Abstract
Objective: We aimed to investigate the relationship between titin (TTN) gene mutations and thyroid cancer (THCA) and to explore the feasibility of the TTN gene as a potential prognostic indicator of THCA.
Methods: From TCGA-THCA cohort, we performed a series of analyses to evaluate the prognostic value and potential mechanism of TTN in THCA. These patients were divided into the mutant-type (MUT) group and the wild-type (WT) group. Differentially expressed genes (DEGs) in the two groups were screened using the ‘DESeq2’ R package. Functional enrichment analysis was performed, and the protein–protein interaction (PPI) network, transcription factor (TF)-target interaction networks, and competitive endogenous RNA (ceRNA) regulatory networks were established for the DEGs. The TIMER database was applied for immune cell infiltration. Survival analysis and Cox regression analysis were used to analyze the potential prognostic value of the TTN gene.
Results: Differential expression analysis showed that 409 genes were significantly up-regulated and 36 genes were down-regulated. Functional enrichment analysis revealed that TTN gene mutations played a potential role in the development of THCA. Analysis of the immune microenvironment indicated that TTN gene mutations were significantly associated with enrichment of M0 macrophages. Survival analysis showed that the MUT group predicted poorer prognosis than the WT group. Cox regression analysis demonstrated that TTN gene mutations were an independent risk factor for THCA. Nomograms also confirmed the prognostic values of the TTN gene in THCA.
Conclusions In summary, our results demonstrated that TTN gene mutations predict poor prognosis in patients with THCA. This is the first study to research TTN gene mutations in THCA and to investigate their prognostic value in THCA.
Keywords: bioinformatics analysis, gene mutations, poor prognosis, thyroid cancer, TTN gene
Introduction
Thyroid cancer (THCA) is the most common endocrine malignancy, and its incidence is increasing rapidly worldwide [1]. Implementation of conservative diagnostic practices did not result in a decline in THCA incidence in men [2]. The follicular-derived thyroid cancers are divided into well-differentiated papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), poorly differentiated thyroid cancer (PDTC), and anaplastic thyroid cancer (ATC). PTC is one of the most common forms of THCA [3]. Although well-differentiated thyroid cancers, such as PTC, have a good prognosis, the mortality rates of PDTC and ATC are still relatively high [4]. To date, the standard treatment for THCA includes surgery, radioactive iodine (RAI) therapy, and thyroid-stimulating hormone (TSH) inhibition. However, many patients still show recurrence or progression. Notably, as more gene mutations are being recognized and becoming drug targets, more novel treatments are being studied in THCA, including the BRAF inhibitors, vemurafenib [5] and dabrafenib [6]. Although the role of gene mutations in THCA has been partially clarified, there are no clear drugs for targeted therapy, and the pathogenesis and molecular mechanisms of THCA remain to be elucidated. The treatment of refractory thyroid cancer also remains a major challenge, and intensive efforts have been made to find an effective treatment for refractory thyroid cancer [7]. Thus, the identification of new potential biomarkers to guide targeted clinical therapy is urgently needed.
In recent years, with the rapid development of molecular biology, it has been found that the occurrence and development of cancer are related to gene mutation and methylation. Many genes and biomarkers related to THCA have been reported. Currently, BRAF mutation, NTRK3 fusion, NRAS mutation, and RET fusion are of concern [8,9]. The discovery of new biomarkers has significantly improved the understanding of the molecular pathogenesis of THCA, thus providing more personalized treatment for patients with THCA. Despite significant advances in molecular therapy, there is still a lack of effective therapies for advanced and radioactive iodine-refractory differentiated thyroid cancer [10]. Good diagnostic and prognostic biomarkers still have not been found.
Studies have shown that titin (TTN) gene mutations are related to the occurrence and development of tumors. TTN genes, also known as TMD, CMH9, CMD1G, CMPD4, and EOMFC, encode proteins in striated muscle. The 127 TTN-coding sequence variants associated with human conditions are distributed as follows: 33 nonsense mutations, 32 frameshifts, 36 missense variants, and 22 splice-site variants [11]. Recent studies have indicated that mutations in the TTN gene are among the most common genomic aberrations in ocular surface squamous neoplasia (OSSN) [12]. TTN was identified as the most frequently mutated gene within the pancancer cohort, and its mutation number was best correlated with tumor mutation burden (TMB) [13]. Yang et al. found that TTN mutations were associated with prognosis and higher TMB in gastric cancer [14]. This evidence supports the key role of the TTN gene in the carcinogenesis process. However, no studies have reported the relationship between TTN gene mutations and THCA. The present study focused on TTN gene mutations in THCA and explored the molecular mechanisms related to TTN gene mutations through bioinformatics analysis, which has not yet been reported.
In the present study, we aimed to explore the relationship between TTN gene mutations and THCA and to explore the feasibility of the TTN gene as a potential prognostic indicator of THCA. The results suggested that TTN gene mutations were closely related to patients with THCA and that the TTN gene was a potential clinical biomarker of THCA.
Methods
Data sources
‘Masked Somatic Mutation’ data can be obtained from the TCGA GDC website and was defined as somatic mutation data in patients with THCA in the present study. Data were processed using VarScan software, and visualization of somatic mutations was performed using the MAFTools R package [15]. Gene expression sequencing data (HTseq counts) of patients with THCA (n=568) were downloaded. In addition, clinical data of the control group (n=507) were downloaded using TCGA GDC software, including age, survival status, follow-up time, clinical stage, etc. Due to the exclusion of data with no survival information and incomplete TNM-staging information, 495 patients were retained for further analysis of clinical baseline data. These data were divided into the TTN gene mutation (mutant-type, MUT) group and the wild-type (WT) group.
