Abstract
In this era of personalized medicine, targeted immunotherapies like immune checkpoint inhibitors (ICI) blocking the programmed death‐1 (PD‐1)/program death ligand‐1 (PD‐L1) axis have become an integral part of treating advanced stage non‐small cell lung carcinoma (NSCLC) and many other cancer types. Multiple monoclonal antibodies are available commercially to detect PD‐L1 expression in tumor cells by immunohistochemistry (IHC). As most clinical trials initially required tumor biopsy for PD‐L1 detection by IHC, many of the currently available PD‐1/PD‐L1 assays have been developed and validated on formalin fixed tissue specimens. The majority (>50%) of lung cancer cases do not have a surgical biopsy or resection specimen available for ancillary testing and instead must rely primarily on fine needle aspiration biopsy specimens for diagnosis, staging and ancillary tests. Review of the literature shows multiple studies exploring the feasibility of PD‐L1 IHC on cytological samples. In addition, there are studies addressing various aspects of IHC validation on cytology preparations including pre‐analytical (e.g., different fixatives), analytical (e.g., antibody clone, staining platforms, inter and intra‐observer agreement, cytology‐histology concordance) and post‐analytical (e.g., clinical outcome) issues. Although promising results in this field have emerged utilizing cytology samples, many important questions still need to be addressed. This review summarizes the literature of PD‐L1 IHC in lung cytology specimens and provides practical tips for optimizing analysis.
Keywords: cancer, concordance, cytology, FNA, immunohistochemistry, lung, PD1/PDL1
1. INTRODUCTION
Non‐small cell lung carcinoma (NSCLC) accounts for 80%–85% of cases and primarily comprises adenocarcinoma, squamous cell carcinoma and a not otherwise specified (NOS) category. 1 The overall 5‐year survival rate for NSCLC is only 17%. Current treatment options include surgical resection, neoadjuvant chemotherapy and targeted therapies. 1 , 2 , 3 Along with molecular prognostic biomarkers, programmed death‐1 (PD‐1)/program death ligand‐1 (PD‐L1) immunohistochemistry (IHC) has become an integral part of standard treatment regimens for NSCLC. T‐lymphocytes express PD‐1 and tumor cells express either PD‐L1 or PD‐L2. Binding of PD‐1 to PD‐L1 leads to increased apoptosis of activated tumor reactive T‐cells that, in turn, promotes growth of tumor cells by an immune escape mechanism. 4 , 5 Novel targeted immunotherapies block this pathway, thereby leading to tumor cell death. The use of such immune checkpoint inhibitors (ICIs) for NSCLC has emerged as one of the most promising cancer treatments.
PD‐L1 expression can be detected by IHC on tissue specimens or immunocytochemistry (ICC) on cytology samples, as well as by flow cytometry, fluorescent in‐situ hybridization (FISH) or molecular testing. Of these tests, IHC or ICC remain the most cost‐effective and most commonly used tests for the detection of PD‐L1 expression. Studies have shown better prognosis with anti‐PD‐1/PD‐L1 therapies for certain cancers compared to platinum‐based chemotherapies. 6 As most clinical trials initially required tumor biopsy for PD‐L1 detection by IHC, most of the currently available PD‐1/PD‐L1 assays have been developed and validated on formalin fixed tissue and not alcohol fixed cytology specimens. 7 , 8 , 9
The majority (>50%) of lung cancer cases do not have a surgical biopsy or resection specimen for ancillary testing and instead rely primarily on fine needle aspiration biopsy (FNAB) specimens for diagnosis, staging and ancillary tests. 10 Not surprisingly, a review of the literature shows multiple studies exploring the feasibility of PD‐L1 immunostaining on cytological samples. In addition, there are numerous cytology studies that address pre‐analytical (e.g., different fixatives), analytical (e.g., antibody clone, staining platforms, inter and intra‐observer agreement, cytology‐histology concordance) and post‐analytical (e.g., clinical outcome) issues. 11 , 12 , 13 , 14 , 15 , 16 Although promising results have emerged, many important questions about this field still need to be addressed.
This review summarizes the literature of PD‐L1 immunostaining in lung cytological specimens and provides practical tips for optimizing PD‐L1 analysis.
2. METHODS
A systematic data search was undertaken using PubMed and Embase electronic databases from January 2017 until April 2021. Key words including “cytology” and “PD‐L1” were used for this data search. We were interested in the current status of PD‐L1 testing in lung cytology specimens with respect to pre‐analytic, analytic and post‐analytic aspects of this test. Exclusion criteria included animal studies, PD‐L1 testing other than IHC/ICC such as flow cytometry or molecular methods, studies not dealing with lung carcinoma, or studies without any comparisons like cytology‐histology concordance, inter‐observer comparison or clone comparison. No language restrictions were applied. Different study designs, different antibody clones and staining platforms employed, as well as variations in study population precluded a formal meta‐analysis of these data.
3. RESULTS
After deleting duplicates, 4730 articles were screened. A total of 77 full‐text and 30 abstract articles matched study goals. Studies analyzing PD‐L1 testing on lung carcinoma cytology specimens originated mostly from the USA, followed by publications arising from Italy, Canada, United Kingdom, China, Germany, Japan, India, Turkey and other countries. The Dako clone 22C3 was the most common clone analyzed on cytology specimens, followed by the Ventana SP263 clone. 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 The data collected included the clone type used in each study. Pertinent details covering the technical aspects of the immunostain assay performed were also noted. An attempt was made to exclusively include only cases involving FNAB of lung NSCLC with a PD‐L1 test. Many studies included a subset of samples procured from metastatic disease involving the lymph nodes or effusion specimens. As noted, heterogeneity of the various study designs precluded meta‐analysis of the collected data.
4. PRE‐ANALYTICAL CONSIDERATIONS
4.1. Specimen collection
Newer generation biopsy needles and minimally invasive trans‐bronchial needle aspiration (TBNA) or endoscopic ultrasound‐guided fine needle aspiration biopsy (EBUS‐FNAB) procedures have revolutionized cytology sample procurement. These procedures have become the test of choice for the initial diagnosis, staging and tissue procurement for biomarker testing of lung lesions. Many studies have proven the feasibility of cytology specimens obtained via EBUS or TBNA for PD‐L1 testing. 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 Standard needle sizes used for these procedures range from 21 to 25 G. 48 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76
Sakakibara et al. 66 compared EBUS‐TBNA from lymph nodes to excised lymph nodes and also to the primary tumors. They demonstrated good concordance with an R value of 0.93 for TBNA and lymph node excisions and 0.75 for TBNA versus primary lung tumor resections, for clone AbCam EPR1161. A recent study by Perrotta et al. 22 studied the effect of assessment of PD‐L1 on TBNA samples that they also compared with other sampling methods such as percutaneous FNA, percutaneous core needle biopsy (CNB), thoracoscopy, excisions by using video‐assisted thoracoscopic surgery (VATS), or open thoracotomy. Needle sizes used in their study were 19G, 21G, 22G and 25G. Sample adequacy between these different methods did not show any statistically significant difference. Davidson et al. 69 undertook a prospective study using a 19G needle for lymph node aspirates and demonstrated that the majority (14/17) samples were adequate for the evaluation of PD‐L1. In this study, 42.9% of the samples demonstrated positive PD‐L1 expression. Similarly, Wahidi et al. 31 concluded that the utility of utilizing a 19G needle for PD‐L1 testing and molecular testing without the risk of any increase in adverse events.
There is no published evidence that needle size significantly affects sample adequacy for PD‐L1 testing. 31 , 62 Although, a recent study by Hardy et al. 76 showed that needle size can still affect adequate sample procurement. In this study, PD‐L1 testing failures occurred in 3/5 (60%) 22G needle biopsies, 1/5 (20%) in 21G needle biopsies, and 2/39 (5.1%) in 19G needle biopsies (with a p value of .016). These results are skewed due to the number of samples compared (five samples with 21–22 G vs. 39 samples with 19G needle).
