SUMMARY
The leaf veins of higher plants contain a highly specialized vascular system comprised of xylem and phloem cells that transport water, organic compounds and mineral nutrients. The development of the vascular system is controlled by phytohormones that interact with complex transcriptional regulatory networks. Before the emergence of true leaves, the cotyledons of young seedlings perform photosynthesis that provides energy for the sustainable growth and survival of seedlings. However, the mechanisms underlying the early development of leaf veins in cotyledons are still not fully understood, in part due to the complex cellular composition of this tissue. To better understand the development of leaf veins, we analyzed 14 117 single cells from 3‐day‐old cotyledons using single‐cell RNA sequencing. Based on gene expression patterns, we identified 10 clusters of cells and traced their developmental trajectories. We discovered multiple new marker genes and developmental features of leaf veins. The transcription factor networks of some cell types indicated potential roles of CYCLING DOF FACTOR 5 (CDF5) and REPRESSOR OF GA (RGA) in the early development and function of the leaf veins in cotyledons. These new findings lay a foundation for understanding the early developmental dynamics of cotyledon veins. The mechanisms underlying the early development of leaf veins in cotyledons are still not fully understood. In this study, we comprehensively characterized the early differentiation and development of leaf veins in 3‐day‐old cotyledons based on single‐cell transcriptome analysis. We identified the cell types and novel marker genes of leaf veins and characterized the novel regulators of leaf vein.
Keywords: novel regulators, development, leaf veins, cotyledons, single‐cell RNA‐sequencing, Arabidopsis thaliana
Significance Statement
The mechanisms underlying the early development of leaf veins in cotyledons are still not fully understood. In this study, we comprehensively characterized the early differentiation and development of leaf veins in 3‐day‐old cotyledons based on single‐cell transcriptome analysis. We identified the cell types and novel marker genes of leaf veins, and characterized the novel regulators of leaf vein.
INTRODUCTION
The evolution of vascular tissue that efficiently transports water and organic solutes was critical for the establishment of land‐based plant life (Furuta et al., 2014a). Water is transported upward in the xylem, which is comprised of tracheids and vessel elements (Lucas et al., 2013). Organic solutes are transported bidirectionally in the phloem (Furuta et al., 2014b), which contains living sieve tube elements (SEs) that lack a nucleus but have nucleated companion cells (CCs) associated with them (Fukuda, 2004). Additionally, the vascular system is often surrounded by a bundle sheath (BS) of parenchyma cells.
In Arabidopsis, the shoot apical meristem and root apical meristem give rise to the procambium, whereas procambial cells (PCs) produce the primary phloem and primary xylem during primary growth (Furuta et al., 2014a). A Zinnia (Zinnia elegans L.) TRACHEARY DIFFERENTIATION INHIBITORY FACTOR (TDIF), identified as the CLAVATA3/ESR‐RELATED (CLE) family peptide CLE41/CEL42/CEL44, was found to function as a signaling molecule that both inhibits xylem cell differentiation from PCs and promotes PC proliferation (Ito et al., 2006; Hirakawa et al., 2008, 2010). Its receptor TDR/PXY (TDIF RECEPTOR/PHLOEM INTERCALATED WITH XYLEM) belongs to the XI LRR‐RLK (LEUCINE‐RICH REPEAT RECEPTOR‐LIKE KINASE) family and is expressed in PCs in Arabidopsis (Ito et al., 2006; Fisher and Turner, 2007; Hirakawa et al., 2008; Etchells and Turner, 2010). In Arabidopsis, the TDIF signal activates the expression of WUSCHEL‐related HOMEOBOX 4 (WOX4), WOX14 and HOMEOBOX GENE 8 (ATHB8) in PCs and cambial cells (Hirakawa et al., 2010; Ji et al., 2010; Suer et al., 2011; Etchells et al., 2013). Arabidopsis ATHB8 encodes an HD‐Zip III transcription factor (TF; Baima et al., 2001) whose paralogs modulate the expression of genes involved in auxin biosynthesis and perception (Muller et al., 2016). The plant glycogen synthase kinase 3 proteins (GSK3s) were identified as crucial downstream components of the TDIF signaling pathway that suppress xylem differentiation from PCs in Arabidopsis (Kondo et al., 2014). A recent study revealed a PXY‐mediated transcriptional regulatory network of Arabidopsis, in which WUSCHEL HOMEOBOX RELATED14 (WOX14), TARGET OF MONOPTEROS6 (TMO6) and LATERAL ORGAN BOUNDARIES DOMAIN4 (LBD4) formed a feed‐forward loop to regulate the expression of target genes in response to auxin, cytokinin (CK) and TDIF PXY signaling (Smit et al., 2020). CKs are key regulators of xylem development (Mahonen et al., 2000, 2006; Matsumoto‐Kitano et al., 2008; Bishopp et al., 2011a,b), and their interaction with CLE peptides was shown to regulate xylem differentiation (Kondo et al., 2011).
The formation of vein patterns in leaves and cotyledons undergoing a spatially regulated development proceeds by the progressive recruitment of ground cells into PCs (Carland and Nelson, 2004). The organization of different cell types in veins is under the control of polar auxin transport and the MYB and HD‐ZIP TFs. Auxin has been shown to play a crucial regulatory role in establishing the continuous procambial strands necessary for vascular differentiation. The synthesis, degradation and distribution of auxin are tightly correlated with the differentiation and patterning of the vascular system (Sachs, 2000). The regulation of auxin flow requires symmetrically localized influx and efflux carriers on the plasma membrane, including AUXIN RESISTANT 1 (AUX1)‐like influx and PIN1‐like efflux proteins in the cambium (Marchant et al., 1999; Steinmann et al., 1999). MONOPTEROS (MP), which encodes an Auxin Response Factor (ARF), is involved in regulating auxin‐mediated vascular tissue specification (Hardtke and Berleth, 1998). MP is thought to act as an activator of vascular proliferation in seedlings (Vera‐Sirera et al., 2015) or as a repressor of vascular proliferation in mature plant tissues (Mattsson et al., 2003; Brackmann et al., 2018). In addition to auxins, CKs have been identified as hormonal regulators of vascular patterning (Burkle et al., 2003). The roots of mutants with an impaired histidine kinase signaling system have been shown to contain increased protoxylem cells in vascular cylinders, while other cell types were depleted in this tissue (Dettmer et al., 2009). CKs have also been shown to be required for the maintenance of cambial cell proliferation during secondary growth (Matsumoto‐Kitano et al., 2008).
To identify the upstream components that spatiotemporally regulate xylem and phloem development, a system known as Vascular Cell Induction Culture System Using Arabidopsis Leaves (VISUAL) has been established (Kondo et al., 2015). The phloem SE‐like cells and xylem tracheary elements can be induced by VISUAL, and genetic analysis has confirmed that ALTERED PHLOEM DEVELOPMENT (APL) plays a central role in SE differentiation via VISUAL (Kondo et al., 2015). APL, which encodes a MYB‐type TF, is thought to be required for later steps of SE development and for spatially limiting the differentiation of the xylem (Bonke et al., 2003). The downstream components of APL include NAC DOMAIN‐CONTAINING PROTEIN 45 (NAC045), NAC086 and NAC45/86‐DEPENDENT EXONUCLEASE‐DOMAIN (NENs), which regulate enucleation during phloem differentiation (Furuta et al., 2014b). Based on VISUAL transcriptome data, a co‐expression network that regulates early phloem SE development has also been identified (Kondo et al., 2016).
After seed germination, the cotyledon is not only an important reserve organ, but also the initial organ that performs photosynthesis to provide energy for growth and survival of seedlings. Leaf veins rapidly appear in cotyledons after germination. They form the key channel network for the transport of photosynthetic products and minerals in seedlings. Although considerable effort has been devoted to understanding vein cell differentiation, a full understanding remains elusive due to the limitations of tissue separation techniques (Fukuda, 2004; Furuta et al., 2014a; Kondo et al., 2016). Recently, the single‐cell RNA‐sequencing (scRNA‐seq) technique has been used to determine the mechanisms that regulate root development (Denyer et al., 2019; Ryu et al., 2019; Zhang et al., 2019; Wendrich et al., 2020; Apelt et al., 2021; Gala et al., 2021; Liu et al., 2021; Serrano‐Ron et al., 2021; Wang et al., 2021; Zhang et al., 2021) and phloem cells in mature leaves (Kim et al., 2021). These studies provide a key reference for analyzing the regulatory mechanisms of the early development and differentiation of leaf veins in cotyledons. In this study, we used scRNA‐seq technology (Liu et al., 2020) to analyze 14 117 single cells isolated from 3‐day‐old cotyledons. Our study presents the early developmental dynamics of the leaf veins in cotyledons and also identified core TFs that are involved in regulating the development of leaf vein. Our study also provides new cell‐type‐specific gene expression profiles and potential marker genes for future studies of the development of plant vascular tissue.
RESULTS
Single‐cell transcriptome profiling of leaf vein cell types and gene expression profiles
The development and function of vascular tissues in leaves and roots are very similar but are controlled independently. The vascular system in leaves connects to the mesophyll cells (MPCs) and acts as the first hub that receives the photosynthetic products transported to all tissues through the phloem system (Amiard et al., 2005; Baker et al., 2016). Therefore, the developmental pattern of leaf veins is impacted by photosynthesis (Li et al., 2017). To explore the mechanisms underlying leaf vein development in cotyledons, we investigated the transcriptome profiles of leaf vein cells via scRNA‐seq. First, we isolated protoplasts from the 3‐day‐old cotyledons and obtained 15 170 single cells for scRNA‐seq (Figure S1). After quality control analysis of the raw sequencing data, 14 117 cells were selected for further analysis (Figure S2). The t‐distributed Stochastic Neighbor Embedding (t‐SNE) analysis and the Uniform Manifold Approximation and Projection (UMAP) algorithms were used to visualize the cell clusters (Figures 1a and S3a). A total of 10 and 11 cell clusters were identified by t‐SNE and UMAP analyses, respectively (Figures 1a and S3a). The differentially expressed genes (DEGs) in each cluster were screened as described in Experimental Procedures (Tables S1 and S2). The top 10 genes that were most unique to a given cell cluster were considered as marker genes and are displayed in a heatmap (Figures 1b and S3b). The representative marker genes are shown on the feature plot in Figure 1(c) and Figure S3(c). The correlation analysis of corresponding cell clusters from t‐SNE and UMAP is displayed in Figure S3(d). Gene Ontology (GO) enrichment analysis of DEGs of all cell clusters produced by UMAP analysis was carried out to analyze biological processes that are enriched in each cluster (Figure S3e).
