Abstract
Context. –
The diagnosis and clinical management of patients with diffuse gliomas have evolved rapidly over the past decade with the emergence of molecular biomarkers that are used to classify, stratify risk and predict treatment response for optimal clinical care.
Objective. –
To develop evidence-based recommendations for informing molecular biomarker testing for pediatric and adult patients with diffuse gliomas and provide guidance for appropriate laboratory test and biomarker selection for optimal diagnosis, risk stratification and prediction.
Design. –
The College of American Pathologists convened an expert panel to perform a systematic review of the literature and develop recommendations. A systematic review of literature was conducted to address the overarching question, “What ancillary tests are needed to classify diffuse gliomas and sufficiently inform the clinical management of patients?” Recommendations were derived from quality of evidence, open comment feedback, and expert panel consensus.
Results. –
Thirteen recommendations and 3 good practice statements were established to guide pathologists and treating physicians on the most appropriate methods and molecular biomarkers to include in laboratory testing to inform clinical management of patients with diffuse gliomas.
Conclusions. –
Evidence-based incorporation of laboratory results from molecular biomarker testing into integrated diagnoses of diffuse gliomas provides reproducible and clinically meaningful information for patient management.
Keywords: diffuse glioma, astrocytoma, oligodendroglioma, glioblastoma, diffuse midline glioma, histone H3, IDH, 1p/19q codeletion, ATRX, TP53, EGFR, TERT promoter, CDKN2A/B
INTRODUCTION
Diffuse gliomas (DG) are primary central nervous system (CNS) neoplasms characterized by the widespread infiltration of individual tumor cells displaying cytologic and histologic features of glial differentiation.1 These tumors affect patients of all ages and arise throughout the neuro-axis but are most common in older adults and occur most frequently in the cerebral hemispheres. Clinical course and treatment varies based on tumor type and grade. Since the early 1900s, and until the 2016 revised 4th edition of the World Health Organization (WHO) Classification of Tumours of the Central Nervous System,1 DG were classified based upon the morphologic features of neoplastic cells, with molecular testing playing an ancillary role.
Over the past decade, numerous investigations have uncovered molecular genetic alterations that can be used to reliably and reproducibly classify DG into clinically meaningful subsets, leading the 2016 WHO 4th Edition update to incorporate diagnostic entities based on the integration of morphologic features with molecular biomarkers.1-6 More recent advances in our understanding of the pathogenesis and clinical behavior of specific DG subtypes has led to the inclusion of additional molecular biomarkers into clinical practice and the 2021 WHO 5th Edition relies even more on molecular test results for diagnosis and grading.7-11 DNA methylome profiling continues to identify numerous tumor types with specific methylation patterns that have characteristic genetic alterations and clinical behavior.12 The increasing complexity and rapid pace of change in diagnostic criteria, relevant molecular biomarkers, laboratory testing platforms, and clinical practice warrant the development of evidence-based recommendations on biomarker testing for DG.
DESIGN
This evidence-based guideline was developed following the standards endorsed by the National Academy of Medicine.13 A detailed description of the methods and the systematic review (including the quality assessment and complete analysis of the evidence) used to create this guideline can be found in the supplemental digital content (SDC) (Copy editors should make the text “supplemental digital content” the hyperlink).
PANEL COMPOSITION
The College of American Pathologists (CAP) in collaboration with the American Association of Neuropathologists (AANP), Association for Molecular Pathology (AMP), and Society for Neuro-Oncology (SNO) convened a multidisciplinary expert panel (EP) consisting of 13 practicing pathologists from a variety of specialties, institution types, and other professional groups, two oncologists who were representatives of the American Society of Clinical Oncology (ASCO), one patient advocate, and a research methodologist consultant to develop this guideline. An advisory panel assisted the expert panel at specific key stages in the development of the guideline. All panel members, except for the methodologist consultant, volunteered their time and were not compensated for their involvement. Detailed information about the panel composition can be found in the SDC.
CONFLICT OF INTEREST POLICY
The collaborators agreed upon a conflict of interest (COI) policy (effective June 2017) and members of the expert panel disclosed all financial interests from three years prior to appointment through the development of the guideline. Individuals were instructed to disclose any relationship that could be interpreted as constituting an actual, potential, or apparent conflict. Complete disclosures of the expert panel members are listed in the Appendix. Disclosures of interest judged by the oversight group to be manageable conflicts are as follows: HC – Consultancies with F. Hoffman-La Roche, Ltd (Basel, Switzerland), Genentech USA, Inc. (South San Francisco, California), Upsher-Smith Laboratories, LLC (Grove, Minnesota), Novocure (St. Helier, Jersey), Insys Therapeutics (Phoenix, Arizona), Mateon Therapeutics (formerly OxiGENE) (Agoura Hills, California), CytRx Corp. (Los Angeles, California), Omniox Inc. (San Carlos, California), AbbVie Inc. (North Chicago, Illinois); EH – Consultancies with E.R. Squibb&Sons, LLC (New Brunswick, New Jersey), Honorarium from Arbor Pharmaceuticals, LLC (Atlanta, Georgia); MvdB – Consultancies with Bristol Myers Squibb (New York, New York), Celldex Therapeutics, Inc. (Hampton, New Jersey), Vaximm AG (Basel, Switzerland), Carthera, Nerviano, Agios, Honorarium from Merk Sharp & Dohme Corp (Kenilworth, New Jersey), Research Grants from AbbVie Inc (North Chicago, Illinois).
The majority of the EP (14 of 17 members) were assessed as having no relevant conflicts of interest. The CAP provided funding for the administration of the project; no industry funds were used in the development of the guideline. All panel members volunteered their time and were not compensated for their involvement, except for the contracted methodologist. See the SDC for complete information about the COI policy.
GUIDELINE OBJECTIVES
The expert panel addressed the overarching question, “What ancillary tests are needed to classify DG in order to sufficiently inform the clinical management of patients?” To answer this, a number of more pointed key questions (KQs) were developed:
KQ 1a: What genetic and molecular alterations should be included for optimal classification of DG?
KQ1b: What are the acceptable techniques/methods for molecular genetic testing of DG? What are the expected turn-around-times for individual assays?
KQ1c: What are the acceptable techniques/methods for assessing whole genome copy number alterations?
KQ 2: What are the core molecular tests/findings that provide sufficient classifying information in the setting of discrete clinicopathologic entities?
KQ 3: What are the acceptable techniques/methods/criteria for determining MGMT promoter methylation status?
OUTCOMES OF INTEREST
The panel established outcomes of interest for both clinical and pathology-based studies/articles. The clinical outcomes of interest included survival rates (overall, 1- and 3- year survival, progression free), recurrence rates, response to treatment, and accuracy of diagnosis. The pathologic outcomes of interest included sensitivity, specificity, positive predictive value, negative predictive value, concordance, turnaround time, reproducibility of the various tests, as well as mutation/alteration/deletion status (percent, presence, frequency, and association with other alterations) for the molecular targets of interest.
The target audience of this review are those within the neuro-oncology community who care for patients with DG, including pathologists, neuroradiologists, neurosurgeons, radiation oncologists, neuro-oncologists, medical oncologists and other members of the patient care team. Recommendations will also be highly relevant to brain tumor investigators, epidemiologists and cancer registrars.
LITERATURE SEARCH AND COLLECTION
Literature search strategies were developed in collaboration with a medical librarian for the concepts of DG, molecular markers or gene alterations, and laboratory test methods. In consultation with the expert panel, the search strategies were created using standardized database terms and keywords. Databases searched included Ovid MEDLINE and Embase.com. Additional searches for unindexed literature were conducted in ClinicalTrials.gov, Cochrane Library, Guidelines International Network, National Guideline Clearinghouse, Trip search engine, University of York Centre for Reviews and Dissemination-PROSPERO, and applicable U.S. and international organizational websites. In addition, EP members were surveyed for any relevant unpublished data at the onset of the project. Initial database searches were completed on November 13, 2017 and refreshed on September 3, 2019 and September 23, 2020. All searches were limited to English language and publication dates of January 1, 2008 to the date of search. Case reports, commentaries, editorials, conference abstracts, and letters were excluded. The Cochrane search filter for humans was applied in Ovid MEDLINE and Embase.com.14 MEDLINE and conference abstract records were excluded in the Embase searches. A targeted search was performed in Ovid MEDLINE on July 24, 2020 to ensure that emerging evidence about bithalamic glioma, infantile-type hemisphere glioma, and diffuse pediatric-type high grade glioma and relevant mutations or amplifications was included in anticipation of the WHO 2021 update.7 The search was limited to publication dates 1/1/2016 – 7/24/2020, English language, and human studies. Case reports, commentaries, editorials, and letters were excluded. The Ovid MEDLINE and Embase search strings and Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) diagrams outlining details of the systematic and targeted review are provided in the SDC as Supplemental Figures 1-3.
INCLUSION AND EXCLUSION CRITERIA
Studies were selected for inclusion in the systematic review of evidence if they met the following criteria:
the study included human patients; the study was a systematic review with or without meta-analysis, a randomized control trial (RCT), or a comparative or non-comparative observational study (prospective or retrospective design); the study included a minimum sample size of at least 30, except for studies evaluating B-Raf proto-oncogene (BRAF) alterations, Fibroblast Growth Factor Receptor (FGFR) alterations, and MYB proto-oncogene (MYB) and MYB-like (MYBL1) alterations, for which the expert panel determined all study sizes should be included based on a lower frequency of these alterations that occur in pediatric brain tumors; the study was published in English; the study addressed one of the key questions; the study included measurable data such as accuracy, sensitivity, specificity, negative predictive value (NPV), positive predictive value (PPV), time to appropriate testing, repeat procedures, overall survival, recurrence rates, turnaround time, concordance; and the study addressed DG and included at least one of the following genetic and molecular alterations:
Isocitrate Dehydrogenase (NADP(+))1 (IDH1) and Isocitrate Dehydrogenase (NADP(+))2 (IDH2) mutations
Histone H3 gene mutations
BRAF alterations
ATRX chromatin remodeler (ATRX) alterations
Tumor protein p53 (TP53) alterations
1p/19q co-deletion
Chromosome 7 gain
Chromosome 10 loss
MYB and MYBL1 alterations
Telomerase Reverse Transcriptase (TERT) promoter mutations
FGFR alterations
Epidermal Growth Factor (EGFR) alterations
Platelet Derived Growth Factor Receptor Alpha PDGFR Alpha (PDGFRA) alterations
C-MET (C-MET) alterations
Cyclin Dependent Kinase Inhibitor 2A (CDKN2A) alterations
O-6-Methylguanine-Deoxiribose Nucleic Acid Methlytransferase (MGMT) promoter alterations
Phosphatase and Tensin Homolog (PTEN) alterations
Neurofibromin 1 (NF1) alterations
Microsatellite instability (MSI) status
MDM2 Proto Oncogene (MDM2) alterations
Cyclin Dependent Kinase 4 (CDK4) alteration
Articles were excluded from the systematic review if they were: meeting abstracts; case reports, consensus documents, editorials, commentaries, or letters; cell line or animal model studies; full-text articles not available in English; studies that didn’t address at least one of the key questions with outcomes of interest as agreed upon.
QUALITY ASSESSMENT
A risk of bias assessment was performed for all fully published studies meeting inclusion criteria by the research methodologist. The methodologist assessed key indicators based on study design and methodological rigor and a rating for the quality of evidence was designated. See Supplemental Tables 1-4 for the quality assessment for included studies by study design. See Supplemental Table 5 for the Quality of Evidence definitions. Using the Grading of Recommendations Assessment, Development and Evaluation (GRADE) approach,15 an overall GRADE rating was given for each recommendation by outcome (Supplemental Table 6). Refer to the SDC for further details.
ASSESSING THE STRENGTH OF RECOMMENDATIONS
Following the quality of evidence assessment, completion of the GRADE Evidence to Decision framework,16 and discussion of the definitions and implications of strength of recommendation (Table 1), the expert panel designated the recommendations as either strong or conditional. Refer to the SDC for further details.
Table 1.
Definitions for Strength of Recommendation
| Category | Definition | Implication |
|---|---|---|
| Strong Recommendation | One for which the guideline panel is confident that the desirable effects of an intervention outweigh its undesirable effects (strong recommendation for an intervention) or that the undesirable effects of an intervention outweigh its desirable effects (strong recommendation against an intervention) | Implies that most or all individuals will be best served by the recommended course of action |
| Conditional (Weak) Recommendation | One for which the desirable effects probably outweigh the undesirable effects (weak recommendation for an intervention) or undesirable effects probably outweigh the desirable effects (weak recommendation against an intervention) but appreciable uncertainty exists | Implies that not all individuals will be best served by the recommended course of action. There is a need to consider more carefully than usual the individual patient’s circumstances, preferences, and values. |
Data derived from Grading of Recommendations, Assessment, Development and Evaluation (GRADE) Working Group materials.196
GUIDELINE REVISION
This guideline will be reviewed every four years, or earlier in the event of publication of substantive and high-quality evidence that could potentially alter the original guideline recommendations. If necessary, the entire panel will reconvene to discuss potential changes. When appropriate, the panel will recommend revision of the guideline to the CAP and its collaborators for review and approval.
RESULTS
A total of 4821 studies met the search term requirements. Based on review of these abstracts, 703 articles met the inclusion criteria and continued to full-text review. A total of 188 articles were included for data extraction, and 86 articles informed the recommendations. Excluded articles were available as discussion or background references. Additional information about the systematic review is available in the SDC. Note that only the studies listed in the quality summary were used to inform the recommendations. These are studies from our systematic review of the literature. All other references mentioned in the write-up of each recommendation were brought in to provide context or further support. For most recommendations, the evidence base consisted of studies reporting on the molecular alteration frequency of the recommended gene in defined WHO diffuse glioma subtypes. To highlight this evidence as it relates to establishing an accurate diagnosis while maintaining brevity for publication, each recommendation is accompanied by a table of representative studies. These representative studies chosen for inclusion in the tables are those that the EP believed most important in summarizing the evidence for each recommendation. Refer to the diffuse gliomas guideline web page on www.cap.org for the complete set of data tables.
The expert panel convened 17 times (15 times by teleconference and two in-person meetings) to develop the scope, draft recommendations, review and respond to solicited feedback, and assess the quality of evidence that supports the final recommendations. A nominal group technique was used for consensus decision making to encourage unique input with balanced participation among group members. An initial open comment period was posted on the CAP website (www.cap.org) from September 9-30, 2019, during which the draft recommendations were posted for public feedback. To allow for more responses, the CAP re-opened the comment period from October 11-31, 2019. Refer to the SDC for further details, including a list of organizations encouraged to participate. The expert panel approved the final recommendations with a supermajority vote.
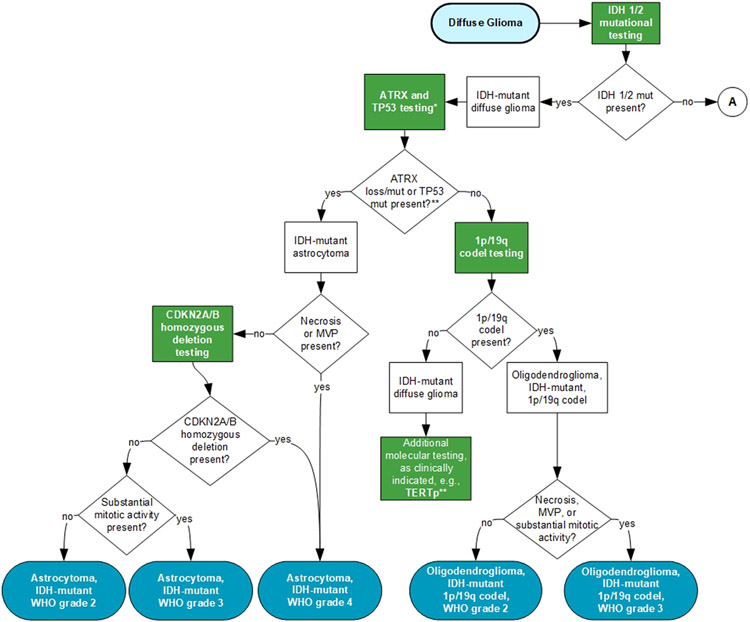
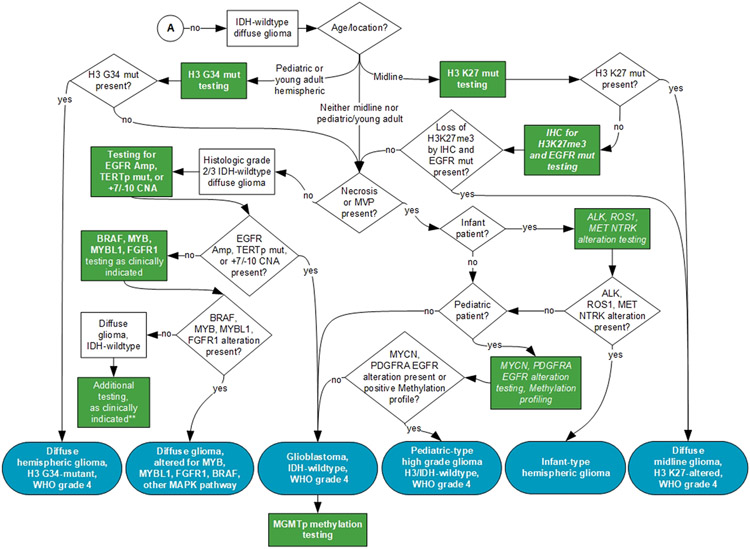
An independent review panel, masked to the expert panel and vetted through the COI process, recommended approval by the CAP Council on Scientific Affairs. The manuscript was also approved by AANP, AMP, and SNO. The final recommendations are summarized in Table 2. In addition, a visual representation of the guideline recommendations is provided to help laboratories and clinicians understand the testing involved that ultimately drives WHO-defined categorization. (Figure 1 shows the testing algorithm where IDH1/2 mutations are present. Figure 2 shows the testing algorithm where IDH1/2 mutations are not present.)
Table 2.
Summary of the Recommendations
| Recommendation | Strength of Recommendation |
|---|---|
| 1. IDH mutational testing must be performed on all diffuse gliomas (DG). | Strong Recommendation |
| 2. ATRX status should be assessed in all IDH-mutant DG unless they show 1p/19q codeletion. | Strong Recommendation |
| 3. TP53 status should be assessed in all IDH-mutant DG unless they show 1p/9q codeletion. | Conditional Recommendation |
| 4. 1p/19q codeletion must be assessed in IDH-mutant DG unless they show ATRX loss or TP53 mutations. | Strong Recommendation |
| 5. CDKN2A/B homozygous deletion testing should be performed on IDH-mutant astrocytomas. | Conditional Recommendation |
| 6. MGMT promoter methylation testing should be performed on all glioblastoma (GBM), IDH-wild type (WT). | Strong Recommendation |
| 7. For IDH-mutant DG, MGMT promoter methylation testing may not be necessary. | Conditional Recommendation |
| 8. TERT promoter mutation testing may be used to provide further support for the diagnosis of oligodendroglioma and IDH-WT GBM. | Conditional Recommendation |
| 9. For histologic grade 2-3 DG that are IDH-WT, testing should be performed for whole chromosome 7 gain/whole chromosome 10 loss, EGFR amplification, and TERT promoter mutation to establish the molecular diagnosis of GBM, IDH-WT, grade IV. | Strong Recommendation |
| 10. H3 K27M testing must be performed in DG that involve the midline in the appropriate clinical and pathologic setting. | Strong Recommendation |
| 11. H3 G34 testing may be performed in pediatric and young adult patients with IDH-WT DG. | Conditional Recommendation |
| 12. BRAF mutation testing (V600) may be performed in DG that are IDH-WT and H3-WT. | Conditional Recommendation |
| 13. MYB/MYBL1 and FGFR1 testing may be performed in children and young adults with DG that are histologic grade 2-3 and are IDH-WT and H3-WT. | Conditional Recommendation |
ATRX, ATRX chromatin remodeler; BRAF, B-Raf proto-oncogene; CDKN2A/B Cyclin Dependent Kinase Inhibitor 2A (CDKN2A)/ Cyclin Dependent Kinase Inhibitor 2B (CDKN2B); EGFR, Epidermal Growth Factor; FGFR1, Fibroblast Growth Factor Receptor 1; H3, histone3; IDH, Isocitrate Dehydrogenase; MGMT, O-6-Methylguanine-Deoxiribose Nucleic Acid Methlytransferase; MYB/MYBL1, MYB proto-oncogene (MYB)/MYB-like; TERT, Telomerase Reverse Transcriptase; TP53, Tumor protein p53
Figure 1. Testing Algorithm for Diffuse Gliomas with IDH1/2 Mutations.
A – refer to Figure 2.
ATRX, ATRX chromatin remodeler; CDKN2A/B Cyclin Dependent Kinase Inhibitor 2A (CDKN2A)/ Cyclin Dependent Kinase Inhibitor 2B (CDKN2B); Codel, codeletion; IDH, Isocitrate Dehydrogenase (NADP(+)); MVP, microvascular proliferation; Mut, mutation; TERTp, Telomerase Reverse Transcriptase promoter; TP53, Tumor protein p53; WHO, World Health Organization.
Blue indicates WHO-defined entities; Green indicates recommended tests; *Some institutions/laboratories may prefer to perform 1p/19q codeletion as the initial step for IDH-mutant gliomas. See recommendations 2-4.**Additional molecular biomarker testing and DNA methylation profiling maybe helpful in establishing a diagnosis for challenging cases.
Figure 2. Testing Algorithm for Diffuse Gliomas Without IDH1/2 Mutations.
A – refer to Figure 1.
