Abstract
A novel quantitative PCR (QPCR) approach, which combines competitive PCR with constant-denaturant capillary electrophoresis (CDCE), was adapted for enumerating microbial cells in environmental samples using the marine nanoflagellate Cafeteria roenbergensis as a model organism. Competitive PCR has been used successfully for quantification of DNA in environmental samples. However, this technique is labor intensive, and its accuracy is dependent on an internal competitor, which must possess the same amplification efficiency as the target yet can be easily discriminated from the target DNA. The use of CDCE circumvented these problems, as its high resolution permitted the use of an internal competitor which differed from the target DNA fragment by a single base and thus ensured that both sequences could be amplified with equal efficiency. The sensitivity of CDCE also enabled specific and precise detection of sequences over a broad range of concentrations. The combined competitive QPCR and CDCE approach accurately enumerated C. roenbergensis cells in eutrophic, coastal seawater at abundances ranging from approximately 10 to 104 cells ml−1. The QPCR cell estimates were confirmed by fluorescent in situ hybridization counts, but estimates of samples with <50 cells ml−1 by QPCR were less variable. This novel approach extends the usefulness of competitive QPCR by demonstrating its ability to reliably enumerate microorganisms at a range of environmentally relevant cell concentrations in complex aquatic samples.
It has become customary during the last two decades to document environmental microbial diversity by cloning and analyzing the sequence of small-subunit rRNA (16S rRNA) genes without prior cultivation of the organisms (2, 9). However, accurate quantification of abundance and dynamics of specific populations has remained difficult despite the importance of such information for increased understanding and prediction of environmental processes. Although hybridization-based approaches, such as quantitative slot-blot hybridization and fluorescent in situ hybridization (FISH), have been used successfully to estimate populations sizes, they require either highly abundant or rapidly growing populations due to their dependence on high target rRNA concentrations for detection (2, 9). Thus, to enhance the sensitivity of detection, quantitative PCR (QPCR) protocols are increasingly being adapted for the enumeration of microbial populations in environmental samples (10, 12, 13, 15, 21, 22, 26, 27). These methods estimate the abundance of specific gene sequences in samples as a proxy for the actual organism possessing the gene and offer the sensitivity, specificity, and ease of use innate to PCR.
Two approaches, competitive QPCR and real-time QPCR, are currently most widely used in microbial ecology applications. Both methods estimate the target gene concentration in a sample by comparison with standard curves constructed from amplifications of serial dilutions of standard DNA. However, they differ substantially in how these standard curves are generated. In competitive QPCR, an internal competitor DNA is added at a known concentration to both serially diluted standard samples and unknown (environmental) samples. After coamplification, ratios of the internal competitor and target PCR products are calculated for both standard dilutions and unknown samples, and a standard curve is constructed that plots competitor-target PCR product ratios against the initial target DNA concentration of the standard dilutions (4, 30). Given equal amplification efficiency of competitor and target DNA, the concentration of the latter in environmental samples can be extrapolated from this standard curve. In the second method, real-time QPCR, the accumulation of amplification product is measured continuously in both standard dilutions of target DNA and samples containing unknown amounts of target DNA. A standard curve is constructed by correlating initial template concentration in the standard samples with the number of PCR cycles (CT) necessary to produce a specific threshold concentration of product. In the test samples, target PCR product accumulation is measured after the same CT, which allows interpolation of target DNA concentration from the standard curve.
Although real-time QPCR permits more rapid and facile measurement of target DNA during routine analyses, competitive QPCR remains an important alternative for target quantification in environmental samples. The coamplification of a known amount of competitor DNA with target DNA is an intuitive way to correct for sample-to-sample variation of amplification efficiency due to the presence of inhibitory substrates and large amounts of background DNA that are obviously absent from the standard dilutions (3). However, many currently used competitive QPCR protocols are hampered by inaccurate and cumbersome post-PCR handling, frequently involving gel quantification of differently sized PCR products. Thus, we adapted a novel combination of competitive QPCR and constant-denaturant capillary electrophoresis (CDCE) for the differentiation and quantification of target and internal competitor PCR products in environmental samples. CDCE was developed for the accurate detection of wild-type and mutant alleles in population studies of diseases caused by low-frequency mutations (as low as 10−6) (11). PCR product separation in CDCE, as in conventional gradient gel electrophoresis, is based on mobility shifts induced in DNA due to partial denaturation of the low-melting-temperature domain of the fragment. However, CDCE can resolve sequences that differ by as little as a single base pair, and quantification of sequences is extremely sensitive due to the use of a laser-based detection system.
Here, we explored the ability of competitive QPCR-CDCE for detecting microbial cells in highly eutrophic coastal water samples, with particular emphasis on the quantification of cells at low concentrations. We used a heterotrophic flagellate, Cafeteria roenbergensis, as a model organism because it provided several advantages over bacterial cells. Complete lysis of the flagellates could be easily achieved, ensuring that the concentration of target DNA in nucleic acid lysates reflected the concentration of cells in the original water samples. The accuracy of cell estimates by QPCR could also be checked by FISH with oligonucleotide probes, an approach that has previously been used to estimate the abundance of heterotrophic flagellates in seawater samples (16, 17). Results demonstrate that competitive QPCR-CDCE allows the use of competitor and target sequences with indistinguishable amplification efficiencies and is capable of accurately enumerating microbial cells in natural samples over a range of environmentally relevant cell numbers. The standard curve extended to a single cell, and as few as 10 cells per ml of seawater could be accurately quantified.