Figure 1.
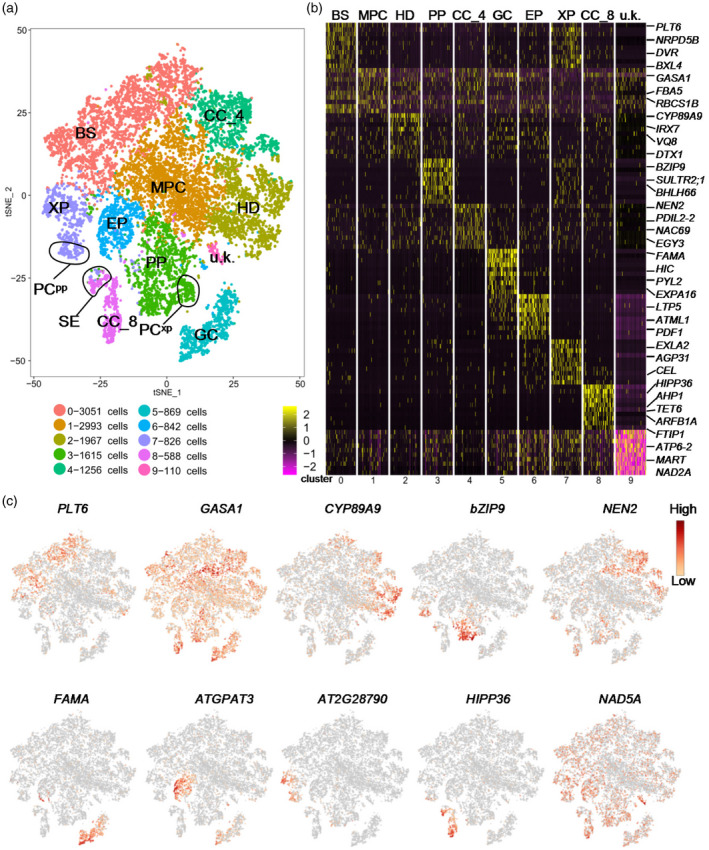
Identification of the cell types of leaf vein in cotyledons.
(a) t‐distributed Stochastic Neighbor Embedding (tSNE) plot of all cells passing the quality control test. Each dot represents a cell colored by clusters labeled by inferred cell types. BS, bundle sheath; MPC, mesophyll cell; PP, phloem parenchyma; HD, hydathode; CC, companion cell; GC, guard cell; EP, epidermis; XP, xylem parenchyma; PCxp, xylem parenchyma cells with features relating to xylem differentiation; PCpp, procambium cells with features relating to phloem differentiation; SE, sieve element; u.k., unknown.
(b) Heatmap of the top 10 genes that were specifically expressed in each of the clusters in the tSNE plot.
(c) Feature plots of the representative marker gene in each cell cluster.
To identify the cell type of each cluster, we first conducted a literature survey of the marker genes of vascular tissues (Table S3). Recent studies have identified APL, SIEVE ELEMENT OCCLUSION B (SEOB), SWEET11 and SWEET12 as marker genes for the phloem parenchyma (PP); SUCROSE‐PROTON SYMPORTER 2 (SUC2) and FT‐INTERACTING PROTEIN 1 (FTIP1) as marker genes for CCs; SCARECROW‐LIKE 23 (SCL23) and SULPHATE TRANSPORTER 2;2 (SULTR2.2) as marker genes for BSs; FAMA, SPEECHLESS (SPCH), TOO MANY MOUTHS (TMM) and MITOGEN‐ACTIVATED PROTEIN KINASE 12 (MPK12) as marker genes for guard cells (GCs); MERISTEM LAYER 1 (ATML1) and CUTICULAR 1 (CUT1) as marker genes for the epidermis (EP); ACAULIS 5 (ACL5) and GLUTAMATE RECEPTOR 3.6 (GLR3.6) as marker genes for the xylem parenchyma (XP); ATHB‐8, REVOLUTA (REV) and ATHB‐15 as marker genes for XP cells with features related to xylem differentiation (PCxp); BETA‐AMYLASE 3 (BAM3) and CLE45 as marker genes for PCs with features related to phloem differentiation (PCpp); PURINE PERMEASE 1 (PUP1), EP3 and NDR1/HIN1‐LIKE1 (NHL1) as marker genes for hydathode (HD); LIGHT HARVESTING CHLOROPHYLL A/B‐BINDING PROTEIN 1.1 LHCB2.1 (LHCB2.1) and RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN 1B (RBCS1B) as marker genes for MPCs (Kim et al., 2021 ). Additionally, SWEET11 has been shown to be expressed in MPCs (Kim et al., 2021). To identify the vein cells, we analyzed the expression of marker genes of the vascular system in each cell cluster on the tSNE (Figure S4) and UMAP plots (Figure S5). This analysis revealed that the cell clusters 0, 1, 2, 3, 4, 5, 6, 7 and 8 belong to BS, MPC, HD, PP, CC_4, GC, EP, XP and CC_8, respectively. Cluster 9 did not display significant expression for any known marker genes and was therefore termed ‘unknown’ (u.k.). Interestingly, genes encoding mitochondrial proteins were strongly expressed in cluster 9, suggesting that mitochondrial activities are very high in cells of this cluster. Based on marker gene analysis, we also identified the cell sub‐clusters for PCpp within XP, PCxp within PP, and SE within CC_8 (Figure 1a). Because more than one cell cluster was identified as CC, we added the serial number of the corresponding cell cluster to distinguish them, as indicated by CC_4 and CC_8, respectively (Figure 1a). The corresponding cell types in UMAP were also identified and annotated according to the highly correlated cell types identified in t‐SNE and marker genes (Figure S3a).
We then investigated the potential biological functions of DEGs in each cell type by GO enrichment analysis (Figure 2; Table S4). As shown in Figure 2, DEGs associated with response to stimulus, metabolic process and localization were enriched in all cell clusters. The DEGs of BS and MPC were found to be primarily associated with photosynthesis and the generation of precursor metabolites and energy (Figure 2a). The DEGs in CC_4 were mainly found to be related to the nucleolus, while the DEGs in CC_8 were mainly related to purine nucleotide metabolic processes and copper ion binding (Figure 2a). The DEGs in PP only share cofactor metabolic processes with other cell clusters, suggesting that PP may perform a relatively unique biological function (Figure 2a). Further analysis showed that DEGs in PP are mainly involved in the response to inorganic substances and to hypoxia (Figure 2b). The DEGs in XP encode proteins primarily localized in the nucleus, membrane protein complexes and chloroplast stroma (Figure 2a). The DEGs in HD are predominantly related to the nucleolus and membrane protein complexes (Figure 2a).
Figure 2.
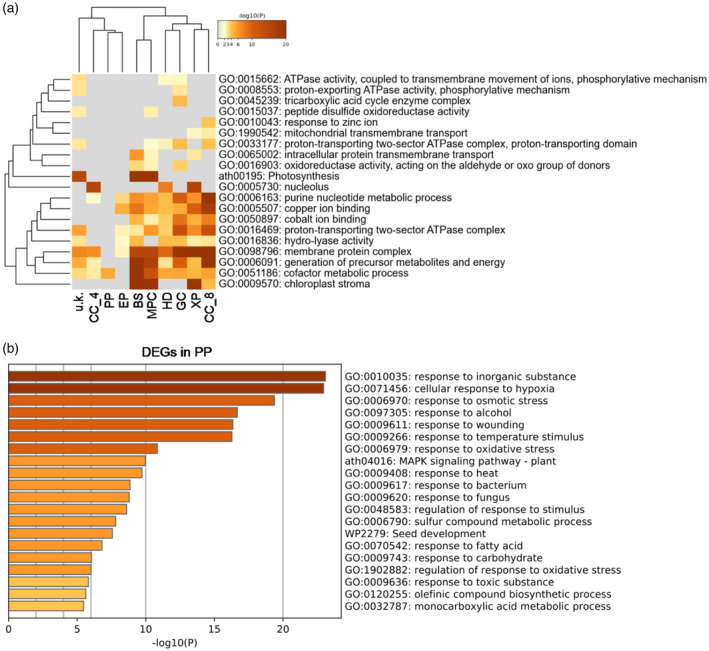
Gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) in each of the cell clusters.
(a) Analysis of the specifically enriched GO terms in each cell cluster. BS, bundle sheath; MPC, mesophyll cell; PP, phloem parenchyma; HD, hydathode; CC, companion cell; GC, guard cell; EP, epidermis; XP, xylem parenchyma; u.k., unknown.
(b) Analysis of the specifically enriched GO terms in PP.