ALK, ALK receptor tyrosine kinase; BRAF, B-Raf proto-oncogene; EGFR, epidermal growth factor receptor; FGFR1, Fibroblast Growth Factor Receptor; H3, histone 3 gene mutation; H3K27me3, H3 K27M trimethylation (H3K27me3) IDH, Isocitrate Dehydrogenase (NADP(+)); IHC, immunohistochemistry; Mut, mutation; TERTp, Telomerase Reverse Transcriptase promoter; CNA, copy number alteration; MAPK, Mitogen-activated protein kinase; MET, mesenchymal epithelial transition factor; MGMTp, O-6-Methylguanine-Deoxiribose Nucleic Acid Methlytransferase promoter; MYB, MYB proto-oncogene (MYB); MYBL1, MYB-like; NTRK, Neurotrophic tyrosine receptor kinase; PDGFRA, platelet derived growth factor receptor alpha; MYCN, N-myc proto-oncogene protein; ROS1, ROS proto-oncogene 1; WHO, World Health Organization.
Blue indicates WHO-defined entities; Green indicates recommended tests; Italic indicates good practice statements. **Additional molecular biomarker testing and DNA methylation profiling maybe helpful in establishing a diagnosis for challenging cases.
RECOMMENDATIONS
1. Strong Recommendation. – IDH mutational testing must be performed on all DG.
The quality of evidence to support this recommendation was assessed as moderate.
Quality Summary: The evidence base informing this recommendation comprises 38 studies.3-6, 17-50 Four studies were genome sequencing studies,5, 6, 21, 22 while the remaining 34 studies were retrospective cohort studies.3, 4, 17-20, 23-50 All genome sequencing studies5, 6, 21, 22 obtained samples retrospectively and were assessed as intermediate-low quality based on risk of selection bias, while no other forms of bias were identified. The retrospective cohort studies all suffered from risk of selection bias and were assessed as low3, 17, 19, 23-26, 28, 31-33, 35-50 and very low quality4, 18, 20, 27, 29, 30, 34 based on risk of selection bias in addition to risk in performance, detection, and reporting domains. The aggregate risk of bias across the entire evidence base was serious but quality of evidence was up-graded based on a strong association between IDH mutational testing and DG diagnostic classification. Refer to the SDC for the quality assessment of included genome sequencing studies, retrospective cohort studies, and the GRADE Quality of Evidence Assessment. See Table 3 for a summary of the mutational status of IDH across all diffuse glioma subtypes.
Table 3.
Representative Studies Reporting on IDH Mutation Testing and Status Across Diffuse Glioma Subtypes
| Study, Study Design |
Number DG Cases |
Testing Method |
Mutation Frequencya |
Study Conclusion |
|---|---|---|---|---|
| Brat et al,6 2015, GSS | n = 293 | Genome sequencing | A: 67.37% (64/95) | Genome-wide data delineated three molecular classes of histologic grade 2/3 DG that were more concordant with IDH, 1p/19q, ATRX and TP53 status than with histologic class and correlated with clinical outcome. |
| OA: 85.13% (63/74) | ||||
| OD: 88.07% (96/109) | ||||
| GBM: NR | ||||
| Ceccarelli et al,5 2016, GSS | n = 1132 | Genome sequencing | A: 69.05% (116/168) | DG classification based on IDH and 1p/19q status was further refined using DNA methylation profiles to identify clinically relevant genetic subsets. |
| OA: 86.84% (99/114) | ||||
| OD: 89.02% (154/173) | ||||
| GBM: 7.51% (34/453) | ||||
| Chan et al,23 2015, RCS | n = 237 | Sequencing | A: 58.33% (98/168) | When combined with IDH and 1p/19q status, TERTp mutation contributed to prognostic subgroups of lower-grade DG. |
| OA: 75.00% (36/48) | ||||
| OD: 95.24% (20/21) | ||||
| GBM: NR | ||||
| Killela et al,34 2014, RCS | n = 473 | Sequencing | A: 78.41% (69/88) | Genetic signatures of DG based on TERTp and IDH status stratifies patients into prognostically distinct cohorts |
| OA: 86.21% (50/58) | ||||
| OD: 96.55% (84/87) | ||||
| GBM: 10.00% (24/240) | ||||
| Mellai et al,39 2011, RCS | n = 287 | Sequencing, IHC | A: 38.89% (7/18) | IDH mutation was specific for DG, was correlated with MGMTp methylation and was anti-correlated with EGFR amp. |
| OA: 100% (4/4) | ||||
| OD: 53.22% (33/62) | ||||
| GBM: 10.22% (19/186) | ||||
| Sanson et al,47 2009, RCS | n = 404 | Sequencing | A: 63.33% (19/30) | IDH1 mutation was closely linked to prognosis, MGMTp status and molecular profile of DG, grades 2-4. |
| OA: 68.18% (60/88) | ||||
| OD: 63.11% (65/103) | ||||
| GBM: NR | ||||
| Labussiere et al,35 2010, RCS | n = 764 | Sequencing | A: NR | IDH1/IDH2 mutation was tightly linked to the finding of t(1;19) translocation in DG, but not with other focal 1p and 19q losses. |
| OA: NR | ||||
| OD: NR | ||||
| GBM: NR | ||||
| Lee et al,36 2017, RCS | n = 168 | Sequencing, MSP | A: 60.53% (23/38) | TERTp mutation strongly correlated with poor survival outcome in patients with IDH-WT GBM |
| OA: NR | ||||
| OD: 100.0% (65/65) | ||||
| GBM: 20.00% (13/65) | ||||
| Eckel-Passow et al,4 2015, RCS | n = 615 | Sequencing | A: NR | DG can be classified into five clinically relevant groups on the basis of IDH, TERT, and 1p/19q |
| OA: NR | ||||
| OD: NR | ||||
| GBM: NR | ||||
| Korshunov et al,3 2015, RCS | n = 202 | 450K BeadChip array | A: NR | Pediatric GBM showed low frequency of IDH1 mutation and high frequency of histone H3 mutations, leading to refined prognostic model. |
| OA: NR | ||||
| OD: NR | ||||
| GBM: 4.95% (10/202) | ||||
| Ebrahimi et al,26 2016, RCS | n = 1064 | IHC | A: 54.93% (156/284) | ATRX is a potential marker for predicting IDH/H3F3A mutations and substratification of DG into prognostic groups |
| OA: 92.96% (66/71) | ||||
| OD: 97.59% (81/83) | ||||
| GBM: 7.41% (27/364) | ||||
| Boots-Sprenger et al,20 2013, RCS | n = 561 | MLPA | A: 72.37% (55/76) | IDH, 1p/19q and MGMTp status were correlated with patient outcome in DG, yet some exceptions were noted. |
| OA: NR | ||||
| OD: NR | ||||
| GBM: 15.92% (36/226) | ||||
| Dubbink et al,25 2016, RCS | n = 133 | NGS | A: 100.0% (20/20) | DG can be subclassified into prognostic groups based on the molecular status of IDH, 1p/19q, TERTp, 7+/10q−, and H3F3A. |
| OA: 0.0% (0/21) | ||||
| OD: 97.96% (48/49) | ||||
| GBM: 0.0% (0/55) |
Mutational frequencies refer to histologic classifications. See full data tables in the supplemental digital content for all outcomes reported by included studies
Abbreviations: 7+/10q−, chromosome 7 gain/chromosome 10 q loss; amp, amplification; A, astrocytoma histology; ATRX, ATRX chromatin remodeler; BRAF, B-Raf proto-oncogene; DG, diffuse glioma; EGFR, Epidermal Growth Factor; GBM, glioblastoma; GSS, genome sequencing study; mut, mutation; H3, histone3; IDH1, Isocitrate Dehydrogenase (NADP(+))1; IDH2, Isocitrate Dehydrogenase (NADP(+))2; IHC, immunohistochemistry; MGMT, O-6-Methylguanine-Deoxiribose Nucleic Acid Methlytransferase; MGMTp, MGMT promoter; MLPA, multiplex ligation-dependent probe amplification; MSP, methylation specific polymerase chain reaction; NGS, next generation sequencing; NR, not reported; OA, oligoastrocytoma histology; OD, oligodendroglioma histology; RCS, retrospective cohort study, t(1;19), translocation (1;19); TERT, Telomerase Reverse Transcriptase; TERTp, TERT promoter; TP53, Tumor protein p53; WT, wild-type.
The identification of an IDH mutation within a DG is required for the diagnosis of specific neoplastic types recognized by the World Health Organization (WHO), including astrocytoma, IDH-mutant and oligodendroglioma, IDH-mutant, 1p/19q codeleted.1, 7 More than 70% of histologic grades 2 and 3 infiltrating astrocytomas in adults are IDH-mutated. By definition, all oligodendrogliomas IDH-mutant, 1p/19q codeleted harbor IDH mutations.4-6, 34 IDH-mutant DG are distinct diseases that have specific genetic profiles, clinical courses and therapeutic options that differ from other forms of DG, such as IDH-wild type (WT) and histone H3-mutant DG.3-6, 18, 20, 23, 25, 35, 36, 47 Therefore, testing for IDH mutations must be performed on DG in order to diagnose these tumor types for proper clinical care. Testing may not be necessary if a definitive diagnosis of an IDH-mutant glioma or another diagnosis that is mutually exclusive with IDH-mutant glioma has been previously established. IDH-mutant gliomas are uncommon in pediatric patients and testing may not be necessary in younger children.51 Testing for IDH mutations also may not be necessary if genetic alterations that are mutually exclusive with IDH mutations have been identified in a DG, such as histone H3 mutations. It should be noted that EGFR amplifications, gain of chromosome 7/loss of chromosome 10, and TERT promoter mutations have been documented in IDH-mutant astrocytomas, albeit at much lower frequency than GBM, IDH-WT, and their identification in a DG should not automatically rule out IDH testing.3, 26, 39, 47, 52 Other clinical or pathologic settings may be encountered in which the diagnosis of an IDH-mutant DG can be formally excluded without testing for the mutation.
IDH1 and IDH2 mutations result in a substitution for a key arginine at codons R132 and R172, respectively.53 The most frequent IDH1 mutation is R132H, which accounts for 89-93% of all IDH1 and IDH2 mutations.5, 6, 27, 35, 47, 53 A highly sensitive and specific monoclonal antibody that recognizes the IDH1-R132H mutant protein is widely used in diagnostic practice. Immunohistochemistry (IHC) for IDH1-R132H is a cost-effective and reliable first line test for IDH1 mutation in supratentorial DG.39, 50, 54-57 IDH1 R132H mutations are followed in frequency by R132C, R132S, R132G and R132L. IDH2 mutations represent approximately 3% of all IDH mutations and are more frequent in oligodendrogliomas than IDH-mutant astrocytomas. R172K is the most frequent, followed by R172M, and R172W. Testing for these non-IDH1-R132H mutations is accomplished by DNA sequence analysis and is necessary when IHC for IDH1-R132H is negative in appropriate settings, with specific exceptions (see above). Recent studies of primary infratentorial IDH-mutant astrocytomas have shown that they have a different spectrum of IDH-mutations, with about 80% harboring non-IDH R132H mutations,58 suggesting that IDH sequencing may be required more frequently in this clinical setting.
Sequence analysis for IDH mutations can be performed by pyrosequencing, Sanger sequencing, polymerase chain reaction (PCR), or next generation sequencing (NGS).27, 34, 59-61 Since patients older than 55-years only rarely develop de novo glioblastomas (GBMs) that are IDH-mutant (4-7%) and the IDH1-R132H antibody recognizes the large majority of IDH mutations, the WHO has recommended that testing for non-IDH1-R132H mutations by sequence analysis may not be necessary in patients older than 55-years whose tumors are negative for IDH1-R132H by IHC.1, 26, 27, 32, 43, 55 However, non-R132H mutations are sufficiently common in DG of all grades under age 55, such that testing for these mutations is indicated when IHC does not detect the R132H mutation. As above, IDH testing may also be unnecessary if other mutually exclusive genetic alterations are identified.
The finding of an IDH mutation in the setting of glial proliferation strongly supports the diagnosis of a DG since this event does not occur in non-neoplastic diseases and is only rarely, if ever, found in CNS neoplasms other than DG.27, 46, 62-64 IDH mutations are almost always stable and persist through the course of disease, which can be exploited to evaluate residual/progressive tumor33, 65, 66; however, loss of the IDH-mutated allele has been documented in some cases at the time of recurrence and may be associated with greater cell proliferation or high grade behavior.67, 68
Public comment response to Recommendation 1: There were 92 respondents, of whom 91.31% (n=84) agreed or agreed with modifications, 6.52 % (n=6) disagreed, and 2.17% (n=2) were neutral. There were 31 written comments, including a number that suggested that the phrase “appropriate clinical and pathologic setting” needed to be further explained. There were also suggestions that the statement should recommend that IDH testing “should” be performed rather than “must” be performed. Others indicated that the methods of testing (eg, IHC versus sequencing) should be described. These comments were taken into consideration. While the recommendation remained the same, the comments were addressed in the text above.
2. Strong Recommendation. – ATRX status should be assessed in all IDH-mutant DG unless they show 1p/19q codeletion.
3. Conditional Recommendation. – TP53 status should be assessed in all IDH-mutant DG unless they show 1p/19q codeletion.
4. Strong Recommendation. – 1p/19q codeletion must be assessed in IDH-mutant DG unless they show ATRX loss or TP53 mutation.
Recommendations 2-4 are tightly linked to one another and should be considered together. Once the initial testing of a DG has revealed that the tumor is IDH-mutant, further testing for ATRX, TP53 and 1p/19q support the diagnosis of IDH-mutant astrocytoma or oligodendroglioma, IDH-mutant, 1p/19q codeleted. Testing algorithms and workflows differ across practices, institutions and countries. Some perform IHC for ATRX and p53 first, with follow-up studies for 1p/19q only if these tests are negative. Others perform 1p/19q testing as the initial step, followed by ATRX and/or p53 testing if the test is negative. Multigene panels represent another approach that provides test results simultaneously; proponents assert that it provides information on other biomarkers relevant to classification and grade.69 Since positive test results from ATRX/p53 and 1p/19q testing are nearly mutually exclusive, it is not necessary to perform additional testing if the initial test results are sufficient to establish a diagnosis. Recommendations 2-4 are written in a manner to reflect these considerations.
The quality of evidence to support Recommendations 2 and 4 is moderate, the quality of evidence to support Recommendation 3 was assessed as low.
Quality Summary: Recommendation 2 was informed by 12 retrospective cohort studies.24-26, 49, 50, 70-76 Eight of these studies were assessed as low quality,24-26, 49, 50, 70, 74, 75 and four were assessed as very low quality.71-73, 76 All included studies were limited by a critical risk of selection bias, plus individual studies were further limited by risk of bias in performance,26, 72, 73 detection,24, 26, 71-76 and reporting,50, 71-74, 76 domains. Although the aggregate risk of bias across the evidence base was very serious, the evidence was upgraded based on a strong association between ATRX assessment and DG WHO classification. Recommendation 3 focuses on the need for TP53 assessment and was informed by two genome sequencing studies,6, 22 two prospective cohort studies,77, 78 and 15 retrospective cohort studies.19, 24, 25, 29, 42, 49, 71, 75, 79-85 The retrospective cohort studies were assessed as low19, 24, 25, 42, 49, 75, 79-81, 83-85 and very low quality29, 71, 82 based on risk of bias in selection,19, 24, 25, 29, 42, 49, 71, 75, 79-85 performance,19, 29, 79 detection,24, 29, 42, 71, 75, 79-82, 84, 85 and reporting29, 42, 71, 80, 82-85 domains. The aggregate risk of bias for the evidence base was very serious and evidence was not further downgraded or upgraded based on any domain. The evidence base supporting Recommendation 4 comprises one genome sequencing study6 and 11 retrospective cohort studies.4, 20, 42, 49, 50, 63, 74-76, 82, 86 All retrospective cohort studies carry a critical risk of selection bias, plus individual studies were further limited by risk of bias in performance,4 detection,4, 20, 42, 63, 74-76, 86 and reporting4, 20, 42, 50, 63, 74, 76, 82, 86 domains. Although the aggregate risk of bias across the evidence base was very serious, the evidence was upgraded based on a strong association between 1p/19q codeletion status and DG WHO classification. Refer to the SDC for the quality assessment of included genome sequencing studies, retrospective cohort studies, and the GRADE Quality of Evidence Assessment. See Table 4 for a summary of the mutational status of ATRX, TP53, and 10/19q across all diffuse glioma subtypes.
Table 4.