Copy number variation analysis
To analyze the change in TTN gene copy-number variations in TCGA-THCA, the ‘TCGAbiolinks’ R package [16] was used to download the ‘Masked Copy Number Segment’ data. Copy number variations (CNVs) between the MUT group and the WT group were explored by GISTIC 2.0 [17] analysis of gene copy numbers from GenePattern (https://cloud.genepattern.org) [18].
The total number of somatic mutations detected in the tumor was defined as the TMB [19]. The Wilcoxon rank-sum test was applied to compare the overall difference in the level of TMB between the two groups.
Gene expression analysis
Differential gene expression refers to alterations in the expression (counts) of the sum of each of the isoforms that are encoded by a gene. The Wilcoxon rank-sum test was used to compare gene expression levels. In addition, differentially expressed genes (DEGs) in the two groups were screened by the ‘DESeq2’ R package [20]. The results are presented in the form of heatmaps and volcanoes. Adjusted P<0.05 and |log2fold change (FC)| > 1 were chosen as the cutoff criteria.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) is designed to analyze the association between gene sets and biological signals in one dataset [21]. To study the differences in biological processes between the two groups, GSEA was performed. The ‘c2.cp.v7.2.symbols [Curated]’ gene set was downloaded from the MSigDB database [22] for GSEA. Then, genetic ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed. GO analysis is a powerful bioinformatics tool used to identify biological processes (BPs), cellular components (CCs), and molecular functions (MFs) [23]. KEGG is a widely used database that stores information about genomes, biological pathways, diseases, and drugs [24]. The ‘ClusterProfiler’ R package [25] was used for GO enrichment analysis and KEGG pathway enrichment analysis. Adjusted P<0.05 and false discovery rate (FDR) < 0.25 were considered statistically significant.
Protein–protein interaction and hub gene identification
Protein–protein interaction (PPI) information was generated using STRING, which is an online database that identifies functional associations between proteins [26]. Genes (score > 0.9) were selected to construct the network model, and the results were visualized by Cytoscape (V3.7.2) software. Then, the maximal clique centrality (MCC) of the node genes in the PPI network was calculated by CytoHubba [27], and the top ten genes were selected as hub genes. Furthermore, we predicted the transcription factor (TF) and competitive endogenous RNA (ceRNA) networks interacting with DEGs using the Cistrome database (http://www.cistrome.org/) [28] and miRTarBase database (https://mirtarbase.cuhk.edu.cn/) [29], respectively. The corresponding interactive diagrams were generated using Cytoscape software.
Immune cell infiltration
CIBERSORT was used to quantify the abundance of immune cells in THCA samples, and the LM22 (10 of 22 cell subsets) signature was performed to distinguish 22 phenotypes of human-infiltrating immune cells [30]. To quantify the proportion of different immune cells in the THCA sample, we used the TIMER database to analyze the correlation between TTN gene mutations and immune cell infiltration in the TCGA-THCA dataset. The TIMER database [31], a web server for the comprehensive analysis of tumor-infiltrating immune cells, provides a user-friendly web interface for dynamic analysis and visualization.
Construction and validation of the clinical prediction model
Survival analysis was performed in both the MUT group and the WT group using the ‘survival’ R package [32]. To further investigate the influence of TTN gene mutations combined with clinicopathological features on the prognosis of patients, univariate and multivariate Cox regression analyses were applied to analyze the independent predictive ability of the risk score for patients with THCA. Subsequently, the TTN gene mutations and clinical information in the TCGA-THCA dataset were used to construct a nomogram that can predict the prognosis of THCA.
TTN gene mutations and drug sensitivity
The Genomics of Drug Sensitivity in Cancer (GDSC) database (http://www.cancerRxgene.org) is the largest public resource for information on drug sensitivity in cancer cells and molecular markers of drug response [33]. The PRRophetic algorithm [34] was used to construct a regression model based on the expression profile of the GDSC cell line and TCGA-THCA gene. IC50 analysis was used to predict the sensitivity of THCA samples to anticancer drugs.
Statistical analysis
Data processing and analysis were mostly achieved using R language, version 4.0.2. Most of the statistical analyses were performed using the bioinformatic tools mentioned above. IBM SPSS statistics 26.0 software was utilized for statistical analysis, and P<0.05 was considered to indicate a statistically significant difference.
Results
Analysis of mutation level and chromosome CNV in patients with THCA
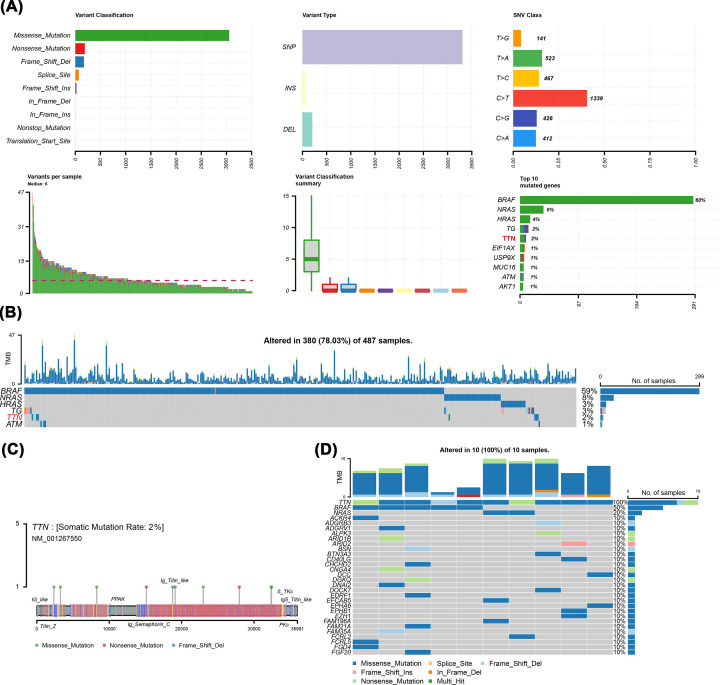
To analyze the impact of gene mutations on THCA, the characteristics of 495 patients with THCA including clinical and gene mutation data were collected from TCGA database. The baseline characteristics of the TCGA-THCA cohort are summarized in Table 1. The results showed that missense mutation was the main mutation type and the frequency of single-nucleotide polymorphisms was higher than that of insertions or deletions, and C>T was most common in single-nucleotide variants (SNVs) in THCA (Figure 1A). Furthermore, among all patients with THCA, the top five mutated genes were BRAF, NRAS, HRAS, TTN, and TG (Figure 1B). In recent years, there have been many reports on the study of BRAF and RAS gene mutations in THCA [35–37], but the role and mechanism of TTN gene mutations in THCA are not clear, and TTN gene mutations in THCA are rarely studied. Consequently, the present study focused on TTN gene mutations in THCA and thoroughly explored TTN mutation-related genes and potential mechanisms through bioinformatics methods. According to the mutation sites, we plotted the mutational landscapes of the TTN gene (Figure 1C). We further made the mutation panorama using the mafTools package (Figure 1D). In addition, we analyzed CNV differences between the two groups. The results indicated that compared with the WT group, the MUT group did not have a significant copy number amplification but did have a significant copy number deletion (Supplementary Figure S1).
