Abstract
Although hundreds of knockout mice show infertility as a major phenotype, the causative genic mutations of male infertility in humans remain rather limited. Here, we report the identification of a missense mutation (D136G) in the X-linked TAF7L gene as a potential cause of oligozoospermia in men. The human aspartate (D136) is evolutionally conserved across species, and its change to glycine (G) is predicted to be detrimental. Genetic complementation experiments in budding yeast demonstrate that the conserved aspartate or its analogous asparagine (N) residue in yeast TAF7 is essential for cell viability and thus its mutation to G is lethal. Although the corresponding D144G substitution in the mouse Taf7l gene does not affect male fertility, RNA-seq analyses reveal alterations in transcriptomic profiles in the Taf7l (D144G) mutant testes. These results support TAF7L mutation as a risk factor for oligozoospermia in humans.
Keywords: infertility, oligozoospermia, spermatogenesis, X chromosome, TAF7L, transcription factor, Characterization of missense mutation in TAF7L gene
A sequencing screen of infertile men identifies a missense mutation in the human TAF7L gene.
Introduction
An estimated 15% of couples suffer from infertility worldwide, half of which is attributed to male factors [1–4]. Spermatogenic failure represents a common cause of male infertility [5]. About 50% of male infertility cases are idiopathic, and idiopathic male infertility is believed to be largely genetic in etiology [6, 7]. Studies of mouse models have defined many pathways that regulate reproduction in mammals [7, 8]. At least 2000 genes are preferentially expressed in mouse spermatogenesis [2], and more than 400 of them are known to regulate mouse fertility [3, 4]. Exome-sequencing of infertile patients has identified mutations in a number of genes in humans as the cause of male sterility [9, 10]. Despite these recent advances, our knowledge about the genetic etiology underlying male infertility in humans remains rather limited.
The X chromosome is enriched for spermatogonially expressed testis-specific genes in mouse [11]. Taf7l (TBP associated factor 7 like) is an X-linked testis-specific homologue of TBP associated factor 7 (Taf7) [11]. Taf7 is an autosomal retrogene and encodes a ubiquitously expressed component of the transcription factor transcription factor IID (TFIID) that is required for the transcription of protein-coding genes by RNA polymerase II [12]. Like TAF7 in somatic tissues, TAF7L interacts with TBP associated factor 1 (TAF1) and is associated with TATA box-binding protein (TBP) in the testes, indicating that TAF7L is a bona fide TBP-associated factor (TAF) [13]. Moreover, biochemical studies have shown that TAF7L replaces TAF7 in the TFIID complex to modulate the transcription in spermatogenesis [13]. Inactivation of Taf7l in mice causes a reduction in the sperm count, sperm motility, testis weight, and male fertility [14]. A backcross of Taf7l knockout mice to the C57BL/6J background produced knockout males that are essentially sterile [15]. The deletion of testis-expressed gene 11 (Tex11), an X-linked germ cell-specific gene, leads to meiotic arrest in males [16]. Interestingly, the Taf7l Tex11 double mutant males exhibit much severe defects in meiosis than either single mouse mutant, suggesting the synergistic regulation of spermatogenesis by these two X-linked genes [17]. In addition to its role in spermatogenesis, TAF7L acts as a transcription regulator in adipocyte differentiation [15, 18].
Based on the phenotype of Taf7l knockout mice, we reasoned that mutations in human TAF7L gene would most likely cause reduced sperm count—oligozoospermia. Sequence variants in TAF7L were previously reported in oligozoospermic men [19]. However, these TAF7L variants were either found in fertile controls or not verified [20], leaving their functional consequence unknown [7]. To determine if TAF7L plays a role in human fertility, we screened 113 infertile men and identified a single amino acid substitution (D136G) in TAF7L in an infertile man with low sperm count. This aspartic residue is conserved in TAF7 from yeast to human and in TAF7L in mammals. We genetically characterized the functional impact of this amino acid substitution in TAF7 and TAF7L in yeast and mouse, respectively.
Materials and methods
Ethics statement
The human study was approved by the Institutional Review Board of Massachusetts Institute of Technology. Informed consent was obtained from all participants. Mice were maintained under veterinarian care from the University Laboratory Animal Resources at the University of Pennsylvania or Nanjing Medical University. All animal experimental procedures were approved by the Institutional Animal Care and Use Committees at University of Pennsylvania and Nanjing Medical University.
Patients and controls
In this study, 113 patients with severe oligozoospermia (sperm count <5 × 106/ml) were included. We excluded patients with any of the following conditions or treatments that cause or predispose to spermatogenic failure: Y-chromosomal deletions [21–25]; a 47, XXY karyotype [26]; cryptorchidism; orchitis; radiotherapy; or chemotherapy. We also studied 175 controls, consisting of men known to be fertile (n = 93) and men of unknown fertility selected to represent worldwide genetic diversity (n = 82, samples from the National Institutes of Health polymorphism discovery panel, Coriell Cell Repositories, and from our own collection) [27]. This collection of 175 control men was previously used in the TEX11 genetic study [28]. Genomic DNA was prepared from peripheral blood leukocytes or EBV-transformed lymphoblastoid cell lines.
Mutation screening
The TAF7L exon/intron structure was determined by the alignment of human TAF7L cDNA sequence (NM_024885) with the genomic sequence [11]. We polymerase chain reaction (PCR)-amplified the TAF7L coding exons (exons 2 through 13) with 12 primer pairs (Supplementary Table S1). PCR was performed in a 25-μl reactor with 12.5 ng of genomic DNA (94 °C, 30 s; 56 °C, 30 s; 72 °C, 90 s; 35 cycles). PCR products were purified by Sephadex S-300 gel filtration. Then, 12.5 μl of the purified PCR product was sequenced in a 25-μl reactor using one of the PCR primers and ABI BigDye on an ABI 3700 sequencer according to the manufacturer’s instructions. Sequence data were analyzed using the Sequencher software (Gene Codes Corporation, Ann Arbor, USA), and sequence variants were identified by an inspection of the aligned sequences in Sequencher.
Pathogenicity prediction of the human TAF7L D136G substitution
Four algorithms, Mutation Taster [29], PolyPhen2 [30], PROVEAN [31], and SIFT [32], were used to predict the possible effects of the missense mutation D136G found in the human TAF7L gene. The web links are as follows: Mutation Taster (http://www.mutation-taster.org); PolyPhen2 (http://genetics.bwh.harvard.edu/pph2); PROVEAN (http://provean.jcvi.org/index.php); and SIFT (https://sift.jcvi.org/).