Representative Studies Reporting on ATRX, TP53, and 1p/19q Status Across Diffuse Glioma Subtypes
| Study, Study Design |
Number DG Cases |
Marker | Testing Method |
Mutation Frequencya |
Study Conclusion |
|---|---|---|---|---|---|
| Brat et al,6 2015, GSS | n=293 | 1p19q, IDH, TP53, ATRX | Genome sequencing | A: IDH + 1p19q, 2.11% (2/95) | Genomewide data delineated three molecular classes of histologic grade 2/3 DG that were more concordant with IDH, 1p/19q, ATRX and TP53 status than with histologic class and correlated with clinical outcome. |
| OA: IDH + 1p19q, 17.57% (13/74) | |||||
| OD: IDH + 1p19q, 63.30% (69/109) | |||||
| GBM: NR | |||||
| McClendon et al,22 2008, GSS | n=91 | TP53 | Genome sequencing | A: NR | Highlighted core molecular pathways consistently altered in GBM, including p53, RTK and RB networks. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR | |||||
| Cai et al,70 2014, RCS | n=169 | ATRX | Sequencing | A: ATRX, 61.76% (42/68) | ATRX expression is tightly correlated with IDH1/2 mutation and can be used to define prognostic DG subgroups. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: ATRX, 41.58% (42/101) | |||||
| Rajmohan et al,74 2016, RCS | n=91 | ATRX, IDH, 1p19q | Sequencing, IHC, FISH | A: ATRX, 83.33% (15/18) 1p19q, 5.56% (1/18) | Histologic grade 3 DG can be stratified into prognostic groups based on IDH, 1p/19q and ATRX status |
| OA: ATRX, 24.24% (8/33) 1p19q, 57.58% (19/33) | |||||
| OD: ATRX, 10.00% (4/40) 1p19q, 70.00% (28/40) | |||||
| GBM: NR | |||||
| Mukasa et al,42 2012, RCS | n=250 | TP53, 1p19q, IDH | Sequencing | A: 1p19q, 6.90% (4/58) TP53, 39.66% (23/58) | IDH mutation in DG was tightly correlated with 1p/19q codeletion and the combination was associated with prolonged survival. |
| OA: 1p19q, 8.33% (1/12) TP53, 41.67% (5/12) | |||||
| OD: 1p19q, 72.50% (29/40) TP53, 7.50% (3/40) | |||||
| GBM: 1p19q, 0.82% (1/122) TP53, 20.49% (25/122) | |||||
| Sahm et al,82 2014, RCS | n=43 | IDH, ATRX, TP53, 1p19q | Sequencing, IHC | A: NR | Histologically defined oligoastrocytomas can be reclassified as astrocytoma or oligodendroglioma based on IDH, TP53, ATRX, and 1p19q |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR | |||||
| Eckel-Passow et al,4 2015, RCS | n=615 | IDH, TERT, 1p19q | Sequencing | A: NR | DG can be classified into five clinically relevant groups on the basis of IDH, TERTp, and 1p/19q |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR | |||||
| Shao et al,75 2016, RCS | n=135 | ATRX, TP53, 1p/19q, IDH | IHC | A: ATRX, 32.79% (20/61) TP53, 50.88% (29/57) 1p19q, 16.00% (4/25) | DG with IDH mutations and ATRX loss also had p53 overexpression and MGMTp methylation, and were mutually exclusive with 1p/19 codeletion. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: ATRX, 20.00% (2/10) TP53, 54.54% (6/11) 1p19q, 0.00% (0/4) | |||||
| Cryan et al,71 2014, RCS | n=108 | ATRX, TP53 | IHC, NGS | A: ATRX, 53.57% (15/28) TP53, 71.41% (20/28) | Oligodendroglioma and astrocytoma can be distinguished by their IDH, 1p/19q, ATRX and TP53 profile. Histologic oligoastrocytomas harbored TP53 and ATRX mutations at frequencies similar to astrocytomas. |
| OA: ATRX, 60.00% (9/15) TP53, 80.00% (12/15) | |||||
| OD: ATRX, 3.08% (2/65) TP53, 6.15% (4/65) | |||||
| GBM: NR | |||||
| Ebrahimi et al,26 2016, RCS | n=1064 | ATRX, IDH | IHC | A: ATRX, 47.89% (136/284) | ATRX is a potential marker for predicting IDH/H3F3A mutations and substratification of DG into prognostic groups |
| OA: ATRX, 32.39% (23/71) | |||||
| OD: ATRX, 1.20% (1/83) | |||||
| GBM: ATRX, 7.42% (27/364) | |||||
| Ikemura et al,73 2016, RCS | n=193 | ATRX, IDH, TP53, 1p19q | IHC | A: ATRX, 54.45% (24/44) TP53, 40.63% (13/32) 1p19q, 10.00% (3/30) | In grade 2/3 DG, ATRX loss was strongly correlated with IDH mutation and p53 overexpression, while mutually exclusive of 1p/19q co-deletion. |
| OA: ATRX, 30.77% (4/13) TP53, 22.22% (2/9) 1p19q, 63.64% (7/11) | |||||
| OD: ATRX, 0.00% (0/18) TP53, 0.00% (0/15) 1p19q, 94.44% (17/18) | |||||
| GBM: ATRX, 12.71% (15/118) TP53, 21.95% (18/82) 1p19q, 3.53% (3/85) | |||||
| Wiestler et al,76 2013, RCS | n=133 | ATRX, 1p19q | IHC | A: ATRX, 44.62% (29/65) | Among patients with anaplastic DG, ATRX loss strongly correlated with IDH mutations, anti-correlated with1p/19q co-deletion, and provided prognostic value. |
| OA: ATRX, 27.08% (13/48) | |||||
| OD: ATRX, 20.00% (2/10) | |||||
| GBM: NR | |||||
| Hewer et al,72 2016, RCS | n=54 | ATRX, IDH, 1p19q | IHC | A: NR | Histologic oligoastrocytomas that had IDH mutations showed either ATRX loss or 1p/19q LOH, but not both alterations. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR | |||||
| Bienkowski et al,49 2018, RCS | n=165 | ATRX, IDH, TP53, 1p19q | IHC, PCR. MLPA | A: ATRX, 30.77% (24/78) TP53, 47.50% (38/80) 1p19q, 1.47% (1/68) | For assessing IDH, 1p/19q and TERTp status in DG, a combination of IHC, direct sequencing, and MLPA is a practical approach. |
| OA: ATRX, 47.17% (25/53) TP53, 56.60% (30/53) 1p19q, 32.08% (17/53) | |||||
| OD: ATRX, 9.68% (3/31) TP53, 29.03% (9/31) 1p19q, 62.07% (18/29) | |||||
| GBM: NR | |||||
| Alentorn et al,19 2014, RCS | n=126 | TP53 | IHC | A: TP53, 35.00% (7/20) | Among IDH-mutant DG, p53 overexpression is mutually exclusive with 1p19q co-deletion. |
| OA: TP53, 38.78% (19/49) | |||||
| OD: TP53, 15.69% (8/51) | |||||
| GBM: NR | |||||
| Wang et al,83 2016, RCS | n=670 | TP53, IDH | IHC | A: TP53, 64.62% (42/65) | IDH mutation was strongly correlated with high p53 expression among histologically defined astrocytic neoplasms. |
| OA: TP53, 62.04% (85/137) | |||||
| OD: TP53, 13.75% (11/80) | |||||
| GBM: TP53, 1.90% (57/300) | |||||
| Rageswarie et al,50 2018, RCS | n=449 | IDH, ATRX, 1p19q | IHC, FISH | A: NR | In a resource-limited set-up, histology with IHC for IDH1-R132H and ATRX form the baseline testing to derive DG subgroups. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR | |||||
| Dubbink et al,25 2016, RCS | n=133 | ATRX, 1p19q, IDH | NGS | A, 1p19q intact: ATRX, 65.00% (13/20) TP53, 95.00% (19/20) | DG can be subclassified into prognostic groups based on the molecular status of IDH, 1p/19q, TERTp, 7+/10q−, and H3F3A. |
| OA: NR | |||||
| OD, 1p19q co-del: ATRX, 2.04% (1/49) TP53, 8.16% (4/49) | |||||
| GBM: ATRX, 0.00% (0/55) TP53, 20.00% (11/55) | |||||
| Chaurasia et al,24 2016 | n=163 | TP53 | NR | A: NR | IHC for ATRX, IDH1, and p53 can be used to stratify GBM patients as individual markers and in combination. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: ATRX, 15.34% (25/163) p53, 49.08% (80/163) |
Mutational frequencies refer to histologic classifications. See full data tables in the supplemental digital content SDC for all outcomes reported by included studies
Abbreviations: 7+/10q−, chromosome 7 gain/chromosome 10 q loss; A, astrocytoma histology; ATRX, ATRX chromatin remodeler; co-del, co-deletion; DG, diffuse glioma; FISH, Fluorescence in situ hybridization; GBM, glioblastoma; GSS, genome sequencing study; H3, histone3; IDH1, Isocitrate Dehydrogenase (NADP(+))1; IDH2, Isocitrate Dehydrogenase (NADP(+))2; IHC, immunohistochemistry; LOH, loss of heterozygosity; mut, mutation; MGMTp, O-6-Methylguanine-Deoxiribose Nucleic Acid Methlytransferase promoter; MLPA, multiplex ligation-dependent probe amplification; NGS, next generation sequencing; NR, not reported; OA, oligoastrocytoma histology; OD, oligodendroglioma histology; PCR, polymerase chain reaction; RB, radial basis; RCS, retrospective cohort study; RTK, receptor tyrosine kinase; TERTp, Telomerase Reverse Transcriptase promoter; TP53, Tumor protein p53
Recommendation 2
As with all cancers, DG must employ a mechanism to prevent telomeres from shortening in order to escape cellular senescence. In the case of IDH-mutant astrocytomas (WHO grades 2-4), this is primarily accomplished by alternative lengthening of telomeres (ALT) via inactivation of the ATRX gene.87 Most oligodendrogliomas, IDH-mutant, 1p/19q-codeleted and IDH-WT GBMs accomplish this via overexpression of telomerase resulting from activating TERT promoter mutations. Due to these associations, loss of nuclear ATRX protein expression in tumor cells of an IDH-mutant DG as determined by IHC serves as a relatively (albeit not completely) sensitive and specific surrogate marker for astrocytic lineage.24-26, 49, 50, 70, 71, 73-75 Since strong and diffuse nuclear p53 immunoreactivity is also commonly encountered in IDH-mutant astrocytomas, the ATRX immunostain is often run simultaneously with the IDH1 R132H (recommendation 1) and p53 stains (discussed in recommendation 3). Less common astrocytoma subtypes, which are more common in children, may also show loss of ATRX expression, including diffuse midline gliomas with H3 K27M mutation (~15% of cases), diffuse hemispheric gliomas with H3 G34 mutation (nearly all cases), and anaplastic astrocytoma with piloid features.26, 88-93 Consistent with their associations with glial lineage, there is a strong inverse relationship among IDH-mutant DG between ATRX loss and the presence of 1p/19q codeletion (ie, molecularly defined oligodendrogliomas).49, 50, 72, 76, 94 Only rare examples of “dual genotype” IDH-mutant DG have been described in which ATRX loss, TP53 mutation and 1p/19q codeletion were identified in the same neoplasm.95-97 For this reason, it has been recommended that 1p/19q testing need not be pursued in IDH-mutant gliomas with immunohistochemically identified ATRX loss or p53 overexpression.98 By the same token, ATRX testing is not considered necessary for IDH-mutant DG with established codeletion of chromosome 1p/19q.
Beyond these basic testing recommendations, a few practical issues are worth highlighting. Firstly, the ATRX immunostain can be technically challenging and appropriate positive and negative controls are therefore critical for accurate assessment. Internal positive controls, such as entrapped neurons, nonneoplastic glia, and endothelial cells, should show retained expression within the same regions where tumor nuclei are immunonegative. If all nuclei (including nonneoplastic cells) are immunonegative, this should be interpreted as a technical failure of the immunostain rather than true loss of expression in the tumor. A mosaic reactivity pattern, wherein some tumor nuclei appear positive and others negative, often represents a technical failure as well and as such, should not be interpreted as loss of expression within just a subset of tumor cells. Regardless, in the case of equivocal immunostaining or in diagnostic practices that do not initially screen for ATRX inactivation using IHC, ATRX gene alterations can be identified instead using other molecular techniques (eg, NGS). Secondly, since only 70-80% of IDH-mutant astroctyomas (WHO grades 2-4) show ATRX inactivation, retained ATRX immunoexpression does not exclude the possibility of this diagnosis. Infratentorial IDH-mutant astrocytomas show loss of ATRX in only about 50% of cases.58 For cases suspected to be IDH-mutant astrocytoma based on morphology, but showing retention of ATRX on IHC, evidence of astrocytic lineage should depend instead on other findings, such as p53 protein overexpression, TP53 gene mutation, or a lack of 1p/19q codeletion. Lastly, ATRX loss may also be encountered in rare IDH-WT GBMs of older adults and in several pediatric/young adult IDH-WT astrocytoma subtypes, as already discussed. As such, ATRX loss is not entirely specific for IDH-mutant astrocytomas and this finding should not be interpreted in isolation, but rather in the context of other alterations, including IDH mutation. DNA methylation profiling has rapidly emerged as a platform that is capable of reproducibly identifying IDH-mutant astrocytomas, as well as subtypes that are infratentorial, supratentorial low grade and supratentorial high grade. Although the technology is not widely available at this time, methylation profiling may be useful in diagnostically challenging cases and will likely emerge as an important diagnostic platform.12, 58
Public comment response to Recommendation 2: There were 92 respondents, of whom 84.78% (n=78) agreed or agreed with modification, 13.05% (n=12) disagreed, and 2.17% (n=2) were neutral. There were 37 written comments, including a number that requested clarification of recommended testing methodologies and a specific algorithm outlining the recommended order of testing, including the relation of ATRX to 1p/19q, p53, histone H3, or TERT promoter analyses. One respondent commented that ATRX status is not needed if 1p/19q is done routinely, whereas another stated that 1p/19q results can be misleading and as such, ATRX data provide useful quality assurance. Although some of these issues are beyond the scope of this paper, a few practical guidelines for testing were nonetheless discussed in the prior paragraph. A few respondents also commented that they did not believe ATRX testing was sufficiently cost effective or diagnostically informative to warrant its routine use, whereas a few felt that Recommendation 2 did not go far enough, since ATRX testing could be potentially useful for all gliomas, including nondiffuse or pediatric astrocytoma variants. These useful comments were taken into consideration and while the final recommendation remained the same, some of these specific concerns were addressed in the text above.
Recommendation 3
Among IDH-mutant DG, TP53 mutation and p53 overexpression are associated with astrocytic lineage and test results for these biomarkers are used to support the diagnosis of IDH-mutant astrocytomas, especially in the setting of ATRX loss or mutation.22, 49, 75, 78, 82, 85, 99 IDH-mutant gliomas that have whole arm 1p/19q codeletion (ie, oligodendroglioma, IDH-mutant, 1p/19q codeleted) only rarely (2-3%) exhibit TP53 mutation or strong p53 overexpression.19, 42, 71, 83
In one large series, TP53 mutations were noted in 94% of IDH-mutant DG that did not demonstrate 1p/19q codeletion.6 Thus, 1p/19q codeletion and TP53 mutations are nearly, but not entirely, mutually exclusive in IDH-mutant DG. In IDH-mutant DG with TP53 mutations, the 17p-arm typically exhibits copy neutral loss of heterozygosity (cnLOH); thus, there are generally two identical copies of the mutant TP53 gene.4-6, 65, 100 In a small percentage of IDH-mutant astrocytomas there are two distinct TP53 mutations; one involves each of the TP53 alleles on the two 17 p-arms.4-6, 65, 100 TP53 mutations that occur in IDH-mutant astrocytomas tend to activate the expression and/or enhance the stability of the p53 protein. Thus, p53 IHC is an excellent surrogate of TP53 mutation in IDH-mutant gliomas—especially if the expression of ATRX is not retained.101, 102 Unlike IHC for ATRX, however, p53 expression is more of a continuum, and the interpretation may not be straightforward. For higher sensitivity and specificity, >10% of tumor cells must show strong nuclear positivity, although most TP53 mutant gliomas show even more widespread (>50%) p53 expression.102 Using this threshold, only a small subset of DG with strong p53 immunoreactivity in >10% of nuclei were found to be TP53 WT by sequencing.102 If tumor DNA sequencing is performed, the combination of IDH, TP53 and ATRX mutations and the associated copy number patterns are distinctive for IDH-mutant astrocytomas.4-6, 65, 100 It should be noted that TP53 mutation is also encountered in other DG, especially IDH-WT GBMs.24, 29, 41, 42, 86, 103 In IDH-WT gliomas, one TP53 gene typically undergoes mutation, and the WT TP53 gene is lost through deletion. In IDH-WT gliomas, TP53 mutation less predictively generates p53 overexpression.75, 83, 104 Nevertheless, p53 overexpression is encountered in up to 30% of IDH-WT GBM, so this finding should not be used as support for IDH-mutant status in isolation.
Public comment response to Recommendation 3: There were 87 respondents, of whom 73.56% (n=64) agreed or agreed with modifications, 18.40% (n=16) disagreed, and 8.04% (n=7) were neutral. There were 35 written comments received during the public comment period. These comments could be classified as follows: 1) The word “should” could be replaced with “must” or “may.” The “should” language of the recommendation was retained based on the strength of the evidence in the literature. 2) The method of TP53 mutation assessment should be addressed. As described in the text above, IHC can be used to assess the expression of p53 protein in the context of IDH-mutant gliomas based on its correlation with TP53 mutation; sequencing methods can also be employed. 3) Some commented that TP53 status should be evaluated in all IDH-mutant DG. Based on the evidence, it was concluded that this is not necessary if whole-arm 1p/19q deletion is known to be present. All comments were taken into consideration and the recommendation remained as initially stated.
Recommendation 4
Confirmation of whole arm 1p/19q codeletion in an IDH-mutant DG is essential to render the diagnoses of oligodendroglioma, IDH-mutant and 1p/19q codeleted. Landmark genomic analyses have firmly established that whole arm 1p/19q codeletion, the product of an unbalanced translocation involving chromosomes 1 and 19, is entirely restricted to gliomas that also harbor mutations in either IDH1 or IDH2.4, 6 Therefore, assessment of 1p/19q codeletion status is only relevant in IDH-mutant DG. 1p/19q testing is not recommended in IDH-WT gliomas, as false positive test results (see below) may complicate effective diagnostic categorization and the results have no bearing on patient outcome or treatment.105, 106
As described above, 1p/19q codeletion arises with near mutual exclusivity with respect to inactivating alterations of TP53 and ATRX.4, 6 Multiple studies have concluded that loss of nuclear ATRX expression by IHC is strongly predictive of a 1p/19q non-codeleted status.74, 76, 82 Confirmed TP53 mutation or p53 overexpression is similarly inversely associated with 1p/19q codeletion.74, 75 Loss of ATRX expression or TP53 mutation has been noted in a small minority of IDH-mutant gliomas that also exhibit 1p/19q codeletion (2-8%).4, 6, 74, 76, 82 The extent to which these cases reflect false positive testing for 1p/19q is unclear. Notably, TCGA analysis, which employed cytogenomic arrays to confirm whole arm 1p/19q codeletion, revealed low rates of coinciding ATRX or TP53 mutations (2-3%),6 suggesting that under strict testing criteria, the mutual exclusivity between ATRX/TP53 mutations and 1p/19q codeletion is near absolute. Nevertheless, the small degree of overlap between these biomarkers may justify 1p/19q codeletion assessment in some IDH-mutant gliomas with established ATRX loss or TP53 mutation/p53 overexpression if oligodendroglioma is suspected based on histopathologic features. As mentioned above, rare cases of carefully documented “dual genotype” IDH-mutant DG have demonstrated the co-occurrence of ATRX loss, TP53 mutation and whole arm 1p/19q codeletion.95-97 Nonetheless, for most cases, 1p/19q testing is no longer needed if an IDH-mutant DG demonstrates ATRX loss and TP53 mutation, as highlighted in cIMPACT-NOW update 2.98
1p/19q codeletion is optimally assessed by cytogenomic array or NGS methodologies capable of definitively demonstrating deletion of whole chromosomal arms of 1p and 19q.35, 107 Other approaches, such as fluorescence in situ hybridization (FISH) and PCR-based testing, have also been effectively applied to 1p/19q analysis4, 74, 76, 82 and have lower cost and quicker turnaround time than their genomic counterparts. DNA methylation profiling is capable of identifying a methylome highly characteristic of oligodendroglioma and has the added benefit of providing copy number alterations that can directly inform the status of 1p/19q codeletion.12
FISH is the most widely used test and has the advantage of assessing a limited sample of tissue (2 slides; a minimum of 50 tumor nuclei required per slide), which is often the case in small biopsy samples. One must take into account the possibility of false positive results, attributable to partial arm deletions that are detected by FISH, yet do not represent whole arm deletions associated with the unbalanced translocation. False positive test results on 1p/19q testing by FISH are also noted in IDH-WT glioblastomas, since they are genomically unstable,105, 106 underlying our recommendation that 1p/19q assessment be exclusively restricted to IDH-mutant gliomas.
Public comment response to Recommendation 4: There were 89 respondents, of whom 85.39% (n=76) agreed or agreed with modifications, 11.24% (n=10) disagreed, and 3.71% (n=3) were neutral. Thirty comments were received. One comment suggested that the term “codeletional status” used in the prior version of the recommendation be removed and replaced with “codeletion” for simplicity. The text was modified in response to this recommendation. Multiple comments suggested that 1p/19q testing be performed more broadly, on all DG or on all IDH-mutant DG. The panel did not reach these conclusions based on review of the evidence, and the detailed reasoning is explained in the preceding paragraph. A related series of comments was made regarding the importance of p53 IHC or TP53 mutational analysis in the classification of DG. One suggested using the phrase “ATRX loss with or without TP53 mutations” instead of “ATRX loss or TP53 mutations”. While it was acknowledged that IHC for p53 has limitations, and that TP53 mutations may rarely co-occur with 1p/19q codeletion in IDH-mutant gliomas (see above), the evidence suggests that TP53 mutations are one of the defining genetic characteristics of IDH-mutant astrocytoma (see preceding sections) and its inclusion is justified in the diagnostic workup of IDH-mutant DG. Multiple comments argued for the formal incorporation of histopathological criteria in addition to or in lieu of biomarkers for the classification of oligodendroglioma. Based on our review of the evidence, the designation of glioma subclasses based on IDH mutation and 1p/19q codeletion is more robust and clinically meaningful than histopathology alone.4, 6, 74, 76, 82 There is also considerable evidence showing that the interpretation of histopathological features varies considerably, even among experienced pathologists. Multiple comments suggested that pediatric gliomas be explicitly excluded from this recommendation. The sentiment underlying this suggestion is correct, given that oligodendroglioma, IDH-mutant, 1p/19q codeleted is rare in children. However, since IDH mutations occur predominantly in adults and our recommendation for 1p/19q codeletion assessment is only for IDH-mutant DG, testing in pediatric gliomas is essentially ruled out in the vast majority of cases. Finally, multiple comments urged specific recommendations regarding the most appropriate testing modalities for 1p/19q assessment. This point was addressed in the preceding paragraphs.
5. Conditional Recommendation. – CDKN2A/B homozygous deletion testing should be performed on IDH-mutant astrocytomas.
The quality of evidence to support this recommendation was assessed as moderate.
Quality Summary: The evidence base informing this recommendation comprises two prospective cohort studies77, 78 and nine retrospective cohort studies.41, 84, 85, 99, 108-112 The included studies were assessed as intermediate,77 intermediate-low,78 low,41, 84, 85, 99, 110-112 and very low quality108, 109 based on risk of bias in selection,41, 77, 78, 84, 85, 99, 108-112 performance,78, 99, 108, 112 detection,41, 77, 84, 85, 108-110, 112, and reporting41, 77, 78, 84, 85, 99, 108-112 domains. Additionally, two of the studies reported statistical analyses that were underpowered,109, 112 and one did not report on sources of funding.109 Although the aggregate risk of bias across the evidence base was very serious, the evidence was up-graded based on a strong association between CDKN2A/B deletion and diffuse glioma WHO classification. Refer to the SDC for the quality assessment of included retrospective cohort studies and the GRADE Quality of Evidence Assessment. See Table 5 for a summary of the mutational status of CDKN2A/B across all diffuse glioma subtypes.
Table 5.
Representative Studies Reporting on CDKN2A Status Across Diffuse Glioma Subtypes
| Study, Study Design |
Number DG Cases |
Testing Method |
CDKN2A Mutation Frequencya |
Study Conclusion |
|---|---|---|---|---|
| Reis et al,99 2015, RCS | n=270 | FISH | A: Loss, 48.15% (52/108) | CDKN2A loss was associated with shorter survival among grade 2/3 IDH-mutant DG that had TP53 mutations and ATRX loss. |
| OA: Loss, 38.78% (19/49) | ||||
| OD: Loss, 23.96% (23/96) | ||||
| GBM: NR | ||||
| Purkait et al,109 2013, RCS | n=67 | FISH | A: NR | CDKN2A deletion was observed GBMs and showed strong correlation with loss of p16 expression by IHC. |
| OA: NR | ||||
| OD: NR | ||||
| GBM: Loss, 43.1%b | ||||
| Collins et al,108 2014, RCS | n=267 | CGH array | A: Hemizygous loss, 30.56% (11/36) Homozygous loss, 2.78% (1/36) | Homozygous deletion of CDKN2A was seen at greater frequency in DG of higher histologic grade, especially GBM. |
| OA: Hemizygous loss, 33.33% (2/6) Homozygous loss, 0.00% (0/6) | ||||
| OD: NR | ||||
| GBM: Hemizygous loss, 29.78% (67/225) Homozygous loss, 31.56% (71/225) | ||||
| Molenaar et al,41 2014, RCS | n=98 | MLPA | A: NR | CDKN2A alteration was not associated with survival in patients with histologically diagnosed GBM. |
| OA: NR | ||||
| OD: NR | ||||
| GBM: Hemizygous loss, 27.6% Homozygous loss, 43.9% | ||||
| Shirahata et al,110 2018, RCS | n=211 | 450k array | A: NR | CDKN2A/B homozygous deletion significantly associated with overall survival in IDH mutant astrocytomas. |
| OA: NR | ||||
| OD: NR | ||||
| GBM: NR | ||||
| Yang et al,85 2020, RCS | n=160 | FISH | A: 15.00% (24/160) | CDKN2A deletion can be used to risk stratify grade 2/3 astrocytomas. |
| OA: NR | ||||
| OD: NR | ||||
| GBM: NR |
Mutational frequencies refer to histologic classifications. See full data tables in the supplemental digital content for all outcomes reported by included studies
Study did not report raw values for CDKN2A loss in primary GBMs.