Screening for marker genes of leaf veins
Many well‐characterized regulators of the development and function of the vascular system are expressed in cotyledon veins, and the identification of marker genes could lead to the discovery of more vascular system regulators (Bonke et al., 2003; Schlereth et al., 2010 ). To this end, we screened for new marker genes by analyzing the top 10 most highly expressed genes in each cluster. SWEET11, SWEET12 and BASIC LEUCINE ZIPPER 9 (bZIP9) were found to be specifically expressed in PP, consistent with previous results (Kim et al., 2021; Figures S4 and S5). In SE, the marker gene SEOB is specifically expressed in living sieve tubes, and is involved in regulating the development of phloem and SE (Froelich et al., 2011; Figures S4 and S5). For CC, we also found expression of several characterized marker genes, such as SUC2 and FTIP1, suggesting that the expression of these genes is conserved in both young cotyledons and mature leaves (Kim et al., 2021; Figures S4 and S5). In addition to these known marker genes, some uncharacterized marker genes are also expressed in specific cell types, such as TETRASPANIN6 (TET6) in CC_8, DNA BINDING WITH ONE FINGER 2.4 (DOF2.4) in CC_8 and SE, IRREGULAR XYLEM 7 (IRX7) in HD, and NHL1, CDF4 and CYP79B3 in PP (Figures S4 and S5). To confirm these new marker genes, we generated transgenic plants expressing the GUS reporter under the control of the promoters (2000 base pair upstream of start code) of the selected marker genes. As expected, the GUS signals of these representative genes of OLIGOPEPTIDE TRANSPORTER 4 (OPT4), EUKARYOTIC RELEASE FACTOR 1–2 (ERF1‐2), AT4G18422, AT1G23200, CYP79B2, CYP79B3, NITRATE TRANSPORTER 1.7 (NRT1.7), NHL1, AT1G04945, BZIP25, AT3G12730, PURPLE ACID PHOSPHATASE 2 (PAP2), DOF2.4, TET6 and CDF4 can be detected in the leaf veins of 3‐day‐old cotyledons (Figure S6). Analysis of the yellow fluorescent protein (YFP) expression of NTR1.7:YFP, NEK6:YFP and BZIP9:YFP indicated that these three genes are expressed in veins of 3‐day‐old cotyledons (Figure S7). These results confirm the expression patterns of these novel marker genes.
We next sought to explore whether the expression of these selected marker genes can be induced during vascular development using the VISUAL system. As shown in Figure 3, the GUS activity of NHL1pro::GUS, DOF2.4pro::GUS, NRT1.7pro::GUS and AT3G12730pro::GUS increased in MPCs compared with that of mock‐treated cells (Figure 3). However, the GUS activity of PAP2pro::GUS and TET6pro::GUS did not significantly change (Figure 3). These results suggest that the expression of some marker genes is activated during the development of veins induced by VISUAL. To explore the potential roles of marker genes, we also generated transgenic plants that over‐express some representative marker genes. As shown in Figure 4, the development of veins in 3‐day‐old cotyledons overexpressing CDF4 (oeCDF4) and oeBZIP9 was impaired compared with wild‐type (WT), suggesting that CDF4 and BZIP9 are involved in regulating the development of veins in cotyledons.
Figure 3.
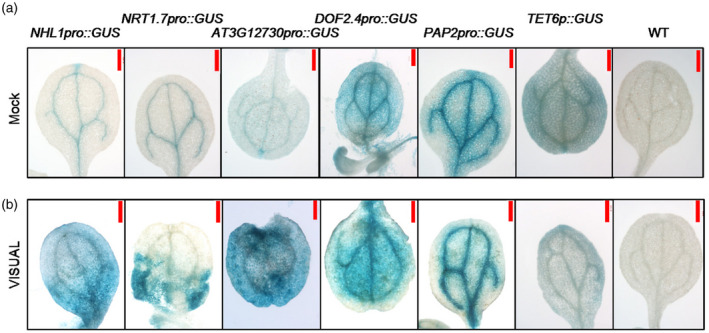
Analysis of the expression patterns of representative vein marker genes.
To detect the expression patterns of representative vein marker genes, we generated transgenic plants expressing the GUS reporter driven by their promoter, wild‐type (WT) was used as negative control.
(a) The GUS signals were detected in the 3‐day‐old cotyledons under mock conditions.
(b) The levels of GUS were detected in the 3‐day‐old cotyledons after Vascular Cell Induction Culture System Using Arabidopsis Leaves (VISUAL) treatment. Scale bar: 500 μm.
Figure 4.
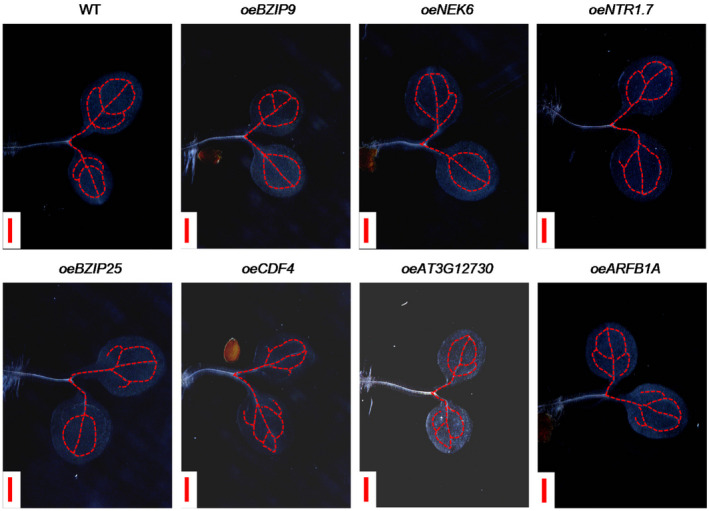
Analysis of the effects of over‐expression of representative marker genes on the developmental pattern of veins in cotyledons.
Detection of the development of the leaf veins in cotyledons of 3‐day‐old seedlings of wild‐type (WT) seedlings and transgenic plants. The leaf veins of cotyledons were marked with red dotted lines. Scale bar: 500 μm.
Tracing of the temporal and spatial patterns of leaf vein marker genes by pseudo_time analysis
To establish the profiles of the marker genes over the course of leaf vein development, we reconstructed the developmental time course and lineage relationships using Monocle analysis (Trapnell et al., 2014). As shown in Figure 5(a,b), the different cell types are distributed in the corresponding regions along the pseudo_time trajectory.
Figure 5.
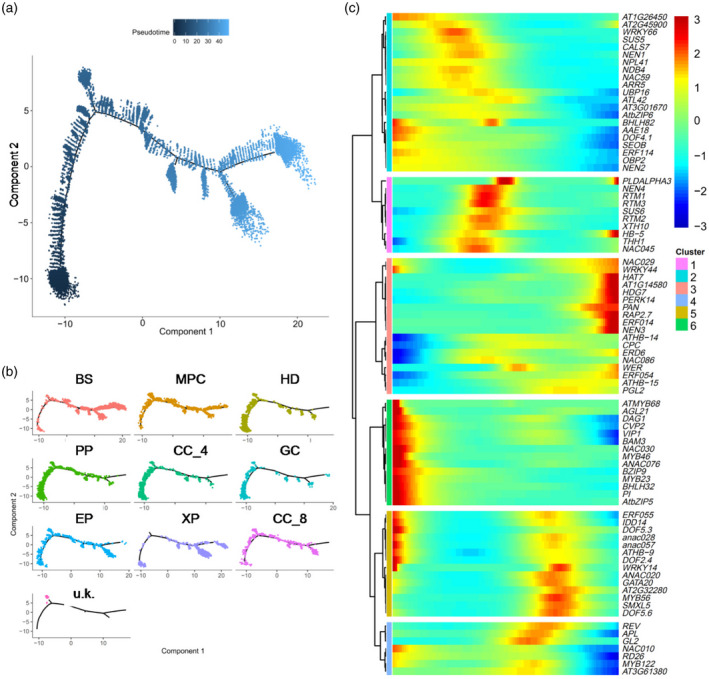
Analysis of the temporal and spatial distribution of cells by pseudo_time analysis.
(a) Distribution of cells of all clusters on the pseudo_time trajectory.
(b) Distribution of cells of each cell type colored according to cluster on the pseudo_time trajectory.
(c) Clustering and expression kinetics of marker genes of vascular along with pseudo_time progression.
Further analysis of the pseudotemporal expression dynamics of specific marker genes of the vascular system indicated that the expression levels of BZIP9, SEOB, BAM3, NAC076 and NAC030 were highest at the beginning of pseudo_time, followed by those of NAC028, NAC057 and DOF2.4 (Figure 5c). This suggests that these TFs play important roles at an early stage of leaf vein development, after which point the expression of NAC045, NAC086, APL, NAC020, REV and RESTRICTED TOBACCO ETCH POTYVIRUS MOVEMENT (RTM3) increases along the developmental trajectory (Figure 5c). Genes involved in the differentiation of CC, phloem and SE, such as NEN1, NEN4, NPL41, RTM1, RTM2 and RTM3, are transiently induced at a later stage of the pseudo_time trajectory (Figure 5c). To further test the pseudotemporal model, we also performed RNA velocity analysis of DEGs for all cell clusters with velocyto (La Manno et al., 2018). Velocyto can be used to analyze changes in transcriptional rate, maturation and turnover of a transcript. This information can then be utilized to predict future up‐ or downregulation of the corresponding transcript (La Manno et al., 2018). RNA velocity was visualized by plotting an arrow for each cell linking the actual state to the estimated future state (Figure 6a; La Manno et al., 2018; Zywitza et al., 2018). As predicted by the pseudotemporal model, the transcriptional profiles of cells in HD, PP, CC_4 and GC changed very little (short or no arrows; Figure 6a), as expected for quiescent and terminally differentiated cells (Svensson and Pachter, 2018; Zywitza et al., 2018). The RNA velocity of cells in clusters BS, XP and CC_8 showed large changes (Figure 6a), as expected for cells that differentiate rapidly. Additionally, analysis of RNA velocity can help to determine the state of gene transcripts (Trapnell et al., 2014). To investigate the specific induction and repression of gene expression along the manifold, we also examined the phase portraits of individual genes. Phase portraits of genes can show the expected deviations from the predicted steady‐state relationship (La Manno et al., 2018). As expected, NAC020 and SEOB are specifically expressed and show positive velocity as indicated by the levels of the spliced mRNA in a similar population of PCpp and SE (Figure 6b). Figure 6(b) further shows that COTYLEDON VASCULAR PATTERN 2 (CVP2) and BAM3 are primarily expressed and show positive velocity in PCpp and SE, while APL and NEN1 are mainly expressed and show positive velocity in CC_8.
Figure 6.