Plasmid constructs and yeast strains
The wild type (WT) Saccharomyces cerevisiae TAF7 gene (scTAF7-N147) was PCR-amplified from the yeast genomic DNA and cloned into the CEN plasmid pRS416 (URA3+). Two TAF7 mutants harboring N147D and N147G point mutations respectively were generated by PCR-based mutagenesis [33] and subcloned into pRS415 (LEU2+) to yield pRS415-scTAF7-N147D and pRS415-scTAF7-N147G plasmids. pRS416-scTAF7-N147 (WT) was transformed into TAF7/taf7∆::kanMX6 heterozygous diploid yeast cells [34, 35], and tetrad analysis was performed to test whether the scTAF7 (WT) plasmid rescues the lethality of the yeast taf7 deletion. Sc taf7∆ segregants containing pRS416-scTAF7-N4147 were viable and plasmid dependency for viability was confirmed by assaying for growth on 5-FOA plates, which counter-selects the URA3-containing pRS416-scTAF7 plasmid [36]. To determine whether scTAF7-N147D or cTAF7-N147G rescued the taf7∆ mutation, haploid taf7∆ cells containing pRS416-scTAF7-N147 (WT) were transformed with pRS415-scTAF7-N147D or pRS415-scTAF7-N147G and the viability of the transformants was assessed after counter-selecting the pRS416-scTAF7-N147 plasmid on 5-FOA plates.
Generation of Taf7l knockin (D144G) mutant mice
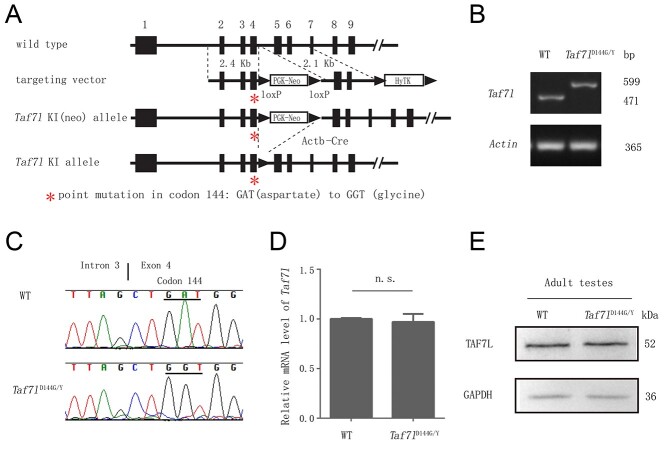
We mutated codon 144 from GAT [aspartate (D)] to GGT [glycine (G)] in the mouse Taf7l gene to generate a mutation analogous to the D136G mutation in the human TAF7L gene. Codon 144 is located in exon 4 (Figure 2A). We generated this point mutation in the targeting vector by an overlapping PCR-mediated mutagenesis strategy as described previously [33]. Using a Taf7l-containing BAC clone (RP22-415C9) as the template, two homologous arms (2.4 and 2.1 kb) were amplified by high-fidelity PCR (Figure 2A). The floxed neomycin selection marker was cloned into intron 4. V6.5 ES cells were electroporated with the linearized targeting vector and were selected for integration in the presence of G418 (350 μg/ml) and gancyclovir (2 μM). In G418-positive clones screened for homologous recombination, 7 out of 184 were homologously targeted. Exon 4 was PCR-amplified from positive ES cell clones and sequenced to confirm the presence of the point mutation. Two ES cells harboring the Taf7l D144G allele were injected into BALB/c blastocysts that were subsequently transferred to the uteri of pseudopregnant females. The resulting male chimeras were bred with C57BL/6J females to obtain germline transmission of the mutant allele. Offspring were genotyped by PCR assays. WT (599 bp) and Taf7l mutant (471 bp) alleles were assayed by PCR with the Taf7l-KI forward and reverse primers (Supplementary Table S2). Taf7l knockin mice were crossed with Actb-Cre to remove the floxed PGK-Neo selection cassette [37]. The Actb-Cre transgene was removed during subsequent breeding. Taf7l knockin mice were backcrossed to the C57BL/6J strain (Jackson Laboratory) for 10 generations. The backcrossed mice were used in this study.
Figure 2.
Generation of Taf7lD144G knockin mice. (A) Schematic of mouse Taf7l gene structure, targeting vector, and various alleles. Taf7l consists of 13 exons. Only the first nine exons are shown in rectangles. Exons 10 to 13 are not shown. The asterisk in Exon 4 denotes the point mutation. PGK-Neo encodes a neomycin selection marker. HyTK enables positive selection with hygromycin (not used) and negative selection with ganciclovir. pOG231 expresses Cre recombinase. (B) Genotyping of Taf7l alleles. (C) Sequencing of the wild type and Taf7lD144G alleles. (D) qRT-PCR analysis of the Taf7l transcript in wild type and Taf7lD144G/Y testes from 3-month-old mice. n.s.: nonsignificant, by Student’s t-test. (E) Western blot analysis of TAF7L in 3-month-old wild type and Taf7lD144G/Y testes. GAPDH serves as a loading control.
Quantitative real-time PCR (qRT-PCR) expression analysis
Total RNA was isolated from WT and Taf7l mutant adult (3 months old) mouse testes using TRIzol reagent (15 596 026, Invitrogen, USA). One microgram of total RNA was reverse-transcribed using the PrimeScript RT Master Mix (RR036A, Takara, Japan). The obtained cDNA was diluted five times and then RT-qPCR was performed using the TB Green Premix Ex Taq II (RR820A, Takara, Japan) on a StepOnePlus RT-PCR machine (4 376 592, Applied Biosystems, USA). The relative expression of target mRNA was determined after normalization to the 36B4 gene. Student t-test was used for statistical analysis.
Western blot analysis
Protein extracts were prepared from fresh testes of adult mice. Testicular protein (5 μg) from each sample was loaded and separated on a 10% SDS–PAGE gel. After transferring onto a polyvinylidene fluoride membrane (1 620 177, Bio-Rad, USA), the blots were incubated with the following primary antibodies: anti-TAF7L (DF2403, Affinity, China; 1:1000) or anti-GAPDH (AC033, Abclonal, Wuhan, China; 1:20 000), and secondary antibodies: HRP goat antimouse IgG (H + L) (AS003, Abclonal, Wuhan, China; 1:10 000) or HRP goat antirabbit IgG (H + L) (AS014, Abclonal, Wuhan, China; 1:10 000).
