Abstract
Crop wild relatives, the closely related species of crops, may harbour potentially important sources of new allelic diversity for (a)biotic tolerance or resistance. However, to date, wild diversity is only poorly characterized and evaluated. Banana has a large wild diversity but only a narrow proportion is currently used in breeding programmes. The main objective of this study was to evaluate genotype‐dependent transpiration responses in relation to the environment. By applying continuous high‐throughput phenotyping, we were able to construct genotype‐specific transpiration response models in relation to light, VPD and soil water potential. We characterized and evaluated six (sub)species and discerned four phenotypic clusters. Significant differences were observed in leaf area, cumulative transpiration and transpiration efficiency. We confirmed a general stomatal‐driven ‘isohydric’ drought avoidance behaviour, but discovered genotypic differences in the onset and intensity of stomatal closure. We pinpointed crucial genotype‐specific soil water potentials when drought avoidance mechanisms were initiated and when stress kicked in. Differences between (sub)species were dependent on environmental conditions, illustrating the need for high‐throughput dynamic phenotyping, modelling and validation. We conclude that the banana wild relatives contain useful drought tolerance traits, emphasising the importance of their conservation and potential for use in breeding programmes.
Keywords: drought, stomata
1. INTRODUCTION
Human selection of plants has mainly focused on improving yields, harvest efficiency and edibility. Such a selection has created genetic bottlenecks as only a limited number of progenitors with beneficial traits were selected (Zhang et al., 2017). Crop wild relatives, the closely related species of cultivated crops, have been omitted in the past because of their poor agronomic traits, but are receiving increasing attention as they contain naturally acquired tolerances and resistances to biotic and abiotic stresses (Bohra et al., 2021; Dempewolf et al., 2017; King et al., 2017; Prohens et al., 2017). Diversity of crop wild relatives was recently screened for drought tolerance in alfalfa (Humphries et al., 2021), wheat (Aberkane et al., 2021; Reynolds et al., 2007), barley (Honsdorf et al., 2014; Naz et al., 2014), sorghum (Cowan et al., 2020; Ochieng et al., 2021) and sunflower (Seiler et al., 2017) amongst many others. However, most crop wild relatives remain poorly characterized and are hardly phenotyped (Castañeda‐Álvarez et al., 2016).
Banana (Musa spp.) production ranks among the top 15 crops worldwide, with annual production of over 158 million tons (FAO, 2019). However, all bananas exported today are still selections from somatic mutants of the Cavendish group and have thereby a very narrow genetic base (Ortiz & Vuylsteke, 1996). In contrast, there is ample diversity in the crop wild relatives of bananas (Mertens, Bawin, et al., 2021), which need to be conserved and characterized (Castañeda‐Álvarez et al., 2016; Eyland et al., 2020; Mertens, Swennen, et al., 2021). Within the Musa acuminata subgroup, 10 different subspecies are commonly described (Christelová et al., 2017). M. acuminata ssp. burmannica/siamea/burmannicoides, ssp. malaccensis, ssp. zebrina and ssp. banksii were suggested to be the main contributors to the A genome in cultivars and parthenocarpy has been linked to M. acuminata ssp. banksii and M. acuminata ssp. errans (Carreel et al., 2002; Martin et al., 2020; Perrier et al., 2011). The most common wild progenitor used in banana breeding is ‘Calcutta 4’ (M. acuminata ssp. burmannicoides) because of its resistance to the black Sigatoka fungal leaf disease and Fusarium wilt race 1 fungal root disease (Gonçalves et al., 2019; Ortiz & Swennen, 2014).
Banana production is sensitive to drought, with estimated yield reductions of 8% per 100 mm shortage in water (van Asten et al., 2011). In its most common agro‐ecological zones, banana requires between 1200 and 2600 mm year−1 evenly distributed throughout the year (Hegde & Srinivas, 1989; Machovina & Feeley, 2013; van Asten et al., 2011). In commercial plantations, the water requirements are met by irrigation (Carr, 2009; Ekanayake et al, 1994), but these plantations only represent 15% of the global production whereas farmer's production is typically rainfed. With climate change, precipitation in banana‐growing regions is expected to shift or decrease, while temperatures and consequently air vapour pressure deficits (VPDs) will continue to increase, thereby causing a major risk for banana production (Calberto et al., 2015; Machovina & Feeley, 2013; Rippke et al., 2016; Varma & Bebber, 2019). Improving banana performance for future climate conditions through breeding is therefore essential (Brown et al., 2020). Wild bananas, with naturally acquired abiotic tolerance, are therefore key (Dempewolf et al., 2017; Hajjar & Hodgkin, 2007). However, until now the diversity in drought tolerance traits within the wild banana populations remains uncharacterized.
The main objective of this study was to evaluate the transpiration responses to soil water deficit within wild banana relatives. Measuring the plant water status in banana is however complex. Typical thermodynamic measurements of leaf water potential are impossible because of its peculiar latex vessel structure (Milburn et al., 1990; Skutch, 1932; Turner et al., 2007). Stomatal conductance or transpiration measurements are described as the most sensitive descriptors (Carr, 2009; Turner & Thomas, 1998). By high‐throughput continuous phenotyping under fluctuating light, air VPD and soil water content we built genotype‐specific transpiration response models and discerned genotype‐specific behaviours. To our knowledge, we present here the first evaluation of the impact of water deficit on wild banana species as a function of the environmental conditions, thereby pinpointing crucial events when drought avoidance mechanisms are initiated and stress is experienced. This knowledge will stimulate the incorporation of crop wild relatives in banana drought breeding programmes.
2. MATERIALS AND METHODS
Two experiments were performed. One high‐throughput phenotyping experiment in the Phenodyn‐PhenoArch platform hosted at M3P, Montpellier Plant Phenotyping Platforms (https://www6.montpellier.inra.fr/lepse/M3P), INRAE. A second validation experiment at the Bioversity phenotyping platform hosted at KU Leuven, confirming three contrasting phenotypes and coupling transpiration measurements to leaf temperature and pressure probe dynamics.
2.1. High‐throughput phenotyping at the Phenodyn platform
2.1.1. Plant material and experiment
A diversity panel of eight wild banana genotypes belonging to six (sub)species (Table 1) were phenotyped for 5 weeks (October–November) at the Phenodyn platform (Sadok et al., 2007). These (sub)species comprise most of the known ancestors of the cultivated banana species (Carreel et al., 2002; Martin et al., 2020; Perrier et al., 2011; Sardos et al., 2021).
Table 1.
Wild banana genotypes screened during the high‐throughput phenotyping experiment at the Phenodyn platform
| Name | Subspecies | ITC codea | Originb | Collection sitec | Collection coordinatesc |
|---|---|---|---|---|---|
| Balbisiana | Musa balbisiana | / | South China, northern Indo‐Burma, Southwest India, Sri Lanka, Philippines and New Guinea | Japan (Amami) | / |
| Banksii_11 | M. acuminata ssp. banksii | / | New Guinea | Papua New Guinea (Madang) | 5° 37′8″S |
| 145° 28′7″E | |||||
| Banksii_17 | M. acuminata ssp. banksii | / | New Guinea | Papua New Guinea (Morobe) | 6° 44′42″S |
| 146° 43′51″E | |||||
| Burmannicoides | M. acuminata ssp. burmannicoides | ITC0249 | Southern Indo‐Burma | / | / |
| Errans | M. acuminata ssp. errans | ITC1028 | Philippines | / | / |
| Malaccensis_33 | M. acuminata ssp. malaccensis | / | Sumatra and Malayan Peninsula | Malaysia (Pahang) | 3° 53′51″N |
| 102° 12′23″E | |||||
| Malaccensis_ITC0074 | M. acuminata ssp. malaccensis | ITC0074 | Sumatra and Malayan Peninsula | / | / |
| Microcarpa | M. acuminata ssp. microcarpa | ITC0253 | Borneo | / | / |
Genotypes without ITC code were collected germplasm not yet available at the International Transit Centre (ITC) collection.
Genotype origin as described by Janssens et al. (2016).
Only location of collected samples is shown.
In vitro banana plants were obtained from the International Transit Center (ITC, Bioversity International, hosted at KU Leuven). Seeds collected in Papua New Guinea, Malaysia and Japan were obtained from the National Agricultural Research Institute of Papua New Guinea (NARI), the Kew Millennium Seed Bank and the Amami Biodiversity Gardens, conditioned by the Standard Material Transfer Agreement (SMTA) of the International Treaty on Plant Genetic Resources for Food and Agriculture. Sixteen replicates per genotype were screened. In vitro plants were acclimated for 6 weeks before starting the experiment. Plants were grown in white 9 L pots filled with a well‐mixed 30:70 (v/v) mixture of fine clay and organic compost. Half of the plants received a water deficit treatment, whereas the other half was maintained under well‐watered conditions. Water deficit plants received no irrigation, while well‐watered plants were irrigated in pulses at night till a specified target weight was reached. Genotypes and treatments were arranged in a randomized complete block design. Air temperature, relative humidity, and light intensity were measured every 5 min at nine points in the greenhouse. The VPD was calculated based on the air temperature and relative humidity. The photoperiod was set to 14 h. Additional light (200 μmol m−2 s−1) was provided when external solar radiation was below 300 W m−2 by using 400 W HPS Plantastar lamps. Soil water potential curves were constructed with soil samples of 100 cm3 following the pressure plate method (Richards, 1948) and fitted according to the van Genuchten (1980) equation.
2.1.2. Image acquisition and analysis
Plants were imaged biweekly in the Phenoarch phenotyping platform (Cabrera‐Bosquet et al., 2016). The daily plant weight and leaf area were estimated from 13 red–green–blue images (12 side views from 30° rotational difference and one top view) (Cabrera‐Bosquet et al., 2016). Top and side views were calibrated using reference objects to convert pixels to mm2.
Calibration curves were constructed by linking image‐based features with destructive ground‐truth measurements of leaf area and fresh biomass of banana plants at different development stages through multiple linear regression (R 2 = 0.99 and 0.97). Plant biomass and leaf area were modelled by a power law equation over time to estimate leaf area and fresh biomass at every time point (Paine et al., 2012):
| (1) |
| (2) |
With and the initial leaf area and plant mass; and c 1, c 2, β 1 and β 2 are coefficients.