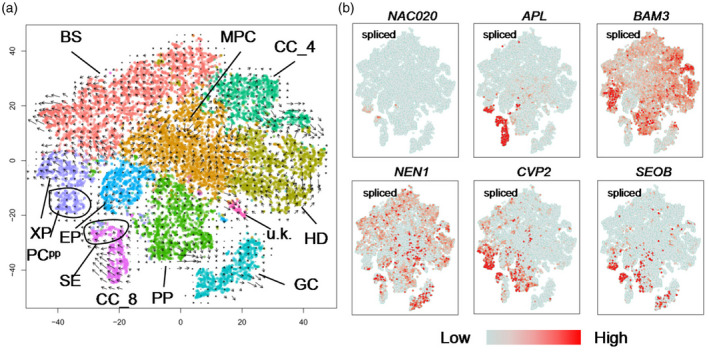
RNA velocity unveils the RNA dynamics of different cell types.
(a) RNA velocity plotted in t‐distributed Stochastic Neighbor Embedding (t‐SNE) space. The direction of state transitions and the extent of change in RNA dynamics are indicated by the vectors (arrows) and their lengths, respectively. Long arrows indicate that cells have either initiated or terminated the transcription of many different genes, while short arrows or no arrows indicate cells with small changes in transcriptional activity.
(b) The plots illustrate phase portraits of representative genes. The signals of spliced mRNA were shown.
Identification of the core TFs of leaf vein
The development of the vascular system is under the control of both phytohormones and a complex transcription regulatory network. To identify the master regulators related to the biogenesis and development of the vascular system, we screened for TFs that are specifically expressed in CC, XP and PP. TF networks for these cell clusters were then generated and the core TFs were identified (Figure 7; Table S5). As shown in Figure 7(a−c), the core TFs are CYCLING DOF FACTER 5 (CDF5) in PP, ETHYLENE‐RESPONSIVE ELEMENT BINDING FACTOR 15 (ERF15) and TFIIIA in CC_4, and REPRESSOR OF GA (RGA) and REPRODUCTIVE MERISTEM (REM19) in XP. CDF5 is an important regulator of circadian rhythm and flowering time (Henriques et al., 2017; Martin et al., 2018, 2020). DNA affinity purification sequencing (DAP‐seq) analysis indicated that CDF5 is significantly enriched on the 5′‐untranslated region (UTR) and promoter of BZIP9, SWEET12 and SULTR2;1 and the coding sequence (CDS) of SWEET11 (Figure 7e−g), suggesting that the expression of these genes can be regulated by CDF5. GO analysis of the target genes of CDF5 indicated that they are enriched for cellular response to hormone stimulus, phloem or xylem histogenesis and circadian rhythm (Figure 7h). To better understand the potential roles of CDF5 in the development of PP, we isolated the cdf5 mutant, characterized the developmental patterns of its cotyledon veins and quantified the expression of potential CDF5 targets. As shown in Figure 7(i), the developmental patterns of veins in the cdf5 mutant are similar to those of WT, but the expression of BZIP9, SWEET11, SWEET12 and SULTR2;1 declined to varying degrees (Figure 7j), suggesting that CDF5 may be involved in regulating the function of leaf veins by regulating the expression of these four target genes. Indeed, they are mainly involved in regulating the function, rather than the development of leaf veins (Maruyama‐Nakashita et al., 2015; Walerowski et al., 2018; Kim et al., 2021), although we found that oeBZIP9 can affect the development of veins (Figure 4).
Figure 7.
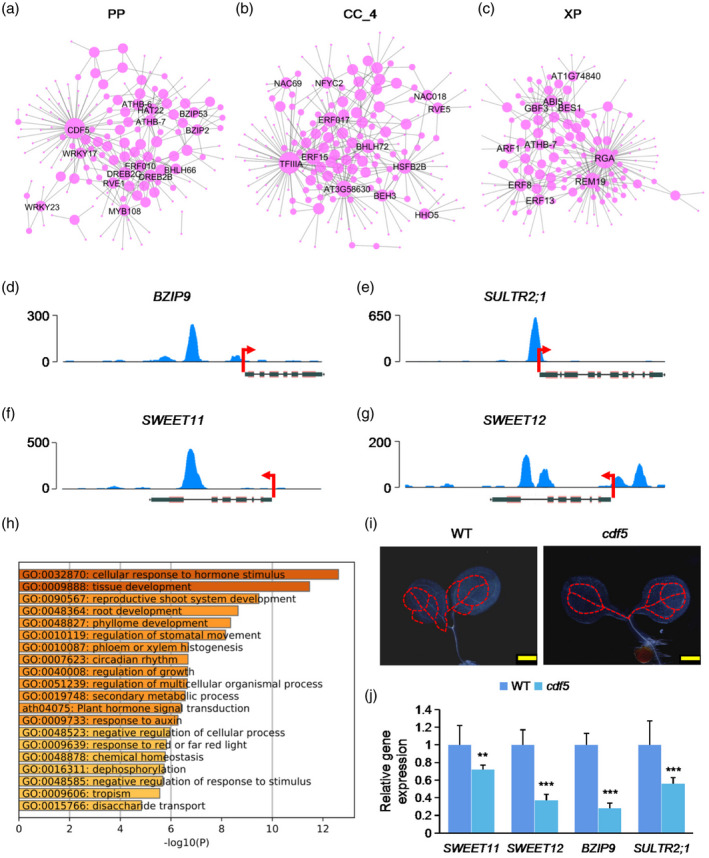
Identification of regulatory networks of transcription factors (TFs) in different cell types.
(a−c) Regulatory network showing potentially core transcriptional regulators for phloem parenchyma (PP), companion cell (CC)_4 and xylem parenchyma (XP) cells. The dot size indicates the number of connections. Gene names of top‐ranked nodes (ranked by the number of connections) are shown.
(d−g) DNA affinity purification sequencing (DAP‐seq) of enrichment of CDF5 in the promoter regions of representative genes. Data are derived from O’Malley et al. (2016).
(h) Gene Ontology (GO) enrichment analysis of target genes of CDF5.
(i) Detection of the developmental patterns of vascular system in cotyledons of 3‐day‐old seedlings of cdf5 mutant and wild‐type (WT) seedlings. Scale bar: 500 μm.
(j) Quantitative polymerase chain reaction (qPCR) analysis of the relative expression of representative target genes of CDF5 in cdf5 mutant and WT. **p < 0.01; ***p < 0.001, Student’s t‐test versus WT.
Recent studies indicate that light can initiate PC formation in VISUAL (Yamazaki et al., 2018). Although light does not elevate endogenous gibberellic acid (GA) content, GA can simulate the effect of light to promote PC formation through degradation of DELLA (GAI, RGA, RGL1, RGL2 and RGL3; Yamazaki et al., 2018). In addition, overexpression of the constitutively active DELLA protein RGA inhibits vascular cell differentiation even under light conditions, suggesting that RGA prevents PC formation during vascular development in VISUAL (Yamazaki et al., 2018). As shown in Figure S8(a−c), feature plots indicate that RGA is highly enriched in PC and PP, suggesting that DELLA signaling may be involved in regulating the development of the leaf veins at a very early stage. To investigate the potential roles of RGA during the early development of the leaf vein in cotyledons, we obtained the oeRGL2 and della quintuple mutant ( gai‐t6 ; rga‐t2 ; rgl1‐1 ; rgl2‐1 ; rgl3‐1 ) from Arabidopsis Biological Resource Center (ABRC). As expected, the developmental pattern of the leaf vein in cotyledons was normal in oeRGL2, but impaired in the della quintuple mutant compared with WT (Figure S8d−f). These results suggest that DELLA is required for the regulation of the early development of the leaf veins in cotyledons.
ATP synthase beta‐subunit is involved in regulating the development of the leaf veins in cotyledons
The vein system is the main transmission pipeline for metabolites generated by photosynthesis and water in leaves, and the development of the leaf vein system is impacted by photosynthetic activity and energy status (Li et al., 2017). Expression profiles of CC_8 and XP are greatly enriched for genes involved in the generation of precursor metabolites and energy (Figure 2a), suggesting that active energy metabolism is required for the early development of the leaf vein in cotyledons. Further analysis indicated that the expression of the mitochondrial marker genes AT5G08670 and AT5G08690, which encode the mitochondrial ATP synthase beta‐subunit, is higher in XP and CC_8 (Figure 8a,b). To further investigate the expression of AT5G08670 and AT5G08690 during the development of the vascular system in cotyledons, we generated corresponding GUS reporter lines. The expression of AT5G08670pro::GUS and AT5G08690pro::GUS was high in the leaf veins in cotyledons (Figure 8c,d). The GUS signals of AT5G08670pro::GUS and AT5G08690pro::GUS were increased in MPCs after VISUAL treatment (Figure 8e,f). To examine whether AT5G08670 and AT5G08690 are involved in the development of leaf veins, we isolated mutants deficient in these genes. Analysis of the development of leaf veins of the single mutants showed that their leaf veins developed normally (data not shown). The mRNA and protein sequences of AT5G08670 and AT5G08690 are highly similar, suggesting that they may be functionally redundant. Because AT5G08670 and AT5G08690 are located close together in the Arabidopsis genome, double mutants could not be generated via crossing. We instead generated the double mutant through the DNA editing technique (Figures 8h and S9). The development of the leaf veins in the cotyledons of the double mutant (here referred to as atpb) was impaired compared with WT (Figure 8g,h), suggesting that AT5G08670 and AT5G08690 are required for the early development of the leaf veins in cotyledons.
Figure 8.
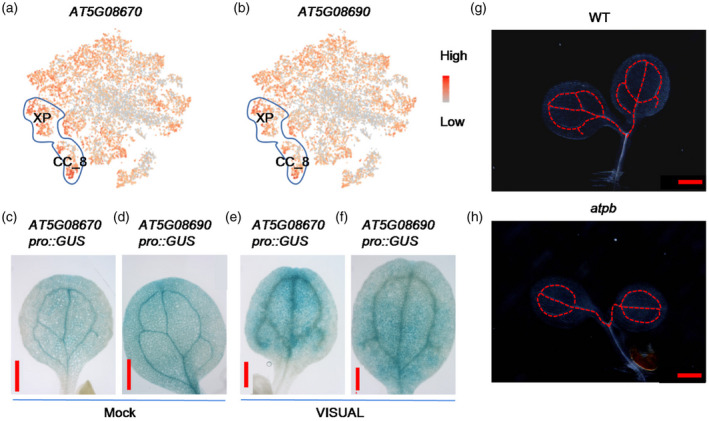
ATP synthase beta‐subunit AT5G08670 and AT5G08690 are involved in regulating the development of veins in cotyledons.