4.2. Role of rapid onsite evaluation
ROSE by a trained cytotechnologist or cytopathologist increases the success rate of tissue procurement and allows for the appropriate triage of ancillary testing of all cancer types. Indeed, studies by Stevenson et al. 77 and Doxtader et al. 78 evaluating ROSE during EBUS procedures confirmed the utility of ROSE to increase the yield of aspirated sample for ancillary tests such as PD‐L1.
4.3. Test requisition form
The pathology laboratory should consider providing guidelines for ordering PD‐L1 testing. Test menus accordingly need to ideally incorporate educational material.
4.4. Type of cytological samples and fixatives:
The IASLC (International Association for the Study of Lung Cancer) 11 discusses the use of a variety of cytology sample types for PD‐L1 immunostaining. Of the different cytological preparations available, the cell‐block (CB) is the most common type of processed specimen material extensively studied for PD‐L1 ICC followed by other preparation types such as direct smears (unstained, air‐dried or alcohol fixed), cell‐transfer, cytospins and liquid‐based preparations. 79 , 80 , 81
Of these specimens, CBs are typically handled similar to formalin fixed, paraffin embedded (FFPE) tissues and thus their use is similar to that of FFPE material. However, one of the major pre‐analytical factors that can affect ICC performance of CBs is the variety of methods in which CBs are prepared and fixatives used prior to cell‐blocking that vary for each laboratory. Cell‐block method preparation was not consistently provided for appropriate analysis. Different fixatives used include alcohol, CytoLyt, CytoRich Red, MicroFix spray and formalin, and RPMI or alcohol followed by formalin. 82 , 83 , 84 , 85 According to some authors, alcohol‐based fixatives might compromise IHC staining. 82 , 83 , 84 However, several studies exploring the effect of different fixatives before cell‐blocking concluded that the type of fixative does not in fact affect PD‐L1 staining. This includes investigations by Wang et al. 35 about alcohol only, formalin only, and both fixatives, as well as the study by Gosney et al. 32 about alcohol‐based fixatives like CytoRich Red or CytoLyt and neutral buffered formalin, and the paper by Lou et al. 28 using CytoLyt. Of the direct smear studies, a study by Lozano et al. 39 demonstrated good concordance between PD‐L1 expression in FFPE tissue, FFPE CBs and alcohol fixed Papanicolaou stained direct smears. Similar results have been confirmed by other researchers. 46 , 49 , 60 Wang et al. 86 further showed that destained Papanicolaou smears are less sensitive than CBs in detecting PD‐L1 for clone SP263. Our literature review supports 10% buffered formalin as the fixative of choice for CBs for PD‐L1 ICC. Vigliar et al. 36 concluded that fixation time affects the performance of a LDT employing the 22C3 PD‐L1 assay, but did not affect the results of a commercially available SP263 assay. Hence, they advocate checking fixation times for LDTs.
4.5. Tissue block age
Ideal surgical pathology specimens used for PD‐L1 testing should not be older than 3 years according to a study conducted by Boothman et al. 87 Studies exploring this aspect on cytology material are lacking.
5. ANALYTICAL CONSIDERATIONS
5.1. Use of appropriate controls
Use of positive and negative controls remain one of the key elements in the analytical stage for optimal PD‐L1 test performance. Placenta and tonsil were the most common external positive controls used for PD‐L1 testing using cytology material. Some researchers used commercial cell‐lines as positive controls while developing protocols for PD‐L1 expression for cytological samples. 88
5.2. Sample adequacy
Multiple studies have assessed the adequacy rate of PD‐L1 evaluation on cytology samples with reported adequacy rates ranging between 50% and 96%. 89 , 90 , 91 , 92 Most studies used a cellularity of less than 100 cells as an exclusion criterion for PD‐L1 ICC. A minimum of 100 viable and well‐preserved cells are required to perform PD‐L1 IHC/ICC. Studies have shown that concordance rates vary in direct proportion to the cellularity of the cytology specimens. 61 In this study, the authors demonstrated that the concordance rate at a cut‐off of 50% was near perfect (>0.80) for cell count of 400 for clone 28–8 and cell count of 500 for clone SP142 compared to moderate concordance at cellularity between 100 and 500 cells.
5.3. Currently available PD‐1/PD‐L1 IHC/ICC assays for NSCLCs
Table 1 provides and overview of the different PD‐L1 clones commercially available for IHC and ICC testing. Due to the availability of these different PD‐L1 assays and different staining platforms, coupled with different cut‐off levels to determine the positivity of PD‐L1 expression, standardize PD‐L1 testing and reporting of results remains challenging.
TABLE 1.
Summary of available commercial monoclonal PD‐L1 antibodies
| Assay clone | Staining platform | Target drug name | Drug target | Cell type and location for assessment |
|---|---|---|---|---|
| 22C3 | Dako | Pembrolizumab | PD1 | Tumor cell membrane |
| 28–8 | Dako | Nivolumab | PD1 | Tumor cell membrane |
| SP263 | Ventana | Durvalumab | PD‐L1 | Tumor cell membrane |
| SP 142 | Ventana | Atezolizumab | PD‐L1 | Tumor cell and/or immune cell membrane |
Abbreviations: PD, programmed death; PD‐L1, programmed death ligand 1.
5.4. PD‐L1 immunocytochemistry interpretation
Table 2 summarizes of interpretation guidelines for cytology material. 11 , 93 , 94 , 95 , 96 , 97
TABLE 2.
PD‐L1 immunocytochemistry interpretation guideline
|
Sample adequacy a : At least 100 viable, well‐preserved, non‐overlapping tumor cells Positive stain result: Complete or partial membranous staining of tumor cells irrespective of the staining intensity Negative stain result: Exclusively cytoplasmic staining, granular cytoplasmic staining or nuclear staining of tumor cells Pitfalls to avoid: Avoid macrophages and inflammatory cells while scoring tumor cells by parallel examination of H&E stained glass or digital slides Cytology specimen considerations: 3‐D clusters (smears), more non‐specific cytoplasmic staining, cellular fragmentation, background cellular debris, inflammatory cell contamination from blood, lack of intact architecture (e.g., cannot count tumor infiltrating lymphocytes) Consideration of repeat testing: Too much background staining, weak staining of control How to improve interpretation: Consider providing training for PD‐L1 interpretation or participation in proficiency testing |
| Sample report format of PD‐L1 on lung cytology specimens: |
| The cytology report should include the following information: |
| PD‐L1 ICC parameters: |
| a. Cytology sample type |
| b. Clone |
| c. Staining platform utilized |
| d. Laboratory developed test (LTD): Yes or No |
| PD‐L1 ICC scoring: |
| Scoring system used |
| Exact Score: e.g., Tumor proportion score for Dako 22C3 clone |
Abbreviations: ICC, immunocytomistry; PD, programmed death; PD‐L1, programmed death ligand 1.
A disclaimer should be included if cell‐block cellularity has <100 cells and that repeat sampling may be considered if clinically indicated.
In brief, a literature review indicates that complete or partial membranous staining (Figure 1), irrespective of stain intensity, was considered positive in most studies. Exclusive cytoplasmic staining or granular cytoplasmic staining should be considered as negative. Similarly, nuclear staining should be considered as artifactual and interpreted as negative.
FIGURE 1.