Transpiration efficiency (TE; g g−1) was calculated as the estimated growth over the whole experiment divided by the cumulative plant water use over the whole experiment:
| (3) |
With and , the modelled plant mass at the start and end of the experiment; and WU as the daily absolute plant water use.
2.1.3. Real‐time transpiration phenotyping
Plant transpiration was measured by keeping plants on high‐precision balances. These balances provided weight measurements every 5 min. This total weight (m tot) was broken down to continuously quantify evapotranspiration and soil water content. m tot consisted of the plant weight (m plant), the dry soil weight (m soil,dry), the amount of water retained in the soil (m water) and the plastic pot and tray (m plastic).
| (4) |
The plastic pot and tray (m plastic) were fixed weights, measured before the experiment. The plant mass (m plant) was retrieved from image analysis (Equation 2). The soil dry weight (m soil,dry) of each pot was calculated by weighing the soil after the initial filling of the pots (m soil,start) and taking a representative sample of which the ratio of dry to fresh weight was calculated by comparing the initial fresh weight with the soil weight after 2 weeks of drying:
| (5) |
The gravimetric soil water content (SWC; g g−1) was calculated by solving Equation (4) for and dividing by as calculated from Equation (4).
| (6) |
Soil evaporation (E soil; g h−1) was calculated using six pots on balances carrying no plant, but an artificial plastic plant mimicking the canopy soil cover.
| (7) |
Half of these empty pots were well‐watered, while the other part were exposed to water deficit. The soil evaporation of a specific pot with plant was estimated based on the soil evaporation of the three empty pots under the same treatment.
Continuous transpiration rates (E rate; g cm− 2 h−1) were calculated by differentiating subsequent weight measurements, subtracting calculated soil evaporation for similar period and dividing by the calculated leaf area (Equation 1) and time between measurements.
| (8) |
Transpiration rates were averaged over 1‐h segments to take into account the balance accuracy (±1 g).
2.1.4. Jarvis–Stewart model
A modified Jarvis–Stewart model was applied (Jarvis, 1976; Whitley et al., 2009) to estimate individual plant transpiration rates in relation to light intensity, VPD and soil water content. Individual plant light irradiance and VPD was interpolated by inverse distance weighting between the environmental sensors using the Shepard method (Shepard, 1968). Light interception was assumed to be uniform and the plant canopy was hypothesized to be well‐coupled with the atmosphere. Transpiration rate was expressed as a function of scaling terms:
| (9) |
where represents a scaling factor for the transpiration rate. With f 1 an asymptotic saturating function:
| (10) |
With Q in the incoming radiation (W m− 2), Q max the maximum radiation observed during the experiment and k 1 describing the sensitivity of the light response.
f 2 describes the VPD response:
| (11) |
where f 2 was scaled between 0 and 1 by dividing by its maximum and k 2 characterizes the curvature of the transpiration response towards increasing VPD.
f 3 describes the transpiration response towards the soil water content:
| (12) |
SWCcrit represents the threshold soil water content at which stomatal closure under progressive soil drying initiates and SWCwilt the soil water content at which water uptake and consequently transpiration is halted. The model was parameterized by minimising the root‐mean‐square error (RMSE) based on the Fletcher variable metric method across 1000 distinct initial values (Nash, 2018). 95% confidence intervals were calculated by bootstrapping (Canty & Ripley, 2019). Parameter values were considered significantly different if bootstrapped confidence intervals did not overlap.
2.1.5. Leaf gas exchange measurements
Measurements were taken from the middle of the second youngest fully developed leaf using an LI‐6800 infra‐red gas analyser (LI‐COR). Measurements were taken between 9:00 and 13:00 to avoid diurnal stomatal regulations (Eyland et al., 2021). Photosynthetic efficiency measurements were performed on well‐watered plants (SWC 1.4 g g−1, −0.006 MPa) and water deficit plants (SWC 0.75 g g−1, −0.13 MPa). The leaf cuvette was maintained at a CO2 concentration of 400 μmol mol−1, a saturating light intensity of 1500 µmol m−2 s−1, a leaf temperature of 26°C and a VPD of 1 kPa. Maximum stomatal conductance (g s,max) and photosynthetic rate (A max) were defined as the steady‐state stomatal conductance (g s) and photosynthesis rate (A) reached under these saturating light conditions. After reaching steady‐state stomatal conductance (g s) and photosynthesis rate (A), the ambient CO2 concentration (Ca) was sequentially decreased to 300, 200, 100, 50 and 0 µmol mol−1 and increased again to 400, 550, 700, 1000 and 1200 µmol mol−1. The maximum velocity of Rubisco for carboxylation (V cmax) and the maximum rate of electron transport (J max) were derived according to Sharkey et al. (2007).
Gas exchange measurements at 200 and 1500 µmol m2 s−1 photosynthetic photon flux density were performed on well‐watered plants. The leaf cuvette was maintained at a CO2 concentration of 400 μmol mol−1, a leaf temperature of 26°C and a VPD of 1 kPa. Both A and g s were allowed to stabilize for a minimum of 30 min before taking measurements. Impression was made by applying dental polymer according to the protocol of Weyers and Johansen (1985), followed by covering the polymer with nail varnish and placement on a microscope slide. Stomatal anatomy was quantified using an EVOS digital inverted microscope.
2.1.6. Blind hierarchical transpiration rate clustering
A database of modelled transpiration rates as a function of light, VPD and soil water content was constructed for each genotype. The data set was composed of all combinations of VPDs (0–2.9 kPa, stepsize 0.1 kPa), light intensities (0–350 W m−2, stepsize 10 W m−2) and soil water contents (0.3–1.4 g g−1, stepsize 0.05 g g−1) measured during the experiment. The optimal number of clusters was determined by the Dunn index, obtained from the Euclidean distance matrix of the complete set of modelled transpiration rate values. Four clusters were proposed (Dunn index of 0.68).
2.2. Validation experiment at the Bioversity phenotyping platform
2.2.1. Plant material and experiment
During the second phenotyping experiment, three contrasting genotypes (Banksii_11, Malaccensis_33 and Balbisiana) were phenotyped for 8 weeks at the Bioversity phenotyping platform hosted at KU Leuven. For each genotype, four plants remained under well‐watered conditions and four were water deficit. After 33 days of drying, the four water deficit plants were replaced with four new ones. The in vitro plants were acclimated for 8 weeks before starting the experiment. Plants were grown in 10 L containers filled with peat‐based compost. Air temperature and relative humidity were measured every 5 min at eight points in the greenhouse. Light intensity was measured in the middle of the greenhouse. The photoperiod was set to 12 h. Supplemental lighting of 14 W m−2 at plant level was provided when solar radiation was lower than 250 W m−2. Soil water potential curves were constructed by soil samples of 100 cm3 following the pressure plate method (Richards, 1948) and fitted according to the van Genuchten (1980) equation.
2.2.2. Image acquisition and transpiration phenotyping
Topview images were taken weekly and correlated to destructive measurements of leaf area and fresh biomass through multiple linear regression (R 2 = 0.92 and 0.95). Plant biomass and leaf area were modelled by Equations (1) and (2). Similar to the high‐throughput phenotyping experiment at the Phenodyn platform, plant transpiration was measured by keeping the plants on high‐precision balances providing continuous weight measurements. The soil was covered by plastic to avoid evaporation and ensure only water loss through transpiration. Transpiration rate was modelled by the Jarvis‐Stewart model (Equations (9), (10), (11), (12)).
2.2.3. Leaf temperature measurements
On September 15, 2020, leaf temperature was measured hourly between 09:00 h and 19:00 h. Temperature was measured at the middle of the second youngest fully developed leaf with a handheld infra‐red thermometer (BOSCH 1000 C; ±0.1°C precision).
2.2.4. Leaf patch pressure probes
Pressure probes with a magnetic pressure adjustment (YARA ZIM Plant Technology GmbH) were used as a proxy for turgor pressure development (Zimmermann et al., 2008). These non‐invasive probes have been shown to be effective in measuring changes in the plant water status in banana (Zait et al., 2017; Zimmermann et al., 2010). The probes magnetically generate a pressure on the leaf patch and determine the attenuated output pressure (P p) which is non‐linearly and inversely coupled to the cell turgor pressure (Rüger et al., 2010; Zimmermann et al., 2008, 2010). Probes were connected to the middle of the second youngest fully developed leaf and removed weekly for plant imaging. The first 24 h were not considered to avoid relaxation of the clamped patches (Ache et al., 2010). Absolute output values varied from leaf to leaf depending on the compressibility of the clamped leaf area and the force applied via the probe (Zimmermann et al., 2008). As a result, relative P p pattern changes were analysed. These relative patterns indicate increases or decreases in the leaf turgor rather than absolute changes. Continuous pressure probe measurements were combined with continuous environmental and transpiration rate data. A repeated measures correlation matrix was constructed based on individual daily plant measurements from one hour before sunrise till one hour after sunset, using the rmcorr package in R (Bakdash & Marusich, 2017). The time needed for nocturnal P p recovery was calculated by fitting an exponential function on the P p data:
| (13) |
where P p is the output leaf patch pressure, a is the P p at the start of the night, b the steady‐state P p obtained throughout the night, ‘t’ the time (min) and τ the time constant for rapidity of turgor recovery (min). Nocturnal P p recovery was modelled between the onset of the night until 1 h after a minimal output leaf patch pressure was observed. Output leaf patch pressure was measured every 5 min and smoothed with a rolling window of 13. Data having an R 2 lower than 0.70 (6% of the data) and the first day after pressure probe attachment were removed.
2.3. Genotyping
Total genomic DNA from silica‐dried leaf material was isolated using an optimized CTAB protocol (Tel‐Zur et al., 1999). Amplification reactions of rps16, atpB‐rbcL, trnL‐F and ITS followed the protocols of Oxelman et al. (1997), Chiang et al. (1998), Taberlet et al. (1991) and White et al. (1990), respectively. A GeneAmp PCR system 9700 (Applied Biosystems) was used for amplification reactions. Sequencing reactions were carried out by Macrogen, Inc. All newly obtained sequences in this study were submitted to GenBank.
