Abstract
We are using directed evolution to extend the range of dioxygenase-catalyzed biotransformations to include substrates that are either poorly accepted or not accepted at all by the naturally occurring enzymes. Here we report on the oxidation of a heterocyclic substrate, 4-picoline, by toluene dioxygenase (TDO) and improvement of the enzyme's activity by laboratory evolution. The biotransformation of 4-picoline proceeds at only ∼4.5% of the rate of the natural reaction on toluene. Random mutagenesis, saturation mutagenesis, and screening directly for product formation using a modified Gibbs assay generated mutant TDO 3-B38, in which the wild-type stop codon was replaced with a codon encoding threonine. Escherichia coli-expressed TDO 3-B38 exhibited 5.6 times higher activity toward 4-picoline and ∼20% more activity towards toluene than wild-type TDO. The product of the biotransformation of 4-picoline is 3-hydroxy-4-picoline; no cis-diols of 4-picoline were observed.
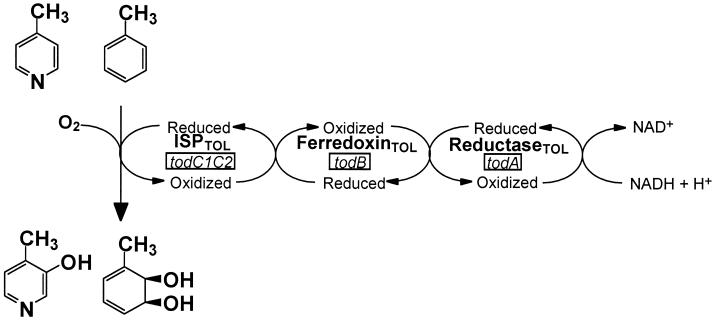
Dioxygenase enzymes involved in the catabolism of aromatic hydrocarbons by soil microorganisms nicely illustrate nature's ability to adapt to different carbon sources through evolution of the substrate specificity of the biodegradation enzymes (35). The biodegradation pathway often begins with the dioxygenase-catalyzed regio- and enantio-specific introduction of molecular oxygen into aromatic compounds to form the corresponding arene cis-diols, with consumption of NADH (13, 36) (Fig. 1).
FIG. 1.
TDO system. TDO catalyzes the insertion of molecular oxygen into toluene and 4-picoline to form toluene cis-dihydrodiol and 3-hydroxy-4-picoline, respectively, with consumption of NADH.
It has been noted that the chiral products of the dioxygenase reaction can be converted in a variety of synthetic reactions to advanced intermediates for natural-product synthesis (8, 15). Toluene dioxygenase (TDO) from Pseudomonas putida readily dihydroxylates aromatic carbocycles with one or two small hydrophobic substituents (6, 33). Activity towards much larger, fused-ring substrates or substrates with polar or bulky substituents is considerably reduced or nonexistent (33).
We are attempting to extend the utility of dioxygenase-catalyzed biotransformations by engineering the catalysts to accept a wider range of substrates, including small heterocyclic compounds and polar-substituted carbocyclic aromatics. N-Heterocyclic compounds are useful for the synthesis of biologically active compounds; recent reports describe the regio- and/or stereo-controlled biooxidation of N-heterocycles (20, 26). Bicyclic compounds such as quinolines and benzofurans are accepted by TDO, with oxygen insertion occurring primarily in the carbocylic rings (3–5).
To date, reactions on single-ring heterocycles have not been reported. We have found that TDO catalyzes the oxygenation of 4-picoline (4-methylpyridine), although at a much slower rate than on its preferred substrate toluene.
A strategy of DNA shuffling to create libraries of hybrid genes and screening has been used with some success to extend the substrate range of dioxygenases that degrade environmental pollutants such as polychlorinated biphenyls (7, 21). An alternative approach is to fine tune a single gene by accumulating beneficial mutations identified by screening randomly mutated libraries (1). We have used this latter approach to investigate how readily TDO adapts to the heterocyclic substrate 4-picoline.
MATERIALS AND METHODS
Materials.