Table 1. The baseline characteristics of the TCGA-THCA cohort.
| Characteristics | N (%) |
|---|---|
| Gender | |
| Female | 361 (73) |
| Male | 134 (27) |
| Age | |
| <60 | 378 (76) |
| ≥60 | 117 (24) |
| Stage | |
| Stage I | 279 (56) |
| Stage II | 52 (11) |
| Stage III | 111 (22) |
| Stage IV | 53 (11) |
| TTN mutation | |
| Wild-type | 485 (98) |
| Mutate-type | 10 (2) |
Figure 1. Analysis of the level of TTN mutations in patients with THCA.
(A) Panoramic images of gene mutations. (B) The top five mutated genes in the TCGA-THCA dataset. (C) Amino acid changes of TTN protein in TCGA-THCA dataset. (D) Analysis of overall mutation signature in the MUT group.
TTN mutations and biological characteristics
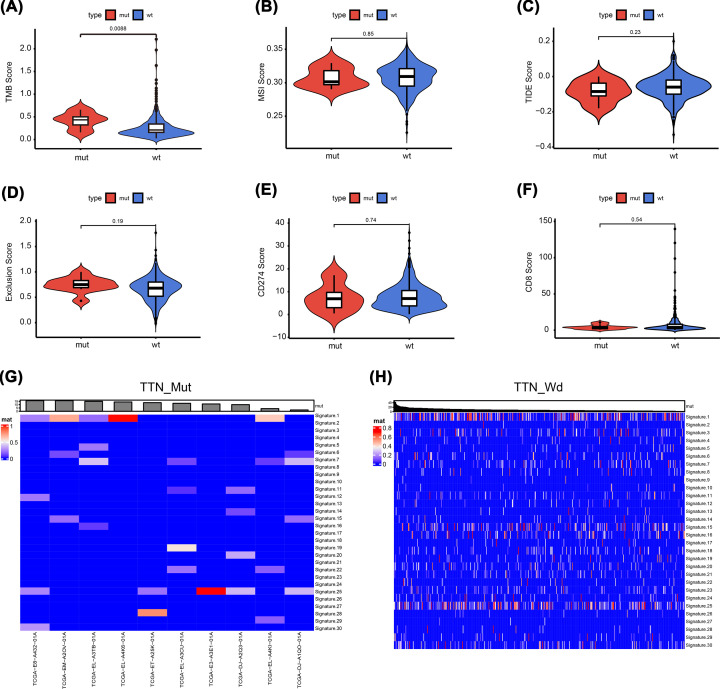
We further analyzed the correlation between TTN gene mutations and different biological characteristics. The results showed that TMB levels were significantly increased in the MUT group compared with the WT group (P=0.0088), but there were no significant differences in microsatellite instability (MSI), TIDE score, immune escape score, CD274 score, or CD8 score between the two groups (Figure 2A–F). In addition, the spectrum of 96 mutations was decomposed into 30 different signatures and combined with biological characteristics and somatic mutation characteristics. The results showed that there was no significant change in signature distribution between the two groups (Figure 2G,H).
Figure 2. Analysis of biological characteristics of TTN mutations in patients with THCA.
(A) TMB levels were significantly increased in the MUT group (P<0.001). (B–F) No significant differences in MSI, TIDE score, immune escape score, CD274 and CD8 scores between the two groups. (G,H) Heat map analysis between the two groups.