Sequence assembly was carried out in Geneious v10.1 (Biomatters). MAFFT (Katoh et al., 2002) implemented in Geneious, was used for automated alignment with the following parameters (E‐INS‐i algorithm, 100PAM/k = 2 scoring matrix, 1.3 gap open penalty and 0.123 offset value). The automatically aligned data set was manually optimized. jModelTest 2.1.4 (Posada, 2008) was used to select the best‐fitting nucleotide substitution model for each data set under the Akaike information criterion (AIC). The GTR+I+G model was determined as best fit for rps16 and trnL‐F, GTR+G for atpB‐rbcL and F81+I for ITS.
Possible topological conflicts between plastid and nuclear data matrices were visually inspected, by searching for conflicting relationships with a bootstrap support value ≥ 70% (Johnson & Soltis, 1998; Pirie, 2015). For this, a Maximum Likelihood (ML) tree of the plastid and nuclear matrix were created using the RAxML search algorithm (Stamatakis et al., 2005). Since no supported topological conflict was detected among individual gene trees, we used the concatenated DNA matrix to make an ultrametric tree using BEAST 1.8.0 (Drummond & Rambaut, 2007). BEAUti was used to prepare the xml‐file as input for the dating analysis in BEAST. A partitioned Bayesian MCMC analysis was performed using the Yule branching model and a relaxed lognormal clock. Data partitions were unlinked for the model of nucleotide evolution, allowing different partitions to adopt different parameter values, according to the different models selected for each. Root prior was set to 1. The analysis ran for 20 million generations and was sampled every 5000 generations. TRACER v.1.6 (Rambaut et al., 2018) was used to evaluate the effective sampling size (ESS) of the posteriors (which should be >200). The maximum clade credibility (MCC) tree was calculated using TreeAnnotator v.1.8.0 (Rambaut & Drummond, 2007). Posterior probabilities were calculated by TreeAnnotator v.1.8.0. (Rambaut & Drummond, 2007) using the raw trees as input and a threshold of 0.5.
2.4. Statistics
All data processing and statistical analysis were carried out in R (V3.6.2). Plant‐specific and date‐specific factors were included as random effects in a linear mixed model to account for repeated measurements. Genotypic differences were tested by applying analysis of variance (ANOVA) with a post hoc Benjamini and Hochberg correction. Segmented regression was performed on the whole‐plant transpiration between one hour before and after the onset of light on transpiration rate data measured every 15 min. Data with no significant segmented regression (p value Davies Test < 0.05, segmented R package) and negative second slopes were removed (3%). Segmented regression was performed for every genotype on the time constant τ describing the rapidity of nocturnal P p recovery.
3. RESULTS
3.1. Significant differences in leaf area, cumulative transpiration and transpiration efficiency across wild banana genotypes
A typical time course of transpiration rate in fluctuating climatic conditions is presented in Figure 1. VPDs and light intensity fluctuated throughout the day (Figure 1a,b). Transpiration rates were calculated from total weight measurements of the lysimeter set‐up and fluctuated in response to the environmental triggers (Equations (4), (5), (6), (7), (8), Figure 1c,d). The transpiration rate reduction of non‐irrigated plants compared to well‐watered plants varied with the occurring environmental conditions, strengthening the need for high‐throughput dynamic phenotyping (Figure 1c). A large range of environmental conditions was measured over the time course of the experiments (Figures S1 and S2).
Figure 1.
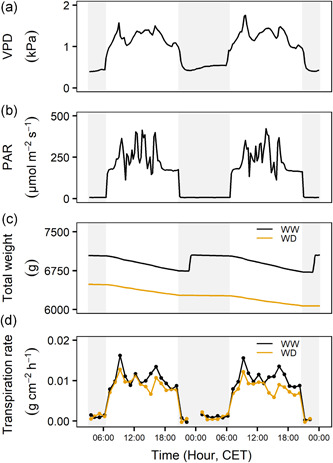
Time courses of (a) vapour pressure deficit (VPD), (b) photosynthetic active radiation (PAR), (c) raw weight measurements for a representative well‐watered (WW) and water deficit (WD) plant of the Banksii_11 genotype and (d) calculated transpiration rates per unit leaf area. The transpiration rate was calculated every hour, the VPD, PAR and total weight are illustrated every 5 min. Grey areas indicate times of darkness [Color figure can be viewed at wileyonlinelibrary.com]
The leaf area differed significantly across wild banana genotypes under both well‐watered and water deficit conditions (p < 0.05). Water deficit resulted in a significant reduction of leaf area and water use (Table S1). Daily water use followed the leaf area trend and was strongly correlated to the final leaf area across genotypes (R 2 = 0.84; p < 0.001). The M. acuminata ssp. banksii genotypes were characterized by a high leaf area and consequently cumulative water loss, while M. acuminata ssp. errans and M. balbisiana genotypes showed a low leaf area and reduced cumulative water loss (Table S1). The smaller leaf area of M. balbisiana genotypes was confirmed during the validation experiment at the Bioversity platform (Table S2). The transpiration efficiency of M. acuminata ssp. burmannicoides genotype was the lowest in both well‐watered and water deficit conditions, while M. acuminata ssp. errans showed the highest transpiration efficiency (Table S1). During the high‐throughput phenotyping experiment water deficit resulted in a significant increase in transpiration efficiency. While the average increase was 34%, the M. acuminata ssp. errans genotype displayed a 69% increase (Table S1).
3.2. Maximum and daily transpiration rates differ under well‐watered conditions
The observed daily maximum transpiration rate under well‐watered conditions differed significantly between genotypes (p < 0.001). Highest maximum transpiration rates were observed for the M. balbisiana and M. acuminata ssp. microcarpa genotypes, in comparison with M. acuminata ssp. malaccensis (Figure S3). The observed daily transpiration rate was strongly correlated to the observed maximal transpiration rate (R 2 = 0.68, p < 0.001) and similar genotype‐specific transpiration rates were observed (Figure S3). Maximum stomatal conductance (g s,max) and photosynthesis (A max) across genotypes were positively correlated to the observed maximum transpiration rate (R2 = 0.77 and 0.85; p < 0.05; Figure S4) and daily transpiration rate (R 2 = 0.80 and 0.87; p < 0.05). During the validation experiment, similar significant differences in maximum and daily transpiration rates were observed with the lowest values observed for the M. acuminata ssp. malaccensis genotypes (Figure S5). Similar to the differences in maximal transpiration rate (E max; Figure S3a), g s,max differed significantly across genotypes under well‐watered conditions with M. balbisiana having a significantly higher g s,max (Figure S6a). M. acuminata ssp. errans and M. acuminata ssp. malaccensis genotypes showed the lowest g s,max.
3.3. Drought impacted transpiration rate and photosynthesis in a genotype‐specific way
When the soil water content was above a certain threshold water status, the maximum transpiration rate of non‐irrigated plants did not differ significantly from the transpiration rate of well‐watered plants (Figure S7a). Below this critical threshold (SWCcrit), a water deficit was experienced and the transpiration rate declined sharply (Figure S7a). This threshold differed significantly between genotypes, as well as the slope of the change in transpiration rate below the threshold, thereby suggesting different levels of stomatal control (Figure S7b,c).
To disentangle environmental impacts, we modelled the individual plant transpiration rate in terms of net radiation, VPD and soil water content based on the Jarvis‐Stewart model (Equations (9), (10), (11), (12); Table S3). For the high‐throughput phenotyping experiment, the RMSE of the model ranged between 0.0018 and 0.0025 g cm−2 h−1 and the R 2 of the observed transpiration rates and modelled rates was for all genotypes higher than 0.80 (Figure S8). RMSE ranged between 0.0015 and 0.0021 g cm−2 h−1 during the second validation experiment with R 2 between 0.83 and 0.90. High correlations were obtained between the modelled critical soil water content (Figure 2) and the critical soil water content of the maximum transpiration rate response (Figure S7; R 2 = 0.87). Similarly, the slope of transpiration rate reductions was tightly related between the modelled and maximum transpiration rate (R 2 = 0.84). The threshold soil water content was significantly higher in the M. acuminata ssp. errans genotype, indicating a sensitive response to a decreasing soil water potential (SWC 1.38 g g−1, −0.008 MPa; Figure 2). The lowest threshold soil water content was observed in the M. balbisiana genotypes (SWC 1.10 g g−1, −0.022 MPa), while the other M. acuminata subspecies showed decreased transpiration rate responses between −0.016 and −0.012 MPa (SWC 1.18 and 1.27 g g−1, Figure 2). The genotypic transpiration behaviour was confirmed in the validation experiment for M. balbisiana compared to M. acuminata ssp. malaccensis and ssp. banksii (Figures S9 and S10).
Figure 2.