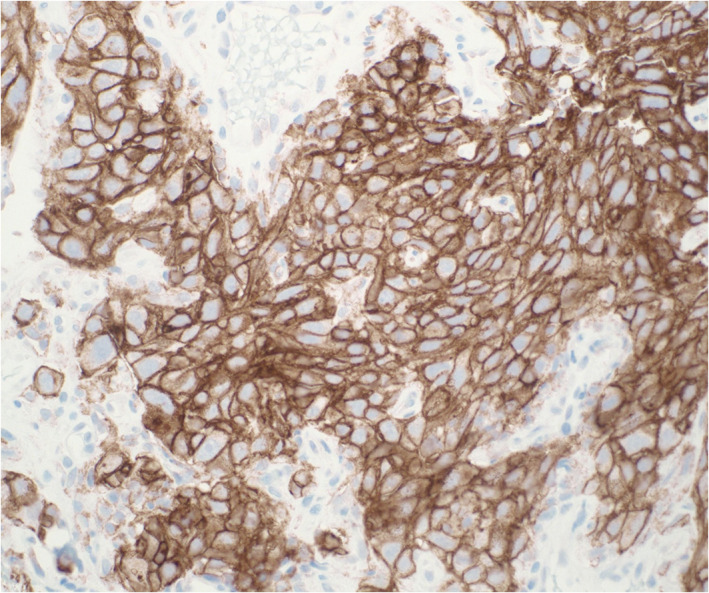
Program death ligand‐1 (PD‐L1) immunostain performed on a cell block section of a non‐small cell lung carcinoma showing diffuse circumferential expression (×200 magnification, clone 22C3, Dako) [Colour figure can be viewed at wileyonlinelibrary.com]
For most assays, only tumor cells are scored whereas for assay SP142 tumor cells and immune cells need to be scored. The SP142 clone is rarely used in cytology due to the inability to establish a true relationship between tumor cells and inflammatory cells in cytology material. The most common scoring system used in the literature includes a 3‐tiered scoring system based on proportion (number of PD‐L1 stained tumor cells divided by the total number of viable tumor cells, multiplied by 100) of tumor cells staining with negative cases (TPS = tumor proportion score < 1), low positive cases (TPS score ≥ 1–49) and high positive cases (TPS score ≥ 50). Caution needs to be taken to exclude inflammatory cells like macrophages while assessing tumor cells for accurate PD‐L1 scoring. Identifying macrophages can be a challenging task for cytology preparations due to the 3‐dimensional nature (applies to aspirate smears only) of cytology samples. 60 Gagne et al. 38 specifically analyzed the level of difficulty encountered while scoring PD‐L1 on cytology preparations. They divided the difficulty level into three categories based on the efforts and magnification needed to interpret PD‐L1 staining as positive or negative. They concluded that PD‐L1 scoring on cytology preparations is more cumbersome compared to surgical pathology preparations. Some of the reasons for increased difficulty in scoring PD‐L1 on cytology preparations included the presence of 3‐D clusters leading to overlapping cells on smears that creates difficulty in assessing complete membranous staining, difficulty in separating tumor cells from inflammatory cells (especially pleural effusion specimens), and more cytoplasmic staining or background staining. Similarly, inflammatory cell contamination from aspirated blood and loss of intact tissue architecture remains limiting factors for PD‐L1 evaluation in cytology samples. 13 Factors to consider for improving the interpretation of PD‐L1 in cytology material include comparing cellular material to the original H&E stained slide of the cell‐block or scanned digital images in parallel during evaluation. Gilani et al. 98 demonstrated that double staining (TTF1 and PD‐L1; p40 and PD‐L1) was very helpful, easy to perform, and more efficient in evaluating PD‐L1 in cytologic preparations. An important limitation to keep in mind while using double staining with TTF‐1 and PD‐L1 is that >30% of lung adenocarcinomas can be negative for TTF‐1. Although many authors 64 consider using similar cut‐offs for starting anti‐PD therapy in cytology material to those employed in surgical/excision specimens, this issue warrants further clinical trials exploring clinical outcomes based on PD‐L1 testing primarily performed on cytology specimens using similar versus modified cut‐offs to surgical specimens.
5.5. Validation and optimization of laboratory developed tests
The CAP provides guidelines for validation of immunohistochemical stains. 99 , 100 Appropriate validation of LTDs and optimization of protocols should be undertaken for achieving diagnostic accuracy comparable to the reference gold standard. The CAP recommends the use of at least 10 positive and 10 negative cases for initial validation and achieving a concordance rate of >90% between the new IHC and comparator IHC. Specific CAP guidelines specific to PD‐L1 IHC are in progress. Illie et al. 41 developed a 22C3 Antibody concentrate‐based LDT that showed a high concordance rate between tissue biopsy (approximately 100%) and cytology (nearly 95%) specimens when compared to PD‐L1 IHC expression determined using the PD‐L1 IHC 22C3 companion assay at both TPS cut points (≥1%, ≥50%). Skov et al. 21 explored the effect of different staining platforms (Autostainer and Omnis) for clone Dako 22C3. This demonstrated concordance of 0.99 between the different staining platforms used for clone 22C3.
5.6. Cytology‐histology correlation
Many investigators study design included matched or un‐matched cytology‐histology concordance in addition to other end points to explore the feasibility of minimally invasive, rapid cytology samples for PD‐L1 testing compared to recommended more invasive biopsy or resection specimens. 23 , 101 , 102 , 103 , 104 , 105 , 106 , 107 , 108 , 109 , 110 , 111 Concordance rates and k‐values were variably provided, including separate concordance for different cut‐offs and/or overall concordance. Sample size varied from 21 to 247 cases, and the concordance rates varied from 53 17 , 23 to almost 100% 46 (Table 3). Some of the reasons for variable concordance reported in the literature include intra‐tumoral heterogeneity of PD‐L1 expression, variable cellularity, and more 3‐dimensional cell clusters in cytology samples. A study conducted by Shen and Li 101 analyzed associations between different specimen types and histopathological characteristics and demonstrated significant heterogeneity not in different tumor subtypes, but between primary and metastatic sites and different sample types attributed to intratumoral heterogeneity. A review article by Gosney et al. 12 evaluated this concordance rate using nine studies, with a total 428 specimens, to demonstrate concordance rates of 88.3% and 89.7% for a TPS cut off of >1% and ≥50%. Overall, these studies confirm the feasibility of cytology specimens for reliable PD‐L1 evaluation. Other rare clones studied by Sakakibara and Russel‐Goldman showed similar results. 66 , 67
TABLE 3.
Summary of published studies assessing cytology‐histology concordance for PD‐L1 testing of non‐small cell lung carcinoma patients
| Reference | Number of specimens | Antibody clone | Cytology‐histology concordance rate (kappa) |
|---|---|---|---|
| Ambrosini et al. 17 | 26 | 22C3 | 53.8% (k = 0.31) |
| Koomen et al. 18 | 47 | 22C3, SP263 | 57% (k = 0.49; 67% (k 0.590 |
| Kuempers et al. 23 | 247 | 22C3 | 74.1% |
| Jug et al. 25 | 53 | 22C3 | 81.5% for adenocarcinoma, 76% for squamous cell carcinoma (k = 0.45) |
| Lou et al. 28 | 81 | 22C3 | 63% (k = 0.68) |
| Bortolloto et al. 29 | 20 | 22C3 | 90% |
| Wang et al. 35 | 34 | 22C3 | 91.2% (34 samples from different sites) and 100% (16 samples from same anatomic site) |
| Tsunoda et al. 37 | 30 | 22C3 | 86.7% |
| Lozano et al. 39 | 113 | 22C3 | 97.3% |
| Ilie et al. 41 | 70 | 22C3 | 97% |
| Xu et al. 42 | 52 | 22C3 | k = 0.54 (adenocarcinoma), k = 0.34 (squamous cell carcinoma) |
| Wang et al. 43 | 29 | 22C3 | Pearson correlation 0.925 |
| Noll et al. 46 | 28 smears and nine cell block | 22C3 | High |
| Arriola et al. 47 | 30 | 22C3 | 80% (smears), 94.4% (cell blocks) and 62% (cell transfer from Pap stained smears) |
| Biswas et al. 48 | 50 |
22C3 28–8 SP263 |
k = 0.554 k = 0.698 k = 0.908 |
| Capizzi et al. 49 | 21 | 22C3 | 93% |
| Chauhan et al. 51 | 40 | SP263 | 82% |
| Gagne et al. 52 | 46 | SP263 | k = 0.56–0.82 |
| Bozzetti et al. 53 | 52 | SP263 | 92.3% (k = 0.731) |
| Ricci et al. 54 | 150 | SP263 | k = 0.534 (cut off of 1%) and k = 0.740 (>50% cut off) |
| Pak, Roh | 58 | SP263 | 94.34 |
| Daverio et al. 56 | 138 | SP263 | k = 0.564 |
| Hendry et al. 57 | 58 | SP263 | k = 0.39 |
| Munari et al. 59 | 55 | SP263 | 90.6 (50% cut off), 81.1% (1% cut off) |
| Jain et al. 60 | 26 | SP263 | 88.4% |
| Dong et al. 61 | 112 | 28–8 | k = 0.377–0.686 |
| Skov 64 | 86 |
28–8 22C3 |
90% (50% cut off), 87% (1% cut off); 94% (50% cut off), 85% (1% cut off) |
Abbreviation: PD‐L1, programmed death ligand 1.