MATERIALS AND METHODS
Cultures.
C. roenbergensis clone SR6 was originally isolated from Sakonnet River, Rhode Island (19). Cells were maintained at room temperature on the bacterium Halomonas halodurans, which was grown in sterile seawater from Vineyard Sound, Massachusetts, supplemented with 0.005% yeast extract. The concentration of C. roenbergensis cells in stationary-growth-phase cultures was determined by direct epifluorescence cell counts following staining with DAPI (4′,6′-diamidino-2-phenylindole) (24) and in situ hybridization with species-specific oligonucleotide probes (see below).
Field samples.
Surface-water samples were collected in sterile bottles in January 1999 and June 2000 from Eel Pond, Woods Hole, Mass. The first sample was used for developing and testing the competitive QPCR protocol, and the second was used in experiments to compare the enumeration of C. roenbergensis by QPCR with that by in situ hybridization. In all cases, 100-ml samples were amended with small amounts of stationary-growth-phase cultures to achieve a known cell density in the water samples. Final concentrations of C. roenbergensis in the different samples were 13, 39, 65, 130, 6,500, and 13,000 cells per ml for the January samples and 16, 43, 75, 160, 7,460, and 16,000 cells per ml for the June samples. One unamended sample from each sampling date served as the control. A set of subsamples from the June 2000 spiked seawater samples was preserved with formaldehyde at a final concentration of 3.7% for in situ hybridization with oligonucleotide probes.
Primers and probes.
Five oligonucleotides served as primers for amplification of the C. roenbergensis 18S ribosomal DNA (rDNA) (Table 1). Primers 187F-GC and 239R were used for QPCR amplifications, while primer pairs AF-219mut and AF-451R were used for construction of the internal competitor (see below). In addition, three probes, CROE239, CROE451, and CROE638, were designed for species-specific in situ hybridization (Table 1); CROE239 and CROE451 were identical in sequence to primers 239R and 451R, respectively. The specificity of all the primers and probes to the C. roenbergensis 18S rDNA except primer AF, which targets all eukaryotic 18S rDNAs (8), was checked against sequences in GenBank and the Ribosomal Database Project II using BLAST and Check_Probe, respectively (1, 20).
TABLE 1.
Primers and hybridization probes used in this study
| Primer or probe | Sequence (5′→3′) | Targeta |
|---|---|---|
| Primersb | ||
| AF | AACCTGGTTGATCCTGCCAGT | 1–21, Eukaryotic 18S rDNA |
| 187Fc | CGAAGTCCGGATCCCTCGG | 168–187, C. roenbergensis 18S rDNA |
| 219Rmutd | TCTATCGAGGTTCGCTCGGTTATTATTATTCACCTGTGT | 219–257, C. roenbergensis 18S rDNA |
| 239R | TCTATCGAGGTTCGCTCGG | 239–257, C. roenbergensis 18S rDNA |
| 451R | CGACATTAAGTCACCCGTC | 451–469, C. roenbergensis 18S rDNA |
| Probese | ||
| CROE 239 | TCTATCGAGGTTCGCTCGG | 239–257, C. roenbergensis 18S rDNA |
| CROE 451 | CGACATTAAGTCACCCGTC | 451–469, C. roenbergensis 18S rDNA |
| CROE 638 | GGATGAACTTGCGGAGTGGT | 638–657, C. roenbergensis 18S rDNA |
Numbers refer to nucleotide positions of the C. roenbergensis 18S rRNA gene.
Forward primers (F) are 18S rRNA-like; reverse primers (R) are complementary to the rRNA.
The 168F primer was synthesized with a GC clamp consisting of the sequence 5′-GCCCGCCCCCGCCGCCCTGCCCGCGCCCCGCGCCGCCCGC-3′ at the 5′ terminus. FITC was conjugated to the 5′ terminus of the clamp.
The underlined nucleotide indicates the G-to-T substitution introduced at position 231 for construction of the internal standard.
The 5′ termini of the probes were biotinylated.
The annealing temperature for the 187F-GC–239R primer pair, which yielded maximum PCR product, was empirically established to be 62°C. The specificity of amplification was further tested using plankton lysates prepared from Boston Harbor seawater samples and analyzing the PCR products by CDCE, which is able to discriminate among related sequences differing by as little as a single-base-pair substitution. For in situ hybridization, wash temperatures were adjusted to optimize signal strength and specificity using two other clones of C. roenbergensis, three other heterotrophic flagellates (Paraphysomonas imperforata, Spumella sp., and Pteridomonas danica), and varied protist assemblages in environmental samples as controls.
Construction of internal competitor for competitive QPCR.