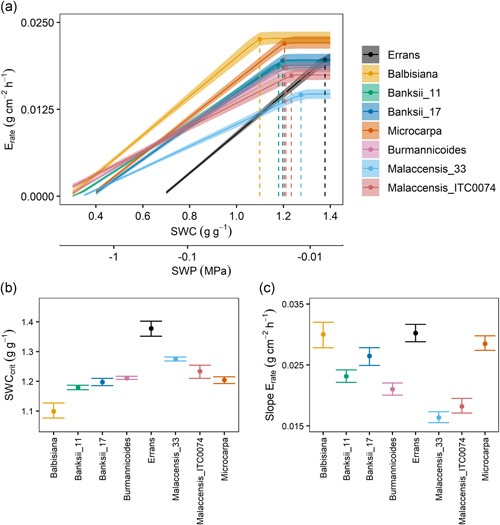
Modelled transpiration rate (E rate; Equation 12) as a function of soil water content (SWC) and soil water potential (SWP) for eight wild banana genotypes. (a) E rate did not differ from well‐watered conditions until a critical soil water content threshold (SWCcrit ) was reached. After this SWCcrit , water content becomes limiting and E rate decreased strongly. Data represent modelled response according to Equation (12) with a 95% confidence interval at VPD = 1.5 kPa and PAR = 500 µmol m−2 s−1. Dotted line represents the SWCcrit for each genotype. Note the logarithmic scale of the soil water potential. (b) SWCcrit at which E max started to decrease. Data represent the modelled parameter with a 95% confidence interval. (c) Slope of E rate decreases after the SWCcrit was reached. Data represent the modelled parameter with a 95% confidence interval. PAR, photosynthetic active radiation; VPD, vapour pressure deficit [Color figure can be viewed at wileyonlinelibrary.com]
In the same way as the reductions in transpiration rate with decreasing soil water content (Figure 2), the maximal stomatal conductance (g s,max) was significantly reduced when measured at a soil water potential of −0.13 MPa (36%–64% reduction) (Figure S6a). Likewise, maximum photosynthesis (A max) and intrinsic water use efficiency (iWUE) were significantly reduced and increased respectively (Figure S6b,c). The M. acuminata ssp. malaccensis genotypes had a low A max (Figure S6b). iWUE across genotypes was significantly correlated to whole‐plant transpiration efficiency (TE) over the complete experiment (R²WW = 0. 92 and R²WD = 0.88). iWUE was highest for the M. acuminata ssp. errans genotype under both well‐watered and water deficit conditions (Figure S6C). Under water deficit conditions g s,max remained highest in M. balbisiana (Figure S6a), comparable to the decrease in E max under decreasing soil water content (Figure S6). The maximum rate of Rubisco activity (V c,max) and the potential rate of electron transport (J max) did not differ significantly between well‐watered and water deficit plants (Figure S6d,e). The photosynthetic capacities were highest in the M. balbisiana genotypes under both well‐watered and water deficit conditions (Figure S6d,e). M. acuminata ssp. errans and ssp. malaccensis genotypes showed the lowest Vc, max and Jmax (Figure S6d,e).
Consistent with the above results, leaf temperature measured during the validation experiment was significantly lower at most time points in M. balbisiana under water deficit conditions compared to M. acuminata ssp. malaccensis and banksii (Figure S11). The leaf temperature was at every time point significantly higher under water deficit compared to well‐watered conditions for both M. acuminata ssp. malaccensis and ssp. banksii (p < 0.05, Figure 3, the soil potential of water deficit treated M. acuminata ssp. malaccensis and ssp. banksii plants was −0.062 ± 0.011 and −0.033 ± 0.004 MPa, respectively). The temperature differences observed for M. balbisiana were not significant (the soil potential of water deficit treated plants was −0.026 ± 0.002 MPa). Similarly, transpiration rates differed only significantly for both M. acuminata ssp. malaccensis and banksii (p < 0.05, Figure 3).
Figure 3.
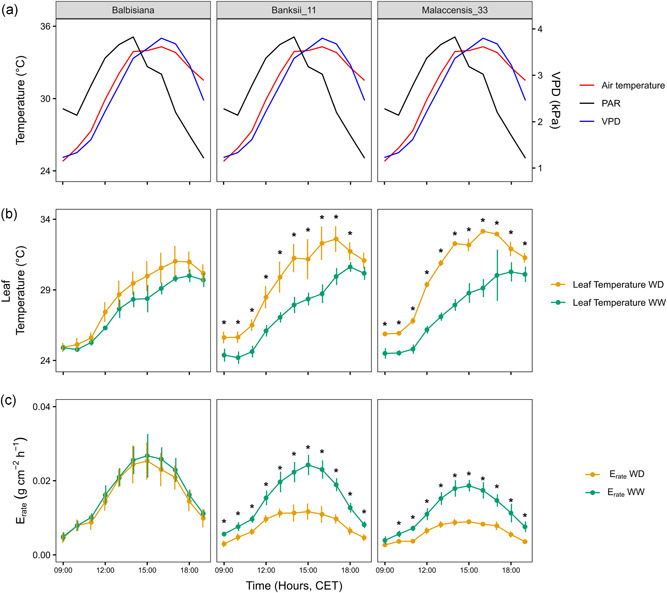
Environmental conditions, leaf temperature and transpiration rate for three wild banana genotypes during 1 day of the validation experiment. (a) Air temperature (red), photosynthetic active radiation (PAR; black) and vapour pressure deficit (VPD; blue) throughout the measurement day. (b) Leaf temperature development for well‐watered (WW; green) and water deficit (WD; orange) plants. Data represent the mean ± stdev (n = 4 per treatment and genotype, * for p < 0.05). (c) Transpiration rate evolution of WW (green) and WD (orange) plants. Data represent the mean ± stdev (n = 4 per treatment and genotype, * for p < 0.05). [Color figure can be viewed at wileyonlinelibrary.com]
3.4. Light and VPD triggered genotype‐specific transpiration rate responses
The continuous transpiration rate response to light and VPD was estimated by the Jarvis‐Stewart model (Equations 10 and 11). At low light intensities, transpiration rates were alike across genotypes, but responses to higher light intensities differed significantly (Figure 4). Both M. acuminata ssp. malaccensis genotypes maintained the lowest transpiration rates with increasing light intensity, while M. acuminata ssp. microcarpa and M. balbisiana achieved higher transpiration rates. During the validation experiment, similar light responses were observed with the highest transpiration rate in M. balbisiana and the lowest in M. acuminata ssp. malaccensis genotype (Figure S12). The estimated transpiration rates at a light intensity of 200 and 1500 µmol m−2 s−1 were significantly correlated to g s measurements of four representative genotypes at these light levels (Figure S13).
Figure 4.
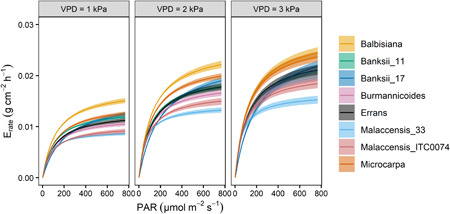
Modelled transpiration rate response of eight wild banana genotypes as a function of light intensity (PAR; photosynthetic active radiation) under well‐watered conditions at three VPD levels (1, 2 and 3 kPa). Data represent the modelled response according to Equation (10) with a 95% confidence interval. VPD, vapour pressure deficit [Color figure can be viewed at wileyonlinelibrary.com]
Transpiration rate responses to morning light intensity have been shown to be a good proxy of light‐induced stomatal speeds (Eyland et al., 2021). Light intensity increased from 0 to 200 µmol m−2 s−1 in the morning. Upon the onset of light in the morning, transpiration rate increased significantly and a breakpoint in transpiration rate increase was determined (Figure S14). This responsiveness of transpiration differed significantly across genotypes, with rapid increases in transpiration rate in M. acuminata ssp. banksii, ssp. errans and ssp. microcarpa genotypes, while M. balbisiana, M. acuminata ssp. burmannicoides and ssp. malaccensis genotypes showed postponed responses (Figure S14).
The transpiration rate continued to increase with increasing VPD in all genotypes (Figure 5). Absolute differences in transpiration rate across genotypes were more expressed under high VPD. M. balbisiana, followed by M. acuminata ssp. microcarpa showed the highest transpiration rate with increasing VPDs (Figure 5). The M. acuminata ssp. malaccensis and ssp. burmannicoides genotypes showed the lowest increase in transpiration rates with VPD. Under higher VPD levels present during the validation experiment (VPDmax 3.8 kPa) the (sub)species differences were confirmed: M. balbisiana showed the highest transpiration rates as a function of VPD while the M. acuminata ssp. malaccensis genotype was characterized by the lowest transpiration rates (Figure S15).
Figure 5.
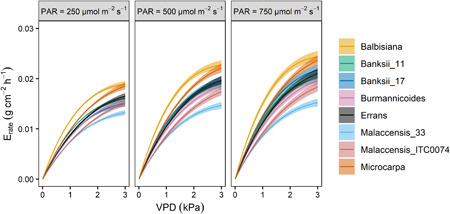
Modelled transpiration rate response of eight wild banana genotypes as a function of vapour pressure deficit (VPD) under well‐watered conditions at three light levels (photosynthetic active radiation [PAR] 250, 500 and 750 µmol m−2 s−1). Data represent the modelled response according to Equation (11) with a 95% confidence interval [Color figure can be viewed at wileyonlinelibrary.com]
3.5. Failure of nocturnal P p recovery under low water potentials was genotype‐dependent
P p dynamics were closely coupled to the transpiration rate patterns under well‐watered conditions and showed significant correlations (Figures 6 and S16). With decreasing soil water content, the correlation decreased (R²WW = 0.58 and R²WD = 0.37). Both the transpiration rate and P p dynamics were also closely correlated to the environmental dynamics, with decreases in light intensity resulting in immediate decreases in transpiration rate and P p values (Figures 6 and S16).
Figure 6.
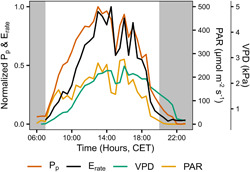
Diurnal dynamics of transpiration rate (E rate, black), leaf patch clamp pressure (Pp, red), vapour pressure deficit (VPD, green) and photosynthetic active radiation (PAR, orange) of a representative well‐watered plant of Musa acuminata ssp. malaccensis. The Pp values have been shown to be inversely coupled to the cell turgor pressure (Zimmermann et al., 2008). Grey areas indicate times of darkness [Color figure can be viewed at wileyonlinelibrary.com]
Under severe water deficit M. acuminata ssp. banksii and ssp. malaccensis showed an increase in the time needed for nocturnal P p recovery (Figures 7 and 8). The time constant of the exponential decrease was significantly increased (p < 0.001) from SWC 1.15 and 0.91 g g−1 (−0.16 and −0.50 MPa) for M. acuminata ssp. banksii and ssp. malaccensis genotypes, respectively (Figure 8). In this water potential range, M. balbisiana showed no significant increase in the time constant.
Figure 7.
