Abstract
A new insertion sequence (IS), IS1405, was isolated and characterized from a Ralstonia solanacearum race 1 strain by the method of insertional inactivation of the sacB gene. Sequence analysis indicated that the IS is closely related to the members of IS5 family, but the extent of nucleotide sequence identity in 5′ and 3′ noncoding regions between IS1405 and other members of IS5 family is only 23 to 31%. Nucleotide sequences of these regions were used to design specific oligonucleotide primers for detection of race 1 strains by PCR. The PCR amplified a specific DNA fragment for all R. solanacearum race 1 strains tested, and no amplification was observed with some other plant-pathogenic bacteria. Analysis of nucleotide sequences flanking IS1405 and additional five endogenous IS1405s that reside in the chromosome of R. solanacearum race 1 strains indicated that IS1405 prefers a target site of CTAR and has two different insertional orientations with respect to this target site. Restriction fragment length polymorphism (RFLP) pattern analysis using IS1405 as a probe revealed extensive genetic variation among strains of R. solanacearum race 1 isolated from eight different host plants in Taiwan. The RFLP patterns were then used to subdivide the race 1 strains into two groups and several subgroups, which allowed for tracking different subgroup strains of R. solanacearum through a host plant community. Furthermore, specific insertion sites of IS1405 in certain subgroups were used as a genetic marker to develop subgroup-specific primers for detection of R. solanacearum, and thus, the subgroup strains can be easily identified through a rapid PCR assay rather than RFLP analysis.
Ralstonia solanacearum (synonym Pseudomonas solanacearum E. F. Smith) (39) causes bacterial wilt, which is one of the most important and widely spread bacterial diseases of crops in the world. The presence of this pathogen on several hundred species representing 44 families of plants has been recorded (4). In Taiwan, the pathogen has been reported to occur on tomato, tobacco, potato, sweet pepper, eggplant, peanut, bird-of-paradise (40), strawberry, perilla (Perilla crispa) (22), castor bean, sesame, radish (26), anthurium (37), and eustoma (Eustoma grandiflorum L.) (5) plants. R. solanacearum strains represent a heterogeneous group that has been subdivided into five races on the basis of host range (3; I. W. Buddenhagen, L. Sequeira, and A. Kelman, abstr., Phytopathology 52:726, 1962) or five biovars based on the ability to oxidize certain disaccharides and hexose alcohols (19, 20). Races and biovars usually do not correspond, although strains of race 3 always belong to biovar 2 (3, 19). Moreover, there is considerable genetic variation among strains within each race or biovar (9, 31). Although strains of R. solanacearum in Taiwan are all race 1, genetic diversity and variation of aggressiveness in race 1 strains in Taiwan have been demonstrated (23, 25). The genetic and pathogenic variations make the development of diagnostic, detection, and control measures of R. solanacearum more difficult and complex.
Several molecular bacterial typing methods have been developed to identify genetic diversity and subdivide races or biovars of R. solanacearum for strain characterization. These methods include restriction fragment length polymorphism (RFLP) patterns (9), genomic fingerprinting (15), pulsed-field gel electrophoresis (36), and PCR-RFLP (31). Rep-PCR and random amplified polymorphic DNA are also used to differentiate strains of R. solanacearum race 1 (25, 27). In addition, insertion sequences (IS) have been reported and proved to be a valuable tool for molecular typing and strain characterization of some bacteria (1, 21, 29). Genomic mutation and rearrangement caused by IS is one of important mechanisms for generating genetic diversity (12, 28). Molecular analysis using IS can detect not only the existence, copy number, and location of IS but also the overall genomic organization of a bacterial strain.