5.7. Intra‐ and inter‐observer reproducibility
Very few studies exclusively analyzed intra‐ and/or inter‐observer reproducibility on paired cytology‐histology specimens and reported good reproducibility (Table 4). Of these studies, Munari et al. 59 reported good inter‐observer concordance (90.5%) and excellent intra‐observer concordance (98.1%) for the SP263 clone. However, Keumpers et al. 23 reported higher inter‐observer variability in cytology samples compared to histology. Few studies reported better concordance with a cut‐off of TPS ≥50% compared to a cut‐off of 1%. Certain studies explored the impact of different assay types. 18 Reasons for the variable interpretation are discussed in detail in the analytic considerations, part 4 interpretation of PDL1 ICC of this manuscript.
TABLE 4.
Summary of published studies assessing inter‐observer agreement/concordance for PD‐L1 testing of non‐small cell lung carcinoma patients
| Reference | Preparation type | Number of specimens | Number of pathologists | Antibody clone | Interobserver concordance |
|---|---|---|---|---|---|
| Koomen et al. 18 | Cell block | 47 | 2 | 22C3 | High |
| Sinclair et al. 19 | Cell block | 86 | 5 | 22C3 | Fleiss' kappa (0.74–0.79) and Cohen's kappa (0.49–0.83 to 0.63–0.90) |
| Hernandez et al. 26 | Cell block | 54 |
3 (cytopathologists with added pulmonary pathology expertise) 4 (without pulmonary expertise) |
22C3 | 42.8% and 61.9% concordance for 21 samples by 7 observers and 3 observers with added pulmonary expertise |
| Veroceq et al. 27 | Cell block | NA | 2 | 22C3 | Discordance rate 16–17.5% |
| Lou et al. 28 | Cell block | 81 | 2 | 22C3 | 0.93–0.97 |
| Krovstov O et al. 30 | Cell block | 50 | 3 | 22C3 | Fliess's kappa 0.66 |
| Heyman et al. 50 | Cell block | Not provided | 22C3 | 93% | |
| Gagne et al. 52 | Cell block | 46 | 4 | SP263, 28–8 | Fliess's kappa 0.74 to 0.82 |
| Daverio et al. 56 | Cell block | 40 | 2 | SP263 | 0.450 |
| Munari et al. 59 | Cell block | 47 | 2 | SP263 | 90.5% concordance, k 0.69 |
| Tsao et al. 102 | Cell block | 22 | 24 | 22C3, 28–8, SP142, SP263, 73–10 |
ICC = 0.78–0.85 Fliess's kappa 0.6–0.85 |
| Russel‐Goldman et al. 67 | Cell block | 56 | 2 | E1L3N | ICC 0.96 |
Abbreviations: ICC, interclass correlation; PD‐L1, programmed death ligand 1.
5.8. Concordance among assays
Studies exploring the effect of different clones affecting PD‐L1 interpretation in cytology material are limited. Sapalidis et al., 34 Lozano et al. 39 and Ilie et al. 41 have all reported excellent concordance rates of 99 (Dako 22C3 and Biocare CAL10), 92.7 (22C3 and SP263) and 99 (22C3 antibody concentrate and Dako 22C3 clone), respectively. Gagne et al. 52 showed a k value of 0.59 (1% cut off) and 0.73 (50% cut off) for clones Ventana SP263 and Dako 28–8. Similar results were shown by Skov et al. 21 for clones Dako 22C3 and 28–8 with a correlation of 0.95. These findings are further supported by a Blueprint PD‐L1 IHC Comparability Project 102 in which 24 pulmonary pathologists from 15 countries scored 22 cytology specimens that showed an ICC of 0.78 (cytology glass slides) and 0.85 (cytology digital slides) for different IHC assays and an ICC of 0.89 (surgical glass slides) and 0.93 (surgical digital slides). More dedicated studies using cytology material are needed to address PD‐L1 assay interchangeability for clinical purposes.
5.9. Clinical correlates
Some studies compared PD‐L1 expression to clinical characteristics such as age, gender, smoking history, specimen anatomic site (primary versus metastatic), type of NSCLC, cancer stage at diagnosis, and molecular findings. They demonstrated variable associations in the different studies. A complete discussion is beyond the scope of this article. However, Wang et al. 112 study showed that NSCLC in higher stages is more likely to express PD‐L1, especially in metastases, for clone 22C3. Sakata et al. 44 compared the effect of neoadjuvant therapy versus no neoadjuvant therapy effect on PD‐L1 and demonstrated a concordance rate of 84% between those groups at a cut off of >50%.
5.10. Role of digital pathology
In recent years, we have witnessed the rise of digital pathology and an increasing adoption of this technology in medical centers. This has given researchers and industry the opportunity to explore different image analysis algorithms to address several issues related to manual quantitative PD‐L1 scoring. 19 , 113 However, only a few computer applications have been considered for PD‐L1 scoring in cytology until now, and we expect an increase in development of digital automated scoring in the near future, which may help standardize the interpretation of PDL1 in cytology.
6. POST‐ANALYTIC CONSIDERATIONS
6.1. Correlation with clinical outcomes
There are limited studies 114 , 115 , 116 , 117 analyzing the actual impact of PD‐L1 analysis on cytology specimens with specific cut‐offs with any particular immunotherapy drug and correlation with clinical outcomes. Studies by Torous et al. 45 and Stanowska et al. 117 compared clinical outcome based on cytology specimens and compared their findings to surgical specimens using similar cut offs. They found no significant difference in disease control rates between cytology and surgical specimens. Lozano et al. 116 also showed a variable response rate to immunotherapy based on cytology samples. Kovacevic et al. 115 in their study concluded that cytological samples from metastases have the poorest predictive value for assessment of immunotherapy treated patients. Additional studies exploring the effect of using cytology as surrogate material for clinical decision making to start anti‐PD‐L1 therapy are clearly needed.
6.2. External quality assurance for inter‐laboratory test performance
Validation of LDTs remain challenging for variety of PD‐L1 assays. External quality assurance for inter‐laboratory test performance may be considered if feasible. 118 , 119
7. CONCLUSION
Our literature review demonstrates the feasibility of PD‐L1 testing utilizing lung cancer cytology specimens. Published studies report a moderate degree of cytology‐histology concordance, as well as good concordance among different assays. This may likely be attributed to the fact most evaluations of PD‐L1 used cell block preparations. Nevertheless, cytological fixatives do not appear to compromise PD‐L1 staining which further supports the utility of employing minimally invasive, rapid, cost‐effective cytology specimen procurement for biomarker assessment. Reasons that account for variable concordance between cytology and surgical specimens include intra‐tumoral heterogeneity of PD‐L1 expression, cellularity, and more 3‐dimensional cell clusters in cytology samples. Unfortunately, different study designs precluded a robust comparison of results to perform a formal meta‐analysis and thereby draw stronger conclusions. One of the limitations of this review article is that some of the articles were only abstracts that were non‐peer reviewed. Further work still needs to be done to establish standard guidelines for PD‐L1 testing of cytology specimens that address pre‐analytical, analytical, and post‐analytical parameters.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
Concept and design: Liron Pantanowitz and Albino Eccher. Data collection and analysis: Swati Satturwar and Ilaria Girolami under supervision of Liron Pantanowitz. Writing first draft: Swati Satturwar. Manuscript review and editing: Ilaria Girolami, Enrico Munari, Francesco Ciompi, Albino Echher and Liron Pantanowitz.