Abbreviations: A, astrocytoma histology; ATRX, ATRX chromatin remodeler; CDKN2A, Cyclin Dependent Kinase Inhibitor 2A; CDKN2B, Cyclin Dependent Kinase Inhibitor 2B; CGH, comparative genomic hybridization; DG, diffuse glioma; FISH, Fluorescence in situ hybridization; GBM, glioblastoma; IDH, Isocitrate Dehydrogenase; IHC, immunohistochemistry; MLPA, multiplex ligation-dependent probe amplification; mut, mutation; NR, not reported; OA, oligoastrocytoma histology; OD, oligodendroglioma histology; RCS, retrospective cohort study; TP53, Tumor protein p53; w/o, without.
CDKN2A lies adjacent to CDKN2B in a region of chromosome 9 that is frequently mutated and deleted in a wide variety of human cancers. CDKN2A/B deletion has been shown by multiple investigations to be an adverse prognostic factor in IDH-mutant astrocytomas.41, 85, 99, 108, 109, 111, 112 Subsequent analyses of multiple cohorts of IDH-mutant astrocytomas showed that homozygous deletion of CDKN2A/B was the most relevant adverse prognostic indicator.110 Patients with histologic grade 3 IDH-mutant astrocytomas that harbored homozygous deletion of CDKN2A/B had shorter survivals than patients with histologic grade 4 tumors that did not have CDKN2A/B homozygous deletion. Since homozygous deletion of CDKN2A/B can be observed in other types of primary brain tumors that have highly variable clinical outcomes, including IDH-WT glioblastoma, pleomorphic xanthoastrocytoma, and anaplastic astrocytoma with piloid features, this recommendation pertains specifically to IDH-mutant astrocytomas.
In addition to studies captured in our systematic review, other studies1, 113 have shown similar associations between CDKN2A/B homozygous deletion and shorter survival among patients with IDH-mutant astrocytomas. Such findings culminated in the cIMPACT-NOW recommendation that CDKN2A/B homozygous deletion could be used as a grade 4 criterion in a histologic grade 2 or 3 IDH-mutant astrocytoma11 and led to its inclusion as a grade 4 criterion within the WHO, 5th Edition.7 While not a part of the current recommendations, CDKN2A/B homozygous deletion has also been shown to be a marker of poor prognosis in oligodendroglioma, IDH-mutant, 1p/19q codeleted, which may be relevant to future recommendations or grading schemes for these tumors.111
Public comment response to Recommendation 5: Of the 86 total respondents to this recommendation, 76.74% (n=66) agreed or agreed with modification, 9.30% (n=8) disagreed, and 13.95% (n=12) were neutral. (Total does not equal 100% due to rounding.) Twenty-three written comments were received. Some respondents expressed a desire to strengthen the language of the recommendation from “may be performed” to “should be performed.” In response to this suggestion, the recommendation was strengthened to “should be performed” based on review of the evidence. Others requested advice regarding methods, desired reconciliation with subsequent cIMPACT-NOW 5 statements, or underscored the importance of the distinction between heterozygous and homozygous loss of CDKN2A/B. The evidence suggests that only homozygous deletion of CDKN2A/B is strongly and reproducibly associated with decreased survival that is independent of tumor grade. These comments were taken into consideration and specific concerns were addressed and clarified in the preceding paragraphs.
6. Strong Recommendation. – MGMT promoter methylation testing should be performed on all GBM IDH-WT.
The quality of evidence to support this recommendation was assessed as moderate.
Quality Summary: This recommendation was informed by two meta-analyses,114, 115 three genome sequencing studies,5, 21, 22 and five retrospective cohort studies.86, 108, 116-118 The included meta-analyses were assessed as high115 and high-intermediate114 quality. Neither of these two studies reported on using publication status as a study selection inclusion criteria,114, 115 and one did not report on conflict of interest or sources of funding.114 The genome sequencing studies were all assessed as intermediate-low quality based on retrospective acquisition of samples in all of them,5, 21, 22 plus individual moderate risk of reporting21 and detection bias.5, 21 Finally, the retrospective cohort studies were assessed as low86, 116, 117 and very low quality108, 118 based on risk of bias. The aggregate risk of bias of the evidence base was serious and the evidence was not further up- or downgraded for any domain. Refer to the SDC for the quality assessment of genome sequencing studies, retrospective cohort studies, meta-analyses, and the GRADE Quality of Evidence Assessment. See Table 6 for a summary of the promoter methylation status of MGMT in glioblastomas.
Table 6.
Representative Studies Reporting on MGMT Promoter Methylation in Glioblastomas
| Study, Study Design |
Number GBM Cases |
Testing Method |
Promoter Methylation Frequencya |
Study Conclusion |
|---|---|---|---|---|
| Houdova Megova et al,31 2017, RCS | n=145 | MSP | IDHwt GBM: 47.78% (43/90) | IDH1 mutations are closely associated with MGMTp methylation in DG. IDH1 mutations in GBMs were a stronger marker of overall patient survival than MGMTp methylation |
| IDHmut GBM:87.50% (7/8) | ||||
| Lee et al,36 2017, RCS | n=65 | Sequencing, MSP | IDHwt GBM: 50.00% (26/52) | MGMTp methylation was much more common in IDH-mutant DG than in IDH-WT DG. |
| IDHmut GBM: 76.92% (10/13) | ||||
| Mulholland et al,143 2012, RCS | n=172 | Pyrosequencing, MSP, RT-PCR | GBM: 52.33% (90/172) | Nearly all IDH-mutant DG (127 of 129) showed MGMTp methylation by quantitative analysis of CpG sites following pyrosequencing. |
| Bady et al,116 2012, RCS | n=174 | BeadChip array | GBM: NR | Two sites within MGMTp were critical for gene silencing. A strong link was noted between MGMTp methylation and G-CIMP status. |
Mutational frequencies refer to histologic classifications. See full data tables in the supplemental digital content for all outcomes reported by included studies
Abbreviations: DG, diffuse glioma; GBM, glioblastoma; G-CIMP, Glioma-CpG island methylator phenotype; IDH1, Isocitrate Dehydrogenase (NADP(+))1; MGMT, O-6-Methylguanine-Deoxiribose Nucleic Acid Methyltransferase; MGMTp, O-6-Methylguanine-Deoxiribose Nucleic Acid Methyltransferase promoter; MSP, methylation-specific polymerase chain reaction; mut, mutation; NR, not reported; RCS, retrospective cohort study; RT-PCR – real time polymerase chain reaction; wt, wild type
The MGMT (O-6-Methylguanine-DNA Methyltransferase) protein, encoded by the MGMT gene, binds to DNA and repairs mutations that occur during DNA replication. Through these actions, it is a key mediator of resistance to alkylating chemotherapy in the treatment of DG. Randomized studies have shown that the clinical benefit of adding temozolomide to radiotherapy is predominantly among patients with MGMT promoter methylated GBM (“predictive marker”).119, 120 Temozolomide monotherapy in newly diagnosed GBM is only effective in patients with tumors that have MGMT promoter methylation.121, 122 In GBM patients treated with combined temozolomide chemo-irradiation, MGMT promoter methylation is an established prognostic parameter. In a large randomized trial on temozolomide dose intensification, MGMT promoter methylation was associated with improved OS (21.2 months versus 14 months; HR, 1.74; P < .001).123 Clinical benefit of treatment with temozolomide or lomustine at the time of tumor progression after initial therapy is particularly observed in patients with methylated MGMT promoter at the time of first diagnosis.124-126 Assessment of MGMT promoter methylation can be useful for making treatment decisions for patients when either radiotherapy or chemotherapy is considered contra-indicated, and in patients with recurrent tumors for whom second line alkylating chemotherapy is unlikely to be successful.
The expression of the MGMT protein is inhibited by methylation of specific CpG sites in the MGMT promoter and laboratory testing is therefore focused on these regions.116, 127 Despite the overall correlation between MGMT promoter methylation and clinical outcome, test characteristics remain suboptimal. MGMT promoter methylation results are affected by the choice of CpG islands and promoter regions that are tested, as well as the platform employed. Techniques used to assess MGMT promoter methylation status include qualitative or quantitative methylation specific PCR,128 pyrosequencing,129 multiplex ligation-dependent probe amplification,130 genome wide methylation analysis,116 and IHC.131 With any of these available assays, the determination of the optimal technical cut off between methylated and unmethylated remains a challenge.132 In one study that compared 5 techniques for measuring MGMT promoter methylation on a series of 100 GBMs, the percentage of tumors with promoter methylation varied between 30% and 60%, a difference that was attributed to both variations between techniques and between laboratories.123, 133, 134
A major obstacle for current studies on the interpretation and optimization of these techniques is the lack of randomized trials with an untreated control arm that would allow assessment of the predictive value of MGMT promoter methylation. Current studies therefore assess the prognostic significance of various MGMT promoter methylation tests in GBM patients being treated with chemo-irradiation with temozolomide134-136 or the concordance of results between various techniques.129 Individual detection techniques show only modest correlation when compared to one another.137, 138 MGMT protein expression by IHC was found unreliable in one series due to large interobserver variation, yet another study concluded that IHC results correlated with methylation-specific-PCR.131, 139, 140 Also, in some studies MGMT promoter status was found to vary between paired samples obtained at the time of first diagnosis and progression.141 With a quantitative methylation-specific-PCR technique, analysis within cohorts have revealed a clinically relevant grey zone between MGMT promoter clearly methylated and unmethylated tumors. Establishing an appropriate threshold within this test grey zone has substantial implications for patients in clinical trials in which temozolomide is excluded from therapy based on MGMT methylation status.142
Public comment response to Recommendation 6: Of the 86 respondents, 76.74% (n=66) agreed or agreed with modifications, 9.30% (n=8) disagreed, and 13.95% (n=12) were neutral. (Total does not equal 100% due to rounding.) Twenty-five comments were received. Comments emphasized that patients with GBM that had an unmethylated MGMT promoter may also benefit from the addition of temozolomide to radiotherapy, assumed the impact was mainly prognostic and stated that the need for testing depended on clinical decision making and treatment planning. While most of the statement remained unchanged, the panel clarified that the recommendation applies to all GBM IDH-WT.
7. Conditional Recommendation. – For IDH-mutant DG, MGMT promoter methylation testing may not be necessary.
The quality of evidence to support this recommendation was assessed as low. Quality Summary: The evidence base informing this recommendation comprises one genome sequencing study5 and four low quality retrospective cohort studies.31, 36, 40, 143 The genome sequencing study5 was assessed as intermediate-low quality based on a serious risk of selection bias and a moderate risk of detection bias. All retrospective cohort studies were limited by critical risk of selection bias.31, 36, 40, 143 Individual studies were further limited by risk of bias in performance,31, 40 reporting,31, 36, 40, 143 and detection domains,36, 143 as well as underpowered statistical analyses in two,31, 143 and a lack of reporting funding sources in one.40 The aggregate risk of bias across these studies was very serious but evidence was not further downgraded for any domain. Refer to the SDC for the quality assessment of included genome sequencing studies, retrospective cohort studies, and the GRADE Quality of Evidence Assessment.
In the section above, data supporting MGMT promoter methylation testing in GBM, IDH-WT were presented. The clinical relevance of testing for MGMT promoter methylation in IDH-mutant DG is not as firmly established. IDH mutations lead directly to global DNA hypermethylation and the establishment of the CpG island methylator phenotype (G-CIMP), as demonstrated experimentally and in numerous investigations of clinical specimens.5, 144, 145 Therefore, the prevalence of MGMT promoter methylation in IDH-mutant DG is very high .40, 144, 146, 147
Data supporting this recommendation from our systematic review documents the tight correlation between IDH-mutant DG and MGMT promoter methylation.31, 36, 40, 143, 148, 149 Using bisulfite treatment, pyrosequencing, and focusing on 16 selected CpG sites, Mulholland et al found that 127/129 (98.45%) gliomas with IDH mutations were MGMT methylated, irrespective of pathologic group.143 A strong association between IDH mutation and MGMT promoter methylation has also been demonstrated by others.31, 40 Thus, the high correlation between IDH-mutation and MGMT promoter methylation suggests that testing for MGMT promoter methylation in IDH-mutant DG may not be necessary.
Public comment response to Recommendation 7: There were 89 respondents, of whom 73.03% (n=65) agreed or agreed with modifications, 16.85% disagreed (n=15), and 10.11% (n=9) were neutral. (Total does not equal 100% due to rounding.) There were 17 written comments, many of which stated that MGMT testing decisions should be left to the treating oncologist and questioned whether it is worthwhile to issue a recommendation for every test that should not be performed. We have carefully taken these comments into consideration and have kept the recommendation as is, since MGMT promoter methylation testing is one of the most frequently requested tests for DG, and we did not find sufficient evidence for reflexively ordering it in the group of IDH-mutant DG. The strong association between IDH mutation and MGMT promoter methylation supports a role for IDH mutation (when present) resulting in MGMT promoter methylation, with the limitations of predominantly retrospective evidence. The recommendation is listed as conditional, which leaves room for individual and institutional treatment decisions.
8. Conditional Recommendation. – TERT promoter mutation testing may be used to provide further support for the diagnosis of oligodendroglioma and IDH-WT GBM.
The quality of evidence to support this recommendation was assessed as low.
Quality Summary: This recommendation was informed by two genome sequencing studies5, 6 and 11 retrospective cohort studies.4, 18, 23, 34, 49, 84, 150-154 The genome sequencing studies5, 6 were assessed as intermediate-low quality while the retrospective cohort studies were assessed as low23, 49, 84, 150, 151, 153, 154 and very low quality.4, 18, 34, 152 Included studies suffered from risk of bias in selection,4-6, 18, 23, 34, 49, 84, 150-154 performance,4, 18, 34, 150 reporting,4, 18, 23, 34, 84, 150-152, 154 and detection4, 5, 23, 34, 84, 150-152, 154 domains. The aggregate risk of bias across these studies was very serious but evidence was not further downgraded for any domain. Refer to the SDC for the quality assessment of included genome sequencing studies, retrospective cohort studies, and the GRADE Quality of Evidence Assessment. See Table 7 for a summary of the mutational status of the TERT promoter across all diffuse glioma subtypes.
Table 7.
Representative Studies Reporting on TERT Promoter, EGFR, Chromosome 7, and Chromosome 10 Status Across Diffuse Glioma Subtypes
| Study, Design |
Number DG Cases |
Marker(s) | Testing Method |
Mutation Frequencya |
Study Conclusion |
|---|---|---|---|---|---|
| McLendon et al,22 2008, GSS | n=91 | EGFR | Genome sequencing | A: NR | Highlighted core molecular pathways consistently altered in GBM, including p53, RTK and RB networks. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR | |||||
| Brat et al,6 2015, GSS | n=293 | IDH, TP53, ATRX, 1p/19q, EGFR | Genome sequencing | A: NR | IDH-WT lower grade DG had genomic alterations and clinical behavior like IDH-WT GBM. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR | |||||
| Brennan et al,21 2013, GSS | n=543 | Chr 7, Chr 10, TERTp, EGFR | Genome sequencing | A: NR | EGFR amp, TERTp mutation, Chr 7 gains and Chr 10 losses were frequent events in IDH-WT GBM. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: TP53, 27.9%b TERT, 84.0%b | |||||
| Chan et al,23 2015, RCS | n=237 | TERT | Sequencing | A: TERT, 19.05% (32/168) | Among histologic grade 2/3 IDH-WT DG, TERTp mutation was associated with short survival, similar to IDH-WT GBM. |
| OA: TERT, 39.58% (19/48) | |||||
| OD: TERT, 76.19% (16/21) | |||||
| GBM: NR | |||||
| Gao et al,150 2016, RCS | n=389 | TERT | Sequencing, Pyrosequencing | A: TERT, 26.72% (66/247) | TERTp mutations a prognostic factors among DG and inversely associated with ATRX inactivation. |
| OA: TERT, 41.30% (19/46) | |||||
| OD: TERT, 22.73% (10/44) | |||||
| GBM: TERT, 32.69% (17/52) | |||||
| Chan et al,160 2015, RCS | n=214 | TERT, EGFR | Sequencing, FISH | A: NR | Among patients with grade 2/3 IDH-WT DG, EGFR amp and TERTp mutation was associated with shorter survival, similar to IDH-WT GBM. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR | |||||
| Eckel-Passow et al,4 2015, RCS | n=615 | TERT | Sequencing | A: NR | DG classified on the basis of IDH, TERTp, and 1p/19q showed that those with TERTp mutation only had short overall survival. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR | |||||
| Labussiere et al,151 2014, RCS | n=807 | TERT | Array, PCR | A: TERT, 30.23% (13/43) | Among IDH-WT GBM, the presence of TERTp mutation or EGFR amp was associated with shorter survival. |
| OA: TERT, 31.95% (54/169) | |||||
| OD: TERT, 61.00% (122/200) | |||||
| GBM: TERT, 75.70% (299/395) | |||||
| Collins et al,108 2014, RCS | n=267 | Chr 7, Chr 10, EGFR | CGH array | A: Chr7 tri/poly, 5.56% (2/36); mono, 0.00% (0/36) Chr10 mono, 11.11% (4/36); 10q partial del, 38.89% (14/36) EGFR amp, 11.11% (4/36) | Among patients with recurrent grade 3/4 astrocytoma, EGFR amp Chr 7 gains and Chr 10 losses were associated with shorter survival. |
| OA: Chr7 tri/poly, 0.00% (0/6); mono, 0.00% (0/6) Chr10 mono, 0.00% (0/6); 10q partial del, 33.33% (2/6) EGFR amp, 0.00% (0/6) | |||||
| OD: NR | |||||
| GBM: Chr7 tri/poly, 63.56% (143/225); mono, 0.44% (1/225) Chr10 mono, 64.89% (146/225); 10q partial del, 20.00% (45/225) EGFR amp, 44.44% (100/225) | |||||
| Little et al,162 2012, RCS | n=342 | EGFR | FISH | A: EGFR amp, 15.38% (2/13) | Detailed FISH analysis of GBMs showed that 44% have EGFR amp, 21% have PDGFRA amp and 9% contain both EGFR amp and PDGFRA amp. |
| OA: NR | |||||
| OD: EGFR amp, 9.76% (4/41) | |||||
| GBM: EGFR amp, 44.17% (106/240) | |||||
| Guillaudeau et al,161 2009, RCS | n=35 | EGFR | FISH, IHC | A: EGFR amp, 0% (0/8) | EGFR amp was associated with increased protein expression of EGFR by IHC in DG |
| OA: NR | |||||
| OD: NR | |||||
| GBM: EGFR amp, 72.73% (8/11) | |||||
| Houdova Megova et al,31 2017, RCS | n=145 | EGFR | Not reported | A: EGFR amp, 7.69% (2/26) | EGFR amp and Chr 10 losses are more frequent in IDH-WT than IDH-mut DG and are most frequent in IDH-WT GBM. |
| OA: EGFR amp, 0% (0/4) | |||||
| OD: EGFR amp, 0% (0/7) | |||||
| GBM: EGFR amp, 36.56% (30/82) | |||||
| Aibaidula et al,18 2017, RCS | n=166 | TERT | Patient records | A: NR | Among adults with IDH-WT grade 2/3 DG, EGFR amp and mutations of TERTp or H3F3A K27M were associated with shorter survival. |
| OA: NR | |||||
| OD: NR | |||||
| GBM: NR |
Mutational frequencies refer to histologic classifications. See full data tables in the supplemental digital content for all outcomes reported by included studies
Study did not report raw values for mutation frequency.
Abbreviations: A, astrocytoma histology; amp, amplification; ATRX, ATRX chromatin remodeler; Chr, chromosome; del, deletion; DG, diffuse glioma; EGFR, epidermal growth factor receptor; FISH, Fluorescence in situ hybridization; GBM, glioblastoma; GSS, genome sequencing study; H3, histone3; IDH, Isocitrate Dehydrogenase; IHC, immunohistochemistry; mono, monopoly; mut, mutation; NR, not reported; OA, oligoastrocytoma histology; OD, oligodendroglioma histology; PCR, polymerase chain reaction; PDGFRA, platelet derived growth factor receptor alpha; poly, polysomy; RCS, retrospective cohort study; TERT, Telomerase Reverse Transcriptase; TERTp, TERT promoter; TP53, Tumor protein p53; tri, trisomy; wt, wild-type.
As discussed for ATRX (Recommendation 2), cancerous proliferation requires escape from cellular senescence. In oligodendrogliomas, IDH-mutant, 1p/19q codeleted and IDH-WT GBM this predominantly occurs via point mutations in the TERT promoter, which leads to enhanced TERT expression. Point mutations occur in two hotspots located 124bp and 146bp upstream of the translation start site and are referred to as C228T and C250T.34, 151 Testing for promoter mutation is performed by DNA sequence analysis, either by Sanger methods, pyrosequencing, PCR or targeted NGS.25, 34, 84, 150, 154, 155
In contrast to oligodendrogliomas and IDH-WT GBM, IDH-mutant astrocytomas primarily use the alternative lengthening of telomeres (ALT) pathway to escape senescence, which is driven by inactivation of ATRX. Notably, TERT promoter mutations and ATRX loss/mutations are mutually exclusive,4, 6, 150 consistent with our understanding that these two genetic mechanisms confer equivalent selective growth advantages. Thus, in the setting of IDH-mutation, TERT promoter mutations can provide additional support for the diagnosis of oligodendroglioma, IDH-mutant, 1p/19q codeleted, WHO grade 2 or 3. It can also provide diagnostic support in cases in which immunostaining for ATRX is equivocal or was not performed. By the same logic, TERT promoter sequencing may not be necessary in IDH-mutant gliomas with sufficient evidence of ATRX loss. Similarly, if there is laboratory evidence that clearly establishes the diagnosis of oligodendroglioma (IDH-mutant and 1p/19q codeleted), then TERT promoter testing may not be necessary.