Differential gene expression analysis in patients with TTN mutations
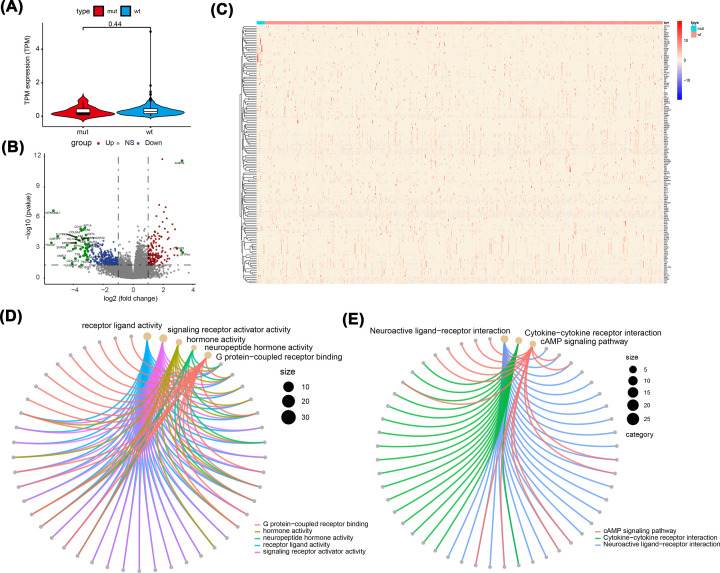
To analyze the effect of TTN gene mutations on the development of THCA, samples with THCA from the TCGA database were divided into the MUT group and the WT group. TTN gene expression levels are shown in Figure 3A. Subsequently, differential expression analysis was conducted between the two groups. The results showed that 409 genes were significantly up-regulated in the MUT group, and 36 genes were significantly down-regulated in the MUT group (Figure 3B,C). Next, we performed functional enrichment analysis of the DEGs. GO enrichment analysis showed that the DEGs were closely related to the biological processes of receptor ligand activity, signalling receptor activator activity, and hormone activity (Figure 3D). KEGG pathway enrichment analysis suggested that the DEGs mainly affected neuroactive ligand–receptor interactions, cytokine–cytokine receptor interactions, and cAMP signalling pathways (Figure 3E).
Figure 3. Differential expression analysis and functional enrichment analysis.
(A) TTN gene expression level between the two groups (P=0.053). (B,C) The volcano map and heat map between the two groups. (D) GO enrichment analysis. (E) KEGG pathway enrichment analysis.
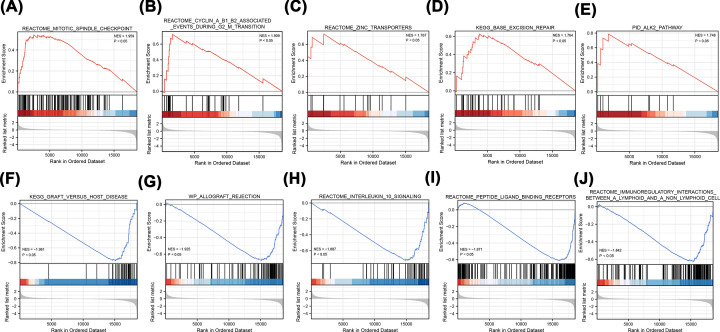
Meanwhile, based on the gene expression profile in the TCGA-THCA dataset, GSEA enrichment analysis results showed that mitotic spindle checkpoint-, cyclin A/B1/B2-associated events during the g2 m transition, zinc transporters, and KEGG base excision repair pathways were significantly enriched in the MUT group. KEGG analysis showed that genes involved in graft versus host disease, allograft rejection, and interleukin-10 signalling and peptide ligand-binding receptor genes were significantly enriched in the WT group (Figure 4A–J).
Figure 4. GSEA enrichment analysis.
(A–D) Based on NES values, the top five enriched pathways in the MUT group were shown. (E–J) Based on NES values, the top five enriched pathways in the WT group were shown. NES value represents the enrichment score after normalization. The higher the NES value, the more genes were enriched in the pathway.
PPI and hub gene identification
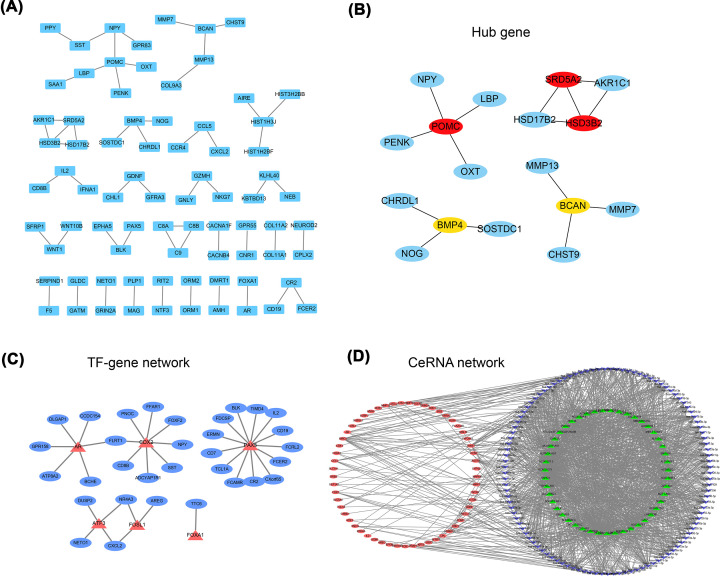
A PPI network between DEGs was established through the STRING database (Figure 5A). Through the CytoHubba plug-in in Cytoscape, the MCC algorithm was used to select the top ten genes, which were called hub genes (Figure 5B). TF-target interaction networks and ceRNA regulatory networks were obtained from the Cistrome database and miRTarBase database (Figure 5C,D), respectively. These data were visualized using Cytoscape software.
Figure 5. Construction of PPI, TF-target interaction networks, and ceRNA regulatory networks.
(A) PPI networks between DEGs was established through STRING database. Each node represents a different gene. (B) Hub gene. (C) TF-target interaction networks. Red represents TF and blue represents target genes. (D) ceRNA regulatory networks. Green, blue, and red represent lncRNA, miRNA, and mRNA, respectively.