PRACTITIONER POINTS
This review demonstrates the feasibility of PD‐L1 testing utilizing lung cancer cytology specimens with a moderate degree of cytology‐histology concordance and good concordance among different assays. Cytological fixatives do not appear to compromise PD‐L1 staining which further supports the utility of employing minimally invasive, rapid, cost‐effective cytology specimen procurement for biomarker assessment.
Satturwar S, Girolami I, Munari E, Ciompi F, Eccher A, Pantanowitz L. Program death ligand‐1 immunocytochemistry in lung cancer cytological samples: A systematic review. Diagnostic Cytopathology. 2022;50(6):313‐323. doi: 10.1002/dc.24955
DATA AVAILABILITY STATEMENT
Data will be available upon request to the corresponding author.
REFERENCES
- 1. Nicholson AG, Tsao MS, Beasley MB, et al. The 2021 WHO classification of lung tumors: impact of advances since 2015. J Thorac Oncol. 2022;17:362‐387. [DOI] [PubMed] [Google Scholar]
- 2. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553:446‐454. [DOI] [PubMed] [Google Scholar]
- 3. Patel SA, Weiss J. Advances in the treatment of non‐small cell lung cancer: immunotherapy. Clin Chest Med. 2020;41:237‐247. [DOI] [PubMed] [Google Scholar]
- 4. Han Y, Liu D, Li L. PD‐1/PD‐L1 pathway: current researches in cancer. Am J Cancer Res. 2020;10:727‐742. [PMC free article] [PubMed] [Google Scholar]
- 5. Velcheti V, Schalper K. Basic overview of current immunotherapy approaches in cancer. Am Soc Clin Oncol Educ Book. 2016;35:298‐308. [DOI] [PubMed] [Google Scholar]
- 6. Doroshow DB, Sanmamed MF, Hastings K, et al. Immunotherapy in non‐small cell lung cancer: facts and hopes. Clin Cancer Res. 2019;25:4592‐4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bodor JN, Boumber Y, Borghaei H. Biomarkers for immune checkpoint inhibition in non‐small cell lung cancer (NSCLC). Cancer. 2020;126:260‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callea M, Pedica F, Doglioni C. Programmed death 1 (PD‐L1) and it's ligand as a new frontier in cancer immunotherapy and challenges for the pathologists: state of the art. Pathologica. 2016;108:48‐58. [PubMed] [Google Scholar]
- 9. Yatabe Y, Kerr KM, Utomo A, et al. EGFR mutation testing practices within the Asia Pacific region: results of a multicenter diagnostic survey. J Thorac Oncol. 2015;10:438‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takigawa N, Ochi N, Yamane H. Histology versus cytology: PD‐L1 testing in non‐small cell lung cancer. Transl lung. Cancer Res. 2018;7(Suppl 3):S225‐S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lantuejoul S, Sound‐Tsao M, Cooper WA, et al. PD‐L1 testing for lung cancer in 2019: perspective from the IASLC pathology committee. J Thorac Oncol. 2020;15:499‐519. [DOI] [PubMed] [Google Scholar]
- 12. Gosney JR, Boothman AM, Ratcliffe M, Kerr KM. Cytology for PD‐L1 testing: a systematic review. Lung Cancer. 2020;141:101‐106. [DOI] [PubMed] [Google Scholar]
- 13. Clark DP. Biomarkers for immune checkpoint inhibitors: the importance of tumor topography and the challenges to cytopathology. Cancer Cytopathol. 2018;126:11‐19. [DOI] [PubMed] [Google Scholar]
- 14. Kim H, Chung JH. PD‐L1 testing in non‐small cell lung cancer: past, present, and future. J Pathol Transl Med. 2019;53:199‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang M, Wang S, Trapani JA, Neeson PJ. Challenges of PD‐L1 testing in non‐small cell lung cancer and beyond. J Thorac Dis. 2020;12:4541‐4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tejerina E, Garca Tobar L, Echeveste JI, de Andrea CE, Vigliar E, Lozano MD. PD‐L1 in cytological samples: a review and a practical approach. Front Med (Lausanne). 2021;8:668612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ambrosini‐Spaltro A, Dubini A, Pieri F, Ravaglia C, Delmonte A, Poletti V. PD‐L1 in NSCLC: role of cell‐blocks and concordance between samples. Diagn Cytopathol. 2021;49:303‐310. [DOI] [PubMed] [Google Scholar]
- 18. Koomen BM, van der Starre‐Gaal J, Vonk JM, et al. Formalin fixation for optimal concordance of programmed death‐ligand 1 immunostaining between cytologic and histologic specimens from patients with non‐small cell lung cancer. Cancer Cytopathol. 2021;129:304‐317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sinclair W, Kobalka P, Ren R, et al. Interobserver agreement in programmed cell death‐ligand 1 immunohistochemistry scoring in nonsmall cell lung carcinoma cytologic specimens. Diagn Cytopathol. 2021;49:219‐225. [DOI] [PubMed] [Google Scholar]
- 20. Casale S, Bortolotto C, Stella GM, et al. Recent advancement on PD‐L1 expression quantification: the radiologist perspective on CT‐guided FNAC. Diagn Interv Radiol. 2021;2:214‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skov BG. Comparison of PD‐L1 expression using 2 validated PD‐L1 IHC 22C3 pharmDx methods in non‐small cell lung cancer in a routine hospital setting. Appl Immunohistochem Mol Morphol. 2021;29:49‐55. [DOI] [PubMed] [Google Scholar]
- 22. Perrotta F, Nankivell M, Adizie JB, et al. Endobronchial ultrasound‐guided Transbronchial needle aspiration for PD‐L1 testing in non‐small cell lung cancer. Chest. 2020;158:1230‐1239. [DOI] [PubMed] [Google Scholar]
- 23. Kuempers C, van der Linde LIS, Reischl M, et al. Comparison of PD‐L1 expression between paired cytologic and histologic specimens from non‐small cell lung cancer patients. Virchows Arch. 2020;476:261‐271. [DOI] [PubMed] [Google Scholar]
- 24. Evans M, O'Sullivan B, Hughes F, et al. The clinicopathological and molecular associations of PD‐L1 expression in non‐small cell lung cancer: analysis of a series of 10,005 cases tested with the 22C3 assay. Pathol Oncol Res. 2020;26:79‐89. [DOI] [PubMed] [Google Scholar]
- 25. Jug R, Giovacchini CX, Liu B, et al. EBUS‐FNA cytologic‐histologic correlation of PD‐L1 immunohistochemistry in non‐small cell lung cancer. J Am Soc Cytopathol. 2020;9:485‐493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hernandez A, Brandler TC, Chen F, et al. Scoring of programmed death‐ligand 1 immunohistochemistry on cytology cell block specimens in non‐small cell lung carcinoma. Am J Clin Pathol. 2020;154:517‐524. [DOI] [PubMed] [Google Scholar]
- 27. Verocq C, Decaestecker C, Rocq L, et al. The daily practice reality of PD‐L1 (CD274) evaluation in non‐small cell lung cancer: a retrospective study. Oncol Lett. 2020;19:3400‐3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lou SK, Ko HM, Kinoshita T, et al. Implementation of PD‐L1 22C3 IHC pharmDxTM in cell block preparations of lung cancer: concordance with surgical resections and technical validation of CytoLyt® Prefixation. Acta Cytol. 2020;64:577‐587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bortolotto C, Maglia C, Ciuffreda A, et al. The growth of non‐solid neoplastic lung nodules is associated with low PD L1 expression, irrespective of sampling technique. J Transl Med. 2020;18:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kravtsov O, Hartley CP, Sheinin Y, et al. Utility of PD‐L1 testing on non‐small cell lung cancer cytology specimens: an institutional experience with interobserver variability analysis. Ann Diagn Pathol. 2020;48:151602. [DOI] [PubMed] [Google Scholar]