Toluene was from Fisher Scientific (Pittsburgh, Pa.). Isopropyl-β-d-thiogalactopyranoside (IPTG) was from ICN Biomedicals (Aurora, Calif.). 4-Picoline and 2,6-dichloroquinone chloroimide (Gibbs reagent) were from Aldrich (Milwaukee, Wis.). Bugbuster was purchased from Novagen (Madison, Wis.). NuPAGE materials, SeeBlue plus 2 size markers, and Novex colloidal blue stain kit were purchased from Invitrogen (Carlsbad, Calif.). Ambersorb XEN-575 and ampicillin were from Sigma (St. Louis. Mo.). cis-(1S,2R)-Dihydroxy-3-methylcyclohexa-3,5-diene (cis-toluene dihydrodiol) was purchased from QuChem (Belfast). Plasmid pTrc99A was from Amersham Pharmacia Biotech (Piscataway, N.J.). Restriction enzymes were from New England Biolabs (Beverly, Calif.), and T4 DNA ligase was from Roche Diagnostics (Indianapolis, Ind.). Amplitaq DNA polymerase was purchased from Perkin-Elmer (Norwalk, Conn.), and Pfu DNA polymerase was from Stratagene (La Jolla, Calif.). The 96-well plates were purchased from Rainin (Emeryville, Calif.).
Bacterial strains and plasmids.
E. coli DH5α and BL21(DE3) were used for cloning and expression, respectively. pDTG601 carrying the todC1C2BA cistron was kindly provided by D. T. Gibson (37). A 2.2-kbp DNA fragment including todC2BA was PCR amplified using primers TDO9F (5′-TTGGATCCGGTGGACCTTGTCCATTTG-3′) and TDO10R (5′-GCTCTAGATCACGTTAGGTCTCCTTCATTCG-3′) and cloned into the BamHI and XbaI sites of pTrc99A to yield plasmid pXTD8. Primers TDO12F (5′-CGGAATTCTAGGAAACAGACCATG-3′) and TDO13R (5′-CCGGATCCAACCTGGGTCGAAGTCAAATG-3′) were used for PCR amplification of a 1.4-kbp DNA fragment carrying todC1, with substitution of a ribosome-binding-site sequence (GAGAA) with a sequence (AGGAA) designed for higher expression of todC1. Following double digestion with EcoRI and BamHI, the DNA fragment was ligated to pXTD8 double-digested with EcoRI and BamHI, which yielded plasmid pXTD12 (wild type).
Random mutagenesis of todC1.
Error-prone PCR was used to introduce mutations into the todC1 structural gene and 45 downstream nucleotides. Reaction mixture (50 μl) contained 66.5 pg of pXTD12 (wild type), 20 pmol each of primers TDO12F and TDO13R, 100 nmol of each deoxynucleoside triphosphate (dNTP), 350 nmol of MgCl2, 30 nmol of MnCl2, and 1.25 U of AmpliTaq DNA polymerase. PCR was carried out with an MJ research PTC-200 thermal cycler (Watertown, Mass.) under the following conditions: 3 min at 94°C, 30 cycles of 30 s at 94°C, 30 s at 50°C, and 1 min at 72°C, and 3 min at 72°C. Following double digestion of the PCR product, the resulting DNA fragment was ligated back to pXTD8 at the EcoRI and BamHI sites.
Saturation mutagenesis.
To minimize point mutations, Pfu DNA polymerase was used in the following PCR. A 50-ul PCR mixture contained 0.5 ng of template DNA, 20 pmol of each primer, 10 nmol of each dNTP, and 1.25 U of Pfu DNA polymerase. PCR was performed using an MJ research PTC-200 thermal cycler as follows: 3 min at 94°C, 30 cycles of 30 s at 94°C, 30 s at 50°C, and 2 min at 72°C, and 5 min at 72°C. Two degenerate primers, TDO32F (5′-AAGGCGACANNNNNNATCCAGAGACAG-3′) and TDO33R (5′-TCTCTGGATNNNNNNTGTCGCCTTCAG-3′) were designed to randomize positions 450 and 451 in the amino acid sequence (Arg and stop codons in the wild-type DNA sequence). Plasmid DNA was purified from mutants 8A (containing mutation c1242t) and 16A (containing mutations t1351c and g1376t). An equimolar mixture served as the template in the following two-step saturation mutagenesis. In the first step, the first 1,400 nucleotides of todC1 were amplified by PCR using primers TDO12F and TDO33R, and in the second step, 100 nucleotides were amplified using primers TDO32F and TDO13R. After the second step, the two PCR products were combined at a 1:1 molar ratio and used as the template in a PCR using TDO12F and TDO13R to obtain the full-length todC1 fragment. This product, which contains randomized positions and recombinations of the mutations in 8A and 16A, was cloned back into pXTD8 to produce a second-generation mutant library of todC1. Screening of ∼2,200 clones on 4-picoline identified seven higher-activity clones. An equimolar-ratio mixture of plasmid DNA from those seven was used for saturation mutagenesis at amino acid position 352. Two degenerate primers, TDO34F (5′-CTGATGCTCCTNNSGATATCAAGA-3′) and TDO35R (5′-CTTGATATCSNNAGGAGCATCA-3′) were designed to randomize this position. The two-step PCR was performed as described above, and todC1 fragments obtained were cloned into pXTD8 to produce the third-generation mutant todC1 library.