TERT promoter mutations were initially noted in the majority of histologically defined GBMs.156, 157 Subsequently, IDH mutations and TERT promoter mutations were shown to be mutually exclusive in GBMs, with those GBMs harboring TERT promoter mutations associated with shorter survivals compared to those with IDH mutations.34, 151, 155 Further comprehensive investigations demonstrated that those adult DG that harbored TERT promoter mutations but did not have IDH-mutations, were tightly associated with genetic signatures of IDH-WT GBM and short overall patient survivals.4, 5, 158 TERT promoter mutations are not specific for IDH-WT GBM, since they have been documented in other forms of CNS neoplasia in addition to oligodendroglioma, IDH-mutant, 1p/19q codeleted.156, 158, 159 However, in the proper clinical, radiological and neuropathological setting, TERT promoter mutations may be used to support the diagnosis of IDH-WT GBM.
Public comment response to Recommendation 8: There were 88 respondents, of which 73.86% (n=69) agreed or agreed with modifications, 17.05% (n=9) disagreed, and 10.23% (n=10) were neutral. (Total does not equal 100% due to rounding.) There were 15 comments, including several that reiterated the importance of interpreting TERT promoter mutations in the context of IDH-status, as well as its established relationships with ATRX mutations and 1p/19q codeletions, as discussed above. The recommendation was unchanged.
9. Strong Recommendation. – For histologic grade 2-3 DG that are IDH-WT, testing should be performed for whole chromosome 7 gain/whole chromosome 10 loss, EGFR amplification, and TERT promoter mutation to establish the molecular diagnosis of IDH-WT GBM, grade 4.
The quality of evidence to support this recommendation was assessed as moderate.
Quality Summary: The evidence base informing this recommendation includes studies evaluating testing of chromosome 7, chromosome 10, EGFR and TERT promoter. One genome sequencing study21 and one retrospective cohort study108 comprise the evidence base for chromosome 7, one retrospective cohort study was included for chromosome 10,108 two genome sequencing studies,21, 22 and eight retrospective cohort studies18, 31, 49, 108, 160-163 for EGFR, and one genome sequencing study5 and eight retrospective cohort studies for TERT.4, 18, 23, 49, 150-153 Included studies were assessed as intermediate-low through very low quality and suffered from risk of bias in selection,4, 5, 18, 21-23, 31, 49, 108, 150-153, 160-163 performance,4, 18, 31, 108, 150, 163 reporting,4, 18, 21, 23, 31, 108, 150-152, 160, 161, 163 and detection4, 5, 21, 23, 108, 150-152, 160-162 domains. Quality of evidence was assessed for each target individually and overall for the recommendation. For both chromosome 7 and EGFR testing, the aggregate risk of bias for studies included in the evidence base was serious and evidence was not further downgraded, resulting in moderate quality of evidence. For chromosome 10 the risk of bias of the one included study was very serious, thus carrying a low quality of evidence. Finally, the quality of evidence for TERT was also low based on a very serious aggregate risk of bias and no further downgrading of quality. The overall recommendation was assessed as moderate. Refer to the SDC for the quality assessment of included genome sequencing studies, retrospective cohort studies, and the GRADE Quality of Evidence Assessment. See Table 8 for a summary of the mutational status of EGFR, TERT promoter, chromosome 7, and chromosome 10 across all diffuse glioma subtypes.
Table 8.
Representative Studies Reporting on Histone H3 Mutation Frequency Across Diffuse Glioma Subtypes
| Study, Study Design |
Number DG Cases |
Marker(s) | Testing Method |
Mutation Frequencya |
Study Conclusion |
|---|---|---|---|---|---|
| Castel et al,168 2015, RCS | n=62 | H3K27M | Sequencing, IHC | Midline Glioma: H2K27M, 95.16% (59/62) | There was a high frequency of H3-K27M mutations in DIPGs and the correlation between Sanger sequencing and IHC for H3-K37M mutations was strong. |
| Khuong-Quang et al,88 2012, RCS | n=42 | H3K27M | Sequencing | Midline Glioma: H3K27M, 71.43% (30/42) | H3-K27M mutations were found in high frequency in DIPG and had a poor prognosis compared to DIPG without this mutation. TP53 mutations co-occurred with H3-K27M mutations with high frequency. |
| Gessi et al,171 2013, RCS | n=123 | H3G34, H3G27 | Pyrosequencing, MLPA | GBM: H3G34/ H3K27 31.71% (39/123) | Histone H3-G34R mutations were identified in a subset of pediatric tumors histologically classified as GBM and as CNS-PNET. |
| CNS-PNET: H3G34/ H3K27, 12.12% (4/33) | |||||
| Korshunov et al,3 2015, RCS | n=202 | H3G34, H3K27M | 450k BeadChip array | GBM: H3G34, 11.88% (24/202) H3G34/ H3K27M, 46.04% (93/202) | Pediatric tumors diagnosed histologically as GBM had specific genetic and clinical subsets, defined by mutations in H3-K27M, H3-G34R and IDH1. |
Mutational frequencies refer to histologic classifications. See full data tables in the supplemental digital content for all outcomes reported by included studies
Abbreviations: CNS-PNET, pediatric primitive neuroectodermal tumors of the central nervous system; DG, diffuse glioma; DIPG, diffuse intrinsic pontine glioma; GBM, glioblastoma; IDH1, Isocitrate Dehydrogenase (NADP(+))1; IHC, immunohistochemistry; MLPA, multiplex ligation-dependent probe amplification; RCS, retrospective cohort study; TP53, Tumor protein p53.
A substantial subset of IDH-WT diffuse astrocytic gliomas in adults are considered grade 2 or 3 based on histologic criteria (no microvascular proliferation or necrosis), yet have an aggressive clinical course, with overall survival times similar to patients with IDH-WT GBM. Numerous studies have attempted to identify molecular genetic biomarkers that reliably identify histologic grade 2 and 3 tumors that behave most aggressively. The strongest evidence indicates that the following markers identify IDH-WT diffuse astrocytic gliomas with grade 4 clinical behavior: 1) whole chromosome 7 gain together with whole chromosome 10 loss (+7/−10); 2) EGFR amplification; or 3) TERT promoter mutation. Studies that have assessed the frequency of +7/−10, EGFR amplification and TERT promoter mutations have demonstrated higher percentages of these alterations in histologically defined GBMs as compared to histologic grade 2 or 3 DG, lending support to the association between these markers and the molecular signature of IDH-WT GBM.18, 21, 22, 31, 108, 151, 161, 162
IDH-WT diffuse astrocytic gliomas of histologic grade 2 or 3 that carry EGFR amplification are associated with significantly shorter patient survival and have outcomes similar to patients with histologically defined IDH-WT GBM.5, 6, 18, 158, 160 EGFR amplification has excellent specificity for gliomas with aggressive behavior and is not present in other glioma subtypes that display a more indolent clinical course. It is typically accompanied by gain of chromosome 7 and loss of chromosome 10 in adult DG, as well as in a smaller subset of pediatric high grade gliomas.158, 164
Similarly, those histologic IDH-WT diffuse astrocytic gliomas of histologic grade 2 or 3 with the +7/−10 signature are associated with short patient survival, similar to histologically defined IDH-WT GBM.5, 158, 165 The +7/−10 signature also has excellent specificity for DG, with the rare exception of pleomorphic xanthoastrocytomas (PXAs).158 Most investigations have focused on the prognostic role of whole gains of chromosome 7 and whole losses of chromosome 10 (+7/−10), since these are most frequent; the prognostic association of other, far less common combinations of imbalances, such as +7q/−10q, +7/−10q or +7q/−10, await further evaluation.
TERT promoter mutation in IDH-WT diffuse astrocytic gliomas of histologic grade 2 or 3 is associated with significantly shorter patient survival.4, 18, 23, 160 However, other types of IDH-WT glial neoplasms without WHO grade 4 histology or aggressive behavior also occasionally harbor TERT promoter mutations.156, 158, 159 Therefore, TERT promoter mutations can be considered a marker for grade 4 behavior if the clinical, radiologic and histopathologic features are those of diffusely infiltrative astrocytic glioma. Further, more recent investigations have concluded that a subset of histologic grade 2 IDH-WT DG that harbor only TERT promoter mutations (no other genetic alterations involving canonical IDH-WT GBM pathways) have longer clinical outcomes than expected for a grade 4 neoplasm.166, 167 The combination of TERT promoter mutation with other markers, such as EGFR amplification and +7/−10 adds specificity as a marker of grade 4 behavior.158
cIMPACT-NOW update 3 has recommended that IDH-WT DG of histologic grade 2 or 3 that have +7/−10, EGFR amplification or TERT promoter mutation should be considered grade 4 and more recently, these molecular alterations have been added as criteria for the diagnosis of GBM, IDH-WT, WHO grade 4 by the WHO 5th edition.7, 10 Clinical implications for the use of these biomarkers to establish the molecular diagnosis of IDH-WT GBM, grade 4 include the use of combined chemo- and radiotherapy and expanded clinical trial inclusion.
Public comment response to Recommendation 9: Of 89 respondents, 86.52% (n=77) agreed or agreed with modifications, 6.74% (n=6) disagreed, and 6.74% (n=6) were neutral. Twenty written comments were received, many expressing concern about the use of “grade IV astrocytic glioma.” As “astrocytic glioma” is not currently an official WHO class, respondents requested that the statement be changed to “molecular diagnosis of GBM.” Additional comments requested that the recommendation include ‘and’ instead of ‘or’ for the target testing to reduce turnaround time. These suggestions were incorporated into the final recommendation.
10. Strong Recommendation. – H3 K27M testing must be performed in DG that involve the midline in the appropriate clinical and pathologic setting.
The quality of evidence to support this recommendation was assessed as moderate.
Quality Summary: This recommendation was informed by two retrospective cohort studies.88, 168 Both retrospective studies were limited by risk of bias in selection, performance, reporting, and detection domains.88, 168 In addition, one study reported underpowered statistical analyses.168 The aggregate risk of bias for the evidence base was very serious and the quality of evidence was up-graded based on a strong association between H3 K27M testing and DG diagnosis in the WHO classification. Refer to the SDC for the quality assessment of included retrospective cohort studies and the GRADE Quality of Evidence Assessment. See Table 8 for a summary of the mutational status of histone H3 in glioblastoma, midline glioma, and pediatric primitive neuroectodermal tumors of the central nervous system.
Diffuse midline glioma with histone H3 K27M mutation was a newly recognized diagnostic entity in the 4th Edition update of the WHO Classification of Tumours of the Central Nervous System (2016).1 The diagnosis relies on the histological identification of diffusely infiltrating glioma within midline CNS structures such as spinal cord, brainstem, or thalamus and the identification of K27M mutation in either H3F3A or HIST1H3B/C using anti-H3K27M IHC or by sequencing methods.90, 169 By histology, tumors can show a large degree of histo- and cytopathological variability, ranging from a low density infiltrate to an undifferentiated, high grade neoplasm. Regardless of histologic features, the presence of an H3 K27M mutation in a diffusely infiltrative glioma involving the midline most often predicts clinically aggressive behavior and poor prognosis, leading to its designation of WHO grade 4.88, 168 While the number of studies investigating this particular mutation is small, the implications for diagnosis, prognosis and treatment are critically important. This underlies the strong recommendation that H3 K27M testing must be performed in DG that involve the midline in the appropriate clinical and pathologic setting. As most of these tumors occur in the pediatric population, this recommendation for additional testing is essential for children and young adults. For older adults there is increasing evidence that the midline location of a DG is also tightly linked with H3 K27M mutation, warranting testing of all patients with midline gliomas in order to guide appropriate care.91
H3 K27M testing can be performed using anti-H3 K27M IHC, which displays strong nuclear staining with relatively high sensitivity for the mutation, ranging from approximately 71% to 100% when compared with sequencing methods.88, 168, 170-172 IHC can be particularly useful in small biopsy samples, when tissue is limited. Several anti-H3 K27M antibodies are commercially available for on-site laboratory development, and reference laboratories also offer testing. Importantly, the staining pattern of a positive test is nuclear, and therefore staining of cytoplasmic and/or cellular processes should not be interpreted as positive. Confirmatory molecular sequencing may be of utility in equivocal cases. DG that have H3 K27M mutations also show loss of H3 K27M trimethylation (H3K27me3), which can be detected by loss of nuclear immunoreactivity for H3 K27me3 by IHC.173 A smaller subset of diffuse midline gliomas that show loss of H3 K27me3 do not have H3 K27M mutations, but rather have EGFR mutations (see Good Practice Statement 1) or overexpression of EZHIP.172, 174, 175
Public comment response to Recommendation10: There were 88 respondents, of whom 96.59% (n=85) agreed or agreed with modifications and 3.37% (n=3) disagreed. There were 24 written comments. Originally, the statement included that the recommendation applied to “the appropriate clinical and pathologic setting.” Some commented that this phrase needed to be further explained. There were also suggestions that the statement should recommend that the H3 K27M test phrasing of “should” be performed rather than “must” be performed. Others indicated that the methods of testing (eg, IHC versus sequencing) should be described. These comments were taken into consideration. While the recommendation remained the same, these comments have been addressed in the preceding paragraphs.
11. Conditional Recommendation. – H3 G34 testing may be performed in pediatric and young adults with IDH-WT DG.
The quality of evidence to support this recommendation was assessed as low.
Quality Summary: The evidence base informing this recommendation comprises two low quality retrospective cohort studies.3, 171 In addition to suffering from critical risk of selection bias, both studies were limited by a moderate risk of reporting bias3, 171 and one was also limited by risk of performance and detection bias.3 The aggregate risk of bias for the studies was very serious and the evidence was not further downgraded for any domain. Refer to the SDC for the quality assessment of included retrospective cohort studies and the GRADE Quality of Evidence Assessment. See Table 8 for a summary of the mutational status of histone H3 in glioblastoma, midline glioma, and pediatric embryonal tumors of the central nervous system.
A hotspot mutation in the histone gene H3F3A found within a subset of pediatric and young adult high-grade gliomas confers either a G34R or G34V substitution in the gene product.3, 171, 176, 177 G34R/V mutant diffuse gliomas occur in a somewhat older age group in comparison with K27M cases (median age, 19-years for G34R/V mutation; median age 6 for K27M mutation) and tend to involve the cerebral hemispheres rather than midline locations.176 Prognosis for G34R/V-mutant gliomas is poor but somewhat better than the K27M-mutant gliomas (median survival of 12 months for K27M and 24 months for G34R or G34V). “Diffuse hemispheric glioma H3 G34-mutant” was recently introduced as a distinct entity in the WHO 5th Edition, underscoring the importance of testing for this molecular alteration.7 Importantly, these DG with G34R/V mutations should not be lumped together with other IDH WT gliomas, as they carry a disease defining genetic alteration that directs aggressive behavior corresponding to a WHO grade 4 neoplasm regardless of histologic appearance.3, 171 Testing by sequencing or with mutation-specific (H3 G34R) antibodies is recommended for non-midline DG in pediatric and young adult populations.
Public comment response to Recommendation 11: There were 89 respondents, of whom 83.15% (n=74) agreed or agreed with modification, 4.49% (n=4) disagreed, and 12.36% (n=11) were neutral. There were several written comments, including one that suggested that the age of the patient should be considered but that older adults might also benefit from testing for some cases. Others indicated that the methods of testing (ie, IHC versus sequencing) should be described. These comments were taken into consideration. While the recommendation remained the same, the comments were addressed in the text above.
12. Conditional Recommendation. – BRAF mutation testing (V600) may be performed in DG that are IDH-WT and H3-WT.
The quality of evidence to support this recommendation was assessed as low.
Quality Summary: The evidence base informing this recommendation consists of one intermediate quality prospective cohort study77 and four retrospective cohort studies.18, 178-180 Two included retrospective studies were assessed as very low quality based on serious or critical risk of bias across more than one domain.18, 178 The other two retrospective studies were also assessed as very low quality and this was based on critical risk of selection bias, moderate risk of bias in other domains, underpowered statistical analyses, and a lack of funding source reporting.179, 180 The aggregate risk of bias for the studies was very serious but evidence was not further downgraded for any domain. Refer to the SDC for the quality assessment of retrospective cohort studies and the GRADE Quality of Evidence Assessment. See Table 9 for a summary of the mutational status of BRAF, MYB/MYBL1, and FGFR1 across diffuse glioma subtypes.
Table 9.
Representative Studies Reporting on BRAF, MYB/MYBL1, and FGFR1 Status Across Diffuse Glioma Subtypes
| Study, Study Design |
Number DG Cases |
Marker | Testing Method |
Mutation Frequencya |
Study Conclusion |
|---|---|---|---|---|---|
| Korshunov et al,179 2017, RCS | n=64 | BRAF V600E | 450k array | E-GBM: BRAF, 56.25% (36/64) | Epithelioid GBMs resolve into three genetic entities. Those clustering with PXA and IDHwt GBM by methylation are enriched for BRAF V600E mutations. |
| Kleinschmidt-DeMasters et al,178 2013, RCS | n=24 | BRAF V600E | IHC, PCR | E-GBM: BRAF, 53.85% (7/13) | BRAF V600E mutations were noted in a substantial percentage of epithelioid GBMs, but not rhabdoid or giant cell GBMs. |
| Nakajima et al,180 2017, RCS | n=14 | BRAF V600E | Sequencing, CGH array | E-GBM: BRAF, 92.86% (13/14) | Epithelioid GBMs are characterized by BRAF V600E mutations, TERTp mutations, and CDKN2A/B deletions, most often in combination. |
| Aibaidula et al,18 2017, RCS | n=166 | BRAF V600E, MYB | Sequencing, FISH | IDHwt (all DGs): BRAF, 6.02% (10/166) MYB, 19.88% (33/166) | Subset of adult IDH wild-type histologic grade 2/3 DGs harbor clinically relevant BRAF V600E mutations and MYB amps. |
| Tatevossian et al,183 2010, RCS | n=77 | MYB | SNP array | A: MYB amp, 14.29% (2/14); poly, 7.14% (1/14) | MYB amp was described in a subset of pediatric low grade DA, leading to increased gene and protein expression. Focal truncating deletion of MYB was noted in AG. |
| Ramkisson et al,184 2013, RCS | n=44 | MYB | Copy number analysis | A: MYBL1 rearrangement, 26.32% (5/19) | Recurrent truncating duplications of MYBL1 were present in a subset of low grade pediatric DA. Focal truncating deletion of MYB was noted in AG. |
| Qaddoumi et al,2 2016, RCS | n=91 | FGFR1 | Sequencing | A: FGFR1/ 3, 23.53% (4/17) | Alterations in BRAF, FGFR1, or MYB/MYBL1 were solitary pathogenic alterations in subsets of low grade pediatric DG, often corresponding to histologic class. |
| OD: FGFR1/ 3, 61.90% (26/42) |
Mutational frequencies refer to histologic classifications. See full data tables in the supplemental digital content for all outcomes reported by included studies
Abbreviations: A, astrocytoma histology; AG, angiocentric glioma; amp, amplification; BRAF, B-Raf proto-oncogene; CDKN2A, Cyclin Dependent Kinase Inhibitor 2A; CDKN2B, Cyclin Dependent Kinase Inhibitor 2B; CGH, comparative genomic hybridization; DA, diffuse astrocytoma; DG, diffuse glioma; E-GBM, epithelioid glioblastoma histology; FGFR1, Fibroblast Growth Factor Receptor 1; FISH, Fluorescence in situ hybridization; GBM, glioblastoma; OD, oligodendroglioma histology; IDH, Isocitrate Dehydrogenase; IHC, immunohistochemistry; MYB, MYB proto-oncogene; MYB1, MYB proto-oncogene like 1; NR, not reported; PCR, polymerase chain reaction; poly, polysomy; PXA, pleomorphic xanthoastrocytomas; RCS, retrospective cohort study; SNP, single nucleotide polymorphism; TERTp, Telomerase Reverse Transcriptase promoter; wt, wild-type.
BRAF V600E mutation is a driver mutation that is present in a wide variety of neoplasms, including melanoma, hairy cell leukemia, papillary thyroid carcinoma, colorectal carcinoma, lung carcinoma, Langerhans cell histiocytosis, Erdheim Chester disease, papillary craniopharyngioma, and ameloblastoma. It is present in a minority of DG, but prevalence is enriched in epithelioid GBMs178-180 and other IDH- and H3-WT DG.2, 18 Of note, BRAF V600E mutation is distinct from the KIAA1549:BRAF fusion and other types of BRAF fusion, which are especially frequent in cerebellar pilocytic astrocytomas.
In addition to these studies, the recommendation was influenced by the availability of targeted therapy181 and the occurrence of BRAF V600E within a variety of brain tumors other than DG, where the mutation is more prevalent; these include PXAs, ganglioglioma (GG), and a minority of pilocytic astrocytomas.2, 182 Although BRAF V600E mutation occurs in a higher proportion of these generally circumscribed neuroepithelial tumors, PXA, GG, and pilocytic astrocytoma fell outside of the scope and search criteria used to develop these guidelines. However, histologic overlap between these entities can be considerable; given the presence of targeted therapy, a liberal approach to testing was favored in borderline or ambiguous cases where the distinction between a diffuse and circumscribed neoplasm could not be made with certainty.