(a,b) Feature plots of the expression of AT5G08670 and AT5G08690. The cells of xylem parenchyma (XP) and companion cell (CC)_8 are marked with blue line circles.
(c,d) The expression of GUS of AT5G08670pro::GUS and AT5G08690pro::GUS is specifically enriched in leaf veins of cotyledon.
(e,f) The levels of GUS of AT5G08670pro::GUS and AT5G08690pro::GUS were increased in mesophyll cells (MPCs) with the Vascular Cell Induction Culture System Using Arabidopsis Leaves (VISUAL). Scale bar: 500 μm (c−f).
(g,h) Detection of the developmental patterns of leaf veins in cotyledons of 3‐day‐old seedlings of wild‐type (WT) seedlings and atpb double mutant (double mutant of AT5G08670 and AT5G08690). Scale bar: 500 μm (g,h).
DISCUSSION
Transcriptome and novel marker genes of the leaf vein in cotyledons
Leaf veins provide an important structural support and transportation system for the transmission of organic matter, minerals and water in leaves. The role of leaf vein formation in the early development of cotyledons still remains poorly understood. Xylem and phloem cells sorted from VISUAL cultured samples by fluorescence‐activated cell sorting (FACS) technology have provided important insights into the molecular mechanisms underlying sequential differentiation of sieve element‐like cells (Kondo et al., 2016). However, only a few cell types can be obtained by FACS, and it is difficult to elucidate the early developmental regulation of leaf vein under conditions of the VISUAL culture system (Kondo et al., 2016). The application of 10X Genomics in the plant scRNA‐seq research field has demonstrated its technical advantages in high cell throughput and its ability to analyze the heterogeneity of single‐cell gene expression patterns. This approach has made it feasible to analyze the molecular mechanisms of the early development of leaf veins in cotyledons. We employed 10X Genomics technology to investigate the global cell atlas of 3‐day‐old cotyledons. We analyzed 14 117 single cells, identified 10 cell clusters and annotated the corresponding cell types. GO analysis of DEGs specifically expressed in different cell types provides a reference for in‐depth and systematic study of the biological functions of various leaf vein cells. The DEGs in BSs and MPCs were found to be related to photosynthesis (Figure 2a), which is consistent with the presence of chloroplasts in these cells (Barton et al., 2016). Interestingly, we found that GO terms enriched in PP share little overlap with those of other cell types, and are mainly involved in responses to biotic and abiotic stimulations (Figure 2a,b). Based on GO analysis, we also found that the DEGs of XP and CC_8 are significantly enriched for energy metabolism in mitochondria (Figure 2a). The representative marker genes AT5G08670 and AT5G08690 were also found to be highly expressed in XP and CC_8 (Figure 8a,b). Development of the leaf vein pattern was impaired in the cotyledons of the atpb mutant, suggesting that energy metabolism in mitochondria is required for the early development of the leaf vein (Figure 2). In addition to the known regulators of the leaf vein, we also found novel marker genes, such as HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 36 (HIPP36), ARABIDOPSIS THALIANA HISTIDINE‐CONTAINING PHOSPHOTRANSMITTER 1 (AHP1), TET6 and PHL12 for CC, ARABINOGALACTAN PROTEIN 31 (AGP31) and CELLULASE 1 (CEL1) for XP, DOF2.4 for SE and IRX7, CDF4, SULTR2;1 and BZIP9 for PP (Figures 1 and S4). The low‐affinity sulfate transporter SULTR2;1 is predominantly expressed in the XP and pericycle cells in Arabidopsis thaliana roots when sulfate is depleted (Maruyama‐Nakashita et al., 2015). However, the expression of SULTR2;1 in the shoot vascular system is not induced by sulfur starvation (Maruyama‐Nakashita et al., 2015). AGP31 and FTIP1 have also been shown to play roles in the regulation of vascular system development (Liu and Mehdy, 2007; Liu et al., 2012; Abe et al., 2015). Analysis of the expression of GUS driven by promoters of marker genes further confirmed their presence in leaf veins of cotyledons (Figure S6). These newly identified marker genes can be used as references in future studies.
Spatial and temporal patterns of the leaf vein transcriptome
The development of the leaf vein is a sequential process that is accompanied by significant changes in transcriptome, metabolism and cell morphology. The results of the pseudo_time analysis indicated that all cells could be ordered along one main developmental trajectory (Figure 5a). Analysis of the distribution of different cell types indicated that the cells of different cell clusters are distributed relatively independently on the pseudo_time trajectory (Figure 5b). Previous studies have shown that key regulators of vein development, such as NAC020, SEOB and APL, are expressed at specific developmental stages of leaf veins (Kondo et al., 2016). Heatmap analysis of representative marker genes in our study yielded patterns that were consistent with previous data (Kondo et al., 2016; Figure 5c), and the dynamic expression patterns of representative marker genes along the time trajectory are compatible with their known functions at particular developmental stages of leaf veins (Figure 5c). These results suggest that the pseudo_time analysis is an effective tool to determine the dynamics of gene expression at different developmental stages. Further RNA velocity analysis showed that RNA metabolism activity in PP, XP and EP cells is relatively low, which is compatible with their nature as quiescent and terminally differentiated cells. In contrast, BSs, MPCs and CC_8 were shown to have high RNA metabolic activity, indicating that these cells are highly differentiated (Figure 6a).
Identification of the core transcriptional factors regulating the development of the leaf veins in cotyledons
The development of the vascular system is regulated by a signaling cascade composed of CLE41/42‐TDR‐WOX4/14, as well as other TFs (Etchells and Turner, 2010; Hirakawa et al., 2010; Suer et al., 2011; Etchells et al., 2013; Kondo et al., 2014). Recently, several important TFs involved in the regulation of vascular system function have been characterized, such as SUPPRESSOR OF MAX2 1‐LIKE 5 (SMXL5), SHORTROOT and WRKY15 (Ge et al., 2020; Kim et al., 2020; Smit et al., 2020; Wallner et al., 2020). Our scRNA‐seq results indicate that different cell types have distinct DEGs related to specific functions during development of the leaf veins (Figure 2). We screened for TFs that were specifically enriched in PP, CC and XP (Figure 7a−c). DAP‐seq and phenotype analysis indicated that CDF5 may be involved in regulating the function rather than the development of veins by regulating the expression of SWEET11, SWEET12, BZIP9 and SULTR2;1 (Figure 7d−j). The identification of these core regulators also indicated the potential roles of GA signaling in the regulation of the development of the leaf veins in cotyledons. For example, we found that the TFs RGA, RGA‐LIKE 1 (RGL1) and RGL2 are highly expressed in the XP and CC_8, and the development of leaf veins in the cotyledons of the della quintuple mutant was impaired (Figure S8). In the past, the roles of jasmonic acid (JA) and indole acetic acid (IAA) in the regulation and regeneration of the vascular system have been demonstrated (Reinhardt, 2003; Scarpella et al., 2006; Foyer et al., 2015; Zhou et al., 2019a). ERF109 and ERF115 were shown to act in response to JA signaling during regeneration of the vascular system (Zhou et al., 2019b), and our results provide new insights into the role of GA in the development of the leaf veins in cotyledons.
Taken together, our results provide new insights into the early transcriptome profiles of leaf veins in cotyledons, and lead to the identification of new marker genes for leaf vein cells. The dynamic expression patterns of these new marker genes were further confirmed with GUS reporter fusions, providing new information about the mechanisms underlying leaf vein development. A more complete picture of the mechanisms underlying the development of leaf veins will be possible by employing these new markers in future studies.
EXPERIMENTAL PROCEDURES
Screening and verification of mutants
The T‐DNA insertion mutants were obtained from ABRC (Table S6). Mutant lines homozygous for the T‐DNA insertion were identified by polymerase chain reaction (PCR) analysis using gene‐specific and T‐DNA‐specific primers (Figure S10; Table S7).
Constructs for plant transformation
To generate the pBGWFS7‐promoter (of marker genes) constructs, the upstream 2000 bp of the start codon ATG of selected marker genes were PCR‐amplified using the primer pairs as described in Table S7. Then the PCR products were purified and first cloned into pDNOR201 by BP Clonase reactions (GATEWAY Cloning; Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions to generate the pDNOR‐promoter (of marker genes). The resulting plasmids were recombined into pBGWFS7 using LR Clonase reactions (GATEWAY Cloning; Invitrogen) to generate the final constructs. To generate the overexpression constructs, the full‐length cDNA fragments of marker genes were PCR‐amplified using the primer pairs described in Table S7. Then the PCR products were purified and first cloned into pDNOR201 by BP Clonase reactions (GATEWAY Cloning; Invitrogen) according to the manufacturer’s instructions to generate the pDNOR‐cDNA vectors. The resulting plasmids were recombined into pB7YWG2.0 using LR Clonase reactions to generate the final constructs.
Plant transformation
The pBGWFS7‐promoter constructs were transformed into Agrobacterium tumefaciens strain GV3105 via electroporation. Then the A. tumefaciens that contained the constructs of pBGWFS7‐promoter was introduced into WT. The resulting T1 transgenic plants were selected by BASTA as described previously (Sun et al., 2016). Homozygous transgenic plants were used in all experiments.