In this study, we attempted to use IS to understand genetic diversity and for strain characterization of R. solanacearum race 1 strains in Taiwan. An insertion sequence, IS1405, was isolated from an R. solanacearum race 1 strain. The 5′ and 3′ noncoding regions of IS1405 were used to design specific oligonucleotide primers for the detection of race 1 strains by PCR. IS1405 was further used as a probe to detect the genetic diversity of R. solanacearum race 1 in Taiwan by RFLP pattern analysis, which allows us to classify race 1 strains and to track strains through a host plant community. In addition, several endogenous insertion sites of IS1405 were analyzed, and certain sites were used as genetic markers for identification.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains of R. solanacearum, Pseudomonas, Xanthomonas, and Erwinia that were used in this study and their pathovar designations are listed in Tables 1 and 2. Plasmid pCPP46-sacB was constructed by digesting pUCD800 (13) with PstI and BamHI and cloning the resulting 2.6-kb fragment containing the levansucrase gene (sacB) into the wide-host-range plasmid pCPP46 (8). Whenever an Escherichia coli host was necessary, strain DH5α (Gibco-BRL Life Technologies, Inc., Gaithersburg, Md.) or S17-1 (35) was used. R. solanacearum was cultured in BG media (2) at 30°C. E. coli DH5α, Xanthomonas spp., Erwinia spp., and Pseudomonas spp. were cultured in Luria-Bertani agar or broth medium (33) at 37°C for E. coli and at 28°C for the other species. Ampicillin (50 μg ml−1) was added as necessary to maintain selection of the resistance marker in pBluescript SK(+) (Stratagene Co., La Jolla, Calif.).
TABLE 1.
Bacterial strains tested, plant hosts, and grouping of R. solanacearum race 1 strains in Taiwan
| PCR-positive hosts | R. solanacearum subgroup(s) and PCR-positive strain(s)a |
|---|---|
| Solanaceous hosts | |
| Tomato | A1, PS21, PS68, PSS4; A2, PSL1, PSL3; A3, PSS180; B1, PSS240; B2, PS95, PS186; B3, PS72, B4, PSS190; B5, PSS197; B6, PSS81 |
| Tobacco | A1, PS31 |
| Sweet pepper | B1, PS-PER1, PS-PER2; B2, PS96 |
| Nonsolanaceous hosts | |
| Anthurium | A1, PS-AU17; A4, PS-AU5, PS-AU10 |
| Cat tail willow | A2, PS-CTW20; A5, PS-CTW9; B1, PS-CTW25, PS-CTW31 |
| New Zealand Flax | B1, PS-PDN2, PS-PDN3 |
| Eustoma | B1, PS-ES17; B7, PS-ES1, PS-ES8, PS-ES10 |
| Rose | A1, PS-RO1, PS-RO5 |
The positive strains had a 1,070-bp DNA fragment amplified by PCR using PS-IS-F and PS-IS-R as primers. The negative strains had no PCR-amplified product. Strains were obtained from Department of Plant Pathology, National Chung Hsing University, Asian Vegetable Research and Development Center, and Department of Pesticide Application, Taiwan Agricultural Chemicals and Toxic Substances Research Institute, Taiwan. The groupings are based on RFLP patterns as described in the text.
TABLE 2.
PCR-negative species tested
| PCR-negative species | Strainsa |
|---|---|
| Erwinia spp. and strains | E. amylovora EA169, E. carotovora subsp. atroseptica UCBPP149, E. carotovora subsp. betavasculorum UCBPP 162, E. carotovora subsp. carotovora Erc1, E. chrysanthemi pv. dieffenbachiae ED 104, E. cypripedii EC155, E. nigrifluens EN 105, E. quercina EQ101, E. rhapontici ER102, E. rubrifaciens ER103 |
| Pseudomonas spp. and strains | P. cepacia PC22, P. gladioli UCBPP550, P. syringae pv. apii PA102, P. syringae pv. tabaci PT124, P. syringae pv. erobotryae UCBPP258, P. syringae pv. atrofaciens UCBPP260, P. syringae pv. coronafaciens UCBPP470, P. syringae pv. glycinea UCBPP527 |
| Xanthomonas spp. and pathovars | X. arboricola pv. corylina XC5, X. arboricola pv. juglandis XJ123, X. arboricola pv. pruni XP10, X. axonopodis pv. begoniae XB110, X. axonopodis pv. cassiae XC112, X. axonopodis pv. manihotis XM125, X. axonopodis pv. tamarindi XT105, X. axonopodis pv. vignicola XV18, X. axonopodis pv. lespedezae XL1, X. campestris pv. campestris XCC1-1, X. hortorum pv. pelargonii XP15 |
The positive strains had a 1,070-bp DNA fragment amplified by PCR using PS-IS-F/PS-IS-R as primers. The negative strains had no PCR-amplified product. The strains were obtained from the International Collection of Phytopathogenic Bacteria, University of California, Berkeley, and American Type Culture Collection, Manassas, Va.
Isolation of insertion sequence in R. solanacearum.