This recommendation is most relevant to IDH- and H3-WT DG in which BRAF V600E mutations have been shown to be enriched, including: epithelioid GBMs and cases overlapping with PXA or anaplastic PXA; pediatric DG or tumors overlapping with GG or pilocytic astrocytoma; and histologic grade 2 and 3 DG lacking a “GBM molecular signature”.10 Evidence for BRAF V600E testing in the setting of an IDH-WT GBM with typical histologic features or with canonical genetic alterations in older adults is currently lacking.
While many of the markers considered in this guideline pertain principally to DG in adults, it should be noted that a small number of pediatric DG with BRAF V600E mutation have been identified,2 and that BRAF V600E testing should be considered in the context of IDH- and H3-WT DG in children.9 This is particularly important given relatively common histologic overlap between circumscribed and DG in children.
Public comment response to Recommendation 12: Of the 89 respondents to this recommendation, 84.27% (n=75) agreed or agreed with modifications, 11.23% (n=10) disagreed, and 4.49% (n=4) were neutral. (Total does not equal 100% due to rounding.) Of 24 written comments, 7 solicited advice as to the best method of testing, and 5 emphasized the importance of clinical and pathological correlation. The remaining 12 comments expressed diverse thoughts, both supportive and skeptical, including concern about the difference between the strength of the recommendation and the level of evidence (2 comments). These helpful comments were taken into account and the initial recommendation was streamlined to encompass all IDH- and H3-WT DG (within the appropriate clinicopathologic context) instead of epithelioid GBM and “other” IDH- and H3-WT DG. We also included a more specific description of the appropriate clinicopathologic context for testing in the text above.
13. Conditional Recommendation. – MYB/MYBL1 and FGFR1 testing may be performed in children and young adults with DG that are histologic grade 2-3 and are IDH-WT and histone H3-WT.
The quality of evidence to support this recommendation was assessed as low.
Quality Summary: The evidence base informing this recommendation includes three retrospective cohort studies evaluating MYB/MYBL1 testing18, 183, 184 and one retrospective cohort study evaluating FGFR1 testing.2 The four studies were assessed as low183, 184 and very low quality2, 18 based on risk of bias in selection,2, 18, 183, 184 performance,18 reporting,2, 18, 183, 184 and detection2, 183, 184 domains. Quality of evidence was assessed for each target individually and then overall for the entire statement. The aggregate risk of bias for MYB/MYBL1 studies was very serious but evidence was not further downgraded for any domain. The risk of bias for the one FGFR1 study was very serious. As only one study was identified, quality of evidence is dependent only on the risk of bias. The quality of evidence for the entire recommendation was assessed as low. Refer to the SDC for the quality assessment of included retrospective cohort studies and the GRADE Quality of Evidence Assessment. See Table 9 for a summary of the mutational status of BRAF, MYB/MYBL1, and FGFR1 across diffuse glioma subtypes.
Recent studies have identified subsets of DG in children and young adults that lack alterations in IDH1, IDH2 or histone H3 genes. In addition to BRAF, mentioned above, genes that are recurrently altered in this group include v-Myb avian myeloblastosis viral oncogene homolog (MYB), v-myb avian myeloblastosis viral oncogene homolog-like 1 (MYBL1), and FGFR1. These tumors are technically IDH- and H3- WT, but in retrospective studies they seem to exhibit an indolent clinical course, especially when compared to most DG of adults.18 Genetic mechanisms of activation of these drivers are heterogeneous, which may affect the availability of specific assays for detection. These include duplications, truncations, amplifications and rearrangements resulting in fusion genes (MYB and MYBL1) and internal tandem duplications of tyrosine kinase domains and single nucleotide variants (FGFR1). The prevalence of these alterations is difficult to assert with certainty, since most of the early data was from small retrospective studies. Integrated molecular and clinical analysis of a large cohort of pediatric low grade gliomas provide insight into the spectrum and frequency of these alterations.185 Specific genetic alterations seem to correlate with glioma subtypes, with MYBL1 rearrangements being reported in up to 50% pediatric diffuse astrocytoma,184 MYB amplifications/copy number gains in 2/14 (14%) pediatric diffuse astrocytoma183 and 33/166 (20%) adult IDH WT lower grade gliomas.18 Conversely, in one study FGFR1 alterations were frequent in rare low-grade neoplasms with ‘oligodendroglial’ phenotype affecting children and young adults, 39/42 (92.86%).2 In addition, MYB-QKI fusions are found in nearly all angiocentric glioma (WHO grade 1), a low grade DG often associated with epilepsy.186 Given the diagnostic and prognostic implications for these genetic drivers in gliomas of children and young adults, testing for them may be advisable depending on laboratory capabilities, but the quality of evidence is lower than that supporting testing for other more commonly altered genes (eg, IDH1/2 and BRAF).
Public comment response to Recommendation 13: There were 88 respondents, of whom 75% (n=66) agreed or agreed with modifications, 9.09% (n=8) disagreed, and 15.91% (n=14) were neutral. There were 19 written comments, including a number that suggested that evidence for the recommendation was not strong and the targets are not actionable; questions about testing methods; requests to clarify what is meant by “in the appropriate clinical setting” and inclusion of other genetic drivers (ie, panels). These comments were taken into consideration and the recommendation was revised (may instead of should) or addressed by the other recommendations (eg, testing for BRAF).
GOOD PRACTICE STATEMENTS
During the period of the systematic literature review and drafting of recommendations, several clinically relevant investigations were published on the topic of biomarker testing for DG subtypes not originally covered by the Patient/population, Intervention, Comparator, and Outcome (PICO) frameworks. In order to include guidance on these subtypes, we performed a focused literature review and included Good Practice Statements based on this second review. A description of the methods used to formulate these statements is included in the SDC.
The targeted search included alterations of ALK receptor tyrosine kinase (ALK), EGFR, ROS proto-oncogene 1 (ROS1), Neurotrophic tyrosine receptor kinase (NTRK), MET, PDGFRA, and MYCN proto-oncogene (MYCN) within bithalamic gliomas, infantile-type hemisphere gliomas, or diffuse pediatric-type high grade gliomas. The panel viewed the statements below as important, but there was not a formal rating for the certainty of evidence. The decision to form these statements was based on 1) emerging evidence with limited number of studies and samples and 2) anticipated updates to the WHO occurring at the time that this manuscript was being written. The panel believed that the tests would provide prognostic or treatment-related information for clinicians to consider. Future updates to the guideline will address these tests using a formal rating of the evidence and will include a grade for the strength of recommendation. Refer to Table 10 for the list of Good Practice Statements.
Table 10.
Summary of the Good Practice Statements (GPS)
| GPS1. EGFR mutation testing may be performed on diffuse gliomas that involve the midline. |
| GPS2. Testing for alterations in ALK, ROS1, MET and NTRK genes may be performed on cerebral hemispheric diffuse gliomas of infancy that are wildtype for IDH and histone H3. |
| GPS3. Testing for DNA methylation class or for alterations in PDGFRA, EGFR, MYCN may be performed in pediatric high grade diffuse gliomas that are wildtype for IDH and histone H3. |
Statement 1. EGFR mutation testing may be performed on diffuse gliomas that involve the midline.
A small subset of diffusely infiltrating midline gliomas that harbor EGFR mutations have a distinct methylation profile.174, 187 While most of these DG with EGFR mutations are histone H3-wildtype, some contain H3 K27M mutations. Similar to H3-K27 mutant gliomas involving the midline, EGFR-mutant DG in this location that are histone H3-wildtype typically show loss of H3 K27M trimethylation (H3K27me3), which can be detected by IHC. Small in-frame insertions/duplications of the tyrosine kinase domain and point mutations of the extracellular domain are detected by NGS and are potential therapeutic targets.
Statement 2. Testing for alterations in ALK, ROS1, MET and NTRK genes may be performed on cerebral hemispheric diffuse gliomas of infancy that are wildtype for IDH and histone H3.
DG that occur in infancy are usually molecularly distinct from those of older childhood and adults. These large cerebral hemispheric tumors are typically wildtype for IDH and histone H3 and often harbor fusions (rarely other activating alterations) involving ROS1, ALK, MET or the NTRK family.188, 189 The identification of such fusions or other activating alterations could lead to targeted therapy. Fusions can be identified by NGS and by FISH if appropriate probes are available.
Statement 3. Testing for DNA methylation class or for alterations in PDGFRA, EGFR, MYCN may be performed in pediatric high grade diffuse gliomas that are wildtype for IDH and histone H3.
Among pediatric diffuse high grade gliomas that are IDH-wildtype and histone H3-wildtype, three molecular subclasses can be identified by DNA methylation profiling that have differing molecular genetic alterations and clinical outcomes. One class is generally characterized by PDGFRA amplifications or mutations; a second class enriched for EGFR amplifications is associated with the longest survival; and the third often harbors MYCN amplifications and is associated with the shortest survival.164, 190
FUTURE CONSIDERATIONS
The recommendations provided are based on evidence available at the time of literature review. It is expected that additional studies and new technologies will emerge that will need to be incorporated into future guidelines. In the category of IDH-WT diffuse astrocytic gliomas of adults, it has been recommended that histologic grade 2 and 3 DG should be tested for chromosome 7 gain/whole chromosome 10 loss, EGFR amplification, and TERT promoter mutation to establish the molecular diagnosis of GBM, grade 4. It might be expected that additional biomarkers or combinations of biomarkers will be identified and validated in histologic grade 2 and 3 DG that are also capable of predicting grade 4 clinical behavior, including alterations in PDGFRA, FGFR3, c-Met, TP53, NF1, PTEN, among others.10 For IDH-mutant astrocytomas of histologic grade 2 and 3, future studies could uncover and validate additional markers that can more optimally stratify risk or predict grade 4 clinical behavior in addition to CDKN2A/B homozygous deletion, such as PDGFRA, MYCN or CDK4 amplifications, mutations in CDKN2A/B or Rb1, among others.11, 65, 85 CDKN2A/B homozygous deletion have recently been shown to be a marker of poor prognosis in oligodendroglioma, IDH-mutant, 1p/19q codeleted, which along with other biomarkers, may be relevant to future recommendations or grading schemes.111 Similarly, it is likely that genetic or epigenetic markers will be uncovered and validated that are capable of stratifying risk among patients with diffuse gliomas driven by histone H3, Mitogen-activated protein kinase (MAPK) pathway or MYB/MYBL1 genes.9, 191
The laboratory findings of tumor mutational burden (TMB), mismatch repair (MMR) loss and microsatellite instability (MSI) are of interest in multiple forms of cancer, since the findings from these tests predict response to immunotherapies, including immune checkpoint inhibitors in some cancer types.192-194 Initial randomized clinical trials of checkpoint inhibitors in newly diagnosed and recurrent glioblastoma have failed to demonstrate improved survival.195 Guidelines for testing of TMB, MMR or MSI in DG will be developed if and when clinical care warrants them.
Whole genome DNA methylation profiling represents a robust and reproducible method for precisely segregating tumor types that have similar genetic alterations, and epigenetic signatures and clinical behaviors.5, 12 IDH-wildtype GBM, IDH-mutant astrocytomas, oligodendrogliomas, IDH-mutant, 1p/19q codeleted, and histone H3 mutant gliomas cluster tightly within their own class with little overlap based on DNA methylation profiling.5, 6, 22 Other DG, such as those that occur in children and are driven by MAPK pathway or MYB/MYBL1 genomic alteration also cluster together, indicating a high degree of specificity for each signature.164, 190 While the literature indicates this method is superior for classification purposes and could have a role in future classification and grading, it currently lacks widespread clinical implementation due to regulatory challenges, prevailing practice patterns and uncertainties in reimbursement. Currently, DNA methylation profiling is an attractive alternative for identifying DG classes that will likely be implemented into clinical practice in the near future.
LIMITATIONS
We were unable to answer KQs 1b. (What are the acceptable techniques/methods for mutation testing of DG and what are the expected turn-around-times for individual assays?) While we collected the various methods of testing, none performed poorly in our assessment. Each have varying strengths and weaknesses. For the latter question, no studies from the literature search reported turn-around time data and so, no recommendation was made. It is up to each laboratory to determine the best methodology and acceptable turn-around-times. Although the individual DG entities have prognostic characteristics that have been well defined in the literature,2-7 a majority of the studies that comprised the evidence base for the recommendations correlated the reported outcomes to DG entities and not to survival outcomes. Additional studies that investigate outcomes related to biomarkers and DG subtypes would increase certainty of the evidence base and lead to stronger recommendations.
CONCLUSIONS
The 13 evidence-based recommendations and three good practice statements that are provided for biomarker testing of DG, together with their explanations and justifications, are intended to guide practice and improve the clinical care of patients with these diseases. For more than a century, the diagnosis of DG was based primarily on histologic appearance, yet recent clinically relevant molecular genetic discoveries have forced a re-evaluation of diagnostic definitions and criteria. We are now firmly within the molecular era in which integrated diagnoses are formulated by directly incorporating molecular biomarker test results. The complexity of establishing an integrated diagnosis has increased and pace of change has been rapid, warranting the recommendations that we have provided. Neuropathologists, molecular pathologists and treating physicians will continue to work closely together to provide optimal clinical care by utilizing molecular biomarkers for diagnostic and treatment purposes.
Supplementary Material
Acknowledgements
We thank the collaborating medical societies and their staff involved in the development of this guideline: American Association of Neuropathologists, Association for Molecular Pathology, Society for Neuro-Oncology. We thank the American Society of Clinical Oncology for identifying representatives to serve on the expert panel. We also thank the advisory panel members representing these organizations for their guidance throughout the development of the guideline and for their thoughtful review of this work: Tracy T. Batchelor, MD, David W. Ellison, MD, PhD, Matthew Foster, MD, Azra H. Ligon, PhD, Keith L. Ligon, MD, PhD, Morgan Mosky, patient advocate, Joanna J. Phillips, MD, PhD, Gordana Raca, MD, PhD, Sharleen Rapp, BS, MB(ASCP)cm, Michelle Stoffel, MD, PhD, Mary Kay Washington, MD, PhD.
Appendix - Disclosed Interests and Activities from July 2017 to April 2021
Daniel J. Brat, MD, PhD
Position of Influence: Society for Neuro-Oncology, American Association of Neuropathologists
Julia A. Bridge, MD
Research Grant: Cepheid
Consulting fees or Advisory Board: Bayer, Bristol Myers Squibb/National Comprehensive Cancer Network, Epizyme, Merck, Moffitt Cancer Center, Translational Genomics Research Institute, VENTANA/Roche
Position of Influence: United States & Canadian Academy of Pathology
Howard Colman, MD, PhD
Consulting fees or Advisory Board: AbbVie, Adastra Pharmaceuticals, Bayer, Best Doctors, CytRx Corporation, F. Hoffman-La Roche, Forma Therapeutics, Foundation Medicine, Genentech, Insys Therapeutics, Karyopharm Therapeutics, NewLink Genetics, Novocure, Omniox, Orbus Therapeutics, OXiGENE, Private Health, Upsher-Smith
Speakers' bureau/lecture fees/honoraria: Merck
Institutional contracts/clinical trials: AbbVie, Array BioPharma, Bayer, BeiGene, Bristol Myers Squibb, DNAtrix, Forma Therapeutics, Global Coalition for Adaptive Research, Kadmon Holdings, Karyopharm Therapeutics, Merck, NewLink Genetics, Nuvation Bio, Orbus, Plexxikon,
Meera R. Hameed, MD
Consulting fees or Advisory Board: Novartis
Eyas M. Hattab, MD, MBA
Consulting fees or Advisory Board: Arbor Pharmaceuticals, Cell Marque, E.R. Squibb&Sons, Bristol Myers Squibb, Ventana Medical System/Roche Diagnostics
Speakers' bureau/lecture fees/honoraria: Arbor Pharmaceuticals, Roche Diagnostics
Position of Influence: College of American Pathologists, United States & Canadian Academy of Pathologists
Other: International Collaboration on Cancer Reporting member
Jason T. Huse, MD, PhD
Research Grants: Taiho Oncology, Inc.
Martin J. van den Bent, MD, PhD
Consulting fees or Advisory Board: AbbVie, Agios, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, CarThera, Celgene, Celldex Therapeutics, Genenta Science, Karyopharm Therapeutics, Nerviano Medical Sciences, VAXIMM
Speakers' bureau/lecture fees/honoraria: Merck Sharp & Dohme Corp
Research Grants: AbbVie
No Disclosures to Report
Arie Perry, MD; Brent T. Harris, MD, PhD; Michelle Hawks Yount, MS; Fausto J. Rodriguez, MD; Robert B. Jenkins, MD, PhD; William C. McDonald, MD; Kenneth Aldape, MD; Peter Canoll, MD, PhD; Dolores H. Lopez-Terrada, MD, PhD; Lesley H. Souter, PhD; Carol Colasacco, MLIS, SCT(ASCP); Nicole E. Thomas, MPH, CT(ASCP)cm
Footnotes
Disclaimer
The College of American Pathologists (CAP) developed the Pathology and Laboratory Quality Center for Evidence-based Guidelines as a forum to create and maintain laboratory practice guidelines (LPGs). Guidelines are intended to assist physicians and patients in clinical decision-making and to identify questions and settings for further research. With the rapid flow of scientific information, new evidence may emerge between the time an LPG is developed and when it is published or read. LPGs are not continually updated and may not reflect the most recent evidence. LPGs address only the topics specifically identified therein and are not applicable to other interventions, diseases, or stages of diseases. Furthermore, guidelines cannot account for individual variation among patients and cannot be considered inclusive of all proper methods of care or exclusive of other treatments. It is the responsibility of the treating physician or other health care provider, relying on independent experience and knowledge, to determine the best course of treatment for the patient. Accordingly, adherence to any LPG is voluntary, with the ultimate determination regarding its application to be made by the physician in light of each patient’s individual circumstances and preferences. The CAP and its collaborators make no warranty, express or implied, regarding LPGs and specifically excludes any warranties of merchantability and fitness for a particular use or purpose. The CAP and its collaborators assume no responsibility for any injury or damage to persons or property arising out of or related to any use of this statement or for any errors or omissions.
Contributor Information
Daniel J. Brat, Department of Pathology, Northwestern University Feinberg School of Medicine, Chicago, IL.
Kenneth Aldape, Laboratory of Pathology, National Cancer Institute, Bethesda, MD.
Julia A. Bridge, Department of Pathology and Microbiology, University of Nebraska Medical Center, Omaha, NE; Cytogenetics, ProPath, Dallas, TX.
Peter Canoll, Department of Pathology and Cell Biology, Columbia University Medical Center, New York, NY.
Howard Colman, Department of Neurosurgery and Huntsman Cancer Institute, University of Utah, Salt Lake City, UT.
Meera R. Hameed, Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, NY.
Brent T. Harris, Department of Neurology and Pathology, MedStar Georgetown University Hospital, Washington, DC.
Eyas M. Hattab, Department of Pathology and Laboratory Medicine, University of Louisville, Louisville, KY.
Jason T. Huse, Departments of Pathology and Translational Molecular Pathology, University of Texas MD, Anderson Cancer Center, Houston, TX.
Robert B. Jenkins, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN.
Dolores H. Lopez-Terrada, Departments of Pathology and Pediatrics, Baylor College of Medicine and Texas Children’s Hospital, Houston, TX.
William C. McDonald, Pathology Department, Abbott Northwestern Hospital, Minneapolis, MN.
Fausto J. Rodriguez, Department of Pathology, The Johns Hopkins Hospital, Baltimore, MD.
Lesley H. Souter, private practice, Wellandport, Ontario, Canada.
Carol Colasacco, Surveys, College of American Pathologists, Northfield, IL.
Nicole E. Thomas, Surveys, College of American Pathologists, Northfield, IL.
Michelle Hawks Yount, Patient Advocate, Brookfield, WI.
Martin J. van den Bent, Brain Tumor Center at Erasmus MC Cancer Institute University Medical Center Rotterdam, Rotterdam, The Netherlands.
Arie Perry, Departments of Pathology and Neurological Surgery University of California San Francisco School of Medicine, San Francisco, CA.
