation for the production of new silk-related genes with control of size,28 and concatemerization for the construction of gene libraries.29 The main purification method used for recombinant silk proteins has been Ni-NTA affinity chromatography,11 while inverse temperature cycling is preferred for some silk constructs with elastin domains.24 The key advantages of genetically engineered silk proteins include the tailorability of sequence, versatility in protein chemistry, and the control of protein size. Recombinant DNA methods provide tools to study sequence-function relationships, yet remain limiting when generating high molecular weight silk, when cloning the full-length silk genes, or when large-scale production of silk proteins is needed.17
2.2. POST BIOSYNTHESIS PROCESSING
As summarized in Figure 1A, Microfluidic14 and wet spinning techniques11, which mimic the natural silk spinning process, have been exploited to fabricate recombinant silk fibers and further the understanding of the natural assembly process. Chemical and physical modification, including methanol vapor treatment and water annealing for the induction of β-sheet secondary structures, and enzymatic crosslinking for the formation of dityrosine networks, have also been utilized to facilitate the assembly of the polymer chains into defined secondary structures or polymer networks for enhanced material properties.24,26 Modifying silk processing parameters, together with protein sequence alterations, allows for the exploration of a large design space to develop new functional polymeric materials.
2.3. MULTISCALE MODELING AND VALIDATION
As summarized in Figure 1B, Common approaches to model material features include molecular dynamics (MD) for small peptides and accelerated sampling methods such as replica exchange molecular dynamics (REMD) for protein polymers and multi-molecular systems on the nanoscale,30 coarse-grained modeling (CG) on the mesoscale,11,24,26 and continuum methods on larger scales. Implicit solvent temperature replica exchange molecular dynamics followed by explicit solvent conventional molecular dynamics have been used to investigate dynamics and structural properties of the systems with atomic resolution.24,26 New mesoscale techniques have also been developed to access larger temporal and spatial scales, which currently exceed the computational capacity of full atomistic molecular dynamics simulations. Classical dissipative particle dynamics (DPD) was used, with two new terms added that describe the hydrogen bonds and covalent bonds in polymer chains. The LAMMPS code was modified to include the new terms in the integration of classical DPD simulations. This new coarse-grained model allows for the simulation of soft biopolymers such as silk proteins and captures the formation of micelles and polymer networks.11 Multiscale modeling provides a useful guide to describe material features starting from fundamental laws of physics, yet it is currently not possible to study complex materials with a single computational method on all scales simultaneously. There are efforts to propose new modeling schemes with tunable resolution to balance accuracy and efficiency,31 but these approaches are not yet in routine use and require further development and testing. New sampling methods, such as replica exchange with solute tempering,32 software developments, and hardware to facilitate high performance computing and data storage help to improve the computational capacity and utility of simulations.
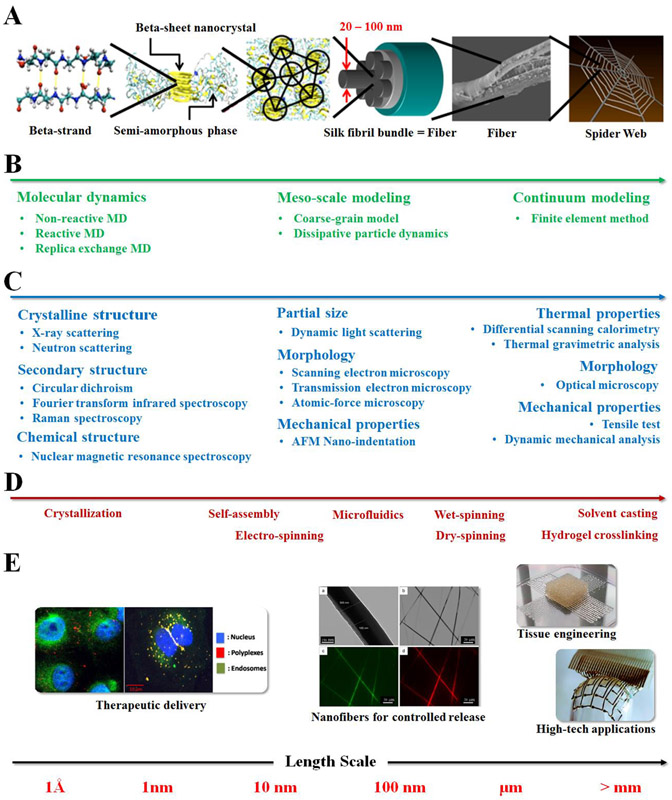
As summarized in Figure 1C, Experimental characterization, coupled with simulation, are routinely used to validate outcomes and provide feedbacks to revise current simulation models at multiple scales. Common experimental methods that are used to assess the protein properties and validate the simulation outcomes are listed in Figure 2. At atomic or nanoscale, experimental methods that are used to measure crystalline, secondary or chemical structures provide information to identify atomic properties.11,13 At the larger scales, experimental methods that measure thermal, optical and mechanical properties provide information about the macroscopic physical properties.13,24 These experimental results served as a feedback loop to revise the simulation models during the iteration.
Figure 2.