A 451-bp fragment of the C. roenbergensis 18S rDNA was mutagenized so that it differed from the wild type by the substitution of a single internal base pair. First, a PCR was carried out with primer AF and a mutagenesis primer, 219Rmut, which created a 257-bp product and changed position 231 from a CG to an AT. This product was gel purified and used as a forward primer in a second PCR together with primer 451R. The resulting 469-bp PCR product was gel purified, reamplified with primers AF and 451R, and gel purified once again to generate a concentrated stock of internal competitor DNA.
Cell lysis and nucleic acid preparation.
For QPCR, total eukaryotic plankton, including added C. roenbergensis cells, was harvested by filtering each of the 100-ml samples through a 0.8-μm-pore-size polycarbonate filter. Immediately after filtration, the filters were put into 1.5-ml microcentrifuge tubes containing 1 ml of 1× PCR buffer (Promega, Madison, Wis.) and frozen at −20°C. To test whether the filtration process led to loss of nucleic acids through cell lysis, a culture of C. roenbergensis in the exponential phase of growth was filtered, and the filtrate was analyzed by PCR.
To release nucleic acids from the cells for QPCR, filters previously frozen in 1× PCR buffer were thawed at room temperature and incubated at 95°C for 15 min. Microscopic observation of cell pellets from pure cultures subjected to this method of cell lysis revealed complete lysis (data not shown). This method of cell lysis is also effective for a variety of naked heterotrophic flagellate species (19). The lysates were then centrifuged at 6,000 rpm for 10 min to pellet cell debris. Internal competitor DNA was added to these cell lysates, and 2 μl of the supernatant was used as the template in QPCR analyses except for the samples that were amended with 6,500 and 13,000 cells ml−1; these samples were diluted 100-fold and amended with internal competitor DNA before use in QPCR analyses.
QPCR.
The primer pair 168F-GC and 239R was used to coamplify the target and internal competitor DNA in all QPCRs. Amplifications were performed in a total volume of 20 μl containing 200 μM each of the four deoxynucleoside triphosphates, 2 mM MgCl2, 1× buffer, 100 nM each primer, and 0.025 U of cloned Pfu polymerase (Stratagene, La Jolla, Calif.) per μl on a thermal cycler (Robocycler; Stratagene). A total of 2 μl of cell lysate was used as the template in each PCR (see below for description of template composition). The amplification program consisted of initial denaturation at 95°C for 180 s, followed by 25 cycles of denaturation at 95°C for 90 s, annealing at 62°C for 60 s, and extension at 72°C for 120 s, with an additional 10-min extension step at 72°C after the last cycle.
A two-stage amplification protocol was carried out to ensure that sufficient product accumulated for CDCE detection while the kinetics of product accumulation remained in the exponential phase. This condition was crucial because one of the goals of this study was to test the ability of QPCR to enumerate very small numbers of microbial cells in environmental samples. The samples were subjected to 25 cycles in the first stage of amplification, and 2 μl from each first-stage reaction was subsequently diluted into fresh reaction mixture and amplified for an additional 15 cycles. The resulting 130-bp PCR products were separated and quantified by CDCE.
The amplification rates of C. roenbergensis template and internal competitor DNAs over a range of PCR cycles were determined to test whether both templates were amplified with equal efficiency when added together to Eel Pond seawater. This experiment also served as a trial for determining the range of PCR cycles during which amplification proceeded at an exponential rate. Reactions contained Eel Pond plankton lysates and C. roenbergensis, and internal competitor DNA equivalent to 50 cells and approximately 40 copies, respectively. These template concentrations were chosen to represent the upper limit of target cells with which we generated the standard curve. Accumulation of target and internal competitor DNA PCR products was quantified at two cycle intervals from 10 to 24 cycles during the second-stage PCR (total of 35 to 49 cycles) by CDCE analysis.
To test the accuracy of the QPCR-CDCE method for quantifying microbial cells, standard curve samples and environmental samples were assembled. The standard curve samples were prepared using pure C. roenbergensis cell lysates equivalent to 1, 2, 4, 8, 10, 20, and 30 cells, while the environmental samples consisted of lysates of Eel Pond seawater which had been spiked with known numbers of C. roenbergensis prior to cell lysis (see above under Field samples). Both were also amended with approximately 40 copies of the internal competitor DNA. Amplifications for the first and second stage were performed in duplicate. The PCR products were run on the CDCE instrument, and the areas of target and internal competitor DNA peaks were quantified (see below). The standard curve was constructed as a plot of the logarithm of the ratio of the target and internal competitor DNA peak areas against the logarithm of the number of C. roenbergensis cells in the standard reactions. The concentration of C. roenbergensis cells in the Eel Pond samples was estimated based on the ratio of the target and internal competitor DNA peak areas and by interpolation from the standard curve.
CDCE.