- 31. Wahidi MM, Davidson K, Shofer S, et al. Pilot study of the performance of 19‐G needle in endobronchial ultrasound‐guided transbronchial aspiration for the diagnosis and testing of molecular markers in lung cancer. J Bronchology Interv Pulmonol. 2021;28:209‐214. [DOI] [PubMed] [Google Scholar]
- 32. Gosney JR, Haragan A, Chadwick C, et al. Programmed death ligand 1 expression in EBUS aspirates of non‐small cell lung cancer: is interpretation affected by type of fixation? Cancer Cytopathol. 2020;128:100‐106. [DOI] [PubMed] [Google Scholar]
- 33. Smith A, Wang H, Zerbo A, et al. Programmed death ligand 1 testing of endobronchial ultrasound‐guided Transbronchial needle aspiration samples acquired for the diagnosis and staging of non‐small cell lung cancer. J Bronchology Interv Pulmonol. 2020;27:50‐57. [DOI] [PubMed] [Google Scholar]
- 34. Sapalidis K, Zarogoulidis P, Petridis D, et al. EBUS‐TNBA 22G samples: comparison of PD‐L1 expression between DAKO and BIOCARE®. J Cancer. 2019;10:4739‐4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang G, Ionescu DN, Lee CH, et al. PD‐L1 testing on the EBUS‐FNA cytology specimens of non‐small cell lung cancer. Lung Cancer. 2019;136:1‐5. [DOI] [PubMed] [Google Scholar]
- 36. Vigliar E, Malapelle U, Iaccarino A, et al. PD‐L1 expression on routine samples of non‐small cell lung cancer: results and critical issues from a 1‐year experience of a centralised laboratory. J Clin Pathol. 2019;72:412‐417. [DOI] [PubMed] [Google Scholar]
- 37. Tsunoda A, Morikawa K, Inoue T, et al. A prospective observational study to assess PD‐L1 expression in small biopsy samples for non‐small‐cell lung cancer. BMC Cancer. 2019;19:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gagné A, Wang E, Bastien N, et al. Impact of specimen characteristics on PD‐L1 testing in non‐small cell lung cancer: validation of the IASLC PD‐L1 testing recommendations. J Thorac Oncol. 2019;14:2062‐2070. [DOI] [PubMed] [Google Scholar]
- 39. Lozano MD, Abengozar‐Muela M, Echeveste JI, et al. Programmed death‐ligand 1 expression on direct pap‐stained cytology smears from non‐small cell lung cancer: comparison with cell blocks and surgical resection specimens. Cancer Cytopathol. 2019;127:470‐480. [DOI] [PubMed] [Google Scholar]
- 40. Mei P, Shilo K, Wei L, et al. Programmed cell death ligand 1 expression in cytologic and surgical non‐small cell lung carcinoma specimens from a single institution: association with clinicopathologic features and molecular alterations. Cancer Cytopathol. 2019;127:447‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ilie M, Juco J, Huang L, Hofman V, Khambata‐Ford S, Hofman P. Use of the 22C3 anti‐programmed death‐ligand 1 antibody to determine programmed death‐ligand 1 expression in cytology samples obtained from non‐small cell lung cancer patients. Cancer Cytopathol. 2018;126:264‐274. [DOI] [PubMed] [Google Scholar]
- 42. Xu H, Bratton L, Nead M, Russell D, Zhou Z. Comparison of programmed death‐ligand 1 (PD‐L1) immunostain for nonsmall cell lung carcinoma between paired cytological and surgical specimens. Cytojournal. 2018;15:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang H, Agulnik J, Kasymjanova G, et al. Cytology cell blocks are suitable for immunohistochemical testing for PD‐L1 in lung cancer. Ann Oncol. 2018;29:1417‐1422. [DOI] [PubMed] [Google Scholar]
- 44. Sakata KK, Midthun DE, Mullon JJ, et al. Comparison of programmed death Ligand‐1 Immunohistochemical staining between Endobronchial ultrasound Transbronchial needle aspiration and resected lung cancer specimens. Chest. 2018;154:827‐837. [DOI] [PubMed] [Google Scholar]
- 45. Torous VF, Rangachari D, Gallant BP, Shea M, Costa DB, VanderLaan PA. PD‐L1 testing using the clone 22C3 pharmDx kit for selection of patients with non‐small cell lung cancer to receive immune checkpoint inhibitor therapy: are cytology cell blocks a viable option? J Am Soc Cytopathol. 2018;7:133‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Noll B, Wang WL, Gong Y, et al. Programmed death ligand 1 testing in non‐small cell lung carcinoma cytology cell block and aspirate smear preparations. Cancer Cytopathol. 2018;126:342‐352. [DOI] [PubMed] [Google Scholar]
- 47. Arriola AGP, Bashover E, Joseph C, Staerkel G, Wang WL, Roy‐Chowdhuri S. The usefulness of various cytologic specimen preparations for PD‐L1 immunostaining in non‐small cell lung carcinoma. J Am Soc Cytopathol. 2018;7:324‐332. [DOI] [PubMed] [Google Scholar]
- 48. Biswas A, Leon ME, Drew P, et al. Clinical performance of endobronchial ultrasound‐guided transbronchial needle aspiration for assessing programmed death ligand‐1 expression in nonsmall cell lung cancer. Diagn Cytopathol. 2018;46:378‐383. [DOI] [PubMed] [Google Scholar]
- 49. Capizzi E, Ricci C, Giunchi F, et al. Validation of the immunohistochemical expression of programmed death ligand 1 (PD‐L1) on cytological smears in advanced non small cell lung cancer. Lung Cancer. 2018;126:9‐14. [DOI] [PubMed] [Google Scholar]
- 50. Heymann JJ, Bulman WA, Swinarski D, et al. PD‐L1 expression in non‐small cell lung carcinoma: comparison among cytology, small biopsy, and surgical resection specimens. Cancer Cytopathol. 2017;125:896‐907. [DOI] [PubMed] [Google Scholar]
- 51. Chauhan A, Siegel L, Freese R, Racila E, Stewart J 3rd, Amin K. Performance of Ventana SP263 PD‐L1 assay in endobronchial ultrasound guided‐fine‐needle aspiration derived non‐small‐cell lung carcinoma samples. Diagn Cytopathol. 2021;49:355‐362. [DOI] [PubMed] [Google Scholar]
- 52. Gagne A, Orain M, Ionescu D, Tsao MS, Joubert D, Joubert P. Comprehensive assessment of PD‐L1 immunohistochemistry on paired tissue and cytology specimens from non‐small cell lung cancer. Lung Cancer. 2020;146:276‐284. [DOI] [PubMed] [Google Scholar]
- 53. Bozzetti C, Squadrilli A, Nizzoli R, et al. Optimizing PD‐L1 evaluation on cytological samples from advanced non‐small‐cell lung cancer. Immunotherapy. 2020;12:183‐193. [DOI] [PubMed] [Google Scholar]
- 54. Ricci C, Capizzi E, Giunchi F, et al. Reliability of programmed death ligand 1 (PD‐L1) tumor proportion score (TPS) on cytological smears in advanced non‐small cell lung cancer: a prospective validation study. Ther Adv Med Oncol. 2020;12:1758835920954802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pak MG, Roh MS. Cell‐blocks are suitable material for programmed cell death ligand‐1 immunohistochemistry: comparison of cell‐blocks and matched surgical resection specimens in lung cancer. Cytopathology. 2019;30:578‐585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Daverio M, Patrucco F, Gavelli F, et al. Comparative analysis of programmed death ligand 1 expression in paired cytologic and histologic specimens of non‐small cell lung cancer. Cancer Cytopathol. 2020;128:580‐588. [DOI] [PubMed] [Google Scholar]
- 57. Hendry S, Byrne DJ, Christie M, et al. Adequate tumour cellularity is essential for accurate PD‐L1 immunohistochemistry assessment on cytology cell‐block specimens. Cytopathology. 2020;31:90‐95. [DOI] [PubMed] [Google Scholar]