Screening procedure.
An aliquot of the ligation mixture was used to transform E. coli BL21(DE3) by the CaCl2 method (29). At the same time, two control plasmids, pXTD12 (wild type) as a positive control and pXTD8 as a negative control, were used to transform E. coli BL21(DE3) separately. Transformants were grown on Luria-Bertani (LB) agar plates (29) containing ampicillin (100 μg/ml) at 37°C. Each colony was inoculated into 100 μl of LB medium containing ampicillin (100 μg/ml) in separate wells of a 96-well plate. Each 96-well plate accommodated 94 mutants plus one positive and one negative control. Plates were shaken at 270 rpm at 37°C overnight with a New Brunswick Scientific Innova 4000 incubator shaker (Edison, N.J.), and 10 μl of the culture was transferred to new wells containing 100 μl of M9 medium (29) supplemented with 0.4 % glucose, 0.5 mM IPTG, and ampicillin (100 μg/ml). Following 5 h of incubation at 30°C, the culture was mixed with reaction medium containing 50 mM sodium phosphate buffer (pH 7.4), 0.2% glucose, and toluene or 4-picoline to give a final concentration of 5 mM substrate and a total volume of 100 μl. Biotransformation was carried out at room temperature for 30 min (toluene) or 1 h (4-picoline).
A Beckman Instruments Multimek 96 automated multichannel pipettor (Fullerton, Calif.) was used for liquid handling in the following protocol. Activities were determined by adding 100 μl of 0.1 M HCl to 100 μl of biotransformation mixture to reduce the pH below 3.0. Following centrifugation at 1,700 × g for 5 min, 100 μl of supernatant was removed and incubated at 37°C for 30 min. Then, 20 μl of 1 M Tris-HCl buffer (pH 8.5) was added, followed by 25 μl of 0.4% Gibbs' reagent ethanol solution. Absorbance was measured at 590 nm for toluene or 600 nm for 4-picoline with a Molecular Devices Spectramax 250 plate reader (Sunnyvale, Calif.) after blue color developed.
Scaled-up biotransformation.
A New Brunswick Scientific Innova 4000 incubator shaker was used for shaking in the following cultivation and biotransformation. Frozen cell culture of E. coli BL21(DE3) carrying the plasmid in question (10 μl) was inoculated into 1 ml of LB medium containing ampicillin (100 μg/ml) and shaken overnight at 270 rpm at 37°C. Then 250 μl of the resulting seed culture was transferred into 25 ml of fresh medium in a 250-ml flask and shaken at 270 rpm at 37°C. When the optical density at 600 nm of the culture reached 0.5 to 0.7, 100 mM IPTG was added to a final concentration of 1 mM. At this point, the growth temperature was shifted to 30°C and incubation proceeded for an additional 3 h. Then 2.5 ml of the culture was removed into a test tube, and cells were spun down at 1,700 × g for 5 min. Collected cells were washed once with 2.5 ml of 50 mM sodium phosphate buffer (pH 7.4) and suspended with 10 ml of reaction medium containing 50 mM sodium phosphate buffer (pH 7.4) and 0.2% glucose. Biotransformation was started by adding substrate to a final concentration of 5 mM. The mixture was shaken at 270 rpm at 30°C, and 100 μl of sample was removed every 30 min for 1.5 h. Product accumulation was monitored using the Gibbs assay, as described above for high-throughput screening.
Protein extraction and expression analysis of mutant clones.
Frozen cell cultures of BL21(DE3) carrying mutant plasmids were grown and induced according to the conditions of the scaled-up biotransformation. After induction with 1 mM IPTG, 20 ml of cells was spun down at 2,060 × g for 10 min and resuspended in 1.5 ml of Bugbuster protein extraction reagent. The cells were broken with shaking at room temperature for 15 min. Insoluble cell debris was removed by centrifugation at 20,200 × g for 20 min at 4°C, and the cell extract was removed to a fresh tube.
To 13 μl of cell extract was added 13 μl of water, 10 μl of NuPAGE lithium dodecyl sulfate sample buffer, and 4 μl of NuPAGE sample reducing agent. The sample was heated for 10 min at 70°C, and 10 μl was loaded into a NuPAGE 10% Bis-Tris gel (1.0 mm × 15) and run according to the manufacturer's instructions. As a size standard, 10 μl of SeeBlue Plus 2 was added to two wells. The gel was stained using the Novex colloidal blue stain kit.