Histology
The testes and epididymides from 3-month-old mice were fixed in Bouin’s solution (HT10132-1 L, Sigma, USA) overnight. The tissues were dehydrated through gradient ethanol, embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin.
Mating test
Each WT or Taf7lD144G/Y male (five males per genotype) was housed with two Taf7l+/D144G females for 10 months. Mice were 2 months old at the start of the mating test. The date and size of litters were recorded.
Sperm count
Cauda epididymides were dissected. Each epididymis was minced in 4 ml 1× phosphate-buffered saline (PBS) solution to disperse the sperm and fixed with 2 ml of 4% paraformaldehyde (PFA) for 10 min. Sperms were counted using a hemocytometer.
Computer-assisted sperm analysis (CASA)
Three small openings were cut on the freshly harvested cauda epididymis from adult mice. The epididymis was placed in 37 °C preheated sperm culture medium (10% fetal bovine serum (FBS) in DMEM) and incubated for 5 min to allow the sperms to swim out. Then, 10 μl of sperm suspension was subjected to CASA analysis (IVOS, Hamilton Thorne, USA).
Transfection and coimmunoprecipitation
HEK293T cells were transfected with plasmids for the expression of HA-TBP and FLAG-tagged TAF7L (WT or KI). Then, 48 h after transfection, the cells were harvested with lysis buffer (20 mM Tris pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 0.5% sodium deoxycholate, 1 mM dithiothreitol (DTT), protease inhibitor). Approximately 10 μg of the indicated antibody was added into cell lysates with constant agitation at 4 °C overnight; 50 μl of protein G dynabeads slurry (Invitrogen; 10004D) was added to pull down the immunocomplexes. Beads were collected with a magnetic grate and washed seven times with cold buffer (20 mM Tris pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA). 50 μl of 2× SDS loading buffer was added and the sample was boiled at 95 °C for 10 min. Protein samples were subjected to immunoblotting with appropriate antibodies.
Isolation of germ cell populations
Pachytene spermatocytes (pacSC) and round spermatids (rST) were isolated twice from adult testes using a STA-PUT apparatus within a 4 °C refrigerator [38]. For each isolation, three pairs of Taf7l WT or KI testes were subjected to first enzymatic digestion with 1 mg/ml collagenase IV (Gibco) in a 37 °C water bath for 15 min, followed by a second enzymatic digestion with 1 mg/ml trypsin (Sigma) and 1 mg/ml DNase I (Sigma) in a 37 °C water bath for 8 min. Then cells were resuspended in 25 ml of 0.5% BSA solution and filtered through a 100 mm mesh to remove cell aggregates and a 40 mm mesh to remove sperm tails. Next, 25 ml of cell suspensions was loaded and stirred gently. The stirrer worked under the 2% BSA solution and artery clips were removed so that both 2 and 4% BSA solutions were introduced to the chamber (~30 min). Artery clips were replaced and the stirrers were switched off, and a gradient of BSA was formed after 2 h of sedimentation in the standard cell sedimentation chamber. In order to collect different cell types with 15-ml centrifuge tubes, the first fraction collected was designated as 1 and the rest were numbered up to 100. The cells in each fraction were collected by centrifugation at 500 g for 5 min. The cells were resuspended in 0.2 ml of cold PBS. An aliquot of each fraction was examined under a DIC microscope to assess cellular integrity and identify the cell types. Fractions containing cells of similar size and morphology were pooled together and prepared for RNA extraction and RNA-seq analysis.
RNA-seq analysis
Total RNA was extracted from 3-month-old WT and Taf7lD144G/Y mouse testes or isolated spermatogenic cells using TRIzol reagent (15 596 026, Invitrogen, USA). The RNA quality was assessed using an Agilent 2100 RNA 6000 Nano kit. Strand-specific RNA-seq libraries (three biological replicates per genotype) were constructed using the TruSeq stranded total RNA sample preparation kit (Illumina, USA) according to the manufacturer’s instructions and sequenced on the Illumina HiSeqTM 4000 platform at Genedenovo Biotechnology Co., Ltd (Guangzhou, China). Low-quality read filtering and adaptor trimming were performed using fastP [39]. High-quality reads were mapped to the mouse genome (mm10) using HISAT2 [40] with a GTF file downloaded from the Ensembl genome browser 104. First, we used StringTie [41] to reconstruct the transcript and calculate the mRNA expression level of all genes in every sample. Second, CPC2 [42] and CNCI [43] were used to predict the coding ability of new transcripts. Transcripts without coding potential were classified as long noncoding RNAs (lncRNAs). Finally, unmapped reads with high quality were processed using the find_circ [44] pipeline to identify circular RNAs (circRNAs). For differential gene expression analysis of mRNAs and lncRNAs, we used DESeq2 [45] to normalize the read count and calculate the P-value and false discovery rate value but used edgeR [46] for circRNA. Significant differentially expressed genes were identified as those with P-value <0.05 and fold-change >1.5.
Statistical analysis
Statistical analysis was performed using the two-tailed unpaired Student’s t-test (ns: not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001).
Results
Mutation screening in the TAF7L gene in infertile men
TAF7L is a component of the transcription factor TFIID complex. Taf7l is an X-linked single-copy testis-specific gene in both mice and humans [11]. Targeted disruption of Taf7l in mice causes significant reduction in sperm count and sperm motility [14]. The most common form of male infertility is low sperm count. As the X chromosome is in the hemizygous state in men, we hypothesized that mutations in TAF7L could cause oligozoospermia (reduced sperm count) in humans. To screen for TAF7L mutations in infertile men, we sequenced 12 exons of TAF7L in 113 severely oligozoospermic men (sperm count <5 × 106/ml) and 175 control men. We found a total of 10 sequence variants (Table 1). Six of these occurred in two or more samples with similar frequencies in both affected and control men and thus were likely to be polymorphisms. Among the six polymorphisms, two resulted in amino acid changes in exons 9 and 13, respectively, and another variant led to a deletion of two amino acids in exon 10 (Table 1). These three missense variants were also identified in infertile and fertile men in previous studies of nonobstructive azoospermic men and thus were not associated with infertility [19, 20].
Table 1.