- 58. KulaÇ İ, Aydin A, Bulutay P, Firat P. Efficiency of cytology samples for PD‐L1 evaluation and comparison with tissue samples. Turk Patoloji Derg. 2020;36:205‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Munari E, Zamboni G. Sighele expression of programmed cell death ligand 1 in non‐small cell lung cancer: comparison between cytologic smears, core biopsies, and whole sections using the SP263 assay. Cancer Cytopathol. 2019;127:52‐61. [DOI] [PubMed] [Google Scholar]
- 60. Jain D, Sukumar S, Mohan A, Iyer VK. Programmed death‐ligand‐1 immunoexpression in matched biopsy and liquid‐based cytology samples of advanced stage non‐small cell lung carcinomas. Cytopathology. 2018;29:550‐557. [DOI] [PubMed] [Google Scholar]
- 61. Dong Z, Liu Y, Jiang T, et al. Cell block as a surrogate for programmed death‐ligand 1 staining testing in patients of non‐small cell lung cancer. J Cancer. 2020;11:551‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stoy SP, Rosen L, Mueller J, Murgu S. Programmed death‐ligand 1 testing of lung cancer cytology specimens obtained with bronchoscopy. Cancer Cytopathol. 2018;126:122‐128. [DOI] [PubMed] [Google Scholar]
- 63. Young S, Griego‐Fullbright C, Wagner A, Chargin A, Patterson BK, Chabot‐Richards D. Concordance of PD‐L1 expression detection in non‐small cell lung cancer (NSCLC) tissue biopsy specimens between OncoTect iO lung assay and immunohistochemistry (IHC). Am J Clin Pathol. 2018;150:346‐352. [DOI] [PubMed] [Google Scholar]
- 64. Skov BG, Skov T. Paired comparison of PD‐L1 expression on cytologic and histologic specimens from malignancies in the lung assessed with PD‐L1 IHC 28‐8pharmDx and PD‐L1 IHC 22C3pharmDx. Appl Immunohistochem Mol Morphol. 2017;25:453‐459. [DOI] [PubMed] [Google Scholar]
- 65. Bubendorf L, Conde E, Cappuzzo F, et al. A noninterventional, multinational study to assess PD‐L1 expression in cytological and histological lung cancer specimens. Cancer Cytopathol. 2020;128:928‐938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sakakibara R, Inamura K, Tambo Y, et al. EBUS‐TBNA as a promising method for the evaluation of tumor PD‐L1 expression in lung cancer. Clin Lung Cancer. 2017;18:527‐534. [DOI] [PubMed] [Google Scholar]
- 67. Russell‐Goldman E, Kravets S, Dahlberg SE, Sholl LM, Vivero M. Cytologic‐histologic correlation of programmed death‐ligand 1 immunohistochemistry in lung carcinomas. Cancer Cytopathol. 2018;126:253‐263. [DOI] [PubMed] [Google Scholar]
- 68. Stoy S, Rosen L, Murgu S. The use of endobronchial ultrasound‐guided transbronchial needle aspiration cytology specimens for programmed death ligand 1 immunohistochemistry testing in non‐small cell lung cancer. J Bronchol Interv Pulmonol. 2017;24:181‐183. [DOI] [PubMed] [Google Scholar]
- 69. Davidson KR, Cheng GZ, Mahmood K, et al. Prospective pilot study of endobronchial ultrasound trasnbronchial needle aspiration with 19‐gauge needle to detect PD‐L1 in lung cancer. Am J Respir Crit Care Med. 2019;199:9.30130137 [Google Scholar]
- 70. Um SW, Kim HK, Jung SH, et al. Endobronchial ultrasound versus mediastinoscopy for mediastinal nodal staging of non‐small‐cell lung cancer. J Thorac Oncol. 2015;10:331‐337. [DOI] [PubMed] [Google Scholar]
- 71. Navani N, Nankivell M, Lawrence DR, et al. Lung cancer diagnosis and staging with endobronchial ultrasound‐guided transbronchial needle aspiration compared with conventional approaches: an open label, pragmatic, randomised controlled trial. Lancet Respir Med. 2015;3:282‐289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hameed F, Moore A, Sykes A, Wrightson J. The utility of Endobronchial ultrasound‐transbronchial needle aspiration (EBUS‐TBNA) in programmed death Ligand‐1 (PD‐L1) analysis of non‐small cell lung cancer Eur. Respir J. 2019;54. [Google Scholar]
- 73. Lee JM, Kim JS, Heymann JJ, et al. Feasibilty of PD‐L1 expression in non‐small cell lung cancer from EBUS‐TBNA samples. Am J Respir Crit Care Med. 2017;195. [Google Scholar]
- 74. Martin‐Deleon R, Teixido C, Reyes R, et al. Feasibility of EBUS‐TBNA cytologies or an extensive assessment of predictive biomarkers in lung cancer. J Thorac Oncol. 2019;14(10):S927‐S928. [Google Scholar]
- 75. Somalaraju S, Naqvi S, Sood R, et al. Adequacy of endobronchial ultrasound (EBUS) for PD‐L1 expression testing. Am J Respir Crit Care Med. 2019;199:9.30130137 [Google Scholar]
- 76. Hardy J, Bhatt N, Medford ARL. Suitability of endobronchial ultrasound‐guided transbronchial needle aspiration samples for programmed death ligand‐1 testing in non‐small cell lung cancer, the Bristol experience. Asia Pac J Clin Oncol. Published online April 18, 2021. doi: 10.1111/ajco.13549 [DOI] [PubMed] [Google Scholar]
- 77. Stevenson T, Powari M, Bowles C. Evolution of a rapid onsite evaluation (ROSE) service for endobronchial ultrasound guided (EBUS) fine needle aspiration (FNA) cytology in a UKHospital: a 7‐year audit. DiagnCytopathol. 2018;46:656‐662. [DOI] [PubMed] [Google Scholar]
- 78. Doxtader E, Yachimiak T, Mukhopadhyay S, et al. Rapid on‐site evaluation (ROSE) of endobronchial ultrasound‐guided fine‐needle aspiration (EBUS‐TBNA) optimizes tissue for evaluation of PD‐L1 expression on formalin fixed non‐small cell lung carcinoma. J Am Soc Cytopathol. 2017;6(5):PPS45. [Google Scholar]
- 79. Escario MDL, Abengozar M, Labiano T, et al. PD‐L1 expression in cytological stained smears using two commercially available assays: comparison with cell blocks and resection specimens. Lab Invest. 2018;98:PP158. [Google Scholar]
- 80. Alruwaii F, Idrees M. Programmed death‐ligand 1 (PD‐L1) expression analysis in cytology specimens using 22C3 clone for targeted therapy‐a validation study. J Am Soc Cytopathol. 2017;6(5):S44. [Google Scholar]
- 81. Kovacevic M, Kern I. PD‐L1 expression in different samples of non‐small cell lung cancer. Virchows Arch. 2017;471(1):S104. [Google Scholar]
- 82. Hart NA T, van der Starre‐Gaal J, Vonk JM, Timens W. Essential pre‐analytics in PD‐L1 immunocytochemistry. Histopathology. 2019;74:362‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lloyd IE, Zhou W, Witt BL, Chadwick BE. Characterization of PD‐L1 Immunohistochemical expression in cell blocks with different specimen fixation and processing methods. Appl Immunohistochem Mol Morphol. 2019;27:107‐113. [DOI] [PubMed] [Google Scholar]
- 84. Gruchy JR, Barnes PJ, Dakin Haché KA. CytoLyt® fixation and decalcification pretreatments alter antigenicity in normal tissues compared with standard formalin fixation. Appl Immunohistochem Mol Morphol. 2015;23:297‐302. [DOI] [PubMed] [Google Scholar]
- 85. Zheng X, Bell H, Donovan M, et al. Performance of PD‐L1 (22C3 clone) antibody in cytological preparations using different collection media in non‐small cell lung carcinoma and comparison with staining in paired histology sections. J Am Soc Cytopathol. 2017;6(5):S81. [Google Scholar]
- 86. Wang H, Liu L, Rabie L, et al. Application of PD‐L1 antibody clone sp263 on paired fine needle aspirations, cell blocks and surgical specimens of non‐small cell lung cancer. Lab Invest. 2018;98:PP182. [Google Scholar]