Experiment and computational methods to study the hierarchical structure and function of silk biopolymers at different length scales. (A) Schematic of the hierarchical spider silk structure that ranges from molecular to macroscopic scales. The hydrogen bonded β-strands at the angstrom level create hetero-nanocomposite of β-sheet nanocrystals embedded in soft semi-amorphous matrix at the nano-meter length scale, which self-assemble into silk fiber at the micron scale to create spider orb web structure. (B) Theoretical and simulation methods for self- and assisted- assembly mechanisms per the length scale they address. (C) Analytical characterization tools for resolving the physical properties of silk proteins at different length scale. (D) Processing methods for the formation of the hierarchical structure on different length scales. These methods control the resulting material structure and properties, and lead to (E) various biomedical and technology applications of silk proteins from nano to macroscopic scale. Adapted with permission from ref. 33-37. Copyright 2010 Nature Publishing Group, 2014 WILEY-VCH, 2015 American Chemical Society, and 2016 MDPI AG.
As shown in the examples below, polymer synthesis, here in the form of recombinant DNA technology, is used to modulate the sequence chemistry on the molecular and nanoscale, which in turn has a major impact on higher level structures and final material properties. Polymer processing controls the environmental conditions to influence the assembly of polymer chains into defined secondary structures at the nanoscale and the formation of polymer networks at the macroscale and mesoscale. Computational modeling, together with experimental validation, guides the biopolymer design process by simulating structures and assembly at different length scales. Interactive steps involving experimental validation and modeling also provide a chance to revise the models during the iteration. This synergistic integration of genetic engineering, simulation and experiment provide a facile approach to modulate biopolymers at the nanoscale and control their function utility at the microscopic and macro scales.
EXAMPLE 1: UNDERSTANDING SPIDER SILK DESIGNS FOR MECHANICALLY ROBUST BIOPOLYMERS
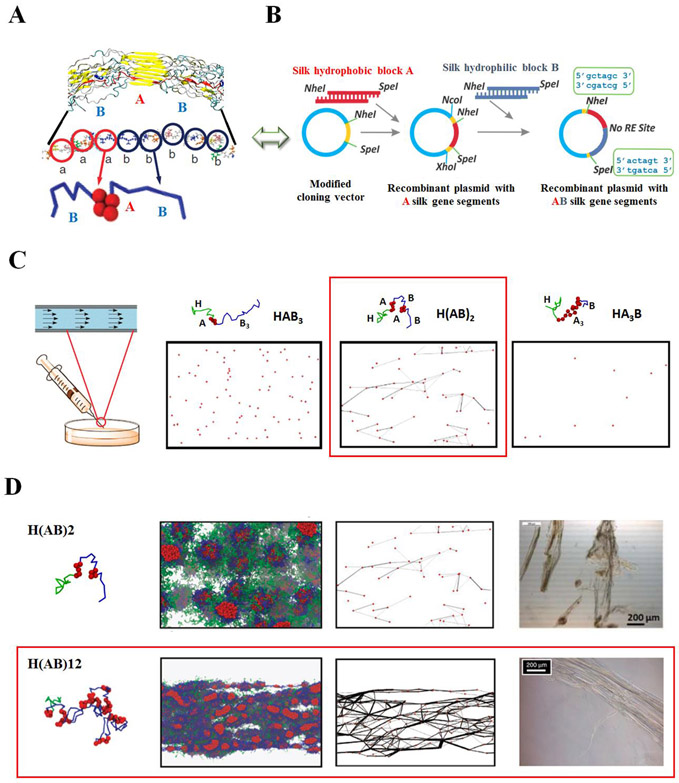
Considering the remarkable mechanical properties of silks, an important challenge remains: the mass production of spider silks and related biomimetic versions. Recombinant DNA approaches provide an alternative route to spider silk synthesis, and give a starting point for developing the fundamental insights into spider silk sequence design, self-assembly mechanisms, and fiber spinning process. To understand sequence-function relationships and harness the self-assembly process for mechanically robust silk-based materials, recombinant spider silks with various molecular weight, domain sizes, domain distributions, and hydrophobic-hydrophilic domain ratios were bio-synthesized via genetic engineering.11,18-22,28,38 The sequence features of the recombinant spider silks were adapted from the representative sequence of MaSp1 in the spider dragline silk of Nephila clavipes, including hydrophobic A domains, GAGAAAAAGGAGTS, and hydrophilic B domains, QGGYGGLGSQGSGRGGLGGQTS.11,18-22,28,38 The recombinant spider silk sequences followed generic design templates, e.g., (ApBq)n, and the secondary structure analysis of recombinant spider silks revealed that the hydrophobic A domains were responsible for the formation of β-sheet crystalline regions, and the hydrophilic B domains were responsible for the formation of the non-crystalline regions.18,38 Studies on the self-assembly of recombinant spider silks revealed that the hydrophobic-hydrophilic domain ratio impacted the formation of micelle structures in solution.20,21,28 Studies of solution parameters suggested that environmental factors, such as pH and ion concentration (e.g., sulfate concentration), influenced the self-assembly of the recombinant spider silks.22
The integration of scalable modelling, biosynthesis and processing also added new insight into the formation of robust silk fibers with a Young’s modulus of 1-8 GPa, similar to native spider silk fibers. This combination approach suggested that a large volume fraction of the hydrophobic A domain in recombinant spider silks tended to have a higher β-sheet content for high mechanical stiffness, and also tended to self-associate rather than forming intermolecular structures necessary for fiber formation.14 Using modified DPD simulations, combined with biomimetic spinning and experimental validation, the results from these studies also suggested that: (i) intermediate ratios of hydrophobic to hydrophilic domains at a 1:1 ratio resulted in well-connected networks, (ii) shear flow increased the connectivity of the polymer networks which translated to improved silk fiber formation, and (iii) stronger polymer networks were formed when longer polymer chains (larger n value) were presented (Figure 3).11 With the above examples of integrating genetic engineering, experiment, processing and modeling, we demonstrate a synergistic path toward more predictable material outcomes, in this case mechanics.