Summary of sequence variants in TAF7L found in oligozoospermic and control men
| Position | Nucleotide changea | Resultant changea | 113 | 175 |
|---|---|---|---|---|
| Exon 4 | c.407A➔G | Missense mutation, D136G | 1b | 0 |
| Exon 5 | c.633G➔A | Silent | 1 | 0 |
| Intron 2 | –47G➔C | Intronic alteration | 2 | 0 |
| Intron 4 | +42A➔C | Intronic alteration | 1 | 0 |
| Polymorphisms | ||||
| Exon 9 | c.922A➔G | Missense mutation, S222G | 19 | 25 |
| Exon 10 | c.1026del(TGAGGA) | Del(ED) in a stretch of EDED | 22 | 40 |
| Exon 13 | c.1373G➔A | Missense mutation, C370Y | 2 | 4 |
| Intron 7 | –57C➔G | Intronic alteration | 1 | 1 |
| Intron 8 | +52A➔G | Intronic SNP: rs3788764 | 44 | 61 |
| Intron 10 | +91A➔G | Intronic SNP: rs3761514 | 40 | 60 |
However, four variants were observed only in one or two infertile men: one missense mutation (D136G), one silent mutation, and two intronic mutations. Therefore, only the D136G mutation is most likely to be deleterious (Figure 1A). The patient with this point mutation (D136G) (WHT3493, Figure 1A) and his female partner had experienced infertility for several years. The evaluation showed sperm counts ranging 4–6 million/ml, far lower than the normal sperm count of >15 million/ml. His karyotype and Y chromosomal microdeletion assay were both noninformative. Bilateral microsurgical varicocelectomy was performed with no demonstrable change in semen parameters. However, over the next several years, the couple had three naturally conceived pregnancies: two miscarriages on either side of the birth of their healthy daughter. Unfortunately, the patient developed unilateral testis cancer but only required orchiectomy. Subsequent sperm counts settled out at an average of 3 million/ml. The couple did not return for follow-up visits or assessment.
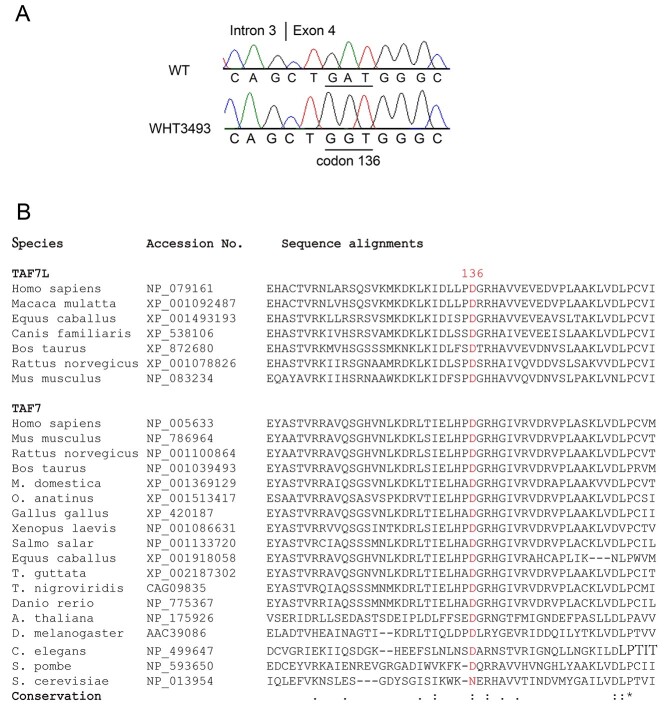
Figure 1.
Identification of a missense mutation (D136G) in human TAF7L gene in an oligozoospermic man. (A) Sequencing chromatograms of human wild type and mutant TAF7L alleles. Patient WHT3759 harbors a missense mutation in exon 4 of TAF7L, resulting in the D136G amino acid substitution. (B) Conservation of the aspartic acid at 136 (D136) in TAF7L and TAF7 proteins among eukaryotes. The position of the conserved aspartic acid/asparagine residue: D136 (human TAF7L), N147 (budding yeast TAF7), and D144 (mouse TAF7L). Sequence alignment was performed with Clustal X.
D136 is highly conserved in the TAF7/TAF7L gene family
Although Taf7l is present only in mammalian species, Taf7, the ubiquitously expressed homologue of Taf7l, is present in all eukaryotes examined [14]. Aspartic acid (D136) is conserved not only in TAF7L but also in TAF7 proteins from diverse species including fission yeast (Schizosaccharomyces pombe), worms, fly, fish, and mammals (Figure 1B). In contrast, the residues adjacent to D136 are not as well conserved. TAF7 and TAF7L are components of the basal transcription factor TFIID complex that interacts with transcription activators to activate transcription. The residue D136 is located in the TAF7/TAF7L domain that is known to interact with transcriptional activators [12]. Therefore, the D136G mutation could disrupt the interaction between TAF7L/TAF7 and transcriptional activators, thereby inhibiting downstream transcription programs necessary for sperm production. The evolutionary conservation of D136 in TAF7L and TAF7 suggests that it is a functionally important residue.
The D136G substitution in TAF7L is predicted to be deleterious
To gain more insights before direct functional studies, we used bioinformatic tools to evaluate the possible impact of the D136G substitution on the function of TAF7L. Four bioinformatics tools including SIFT, MutationTaster, PolyPhen-2, and PROVEAN were employed. Among them, the SIFT software is based on sequence homology and physical properties of amino acids. PolyPhen-2, MutationTaster, and PROVEAN predict the impact of an amino acid change on the structure and function of the protein based on a large number of sequences, evolutionary relationships, and structural features. All four analyses predicted that the D136G substitution was potentially detrimental to the function of TAF7L (Table 2).
Table 2.
Bioinformatic prediction of possible functional impacts of the missense mutation (c. 407A>G; p.D136G) in the human TAF7L gene
| Bioinformatic tools | Prediction |
|---|---|
| Mutation Taster | Disease causing |
| PolyPhen-2 | Probably damaging |
| PROVEAN | Deleterious |
| SIFT | Deleterious |
The conserved D/N residue is required for the TAF function in budding yeast
The D136 (human TAF7L) is conserved in TAF7L and TAF7 in all species examined except S. cerevisiae (budding yeast) (Figure 1B). Budding yeast is the only species that we found to contain asparagine (N) instead of aspartic acid (D) at this position. Notably, asparagine and aspartic acid are structurally similar amino acids. We found that TAF7-null yeast mutants were not viable, showing that TAF7 is an essential gene in budding yeast (Table 3). We next performed tetrad analysis to determine whether the N147 in yeast TAF7 (corresponds to D136 in human TAF7L) is required for its function. We cloned WT yeast TAF7 (scTAF7-N147) into a low-copy CEN plasmid pRS416. As expected, the pRS416-scTAF7-N147 rescues the lethality of TAF7-null yeast cells (Table 3). We made two TAF7 mutants harboring N147D and N147G point mutations respectively, resulting in two constructs pRS415-scTAF7-N147D and pRS415-scTAF7-N147G. We found that scTAF7-N147D, like the WT TAF7-N147, rescued the lethality of TAF7-null yeast cells. This was expected because the aspartate residue is conserved in all other species, including the fission yeast (S. pombe). Notably, however, the scTAF7-N147G (analogous to the infertility-associated human mutation) did not rescue the lethality caused by TAF7 deletion in yeast (Table 3), showing that this conserved D/N residue is required for the function of TAF7.