Cotyledon collection and protoplast preparation
We isolated protoplasts from cotyledons of 3‐day‐old Arabidopsis seedlings as described previously (Yoo et al., 2007) with slight modifications to adjust to the cotyledon tissue. Briefly, the cotyledons were harvested from seedlings and cut into 2‐mm sticks and submerged in a solution [0.5 mm CaCl2, 0.5 mm MgCl2, 5 mm MES, 1.5% Cellulase RS, 0.03% Pectolyase Y23, 0.25% BSA, actinomycin D (33 mg L−1) and cordycepin (100 mg L−1), pH 5.5] by vacuum infiltration for 10 min. The samples were then incubated for 4 h to isolate protoplasts. Afterwards, the isolated cells were washed three times with 8% mannitol buffer to remove Mg2+. Cells were then filtered with a 40‐μm cell strainer. Cell activity was detected by trypan blue staining and cell concentration was measured with a hemocytometer.
scRNA‐seq library preparation
We prepared scRNA‐seq libraries with Chromium Single Cell 3′ Gel Beads‐in‐emulsion (GEM) Library & Gel Bead Kit v3 according to the user manual supplied by the kit.
scRNA‐seq data preprocessing
The raw scRNA‐seq data preprocessing was performed as described (Liu et al., 2020). Firstly, the preliminary quality control of raw data was carried out by using CellRanger (V 3.0.0) software. The average Q30 quality of RNA reads was 93.1%, the median UMI value of each cell was 24 650, and the median gene value of each cell was 2321. On the basis of CellRanger’s preliminary quality control, Seurat software package (v 2.3.4) was used for further quality control of the data. Theoretically, the number of genes (nGene), number of UMI (nUMI), mitochondrial genes (percent.mito) and chloroplast genes (percent.pt) expressed by most cells will be concentrated in a certain region. A very low number of genes or UMI is considered as low‐quality cell, and a too high number may be considered as double cells or multicellular. Therefore, cell filtration was analyzed according to the distribution of four indexes of nUMI, nGene, percent.mito and percent.pt. To remove low‐quality cells and likely multiplet captures, which is a major concern in microdroplet‐based experiments, we further applied a criteria to filter out cells with UMI/gene numbers beyond the limit of the mean value ± twofold of standard deviations, assuming a Gaussian distribution of each cell’s UMI/gene number. Following visual inspection of the distribution of cells by the fraction of mitochondrial genes and chloroplast genes expressed, we further discarded low‐quality cells where > 10% of the counts belonged to mitochondrial genes and > 40% of the counts belonged to chloroplast genes. After quality control, 14 117 cells in total remained and were used in the downstream analyses. Library size normalization was performed in Seurat on the filtered matrix to obtain normalized counts.
Genes with the highest variable expression amongst single cells were identified using the method described previously (Macosko et al., 2015). The average expression and dispersion were briefly calculated for all genes, which were subsequently placed into 10 bins based on expression. Principal component analysis (PCA) was performed to reduce the dimensionality on the log‐ transformed gene‐barcode matrices of the most variable genes. Cells were clustered via a graph‐based approach and visualized in two dimensions using tSNE. A likelihood ratio test, which simultaneously tests for changes in mean expression and percentage of cells expressing a gene, was used to identify significantly DEGs between clusters. We used the FindAllMarkers function (test.use = bimod, logfc.thresold = 0,min.pct = 0.25) in Seurat to identify DEGs of each cluster. For a given cluster, FindAllMarkers identified positive markers compared with all other cells.
To confirm the identification of cell clusters by tSNE, we also performed the UMAP analysis (Becht et al., 2019). For PCA, the scaled data were reduced into 30 approximate PCs depending on the 5965 highly variable genes (set npcs = 30). Clusters were identified using the Seurat function ‘FindClusters’ with ‘resolution = 0.4’. The data structures were separately visualized and explored by UMAP (run the ‘RunUMAP’ function with ‘n.neighbors=30, metric = correlation and min.dist=0.3’).
Pseudo_time trajectory analysis of single‐cell transcriptomes was conducted using Monocle 2 (Trapnell et al., 2014). For pseudo_time analysis, the raw count in Seurat object was first converted into the CellDataSet with the importCDS (object, import_all = F) function in Monocle 2. Then we used estimateSizeFactors() and estimateDispersions() functions to pre‐calculate some parameters about the data. Specifically, size factors helped us normalize the differences in mRNA recovered across cells, and ‘dispersion’ values helped us perform the differential expression analysis later. We used differential GeneTest function (fullModelFormulaStr = ‘~clusters’) of the Monocle 2 package for ordering genes (qval < 0.01) that were likely to be informative for ordering cells along the pseudo‐time trajectory. The ordered genes were then marked with the setOrderingFilter() function. The dimensional reduction clustering analysis was performed with the reduceDimension() function, max_components = 2, reduction_method = ‘DDRTree’, and then with the trajectory inference (‘orderCells’ function) with default parameters. Gene expression was then plotted as a function of pseudo‐time in Monocle 2 to track changes across pseudo‐time. We also plotted TFs and marker genes along the inferred developmental pseudo‐time. The regulation networks for the TFs and target genes were plotted by Cytoscape according to the PlantTFDB database. Bulk and scRNA‐seqs correlation analysis was performed as described by Rheaume et al. (2018). Differential expression analysis was performed with t.test. Using the t.test function to test the gene expression value in scRNA‐seq and bulk, the significant P‐value was obtained. The difference multiple of log2FC was calculated as follows: log2 ((mean gene expression value in scRNA‐seq) + 0.001)/((mean gene expression value in bulk) + 0.001). Finally, the genes with significant difference were screened according to P < 0.05 and ¦log2FC¦ > 1.
RNA‐seq analysis
Cotyledons of 3‐day‐old seedlings were harvested for extracting total RNA using the mirVana miRNA Isolation Kit (Ambion) following the manufacturer’s protocol. The samples with RNA Integrity Number (RIN) ≥ 7 were subjected to subsequent RNA‐seq analysis. The libraries were constructed using TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten) and 125‐bp/150‐bp paired‐end reads were generated.
GUS staining and histological analysis
Histochemical GUS staining was performed with GUS staining kit according to the manual (G3061, Solarbio, Beijing, China). Samples were fixed in 90% acetone at −20°C, rinsed four times with 0.1 m sodium phosphate buffer (pH 7.4), and then incubated in X‐Gluc solution [0.1 m sodium phosphate (pH 7.4), 3 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 0.5 g L−1 5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐glucuronide cyclohexilammonium salt] at 37°C. After staining, samples were incubated in methanol to remove chlorophyll and then mounted in the clearing solution (a mixture of chloral hydrate, water and glycerol in a ratio of 8:2:1). Observation was performed using a stereomicroscope (MZ16F, Leica Microsystems, Germany) or a microscope equipped with Nomarski optics (BX51, Olympus, Tokyo, Japan). For the observation of vascular patterns, cotyledons were fixed in a mixture of ethanol and acetic acid in a ratio of 9:1, hydrated through a graded series of ethanol, and then mounted with the clearing solution (Konishi and Sugiyama, 2003).
Microscopy
The seedlings were stained with 10 g ml−1 propidium iodide (PI; P4170, Sigma, St Louis, MO, USA) for 1 min before imaging. For confocal microscopy, fluorescence in roots was detected using a confocal laser‐scanning microscope (Zeiss, LSM980, Oberkochen, Germany). PI signal was visualized using wavelengths of 610–630 nm. YFP was observed using wavelengths of 510–530 nm. Images and green fluorescent protein (GFP) intensities were processed using Zeiss Confocal Software.
GO enrichment analysis
The enrichment of GO terms and pathways for the DEGs were analyzed using Metascape (http://metascape.org/; Zhou, Zhou, et al., 2019).
ACCESSION NUMBERS
Sequence data from this study can be found in the Arabidopsis Genome Initiative data library under the following accession numbers: RGL2 (AT3G03450), PIN1(AT1G73590), ARR4 (AT1G10470). scRNA‐seq data are available at the https://dataview.ncbi.nlm.nih.gov/?search=SUB6947465 (https://www.ncbi.nlm.nih.gov).
AUTHOR CONTRIBUTIONS
Conceptualization of the project: X.S. Experimental design: XS and ZL Performance of some specific experiments: ZL, JW, YZ, YZ, AQ, XY, ZZ, RW, XY and CG Data analysis: ZL, JW, YZ, YZ and JR Manuscript drafting: SX Contribution to the editing and proofreading of the manuscript draft: GB, JR and XS All authors have read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
OPEN RESEARCH BADGES
This article has earned an Open Data badge for making publicly available the digitally shareable data necessary to reproduce the reported results. The data are available at https://dataview.ncbi.nlm.nih.gov/?search=SUB6947465 (https://www.ncbi.nlm.nih.gov).
Supporting information
Figure S1. The experimental flow chart used in this study.
Figure S2. Quality control analysis of the raw data of scRNA‐seq.
Figure S3. UMAP analysis of the cells.
Figure S4. Analysis of the expression of known marker genes of different kinds of cell types on tSNE plot.
Figure S5. Analysis of the expression of known marker genes of different kinds of cell types on UMAP plot.
Figure S6. Analysis of the expression of representative vein marker genes.
Figure S7. NTR1.7, NEK6 and BZIP9 were expressed in vein.
Figure S8. DELLA proteins are required for the regulation of the patterns of vascular systems in cotyledons.
Figure S9. Vector information of gRNA CRISPR/Cas9 and double mutant verification.
Figure S10. Verification of the T‐DNA insertion of mutants.
Table S6. List of mutant lines used in this study
Table S7. List of oligonucleotides used in this study
Table S1. Differentially expressed genes in each of cell cluster of tSNE analysis
Table S2. Differentially expressed genes in each of cell cluster of UMAP analysis
Table S3. The marker genes of vascular tissues
Table S4. GO enrichment analysis of DEGs of each of cell cluster of tSNE analysis
Table S5.Transcriptional factor network analysis
ACKNOWLEDGEMENTS
The authors are grateful to ABRC for the Arabidopsis seeds. This research was supported by the National Natural Science Foundation of China (31670233).
Linked article: This paper is the subject of a Research Highlight article. To view this Research Highlight article visit https://doi.org/10.1111/tpj.15738
DATA AVAILABILITY STATEMENT
All data supporting the findings of this study are available within the paper and within its supplementary data published online.