- 87. Boothman AM, Scott M, Ratcliffe M, et al. Impact of patient characteristics, prior therapy, and sample type on tumor cell programmed cell death ligand 1 expression in patients with advanced NSCLC screened for the ATLANTIC study. J Thorac Oncol. 2019;14:1390‐1399. [DOI] [PubMed] [Google Scholar]
- 88. Boyiddle C, Ruboyianes M, Valle D, et al. Development of IHC staining protocols for assessment of PD‐L1 expression in cytological samples. Cancer Res. 2017;V77:13. [Google Scholar]
- 89. Labiano T, Almudevar E, Lozano MD, et al. Feasibility of cytological samples for PD‐L1 assessment in lung cancer. Cytopathology. 2018;29:10‐11.28913874 [Google Scholar]
- 90. Behtaj M, Rai H, Awadallah A, et al. Programmed death lignad‐1 (PDL1) testing in advanced non‐small cell lung carcinoma (NSCLC). Comparing adequacy of immunohistochemical results between cytology and surgical specimens. Mod Pathol. 2019;32:3. [Google Scholar]
- 91. Torous V, Rangachari D, Costa D, et al. PD‐L1 immunohistochemical testing for lung cancer: cytology and surgical pathology specimens demonstrate similar overall expression patterns. J Am Soc Pathol. 2017;6(5):S43‐S44. [Google Scholar]
- 92. Agulnik J, Kasymjanova G, Wang H, et al. EBUS‐TBNA in assessing PD‐L1 expression in NSCLC. Thorac Oncol. 2018;13(10):S387. [Google Scholar]
- 93. Schidhaus HU. Predictive value of PD‐L1 diagnostics. Pathologe. 2018;39(6):498‐519. [DOI] [PubMed] [Google Scholar]
- 94. Kovacevic M, Kern I, Gabric S. PD‐L1 in NSCLC cytology. Ann Oncol. 2017;28:iii1‐iii2. [Google Scholar]
- 95. Lantuejoul S, Adam J, Girard N, et al. PD‐L1 testing in non‐small cell lung carcinona: guidelines from the PATTERN group of thoracic pathologists. Ann Pathol. 2018;38(2):110‐125. [DOI] [PubMed] [Google Scholar]
- 96. Elshiekh M, Iles S, Hopcroft D, et al. Lung cancer and immunotherapy: how good are we in selecting the right patients to treat? An audit on PD‐L1 immunohistochemistry scoring sytem on cytology samples. J Pathol. 2019;248:S7. [Google Scholar]
- 97. He X, Reynolds J. Programmed death ligand1(PD‐L1) testing on cytology specimens in non‐small cell lung carcinoma: in‐house versus send‐out. J Am Soc Cytopathol. 2020;9(6):S36. [Google Scholar]
- 98. Gilani S, Zomorrodian S, Farooq T, et al. Using double staining immunohistochemistry to accurately evaluate lung tumor PD‐L1 expression in cytology specimens. Lab Invest. 2018;98:144. [Google Scholar]
- 99. Fitzgibbons PL, Bradley LA, Fatheree LA, et al. Principles of analytic validation of immunohistochemical assays: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2014;138:1432. [DOI] [PubMed] [Google Scholar]
- 100. Satturwar S, Malenie R, Sutton A, Dai D, Aly FZ. Validation of immunohistochemical tests performed on cytology cell block material: practical application of the College of American Pathologists' guidelines. Cyto J. 2019;16(6). doi: 10.4103/cytojournal.cytojournal_29_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shen X, Li Y. Heterogeneity of PD‐L1 expression in non‐small cell lung cancer. Mod Pathol. 2020;33(3):1817‐1818. [Google Scholar]
- 102. Tsao MS, Kerr KM, Kockx M, et al. PD‐L1 immunohistochemistry comparability study in real‐life clinical samples: results of blueprint phase 2 project. J Thorac Oncol. 2018;13:1302‐1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ellwood T, Cooper W. Comparative study of PD‐L1 expression in paired fine‐needle aspiration and core biopsy specimens of non‐small cell lung cancer. Patholo J. 2019;51:S76‐S77. [Google Scholar]
- 104. Zhou C, Ionescu D, Hiruki T. PD‐L1 testing on the EBUS‐FNA cytology specimen has results highly concordant to those of surgical specimen. Lab Invesxt. 2018;98:187‐188. [Google Scholar]
- 105. Beech C, Rimm DL, Neumeister V, Cai G. Assessment of PD‐L1 status by immunohistochemistry in cytological samples of non‐small cell lung cancers: correlation with the results of concurrent surgical specimens. Lab Invesr. 2017;97:88A. [Google Scholar]
- 106. Chen Y, Shen X, Wang Y, et al. Comparisons of PD‐L1 expression between paired cytologic and histologic specimens obtained by endobronchial ultrasound‐guided transbronchial needle aspiration in non‐small cell lung cancer patients. Mod Pathol. 2020;33(3):338‐339. [Google Scholar]
- 107. Doxtader E, Mukhopadhyay S, Brainard J, et al. Evaluation of PD‐L1 expression of non‐small cell lung carcinoma on non‐formalin‐fixed cell blocks with comparison to paired formalin‐fixed surgical pathology specimens. Lab Invest. 2019;98:139‐140. [Google Scholar]
- 108. Frigola G, Vega N, Gonzalez‐Carreras A, et al. Validation of PD‐L1 PD‐L1 performance in cytological samples of non‐small cell lung carcinoma obtained from early‐stage resection specimens. Mod Pathol. 2020;33:344‐345.31477813 [Google Scholar]
- 109. Santana B, Hernández‐Bonilla S, Garcia R, et al. PD‐L1 expression in 1051 non‐small cell lung cancer samples of a tertiary hospital in 2017–2018 and concordance in paired samples. ECP. 2019; PS‐13:006. [Google Scholar]
- 110. Wagner CA, Christie M, Irving L, Steinfort D. Accuracy of PD‐L1 tumor staining of cytological and histological samples of lung adenocarcinoma. Eur Respir J. 2018;52. [Google Scholar]
- 111. Bratton L, Russel D, Yong Q, et al. Comparison of PD‐L1 immunostaining for non‐small cell carcinoma of the lung between paired cytological and surgical specimens. J Am Soc Cytopathol. 2016;5(8):S48. [Google Scholar]
- 112. Wang H, Agulnik J, Kasymjanova G, et al. PD‐L1 expression is related to tumor staging in NSCLC. Lab Invest. 2018;98:755.29483622 [Google Scholar]
- 113. Inge LJ, Dennis E. Development and applications of computer image analysis algorithms for scoring of PD‐L1 immunohistochemistry. Immuno‐Oncol Technol. 2020;6:2‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Jackson J, Valentine E, Toy E, et al. PD‐L1 expression in histological and cytological specimens and outcomes following immunotherapy in non‐small cell lung cancer. J Pathol. 2019;248:S11. [Google Scholar]
- 115. Kovacevic M, Ivanovic M, Kern I, Cufer T. PD‐L1 testing and clinical benefit in patients treated with CPI. J Thorac Oncol. 2019;14:S781‐S782. [Google Scholar]
- 116. Lozano M, Tobar LG, Abengozar M, et al. Feasibility of PD‐L1 expression in cytological stained smears: comparison with cell‐blocks and relationship with the outcomes of NSCLC patients treated with check‐point inhibitors. Mod Pathol. 2020;33(3):392. [Google Scholar]
- 117. Stanowska O, Wisniewski P, Knetki‐Wroblewska M, et al. PDL1 (22C3) expression in different samples of non‐small cell lung cancer (NSCLC) in correlation with response to pembrolizumab treatment a single institute experience. Virchows Arch. 2018;473:S116. [Google Scholar]
- 118. Torlakovic E, Albadine R, Bigras G, et al. Canadian multicenter project on standardization of programmed death‐ligand 1 immunohistochemistry 22C3 laboratory‐developed tests for pembrolizumab therapy in NSCLC. J Thorac Oncol. 2020;15:1328‐1337. [DOI] [PubMed] [Google Scholar]
- 119. Dodson A, Parry S, Lissenberg‐Witte B, et al. External quality assessment demonstrates that PD‐L1 22C3 and SP263 assays are systematically different. J Pathol Clin Res. 2020;6:138‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available upon request to the corresponding author.