Table 3.
Yeast (S. cerevisiae) complementation assay
| Plasmid | taf7∆ (null) yeast |
|---|---|
| None | nonviable |
| scTAF7-N147(WT) | viable |
| scTAF7-N147D | viable |
| scTAF7-N147G | nonviable |
Taf7l D144G/Y male mice exhibit apparently normal fertility
To assess if this point mutation in TAF7L contributes to human male infertility, we generated Taf7lD144G knockin mice, which harbor the D144G point mutation (analogous to the human D136G mutation) (Figure 2A). The point mutation was located in the fourth exon of Taf7l (Figure 2A). The WT and knockin Taf7l alleles were assayed by PCR (Figure 2B). Sanger sequencing of PCR products confirmed the point mutation in mice (Figure 2C). The Taf7lD144G allele was backcrossed to the C57BL/6J genetic background for 10 generations. Quantitative RT-PCR and western blot analyses of adult testes (3-month-old) showed no significant difference in mRNA and protein levels of Taf7l between WT (Taf7l+/Y) and Taf7lD144G/Y testes (Figure 2D and E).
Taf7l D144G/Y male mice displayed no overt developmental defects and sired offspring. The testis size of Taf7lD144G/Y males was comparable to that of WT males (Figure 3A). Body weight, testis weight, and epididymis weight were similar between WT and Taf7lD144G/Y males (Figure 3B–D). CASA showed no difference in epididymal sperm count or the percentage of sperm with progressive motility between WT and Taf7lD144G/Y males (Figure 3E and F). All other sperm motility parameters for Taf7lD144G/Y males were normal as well (Supplementary Table S3). Mating tests were performed by mating WT or Taf7lD144G/Y males with Taf7lD144G/+ females for 10 months. The total number of litters sired was similar between WT and Taf7lD144G/Y males (Figure 3G). The average litter size was lower in Taf7lD144G/Y males than in WT males, but the difference was not statistically significant (Figure 3G). Histological analysis of testes revealed apparently normal spermatogenesis in seminiferous tubules from Taf7lD144G/Y testes, which contained the full spectrum of spermatogenic cells: spermatogonia, spermatocytes, and postmeiotic spermatids (Figure 3H). Mature sperm were abundant in the cauda epididymides of WT and Taf7lD144G/Y males (Figure 3H). These results indicated that the fertility of Taf7lD144G/Y males was normal.
Figure 3.
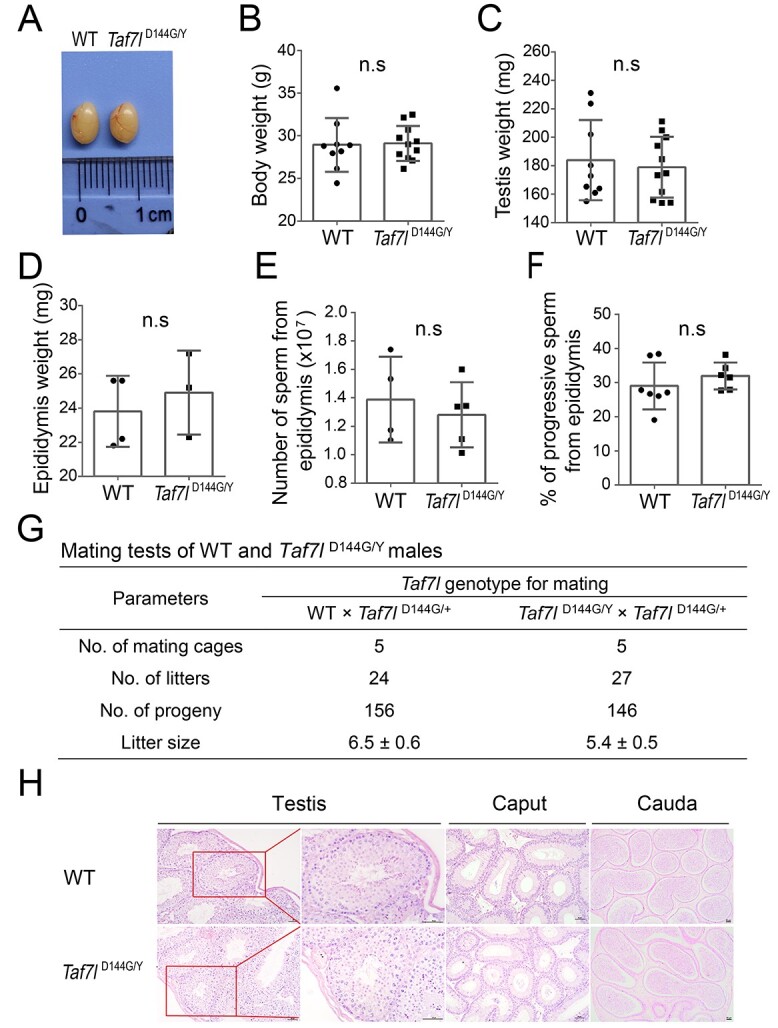
Characterization of the Taf7lD144G/Y male mice. (A) Images of testes from WT and Taf7lD144G/Y males (3 months old). (B–F) Body weight (B), testis weight (C), epididymis weight (D), epididymal sperm count (E), and percentage of epididymal sperm with progressive motility (F) of 3-month-old WT and Taf7lD144G/Y mice. The mean ± SEM is plotted. n, number of mice. n.s.: nonsignificant. (G) Mating test results. Each male was housed with two females for 10 months. (H) Histology of testis, caput epididymis, and cauda epididymis from 3-month-old WT and Taf7lD144G/Y males. Scale bar: 50 μm.
To test whether the D144G substitution in TAF7L affects its interaction with TBP, we constructed expression plasmids for HA-TBP, FLAG-TAF7L (WT), and FLAG-TAF7L (D144G). Following transfection in HEK293T cells, co-immunoprecipitations coupled with the western blot assay showed that the TAF7L (D144G) protein was still able to interact with TBP (Figure S1).