References
- 1.Louis DN, Oghaki H, Wiestler OD, Caenee WK, eds. Who Classification of Tumours of the Central Nervous System. 4th ed. WHO Press; 2016. World Health Organization Classification of Tumours, vol 1. [Google Scholar]
- 2.Qaddoumi I, Orisme W, Wen J, et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016;131(6):833–845. doi: 10.1007/s00401-016-1539-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korshunov A, Ryzhova M, Hovestadt V, et al. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol. 2015;129(5):669–678. doi: 10.1007/s00401-015-1405-4 [DOI] [PubMed] [Google Scholar]
- 4.Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. doi: 10.1056/NEJMoa1407279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brat DJ, Verhaak RGW, Aldape KA, et al. ; Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Classification of Tumours Editorial Board. WHO Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer; 2021. World Health Organization Classification of Tumours, 5th ed.; vol 1. [Google Scholar]
- 8.Louis DN, Ellison DW, Brat DJ, et al. cIMPACT-NOW: a practical summary of diagnostic points from round 1 updates. Brain Pathol. 2019;29(4):469–472. doi: 10.1111/bpa.12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison DW, Hawkins C, Jones DTW, et al. cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAF(v600e) mutation. Acta Neuropathol. 2019;137(4):683–687. doi: 10.1007/s00401-019-01987-0 [DOI] [PubMed] [Google Scholar]
- 10.Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for "diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO Grade IV". Acta Neuropathol. 2018;136(5):805–810. doi: 10.1007/s00401-018-1913-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020;139(3):603–608. doi: 10.1007/s00401-020-02127-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. doi: 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham R, Mancher M, Wolman D, Greenfield S, Steinberg E, eds; Committee on Standards for Developing Trustworthy Clinical Practice Guidelines. Clinical Practice Guidelines We Can Trust. National Academies Press; 2011. [PubMed] [Google Scholar]
- 14.Lefebvre C, Manheimer E, Glanville J. Chapter 6: Searching for studies. In: Higgins JPT, Green S, eds. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0. The Cochrane Collaboration and John Wiley & Sons Inc.; 2011:137–138. [Google Scholar]
- 15.Guyatt G, Oxman AD, Akl EA, et al. Grade guidelines: 1. Introduction: GRADE evidence profiles and summary of findings tables. J Clin Epidemiol. 2011;64(4):383–394. doi: 10.1016/j.jclinepi.2010.04.026 [DOI] [PubMed] [Google Scholar]
- 16.Alonso-Coello P, Schunemann HJ, Moberg J, et al. Grade evidence to decision (EtD) frameworks: a systematic and transparent approach to making well informed healthcare choices. 1: Introduction. BMJ. 2016;353:i2016. doi: 10.1136/bmj.i2016 [DOI] [PubMed] [Google Scholar]
- 17.Ahmadi R, Stockhammer F, Becker N, et al. No prognostic value of IDH1 mutations in a series of 100 WHO grade II astrocytomas. J Neurooncol. 2012;109(1):15–22. doi: 10.1007/s11060-012-0863-y [DOI] [PubMed] [Google Scholar]
- 18.Aibaidula A, Chan AK, Shi Z, et al. Adult IDH wild-type lower-grade gliomas should be further stratified. Neuro Oncol. 2017;19(10):1327–1337. doi: 10.1093/neuonc/nox078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alentorn A, van Thuijl HF, Marie Y, et al. Clinical value of chromosome arms 19q and 11p losses in low-grade gliomas. Neuro Oncol. 2014;16(3):400–408. doi: 10.1093/neuonc/not227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boots-Sprenger SH, Sijben A, Rijntjes J, et al. Significance of complete 1p/19q co-deletion, IDH1 mutation and MGMT promoter methylation in gliomas: use with caution. Mod Pathol. 2013;26(7):922–929. doi: 10.1038/modpathol.2012.166 [DOI] [PubMed] [Google Scholar]
- 21.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLendon R, Friedman A, Bigner D, et al. ; Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan AK, Yao Y, Zhang Z, et al. TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Mod Pathol. 2015;28(2):177–186. doi: 10.1038/modpathol.2014.94 [DOI] [PubMed] [Google Scholar]
- 24.Chaurasia A, Park SH, Seo JW, Park CK. Immunohistochemical analysis of ATRX, IDH1 and p53 in glioblastoma and their correlations with patient survival. J Korean Med Sci. 2016;31(8):1208–1214. doi: 10.3346/jkms.2016.31.8.1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubbink HJ, Atmodimedjo PN, Kros JM, et al. Molecular classification of anaplastic oligodendroglioma using next-generation sequencing: a report of the prospective randomized EORTC Brain Tumor Group 26951 phase III trial. Neuro Oncol. 2016;18(3):388–400. doi: 10.1093/neuonc/nov182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebrahimi A, Skardelly M, Bonzheim I, et al. ATRX immunostaining predicts IDH and H3F3A status in gliomas. Acta Neuropathol Commun. 2016;4(1):60. doi: 10.1186/s40478-016-0331-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felsberg J, Wolter M, Seul H, et al. Rapid and sensitive assessment of the IDH1 and IDH2 mutation status in cerebral gliomas based on DNA pyrosequencing. Acta Neuropathol. 2010;119(4):501–507. doi: 10.1007/s00401-010-0647-4 [DOI] [PubMed] [Google Scholar]
- 28.Fontana L, Tabano S, Bonaparte E, et al. MGMT-methylated alleles are distributed heterogeneously within glioma samples irrespective of IDH status and chromosome 10q deletion. J Neuropathol Exp Neurol. 2016;75(8):791–800. doi: 10.1093/jnen/nlw052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gravendeel LA, Kloosterhof NK, Bralten LB, et al. Segregation of non-p.R132H mutations in IDH1 in distinct molecular subtypes of glioma. Hum Mutat. 2010;31(3):E1186–1199. doi: 10.1002/humu.21201 [DOI] [PubMed] [Google Scholar]
- 30.Hattori N, Hirose Y, Sasaki H, et al. World Health Organization grade II-III astrocytomas consist of genetically distinct tumor lineages. Cancer Sci. 2016;107(8):1159–1164. doi: 10.1111/cas.12969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houdova Megova M, Drabek J, Dwight Z, et al. Isocitrate dehydrogenase mutations are better prognostic marker than O6-methylguanine-DNA methyltransferase promoter methylation in glioblastomas - a retrospective, single-centre molecular genetics study of gliomas. Klin Onkol. 2017;30(5):361–371. doi: 10.14735/amko2017361 [DOI] [PubMed] [Google Scholar]
- 32.Jha P, Suri V, Sharma V, et al. IDH1 mutations in gliomas: first series from a tertiary care centre in India with comprehensive review of literature. Exp Mol Pathol. 2011;91(1):385–393. doi: 10.1016/j.yexmp.2011.04.017 [DOI] [PubMed] [Google Scholar]
- 33.Juratli TA, Kirsch M, Robel K, et al. IDH mutations as an early and consistent marker in low-grade astrocytomas WHO grade II and their consecutive secondary high-grade gliomas. J Neurooncol. 2012;108(3):403–410. doi: 10.1007/s11060-012-0844-1 [DOI] [PubMed] [Google Scholar]
- 34.Killela PJ, Pirozzi CJ, Healy P, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–1525. doi: 10.18632/oncotarget.1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labussiere M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–1890. doi: 10.1212/WNL.0b013e3181e1cf3a [DOI] [PubMed] [Google Scholar]
- 36.Lee Y, Koh J, Kim SI, et al. The frequency and prognostic effect of TERT promoter mutation in diffuse gliomas. Acta Neuropathol Commun. 2017;5(1):62. doi: 10.1186/s40478-017-0465-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewandowska MA, Furtak J, Szylberg T, et al. An analysis of the prognostic value of IDH1 (isocitrate dehydrogenase 1) mutation in Polish glioma patients. Mol Diagn Ther. 2014;18(1):45–53. doi: 10.1007/s40291-013-0050-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li MY, Wang YY, Cai JQ, et al. Isocitrate dehydrogenase 1 gene mutation is associated with prognosis in clinical low-grade gliomas. PLoS ONE. 2015;10(6):e0130872. doi: 10.1371/journal.pone.0130872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mellai M, Piazzi A, Caldera V, et al. IDH1 and IDH2 mutations, immunohistochemistry and associations in a series of brain tumors. J Neurooncol. 2011;105(2):345–357. doi: 10.1007/s11060-011-0596-3 [DOI] [PubMed] [Google Scholar]
- 40.Minniti G, Scaringi C, Arcella A, et al. IDH1 mutation and MGMT methylation status predict survival in patients with anaplastic astrocytoma treated with temozolomide-based chemoradiotherapy. J Neurooncol. 2014;118(2):377–383. doi: 10.1007/s11060-014-1443-0 [DOI] [PubMed] [Google Scholar]
- 41.Molenaar RJ, Verbaan D, Lamba S, et al. The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro Oncol. 2014;16(9):1263–1273. doi: 10.1093/neuonc/nou005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukasa A, Takayanagi S, Saito K, et al. Significance of IDH mutations varies with tumor histology, grade, and genetics in Japanese glioma patients. Cancer Sci. 2012;103(3):587–592. doi: 10.1111/j.1349-7006.2011.02175.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. doi: 10.1158/1078-0432.CCR-09-0715 [DOI] [PubMed] [Google Scholar]
- 44.Olar A, Wani KM, Alfaro-Munoz KD, et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol. 2015;129(4):585–596. doi: 10.1007/s00401-015-1398-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polivka J, Polivka J Jr., Rohan V, et al. Isocitrate dehydrogenase-1 mutations as prognostic biomarker in glioblastoma multiforme patients in West Bohemia. Biomed Res Int. 2014;2014:735659. doi: 10.1155/2014/735659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi ST, Yu L, Lu YT, et al. IDH mutations occur frequently in Chinese glioma patients and predict longer survival but not response to concomitant chemoradiotherapy in anaplastic gliomas. Oncol Rep. 2011;26(6):1479–1485. doi: 10.3892/or.2011.1428 [DOI] [PubMed] [Google Scholar]
- 47.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. doi: 10.1200/JCO.2009.21.9832 [DOI] [PubMed] [Google Scholar]
- 48.Shibahara I, Sonoda Y, Kanamori M, et al. IDH1/2 gene status defines the prognosis and molecular profiles in patients with grade III gliomas. Int J Clin Oncol. 2012;17(6):551–561. doi: 10.1007/s10147-011-0323-2 [DOI] [PubMed] [Google Scholar]
- 49.Bienkowski M, Wohrer A, Moser P, et al. Molecular diagnostic testing of diffuse gliomas in the real-life setting: a practical approach. Clin Neuropathol. 2018;37(4):166–177. doi: 10.5414/NP301110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajeswarie RT, Rao S, Nandeesh BN, Yasha TC, Santosh V. A simple algorithmic approach using histology and immunohistochemistry for the current classification of adult diffuse glioma in a resource-limited set-up. J Clin Pathol. 2018;71(4):323–329. doi: 10.1136/jclinpath-2017-204638 [DOI] [PubMed] [Google Scholar]
- 51.Pollack IF, Hamilton RL, Sobol RW, et al. IDH1 mutations are common in malignant gliomas arising in adolescents: a report from the Children's Oncology Group. Childs Nerv Syst. 2011;27(1):87–94. doi: 10.1007/s00381-010-1264-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong QH, Li KK, Wang WW, et al. Molecular landscape of IDH-mutant primary astrocytoma grade IV/glioblastomas. Mod Pathol. 2021;34(7):1245–1260. doi: 10.1038/s41379-021-00778-x [DOI] [PubMed] [Google Scholar]
- 53.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capper D, Reuss D, Schittenhelm J, et al. Mutation-specific IDH1 antibody differentiates oligodendrogliomas and oligoastrocytomas from other brain tumors with oligodendroglioma-like morphology. Acta Neuropathol. 2011;121(2):241–252. doi: 10.1007/s00401-010-0770-2 [DOI] [PubMed] [Google Scholar]
- 55.DeWitt JC, Jordan JT, Frosch MP, et al. Cost-effectiveness of IDH testing in diffuse gliomas according to the 2016 WHO classification of tumors of the central nervous system recommendations. Neuro Oncology. 2017;219(12):1640–1650. doi: 10.1093/neuonc/nox120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Preusser M, Wohrer A, Stary S, Hoftberger R, Streubel B, Hainfellner JA. Value and limitations of immunohistochemistry and gene sequencing for detection of the IDH1-R132H mutation in diffuse glioma biopsy specimens. J Neuropathol Exp Neurol. 2011;70(8):715–723. doi: 10.1097/NEN.0b013e31822713f0 [DOI] [PubMed] [Google Scholar]
- 57.Pyo JS, Kim NY, Kim RH, Kang G. Concordance analysis and diagnostic test accuracy review of IDH1 immunohistochemistry in glioblastoma. Brain Tumor Pathol. 2016;33(4):248–254. doi: 10.1007/s10014-016-0272-6 [DOI] [PubMed] [Google Scholar]
- 58.Banan R, Stichel D, Bleck A, et al. Infratentorial IDH-mutant astrocytoma is a distinct subtype. Acta Neuropathol. 2020;140(4):569–581. doi: 10.1007/s00401-020-02194-y [DOI] [PubMed] [Google Scholar]
- 59.Carter JH, McNulty SN, Cimino PJ, et al. Targeted next-generation sequencing in molecular subtyping of lower-grade diffuse gliomas: application of the World Health Organization's 2016 revised criteria for central nervous system tumors. J Mol Diagn. 2017;19(2):328–337. doi: 10.1016/j.jmoldx.2016.10.010 [DOI] [PubMed] [Google Scholar]
- 60.Catteau A, Girardi H, Monville F, et al. A new sensitive PCR assay for one-step detection of 12 IDH1/2 mutations in glioma. Acta Neuropathol Commun. 2014;2:58. doi: 10.1186/2051-5960-2-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Zhao YY, Li JF, et al. IDH1 mutation detection by droplet digital PCR in glioma. Oncotarget. 2015;6(37):39651–39660. doi: 10.18632/oncotarget.5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2 [DOI] [PubMed] [Google Scholar]
- 63.Mellai M, Annovazzi L, Senetta R, et al. Diagnostic revision of 206 adult gliomas (including 40 oligoastrocytomas) based on ATRX, IDH1/2 and 1p/19q status. J Neurooncol. 2017;131(2):213–222. doi: 10.1007/s11060-016-2296-5 [DOI] [PubMed] [Google Scholar]
- 64.Zhou YX, Wang JX, Feng M, et al. Analysis of isocitrate dehydrogenase 1 mutation in 97 patients with glioma. J Mol Neurosci. 2012;47(3):442–447. doi: 10.1007/s12031-011-9681-5 [DOI] [PubMed] [Google Scholar]
- 65.Bai H, Harmanci AS, Erson-Omay EZ, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48(1):59–66. doi: 10.1038/ng.3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barthel FP, Johnson KC, Varn FS, et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature. 2019;576(7785):112–120. doi: 10.1038/s41586-019-1775-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lass U, Numann A, von Eckardstein K, et al. Clonal analysis in recurrent astrocytic, oligoastrocytic and oligodendroglial tumors implicates IDH1- mutation as common tumor initiating event. PLoS ONE. 2012;7(7):e41298. doi: 10.1371/journal.pone.0041298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mazor T, Chesnelong C, Pankov A, et al. Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc Natl Acad Sci U S A. 2017;114(40):10743–10748. doi: 10.1073/pnas.1708914114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zacher A, Kaulich K, Stepanow S, et al. Molecular diagnostics of gliomas using next generation sequencing of a glioma-tailored gene panel. Brain Pathol. 2017;27(2):146–159. doi: 10.1111/bpa.12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai J, Yang P, Zhang C, et al. ATRX mRNA expression combined with IDH1/2 mutational status and Ki-67 expression refines the molecular classification of astrocytic tumors: evidence from the whole transcriptome sequencing of 169 samples samples. Oncotarget. 2014;5(9):2551–2561. doi: 10.18632/oncotarget.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cryan JB, Haidar S, Ramkissoon LA, et al. Clinical multiplexed exome sequencing distinguishes adult oligodendroglial neoplasms from astrocytic and mixed lineage gliomas. Oncotarget. 2014;5(18):8083–8092. doi: 10.18632/oncotarget.2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hewer E, Vajtai I, Dettmer MS, Berezowska S, Vassella E. Combined ATRX/IDH1 immunohistochemistry predicts genotype of oligoastrocytomas. Histopathology. 2016;68(2):272–278. doi:doi.org/ 10.1111/his.12743 [DOI] [PubMed] [Google Scholar]
- 73.Ikemura M, Shibahara J, Mukasa A, et al. Utility of ATRX immunohistochemistry in diagnosis of adult diffuse gliomas. Histopathology. 2016;69(2):260–267. doi: 10.1111/his.12927 [DOI] [PubMed] [Google Scholar]
- 74.Rajmohan KS, Sugur HS, Shwetha SD, et al. Prognostic significance of histomolecular subgroups of adult anaplastic (WHO grade III) gliomas: applying the 'integrated' diagnosis approach. J Clin Pathol. 2016;69(8):686–694. doi: 10.1136/jclinpath-2015-203456 [DOI] [PubMed] [Google Scholar]
- 75.Shao LW, Pan Y, Qi XL, et al. ATRX loss in adult supratentorial diffuse astrocytomas correlates with p53 over expression and IDH1 mutation and predicts better outcome in p53 accumulated patients. Histol Histopathol. 2016;31(1):103–114. doi: 10.14670/HH-11-664 [DOI] [PubMed] [Google Scholar]
- 76.Wiestler B, Capper D, Holland-Letz T, et al. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126(3):443–451. doi: 10.1007/s00401-013-1156-z [DOI] [PubMed] [Google Scholar]
- 77.Jonsson P, Lin AL, Young RJ, et al. Genomic correlates of disease progression and treatment response in prospectively characterized gliomas. Clin Cancer Res. 2019;25(18):5537–5547. doi: 10.1158/1078-0432.CCR-19-0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pessoa IA, Amorim CK, Ferreira WAS, et al. Detection and correlation of single and concomitant TP53, PTEN, and CDKN2A alterations in gliomas. Int J Mol Sci. 2019;20(11):2658. doi: 10.3390/ijms20112658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Figarella-Branger D, Bouvier C, de Paula AM, et al. Molecular genetics of adult grade II gliomas: towards a comprehensive tumor classification system. J Neurooncol. 2012;110(2):205–213. doi: 10.1007/s11060-012-0953-x [DOI] [PubMed] [Google Scholar]
- 80.Gorovets D, Kannan K, Shen R, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18(9):2490–2501. doi: 10.1158/1078-0432.CCR-11-2977 [DOI] [PubMed] [Google Scholar]
- 81.Ruano Y, Ribalta T, de Lope AR, et al. Worse outcome in primary glioblastoma multiforme with concurrent epidermal growth factor receptor and p53 alteration. Am J Clin Pathol. 2009;131(2):257–263. doi: 10.1309/AJCP64YBDVCTIRWV [DOI] [PubMed] [Google Scholar]
- 82.Sahm F, Reuss D, Koelsche C, et al. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol. 2014;128(4):551–559. doi: 10.1007/s00401-014-1326-7 [DOI] [PubMed] [Google Scholar]
- 83.Wang PF, Liu N, Song HW, et al. IDH-1R132H mutation status in diffuse glioma patients: implications for classification. Oncotarget. 2016;7(21):31393–31400. doi: 10.18632/oncotarget.8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Higa N, Akahane T, Yokoyama S, et al. A tailored next-generation sequencing panel identified distinct subtypes of wildtype IDH and TERT promoter glioblastomas. Cancer Sci. 2020;111(10):3902–3911. doi: 10.1111/cas.14597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang RR, Shi ZF, Zhang ZY, et al. IDH mutant lower grade (WHO grades II/III) astrocytomas can be stratified for risk by CDKN2A, CDK4 and PDGFRA copy number alterations. Brain Pathol. 2020;30(3):541–553. doi: 10.1111/bpa.12801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hartmann C, Hentschel B, Simon M, et al. Long-term survival in primary glioblastoma with versus without isocitrate dehydrogenase mutations. Clin Cancer Res. 2013;19(18):5146–5157. doi: 10.1158/1078-0432.CCR-13-0017 [DOI] [PubMed] [Google Scholar]
- 87.Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425. doi: 10.1126/science.1207313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124(3):439–447. doi: 10.1007/s00401-012-0998-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pathak P, Jha P, Purkait S, et al. Altered global histone-trimethylation code and H3F3A-ATRX mutation in pediatric GBM. J Neurooncol. 2015;121(3):489–497. doi: 10.1007/s11060-014-1675-z [DOI] [PubMed] [Google Scholar]
- 90.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- 91.Solomon DA, Wood MD, Tihan T, et al. Diffuse midline gliomas with histone H3-k27m mutation: a series of 47 cases assessing the spectrum of morphologic variation and associated genetic alterations. Brain Pathol. 2016;26(5):569–580. doi: 10.1111/bpa.12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olar A, Tran D, Mehta VP, et al. ATRX protein loss and deregulation of PI3K/AKT pathway is frequent in pilocytic astrocytoma with anaplastic features. Clin Neuropathol. 2019;38(2):59–73. doi: 10.5414/NP301105 [DOI] [PubMed] [Google Scholar]
- 93.Rodriguez FJ, Brosnan-Cashman JA, Allen SJ, et al. Alternative lengthening of telomeres, ATRX loss and H3-K27M mutations in histologically defined pilocytic astrocytoma with anaplasia. Brain Pathol. 2019;29(1):126–140. doi: 10.1111/bpa.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reuss DE, Sahm F, Schrimpf D, et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an "integrated" diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2015;129(1):133–146. doi: 10.1007/s00401-014-1370-3 [DOI] [PubMed] [Google Scholar]
- 95.Huse JT, Diamond EL, Wang L, Rosenblum MK. Mixed glioma with molecular features of composite oligodendroglioma and astrocytoma: a true "oligoastrocytoma"? Acta Neuropathol. 2015;129(1):151–153. doi: 10.1007/s00401-014-1359-y [DOI] [PubMed] [Google Scholar]
- 96.Qu M, Olofsson T, Sigurdardottir S, et al. Genetically distinct astrocytic and oligodendroglial components in oligoastrocytomas. Acta Neuropathol. 2007;113(2):129–136. doi: 10.1007/s00401-006-0142-0 [DOI] [PubMed] [Google Scholar]
- 97.Zepeda-Mendoza CJ, Vaubel RA, Zarei S, et al. Concomitant 1p/19q co-deletion and IDH1/2, ATRX, and TP53 mutations within a single clone of "dual-genotype" IDH-mutant infiltrating gliomas. Acta Neuropathol. 2020;139(6):1105–1107. doi: 10.1007/s00401-020-02141-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Louis DN, Giannini C, Capper D, et al. cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol. 2018;135(4):639–642. doi: 10.1007/s00401-018-1826-y [DOI] [PubMed] [Google Scholar]