For separation and quantification, target and competitor PCR products were electrophoresed in fused-silica capillaries (75-μm inner diameter, 350-μm outer diameter; Polymicro Technologies, Inc., Phoenix, Ariz.), coated with 6% linear polyacrylamide in TBE (89 mM Tris-borate, 1 mM EDTA [pH 8.3]). A portion of the capillary was surrounded by a 10-cm water jacket to create a zone of constant temperature for partial denaturation of the DNA fragments. The gel in the capillary was replaced prior to each run with a high-molecular-weight linear polyacrylamide gel (5% linear polyacrylamide in 1× TBE). PCR products (0.3 μl) were diluted 10-fold and electroinjected into the capillary by applying 2 μA of current for 30 s. Samples were electrophoresed for 15 to 20 min by connection to a 30-kV direct current power supply (model CZE, 1000R-2032; Spellman, Hauppauge, N.Y.). Detection and quantification were accomplished by excitation of the fluorescent label by an argon laser (ILT, Salt Lake City, Utah) filtered through a 515-nm narrow-bandpass filter (Corion, Franklin, Mass.) and focused on the separation capillary. Emitted light was collected by a microscope objective (Oriel, Stratford, Conn.) at a right angle to the capillary. This light was directed through two filters (540-nm bandpass and 530-nm long pass) (Corion) into a photo multiplier (Oriel). The signal from the photo multiplier is amplified (107 or 108 V/A) by a current preamplifier (Oriel) and recorded by computer on the Workbench data acquisition system (Strawberry Tree, Inc. Sunnyvale, Calif.).
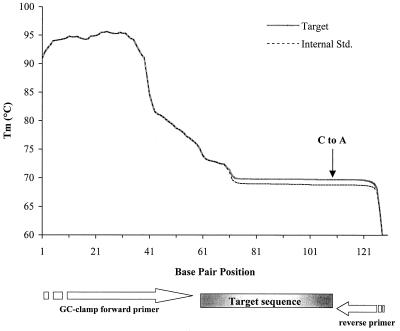
The theoretical separation temperature for the amplified target and internal competitor DNA was determined from melting profiles of the sequences created by the program MacMelt (MedProbe AS, Oslo, Norway) (see Fig. 2). This temperature was based on the Tm of the low-melting-temperature region of the target and internal competitor sequences and was refined by performing test runs with the sequences.
FIG. 2.
Melting profile of the 130-bp target and internal competitor DNA fragments. A substitution was introduced at position 231 of the internal competitor fragment (GC to AT, vertical arrow). Bp 41 on the map corresponds to base position 168 of the C. roenbergensis 18S rRNA gene. The Tm of the low-melting-temperature domain of target sequence is 0.5°C higher than that of the internal competitor. Std., standard.
FISH.
C. roenbergensis cells added to Eel Pond samples collected in June 2000 for QPCR analysis were also enumerated by FISH using biotinylated probes and fluorescein isothiocyanate (FTTC)-labeled avidin. Formaldehyde-preserved samples were vacuum filtered onto 0.4-μm polycarbonate filters of Transwell tissue culture inserts (Costar), dehydrated in a series of ethanol washes, and prehybridized in hybridization buffer (10× Denhardt's solution, 0.1 mg of polyadenylic acid per ml, 5× SET buffer [750 mM NaCl, 100 mM Tris-HCl (pH 7.8), 5 mM EDTA], 0.1% sodium dodecyl sulfate) for at least 45 min at 40°C. Oligonucleotide probes were then added to the samples at a final concentration of 2.5 ng μl−1 and hybridized overnight at 40°C. Following hybridization, samples were washed at 45°C in 0.2× SET buffer (30 mM NaCl, 4 mM Tris-HCl [pH 7.8], 0.2 mM EDTA) for 10 min, incubated with FITC-labeled avidin (20 μg ml−1 in 100 mM NaHCO3-buffered saline [pH 8.2]), and washed with cold NaHCO3-buffered saline to remove unincorporated FITC-avidin. Control hybridizations incubated with FITC-avidin only were also performed for all the samples examined to check for background binding. Filters were cut out of the Transwells and mounted on glass slides for observation by epifluorescence microscopy. Triplicate hybridizations were carried out for each sample. Enumeration of FITC-labeled cells was performed at 1,000× magnification with a Zeiss Axioskop II using a BP450-490 exciter filter and an LP520 barrier filter. Approximately 20 to 120 fields per filter were observed in order to obtain cell counts of C. roenbergensis in the Eel Pond samples.
RESULTS
Evaluation of primer specificity.
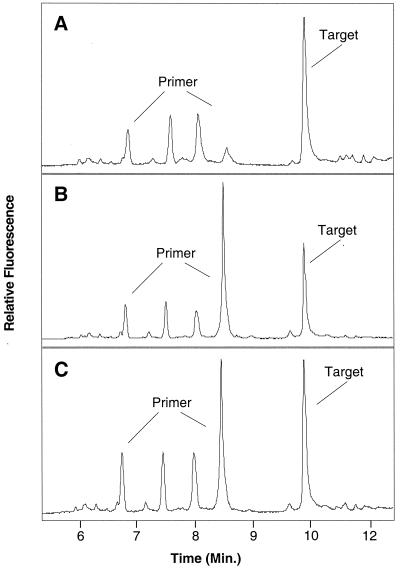
The specificity of the QPCR primer pair 187F-GC and 239R (Table 1) was confirmed by amplification of C. roenbergensis from pure cultures and from a seawater sample collected from Boston Harbor. In both cases, a single 130-bp product was detected by agarose gel electrophoresis (not shown), and only peaks corresponding to the primer and the specific amplification product were evident by CDCE (Fig. 1). The amplified DNA peaks of C. roenbergensis from pure culture (Fig. 1A) and Boston Harbor (Fig. 1B) migrated at the same speed, indicating that the cultured clone was representative of C. roenbergensis in nature. PCR products from both samples were also confirmed to be identical by the presence of a single target peak when they were mixed and electrophoresed together (Fig. 1C).