Purification and identification of biotransformation product.
Biotransformation of 4-picoline was carried out with E. coli BL21(DE3) expressing wild-type TDO. The oxygenated product was bound to Ambersorb XEN-575 (11) and eluted into ethyl acetate-ethanol. (1:1, vol/vol). The concentrated sample was subjected to silica gel column (2.5 by 30 cm) chromatography. The product was eluted with hexane-acetone (1:1, vol/vol), and fractions containing the product, as determined by the Gibbs assay, were collected. Solvent was removed by evaporation, and the product was obtained as slightly yellow crystals.
Analytical.
The fragmentation pattern of the biotransformation product was obtained with a Hewlett-Packard HP5890 series II gas chromatograph and an HP5989A mass spectrometer (Palo Alto, Calif.) equipped with a Quadrex 007-1, 0.1-μm-thickness dimethylpolysiloxane phase (0.18 mm diameter by 3 m) column (Woodbridge, Conn.) 1H nuclear magnetic resonance (NMR) spectra were obtained with a Varian, Inc., Mercury 300-MHz NMR spectrometer (Palo Alto, Calif.) on samples dissolved in chloroform-d (minimum 99.8%).
RESULTS AND DISCUSSION
Biooxidation of 4-picoline by E. coli BL21(DE3) expressing TDO.
Biotransformation using TDO is performed in a whole-cell system because the enzyme comprises multiple components and requires cofactor regeneration (13). Recombinant E. coli cells expressing TDO accumulate relatively large amounts of the biocatalyst and do not have any cis-diol-degrading activity. In a system such as this, total activity is the important figure of merit and reflects the catalyst expression level as well as specific activity.
Gas chromatography-mass spectroscopy (GC-MS) of the product of TDO-catalyzed biotransformation of 4-picoline by E. coli BL21(DE3) cells showed a molecular mass (109 D) and fragmentation pattern indicative of hydroxypicoline. The 1H-NMR spectrum of 2-hydroxy-4-picoline, with peaks at 2.23 ppm (s, 3H), 6.13 ppm (dd, J = 6.7 Hz, 1.6 Hz, 1 H), 6.38 ppm (m, J = 0.8 Hz, 1 H) and 7.25 ppm (d, J = 7.0 Hz, 1 H), differs from that of the product, 2.33 ppm (s, 3H), 7.15 ppm (d, J = 4.8 Hz, 1 H), 7.98 ppm (d, J = 4.8 Hz, 1 H), and 8.24 ppm (s, 1 H), which we have identified as 3-hydroxy-4-picoline. cis-Dihydrodiols of 4-picoline were not detected either by NMR or by GC-MS analysis of the ethyl acetate extract.
Formation of the monohydroxylated product 3-hydroxy-4-picoline likely involves an unstable cis-dihydrodiol which undergoes spontaneous dehydration, as has been proposed to explain the formation of indigo from indole (9), the metabolism of 4-nitrotoluene (28), and analogous monohydroxylated products of the TDO-catalyzed oxidation of quinolines (5). With an initial rate of product formation of 0.22 mmol of 3-hydroxy-4-picoline/h/g of dried cells, TDO is ∼4.5% as active on the heterocyclic substrate as it is towards toluene (5.1 mmol of toluene cis-dihydrodiol/h/g of dried cells). The polar nitrogen-containing heterocycle is not transformed as efficiently as are other nonnatural TDO substrates, such as aromatics with bulky substituents and aliphatic olefins (12, 22, 33)
Directed evolution of TDO.
Oxygenase activity is often determined by monitoring consumption of exogenously added NADH (16). We chose, however, to monitor product formation directly, as we expect mutant TDOs to participate in uncoupling reactions (23) to different extents. The Gibbs reaction has been used for detection of dioxygenase activity (27), but the reported protocol was not sensitive enough to differentiate the small changes in product levels expected during directed evolution. With acid treatment prior to the Gibbs reaction (to catalyze cis-dihydrodiol dehydration to the corresponding phenol) and other modifications, we established a liquid assay system ∼20 times more sensitive than that reported previously (19). A solid-phase version of the assay was also described (19). This assay is suitable for monitoring production of 3-hydroxy-4-picoline. Although this product can be detected without the acidification step, reducing the pH significantly lowers the background absorption that arises from contaminants in the culture medium. We therefore retained this step while screening mutants for activity towards 4-picoline.