Effect of TTN mutations on immune cell infiltration in patients with THCA
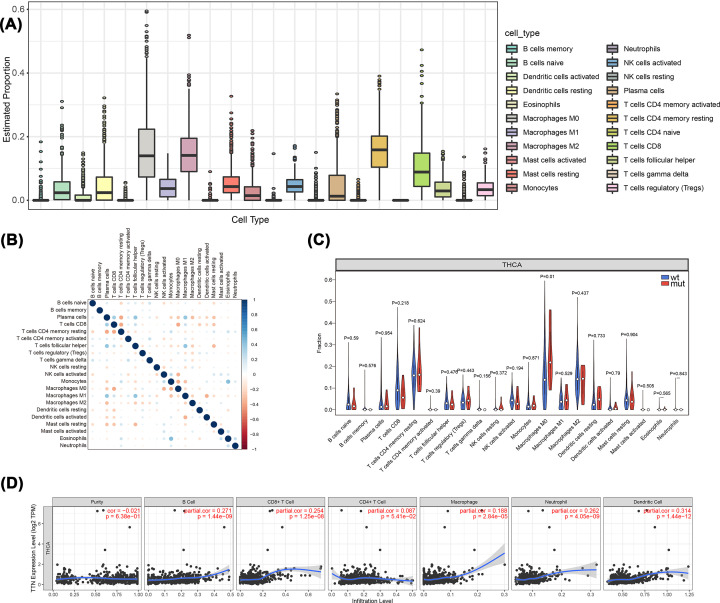
To analyze the relationship between TTN gene mutations and immune cell invasion in the tumor microenvironment, we calculated the proportion of immune cell invasion in the tumor microenvironment using the CIBERSORT algorithm. Figure 6A,B shows the panorama of immune cell infiltration and immune cell score, respectively. Then, the expression differences of immune cells in the two groups were further analyzed, and M0 macrophages were found to have a higher infiltration score in the MUT group than in the WT group (Figure 6C). Correlation analysis based on the TIMER database showed that there were significant differences in the infiltration levels of various immune cells among different TTN gene mutation levels (Figure 6D).
Figure 6. The relationship between TTN mutations and immune cell invasion.
(A) The panorama of immune cell infiltration. (B) Correlation analysis of 22 kinds of immune cells in the TCGA-THCA dataset. (C) Comparison of immune cell infiltration between the two groups. (D) Correlation analysis between TTN and different immune cells.
Correlation analysis of TTN mutations and clinical features
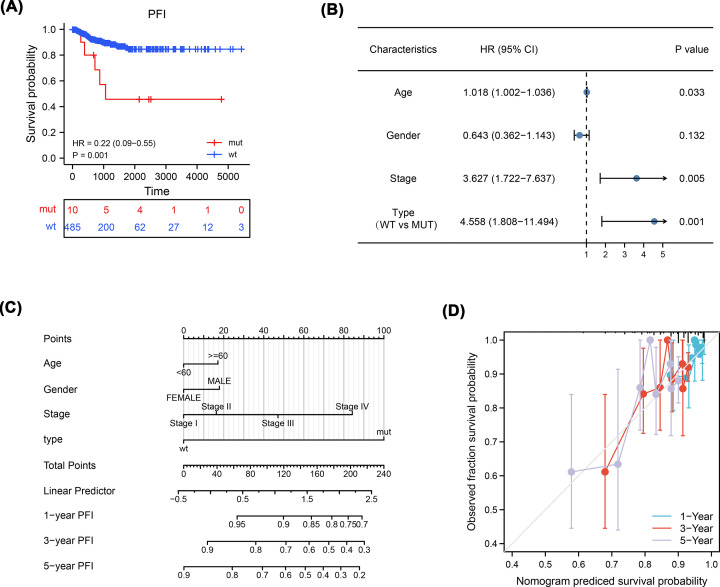
To explore the association between TTN gene mutations and the clinical prognosis of patients with THCA, we conducted a clinical correlation analysis combining clinicopathological features. Survival analysis showed that the MUT group predicted poorer prognosis [progression-free interval (PFI), hazard ratio (HR)=0.22, [95% confidence interval (CI), 0.09–0.55], P=0.001] than the WT group (Figure 7A). Univariate and multivariate Cox regression analyses demonstrated that TTN gene mutations were an independent risk factor for THCA [overall survival (OS), HR=4.558, [CI, 1.808–11.494], P=0.001] (Figure 7B). Moreover, we combined TTN gene mutations with clinicopathological features to construct a nomogram to predict the prognosis of patients with THCA (Figure 7C). The efficiency of the nomogram was verified using the calibration curve, and the results showed that the 1-year nomogram had good consistency with the actual observed OS of patients (Figure 7D).
Figure 7. Effect of TTN mutations on clinicopathological features.
(A) Survival analysis showed that the MUT group predicted poorer prognosis (PFI, HR=0.22, [95% CI, 0.09–0.55], P=0.001) than the WT group. (B) Cox regression analyses demonstrated that TTN gene mutations were an independent risk factor for THCA (OS, HR=4.558, [CI, 1.808–11.494], P=0.001). (C) Construction of a nomogram. (D) Calibration curve of the nomogram.