Alterations in the transcriptome of Taf7lD144G/Y testes
As TAF7L is a transcription factor, we performed RNA-seq analysis of WT and Taf7lD144G/Y testes to address whether the D144G substitution causes transcriptomic changes. Principal component analysis showed that the three biological replicates for each genotype were apparently clustered, except for a relative divergence of one sample per genotype (mRNAs, Figure 4A; lncRNAs, Supplementary S2A; circRNAs, Figure S2B). The intergenotype divergence and intragenotype convergence were clearly shown by the sample correlation analyses by Pearson’s method (mRNAs, Figure 4B; lncRNAs, Supplementary Figure S2C; circRNAs, Supplementary Figure S2D). We identified differentially expressed protein-coding genes (DE genes) in Taf7lD144G/Y testis with the cutoff of >1.5 fold-change and P < 0.05: 165 upregulated genes and 77 downregulated genes (Figure 4C). In addition, our analysis revealed the altered expression of 507 lncRNAs (Supplementary Figure S2E) and 135 circRNAs (Supplementary Figure S2F) in Taf7lD144G/Y testis. Furthermore, we prepared populations of pacSC and rST from WT and Taf7lD144G/Y testes by the STA-PUT method (Supplementary Figure S3), and performed RNA-seq analysis (Supplementary Figures S4 and S5). For both cell types, the intragenotype triplicates were much more uniform than the intergenotype groups (Supplementary Figure S4A–D). We identified 38 upregulated and 227 downregulated genes in Taf7lD144G/Y pacSC (Supplementary Figure S5A and C). In Taf7lD144G/Y rST, 42 genes were upregulated and 80 downregulated (Supplementary Figure S5B and D). A selection of differentially expressed DE genes in Taf7lD144G/Y rST were validated by qRT-PCR (Supplementary Figure S5E and F; Supplementary Table S4). We then analyzed the overlap of DE genes in the Taf7lD144G/Y testes with those reported in the Taf7l knockout testes [14, 15] and found minimal overlaps (Figure 4D and E). These results indicate that the D144G substitution alters the testicular transcriptome, and these alterations are not severe enough to impact fertility in the Taf7lD144G/Y males.
Figure 4.
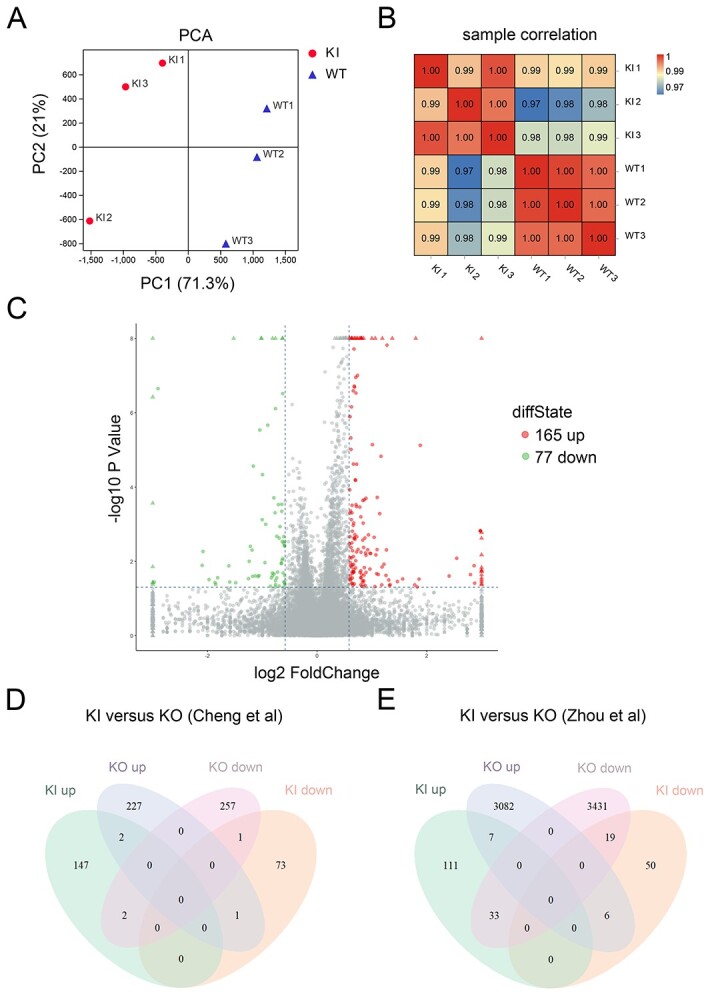
Dysregulation of transcriptomes in Taf7lD144G/Y mouse testes. The RNA-seq data from WT and Taf7lD144G/Y mouse testes (3 months old) were analyzed for protein-coding transcripts. (A) Principal component analysis of RNA-seq samples from WT and Taf7lD144G/Y testes. Three WT samples: WT1, WT2, and WT3; Three knockin samples: KI1, KI2, and KI3. (B) Pearson’s correlation analysis of RNA-seq samples. (C) Volcano plot of transcript levels between WT and Taf7lD144G/Y testes. The differentially expressed genes are highlighted in red (upregulated in Taf7lD144G/Y testis) or green (downregulated in Taf7lD144G/Y testis). (D) Comparison of differentially expressed coding transcripts in Taf7lD144G/Y testis and with those identified in Taf7l knockout testis [14]. (E) Comparison of differentially expressed coding transcripts in Taf7lD144G/Y testis and Taf7l knockout testis [15].
Discussion
Here we identified a missense mutation (D136G) in TAF7L in an oligozoospermic man. This aspartate residue is critical for TAF7 function in budding yeast. Interestingly, a single amino acid substitution (G690D) in TAF1, an X chromosome-encoded TAF that is the largest subunit of TFIID, confers a temperature-sensitive phenotype in the hamster ts13 cell line [47]. TAF1L is an autosomal retrotransposed homologue of TAF1 and is testis-specific in humans [33]. The WT TAF1L rescued the temperature-sensitive lethality of hamster ts13 cells but a mutant TAF1L (G714D; analogous mutation) did not, indicating that TAF1L and TAF1 are functionally equivalent [33]. These studies point to the selection pressure (from meiotic silencing of sex chromosomes in male mammals) on X-linked TAFs and the continuous evolution of testis-specific TAF homologues.