Because random mutagenesis of an enzyme as large as TDO (637 amino acids) generates a large number of potential mutants, it is desirable to target the mutagenesis to sequences that are most likely to affect substrate specificity. TDO substrate specificity is determined by the terminal oxygenase, ISPTOL, which is composed of a larger α subunit and a smaller β subunit (30–32). The α subunit accommodates the Rieske [2Fe-2S] center and the mononuclear iron necessary for catalytic activity and appears to play a major role in determining substrate specificity (2, 7, 10, 25, 34), whereas the β subunit is proposed to have only a structural role (18). Although the β subunit of ISPTOL might also affect interactions with substrate (14, 17), we targeted only the α subunit in this initial study.
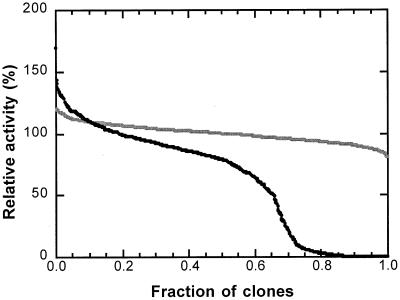
The first mutant library was generated by error-prone PCR of the 1.4-kbp todC1 fragment encoding the α subunit of ISPTOL using 0.6 mM MnCl2. Screening ∼1,000 members of this library for activity on toluene showed ∼20 to 30% inactivation (Fig. 2), which in our experience corresponds to an optimal mutation rate of 2 to 3 nucleotides per sequence. Subsequent DNA sequencing of selected mutants in fact revealed 1 to 3 mutations per 1.4-kbp DNA fragment. We observed a heavy bias for transitions over transversions (7 to 1), however, which limits diversity at the amino acid level.
FIG. 2.
Activities of wild-type TDO and the first-generation mutant TDO library. The activities of each wild-type TDO relative to the average (shaded line) and that of mutant TDO relative to wild-type TDO (black line) are plotted in descending order.
Important for validation of the screening method, the standard deviation of the activity measurement for screening wild-type clones is small (4.5%) compared to the variation in activity in the mutant library (Fig. 2). One mutant, designated 3B, exhibited significantly higher activity towards toluene than the wild type. This mutant also proved superior in a scaled-up biotransformation of toluene (Table 1).
TABLE 1.
Activities and mutations present in TDO variants
| TDO | Amino acid or codon at position:
|
Additional mutation(s) | Relative activity (% of wild type) ± SD
|
|||
|---|---|---|---|---|---|---|
| 352 | 450 | 451 | Toluene | 4-Picoline | ||
| Wild type | D | R | Stop | 100 | 100 | |
| 3B | D | R | R | F181L (t541c), I232V (a694g) | 140 ± 10 | 45 ± 3 |
| 6B | D | H | Stop | R450H (g1349a) | 170 ± 20 | 170 ± 20 |
| 8A | D | R | Stop | [c1242t]a | 190 ± 20 | 170 ± 20 |
| 12E | G | R | Stop | D352G (a1055g) | 140 ± 10 | 170 ± 20 |
| 16A | D | R | R | R459L (g1376t) | 150 ± 10 | 370 ± 30 |
| 3-A69 | D | K | P | 140 ± 10 | 550 ± 40 | |
| 3-B38 | D | R | T | R459L (g1376t) | 150 ± 10 | 560 ± 30 |
| 3-B61 | P | R | P | R459L (g1376t) | 150 ± 10 | 510 ± 40 |
Synonymous mutation.
Approximately 9,000 clones from this library were screened for high activity toward 4-picoline. Selected mutants were tested in a 10-ml scale biotransformation, and those results are summarized in Table 1. The best mutant, 16A, was 3.7 times more active toward 4-picoline than wild-type TDO; activity toward toluene increased 1.5-fold. DNA sequencing of 16A and three different mutant TDOs which showed activity profiles similar to that of 16A revealed that the common substitution of the stop codon (TGA) with Arg (CGA) at amino acid position 451 conferred the improvement in total activity toward 4-picoline and toluene.
The identical stop codon mutation was also observed in mutant 3B, obtained during screening for activity on toluene. Mutants 3B and 16A show the same activity towards toluene, but the activity of 3B toward 4-picoline is only a fraction of that of 16A. The difference in substrate specificity is attributed to either or both of the additional amino acid substitutions in 3B, F181L and I232V.
Synonymous mutation c1242t (in mutant 8A) and the nonsynonymous g1349a mutation in 6B (substitution of R450 by H) and a1055g mutation in 12E (substitution of D352 by G) all increase total activity but have no or little effect on substrate specificity. The only mutation in 8A, c1242t, is located in the middle of a CG stretch of 10 nucleotides. This mutation might facilitate transcription of todC1 by destabilizing the DNA secondary structure.