FIG. 1.
Electropherograms of C. roenbergensis DNA fragments amplified from pure culture (A) and Boston Harbor seawater (B) analyzed by CDCE. PCR products from both amplifications were mixed in equal amounts and electrophoresed (C). The primer peaks correspond to the FITC-labeled 187F primer that remained after PCR. The smaller primer peaks are fractions of labeled primers which disintegrated into shorter pieces during PCR.
CDCE separation of target and internal competitor DNA.
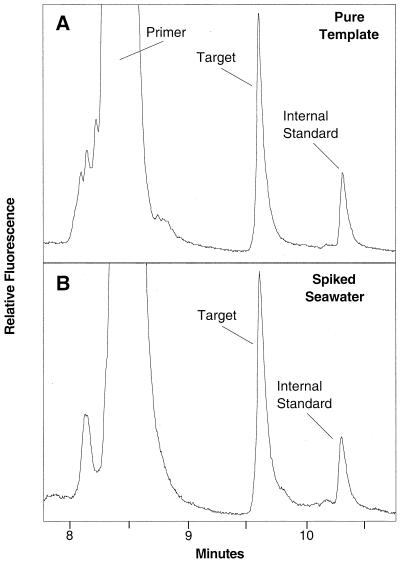
Amplification of the internal competitor generated a 130-bp product that differed from the corresponding wild-type C. roenbergensis product by only a single-base-pair change (Fig. 2). This substitution lowered the calculated Tm of the low-melting-temperature domain of the internal competitor sequence by 0.5°C compared to the target sequence (Fig. 2) and was sufficient to clearly separate the two sequences by CDCE (Fig. 3A). A temperature of 71.8°C was found to provide optimum resolution of competitor and target, which is in good agreement with the calculated temperature. A comparison of CDCE electropherograms of coamplified competitor and target DNA from pure templates and spiked seawater showed only the expected primer, target, and internal competitor peaks (Fig. 3B). This result indicates that amplification of DNA from seawater samples was not adversely affected by the higher complexity of the seawater community or by contaminating substances.
FIG. 3.
Electropherograms of target and internal competitor DNAs separated by CDCE at 71.8°C. The DNA fragments were coamplified from pure target and internal competitor DNA templates (A) and from pure template added to seawater cell lysate (B) at the same concentration as in panel A.
Amplification efficiencies of target and internal competitor sequences.
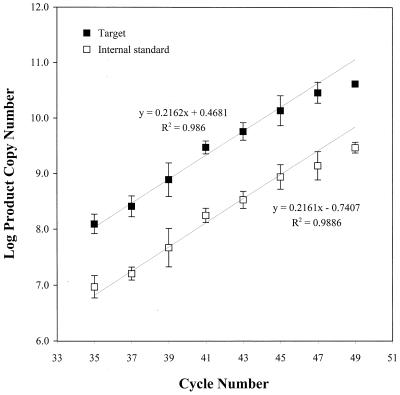
A comparison of the amplification efficiencies of C. roenbergensis and its internal competitor DNA versus cycle number is shown in Fig. 4. The PCR efficiency curves were plotted as log (target copies formed) and log (internal competitor copies formed) versus cycle number (after two-stage amplification), and a linear regression was performed on the data points from cycle 35 to cycle 45. The slopes of both regression lines are essentially identical (0.2161 and 0.2162; R2 = 0.99 for both lines), demonstrating that the amplification efficiencies of the target and internal competitor templates are indistinguishable. Product accumulation proceeded exponentially up to cycle 45, after which it approached the plateau phase (Fig. 4). Based on these data, 40 cycles (25 and 15 cycles for the first and second stages, respectively) were chosen for all QPCR analyses.
FIG. 4.
Comparison of amplification efficiencies in Eel Pond seawater of C. roenbergensis and internal competitor DNA as a function of PCR cycle number (two-stage amplification). PCR products were quantified by integrating the target and internal competitor DNA peaks and multiplying the ratios of the target to total peak area and primer to total peak area by the concentration (copies per microliter) of primer used in the PCR. The error bars represent the standard deviation of the mean of quadruple measurements. Linear regressions were performed on data points from cycles 35 to 45.
Competitive QPCR and FISH estimates of C. roenbergensis in field samples.