Although the Taf7l knockout male mice are subfertile with reduced sperm count [14], the Taf7lD144G/Y males have normal sperm count. The difference in the phenotypes of these two mouse mutants (knockout versus point mutation) could reflect functional compensation by TAF7. TAF7 compensation might be sufficient to restore fertility in Taf7lD144G/Y mice with compromised TAF7L function but not in knockout mice with a complete loss of TAF7L. TAF7 is essential for embryonic development [48]. Therefore, it might be informative to generate Taf7+/−Taf7lD144G/Y mice or Taf7+/−Taf7l-/Y mice in future studies to determine whether Taf7 heterozygosity leads to more severe spermatogenic defects in Taf7l mutant mice.
As exome and whole-genome sequencing has become increasingly cost-effective, a large number of human patients can be screened and as a result, an overwhelming number of sequence variants have been identified. However, the challenge lies in the prediction and interpretation of the functional consequences of these variants. Although the deleterious effect of nonsense mutations is predictable, the functional relevance of missense mutations or intronic variants is difficult to infer. Bioinformatic tools provide predictions as to the possible pathological impact of variants but cannot substitute for experimental validation. Although variants for somatic functions can be tested in cell lines, this becomes more challenging for testing variants in human germ cells because a human in vitro gametogenesis system is yet to be developed [49]. Therefore, animal modeling of sequence variants identified in infertile patients remains the only option for functional studies. We have investigated the functional consequences of the human D136G substitution in TAF7L identified in an oligozoospermic man by modeling this substitution in budding yeast and mouse. While aspartic acid and asparagine are functionally interchangeable in the yeast TAF7 gene, G at this position leads to lethality in yeast (Table 3). We also modeled this mutation in Taf7lD144G/Y mice. Although Taf7lD144G/Y testes exhibit transcriptomic changes, Taf7lD144G/Y mice have normal sperm count, normal sperm motility, and normal fertility. There are several possible explanations for the difference in phenotypes for this amino acid substitution between mouse and human. First, there might be differences in species-specific requirements for TAF7L. Although the human and mouse TAF7 proteins share 90% sequence identity, the human and mouse TAF7L proteins exhibit only 57% sequence identity, showing that the TAF7L proteins are more diverged. Thus, it is possible that human spermatogenesis is more dependent on TAF7L than mouse spermatogenesis. Second, the human patient may have variants in other genes that have synthetic deleterious effects with the TAF7L D136G substitution. Third, given dysregulation of some noncoding RNAs (lncRNAs and circRNAs) in the Taf7lD144G/Y testis, a fertility defect might only be induced under certain stress circumstances [50, 51]. For example, circRNAs protect male fertility under heat stress conditions through the regulation of ubiquitination and degradation of heat shock proteins [52]. Considering that the patient with this point mutation became infertile after fathering a child and that TAF7L plays a crucial role in metabolism in mouse [18], the infertility in this patient could be induced as a response to metabolic distress.
Data availability
The RNA-seq data are available from the National Center for Biotechnology Information’s GEO database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE186296.
Authors’ contributions
K.Z., L.L., F.F.L., P.L.Y., and Q.L.Y. carried out the experiments. R.D.O. and S.S. provided patient samples. P.J.W., H.S., L.G.B., and S.R. carried out the mutation screening. P.J.W., D.C.P., and K.Z. analyzed data. C.K. and F.C.L. performed the yeast experiments. N.A.L. performed blastocyst injection. J.W.Z. performed RNA-seq analyses. P.J.W., K.Z., L.L., and F.F.L. wrote the manuscript.
Supplementary Material
Acknowledgment
We thank Haiying Zhou (University of California, Berkeley) for western blotting analysis.
Contributor Information
Li Ling, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing, China.
Fangfang Li, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing, China.
Pinglan Yang, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing, China.
Robert D Oates, Department of Urology, Boston University Medical Center, Boston, MA, USA.
Sherman Silber, Infertility Center of St. Louis, St. Luke's Hospital, St. Louis, MO, USA.
Cornelia Kurischko, Department of Biomedical Sciences, University of Pennsylvania School of Veterinary Medicine, Philadelphia, PA, USA.
Francis C Luca, Department of Biomedical Sciences, University of Pennsylvania School of Veterinary Medicine, Philadelphia, PA, USA.
N Adrian Leu, Department of Biomedical Sciences, University of Pennsylvania School of Veterinary Medicine, Philadelphia, PA, USA.
Jinwen Zhang, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing, China.
Qiuling Yue, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing, China.
Helen Skaletsky, Howard Hughes Medical Institute, Whitehead Institute, and Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.
Laura G Brown, Howard Hughes Medical Institute, Whitehead Institute, and Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.
Steve G Rozen, Centre for Computational Biology, Duke-NUS Graduate Medical School, Singapore.
David C Page, Howard Hughes Medical Institute, Whitehead Institute, and Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.
P Jeremy Wang, Department of Biomedical Sciences, University of Pennsylvania School of Veterinary Medicine, Philadelphia, PA, USA.
Ke Zheng, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing, China.
Conflict of interest
The authors have declared that no conflict of interest exists.