Further evolution of TDO.
Random mutagenesis highlighted C-terminal positions 450 and 451 in the amino acid sequence as contributing to the specificity and total activity of TDO. We therefore used saturation mutagenesis to explore all possible substitutions at these two consecutive codons. Using the strategy described in Materials and Methods, we simultaneously randomized the codons and recombined mutations c1242t (in 8A) and g1376t (in 16A). Seven superior clones were picked out of 2,200 clones screened, and their plasmid mixture was subjected to another round of saturation mutagenesis at codon 352 (at which 12E had a beneficial mutation). Screening ∼600 clones identified three positive mutants, and the scale-up biotransformation tests revealed further improvements in activity toward 4-picoline (Table 1).
E. coli BL21(DE3)-expressed mutant TDO 3B-38 exhibited a 5.6-fold-enhanced initial reaction rate for oxidation of 4-picoline (1.2 mmol of 3-hydroxy-4-picoline/h/g of dried cells) as well as a slight increase in activity towards toluene (6.9 mmol of toluene cis-dihydrodiol/h/g of dried cells) compared to wild-type TDO. The product of TDO 3B-38 oxidation of picoline is 3-hydroxy-4-picoline, identical to the product of reaction with wild-type TDO.
The results in Table 1 indicate a key role for amino acid 451 in determining activity towards 4-picoline. A proline or threonine in place of arginine at this position increases activity toward 4-picoline 1.5-fold, with no effect on activity toward toluene. The preferred amino acid at 450 is either arginine (wild type) or lysine. The saturation mutagenesis results also show no particular benefit to be gained by substitution at position 352. Furthermore, the synonymous mutation, c1242t, which conferred higher activity in 8A, was not retained in any of the three highest-activity clones, indicating that it provides no additional benefit in the presence of the stop codon mutation.
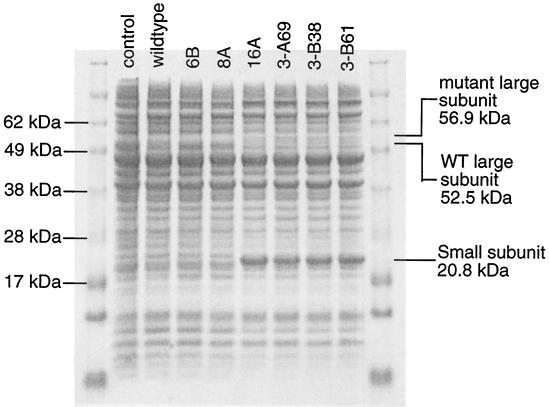
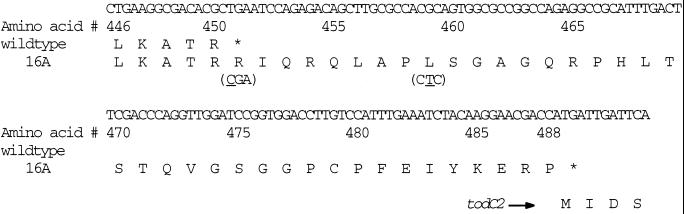
Several of the mutants described in Table 1 were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 3). For four variants containing the stop codon mutation (16A, 3-A69, 3-B38, and 3-B61), the ISPTOL α subunit (the large subunit) migrates as a slightly larger protein. In addition, these variants showed dramatically increased expression of the β (smaller) subunit compared to variants without the stop codon mutation. Presumably, the new stop codon for todC1 in these variants appears just after the start codon of todC2, which would extend the C terminus of the α subunit by 38 amino acids (Fig. 4). This type of overlapping gene structure has also been reported to help expression of the subsequent gene (24), as we have observed in this case for the β subunit. Thus, mutation of the stop codon leads to extension of todC1 and a C-terminal extension of the protein which improve activity toward 4-picoline by increasing expression and altering the enzyme's substrate specificity.
FIG. 3.
SDS-PAGE comparison of cell extracts from mutant and wild-type strains. The lane labeled control contains an extract from BL21(DE3) carrying pTrc99A, the empty vector used to construct the wild-type and mutant strains.
FIG. 4.
Nucleotide and deduced amino acid sequences between the C-terminus of todC1 and the N-terminus of todC2. Amino acids are numbered from the start codon of todC1. The nucleotide substitution found in 16A is underlined in parenthesis. The stop codon of todC1-16A (TGA) overlaps with the start codon of todC2 (ATG).