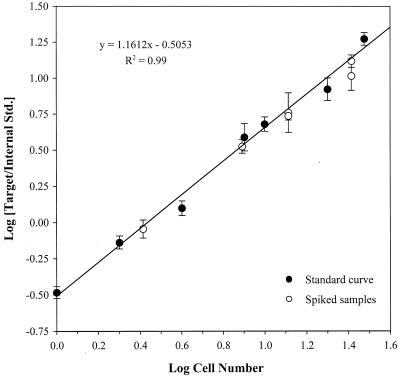
C. roenbergensis was not detected in the unamended Eel Pond water sample. Subsamples of Eel Pond seawater spiked with C. roenbergensis contained cells ranging in concentration from approximately 10 to 104 ml−1, which spans the abundance of eukaryotic microorganisms in coastal seawater. The abundance of C. roenbergensis in these samples was enumerated based on the competitive QPCR standard curve shown in Fig. 5. The lower limit of the standard curve was 1 cell per reaction, equivalent to a detection limit of 5 cells ml−1. The estimates of C. roenbergensis numbers ± 1 standard deviation in all of the January 1999 Eel Pond samples (open circles) fell on the regression line, indicating a 1:1 agreement between the expected and QPCR-estimated concentrations of C. roenbergensis (Fig. 5).
FIG. 5.
QPCR standard curve for C. roenbergensis. The ratio of target peak to internal competitor peak areas was plotted against cell number in 20-μl PCR mixes. All the data points are the mean values of four measurements, and the error bars represent the standard deviation of the mean. The regression (R2 = 0.99) was performed on the standard-curve data points only. Values from the spiked samples which fall on the regression line indicate a 1:1 agreement between the expected and QPCR-estimated concentrations of C. roenbergensis in the spiked samples.
Tables 2 and 3 depict a comparison of the expected and estimated cell concentrations of C. roenbergensis by competitive QPCR alone (Table 2) and in combination with FISH (Table 3). The abundance of C. roenbergensis was calculated by multiplying the number of cells per 20-μl reaction, obtained from the QPCR standard curve, by a factor of 5 (the dilution factor as a result of sample processing). Differences in cell estimates that fall within the average coefficient of variation associated with either the competitive QPCR or FISH assay were considered to be within the counting error of the assay, and thus, those cell counts were considered similar. The QPCR estimates of C. roenbergensis abundance corresponded well with the expected number of C. roenbergensis in all the field samples examined. Given that the coefficient of variation associated with the QPCR assay averaged 14% ± 8% (determined from the data in Table 2), QPCR estimates of C. roenbergensis were found to be similar to the expected cell number in all but one sample. The QPCR-estimated concentration of C. roenbergensis in this sample was, however, the same as the FISH estimate (Table 3).
TABLE 2.
C. roenbergensis cell abundance estimated by competitive QPCRa
| Spiked concn (cells ml−1) | Estimated concn (cells ml−1) ± SD |
|---|---|
| 13 | 13 ± 2 |
| 39 | 39 ± 4 |
| 65 | 63 ± 17 |
| 130 | 104 ± 21 |
| 6.5 × 103 | 5.9 (± 0.3) × 103 |
| 1.3 × 104 | 1.3 (± 0.1) × 104 |
Eel Pond seawater was collected in January 1999 and spiked with C. roenbergensis at the concentrations noted in the first column.
TABLE 3.
Comparison of C. roenbergensis cell abundance estimated by competitive QPCR and FISHa
| Spiked concn (cells ml−1) | Estimated concn (cells ml−1) ± SD
|
|
|---|---|---|
| QPCR | FISH | |
| 16 | 24 | 25 ± 7 |
| 43 | 51 | 40 ± 14 |
| 75 | 85 | 94 ± 2 |
| 160 | 156 | 172 ± 13 |
| 7.5 × 103 | 8.6 × 103 | 1.2 (± 0.1) × 104 |
| 1.6 × 104 | 1.6 × 104 | 2.4 (± 0.3) × 104 |
Eel Pond seawater was collected in June 2000 and spiked with the concentrations of C. roenbergensis noted in the first column.
The ability of the competitive PCR method to provide accurate counts of C. roenbergensis relative to FISH counts was examined with the Eel Pond samples collected in June 2000. The coefficient of variation associated with the FISH counts averaged 16% ± 13%. Competitive QPCR estimates of C. roenbergensis were not different from the FISH counts except for two of the samples (Table 3). In both of these samples, the QPCR estimates were similar to the expected concentration of C. roenbergensis, but the FISH estimates were 50 to 60% higher than the expected C. roenbergensis cell number.
DISCUSSION
The application of the CDCE method to the analysis of microbial communities represents a novel approach in molecular ecological studies. PCR products amplified with primers specific to the target organism could be detected and quantified via the migration distance of specific DNA peaks in CDCE electropherograms. The position of these peaks is sequence dependent and sensitive to as little as a single-base-pair change. Thus, a major advantage of CDCE in analyzing environmental samples of unknown species composition is that nonspecific or nontarget amplification products can be discriminated with high resolution from the target amplification products. In this study, both the forward and reverse PCR primers were designed to specifically amplify a region of the 18S rDNA sequence unique to known strains of the heterotrophic flagellate C. roenbergensis (14). The detection of a single peak, which was identical to cultured C. roenbergensis, in PCR products amplified from Boston Harbor seawater samples confirmed the suitability of the target sequence chosen for the CDCE assay and its specificity for quantifying this flagellate in environmental samples.