Figure 3.
Integrated modeling and experimental approaches for the production of robust recombinant spider silk fibers. Schematic of (A) coarse-graining simulation, (B) recombinant DNA strategy and (C) processing method (e.g., wet spinning) used to study spider silk block copolymers. (C) Coarse-grained representations, and the mesoscopic dissipative particle dynamics (DPD) simulation node-bridge diagram under the shear flow of the recombinant spider silk peptides: HAB3, H(AB)2 and HA3B. Simulation suggested that under shear flow spinning process, only the H(AB)2 sequence (highlighted in red box) with alternating hydrophobic/hydrophilic domains at a 1:1 ratio promote network formation over other domain distributions. (D) Coarse-grained representations, mesoscopic DPD simulation snapshots and the node-bridge diagram under shear flow, and the bright-field microscopic images of spun fibers in bundles in the coagulation bath of the recombinant spider silk peptides: H(AB)2 and H(AB)12. As length is increased from H(AB)2 to H(AB)12, mature networks for robust spider silk fibers formed in H(AB)12 (highlighted in red box) under shear flow. Red beads - hydrophobic ‘A’ beads in the ‘A’ domain, blue lines - hydrophilic ‘B’ beads in the ‘B’ domain and green lines - hydrophilic ‘B’ beads in the ‘H’ domain. Water beads are not shown for clarity. Adapted with permission from ref. 11 and 38. Copyright 2014 Elsevier Inc., and 2015 Nature Publishing Group.
Example 2: Optimization of silk designs for biomaterial interfaces: silk mineralization-peptide fusion proteins
Organic-inorganic interfaces are integral to biomaterial functions in many areas of tissue repair and regeneration.39,40 Fundamental insight into interfacial mineralization of proteins related to calcification of medical materials would permit insights into the fine-tuning of biomaterials to either promote the formation of inorganic-organic hybrid materials as a route to stiffer and stronger materials for bone regeneration or to prevent the mineralization to sustain flexible and dynamic material functions, such as for heart valves and blood vessels.11,24,26
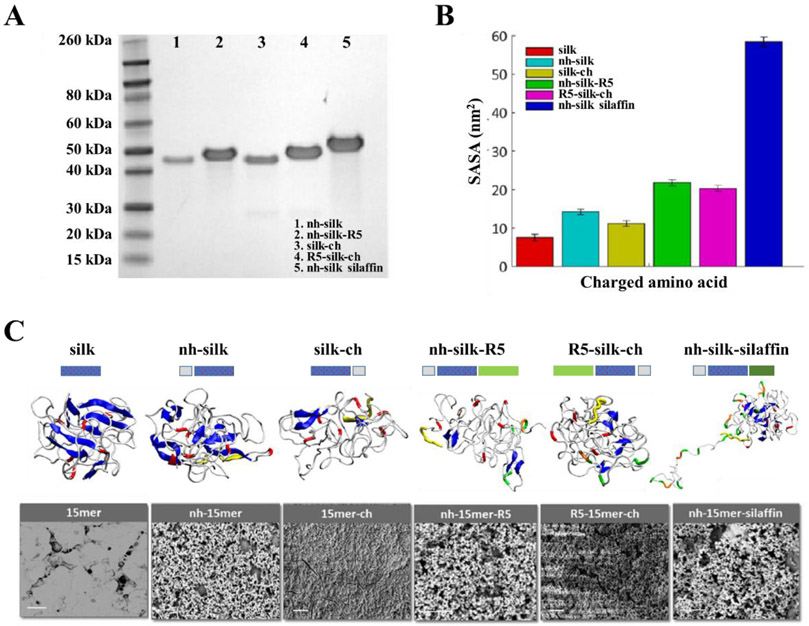
To address the challenge of biomineralization interfaces and the needs derived from hardened/stiffer biomaterial systems, silks were modified via genetic engineering with different functional domains to control biomineralization (Figure 4).41,42 A number of key features were elucidated from these studies, including domain types and distributions related to mineralization, combined with the mechanical properties of the silks where the presence of charged termini on silk protein assembly improved mechanical/functional properties. For instance, the silica-binding peptide R5 (SSKKSGSYSGSKGSKRRIL) derived from Cerithiopsis fusiformis silaffin gene was fused to the N- or C-termini of the silk polymer, (SGRGGLGGQGAGAAAAAGGAGQGGYGGLGSQGT)15, derived from the consensus repeat of N. clavipes dragline silk protein.43 Genetically engineered constructs (nh-silk, nh-silk-R5, silk-ch , R5-silk-ch and nh-silk-sillafin, where nh and ch stand for N- or C-termini fused histidine tag), were produced and processing parameters (solvating agent, β-sheet induction method, temperature) that affected protein secondary structure were defined.26 The presence of positively charged amino acids was essential for supporting biomineralization of the silk fusion proteins, however the process was not amino acid specific. Moreover, the location of the charged domain involved in the biomineralization in the fusion protein affected protein folding and consequently surface exposure of the charged amino acids.26 In another example, artificial silk block copolymers were decorated with a VTKHLNQISQSY (VTK) domain, identified via phage display with preferential adsorption on bone-like mineral and hydroxyapatite, where the silk domain was critical to the material properties and the VTK domain for biomineralization.44,45 Understanding the mechanism by which these key factors impact biomineralization allows for the rational design of silk-based materials to generate new inorganic/organic hybrid systems for biomedical applications.