REFERENCES
- Abe, M. , Kaya, H. , Watanabe‐Taneda, A. , Shibuta, M. , Yamaguchi, A. , Sakamoto, T. et al. (2015) FE, a phloem‐specific Myb‐related protein, promotes flowering through transcriptional activation of FLOWERING LOCUS T and FLOWERING LOCUS T INTERACTING PROTEIN 1. Plant Journal, 83, 1059–1068. [DOI] [PubMed] [Google Scholar]
- Amiard, V. , Mueh, K.E. , Demmig‐Adams, B. , Ebbert, V. , Turgeon, R. & Adams, W.W., 3rd . (2005) Anatomical and photosynthetic acclimation to the light environment in species with differing mechanisms of phloem loading. Proceedings of the National Academy of Sciences of the United States of America, 102, 12968–12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelt, F. , Mavrothalassiti, E. , Gupta, S. , Machin, F. , Olas, J.J. , Annunziata, M.G. et al. (2021) Shoot and root single cell sequencing reveals tissue‐ and daytime‐specific transcriptome profiles. Plant Physiology, 188, 861–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima, S. , Possenti, M. , Matteucci, A. , Wisman, E. , Altamura, M.M. , Ruberti, I. et al. (2001) The arabidopsis ATHB‐8 HD‐zip protein acts as a differentiation‐promoting transcription factor of the vascular meristems. Plant Physiology, 126, 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R.F. , Leach, K.A. , Boyer, N.R. , Swyers, M.J. , Benitez‐Alfonso, Y. , Skopelitis, T. et al. (2016) Sucrose transporter ZmSut1 expression and localization uncover new insights into sucrose phloem loading. Plant Physiology, 172, 1876–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, K.A. , Schattat, M.H. , Jakob, T. , Hause, G. , Wilhelm, C. , McKenna, J.F. et al. (2016) Epidermal pavement cells of Arabidopsis have chloroplasts. Plant Physiology, 171, 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht, E. , McInnes, L. , Healy, J. , Dutertre, C.A. , Kwok, I.W.H. , Ng, L.G. et al. (2019) Dimensionality reduction for visualizing single‐cell data using UMAP. Nature Biotechnology, 37, 38‐+. [DOI] [PubMed] [Google Scholar]
- Bishopp, A. , Help, H. , El‐Showk, S. , Weijers, D. , Scheres, B. , Friml, J. et al. (2011) A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Current Biology, 21, 917–926. [DOI] [PubMed] [Google Scholar]
- Bishopp, A. , Lehesranta, S. , Vaten, A. , Help, H. , El‐Showk, S. , Scheres, B. et al. (2011) Phloem‐transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Current Biology, 21, 927–932. [DOI] [PubMed] [Google Scholar]
- Bonke, M. , Thitamadee, S. , Mahonen, A.P. , Hauser, M.T. & Helariutta, Y. (2003) APL regulates vascular tissue identity in Arabidopsis. Nature, 426, 181–186. [DOI] [PubMed] [Google Scholar]
- Brackmann, K. , Qi, J.Y. , Gebert, M. , Jouannet, V. , Schlamp, T. , Grunwald, K. et al. (2018) Spatial specificity of auxin responses coordinates wood formation. Nature Communications, 9, 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkle, L. , Cedzich, A. , Dopke, C. , Stransky, H. , Okumoto, S. , Gillissen, B. et al. (2003) Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. The Plant Journal, 34, 13–26. [DOI] [PubMed] [Google Scholar]
- Carland, F.M. & Nelson, T. (2004) Cotyledon vascular pattern2‐mediated inositol (1,4,5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. The Plant Cell, 16, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer, T. , Ma, X. , Klesen, S. , Scacchi, E. , Nieselt, K. & Timmermans, M.C.P. (2019) Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high‐throughput single‐cell RNA sequencing. Developmental Cell, 48(840–852), e845. [DOI] [PubMed] [Google Scholar]
- Dettmer, J. , Elo, A. & Helariutta, Y. (2009) Hormone interactions during vascular development. Plant Molecular Biology, 69, 347–360. [DOI] [PubMed] [Google Scholar]
- Etchells, J.P. , Provost, C.M. , Mishra, L. & Turner, S.R. (2013) WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development, 140, 2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells, J.P. & Turner, S.R. (2010) The PXY‐CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development, 137, 767–774. [DOI] [PubMed] [Google Scholar]
- Fisher, K. & Turner, S. (2007) PXY, a receptor‐like kinase essential for maintaining polarity during plant vascular‐tissue development. Current Biology, 17, 1061–1066. [DOI] [PubMed] [Google Scholar]
- Foyer, C.H. , Verrall, S.R. & Hancock, R.D. (2015) Systematic analysis of phloem‐feeding insect‐induced transcriptional reprogramming in Arabidopsis highlights common features and reveals distinct responses to specialist and generalist insects. Journal of Experimental Botany, 66, 495–512. [DOI] [PubMed] [Google Scholar]
- Froelich, D.R. , Mullendore, D.L. , Jensen, K.H. , Ross‐Elliott, T.J. , Anstead, J.A. , Thompson, G.A. et al. (2011) Phloem ultrastructure and pressure flow: sieve‐element‐Occlusion‐related agglomerations do not affect translocation. The Plant Cell, 23, 4428–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, H. (2004) Signals that control plant vascular cell differentiation. Nature Reviews Molecular Cell Biology, 5, 379–391. [DOI] [PubMed] [Google Scholar]
- Furuta, K.M. , Hellmann, E. & Helariutta, Y. (2014) Molecular control of cell specification and cell differentiation during procambial development. Annual Review of Plant Biology, 65, 607–638. [DOI] [PubMed] [Google Scholar]
- Furuta, K.M. , Yadav, S.R. , Lehesranta, S. , Belevich, I. , Miyashima, S. , Heo, J.O. et al. (2014) Plant development. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science, 345, 933–937. [DOI] [PubMed] [Google Scholar]
- Gala, H.P. , Lanctot, A. , Jean‐Baptiste, K. , Guiziou, S. , Chu, J.C. , Zemke, J.E. et al. (2021) A single‐cell view of the transcriptome during lateral root initiation in Arabidopsis thaliana. The Plant Cell, 33, 2197–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, S. , Han, X. , Xu, X. , Shao, Y. , Zhu, Q. , Liu, Y. et al. (2020) WRKY15 suppresses tracheary element differentiation upstream of VND7 during xylem formation. The Plant Cell, 32, 2307–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S. & Berleth, T. (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. The EMBO Journal, 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques, R. , Wang, H. , Liu, J. , Boix, M. , Huang, L.F. & Chua, N.H. (2017) The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. The New Phytologist, 216, 854–867. [DOI] [PubMed] [Google Scholar]
- Hirakawa, Y. , Kondo, Y. & Fukuda, H. (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 Homeobox gene in Arabidopsis. The Plant Cell, 22, 2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa, Y. , Shinohara, H. , Kondo, Y. , Inoue, A. , Nakanomyo, I. , Ogawa, M. et al. (2008) Non‐cell‐autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proceedings of the National Academy of Sciences of the United States of America, 105, 15208–15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, Y. , Nakanomyo, I. , Motose, H. , Iwamoto, K. , Sawa, S. , Dohmae, N. et al. (2006) Dodeca‐CLE peptides as suppressors of plant stem cell differentiation. Science, 313, 842–845. [DOI] [PubMed] [Google Scholar]
- Ji, J.B. , Strable, J. , Shimizu, R. , Koenig, D. , Sinha, N. & Scanlon, M.J. (2010) WOX4 Promotes Procambial Development. Plant Physiology, 152, 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. , Zhou, J. , Kumar, D. , Jang, G. , Ryu, K.H. , Sebastian, J. et al. (2020) SHORTROOT‐mediated intercellular signals coordinate phloem development in Arabidopsis roots. The Plant Cell, 32, 1519–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y. , Symeonidi, E. , Pang, T.Y. , Denyer, T. , Weidauer, D. , Bezrutczyk, M. et al. (2021) Distinct identities of leaf phloem cells revealed by single cell transcriptomics. The Plant Cell, 33, 511–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, Y. , Fujita, T. , Sugiyama, M. & Fukuda, H. (2015) A novel system for xylem cell differentiation in Arabidopsis thaliana. Molecular Plant, 8, 612–621. [DOI] [PubMed] [Google Scholar]
- Kondo, Y. , Hirakawa, Y. , Kieber, J.J. & Fukuda, H. (2011) CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant and Cell Physiology, 52, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, Y. , Ito, T. , Nakagami, H. , Hirakawa, Y. , Saito, M. , Tamaki, T. et al. (2014) Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF‐TDR signalling. Nature Communications, 5, 3504. [DOI] [PubMed] [Google Scholar]
- Kondo, Y. , Nurani, A.M. , Saito, C. , Ichihashi, Y. , Saito, M. , Yamazaki, K. et al. (2016) Vascular cell induction culture system using Arabidopsis leaves (VISUAL) reveals the sequential differentiation of sieve element‐like cells. The Plant Cell, 28, 1250–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, M. & Sugiyama, M. (2003) Genetic analysis of adventitious root formation with a novel series of temperature‐sensitive mutants of Arabidopsis thaliana. Development, 130, 5637–5647. [DOI] [PubMed] [Google Scholar]
- La Manno, G. , Soldatov, R. , Zeisel, A. , Braun, E. , Hochgerner, H. , Petukhov, V. et al. (2018) RNA velocity of single cells. Nature, 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Wu, L. , Foster, R. & Ruan, Y.L. (2017) Molecular regulation of sucrose catabolism and sugar transport for development, defence and phloem function. Journal of Integrative Plant Biology, 59, 322–335. [DOI] [PubMed] [Google Scholar]
- Liu, C. & Mehdy, M.C. (2007) A nonclassical arabinogalactan protein gene highly expressed in vascular tissues, AGP31, is transcriptionally repressed by methyl jasmonic acid in Arabidopsis. Plant Physiology, 145, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Liu, C. , Hou, X.L. , Xi, W.Y. , Shen, L.S. , Tao, Z. et al. (2012) FTIP1 is an essential regulator required for florigen transport. PLoS Biology, 10, e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Liang, Z. , Feng, D. , Jiang, S.J. , Wang, Y.F. , Du, Z.Y. et al. (2021) Transcriptional landscape of rice roots at the single‐cell resolution. Molecular Plant, 14, 384–394. [DOI] [PubMed] [Google Scholar]