Plasmid pCPP46-sacB was first transferred into E. coli S17-1, which supplies RP4 transfer function (35), by the calcium chloride procedure (33), and then mobilized into nalidixic acid-resistant R. solanacearum PS68 by mating between E. coli S17-1 (pCPP46-sacB) and PS68 (Nalr). The mating was carried out by mixing mid-log-phase cultures of PS68 (Nalr) as the recipient with E. coli S17-1 (pCPP46-sacB) as the donor. The mixture was spread onto nutrient yeast extract-glycerol agar plates (11) and incubated at 30°C for 16 h. The transconjugants, PS68 (pCPP46-sacB), were selected and plated on BG medium containing tetracycline (12.5 μg/ml), nalidixic acid (25 μg/ml), and 10% sucrose. After 3 to 4 days of growth, plasmid mutants with an insertion in the sacB gene were identified from survivors on the plates (13). The plasmid from each survivor was isolated and transformed into E. coli DH5α.
General DNA manipulations.
Mini-scale preparations of E. coli plasmid DNA, total genomic DNA isolation of plant pathogenic bacteria, restriction endonuclease treatments, DNA ligation, transformation, and agarose gel electrophoresis were done as described by Sambrook et al. (33). PCR-amplified DNA fragments used as probes were recovered from agarose by using the QIAEX II Gel Extraction Kit (QIAGEN Inc., Chatsworth, Calif.), and labeled with digoxigenin-11-dUTP (DIG) using a PCR DIG probe synthesis kit (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). The PCR for preparation of DIG-labeled DNA probes was performed in an air thermal cycler (Rapidcycler; Idaho Technology) programmed for denaturation at 94°C for 2 min and then for 35 cycles of 2 s at 94°C, 2 s at 50°C, and 35 s at 72°C. Prehybridization, hybridization, and washing for Southern hybridization with DIG-labeled probes were performed at 68°C according to the manufacturer's protocol. Double-stranded sequencing was performed by the dideoxy chain termination method (34) adapted for the AutoRead Sequencing Kit (Pharmacia Biotech, Uppsala, Sweden) with standard universal forward and reverse M13 primers. Sequence data were compiled and analyzed by using the computer programs by the Genetics Computer Group.
Primers, PCR amplification, and sensitivity of PCR detection of R. solanacearum.
For specific amplification of R. solanacearum, two oligonucleotide primers, PS-IS-F and PS-IS-R, were designed from the nucleotide sequence of IS1405 and synthesized with a DNA synthesizer by the Agricultural Biotechnology Laboratories, National Chung Hsing University, Taichung, Taiwan. PS-IS-F and PS-IS-R (Table 3) delineated a 1,070-bp fragment. PCR amplifications were carried out in 10-μl volumes which contained 10 ng of genomic DNA, 3 mM MgCl2, 0.5 μM primer, 0.4 U of Taq DNA polymerase (DyNAzyme II, Finnzymes Oy, Finland), 200 μM (each) deoxynucleoside triphosphate (dNTP) in 50 mM Tris-HCl (pH 8.3), and 500 μg of bovine serum albumin ml−1. PCR conditions were programmed as follows: denaturation at 94°C for 2 min and then 35 cycles of 2 s at 94°C, 2 s at 50°C, and 70 s at 74°C. Amplified DNAs were detected by electrophoresis in 0.8% agarose gels.
TABLE 3.
Primers used for PCRs
| Primers | Oligonucleotide sequence | Paired primer | Product size (bp) |
|---|---|---|---|
| PS-IS-F | 5′-CGCAACGCTGGATGAACCC-3′ | PS-IS-R | 1,070 |
| PS-IS-R | 5′-CAGACGATGCGAAGCCTGAC-3′ | ||
| PS-IS-Louta | 5′-GCCCGCGCTTGCGGGAGTT-3′ | PS-IS-Rout | |
| PS-IS-Rout | 5′-GCTCCGCTTCGATGACTC-3′ | ||
| PS-IS-RA1 | 5′-CACCCTAATCGGCACTAGCG-3′ | PS-IS-F | 1,181 |
| PS-IS-RB1 | 5′-CTCATGCTGACTGGCTACCC-3′ | PS-IS-F | 1,256 |
PS-IS-Lout and PS-IS-Rout primers were used in the inverted PCR. The size of the PCR product depends on the size of eluted DNA as described in the text.