ACKNOWLEDGMENTS
We thank R. W. T. Lee for technical assistance with NMR.
We thank Maxygen Inc. for supporting this research.
REFERENCES
- 1.Arnold F H. Design by directed evolution. Acc Chem Res. 1998;31:125–131. [Google Scholar]
- 2.Beil S, Mason J R, Timmis K N, Pieper D H. Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J Bacteriol. 1998;180:5520–5528. doi: 10.1128/jb.180.21.5520-5528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd D R, Sharma N D, Dorrity M R J, Hand M V, Austin R, McMordie S, Malone J F, Porter H P, Dalton H, Chima J, Sheldrake G N. Structure and stereochemistry of cis-dihydrodiol and phenol metabolites of bicyclic azaarenes from Pseudomonas putida UV4. J Chem Soc Perkin Trans. 1993;1:1065–1071. [Google Scholar]
- 4.Boyd D R, Sharma N D, Boyle R, Malone J F, Chima J, Dalton H. Structures and stereochemical assignments of some novel chiral synthons derived from the biotransformation of 2,3-dihydrobenzofuran and benzofuran by Pseudomonas putida. Tetrahedron Asymmetry. 1993;4:1307–1324. [Google Scholar]
- 5.Boyd D R, Sharma N D, Carroll J G, Malone J F, Mackerracher D G, Allen C C R. Dioxygenase-catalysed cis-dihydrodiol formation in the carbo- and hetero-cyclic rings of quinolines. J Chem Soc Chem Commun. 1998;6:683–684. [Google Scholar]
- 6.Boyd D R, Sheldrake G N. The dioxygenase-catalysed formation of vicinal cis-diols. Natural Product Reports. 1998;15:309–324. [Google Scholar]
- 7.Bruhlmann F, Chen W. Tuning biphenyl dioxygenase for extended substrate specificity. Biotechnol Bioeng. 1999;63:544–551. doi: 10.1002/(sici)1097-0290(19990605)63:5<544::aid-bit4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 8.Buckland B C, Drew S W, Connors N C, Chartrain M M, Lee C, Salmon P M, Gbewonyo K, Zhou W, Gailliot P, Singhvi R, Jr, Olewinski R C, Sun W-J, Reddy J, Zhang J, Jackey B A, Taylor C, Goklen K E, Junker B, Greasham R L. Microbial conversion of indene to indandiol, a key intermediate in the synthesis of CRIXIVAN. Metabolic Eng. 1999;1:63–74. doi: 10.1006/mben.1998.0107. [DOI] [PubMed] [Google Scholar]
- 9.Ensley B D, Ratzkin B J, Osslund T D, Simon M J, Wackett L P, Gibson D T. Expression of naphthalene dioxygenase genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983;222:167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- 10.Erickson B D, Mondello F J. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl Environ Microbiol. 1993;59:3858–3862. doi: 10.1128/aem.59.11.3858-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia A A, Kim D H, Whited G, Kwart L, Anthony W, Downie C. Recovery of a cyclic diol produced via biocatalysis. Isolation Purification. 1994;2:19–25. [Google Scholar]
- 12.Gibson D T, Gschwendt B, Yeh W-K, Kobal V M. Initial reactions in the oxidation of ethylbenzene by Pseudomonas putida. Biochemistry. 1973;12:1520–1528. doi: 10.1021/bi00732a008. [DOI] [PubMed] [Google Scholar]
- 13.Gibson D T, Hensley M, Yoshioka H, Marby T G. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry. 1970;9:1626–1630. doi: 10.1021/bi00809a023. [DOI] [PubMed] [Google Scholar]
- 14.Hirose J, Suyama A, Hayashida S, Furukawa K. Construction of hybrid biphenyl (bph) and toluene (tod) genes for functional analysis of aromatic ring dioxygenases. Gene. 1994;138:27–33. doi: 10.1016/0378-1119(94)90779-x. [DOI] [PubMed] [Google Scholar]
- 15.Hudlicky T, Gonzalez D, Gibson D T. Enzymatic dihydroxylation of aromatics in enantioselective synthesis: expanding asymmetric methodology. Aldrichimica Acta. 1999;32:35–62. [Google Scholar]