Figure 4.
Integrated modeling and experimental approach for the design of biomineralization interfaces. (A) SDS-PAGE of recombinant silk-silica fusion proteins: nh-silk (40 kDa), nh-silk-R5 (43 kDa), silk-ch (40 kDa), R5-silk-ch (43 kDa) and nh-silk-sillafin (55 kDa). (B) Solvent accessible surface area (SASA) of positively charged amino acids in recombinant silk-silica fusion proteins. (C) Schematics of the silk-silica fusion protein designs, REMD simulation of protein folding, and SEM evaluation of the ability to induce silica formation. Correlation between REMD snapshot and SEM images suggested that the folding and exposure of charged functional groups played key roles in biomineralization. Peptide design: his-tag - grey box, spider silk polymer – dotted blue box, and R5 domain - green box. REMD snapshot: β-sheets – blue, random coils - white, histidine - yellow, arginine (in silk domain) - red, arginine (in R5 binding peptide) - orange, and lysine - green. SEM image scale bars are 10 μm. Adapted with permission from ref. 26. Copyright 2016 American Chemical Society.
The integrated modeling-experimental approach allowed for the efficient identification of key parameters in material design to control function, here related to mineralization. Through the integration of modeling, insight was gained into key factors such as protein folding, exposure and alignment of charged units, fostering the fine-tuning of protein sequence and processing parameters to support biomineralization. The computational procedure started with initial predictions of the protein structure from homology modeling. Replica exchange simulations in implicit solvents (water and ethanol) were performed to identify protein folding at ambient (300 K) temperature.26 For each case, the most probable structure was selected using a single linkage clustering algorithm. The representative structures were then refined in explicit solvents and average properties were reported from the last part of the simulation. The results showed that the extent of silicification in the experiments was correlated with the amount of solvent accessible surface area of positively charged amino acids in the folded-assembled structures.
Example 3: Extending silk designs towards dynamic materials: silk-elastin-like proteins
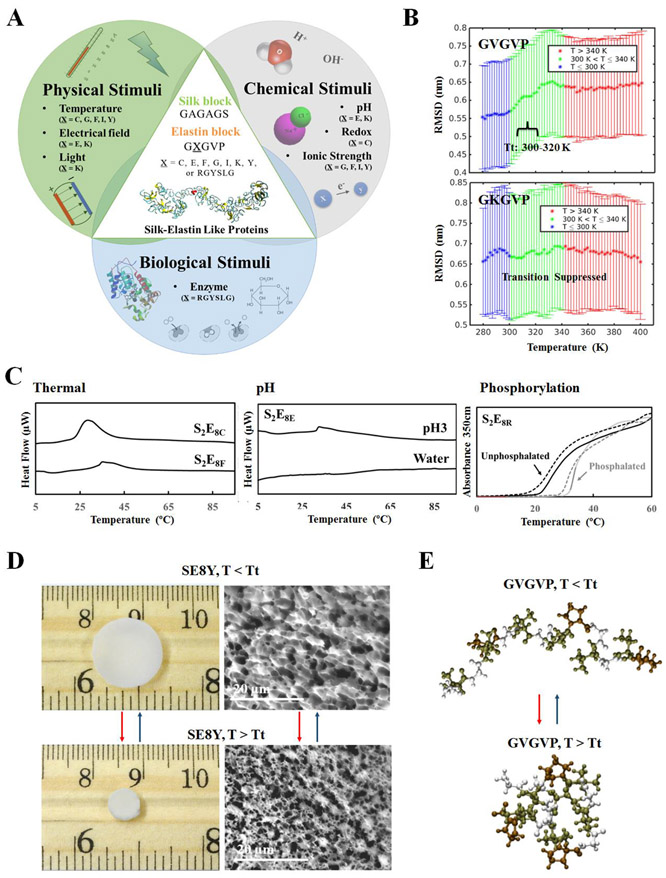
Their impressive material properties render silks as ideal candidate materials for responsive composites. By this motivation, we examine silk-elastin composite materials for controlled mutability design. The representative sequence of Bombyx mori silkworm silk, GAGAGS, was fused with the elastin-like peptide, GXGVP (where X is an interchangeable amino acid), via genetic engineering, to encode and then express silk-elastin-like proteins (SELPs) as templates for stimuli-responsive materials. The robust mechanical properties inherent to silk and the dynamic nature originating from the elastin domains formed the basis for these designs. 46,47
A series of SELPs with different silk-to-elastin ratios,24,47,48 molecular weights29 and protein chemistry24,29,49 were constructed to address sequence-structure-function relationships and control of dynamic properties (Figure 5). The study of self-assembly mechanisms of SELP micelles for drug delivery revealed that the incorporation of a silk domain into SELP facilitated micelle formation.47 Chemical modification of SELPs with retinal for photo-responsive biopolymers suggested that the protein chemistry in the elastin domain can be exploited to expand SELP constructs for new and specific functional responses.50 Actuating properties of SELP hydrogels revealed that elastin chain folding-unfolding at the molecular level during inverse temperature transition (Tt) could be translated into macroscopic reversible materials where the physical properties changed based on crosslinking the elastin domain.24 Studies of the responsive properties of SELP solutions and SELP hydrogels suggested that incorporation of different guest residues in the X position of the elastin domain modulated the protein chemistry and shifted the inverse transition temperature as a function of the silk-to-elastin ratio, chain length, protein concentration and environmental factors, expanding the thermal sensitivity of these materials to a range of other stimuli-responsive properties, including pH, ionic strength, electric field, and enzymatic (phosphorylation) responses.24,29 Though these studies provided further insight into sequence-function relationships of SELPs, fine-tuning of the inverse temperature transitions of SELPs per functional need remains a key issue to be resolved for dynamic biomaterial designs.