To determine the minimum number of R. solanacearum cells that could be detected by PCR, 10-fold serial dilutions of the R. solanacearum PS68 strain were made. In each of six experiments, 2-μl aliquots from each tube of the dilution series were added directly to the PCR mixture. Aliquots from undiluted and serial dilutions of cultures were spread on BG plates to determine titers of bacteria, and CFU were determined.
Isolation of flanking fragments of IS1405 by an inverted PCR.
To determine the flanking sequences of IS1405 in the genome of R. solanacearum, a 0.5-kb PstI internal fragment of IS1405 was used as a probe to hybridize with EcoRI-digested total genomic DNA of different strains of R. solanacearum. Multiple copies were found, and some of them were recovered from the gels, self-ligated by T4 DNA ligase, and cleaved by SalI that is located in IS1405. The resulting DNAs were amplified by an inverted PCR (37) using oligonucleotide primers, PS-IS-Lout and PS-IS-Rout (Table 3), which correspond to 5′ and 3′ regions of IS1405 and were synthesized in an orientation opposite to that for normal PCR. The amplified fragments were cloned into pGEM-T Easy vectors (Promega Corp., Madison, Wis.) and then sequenced.
Isolation of iso-IS1405s from genomes of R. solanacearum.
To isolate extra copies of iso-IS1405 from R. solanacearum cells, DNA fragments containing a homolog of IS1405 were cloned. A 175-bp PstI internal fragment of IS1405 was used as a probe to hybridize EcoRI-digested chromosomal DNA from strains of R. solanacearum. Multiple copies were also found, and some of them were eluted from a gel, ligated to EcoRI-digested pBluescript SK(+) vector, and transformed into E. coli DH5α. The location of the IS1405 homolog in the eluted fragments was determined by restriction mapping and hybridization with the 175-bp PstI fragment, and the subclones of fragments were sequenced.
Nucleotide sequence accession number.
The IS1405 nucleotide sequence has been assigned GenBank accession number AF167984.
RESULTS
Isolation of insertion sequence from R. solanacearum.
R. solanacearum PS68 cells containing pCPP46-sacB were plated on BG medium containing 10% sucrose. The sucrose-viable mutant colonies were individually checked by agarose gel electrophoresis for the presence of pCPP46-sacB containing an insert. Those containing an insertion in the 2.6-kb BamHI-PstI sacB fragment (2 out of 12) were selected, and one of them, designated pCPP46-sacB1, was chosen for further study. Restriction mapping showed that pCPP46-sacB1 contained an insert of approximately 1.2 kb within the KpnI-HindIII fragment of sacB.
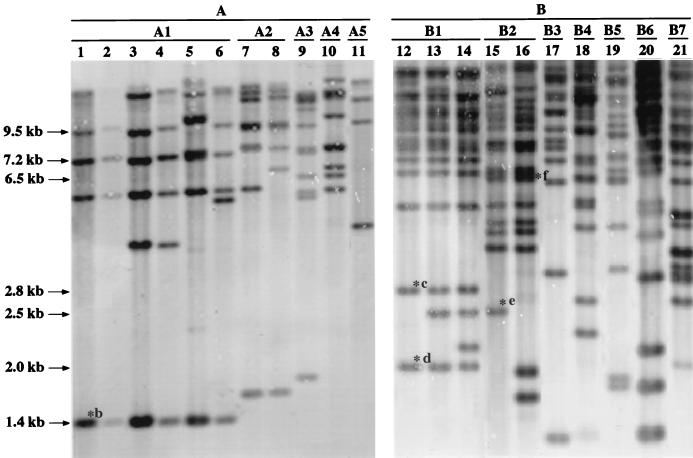
To determine whether the insertion in pCPP46-sacB1 originated from the chromosome of R. solanacearum PS68, a 175-bp PstI internal fragment was used as a probe to hybridize EcoRI-digested chromosomal DNA from R. solanacearum PS68 and E. coli DH5α. The probe hybridized strongly to chromosomal DNA of R. solanacearum PS68 but not to E. coli DH5α. The same probe was then used to hybridize with EcoRI-digested total genomic DNA of from strains of R. solanacearum in Table 1. It was found that the probe hybridized to genomic DNAs of all strains tested. The number of fragments hybridized to the probe varied from 4 to more than 15, indicating that all tested strains of R. solanacearum had multiple copies of insertion in their genomes. Since there were no EcoRI sites within the insertion, the number of hybridized fragments was the minimum number of copies of insertion. The hybridization banding patterns were further used for strain characterization of R. solanacearum in Taiwan as described below. Representative results are shown in Fig. 1.