TTN mutations and drug sensitivity
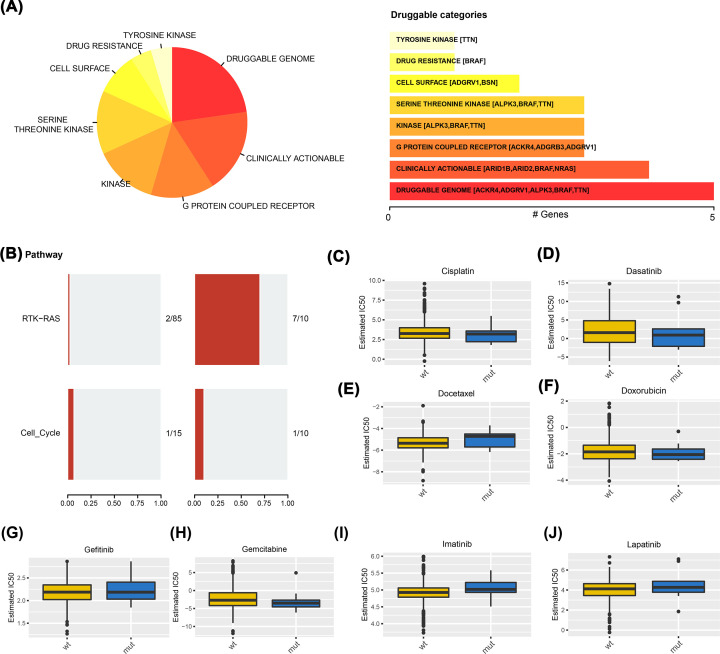
Finally, we analyzed the effect of TTN gene mutations on the drug sensitivity of patients with THCA and found that a variety of drugs were correlated with the level of TTN gene mutations, especially the druggable genome (Figure 8A). Pathway analysis showed that the RTK-RAS pathway was enriched in the MUT group (Figure 8B). Moreover, the IC50 values of antitumor drugs between the MUT group and WT group were analyzed using the GDSC database, and the results showed that cisplatin, docetaxel, and imatinib had different trends (Figure 8C–J).
Figure 8. The relationship between TTN mutations and drug sensitivity.
(A) A variety of drugs were correlated with the level of TTN mutations. (B) Analysis of changes in gene mutation levels in different oncogenic signaling pathways. (C–J) IC50 values of antitumor drugs were analyzed by GDSC database between the two groups.
Discussion
Many molecular alterations represent novel diagnostic and prognostic molecular markers and therapeutic targets for cancer, which provide unprecedented opportunities for further research and clinical development of novel treatment strategies [38]. In recent years, studies have shown that TTN gene mutations may be involved in the development of tumors. TTN is the largest described protein and constitutes the third most abundant type of filament in both cardiac and skeletal human muscle, together with actin and myosin [39]. The entire TTN gene consists of 364 exons and is located on chromosome 2q31 [40]. Single-cell DNA sequencing revealed that the most common mutation in gastroesophageal carcinoma is the TTN gene, which was reported to be associated with TMB in cancer [41]. TTN mutations play an important role in human cancer. However, the relationship between TTN mutations and THCA has not been reported. Hence, we performed a comprehensive bioinformatics analysis of TTN gene mutations and survival prognostic value in THCA. Our results demonstrated that TTN gene mutations predict poor prognosis in patients with THCA and that the TTN gene may serve as a potential prognostic indicator of THCA. This is the first study to research TTN gene mutations in THCA and to investigate their prognostic value in THCA.
TTN mutations associated with human conditions include nonsense mutations, frameshift mutations, missense mutations, and splice site mutations. Studies have reported that TTN missense mutations are correlated with lung squamous cell carcinoma [42,43]. Our study also proved that missense mutations account for the dominant mutations in patients with THCA. Next, to analyze the effect of TTN gene mutations on tumorigenesis in THCA, we divided the patients with THCA into the MUT group and the WT group. Differential expression analysis showed that 409 genes were significantly up-regulated and 36 genes were down-regulated in the MUT group compared with the WT group. Moreover, functional enrichment analysis was performed for the DEGs, including GO enrichment analysis, KEGG pathway enrichment analysis, and GSEA. Functional enrichment analysis revealed that TTN gene mutations played a potential role in the development of THCA. In addition, a PPI network was established. The results showed that TTN gene mutations mainly interact with POMC, SRD5A2, HSD3B2, BMP4, and BCAN. It was recently reported that these genes are involved in the progression of cancer [44–46]. We also explored the molecular mechanism of TTN gene mutations in THCA through TF-target interaction networks and ceRNA regulatory networks.
Increasing evidence has shown that immune infiltration is closely related to cancer [47–49]. Novel-targeted drugs and immunotherapies have developed rapidly over the last decade, bringing new treatments to patients with malignant tumors. Hence, we further analyzed the relationship between the carcinogenic effect of TTN gene mutations and immune infiltration. Analysis of the THCA immune microenvironment indicated that TTN mutation was significantly associated with enrichment of M0 macrophages (P<0.05).
Interestingly, we focused on the clinical significance of TTN gene mutations in THCA and discovered that TTN gene mutations may be a potential prognostic indicator for patients with THCA. First, survival analysis showed that the MUT group had a poorer predicted prognosis (PFI, HR=0.22, [95% CI, 0.09–0.55], P=0.001) than the WT group. Then, Cox regression analysis demonstrated that TTN gene mutations were an independent risk factor for THCA (OS, HR=4.558, [95% CI, 1.808–11.494], P=0.001). More importantly, we combined TTN gene mutations with clinicopathological features to construct a nomogram. The nomogram also confirmed the prognostic value of the TTN gene in THCA. In addition, TTN mutations resulted in increased thyroid cancer sensitivity to eight drugs (P<0.05), especially the druggable genome. These results suggest that the TTN gene plays a unique and crucial role in the prognosis of THCA.
Nevertheless, there are some limitations to our study. First, although bioinformatics analysis is a powerful and efficient tool to aid in understanding of molecular mechanisms and for identifying potential biomarkers underlying THCA, further experimental validations of the TTN gene are needed at the molecular, cellular, and organismal levels. Second, the present study focused on the prognostic value rather than the predictive value of TTN mutations in patients with THCA. Finally, further experimental evidence, such as evidence obtained from real-time PCR, western blot, and immunohistochemistry assays, is required to fully elucidate the role of hub genes and the underlying mechanisms of THCA.
In conclusion, further exploration of the potential value of the TTN gene as a therapeutic target is highly warranted and may ultimately lead to the development of innovative drug therapies for complex and serious diseases. This is the first study to demonstrate TTN gene mutations in THCA and to investigate their prognostic value in THCA. The present study will provide a potential therapeutic strategy for thyroid cancers.