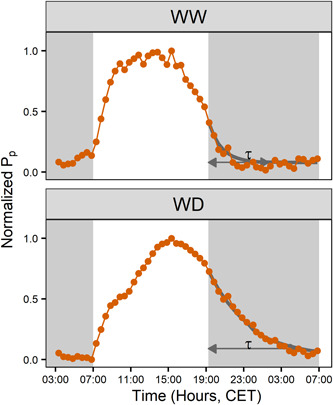
Diurnal pattern of leaf patch clamp pressure (Pp) for a representative well‐watered (WW) and water deficit (WD) plant (SWC 0.63 g g−1) of Musa acuminata ssp. malaccensis on the same day. Nocturnal Pp recovery followed an exponential decrease (grey line) and a time constant (τ) was calculated. The Pp values are inversely coupled to the turgor pressure. Grey areas indicate times of darkness. SWC, soil water content [Color figure can be viewed at wileyonlinelibrary.com]
Figure 8.
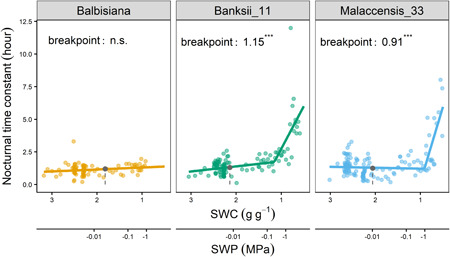
Nocturnal time constant for the exponential decrease in leaf patch clamp pressure at night under decreasing soil water content (SWC) and soil water potential (SWP). A significant increase in the time constant was observed for genotypes Malaccensis_33 and Banksii_11. Grey point and dashed grey line represent the threshold soil water content at which transpiration started to decrease and drought avoidance begins (*** for p < 0.001, n = 93–135). Note the logarithmic scale of the soil water potential [Color figure can be viewed at wileyonlinelibrary.com]
3.6. Four transpiration phenotypes were observed versus three genotypic groups
Clustering of genotypes was performed based on phenotypic similarity of the modelled transpiration rates across a wide range of light, VPD and soil water content conditions. Four major groups were blindly separated via hierarchical clustering (Figure 9). The most distant members of the cluster were M. balbisiana and M. acuminata ssp. errans, characterising groups I and II, respectively. Cluster III consisted of both M. acuminata ssp. banksii genotypes and the M. acuminata ssp. microcarpa genotype. Both M. acuminata ssp. malaccensis genotypes clustered in group IV together with the M. acuminata ssp. burmannicoides genotype.
Figure 9.
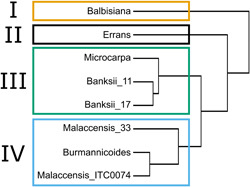
Blind hierarchical clustering of eight wild banana genotypes based on modelled transpiration rates as a function of light, vapour pressure deficit and soil water content. Four major groups (I–IV) were discerned and represented by colours [Color figure can be viewed at wileyonlinelibrary.com]
The phylogenetic relationship between the wild banana genotypes is shown in Figure 10. The phylogeny indicates that the two M. acuminata ssp. banksii accessions are closely related to M. acuminata ssp. errans and ssp. microcarpa, yet with the ssp. microcarpa node not fully supported (Figure 10). Similarly, the two M. acuminata ssp. malaccensis grouped together with M. acuminata ssp. burmannicoides. Musa balbisiana differed most from all the M. acuminata subspecies. Despite that M. acuminata ssp. errans was genetically close to the M. acuminata ssp. microcarpa and banksii (Figure 10), a different phenotype was observed (Figure 9).
Figure 10.
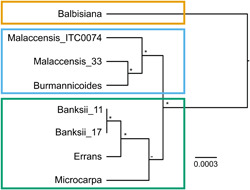
Ultrametric Bayesian maximum clade credibility (MCC) tree of the eight wild banana genotypes. The tree was constructed with BEAST v1.10 from the combined plastid‐nuclear data set. An asterisk indicates nodes that are fully supported (=1), whereas a dash indicates nodes without any support (<0.5). Three groups were discerned and represented by colours [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
Whole‐plant phenotypic responses are the result of a complex interplay between genotype (G), environment (E) and management (M). Phenotyping for drought tolerance under fully controlled conditions ignores G×E interactions (Poorter et al., 2016). The unpredicted G×E interactions often result in a poor translation towards the field, especially under unfavourable conditions (Miflin, 2000; Negin & Moshelion, 2017; Poorter et al., 2016). By measuring transpiration rates continuously across a wide range of environmental conditions and genotypes, we were able to discern genotype‐specific drought‐related traits. A plant's reaction towards water deficit is not solely determined by the genotype, but also by the environment. Radiation, VPD and wind have been shown to be interacting with the effect of soil water potential in banana (Robinson & Bower, 1988). These complex interactions explain why one specific universal soil water content, where a reduction or complete stop in transpiration takes place, cannot be defined. However, the modelling of the transpiration rate responses towards such environmental conditions enabled us to anticipate the fluctuating environment and validate genotypic differences. In literature, many different soil water contents have been reported from diverse environments and mainly for the commercial banana cultivar Cavendish. Robinson and Alberts (1986) are stating that a soil moisture content of −0.023 MPa resulted in a smaller bunch mass. Thomas and Turner (1998) found that a soil water potential of −0.062 MPa stopped gas exchange and mentioned −0.030 MPa for inducing water deficit. Ekanayake et al. (1994) mentioned practices in commercial plantations in which irrigation is applied when the soil water potential exceeds −0.020 MPa. Irrespective of the absolute soil water content and the conditions, it is clear that bananas respond very early to a reduced soil water content. Our results not only confirm the stomatal‐driven ‘isohydric’ drought avoidance behaviour (Eyland et al., 2021; Turner et al., 2007), but also indicate that there is genetic diversity in responsiveness within the Musa genus. All wild banana genotypes reduced their transpiration rate at a relatively high soil moisture content (all below – 0.025 MPa), displaying drought avoidance. At a soil water potential of −0.13 MPa, g s,max and Amax were both significantly reduced compared to well‐watered conditions (Figure S6). At these soil water potentials, the photosynthetic capacities (V cmax and J max) were however not yet affected (Figure S6). Similarly, the drought avoidance behaviour assured that the nocturnal P p recovery was only affected at lower water potentials (Figure 8, −0.16 and −0.50 MPa). This Pp recovery has been shown to be an effective indicator of drought stress, defining the point at which turgor cannot be restored quickly and at which the stress cannot be avoided anymore (Zimmermann et al., 2010).
Despite the fact that stomatal control is the dominant drought avoiding and water‐saving factor in all bananas (Aubert & Catsky, 1970; Eyland et al., 2021; Thomas & Turner, 1998), we described within the wild banana (sub)species a large diversity of growth and transpiration responses. Four diverse transpiration clusters were discerned (Figure 9). These phenotypic clusters were, except for M. acuminata ssp. errans, in agreement with previous genetic characterisations (Figure 10; Martin et al., 2020; Perrier et al., 2009; Sardos et al., 2016, 2021).
As expected from its genetic distance (Figure 10), the M. balbisiana genotype differed phenotypically most strongly from the M. acuminata genotypes. The balbisiana B genome has been frequently associated with improved drought tolerance in triploid cultivars (Ekanayake et al., 1994; Ravi et al., 2013; Thomas et al., 1998; van Wesemael et al., 2019; Vanhove et al., 2012). The M. balbisiana genotype showed in both our experiments a relatively small leaf area (Tables S1 and S2). A reduced leaf area limits the absolute water consumption, thereby avoiding fast water depletion, making it an important drought avoidance trait (Tardieu, 2012; Vadez, 2014). Irrespective of the absolute water consumption, a transpiration rate reduction was observed at a significantly lower soil water content compared to the M. acuminata subspecies (Figures 2, S7, S9, and S10, −0.032 to −0.025 MPa). This maintained stomatal opening is predicted to result in a continued high carbon uptake, which is in line with the higher A max under water‐deficit conditions (Figure S6c). Cultivars containing a large portion of the balbisiana B genome indeed maintain growth under mild water deficit (Kissel et al., 2015; van Wesemael et al., 2019). Despite the transpiration rate reduction at a lower soil water content, M. balbisiana showed together with M. acuminata ssp. errans the most pronounced response to lower soil water contents by strongly decreasing the transpiration rate (Figure 2). In addition to those two drought avoidance strategies (reduced leaf area and strong transpiration rate reduction), M. balbisiana was characterized by the highest transpiration rates, stomatal conductance and photosynthetic capacities (Figures S3 and S6). The M. balbisiana phenotype can be linked to its ecological background as it originates from regions with a very distinct yearly dry season (Janssens et al., 2016; Mertens, Bawin et al., 2021; Perrier et al., 2011). In contrast to the wild M. acuminata, M. balbisiana does also occur in open landscapes in full sunlight, but always close to waterways and can thereby benefit from increased transpiration rates for cooling (Argent, 1976; Eyland et al., 2020; Simmonds, 1956). M. balbisiana was indeed the only genotype to maintain a cool leaf temperature under mild water deficits and high VPDs (Figures 3 and S11).
M. acuminata ssp. banksii genotypes grouped together with M. acuminata ssp. microcarpa in their transpiration behaviour, in accordance to their limited genetic distance (Figures 9 and 10). M. acuminata ssp. banksii is one of the main ancestors of the commonly cultivated banana (Martin et al., 2020) and was characterized by the highest leaf area and thereby with the highest absolute water consumption (Table S1). Together with Musa acuminata ssp. microcarpa, high transpiration rates and stomatal conductance were observed under well‐watered conditions at multiple light and VPD levels (Figures 4, 5 and S6a). This ‘risk taking’ behaviour of these genotypes was further exemplified by fast increases in transpiration rate upon morning irradiation (Figure S14). These fast stomatal opening responses towards light might result in increased carbon uptake for plant growth, but are as well associated with a higher transpirational water loss (Eyland et al., 2021; Lawson & Blatt, 2014).