Figure 5.
Integrated modeling and experimental approach for the design of stimuli-responsive biomaterials. (A) Schematic of the sequence dependent stimuli-responsive features of SELPs. By varying position “X” amino acid in the elastin domains, SELPs can be designed to predictably respond to physical stimuli (e.g., temperature, electric field, light), chemical stimuli (e.g. pH, redox state, ionic strength), and biological stimuli (e.g., enzymatic triggers). (B) Root mean square deviation (RMSD) values for the elastin domain (GVGVP)(GXGVP)(GVGVP) from REMD simulations. RMSD values for X = V indicate a transition from extended to folded conformations. RMSD values for the X = K sequence suggest a suppressed transition in the elastin domain. The analysis suggests that the inverse temperature transition is governed by the “X” residue in elastin domain. (C) Differential scanning calorimetry and turbidity profiles of SELP solution demonstrated that SELPs can be designed to respond to a variety of stimuli (e.g., temperature, pH and phosphorylation). (D) Optical and SEM images of SELP, below and above Tt, exemplified by SE8Y with sequence of [(GAGAGS)(GVGVP)4(GYGVP)(GVGVP)3]14. Result demonstrated that SELPs can be fabricate into hydrogel for stimuli-responsive actuators. (E) Snapshots of the elastin domain (GVGVP)(GXGVP)(GVGVP), from REMD simulation, below and above Tt. An extended conformation is observed below Tt and a folded configuration is present above Tt. Adapted with permission from ref. 24. Copyright 2016 WILEY–VCH.
The incorporation of computational modeling provides a useful tool to reduce the trial-and-error outcomes and accelerate the rational design of stimuli-responsive materials to meet specific functional goals. Multiscale molecular modeling has been employed for the study of elastin-like peptides (ELPs),51 tropoelastin,52 silk protein53 and most recently SELPs. Drawing inspiration from ELP and silk models, SELP models were created to combine the unique mechanical and stimuli-sensitive characteristics of the two main component parts. These models describe molecular mechanisms of structural transitions and shifts in mechanical properties in response to temperature. An important future direction is the scale-up of the phase transition effects to macroscales to guide future design of novel stimuli-responsive SELPs.
6. CONCLUDING REMARKS AND PROSPECTS
Tailored biomaterials with tunable functional properties are desirable for a variety of task-specific applications ranging from drug delivery, tissue engineering, dynamic biomaterial implants, material coatings, components for robotic devices, and implantable devices, among others. Silks, due to the genetic basis of encoded protein sequence control, mechanical properties, biocompatibility and biodegradability, were selected as a prototype model to emulate in the pursuit of the de novo design of functional biomaterials to bridge experimental and modeling strategies in unison. In this Account, we demonstrated the power of the integrated simulation and experimental approaches on biopolymer design through three prominent examples: (i) spider silks to understand sequence-function relationships for mechanically robust biopolymer designs, (ii) silk-silica fusion proteins for functionalization of silk system for biomineralization interfaces, and (iii) silk-elastin-like proteins for stimuli responsive, shape-changing materials. The integrated modeling-experimental approach elucidated several key parameters to address the fundamental challenges in biopolymer designs, including specific insights into the role of molecular weight, hydrophobic/hydrophilic partitioning, and shear stress for silk fiber formation, as well as on protein folding, silk volume fraction and controlling mutability for functional material designs. With the above examples of integrating genetic engineering, experiment, processing and modeling, synergy was demonstrated for improved predictive outcomes for the materials.