Conclusions
In summary, new gene mutations in THCA have been identified through bioinformatics analysis, and these results contribute to the treatment and prognosis of thyroid cancer. This is the first study to demonstrate TTN gene mutations in THCA and to investigate their prognostic value in THCA.
Supplementary Material
Abbreviations
- ATC
anaplastic thyroid cancer
- BP
biological process
- CC
cellular component
- ceRNA
competitive endogenous RNA
- CI
confidence interval
- CNV
copy number variation
- DEG
differentially expressed gene
- FDR
false discovery rate
- FTC
follicular thyroid cancer
- GDSC
Genomics of Drug Sensitivity in Cancer
- GO
genetic ontology
- GSEA
gene set enrichment analysis
- HR
hazard ratio
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MCC
maximal clique centrality
- MF
molecular function
- MSI
microsatellite instability
- MUT
mutant-type
- OS
overall survival
- OSSN
ocular surface squamous neoplasia
- PDTC
poorly differentiated thyroid cancer
- PFI
progression-free interval
- PPI
protein–protein interaction
- PTC
papillary thyroid cancer
- RAI
radioactive iodine
- SNV
single-nucleotide variant
- TF
transcription factor
- THCA
thyroid cancer
- TMB
tumor mutation burden
- TSH
thyroid stimulating hormone
- TTN
titin
- WT
wild-type
Data Availability
Our study used public online database. The data can be accessed by following websites: https://cloud.genepattern.org, https://www.gsea-msigdb.org, http://geneontology.org/, http://www.genome.jp/kegg, https://string-db.org/, http://www.cistrome.org/, https://mirtarbase.cuhk.edu.cn/, and http://www.cancerRxgene.org.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
Support was provided by grants from the National Natural Science Funds of China [grant number 81760168].
CRediT Author Contribution
Xiudan Han: Formal analysis, Funding acquisition, Writing—original draft, Writing—review & editing. Jianrong Chen: Investigation. Jiao Wang: Visualization. Jixiong Xu: Project administration. Ying Liu: Resources, Data curation, Validation, Project administration.
References
- 1.Cabanillas M.E., McFadden D.G. and Durante C. (2016) Thyroid cancer. Lancet 388, 2783–2795 10.1016/S0140-6736(16)30172-6 [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fuchs H.E.et al. (2021) Cancer statistics, 2021. CA Cancer J. Clin. 71, 7–33 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 3.Hitu L., Cainap C., Apostu D.et al. (2020) Skeletal muscle metastasis in papillary thyroid microcarcinoma evaluated by F18-FDG PET/CT. Diagnostics (Basel) 10, 100 10.3390/diagnostics10020100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B. and Ghossein R. (2016) Genomic landscape of poorly differentiated and anaplastic thyroid carcinoma. Endocr. Pathol. 27, 205–212 10.1007/s12022-016-9445-4 [DOI] [PubMed] [Google Scholar]
- 5.Dadu R., Shah K., Busaidy N.L.et al. (2015) Efficacy and tolerability of vemurafenib in patients with BRAF(V600E)-positive papillary thyroid cancer: M.D. Anderson Cancer Center off label experience. J. Clin. Endocrinol. Metab. 100, E77–E81 10.1210/jc.2014-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falchook G.S., Millward M., Hong D.et al. (2015) BRAF inhibitor dabrafenib in patients with metastatic BRAF-mutant thyroid cancer. Thyroid 25, 71–77 10.1089/thy.2014.0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitale G., Pellegrino G., Desiderio E.et al. (2021) Radioiodine-refractory thyroid cancer: a complex challenge. Minerva Med. 112, 686–688 [DOI] [PubMed] [Google Scholar]
- 8.Huang G., Chen J., Zhou J.et al. (2021) Epigenetic modification and BRAF gene mutation in thyroid carcinoma. Cancer Cell Int. 21, 687 10.1186/s12935-021-02405-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macerola E., Proietti A., Poma A.M.et al. (2021) Molecular alterations in relation to histopathological characteristics in a large series of pediatric papillary thyroid carcinoma from a single institution. Cancers 13, 3123 10.3390/cancers13133123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laha D., Nilubol N. and Boufraqech M. (2020) New therapies for advanced thyroid cancer. Front. Endocrinol. (Lausanne) 11, 82 10.3389/fendo.2020.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chauveau C., Rowell J. and Ferreiro A. (2014) A rising titan: TTN review and mutation update. Hum. Mutat. 35, 1046–1059 10.1002/humu.22611 [DOI] [PubMed] [Google Scholar]
- 12.Djulbegovic M.B., Uversky V.N., Karp C.L.et al. (2022) Functional impact of titin (TTN) mutations in ocular surface squamous neoplasia. Int. J. Biol. Macromol. 195, 93–101 10.1016/j.ijbiomac.2021.11.120 [DOI] [PubMed] [Google Scholar]
- 13.Oh J.H., Jang S.J., Kim J.et al. (2020) Spontaneous mutations in the single TTN gene represent high tumor mutation burden. NPJ Genom. Med. 5, 33 10.1038/s41525-019-0107-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., Zhang J., Chen Y.et al. (2020) MUC4, MUC16, and TTN genes mutation correlated with prognosis, and predicted tumor mutation burden and immunotherapy efficacy in gastric cancer and pan-cancer. Clin. Transl. Med. 10, e155 10.1002/ctm2.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayakonda A., Lin D.C., Assenov Y.et al. (2018) Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 28, 1747–1756 10.1101/gr.239244.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colaprico A., Silva T.C., Olsen C.et al. (2016) TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic. Acids. Res. 44, e71 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mermel C.H., Schumacher S.E., Hill B.et al. (2011) GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reich M., Liefeld T., Gould J.et al. (2006) GenePattern 2.0. Nat. Genet. 38, 500–501 10.1038/ng0506-500 [DOI] [PubMed] [Google Scholar]