FIG. 1.
Southern blot analysis of EcoRI-digested total genomic DNA from R. solanacearum race 1 strains, hybridized with a DIG-labeled 175-bp PstI DNA fragment of IS1405. Strains were classified into A and B groups based on the number of hybridized DNA fragments and subsequently into subgroups based on hybridization patterns. ∗b to ∗f, bands containing IS1405b, IS1405c, IS1405d, IS1405e, and IS1405f, respectively, which were eluted for inverted PCR or for cloning as described in Results. Lanes: 1, strain PS68; 2, PSS4; 3, PS31; 4, PS-AU17; 5, PS21; 6, PS-RO1; 7, PS-CTW20; 8, PS-L1; 9, PSS180; 10, PS-AU5; 11, PS-CTW9; 12, PS-CTW31; 13, PS-PER1; 14, PS-PDN3; 15, PS96; 16, PS95; 17, PS72; 18, PSS-190; 19, PSS-197; 20, PSS-81; 21, PS-ES1. The sizes of the bands are on the left.
Sequence analysis and feature of the insertion sequence.
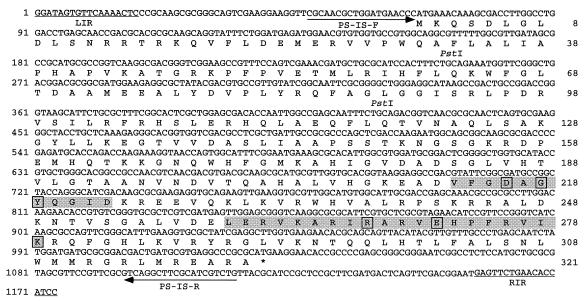
To determine the identity of the insertion in pCPP46-sacB1, the KpnI-HindIII fragment containing the insertion was cloned into pBluescript SK(+) vector and sequenced. The insertion sequence element in the fragment was named IS1405 afterward. The sequence of IS1405 is shown in Fig. 2. The element is 1,174 bp in length and has 18-bp imperfect terminal inverted repeats, (IRs). The overall G+C content of IS1405 is 60.2%. Upon insertion, IS1405 caused a 4-bp target site duplication (Fig. 3). Sequence analysis revealed that IS1405 contains a single open reading frame (ORF), indicated in Fig. 2. No consensus E. coli −10 and −35 promoter sequences (18) were found in the region upstream of this coding region. The actual promoter sequences have not yet been defined. The ORF begins with an ATG at position 67 and ends at position 1030. The ORF could encode a protein of 321 amino acids with a predicted molecular mass of 36.4 kDa and a theoretical isoelectric point of 11.1. This ORF contains an N-terminal region with a D(1)GY signature (called the N3 domain) in amino acids 213 to 223 and a C-terminal region with R(3)E(6)K signature (called the C1 domain) in amino acids 260 to 279 (Fig. 2). These two domains are highly conserved within transposases of members of the IS4 and IS5 families. The major difference between the IS4 and IS5 families is the relative location of the N3 and C1 domains. In the IS4 family, the distance between the N3 and C1 domains is about 100 amino acids, while in the IS5 group, the N3 and C1 domains are adjacent (28, 32). Since these two domains within the putative transposase of IS1405 were separated by 37 amino acids, IS1405 should be assigned to the IS5 family. In addition, IS4 and IS5 families can also be discriminated on the basis of the nucleotide sequence of the IR. Typically, the external trinucleotides of the IR of IS5 family are GGA, GAG or GGC, in contrast to that of the IS4 family, which is typically CAT (28, 32). IS1405 terminated in 5′-GGA-3′ at both ends and should be assigned to the IS5 family.
FIG. 2.
Nucleotide sequence of IS1405. The deduced amino acid sequence of the ORF is given below the nucleotide sequence, and the termination codon is marked (∗). The left and right terminal IR sequences of IS1403, LIR and RIR, are underlined. The N3 domain with the D(1)GY signature in amino acids 213 to 223 and a C1 domain with the R(3)E(6)K signature in amino acids 260 to 279 are shaded. The arrows denote the locations and orientations of the primers, PS-IS-F and PS-IS-R, designed to detect IS1405 of R. solanacearum race 1 strains.