- Liu, Z.X. , Zhou, Y.P. , Guo, J.G. , Li, J.A. , Tian, Z.X. , Zhu, Z.N. et al. (2020) Global dynamic molecular profiling of stomatal lineage cell development by single‐cell RNA sequencing. Molecular Plant, 13, 1178–1193. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J. , Groover, A. , Lichtenberger, R. , Furuta, K. , Yadav, S.R. , Helariutta, Y. et al. (2013) The plant vascular system: evolution, development and functions. Journal of Integrative Plant Biology, 55, 294–388. [DOI] [PubMed] [Google Scholar]
- Macosko, E.Z. , Basu, A. , Satija, R. , Nemesh, J. , Shekhar, K. , Goldman, M. et al. (2015) Highly parallel genome‐wide expression profiling of individual cells using nanoliter droplets. Cell, 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen, A.P. , Bishopp, A. , Higuchi, M. , Nieminen, K.M. , Kinoshita, K. , Tormakangas, K. et al. (2006) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science, 311, 94–98. [DOI] [PubMed] [Google Scholar]
- Mahonen, A.P. , Bonke, M. , Kauppinen, L. , Riikonen, M. , Benfey, P.N. & Helariutta, Y. (2000) A novel two‐component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes & Development, 14, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant, A. , Kargul, J. , May, S.T. , Muller, P. , Delbarre, A. , Perrot‐Rechenmann, C. et al. (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. The EMBO Journal, 18, 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G. , Rovira, A. , Veciana, N. , Soy, J. , Toledo‐Ortiz, G. , Gommers, C.M.M. et al. (2018) Circadian waves of transcriptional repression shape PIF‐regulated photoperiod‐Responsive growth in Arabidopsis. Current Biology, 28(311–318), e315. [DOI] [PubMed] [Google Scholar]
- Martin, G. , Veciana, N. , Boix, M. , Rovira, A. , Henriques, R. & Monte, E. (2020) The photoperiodic response of hypocotyl elongation involves regulation of CDF1 and CDF5 activity. Physiologia Plantarum, 169, 480–490. [DOI] [PubMed] [Google Scholar]
- Maruyama‐Nakashita, A. , Watanabe‐Takahashi, A. , Inoue, E. , Yamaya, T. , Saito, K. & Takahashi, H. (2015) Sulfur‐Responsive elements in the 3 '‐nontranscribed intergenic region are essential for the induction of SULFATE TRANSPORTER 2;1 gene expression in Arabidopsis roots under sulfur deficiency. The Plant Cell, 27, 1279–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto‐Kitano, M. , Kusumoto, T. , Tarkowski, P. , Kinoshita‐Tsujimura, K. , Vaclavikova, K. , Miyawaki, K. et al. (2008) Cytokinins are central regulators of cambial activity. Proceedings of the National Academy of Sciences of the United States of America, 105, 20027–20031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J. , Ckurshumova, W. & Berleth, T. (2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiology, 131, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, C.J. , Valdes, A.E. , Wang, G.D. , Ramachandran, P. , Beste, L. , Uddenberg, D. et al. (2016) PHABULOSA mediates an auxin signaling loop to regulate vascular patterning in Arabidopsis. Plant Physiology, 170, 956–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley, R.C. , Huang, S.S.C. , Song, L. , Lewsey, M.G. , Bartlett, A. , Nery, J.R. et al. (2016) Cistrome and Epicistrome features shape the regulatory DNA landscape. Cell, 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D. (2003) Vascular patterning: more than just auxin? Current Biology, 13, R485–R487. [DOI] [PubMed] [Google Scholar]
- Rheaume, B.A. , Jereen, A. , Bolisetty, M. , Sajid, M.S. , Yang, Y. , Renna, K. et al. (2018) Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nature Communications, 9, 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, K.H. , Huang, L. , Kang, H.M. & Schiefelbein, J. (2019) Single‐cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiology, 179, 1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, T. (2000) Integrating cellular and organismic aspects of vascular differentiation. Plant and Cell Physiology, 41, 649–656. [DOI] [PubMed] [Google Scholar]
- Scarpella, E. , Marcos, D. , Friml, J. & Berleth, T. (2006) Control of leaf vascular patterning by polar auxin transport. Genes & Development, 20, 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth, A. , Moller, B. , Liu, W.L. , Kientz, M. , Flipse, J. , Rademacher, E.H. et al. (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature, 464, 913–U128. [DOI] [PubMed] [Google Scholar]
- Serrano‐Ron, L. , Perez‐Garcia, P. , Sanchez‐Corrionero, A. , Gude, I. , Cabrera, J. , Ip, P.L. et al. (2021) Reconstruction of lateral root formation through single‐cell RNA sequencing reveals order of tissue initiation. Molecular Plant, 14, 1362–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, M.E. , McGregor, S.R. , Sun, H. , Gough, C. , Bagman, A.M. , Soyars, C.L. et al. (2020) A PXY‐mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. The Plant Cell, 32, 319–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T. , Geldner, N. , Grebe, M. , Mangold, S. , Jackson, C.L. , Paris, S. et al. (1999) Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science, 286, 316–318. [DOI] [PubMed] [Google Scholar]
- Suer, S. , Agusti, J. , Sanchez, P. , Schwarz, M. & Greb, T. (2011) WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. The Plant Cell, 23, 3247–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Xu, D. , Liu, Z. , Kleine, T. & Leister, D. (2016) Functional relationship between mTERF4 and GUN1 in retrograde signaling. Journal of Experimental Botany, 67, 3909–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, V. & Pachter, L. (2018) RNA velocity: molecular kinetics from single‐cell RNA‐seq. Molecular Cell, 72, 7–9. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Cacchiarelli, D. , Grimsby, J. , Pokharel, P. , Li, S. , Morse, M. et al. (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology, 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera‐Sirera, F. , De Rybel, B. , Urbez, C. , Kouklas, E. , Pesquera, M. , Alvarez‐Mahecha, J.C. et al. (2015) A bHLH‐based feedback loop restricts vascular cell proliferation in plants. Developmental Cell, 35, 432–443. [DOI] [PubMed] [Google Scholar]
- Walerowski, P. , Gundel, A. , Yahaya, N. , Truman, W. , Sobczak, M. , Olszak, M. et al. (2018) Clubroot disease stimulates early steps of phloem differentiation and recruits SWEET sucrose transporters within developing galls. The Plant Cell, 30, 3058–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner, E.S. , Tonn, N. , Shi, D. , Jouannet, V. & Greb, T. (2020) SUPPRESSOR OF MAX2 1‐LIKE 5 promotes secondary phloem formation during radial stem growth. The Plant Journal, 102, 903–915. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Huan, Q. , Li, K. & Qian, W. (2021) Single‐cell transcriptome atlas of the leaf and root of rice seedlings. Journal of Genetics and Genomics, 48, 881–898. [DOI] [PubMed] [Google Scholar]
- Wendrich, J.R. , Yang, B. , Vandamme, N. , Verstaen, K. , Smet, W. , Van de Velde, C. et al. (2020) Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science, 370, eaay4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, K. , Kondo, Y. , Kojima, M. , Takebayashi, Y. , Sakakibara, H. & Fukuda, H. (2018) Suppression of DELLA signaling induces procambial cell formation in culture. The Plant Journal, 94, 48–59. [DOI] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. & Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols, 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang, T.Q. , Chen, Y. , Liu, Y. , Lin, W.H. & Wang, J.W. (2021) Single‐cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root. Nature Communications, 12, 2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T.Q. , Xu, Z.G. , Shang, G.D. & Wang, J.W. (2019) A single‐cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Molecular Plant, 12, 648–660. [DOI] [PubMed] [Google Scholar]
- Zhou, W. , Lozano‐Torres, J.L. , Blilou, I. , Zhang, X. , Zhai, Q. , Smant, G. et al. (2019) A Jasmonate signaling network activates root stem cells and promotes regeneration. Cell, 177(942–956), e914. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Zhou, B. , Pache, L. , Chang, M. , Khodabakhshi, A.H. , Tanaseichuk, O. et al. (2019) Metascape provides a biologist‐oriented resource for the analysis of systems‐level datasets. Nature Communications, 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zywitza, V. , Misios, A. , Bunatyan, L. , Willnow, T.E. & Rajewsky, N. (2018) Single‐cell transcriptomics characterizes cell types in the subventricular zone and uncovers molecular defects impairing adult neurogenesis. Cell Reports, 25(2457–2469), e2458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The experimental flow chart used in this study.
Figure S2. Quality control analysis of the raw data of scRNA‐seq.
Figure S3. UMAP analysis of the cells.
Figure S4. Analysis of the expression of known marker genes of different kinds of cell types on tSNE plot.
Figure S5. Analysis of the expression of known marker genes of different kinds of cell types on UMAP plot.
Figure S6. Analysis of the expression of representative vein marker genes.
Figure S7. NTR1.7, NEK6 and BZIP9 were expressed in vein.
Figure S8. DELLA proteins are required for the regulation of the patterns of vascular systems in cotyledons.
Figure S9. Vector information of gRNA CRISPR/Cas9 and double mutant verification.
Figure S10. Verification of the T‐DNA insertion of mutants.
Table S6. List of mutant lines used in this study
Table S7. List of oligonucleotides used in this study
Table S1. Differentially expressed genes in each of cell cluster of tSNE analysis
Table S2. Differentially expressed genes in each of cell cluster of UMAP analysis
Table S3. The marker genes of vascular tissues
Table S4. GO enrichment analysis of DEGs of each of cell cluster of tSNE analysis
Table S5.Transcriptional factor network analysis
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary data published online.