The combination of competitive QPCR and CDCE proved highly effective for the enumeration of microorganisms over a broad range of environmentally relevant cell concentrations. As few as 13 cells of the marine flagellate C. roenbergensis per ml could be reproducibly quantified in eutrophic seawater samples. The combined sensitivity and quantitative range of this approach are significant, because accurate identification and enumeration of small flagellates had been virtually impossible due to their generally low abundance (18).
A number of studies have shown the utility of competitive QPCR for estimating the abundance of bacteria in soil, biofilm, and plankton communities (10, 12, 13, 15, 21, 22, 27). For the method to accurately estimate population abundance, the amplification efficiency of the target and internal competitor DNA must be equal, resulting in a linear standard curve with a slope of 1 (5, 25). In practice, the construction of a competitor that can be amplified with the same efficiency as the target for conventional competitive QPCR analysis is often difficult because extensive sequence modification of the competitor is required. For ease of differentiation on agarose gels, competitors which differ substantially in length from the targets are most commonly employed. Furthermore, target and competitor quantification on gels can lead to inaccuracies due to shading effects that quench the fluorescent signal.
The main advantage of CDCE is that it allows the use of a competitor that is virtually identical to the target sequence. Indeed, the amplification efficiency of the target and competitor sequences used in this study, expressed as the slopes of the regressions in Fig. 4, was indistinguishable. The standard curve was also linear (R2 = 0.99), with a close agreement to a slope of 1 (Fig. 5). Thus, the accuracy and precision of the competitive QPCR protocol presented here are in large part due to the sensitivity and high resolution of CDCE, which allowed small changes in DNA concentration to be measured accurately. Another factor that contributed to the high reproducibility of the results was the use of high-fidelity PCR with Pfu polymerase, which gave more consistent DNA peaks and lower background in CDCE than Taq polymerase (data not shown).
The accuracy of the competitive QPCR-CDCE method for enumerating the abundance of C. roenbergensis in field samples was supported by the congruence of the QPCR estimates with the expected cell numbers and most of the FISH cell counts. Cell number estimates obtained by competitive QPCR and FISH were generally within the counting error of the respective assays; however, the FISH counts appeared less reliable than the competitive QPCR estimates in two respects. First, FISH counts of C. roenbergensis were higher than expected in two samples, while the corresponding QPCR estimates matched the expected cell numbers (Table 3). Second, FISH counts of samples with <50 cells ml−1 were more variable than competitive QPCR counts, with coefficients of variation of 32% compared to only 12.5%. High variability associated with FISH counts of cells at low abundance is a general problem. In a previous study, we found that the counting error associated with FISH counts was too high to resolve the seasonal dynamics of the protist Paraphysomonas imperforata, which did not exceed a concentration of about 50 cells ml−1 (18). Other investigators have noted problems with counting error when quantifying single bacterial strains in natural samples by immunofluorescent staining (6, 28). A way to lower the variability is by filtering larger sample volumes to obtain more cells. However, this procedure introduces higher background fluorescence and shading effects which compromise accuracy. Alternatively, a larger area of the filter may be counted, but this approach is extremely time-consuming. In contrast, the QPCR method was not affected by such problems because it was possible to filter 6 to 10 times more sample volume than for FISH.
A problem that is still poorly explored is the effect of environmental inhibitors or background DNA on QPCR estimates. Indeed, a long-recognized cause of PCR variability is the presence of a variety of inhibitors in environmental samples (29). For example, humic substances frequently copurify with environmental DNA and are known to lower amplification efficiency. This may compromise the accuracy of real-time PCR to a larger extent than competitive QPCR because product accumulation is measured on an absolute scale, while in competitive PCR, the ratio of the coamplified target and competitor is determined. An indication of PCR inhibition using the real-time PCR approach is lower than expected target quantification, which was reported in a recent comparison of real-time and competitive QPCR using samples rich in potential inhibitors (7). Similarly, a recent study by Becker et al. (3) exploring real-time QPCR under competitive amplification conditions for the quantification of cyanobacterial ecotypes also highlighted the importance of controlling for amplification efficiency. Although further careful investigation is required, it is possible that competitive QPCR is more robust for variable amplification efficiencies between different samples and thus remains an important alternative for analysis of unknown environmental samples.
In summary, quantitative PCR is increasingly being used for enumeration of microorganisms in environmental samples because of its specificity and sensitivity. Our study has shown that the combination of competitive QPCR and CDCE is a reliable method for accurately enumerating microorganisms over the relevant range of cell concentrations in a complex environmental sample including cell concentrations as low as 10 ml−1.
ACKNOWLEDGMENT
This study was supported partially by a grant from the Department of Civil and Environmental Engineering and by SeaGrant.