- 16.Hurtubise Y, Barriault D, Powlowski J, Sylvestre M. Purification and characterization of the Comamonas testosteroni B-356 biphenyl dioxygenase components. J Bacteriol. 1995;177:6610–6618. doi: 10.1128/jb.177.22.6610-6618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurtubise Y, Barriault D, Sylvestre M. Involvement of the terminal oxygenase β subunit in the biphenyl dioxygenase reactivity pattern toward chlorobiphenyls. J Bacteriol. 1998;180:5828–5835. doi: 10.1128/jb.180.22.5828-5835.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang H, Parales R E, Gibson D T. The α subunit of toluene dioxygenase from Pseudomonas putida F1 can accept electrons from reduced ferredoxinTOL but is catalytically inactive in the absence of the β subunit. Appl Environ Microbiol. 1999;65:315–318. doi: 10.1128/aem.65.1.315-318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joern J M, Sakamoto T, Arisawa A, Arnold F H. A versatile high-throughput screen for dioxygenase activity using solid-phase digital imaging. J Biomol Screening, 2001;6:213–217. doi: 10.1177/108705710100600403. [DOI] [PubMed] [Google Scholar]
- 20.Kiener A. Biosynthesis of functionalized aromatic N-heterocycles. Chemtech. 1995;25:31–35. [Google Scholar]
- 21.Kumamaru T, Suenaga H, Mitsuoka M, Watanabe T, Furukawa K. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat Biotechnol. 1998;16:663–666. doi: 10.1038/nbt0798-663. [DOI] [PubMed] [Google Scholar]
- 22.Lange C C, Wackett L P. Oxidation of aliphatic olefins by toluene dioxygenase: enzyme rates and product identification. J Bacteriol. 1997;179:3858–3865. doi: 10.1128/jb.179.12.3858-3865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K. Benzene-induced uncoupling of naphthalene dioxygenase activity and enzyme inactivation by production of hydrogen peroxide. J Bacteriol. 1999;181:2719–2725. doi: 10.1128/jb.181.9.2719-2725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Normark S, Bergstrom S, Edlund T, Grundstrom T, Jaurin B, Lindberg F P, Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- 25.Parales J V, Parales R E, Resnick S M, Gibson D T. Enzyme specificity of 2-nitrotoluene 2,3-dioxygenase from Pseudomonas sp. strain JS42 is determined by the C-terminal region of the α subunit of the oxygenase component. J Bacteriol. 1998;180:1194–1199. doi: 10.1128/jb.180.5.1194-1199.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen M, Kiener A. Preparation and functionalization of N-heterocycles. Green Chem. 1999;1:99–106. [Google Scholar]
- 27.Quintana M G, Didion C, Dalton H. Colorimetric method for a rapid detection of oxygenated aromatic biotransformation products. Biotechnol Techniques. 1997;11:585–587. [Google Scholar]
- 28.Robertson J B, Spain J C, Haddock J D, Gibson D T. Oxidation of nitrotoluenes by toluene dioxygenase: evidence for a monooxygenase reaction. Appl Environ Microbiol. 1992;58:2643–2648. doi: 10.1128/aem.58.8.2643-2648.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Subramanian V, Liu T-N, Yeh W-K, Gibson D T. Toluene dioxygenase: purification of an iron-sulfur protein by affinity chromatography. Biochem Biophys Res Commun. 1979;91:1131–1139. doi: 10.1016/0006-291x(79)91998-3. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian V, Liu T-N, Yeh W-K, Narro M, Gibson D T. Purification and properties of NADH-ferredoxinTOL reductase. J Biol Chem. 1981;256:2723–2730. [PubMed] [Google Scholar]
- 32.Subramanian V, Liu T-N, Yeh W-K, Serdar C M, Wackett L P, Gibson D T. Purification and properties of ferredoxinTOL. J Biol Chem. 1985;260:2355–2363. [PubMed] [Google Scholar]
- 33.Sheldrake G N. Biologically derived arene cis-dihydrodiols as synthetic building blocks. In: Collins A N, Sheldrake G N, Crosby J, editors. Chirality in industry: the commercial manufacture and application of optically active compounds. Chichester, England: John Wiley & Sons Ltd.; 1992. pp. 127–166. [Google Scholar]
- 34.Tan H-M, Cheng C-M. Substitution of the ISP α subunit of biphenyl dioxygenase from Pseudomonas results in a modification of the enzyme activity. Biochem Biophys Res Commun. 1994;204:912–917. doi: 10.1006/bbrc.1994.2546. [DOI] [PubMed] [Google Scholar]
- 35.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]
- 36.Yeh W-K, Gibson D T, Liu T-N. Toluene dioxygenase: a multicomponent enzyme system. Biochem Biophys Res Commun. 1977;78:401–410. doi: 10.1016/0006-291x(77)91268-2. [DOI] [PubMed] [Google Scholar]
- 37.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]