While silk-mimetic peptides suggested useful insights for predicting function from the molecular building blocks, the mechanical properties of these silks remain inferior to the native materials, in part due to low molecular weight, and the truncation of some critical domains such as the N- and C-termini. The integrated approach to generate predictive outcomes for full length silks, which has not yet been realized, combined with synergistic feedback loops of experiment and computation, should advance the understanding of basic mechanisms and trigger more optimized material designs to address current limitations.
Understanding the assembly of amino acid building blocks into multi-functional structures remains in its infancy. The integrated experimental and modeling approach provides new insights into structure-function relationships. Further understanding and expansion of this approach to add functionality to silk-based materials systems, including copolymers and composites as summarized earlier, will continue to propel utility for these methods. The approach also offers significant implications for other protein materials and eventually nonprotein polymer systems.
ACKNOWLEDGMENTS
The authors acknowledge the financial support of the NIH (U01 EB014976), Tissue Engineering Resource Center (NIH P41 EB002520), ONR (N00014-16-1-2333) and AFOSR (FA9550-11-1-0199).
Biographies
David Kaplan is the Stern Family Endowed Professor of Engineering and Chair of the Department of Biomedical Engineering at Tufts University. His research focus is on biopolymer engineering to understand structure-function relationships, with emphasis on studies related to self-assembly, biomaterials engineering and regenerative medicine.
Markus J. Buehler is McAfee Professor of Engineering and Department Head in the Department of Civil and Environmental Engineering at the Massachusetts Institute of Technology, where he directs the Laboratory for Atomistic and Molecular Mechanics (LAMM). His research focuses on bottom-up modeling of structural and mechanical properties of biological, bioinspired and synthetic materials across multiple scales.
Joyce Y. Wong is a Professor in the Department of Biomedical Engineering and Division of Materials Science and Engineering at Boston University. Her research focuses on using a biomaterials science and engineering approach to develop model systems to understand processes during normal tissue development and disease.
Wenwen Huang is a postdoctoral researcher in Prof. David Kaplan’s group in the Department of Biomedical Engineering at Tufts University. Her research interests include genetic engineering hierarchical and stimuli-responsive proteins, physical and chemical modification of protein-based materials, as well as protein self-assembly mechanism.
Davoud Ebrahimi is a postdoctoral research associate at LAMM, MIT. His research interests include molecular simulation, multiscale modeling, biotechnology and advanced material design.
Nina Dinjaski is a postdoctoral researcher in Prof. David Kaplan’s group in the Department of Biomedical Engineering at Tufts University. Her research interests include genetic engineering strategies to functionalize biomaterials for medical applications.
Anna Tarakanova is a PhD student with Markus J. Buehler at LAMM, MIT. Her research interests include molecular and multiscale modeling of biomaterials, and novel materials design.
REFERENCES
- (1).Blackledge TA; Perez-Rigueiro J; Plaza GR; Perea B; Navarro A; Guinea GV; Elices M: Sequential origin in the high performance properties of orb spider dragline silk. Sci. Rep 2012, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Omenetto FG; Kaplan DL: New Opportunities for an Ancient Material. Science 2010, 329, 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Lewis RV: SPIDER SILK - THE UNRAVELING OF A MYSTERY. Acc. Chem. Res 1992, 25, 392–398. [Google Scholar]
- (4).Shao Z; Vollrath F: Materials: Surprising strength of silkworm silk. Nature 2002, 418, 741–741. [DOI] [PubMed] [Google Scholar]
- (5).Vepari C; Kaplan DL: Silk as a biomaterial. Prog. Polym. Sci 2007, 32, 991–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kundu B; Rajkhowa R; Kundu SC; Wang XG: Silk fibroin biomaterials for tissue regenerations. Adv. Drug Delivery Rev 2013, 65, 457–470. [DOI] [PubMed] [Google Scholar]
- (7).Wenk E; Merkle HP; Meinel L: Silk fibroin as a vehicle for drug delivery applications. J. Controlled Release 2011, 150, 128–141. [DOI] [PubMed] [Google Scholar]