Despite that M. acuminata ssp. errans genotype grouped genetically with the M. acuminata ssp. banksii genotypes (Figure 10), it does have genetic differences (the only wild type with an α mitotype according to Carreel et al. [2002]), and its phenotypic behaviour was contrasting (Figure 9). M. acuminata ssp. errans showed together with M. balbisiana the most divergent phenotype. With the lowest leaf area and lowest absolute water use, the errans genotype was the most ‘conservative’ drought avoiding genotype studied (Table S1). Under water deficit the Musa acuminata ssp. errans* genotype responded by a significantly earlier reduction in transpiration rate (−0.008 MPa, Figures 2 and S7), the greatest transpiration rate reduction (Figure 2c) and the lowest photosynthetic rate (Figure S6b). Both at the stomatal and whole‐plant level it showed the highest WUE (Figure S9c and Table S1). Selecting for a high WUE is however biased towards slow‐growing genotypes because of the nonlinearity between carbon uptake and water loss (Blum, 2009).
Both M. acuminata ssp. malaccensis genotypes were characterized by the lowest daily transpiration rate (Figure S3) and by low stomatal conductance and photosynthesis, even under well‐watered conditions (Figure S6a,b). The Musa acuminata ssp. malaccensis genotypes grouped with ssp. burmannicoides genotype and showed a similar phenotype (Figure 9). In response to increasing light and VPD the genotypes from this cluster showed low transpiration rates (Figures 4 and 5). Similarly, their morning light responsiveness was significantly lower compared to the other M. acuminata subspecies (Figure S14). We deduce that those types do not thrive well in environments with high evaporative demands.
Our findings show that there is significant phenotypic diversity in the wild banana relatives. By high‐throughput continuous phenotyping under variable environmental conditions, we could discern multiple transpiration phenotypes associated with specific drought‐related traits. The effectiveness of a given trait will however depend on the provided drought scenario (Tardieu, 2012; Vadez, 2014). The traits of the M. balbisiana genotype are beneficial under high evaporative demands and relatively short dry seasons. Under well‐watered conditions M. acuminata ssp. banksii and ssp. microcarpa showed the highest growth potential. This assessed diversity in wild banana drought tolerance traits stresses the importance of conserving and characterising crop wild relatives for potential use in breeding programmes.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGEMENTS
The authors would like to thank Edwige Andre and Kevin Longin for the plant propagation and embryo rescue; Hendrik Siongers for measuring the soil water retention; Mathilde Vantyghem for aiding with the leaf temperature measurements; Stéphane Berthézène, Anthony Rosello, Benoît Suard, Vincent Nègre, Hendrik Siongers, Stan Blomme, Loïck Derette, Poi Verwilt, Simon Costers, Eline van Dijk and Bill Smeets for technical assistance during plant growth and phenotyping. The authors acknowledge Matthew Turner for providing the seeds collected in Amami. Julie Sardos, Bart Panis and Janet Paofa for providing seeds collected in Papua New Guinea. The stay of SC and DE in the phenotyping facilities at INRAE was possible through the EU project H2020 731013 (EPPN2020) TNA grant 76: The quest for climate‐smart varieties: phenotyping the banana biodiversity with acronym ‘Bananadyn’. This study was undertaken as part of the initiative ‘Adapting Agriculture to Climate Change: Collecting, Protecting and Preparing Crop Wild Relatives’ which is supported by the Government of Norway. The project is managed by the Global Crop Diversity Trust in partnership with national and international gene banks and plant breeding institutes around the world http://www.cwrdiversity.org/. DE is supported by a scholarship funded by the Global TRUST foundation project ‘Crop wild Relatives Evaluation of drought tolerance in wild bananas from Papua New Guinea’, Grant no: GS15024. The authors thank all donors who supported this study also through their contributions to the CGIAR Fund (http://www.cgiar.org/who-we-are/cgiar-fund/fund-donors-2/), and in particular to the CGIAR Research Programme Roots, Tubers and Bananas (RTB‐CRP) and the Belgian Development Cooperation project ‘More fruit for food security: developing climate‐smart bananas for the African Great Lakes region’. COST Action Phenomenall FA 1306 is gratefully acknowledged for financing the participation at numerous meetings.
Eyland, D. , Luchaire, N. , Cabrera‐Bosquet, L. , Parent, B. , Janssens, S.B. & Swennen, R. et al. (2022) High‐throughput phenotyping reveals differential transpiration behaviour within the banana wild relatives highlighting diversity in drought tolerance. Plant, Cell & Environment, 45, 1647–1663. 10.1111/pce.14310
DATA AVAILABILITY STATEMENT
The data sets used and/or analysed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- Aberkane, H. , Amri, A. , Belkadi, B. , Filali‐Maltouf, A. , Kehel, Z. & Tahir, I.S.A. et al. (2021) Evaluation of durum wheat lines derived from interspecific crosses under drought and heat stress. Crop Science, 61(1), 119–136. Available from: 10.1002/csc2.20319 [DOI] [Google Scholar]
- Ache, P. , Bauer, H. , Kollist, H. , Al‐Rasheid, K.A.S. , Lautner, S. & Hartung, W. et al. (2010) Stomatal action directly feeds back on leaf turgor: new insights into the regulation of the plant water status from non‐invasive pressure probe measurements. Plant Journal, 62(6), 1072–1082. Available from: 10.1111/j.1365-313X.2010.04213.x [DOI] [PubMed] [Google Scholar]
- Argent, G.C.G. (1976) The wild bananas of Papua New Guinea. Notes from the Royal Botanic Garden, Edinburgh, 35, 77–114. Available from: https://www.musalit.org/seeMore.php?id=12231 [Google Scholar]
- Aubert, B. & Catsky, J. (1970) The onset of photosynthetic CO2 influx in banana leaf segments as related to stomatal diffusion resistance at different air humidities. Photosynthetica, 4(3), 254–256. [Google Scholar]
- Bakdash, J.Z. & Marusich, L.R. (2017) Repeated measures correlation. Frontiers in Psychology, 8, 1–13. Available from: 10.3389/fpsyg.2017.00456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, A. (2009) Effective use of water (EUW) and not water‐use efficiency (WUE) is the target of crop yield improvement under drought stress. Field Crops Research, 112(2–3), 119–123. Available from: 10.1016/j.fcr.2009.03.009 [DOI] [Google Scholar]
- Bohra, A. , Kilian, B. , Sivasankar, S. , Caccamo, M. , Mba, C. & McCouch, S.R. et al. (2021) Reap the crop wild relatives for breeding future crops. Trends in Biotechnology, 40, 1–20. Available from: 10.1016/j.tibtech.2021.08.009 [DOI] [PubMed] [Google Scholar]
- Brown, A. , Carpentier, S.C. & Swennen, R. (2020) Breeding climate‐resilient bananas. In: Kole, C. (Ed.) Genomic designing of climate‐smart fruit crops pp. 1–498. Available from: 10.1007/978-3-319-93536-2 [DOI] [Google Scholar]
- Cabrera‐Bosquet, L. , Fournier, C. , Brichet, N. , Welcker, C. , Suard, B. & Tardieu, F. (2016) High‐throughput estimation of incident light, light interception and radiation‐use efficiency of thousands of plants in a phenotyping platform. New Phytologist, 212, 269–281. Available from: 10.1111/nph.14027 [DOI] [PubMed] [Google Scholar]
- Calberto, G. , Staver, C. & Siles, P. (2015) An assessment of global banana production and suitability under climate change scenarios. In: Aziz, E. (Ed.) Climate change and food systems: global assessments and implications for food security and trade. Rome: food Agriculture Organization of the United Nations, pp. 266–291. [Google Scholar]
- Canty, A. & Ripley, B. (2019) boot: Bootstrap R (S‐Plus) functions. Available at: https://cran.r-project.org/web/packages/boot/boot.pdf
- Carr, M.K.V. (2009) The water relations and irrigation requirements of banana (Musa spp.). Experimental Agriculture, 45(3), 333–371. Available from: 10.1017/S001447970900787X [DOI] [Google Scholar]
- Carreel, F. , de Leon, D.G. , Lagoda, P. , Lanaud, C. , Jenny, C. & Horry, J.P. et al. (2002) Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome, 45(4), 679–692. Available from: 10.1139/g02-033 [DOI] [PubMed] [Google Scholar]
- Castañeda‐Álvarez, N.P. , Khoury, C.K. , Achicanoy, H.A. , Bernau, V. , Dempewolf, H. & Eastwood, R.J. et al. (2016) Global conservation priorities for crop wild relatives. Nature Plants, 2(4), 16022. Available from: 10.1038/nplants.2016.22 [DOI] [PubMed] [Google Scholar]
- Chiang, T.Y. , Schaal, B.A. & Peng, C.I. (1998) Universal primers for amplification and sequencing a noncoding spacer between the atpB and rbcL genes of chloroplast DNA. Botanical Bulletin of Academia Sinica, 39(4), 245–250. [Google Scholar]
- Christelová, P. , De Langhe, E. , Hřibová, E. , Čížková, J. , Sardos, J. & Hušáková, M. et al. (2017) Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodiversity and Conservation, 26(4), 801–824. Available from: 10.1007/s10531-016-1273-9 [DOI] [Google Scholar]
- Cowan, M.F. , Blomstedt, C.K. , Norton, S.L. , Henry, R.J. , Møller, B.L. & Gleadow, R. (2020) Crop wild relatives as a genetic resource for generating low‐cyanide, drought‐tolerant Sorghum. Environmental and Experimental Botany, 169, 103884. Available from: 10.1016/j.envexpbot.2019.103884 [DOI] [Google Scholar]
- Dempewolf, H. , Baute, G. , Anderson, J. , Kilian, B. , Smith, C. & Guarino, L. (2017) Past and future use of wild relatives in crop breeding. Crop Science, 57, 1070–1082. Available from: 10.2135/cropsci2016.10.0885 [DOI] [Google Scholar]