We thank David A. Caron (University of Southern California, Los Angeles) for the C. roenbergensis clone SR6 culture.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker S, Böger P, Oehlmann R, Ernst A. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl Environ Microbiol. 2000;66:4945–4953. doi: 10.1128/aem.66.11.4945-4953.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker-André M, Hahlbrock K. Absolute mRNA quantitation using the polymerase chain reaction (PCR): a novel approach by a PCR aided transcript titration assay (PATTY) Nucleic Acids Res. 1989;17:9437–9446. doi: 10.1093/nar/17.22.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandler D P. Redefining relativity: quantitative PCR at low template concentrations for industrial and environmental microbiology. J Ind Microbiol Biotechnol. 1998;21:128–140. [Google Scholar]
- 6.Dahle A B, Laake M. Diversity dynamics of marine bacteria studied by immunofluorescent staining on membrane filters. Appl Environ Microbiol. 1982;43:169–176. doi: 10.1128/aem.43.1.169-176.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desjardin L E, Chen Y, Perkins M D, Teixeira L, Cave M D, Eisenach K D. Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis. J Clin Microbiol. 1998;36:1964–1968. doi: 10.1128/jcm.36.7.1964-1968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 10.Johnsen K, Enger Ø, Jacobsen C S, Thirup L, Torsvik V. Quantitative selective PCR of16S ribosomal DNA correlates well with selective agar plating in describing population dynamics of indigenous Pseudomonas spp. in soil hot spots. Appl Environ Microbiol. 1999;65:1786–1789. doi: 10.1128/aem.65.4.1786-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khrapko K, Hanekamp J S, Thilly W G, Belenkii A, Foret F, Karger B L. Constant denaturant capillary electrophoresis (CDCE): a high resolution appraoch to mutational analysis. Nucleic Acids Res. 1994;22:364–369. doi: 10.1093/nar/22.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurie A D, Lloyd-Jones G. Quantification of phnAc and nahAc in contaminated New Zealand soils by competitive PCR. Appl Environ Microbiol. 2000;66:1814–18117. doi: 10.1128/aem.66.5.1814-1817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S-Y, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultivated soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leipe D D, Wainright P O, Gunderson J H, Porter D, Patterson D J, Valois F, Himmerich S, Sogin M L. The stramenopiles from a molecular perspective: 16S-like rRNA sequences from Labyrinthuloides minuta and Cafeteria roenbergensis. Phycology. 1994;33:369–377. [Google Scholar]
- 15.Leser T D, Boye M, Hendriksen N B. Survival and activity of Pseudomonas sp. strain B13(FR1) in a marine microcosm determined by quantitative PCR and an rRNA-targeting probe and its effect on the indigenous bacterioplankton. Appl Environ Microbiol. 1995;61:1201–1207. doi: 10.1128/aem.61.4.1201-1207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim E L, Amaral L A, Caron D A, DeLong E F. Application of rRNA-based probes for observing marine nanoplanktonic protists. Appl Environ Microbiol. 1993;59:1647–1655. doi: 10.1128/aem.59.5.1647-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim E L, Caron D A, DeLong E F. Development and field application of a quantitative method for examining natural assemblages of protists with oligonucleotide probes. Appl Environ Microbiol. 1996;62:1416–1423. doi: 10.1128/aem.62.4.1416-1423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim E L, Dennett M R, Caron D A. The ecology of Paraphysomonas imperforata based on studies employing oligonucleotide probe identification in coastal water samples and enrichment cultures. Limnol Oceanogr. 1999;44:37–51. [Google Scholar]
- 19.Lim E L, Dennett M R, Caron D A. Identification of heterotrophic nanoflagellates by restriction fragment length polymorphism analysis of small subunit ribosomal DNA. J Eukaryot Microbiol. 2001;48:247–257. doi: 10.1111/j.1550-7408.2001.tb00312.x. [DOI] [PubMed] [Google Scholar]
- 20.Maidak B L, Cole J R, Parker C T J, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michotey V, Méjean V, Bonin V. Comparison of methods for quantification of cytochrome cd1-denitrifying bacteria in environmental marine samples. Appl Environ Microbiol. 2000;66:1564–1571. doi: 10.1128/aem.66.4.1564-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Möller A, Jansson J K. Quantification of genetically tagged cyanobacteria in Baltic Sea sediment by competitive PCR. Biotechniques. 1997;22:512–518. doi: 10.2144/97223rr02. [DOI] [PubMed] [Google Scholar]
- 23.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 24.Raeymaekers L. Quantitative PCR: thoeretical considerations with practical implications. Anal Biochem. 1993;214:582–585. doi: 10.1006/abio.1993.1542. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki M T, Taylor L T, DeLong E F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microbiol. 2000;66:4605–4614. doi: 10.1128/aem.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tay T-L, Hemond H, Krumholz L, Cavanaugh C M, Polz M F. Population dynamics of two toluene degrading species in a contaminated stream assessed by quantitative PCR. Microb Ecol. 2001;41:124–131. doi: 10.1007/s002480000089. [DOI] [PubMed] [Google Scholar]
- 27.Tuomi P, Torsvik T, Heldal M, Bratbak G. Bacterial population dynamics in a meromictic lake. Appl Environ Microbiol. 1997;63:2181–2188. doi: 10.1128/aem.63.6.2181-2188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zachar V, Thomas R A, Goustin A S. Absolute quantitation of target DNA: a simple competitive PCR for efficient analysis of multiple samples. Nucleic Acids Res. 1993;21:2017–2018. doi: 10.1093/nar/21.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]