- (8).Hwang S-W; Tao H; Kim D-H; Cheng H; Song J-K; Rill E; Brenckle MA; Panilaitis B; Won SM; Kim Y-S; Song YM; Yu KJ; Ameen A; Li R; Su Y; Yang M; Kaplan DL; Zakin MR; Slepian MJ; Huang Y; Omenetto FG; Rogers JA: A Physically Transient Form of Silicon Electronics. Science 2012, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wong JY; McDonald J; Taylor-Pinney M; Spivak DI; Kaplan DL; Buehler MJ: Materials by Design: Merging Proteins and Music. Nano Today 2012, 7, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Buehler MJ: Materials by Design-A Perspective From Atoms to Structures. MRS Bull. 2013, 38, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lin S; Ryu S; Tokareva O; Gronau G; Jacobsen MM; Huang W; Rizzo DJ; Li D; Staii C; Pugno NM; Wong JY; Kaplan DL; Buehler MJ: Predictive modelling-based design and experiments for synthesis and spinning of bioinspired silk fibres. Nat. Commun 2015, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ebrahimi D; Tokareva O; Rim NG; Wong JY; Kaplan DL; Buehler MJ: Silk–Its Mysteries, How It Is Made, and How It Is Used. ACS Biomater. Sci. Eng 2015, 1, 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gronau G; Krishnaji ST; Kinahan ME; Giesa T; Wong JY; Kaplan DL; Buehler MJ: A review of combined experimental and computational procedures for assessing biopolymer structure-process-property relationships. Biomaterials 2012, 33, 8240–8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Krishnaji ST; Bratzel G; Kinahan ME; Kluge JA; Staii C; Wong JY; Buehler MJ; Kaplan DL: Sequence-Structure-Property Relationships of Recombinant Spider Silk Proteins: Integration of Biopolymer Design, Processing, and Modeling. Adv. Funct. Mater 2013, 23, 241–253. [Google Scholar]
- (15).Su I; Buehler MJ: Nanomechanics of silk: the fundamentals of a strong, tough and versatile material. Nanotechnology 2016, 27, 15. [DOI] [PubMed] [Google Scholar]
- (16).Tarakanova A; Buehler MJ: A Materiomics Approach to Spider Silk: Protein Molecules to Webs. JOM 2012, 64, 214–225. [Google Scholar]
- (17).Xia XX; Qian ZG; Ki CS; Park YH; Kaplan DL; Lee SY: Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 14059–14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Huang WW; Krishnaji S; Hu X; Kaplan D; Cebe P: Heat Capacity of Spider Silk-Like Block Copolymers. Macromolecules 2011, 44, 5299–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Huang W; Krishnaji S; Tokareva OR; Kaplan D; Cebe P: Influence of Water on Protein Transitions: Thermal Analysis. Macromolecules 2014, 47, 8098–8106. [Google Scholar]
- (20).Huang W; Krishnaji S; Tokareva OR; Kaplan D; Cebe P: Influence of Water on Protein Transitions: Morphology and Secondary Structure. Macromolecules 2014, 47, 8107–8114. [Google Scholar]
- (21).Krishnaji ST; Huang WW; Rabotyagova O; Kharlampieva E; Choi I; Tsukruk VV; Naik R; Cebe P; Kaplan DL: Thin Film Assembly of Spider Silk-like Block Copolymers. Langmuir 2011, 27, 1000–1008. [DOI] [PubMed] [Google Scholar]
- (22).Krishnaji ST; Huang W; Cebe P; Kaplan DL: Influence of Solution Parameters on Phase Diagram of Recombinant Spider Silk-Like Block Copolymers. Macromol. Chem. Phys 2014, 215, 1230–1238. [Google Scholar]
- (23).Huang W; Rollett A; Kaplan DL: Silk-elastin-like protein biomaterials for the controlled delivery of therapeutics. Expert Opin. Drug Delivery 2015, 12, 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Huang WW; Tarakanova A; Dinjaski N; Wang Q; Xia XX; Chen Y; Wong JY; Buehler MJ; Kaplan DL: Design of Multistimuli Responsive Hydrogels Using Integrated Modeling and Genetically Engineered Silk-Elastin-Like Proteins. Adv. Funct. Mater 2016, 26, 4113–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhou S; Huang W; Belton DJ; Simmons LO; Perry CC; Wang X; Kaplan DL: Control of silicification by genetically engineered fusion proteins: Silk-silica binding peptides. Acta Biomater. 2015, 15, 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dinjaski N; Ebrahimi D; Ling S; Shah S; Buehler MJ; Kaplan DL: Integrated Modeling and Experimental Approaches to Control Silica Modification of Design Silk-Based Biomaterials. ACS Biomater. Sci. Eng 2016, Article ASAP, DOI: 10.1021/acsbiomaterials.6b00236. [DOI] [PubMed] [Google Scholar]
- (27).Hu X; Shmelev K; Sun L; Gil E-S; Park S-H; Cebe P; Kaplan DL: Regulation of Silk Material Structure by Temperature-Controlled Water Vapor Annealing. Biomacromolecules 2011, 12, 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Rabotyagova OS; Cebe P; Kaplan DL: Self-Assembly of Genetically Engineered Spider Silk Block Copolymers. Biomacromolecules 2009, 10, 229–236. [DOI] [PubMed] [Google Scholar]
- (29).Wang Q; Xia X; Huang W; Lin Y; Xu Q; Kaplan DL: High Throughput Screening of Dynamic Silk-Elastin-Like Protein Biomaterials. Adv. Funct. Mater 2014, 24, 4303–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chang DK; Urry DW: Molecular-Dynamics Calculations on Relaxed and Extended States of the Polypentapeptide of Elastin. Chem. Phys. Lett 1988, 147, 395–400. [Google Scholar]