- Drummond, A.J. & Rambaut, A. (2007) BEAST: bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7(214), 1–8. Available from: 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekanayake, I. J. , Ortiz, R. & Vuylsteke, D. R. (1994) Influence of leaf age, soil moisture, VPD and time of day on leaf conductance of various Musa genotypes in a humid forest‐moist savanna transition site. Annals of Botany, 74(2), 173–178. Available from: 10.1006/anbo.1994.1106 [DOI] [Google Scholar]
- Eyland, D. , Breton, C. , Sardos, J. , Kallow, S. , Panis, B. & Swennen, R. et al. (2020) Filling the gaps in gene banks: collecting, characterizing, and phenotyping wild banana relatives of Papua New Guinea. Crop Science, 61(1), 137–149. Available from: 10.1002/csc2.20320 [DOI] [Google Scholar]
- Eyland, D. , van Wesemael, J. , Lawson, T. & Carpentier, S.C. (2021) The impact of slow stomatal kinetics on photosynthesis and water use efficiency under fluctuating light. Plant Physiology, 186(2), 998–1012. Available from: 10.1093/plphys/kiab114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO . (2019) FAOSTAT Database. Available at: http://www.fao.org/faostat/
- Gonçalves, Z.S. , Haddad, F. , de Oliveira Amorim, V.B. , Ferreira, C.F. , de Oliveira, S.A.S. & Amorim, E.P. (2019) Agronomic characterization and identification of banana genotypes resistant to Fusarium wilt race 1. European Journal of Plant Pathology, 155(4), 1093–1103. Available from: 10.1007/s10658-019-01837-5 [DOI] [Google Scholar]
- Hajjar, R. & Hodgkin, T. (2007) The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica, 156, 1–13. Available from: 10.1007/s10681-007-9363-0 [DOI] [Google Scholar]
- Hegde, D.M. & Srinivas, K. (1989) Irrigation and nitrogen fertility influences on plant water relations, biomass, and nutrient accumulation and distribution in banana cv. Robusta. Journal of Horticultural Science, 64(1), 91–98. Available from: 10.1080/14620316.1989.11515932 [DOI] [Google Scholar]
- Honsdorf, N. , March, T.J. , Berger, B. , Tester, M. & Pillen, K. (2014) High‐Throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLOS One, 9(5), e97047. Available from: 10.1371/journal.pone.0097047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries, A.W. , Ovalle, C. , Hughes, S. , del Pozo, A. , Inostroza, L. & Barahona, V. et al. (2021) Characterization and pre‐breeding of diverse alfalfa wild relatives originating from drought‐stressed environments. Crop Science, 61(1), 69–88. Available from: 10.1002/csc2.20274 [DOI] [Google Scholar]
- Janssens, S.B. , Vandelook, F. , De Langhe, E. , Verstraete, B. , Smets, E. & Vandenhouwe, I. et al. (2016) Evolutionary dynamics and biogeography of Musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of Southeast Asia. New Phytologist, 210, 1453–1465. Available from: 10.1111/nph.13856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, P.G. (1976) The interpretation of the variations in leaf water potential and stomatal conductance found in canopies in the field. Philosophical Transactions of the Royal Society of London B, 273(927), 593–610. Available from: 10.1098/rstb.1976.0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L.A. & Soltis, D.E. (1998) Assessing congruence: empirical examples from molecular data. Molecular Systematics of Plants II, 297–348. Available from: 10.1007/978-1-4615-5419-6_11 [DOI] [Google Scholar]
- Katoh, K. , Misawa, K. , Kuma, K.I. & Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30(14), 3059–3066. Available from: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J. , Grewal, S. , Yang, C.Y. , Hubbart, S. , Scholefield, D. & Ashling, S. et al. (2017) A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum . Plant Biotechnology Journal, 15(2), 217–226. Available from: 10.1111/pbi.12606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissel, E. , van Asten, P. , Swennen, R. , Lorenzen, J. & Carpentier, S.C. (2015) Transpiration efficiency versus growth: exploring the banana biodiversity for drought tolerance. Scientia Horticulturae, 185, 175–182. Available from: 10.1016/j.scienta.2015.01.035 [DOI] [Google Scholar]
- Lawson, T. & Blatt, M.R. (2014) Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiology, 164, 1556–1570. Available from: 10.1104/pp.114.237107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machovina, B. & Feeley, K.J. (2013) Climate change driven shifts in the extent and location of areas suitable for export banana production. Ecological Economics, 95, 83–95. Available from: 10.1016/j.ecolecon.2013.08.004 [DOI] [Google Scholar]
- Martin, G. , Cardi, C. , Sarah, G. , Ricci, S. , Jenny, C. & Fondi, E. et al. (2020) Genome ancestry mosaics reveal multiple and cryptic contributors to cultivated banana. Plant Journal, 102, 1–18. Available from: 10.1111/tpj.14683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens, A. , Bawin, Y. , Vanden Abeele, S. , Kallow, S. , Toan Vu, D. & Thi Le, L. et al. (2021) Genetic diversity and structure of Musa balbisiana populations in Vietnam and its implications for the conservation of banana crop wild relatives. PLOS One, 16(6), e0253255. Available from: 10.1371/journal.pone.0253255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens, A. , Swennen, R. , Rønsted, N. , Vandelook, F. , Panis, B. & Sachter‐Smith, G. et al. (2021) Conservation status assessment of banana crop wild relatives using species distribution modelling. Diversity and Distributions, 27(4), 729–746. Available from: 10.1111/ddi.13233 [DOI] [Google Scholar]
- Miflin, B. (2000) Crop improvement in the 21st century. Journal of Experimental Botany, 51(342), 1–8. Available from: 10.1093/jxb/51.342.1 [DOI] [PubMed] [Google Scholar]
- Milburn, J. , Kallarackal, J. & Baker, D. (1990) Water relations of the banana. 1. Predicting the water relations of the field‐grown banana using the exuding latex. Australian Journal of Plant Physiology, 17, 57–68. Available from: 10.1071/pp9900057 [DOI] [Google Scholar]
- Nash, J. (2018) Rvmmin: variable metric nonlinear function minimization. Available at: https://cran.r-project.org/package=Rvmmin
- Naz, A.A. , Arifuzzaman, M. , Muzammil, S. , Pillen, K. & Léon, J. (2014). Wild barley introgression lines revealed novel QTL alleles for root and related shoot traits in the cultivated barley (Hordeum vulgare L.). BMC Genetics, 15(1), 1–12. Available from: 10.1186/s12863-014-0107-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negin, B. & Moshelion, M. (2017) The advantages of functional phenotyping in pre‐field screening for drought‐tolerant crops. Functional Plant Biology, 44(1), 107–118. Available from: 10.1071/FP16156 [DOI] [PubMed] [Google Scholar]
- Ochieng, G. , Ngugi, K. , Wamalwa, L.N. , Manyasa, E. , Muchira, N. & Nyamongo, D. et al. (2021) Novel sources of drought tolerance from landraces and wild sorghum relatives. Crop Science, 61(1), 104–118. Available from: 10.1002/csc2.20300 [DOI] [Google Scholar]
- Ortiz, R. & Swennen, R. (2014) From crossbreeding to biotechnology‐facilitated improvement of banana and plantain. Biotechnology Advances, 32(1), 158–169. Available from: 10.1016/j.biotechadv.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Ortiz, R. & Vuylsteke, D. (1996) Recent advances in Musa genetics, breeding and biotechnology. Plant Breeding Abstracts, 66(10), 1355–1363. [Google Scholar]
- Oxelman, B. , Lidén, M. & Berglund, D. (1997) Chloroplastrps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Systematics and Evolution, 206, 393–410. Available from: 10.1007/BF00987959 [DOI] [Google Scholar]
- Paine, C.E.T. , Marthews, T.R. , Vogt, D.R. , Purves, D. , Rees, M. & Hector, A. et al. (2012) How to fit nonlinear plant growth models and calculate growth rates: an update for ecologists. Methods in Ecology and Evolution, 3, 245–256. Available from: 10.1111/j.2041-210X.2011.00155.x [DOI] [Google Scholar]
- Perrier, X. , Bakry, F. , Carreel, F. , Jenny, C. , Horry, J.P. & Lebot, V. et al. (2009) Combining biological approaches to shed light on the evolution of edible bananas. Ethnobotany Research and Applications, 7, 199–216. Available from: 10.17348/era.7.0.199-216 [DOI] [Google Scholar]
- Perrier, X. , De Langhe, E. , Donohue, M. , Lentfer, C. , Vrydaghs, L. & Bakry, F. et al. (2011) Multidisciplinary perspectives on banana (Musa spp.) domestication. Proceedings of the National Academy of Sciences of the United States of America, 108(28), 11311–11318. Available from: 10.1073/pnas.1102001108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirie, M.D. (2015) Phylogenies from concatenated data: is the end nigh? Taxon, 64(3), 421–423. Available from: 10.12705/643.1 [DOI] [Google Scholar]
- Poorter, H. , Fiorani, F. , Pieruschka, R. , Wojciechowski, T. , van der Putten, W.H. & Kleyer, M. et al. (2016) Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytologist, 212(4), 838–855. Available from: 10.1111/nph.14243 [DOI] [PubMed] [Google Scholar]
- Posada, D. (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution, 25(7), 1253–1256. Available from: 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Prohens, J. , Gramazio, P. , Plazas, M. , Dempewolf, H. , Kilian, B. & Díez, M.J. et al. (2017) Introgressiomics: a new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica, 213(7), 1–9. Available from: 10.1007/s10681-017-1938-9 [DOI] [Google Scholar]
- Rambaut, A. & Drummond, A.J. (2007) TreeAnnotator. Available at: https://beast.community/treeannotator