- 99.Reis GF, Pekmezci M, Hansen HM, et al. CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II-III) astrocytomas. J Neuropathol Exp Neurol. 2015;74(5):442–452. doi: 10.1097/NEN.0000000000000188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–468. doi: 10.1038/ng.3273 [DOI] [PubMed] [Google Scholar]
- 101.Gillet E, Alentorn A, Doukoure B, et al. TP53 and p53 statuses and their clinical impact in diffuse low grade gliomas. J Neurooncol. 2014;118(1):131–139. doi: 10.1007/s11060-014-1407-4 [DOI] [PubMed] [Google Scholar]
- 102.Takami H, Yoshida A, Fukushima S, et al. Revisiting TP53 mutations and immunohistochemistry--a comparative study in 157 diffuse gliomas. Brain Pathol. 2015;25(3):256–265. doi: 10.1111/bpa.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mellai M, Monzeglio O, Piazzi A, et al. MGMT promoter hypermethylation and its associations with genetic alterations in a series of 350 brain tumors. J Neurooncol. 2012;107(3):617–631. doi: 10.1007/s11060-011-0787-y [DOI] [PubMed] [Google Scholar]
- 104.Kim B, Myung JK, Seo JH, et al. The clinicopathologic values of the molecules associated with the main pathogenesis of the glioblastoma. J Neurol Sci. 2010;294(1-2):112–118. doi: 10.1016/j.jns.2010.03.019 [DOI] [PubMed] [Google Scholar]
- 105.Clark KH, Villano JL, Nikiforova MN, Hamilton RL, Horbinski C. 1p/19q testing has no significance in the workup of glioblastomas. Neuropathol Appl Neurobiol. 2013;39(6):706–717. doi: 10.1111/nan.12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clark K, Voronovich Z, Horbinski C. How molecular testing can help (and hurt) in the workup of gliomas. Am J Clin Pathol. 2013;139(3):275–288. doi: 10.1309/AJCPFO8IIDNBIJ8Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dubbink HJ, Atmodimedjo PN, van Marion R, et al. Diagnostic detection of allelic losses and imbalances by next-generation sequencing: 1p/19q co-deletion analysis of gliomas. J Mol Diagn. 2016;18(5):775–786. doi: 10.1016/j.jmoldx.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 108.Collins VP, Ichimura K, Di Y, et al. Prognostic and predictive markers in recurrent high grade glioma; results from the BR12 randomised trial. Acta Neuropathol Commun. 2014;2:68. doi: 10.1186/2051-5960-2-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Purkait S, Jha P, Sharma MC, et al. CDKN2A deletion in pediatric versus adult glioblastomas and predictive value of p16 immunohistochemistry. Neuropathology. 2013;33(4):405–412. doi: 10.1111/neup.12014 [DOI] [PubMed] [Google Scholar]
- 110.Shirahata M, Ono T, Stichel D, et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018;136(1):153–166. doi: 10.1007/s00401-018-1849-4 [DOI] [PubMed] [Google Scholar]
- 111.Appay R, Dehais C, Maurage CA, et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol. 2019;21(12):1519–1528. doi: 10.1093/neuonc/noz124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mirchia K, Sathe AA, Walker JM, et al. Total copy number variation as a prognostic factor in adult astrocytoma subtypes. Acta Neuropathol Commun. 2019;7(1):92. doi: 10.1186/s40478-019-0746-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Korshunov A, Casalini B, Chavez L, et al. Integrated molecular characterization of IDH-mutant glioblastomas. Neuropathol Appl Neurobiol. 2019;45(2):108–118. doi: 10.1111/nan.12523 [DOI] [PubMed] [Google Scholar]
- 114.Chen Y, Hu F, Zhou Y, Chen W, Shao H, Zhang Y. MGMT promoter methylation and glioblastoma prognosis: a systematic review and meta-analysis. Arch Med Res. 2013;44(4):281–290. doi: 10.1016/j.arcmed.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 115.Yin AA, Zhang LH, Cheng JX, et al. The predictive but not prognostic value of MGMT promoter methylation status in elderly glioblastoma patients: a meta-analysis. PLoS ONE. 2014;9(1):e85102. doi: 10.1371/journal.pone.0085102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. doi: 10.1007/s00401-012-1016-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brandes AA, Franceschi E, Paccapelo A, et al. Role of MGMT methylation status at time of diagnosis and recurrence for patients with glioblastoma: clinical implications. Oncologist. 2017;22(4):432–437. doi: 10.1634/theoncologist.2016-0254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Reifenberger G, Hentschel B, Felsberg J, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2012;131(6):1342–1350. doi: 10.1002/ijc.27385 [DOI] [PubMed] [Google Scholar]
- 119.Perry JR, Laperriere N, O'Callaghan CJ, et al. Short-course radiation plus temozolomide in elderly patients with glioblastoma. N Engl J Med. 2017;376(11):1027–1037. doi: 10.1056/NEJMoa1611977 [DOI] [PubMed] [Google Scholar]
- 120.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
- 121.Malmstrom A, Gronberg BH, Marosi C, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. doi: 10.1016/S1470-2045(12)70265-6 [DOI] [PubMed] [Google Scholar]
- 122.Wick W, Platten M, Meisner C, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. doi: 10.1016/S1470-2045(12)70164-X [DOI] [PubMed] [Google Scholar]
- 123.Gilbert MR, Wang M, Aldape KD, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J Clin Oncol. 2013;31(32):4085–4091. doi: 10.1200/JCO.2013.49.6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weller M, Tabatabai G, Kastner B, et al. MGMT promoter methylation is a strong prognostic biomarker for benefit from dose-intensified temozolomide rechallenge in progressive glioblastoma: the DIRECTOR trial. Clin Cancer Res. 2015;21(9):2057–2064. doi: 10.1158/1078-0432.CCR-14-2737 [DOI] [PubMed] [Google Scholar]
- 125.Wick W, Gorlia T, Bendszus M, et al. Lomustine and bevacizumab in progressive glioblastoma. N Engl J Med. 2017;377(20):1954–1963. doi: 10.1056/NEJMoa1707358 [DOI] [PubMed] [Google Scholar]
- 126.Taal W, Oosterkamp HM, Walenkamp AM, et al. Single-agent bevacizumab or lomustine versus a combination of bevacizumab plus lomustine in patients with recurrent glioblastoma (BELOB trial): a randomised controlled phase 2 trial. Lancet Oncol. 2014;15(9):943–953. doi: 10.1016/S1470-2045(14)70314-6 [DOI] [PubMed] [Google Scholar]
- 127.Malley DS, Hamoudi RA, Kocialkowski S, Pearson DM, Collins VP, Ichimura K. A distinct region of the MGMT CPG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol. 2011;121(5):651–661. doi: 10.1007/s00401-011-0803-5 [DOI] [PubMed] [Google Scholar]
- 128.Vlassenbroeck I, Califice S, Diserens AC, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008;10(4):332–337. doi: 10.2353/jmoldx.2008.070169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Quillien V, Lavenu A, Sanson M, et al. Outcome-based determination of optimal pyrosequencing assay for MGMT methylation detection in glioblastoma patients. J Neurooncol. 2014;116(3):487–496. doi: 10.1007/s11060-013-1332-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Trabelsi S, Mama N, Ladib M, et al. MGMT methylation assessment in glioblastoma: MS-MLPA versus human methylation 450K BeadChip array and immunohistochemistry. Clin Transl Oncol. 2016;18(4):391–397. doi: 10.1007/s12094-015-1381-0 [DOI] [PubMed] [Google Scholar]
- 131.Lechapt-Zalcman E, Levallet G, Dugue AE, et al. O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation and low MGMT-encoded protein expression as prognostic markers in glioblastoma patients treated with biodegradable carmustine wafer implants after initial surgery followed by radiotherapy with concomitant and adjuvant temozolomide. Cancer. 2012;118(18):4545–4554. doi: 10.1002/cncr.27441 [DOI] [PubMed] [Google Scholar]
- 132.Quillien V, Lavenu A, Ducray F, et al. Clinical validation of the CE-IVD marked Therascreen MGMT kit in a cohort of glioblastoma patients. Cancer Biomark. 2017;20(4):435–441. doi: 10.3233/CBM-170191 [DOI] [PubMed] [Google Scholar]
- 133.Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer. 2012;118(17):4201–4211. doi: 10.1002/cncr.27392 [DOI] [PubMed] [Google Scholar]
- 134.Yuan G, Niu L, Zhang Y, et al. Defining optimal cutoff value of MGMT promoter methylation by ROC analysis for clinical setting in glioblastoma patients. J Neurooncol. 2017;133(1):193–201. doi: 10.1007/s11060-017-2433-9 [DOI] [PubMed] [Google Scholar]
- 135.Christians A, Hartmann C, Benner A, et al. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS ONE. 2012;7(3):e33449. doi: 10.1371/journal.pone.0033449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yamashita S, Yokogami K, Matsumoto F, et al. MGMT promoter methylation in patients with glioblastoma: is methylation-sensitive high-resolution melting superior to methylation-sensitive polymerase chain reaction assay? J Neurosurg. 2018;130(3):780–788. doi: 10.3171/2017.11.JNS171710 [DOI] [PubMed] [Google Scholar]
- 137.Lattanzio L, Borgognone M, Mocellini C, et al. MGMT promoter methylation and glioblastoma: a comparison of analytical methods and of tumor specimens. Int J Biol Markers. 2015;30(2):e208–216. doi: 10.5301/jbm.5000126 [DOI] [PubMed] [Google Scholar]
- 138.Wang L, Li Z, Liu C, et al. Comparative assessment of three methods to analyze MGMT methylation status in a series of 350 gliomas and gangliogliomas. Pathol Res Pract. 2017;213(12):1489–1493. doi: 10.1016/j.prp.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 139.Lalezari S, Chou AP, Tran A, et al. Combined analysis of O6-methylguanine-DNA methyltransferase protein expression and promoter methylation provides optimized prognostication of glioblastoma outcome. Neuro Oncol. 2013;15(3):370–381. doi: 10.1093/neuonc/nos308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Preusser M, Charles Janzer R, Felsberg J, et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. 2008;18(4):520–532. doi: 10.1111/j.1750-3639.2008.00153.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Draaisma K, Chatzipli A, Taphoorn M, et al. Molecular evolution of IDH wild-type glioblastomas treated with standard of care affects survival and design of precision medicine trials: a report from the EORTC 1542 study. J Clin Oncol. 2020;38(1):81–99. doi: 10.1200/JCO.19.00367 [DOI] [PubMed] [Google Scholar]
- 142.Hegi ME, Genbrugge E, Gorlia T, et al. MGMT promoter methylation cutoff with safety margin for selecting glioblastoma patients into trials omitting temozolomide: a pooled analysis of four clinical trials. Clin Cancer Res. 2019;25(6):1809–1816. doi: 10.1158/1078-0432.CCR-18-3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mulholland S, Pearson DM, Hamoudi RA, et al. MGMT CPG island is invariably methylated in adult astrocytic and oligodendroglial tumors with IDH1 or IDH2 mutations. Int J Cancer. 2012;131(5):1104–1113. doi: 10.1002/ijc.26499 [DOI] [PubMed] [Google Scholar]
- 144.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CPG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Christensen BC, Smith AA, Zheng S, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011;103(2):143–153. doi: 10.1093/jnci/djq497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tabatabai G, Stupp R, van den Bent MJ, et al. Molecular diagnostics of gliomas: the clinical perspective. Acta Neuropathol. 2010;120(5):585–592. doi: 10.1007/s00401-010-0750-6 [DOI] [PubMed] [Google Scholar]
- 148.van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the european organization for research and treatment of cancer brain tumor group. Clin Cancer Res. 2010;16(5):1597–604. doi: 10.1158/1078-0432.CCR-09-2902 [DOI] [PubMed] [Google Scholar]
- 149.Wiestler B, Capper D, Hovestadt V, et al. Assessing CPG island methylator phenotype, 1p/19q codeletion, and MGMT promoter methylation from epigenome-wide data in the biomarker cohort of the noa-04 trial. Neuro Oncol. 2014;16(12):1630–1638. doi: 10.1093/neuonc/nou138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao K, Li G, Qu Y, et al. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget. 2016;7(8):8712–8725. doi: 10.18632/oncotarget.6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Labussiere M, Di Stefano AL, Gleize V, et al. TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations. Br J Cancer. 2014;111(10):2024–2032. doi: 10.1038/bjc.2014.538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126(6):931–937. doi: 10.1007/s00401-013-1163-0 [DOI] [PubMed] [Google Scholar]
- 153.Kim HS, Kwon MJ, Song JH, Kim ES, Kim HY, Min KW. Clinical implications of TERT promoter mutation on IDH mutation and MGMT promoter methylation in diffuse gliomas. Pathol Res Pract. 2018;214(6):881–888. doi: 10.1016/j.prp.2018.04.002 [DOI] [PubMed] [Google Scholar]
- 154.Brito C, Azevedo A, Esteves S, et al. Clinical insights gained by refining the 2016 WHO classification of diffuse gliomas with: EGFR amplification, TERT mutations, PTEN deletion and MGMT methylation. BMC Cancer. 2019;19(1):968. doi: 10.1186/s12885-019-6177-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Labussiere M, Boisselier B, Mokhtari K, et al. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology. 2014;83(13):1200–1206. doi: 10.1212/WNL.0000000000000814 [DOI] [PubMed] [Google Scholar]
- 156.Koelsche C, Sahm F, Capper D, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013;126(6):907–915. doi: 10.1007/s00401-013-1195-5 [DOI] [PubMed] [Google Scholar]
- 157.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stichel D, Ebrahimi A, Reuss D, et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018;136(5):793–803. doi: 10.1007/s00401-018-1905-0 [DOI] [PubMed] [Google Scholar]
- 159.Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185 [DOI] [PubMed] [Google Scholar]
- 160.Chan AK, Yao Y, Zhang Z, et al. Combination genetic signature stratifies lower-grade gliomas better than histological grade. Oncotarget. 2015;6(25):20885–20901. doi: 10.18632/oncotarget.4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Guillaudeau A, Durand K, Pommepuy I, et al. Determination of EGFR status in gliomas: usefulness of immunohistochemistry and fluorescent in situ hybridization. Appl Immunohistochem Molecul Morphol. 2009;17(3):220–226. doi: 10.1097/pai.0b013e31818db320 [DOI] [PubMed] [Google Scholar]
- 162.Little SE, Popov S, Jury A, et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res. 2012;72(7):1614–1620. doi: 10.1158/0008-5472.CAN-11-4069 [DOI] [PubMed] [Google Scholar]
- 163.Lee M, Kang SY, Suh YL. Genetic alterations of epidermal growth factor receptor in glioblastoma: the usefulness of immunohistochemistry. Appl Immunohistochem Molecul Morphol. 2019;27(8):589–598. doi: 10.1097/pai.0000000000000669 [DOI] [PubMed] [Google Scholar]
- 164.Korshunov A, Schrimpf D, Ryzhova M, et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 2017;134(3):507–516. doi: 10.1007/s00401-017-1710-1 [DOI] [PubMed] [Google Scholar]
- 165.Wijnenga MMJ, Dubbink HJ, French PJ, et al. Molecular and clinical heterogeneity of adult diffuse low-grade IDH wild-type gliomas: assessment of TERT promoter mutation and chromosome 7 and 10 copy number status allows superior prognostic stratification. Acta Neuropathol. 2017;134(6):957–959. doi: 10.1007/s00401-017-1781-z [DOI] [PubMed] [Google Scholar]
- 166.Berzero G, Di Stefano AL, Ronchi S, et al. IDH-wildtype lower-grade diffuse gliomas: the importance of histological grade and molecular assessment for prognostic stratification. Neuro Oncol. 2021;23(6):955–966. doi: 10.1093/neuonc/noaa258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lin AL, Rosenblum M, Mellinghoff IK, et al. Prognostic and radiographic correlates of a prospectively collected molecularly profiled cohort of IDH1/2-wildtype astrocytomas. Brain Pathol. 2020;30(3):653–660. doi: 10.1111/bpa.12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Castel D, Philippe C, Calmon R, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130(6):815–827. doi: 10.1007/s00401-015-1478-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wu G, Broniscer A, McEachron TA, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. doi: 10.1038/ng.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bechet D, Gielen GG, Korshunov A, et al. Specific detection of methionine 27 mutation in histone 3 variants (H3K27M) in fixed tissue from high-grade astrocytomas. Acta Neuropathol. 2014;128(5):733–741. doi: 10.1007/s00401-014-1337-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gessi M, Gielen GH, Hammes J, et al. H3.3 G34R mutations in pediatric primitive neuroectodermal tumors of central nervous system (CNS-PNET) and pediatric glioblastomas: possible diagnostic and therapeutic implications? J Neurooncol. 2013;112(1):67–72. doi: 10.1007/s11060-012-1040-z [DOI] [PubMed] [Google Scholar]
- 172.Venneti S, Santi M, Felicella MM, et al. A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol. 2014;128(5):743–753. doi: 10.1007/s00401-014-1338-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huang T, Garcia R, Qi J, et al. Detection of histone H3 K27M mutation and post-translational modifications in pediatric diffuse midline glioma via tissue immunohistochemistry informs diagnosis and clinical outcomes. Oncotarget. 2018;9(98):37112–37124. doi: 10.18632/oncotarget.26430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mondal G, Lee JC, Ravindranathan A, et al. Pediatric bithalamic gliomas have a distinct epigenetic signature and frequent EGFR exon 20 insertions resulting in potential sensitivity to targeted kinase inhibition. Acta Neuropathol. 2020;139(6):1071–1088. doi: 10.1007/s00401-020-02155-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Castel D, Kergrohen T, Tauziede-Espariat A, et al. Histone H3 wild-type DIPG/DMG overexpressing EZHIP extend the spectrum diffuse midline gliomas with PRC2 inhibition beyond H3-K27M mutation. Acta Neuropathol. 2020;139(6):1109–1113. doi: 10.1007/s00401-020-02142-w [DOI] [PubMed] [Google Scholar]
- 176.Korshunov A, Capper D, Reuss D, et al. Histologically distinct neuroepithelial tumors with histone 3 G34 mutation are molecularly similar and comprise a single nosologic entity. Acta Neuropathol. 2016;131(1):137–146. doi: 10.1007/s00401-015-1493-1 [DOI] [PubMed] [Google Scholar]
- 177.Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024 [DOI] [PubMed] [Google Scholar]
- 178.Kleinschmidt-DeMasters BK, Aisner DL, Birks DK, Foreman NK. Epithelioid GBMs show a high percentage of BRAF v600e mutation. Am J Surg Pathol. 2013;37(5):685–698. doi: 10.1097/PAS.0b013e31827f9c5e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Korshunov A, Chavez L, Sharma T, et al. Epithelioid glioblastomas stratify into established diagnostic subsets upon integrated molecular analysis. Brain Pathol. 2018;28(5):656–662. doi: 10.1111/bpa.12566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nakajima N, Nobusawa S, Nakata S, et al. BRAF v600e, TERT promoter mutations and CDKN2A/B homozygous deletions are frequent in epithelioid glioblastomas: a histological and molecular analysis focusing on intratumoral heterogeneity. Brain Pathol. 2018;28(5):663–673. doi: 10.1111/bpa.12572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schreck KC, Ranjan S, Skorupan N, et al. Incidence and clinicopathologic features of H3 K27M mutations in adults with radiographically-determined midline gliomas. J Neurooncol. 2019;143(1):87–93. doi: 10.1007/s11060-019-03134-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schindler G, Capper D, Meyer J, et al. Analysis of BRAF v600e mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. doi: 10.1007/s00401-011-0802-6 [DOI] [PubMed] [Google Scholar]
- 183.Tatevossian RG, Tang B, Dalton J, et al. MYB upregulation and genetic aberrations in a subset of pediatric low-grade gliomas. Acta Neuropathol. 2010;120(6):731–743. doi: 10.1007/s00401-010-0763-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ramkissoon LA, Horowitz PM, Craig JM, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci U S A. 2013;110(20):8188–8193. doi: 10.1073/pnas.1300252110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ryall S, Zapotocky M, Fukuoka K, et al. Integrated molecular and clinical analysis of 1,000 pediatric low-grade gliomas. Cancer Cell. 2020;37(4):569–583.e5. doi: 10.1016/j.ccell.2020.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bandopadhayay P, Ramkissoon LA, Jain P, et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet. 2016;48(3):273–282. doi: 10.1038/ng.3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Broniscer A, Hwang SN, Chamdine O, et al. Bithalamic gliomas may be molecularly distinct from their unilateral high-grade counterparts. Brain Pathol. 2018;28(1):112–120. doi: 10.1111/bpa.12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Guerreiro Stucklin AS, Ryall S, Fukuoka K, et al. Alterations in ALK/ROS1/NTRK/MET drive a group of infantile hemispheric gliomas. Nat Commun. 2019;10(1):4343. doi: 10.1038/s41467-019-12187-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Clarke M, Mackay A, Ismer B, et al. Infant high-grade gliomas comprise multiple subgroups characterized by novel targetable gene fusions and favorable outcomes. Cancer Discov. 2020;10(7):942–963. doi: 10.1158/2159-8290.CD-19-1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mackay A, Burford A, Carvalho D, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537.e5. doi: 10.1016/j.ccell.2017.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–450. doi: 10.1038/ng.2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362(6411):eaar3593. doi: 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mandal R, Samstein RM, Lee KW, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. 2019;364(6439):485–491. doi: 10.1126/science.aau0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Omuro A, Vlahovic G, Lim M, et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro Oncol. 2018;20(5):674–686. doi: 10.1093/neuonc/nox208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schuenemann H, Brozek J, Guyatt G, Oxman A; The GRADE Working Group. GRADE handbook for grading quality of evidence and strength of recommendations. Updated October 2013. Accessed March 13, 2021. https://gdt.gradepro.org/app/handbook/handbook.html [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.