- (31).Kirschner DE; Hunt CA; Marino S; Fallahi-Sichani M; Linderman JJ: Tuneable resolution as a systems biology approach for multi-scale, multi-compartment computational models. Wiley Interdiscip. Rev. Syst. Biol. Med 2014, 6, 225–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wang LL; Friesner RA; Berne BJ: Replica Exchange with Solute Scaling: A More Efficient Version of Replica Exchange with Solute Tempering (REST2). J. Phys. Chem. B 2011, 115, 9431–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Keten S; Xu Z; Ihle B; Buehler MJ: Nanoconfinement controls stiffness, strength and mechanical toughness of beta sheet crystals in silk. Nat. Mater 2010, 9, 359–367. [DOI] [PubMed] [Google Scholar]
- (34).Yigit S; Tokareva O; Varone A; Georgakoudi I; Kaplan DL: Bioengineered Silk Gene Delivery System for Nuclear Targeting. Macromol. Biosci 2014, 14, 1291–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhu J; Huang W; Zhang Q; Ling S; Chen Y; Kaplan LD: Aqueous-Based Coaxial Electrospinning of Genetically Engineered Silk Elastin Core-Shell Nanofibers. Materials 2016, 9, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jose RR; Brown JE; Polido KE; Omenetto FG; Kaplan DL: Polyol-Silk Bioink Formulations as Two-Part Room-Temperature Curable Materials for 3D Printing. ACS Biomater. Sci. Eng 2015, 1, 780–788. [DOI] [PubMed] [Google Scholar]
- (37).Kim DH; Viventi J; Amsden JJ; Xiao JL; Vigeland L; Kim YS; Blanco JA; Panilaitis B; Frechette ES; Contreras D; Kaplan DL; Omenetto FG; Huang YG; Hwang KC; Zakin MR; Litt B; Rogers JA: Dissolvable films of silk fibroin for ultrathin conformal bio-integrated electronics. Nat. Mater 2010, 9, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tokareva OS; Lin S; Jacobsen MM; Huang W; Rizzo D; Li D; Simon M; Staii C; Cebe P; Wong JY; Buehler MJ; Kaplan DL: Effect of sequence features on assembly of spider silk block copolymers. J. Struct. Biol 2014, 186, 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Shin H; Jo S; Mikos AG: Biomimetic materials for tissue engineering. Biomaterials 2003, 24, 4353–4364. [DOI] [PubMed] [Google Scholar]
- (40).Hubbell JA: Bioactive biomaterials. Curr. Opin. Biotechnol 1999, 10, 123–129. [DOI] [PubMed] [Google Scholar]
- (41).Plowright R; Dinjaski N; Zhou S; Belton DJ; Kaplan DL; Perry CC: Influence of silk-silica fusion protein design on silica condensation in vitro and cellular calcification. RSC Adv. 2016, 6, 21776–21788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Foo CWP; Patwardhan SV; Belton DJ; Kitchel B; Anastasiades D; Huang J; Naik RR; Perry CC; Kaplan DL: Novel nanocomposites from spider silk-silica fusion (chimeric) proteins. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 9428–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Plowright R; Dinjaski N; Zhou S; Belton DJ; Kaplan DL; Perry CC: Influence of silk-silica fusion protein design on silica condensation and cellular calcification. RSC Adv. 2016, 6, 21776–21788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Addison WN; Miller SJ; Ramaswamy J; Mansouri A; Kohn DH; McKee MD: Phosphorylation-dependent mineral-type specificity for apatite-binding peptide sequences. Biomaterials 2010, 31, 9422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Segvich SJ; Smith HC; Kohn DH: The adsorption of preferential binding peptides to apatite-based materials. Biomaterials 2009, 30, 1287–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Urry DW; Hugel T; Seitz M; Gaub HE; Sheiba L; Dea J; Xu J; Parker T: Elastin: a representative ideal protein elastomer. Philos. Trans. R. Soc. Lond. B. Biol. Scis 2002, 357, 169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Xia XX; Xu QB; Hu X; Qin GK; Kaplan DL: Tunable Self-Assembly of Genetically Engineered Silk-Elastin-like Protein Polymers. Biomacromolecules 2011, 12, 3844–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Xia X-X; Wang M; Lin Y; Xu Q; Kaplan DL: Hydrophobic Drug-Triggered Self-Assembly of Nanoparticles from Silk-Elastin-Like Protein Polymers for Drug Delivery. Biomacromolecules 2014, 15, 908–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lin Y; Xia X; Wang M; Wang Q; An B; Tao H; Xu Q; Omenetto F; Kaplan DL: Genetically Programmable Thermoresponsive Plasmonic Gold/Silk-Elastin Protein Core/Shell Nanoparticles. Langmuir 2014, 30, 4406–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Sun Z; Qin G; Xia X; Cronin-Golomb M; Omenetto FG; Kaplan DL: Photoresponsive Retinal-Modified Silk-Elastin Copolymer. J. Am. Chem. Soc 2013, 135, 3675–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Rousseau R; Schreiner E; Kohlmeyer A; Marx D: Temperature-dependent conformational transitions and hydrogen-bond dynamics of the elastin-like octapeptide GVG(VPGVG): A molecular-dynamics study. Biophys. J 2004, 86, 1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Yeo GC; Tarakanova A; Baldock C; Wise SG; Buehler MJ; Weiss AS: Subtle balance of tropoelastin molecular shape and flexibility regulates dynamics and hierarchical assembly. Sci. Adv 2016, 2, e1501145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Keten S; Buehler MJ: Atomistic model of the spider silk nanostructure. Appl. Phys. Lett 2010, 96, 153701. [Google Scholar]