References
- 1. Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol 2002; 4 (Suppl):s41–49. [DOI] [PubMed] [Google Scholar]
- 2. Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol 2018; 15:369–384. [DOI] [PubMed] [Google Scholar]
- 3. Jiao S-Y, Yang Y-H, Chen S-R. Molecular genetics of infertility: loss-of-function mutations in humans and corresponding knockout/mutated mice. Hum Reprod Update 2021; 27:154–189. [DOI] [PubMed] [Google Scholar]
- 4. Alhathal N, Maddirevula S, Coskun S, Alali H, Assoum M, Morris T, Deek HA, Hamed SA, Alsuhaibani S, Mirdawi A, Ewida N, Al-Qahtani Met al. . A genomics approach to male infertility. Genet Med 2020; 22:1967–1975. [DOI] [PubMed] [Google Scholar]
- 5. Irvine DS. Epidemiology and aetiology of male infertility. Hum Reprod 1998; 13:33–44. [DOI] [PubMed] [Google Scholar]
- 6. Mitchell MJ, Metzler-Guillemain C, Toure A, Coutton C, Arnoult C, Ray PF. Single gene defects leading to sperm quantitative anomalies. Clin Genet 2017; 91:208–216. [DOI] [PubMed] [Google Scholar]
- 7. Vockel M, Riera-Escamilla A, Tüttelmann F, Krausz C. The X chromosome and male infertility. Hum Genet 2021; 140:203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med 2008; 14:1197–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Houston BJ, Riera-Escamilla A, Wyrwoll MJ, Salas-Huetos A, Xavier MJ, Nagirnaja L, Friedrich C, Conrad DF, Aston KI, Krausz C, Tüttelmann F, O'Bryan MKet al. . A systematic review of the validated monogenic causes of human male infertility: 2020 update and a discussion of emerging gene-disease relationships. Hum Reprod Update 2021; 28:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Houston BJ, Conrad DF, O'Bryan MK. A framework for high-resolution phenotyping of candidate male infertility mutants: from human to mouse. Hum Genet 2021; 140:155–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet 2001; 27:422–426. [DOI] [PubMed] [Google Scholar]
- 12. Chiang CM, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 1995; 267:531–536. [DOI] [PubMed] [Google Scholar]
- 13. Pointud J-C, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, Davidson I. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci 2003; 116:1847–1858. [DOI] [PubMed] [Google Scholar]
- 14. Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, Davidson I, Wang PJ. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol 2007; 27:2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou H, Grubisic I, Zheng K, He Y, Wang PJ, Kaplan T, Tjian R. Taf7l cooperates with Trf2 to regulate spermiogenesis. Proc Natl Acad Sci U S A 2013; 110:16886–16891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang F, Gell K, Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Höög C, McLaughlin KJ, Wang PJ. Meiotic failure in male mice lacking an X-linked factor. Genes Dev 2008; 22:682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng K, Yang F, Wang PJ. Regulation of male fertility by X-linked genes. J Androl 2010; 31:79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou H, Kaplan T, Li Y, Grubisic I, Zhang Z, Wang PJ, Eisen MB, Tjian R. Dual functions of TAF7L in adipocyte differentiation. Elife 2013; 2:e00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akinloye O, Gromoll J, Callies C, Nieschlag E, Simoni M. Mutation analysis of the X-chromosome linked, testis-specific TAF7L gene in spermatogenic failure. Andrologia 2007; 39:190–195. [DOI] [PubMed] [Google Scholar]
- 20. Stouffs K, Willems A, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. The role of the testis-specific gene hTAF7L in the aetiology of male infertility. Mol Hum Reprod 2006; 12:263–267. [DOI] [PubMed] [Google Scholar]
- 21. Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta Oet al. . Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet 1995; 10:383–393. [DOI] [PubMed] [Google Scholar]
- 22. Reijo R, Alagappan RK, Patrizio P, Page DC. Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet 1996; 347:1290–1293. [DOI] [PubMed] [Google Scholar]
- 23. Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Köhn FM, Schill WB, Farah S, Ramos C, Hartmann M, Hartschuh Wet al. . Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet 1996; 5:933–943. [DOI] [PubMed] [Google Scholar]
- 24. Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 2001; 29:279–286. [DOI] [PubMed] [Google Scholar]
- 25. Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, Page DC, Rozen S. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet 2002; 71:906–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Green T, Green T, Flash S. Sex differences in psychiatric disorders: what we can learn from sex chromosome aneuploidies. Neuropsychopharmacology 2019; 44:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res 1998; 8:1229–1231. [DOI] [PubMed] [Google Scholar]
- 28. Yang F, Silber S, Leu NA, Oates RD, Marszalek JD, Skaletsky H, Brown LG, Rozen S, Page DC, Wang PJ. TEX11 is mutated in infertile men with azoospermia and regulates genome-wide recombination rates in mouse. EMBO Mol Med 2015; 7:1198–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7:575–576. [DOI] [PubMed] [Google Scholar]
- 30. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013; Chapter 7:Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015; 31:2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gutkin M, Brown RJ, McLean L, Streimer J, Kanaan RA. Shared Individual Formulation Therapy (SIFT): an open-label trial of a new therapy accommodating patient heterogeneity in functional neurological disorder. J Neurol 2021; 268:4882–4889. [DOI] [PubMed] [Google Scholar]
- 33. Wang PJ, Page DC. Functional substitution for TAF(II)250 by a retroposed homolog that is expressed in human spermatogenesis. Hum Mol Genet 2002; 11:2341–2346. [DOI] [PubMed] [Google Scholar]
- 34. Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly Cet al. . Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999; 285:901–906. [DOI] [PubMed] [Google Scholar]
- 35. Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff Aet al. . Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002; 418:387–391. [DOI] [PubMed] [Google Scholar]
- 36. Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol 1991; 194:1–863. [PubMed] [Google Scholar]
- 37. Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol 1997; 62:159–168. [PubMed] [Google Scholar]
- 38. Bryant J, Meyer-Ficca M, Dang V, Berger S, Meyer R. Separation of spermatogenic cell types using STA-PUT velocity sedimentation. J Vis Exp 2013; 80:50648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements, Nat Methods 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads, Nat Biotechnol 2015; 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, Gao G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res 2017; 45:W12–w16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 2013; 41:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold Uet al. . Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 45. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hayashida T, Sekiguchi T, Noguchi E, Sunamoto H, Ohba T, Nishimoto T. The CCG1/TAFII250 gene is mutated in thermosensitive G1 mutants of the BHK21 cell line derived from golden hamster. Gene 1994; 141:267–270. [DOI] [PubMed] [Google Scholar]
- 48. Gegonne A, Tai X, Zhang J, Wu G, Zhu J, Yoshimoto A, Hanson J, Cultraro C, Chen Q-R, Guinter T, Yang Z, Hathcock Ket al. . The general transcription factor TAF7 is essential for embryonic development but not essential for the survival or differentiation of mature T cells. Mol Cell Biol 2012; 32:1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saitou M, Hayashi K. Mammalian in vitro gametogenesis. Science 2021; 374:eaaz6830. [DOI] [PubMed] [Google Scholar]
- 50. Gao F, Cai Y, Kapranov P, Xu D. Reverse-genetics studies of lncRNAs-what we have learnt and paths forward. Genome Biol 2020; 21:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Han X, Luo S, Peng G, Lu JY, Cui G, Liu L, Yan P, Yin Y, Liu W, Wang R, Zhang J, Ai Set al. . Mouse knockout models reveal largely dispensable but context-dependent functions of lncRNAs during development. J Mol Cell Biol 2018; 10:175–178. [DOI] [PubMed] [Google Scholar]
- 52. Gao L, Chang S, Xia W, Wang X, Zhang C, Cheng L, Liu X, Chen L, Shi Q, Huang J, Xu EY, Shan G. Circular RNAs from BOULE play conserved roles in protection against stress-induced fertility decline. Sci Adv 2020; 6:eabb7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are available from the National Center for Biotechnology Information’s GEO database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE186296.