- Rambaut, A. , Drummond, A.J. , Xie, D. , Baele, G. & Suchard, M.A. (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67(5), 901–904. Available from: 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi, I. , Uma, S. , Vaganan, M.M. & Mustaffa, M.M. (2013) Phenotyping bananas for drought resistance. Frontiers in Physiology, 4, 1–15. Available from: 10.3389/fphys.2013.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, M. , Dreccer, F. & Trethowan, R. (2007) Drought‐adaptive traits derived from wheat wild relatives and landraces. Journal of Experimental Botany, 58(2), 177–186. Available from: 10.1093/jxb/erl250 [DOI] [PubMed] [Google Scholar]
- Richards, L.A. (1948) Porous plate apparatus for measuring moisture retention and transmission by soil. Soil Science, 66, 105–110. Available from: 10.1097/00010694-194808000-00003 [DOI] [Google Scholar]
- Rippke, U. , Ramirez‐Villegas, J. , Jarvis, A. , Vermeulen, S.J. , Parker, L. & Mer, F. et al. (2016) Timescales of transformational climate change adaptation in sub‐Saharan African agriculture. Nature Climate Change, 6(6), 605–609. Available from: 10.1038/nclimate2947 [DOI] [Google Scholar]
- Robinson, J.C. & Alberts, A.J. (1986) Growth and yield responses of banana (cultivar‘Williams’) to drip irrigation under drought and normal rainfall conditions in the subtropics. Scientia Horticulturae, 30, 187–202. Available from: 10.1016/0304-4238(86)90097-X [DOI] [Google Scholar]
- Robinson, J.C. & Bower, J.P. (1988) Transpiration from banana leaves in the subtropics in response to diurnal and seasonal factors and high evaporative demand. Scientia Horticulturae, 37, 129–143. Available from: 10.1016/0304-4238(88)90156-2 [DOI] [Google Scholar]
- Rüger, S. , Netzer, Y. , Westhoff, M. , Zimmermann, D. , Reuss, R. & Ovadiya, S. et al. (2010) Remote monitoring of leaf turgor pressure of grapevines subjected to different irrigation treatments using the leaf patch clamp pressure probe. Australian Journal of Grape and Wine Research, 16(3), 405–412. Available from: 10.1111/j.1755-0238.2010.00101.x [DOI] [Google Scholar]
- Sadok, W. , Naudin, P. , Boussuge, B. , Muller, B. , Welcker, C. & Tardieu, F. (2007) Leaf growth rate per unit thermal time follows QTL‐dependent daily patterns in hundreds of maize lines under naturally fluctuating conditions. Plant, Cell and Environment, 30, 135–146. Available from: 10.1111/j.1365-3040.2006.01611.x [DOI] [PubMed] [Google Scholar]
- Sardos, J. , Breton, C. , Perrier, X. , van den Houwe, I. , Paofa, J. & Rouard, M. et al. (2021) Wild to domesticates: genomes of edible diploid bananas hold traces of several genepools. BioRxiv. Available from: 10.1101/2021.01.29.428762 [DOI] [Google Scholar]
- Sardos, J. , Perrier, X. , Doležel, J. , Hřibová, E. , Christelová, P. & Van Den Houwe, I. et al. (2016) DArT whole genome profiling provides insights on the evolution and taxonomy of edible banana (Musa spp.). Annals of Botany, 118(7), 1269–1278. Available from: 10.1093/aob/mcw170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, G.J. , Qi, L.L. & Marek, L.F. (2017) Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Science, 57(3), 1083–1101. Available from: 10.2135/cropsci2016.10.0856 [DOI] [Google Scholar]
- Sharkey, T.D. , Bernacchi, C.J. , Farquhar, G.D. & Singsaas, E.L. (2007) Fitting photosynthetic carbon dioxide response curves for C3 leaves. Plant, Cell and Environment, 30(9), 1035–1040. Available from: 10.1111/j.1365-3040.2007.01710.x [DOI] [PubMed] [Google Scholar]
- Shepard, D. (1968) A two‐dimensional interpolation function for irregularly‐spaced data. Proceedings of the 1968 23rd ACM national conference, pp. 517–524. Available at: 10.1145/800186.810616 [DOI]
- Simmonds, N.W. (1956) Botanical results of the banana collecting expedition, 1954‐5. Kew Bulletin, 11(3), 463–489. Available from: 10.2307/4109131 [DOI] [Google Scholar]
- Skutch, A.F. (1932) Anatomy of the axis of the banana. Botanical Gazette, 93(3), 233–258. Available from: 10.1086/334256 [DOI] [Google Scholar]
- Stamatakis, A. , Ludwig, T. & Meier, H. (2005) RAxML‐III: a fast program for maximum likelihood‐based inference of large phylogenetic trees. Bioinformatics, 21(4), 456–463. Available from: 10.1093/bioinformatics/bti191 [DOI] [PubMed] [Google Scholar]
- Taberlet, P. , Gielly, L. , Pautou, G. , Bouvet, J. & Biologie, L.D. et al. (1991) Plant universal primer. Plant Molecular Biology, 17, 1105–1109. [DOI] [PubMed] [Google Scholar]
- Tardieu, F. (2012) Any trait or trait‐related allele can confer drought tolerance: just design the right drought scenario. Journal of Experimental Botany, 63(1), 25–31. Available from: 10.1093/jxb/err269 [DOI] [PubMed] [Google Scholar]
- Tel‐Zur, N. , Abbo, S. , Myslabodski, D. & Mizrahi, Y. (1999) Modified CTAB procedure for DNA isolation from Epiphytic Cacti of the Genera Hylocereus and Selenicereus (Cactaceae). Plant Molecular Biology Reporter, 17(3), 249–254. Available from: 10.1023/A:1007656315275 [DOI] [Google Scholar]
- Thomas, D.S. & Turner, D.W. (1998) Leaf gas exchange of droughted and irrigated banana cv. Williams (Musa spp.) growing in hot, arid conditions. Journal of Horticultural Science and Biotechnology, 73(3), 419–429. Available from: 10.1080/14620316.1998.11510994 [DOI] [Google Scholar]
- Thomas, D.S. , Turner, D.W. & Eamus, D. (1998) Independent effects of the environment on the leaf gas exchange of three banana (Musa sp.) cultivars of different genomic constitution. Scientia Horticulturae, 75, 41–57. Available from: 10.1016/S0304-4238(98)00114-9 [DOI] [Google Scholar]
- Turner, D.W. , Fortescue, J.A. & Thomas, D.S. (2007) Environmental physiology of the bananas (Musa spp.). Brazilian Journal of Plant Physiology, 19(4), 463–484. Available from: 10.1590/S1677-04202007000400013 [DOI] [Google Scholar]
- Turner, D.W. & Thomas, D.S. (1998) Measurements of plant and soil water status and their association with leaf gas exchange in banana (Musa spp.): a laticiferous plant. Scientia Horticulturae, 77, 177–193. Available from: 10.1016/S0304-4238(98)00168-X [DOI] [Google Scholar]
- Vadez, V. (2014) Root hydraulics: the forgotten side of roots in drought adaptation. Field Crops Research, 165, 15–24. Available from: 10.1016/j.fcr.2014.03.017 [DOI] [Google Scholar]
- van Asten, P.J.A. , Fermont, A.M. & Taulya, G. (2011) Drought is a major yield loss factor for rainfed East African highland banana. Agricultural Water Management, 98(4), 541–552. Available from: 10.1016/j.agwat.2010.10.005 [DOI] [Google Scholar]
- van Genuchten, M.T. (1980) Closed‐form equation for predicting the hydraulic conductivity of unsaturated soils. Soil Science Society of America Journal, 44(5), 892–898. Available from: 10.2136/sssaj1980.03615995004400050002x [DOI] [Google Scholar]
- van Wesemael, J. , Kissel, E. , Eyland, D. , Lawson, T. , Swennen, R. & Carpentier, S.C. (2019) Using growth and transpiration phenotyping under controlled conditions to select water efficient banana genotypes. Frontiers in Plant Science, 10, 1–14. Available from: 10.3389/fpls.2019.00352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhove, A.‐C. , Vermaelen, W. , Panis, B. , Swennen, R. & Carpentier, S.C. (2012) Screening the banana biodiversity for drought tolerance: can an in vitro growth model and proteomics be used as a tool to discover tolerant varieties and understand homeostasis. Frontiers in Plant Science, 3, 1–10. Available from: 10.3389/fpls.2012.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, V. & Bebber, D.P. (2019) Climate change impacts on banana yields around the world. Nature Climate Change, 9(10), 752–757. Available from: 10.1038/s41558-019-0559-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyers, J.D.B. & Johansen, L.G. (1985) Accurate estimation of stomatal aperture from silicone rubber impressions. New Phytologist, 101, 109–115. Available from: 10.1111/j.1469-8137.1985.tb02820.x [DOI] [PubMed] [Google Scholar]
- White, T.J. , Bruns, T. , Lee, S.J.W.T. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a Guide to Methods and Applications, 18(1), 315–322. [Google Scholar]
- Whitley, R. , Medlyn, B. , Zeppel, M. , Macinnis‐Ng, C. & Eamus, D. (2009) Comparing the Penman‐Monteith equation and a modified Jarvis‐Stewart model with an artificial neural network to estimate stand‐scale transpiration and canopy conductance. Journal of Hydrology, 373(1–2), 256–266. Available from: 10.1016/j.jhydrol.2009.04.036 [DOI] [Google Scholar]
- Zait, Y. , Shapira, O. & Schwartz, A. (2017) The effect of blue light on stomatal oscillations and leaf turgor pressure in banana leaves. Plant, Cell & Environment, 40(7), 1143–1152. Available from: 10.1111/pce.12907 [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Mittal, N. , Leamy, L.J. , Barazani, O. & Song, B.H. (2017) Back into the wild—apply untapped genetic diversity of wild relatives for crop improvement. Evolutionary Applications, 10(1), 5–24. Available from: 10.1111/eva.12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, D. , Reuss, R. , Westhoff, M. , Geßner, P. , Bauer, W. & Bamberg, E. et al. (2008) A novel, non‐invasive, online‐monitoring, versatile and easy plant‐based probe for measuring leaf water status. Journal of Experimental Botany, 59(11), 3157–3167. Available from: 10.1093/jxb/ern171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, U. , Rüger, S. , Shapira, O. , Westhoff, M. , Wegner, L.H. & Reuss, R. et al. (2010) Effects of environmental parameters and irrigation on the turgor pressure of banana plants measured using the non‐invasive, online monitoring leaf patch clamp pressure probe. Plant Biology, 12(3), 424–436. Available from: 10.1111/j.1438-8677.2009.00235.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
The data sets used and/or analysed during the current study are available from the corresponding author on reasonable request.