- 19.Merino D.M., McShane L.M., Fabrizio D.et al. (2020) Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J. Immunother. Cancer 8, e000147 10.1136/jitc-2019-000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love M.I., Huber W. and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A., Tamayo P., Mootha V.K.et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberzon A., Birger C., Thorvaldsdóttir H.et al. (2015) The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gene Ontology Consortium (2015) Gene Ontology Consortium: going forward. Nucleic. Acids. Res. 43, D1049–D1056 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M., Furumichi M., Tanabe M.et al. (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic. Acids. Res. 45, D353–D361 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G., Wang L.G., Han Y.et al. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D., Gable A.L., Nastou K.C.et al. (2021) The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic. Acids. Res. 49, D605–D612 10.1093/nar/gkaa1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin C.H., Chen S.H., Wu H.H.et al. (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8, S11 10.1186/1752-0509-8-S4-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng R., Wan C., Mei S.et al. (2019) Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic. Acids. Res. 47, D729–D735 10.1093/nar/gky1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H.Y., Lin Y.C., Li J.et al. (2020) miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic. Acids. Res. 48, D148–D154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman A.M., Liu C.L., Green M.R.et al. (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T., Fan J., Wang B.et al. (2017) TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108–e110 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonabend R., Király F.J., Bender A.et al. (2021) mlr3proba: an R package for machine learning in survival analysis. Bioinformatics 37, 2789–2791 10.1093/bioinformatics/btab039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W., Soares J., Greninger P.et al. (2013) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic. Acids. Res. 41, D955–D961 10.1093/nar/gks1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geeleher P., Cox N. and Huang R.S. (2014) pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 9, e107468 10.1371/journal.pone.0107468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaabouch M., Chahdi H., Azouzi N.et al. (2020) BRAF(V600E) hot spot mutation in thyroid carcinomas: first Moroccan experience from a single-institution retrospective study. Afr. Health Sci. 20, 1849–1856 10.4314/ahs.v20i4.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang E.K., Song D.E., Sim S.Y.et al. (2014) NRAS codon 61 mutation is associated with distant metastasis in patients with follicular thyroid carcinoma. Thyroid 24, 1275–1281 10.1089/thy.2014.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou R., Zhang L., Lu T.et al. (2018) Identification of a novel HRAS variant and its association with papillary thyroid carcinoma. Oncol. Lett. 15, 4511–4516 10.3892/ol.2018.7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing M. (2013) Molecular pathogenesis and mechanisms of thyroid cancer. Nat. Rev. Cancer 13, 184–199 10.1038/nrc3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vikhlyantsev I.M. and Podlubnaya Z.A. (2012) New titin (connectin) isoforms and their functional role in striated muscles of mammals: facts and suppositions. Biochemistry (Mosc.) 77, 1515–1535 10.1134/S0006297912130093 [DOI] [PubMed] [Google Scholar]
- 40.Hackman P., Vihola A., Haravuori H.et al. (2002) Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am. J. Hum. Genet. 71, 492–500 10.1086/342380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duan Q., Tang C., Ma Z.et al. (2021) Genomic heterogeneity and clonal evolution in gastroesophageal junction cancer revealed by single cell DNA sequencing. Front. Oncol. 11, 672020 10.3389/fonc.2021.672020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng X., Yin H., Fu J.et al. (2019) Aggregate analysis based on TCGA: TTN missense mutation correlates with favorable prognosis in lung squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 145, 1027–1035 10.1007/s00432-019-02861-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou S., Ye J., Hu S.et al. (2022) Mutations in the TTN gene are a prognostic factor for patients with lung squamous cell carcinomas. Int. J. Gen. Med. 15, 19–31 10.2147/IJGM.S343259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuo H., Yang D. and Wan Y. (2021) Fam20C regulates bone resorption and breast cancer bone metastasis through osteopontin and BMP4. Cancer Res. 81, 5242–5254 10.1158/0008-5472.CAN-20-3328 [DOI] [PubMed] [Google Scholar]
- 45.van Boxtel W., Verhaegh G.W., van Engen-van Grunsven I.A.et al. (2020) Prediction of clinical benefit from androgen deprivation therapy in salivary duct carcinoma patients. Int. J. Cancer 146, 3196–3206 10.1002/ijc.32795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alvarez-Breckenridge C., Miller J.J., Nayyar N.et al. (2017) Clinical and radiographic response following targeting of BCAN-NTRK1 fusion in glioneuronal tumor. NPJ Precis. Oncol. 1, 5 10.1038/s41698-017-0009-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao X., Xu J., Wang W.et al. (2021) Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. Cancer 20, 131 10.1186/s12943-021-01428-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu C.Q., Xia Q.D., Sun J.X.et al. (2022) Identification and validation of a twelve immune infiltration-related lncRNA prognostic signature for bladder cancer. Aging 14, 1492–1507 10.18632/aging.203889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferrall-Fairbanks M.C., Chakiryan N., Chobrutskiy B.I.et al. (2022) Quantification of T- and B-cell immune receptor distribution diversity characterizes immune cell infiltration and lymphocyte heterogeneity in clear cell renal cell carcinoma. Cancer Res. 82, 929–942 10.1158/0008-5472.CAN-21-1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our study used public online database. The data can be accessed by following websites: https://cloud.genepattern.org, https://www.gsea-msigdb.org, http://geneontology.org/, http://www.genome.jp/kegg, https://string-db.org/, http://www.cistrome.org/, https://mirtarbase.cuhk.edu.cn/, and http://www.cancerRxgene.org.