FIG. 3.
Alignment of flanking nucleotide sequences of IS1405a and five endogenous IS1405s of R. solanacearum race 1 strains. IS1405a is the IS1405 element in pCPP46-sacB1 that originated from R. solanacearum PS68. The sources of other iso-IS1405 are described in Results. The 4-bp target site duplications are underlined. The orientation of each IS1405s is represented by an arrow. Identical nucleotides are in boldface type. Dashes represent gaps introduced for optimal alignment. Consensus nucleotides are listed in the last line, indicating bases that were conserved >60%.
Specific primers for detection of R. solanacearum by PCR.
Comparison of the nucleotide sequence of IS1405 with that of insertion sequences of the IS5 family indicated that the extent of overall nucleotide sequence identity is 51 to 54%. It is noteworthy that the identity of nucleotide sequence in 5′ and 3′ noncoding regions is only 23 to 31% rather than 51 to 60% in the coding region, indicating that the rate of nucleotide mutation is different between noncoding and coding regions. The overall G+C contents of these IS5 subgroup elements are similar and range from 52.7 to 61.7% (Table 4).
TABLE 4.
Comparison of nucleotide sequence of IS1405 with those of members of IS5 family
| IS (accession no.) | 5′ Noncoding region
|
Coding region
|
3′ Noncoding region
|
% G+C | |||
|---|---|---|---|---|---|---|---|
| Length (bp) | Identity (%) | Length (bp) | Identity (%) | Length (bp) | Identity (%) | ||
| IS5 (U95365) | 36 | 24 | 1,029 | 51 | 150 | 30 | 53.2 |
| IS1051 (X70380) | 64 | 30 | 980 | 59 | 114 | 23 | 60.2 |
| IS1384 (AF052751) | 58 | 27 | 981 | 58 | 139 | 23 | 57.0 |
| IS1479a (U56973) | 63 | 30 | 975 | 60 | 115 | 26 | 61.7 |
| IS1646 (AF077016) | 68 | 28 | 981 | 60 | 114 | 26 | 61.7 |
| ISPSMC (AB023075) | 65 | 30 | 978 | 55 | 145 | 22 | 52.7 |
| IS1405 (AF167984) | 66 | 100 | 966 | 100 | 142 | 100 | 60.2 |
Since 5′ and 3′ noncoding regions are more diverse, the regions were used to design a primer set, PS-IS-F and PS-IS-R, specific to IS1405 of R. solanacearum for PCR amplification (Fig. 2 and Table 3). To determine the specificity of these primers, PCRs were carried out with DNAs of all of the strains listed in Tables 1 and 2. The PCR amplification using the PS-IS-F and PS-IS-R primers amplified a 1,070-bp DNA fragment for all R. solanacearum strains tested. No amplification was observed with other plant pathogenic bacteria tested, such as Pseudomonas, Xanthomonas, or Erwinia strains (Table 2).
To determine the sensitivity of detection of the PCR assay with PS-IS-F and PS-IS-R, 2-μl samples of serial dilutions of R. solanacearum PS68 colonies grown on BG medium were used as template for the PCR. The tests were repeated six times, and the sensitivity of detection was determined as 104 CFU per ml of bacterial suspension on the ethidium bromide-stained gel. Since only 2 μl of bacterial suspension was used for PCR, this method was able to detect 20 cells of R. solanacearum.
Characterization of nucleotide sequences flanking IS1405 elements.
To understand whether IS1405 exhibits a target preference, nucleotide sequences flanking five endogenous IS1405s that reside in the chromosome of R. solanacearum race 1 strains were analyzed. The flanking DNAs of three endogenous IS1405s were amplified by an inverted PCR method with the PS-IS-Lout and PS-IS-Rout primer set (Table 3). The eluted 1.4-kb EcoRI fragments of PS68 containing IS1405b, 2.8-kb EcoRI fragments of PS-CTW31 containing IS1405c, and 2.5-kb EcoRI fragments of PS96 containing IS1405e were used as inverted PCR templates, respectively (Fig. 1). The expected 0.2-, 1.6-, and 1.4-kb flanking fragments of three iso-IS1405s were obtained and cloned into pGEM-T Easy vector. In addition, two DNA fragments containing endogenous IS1405 from R. solanacearum race 1 strains were cloned. Also, 2.0-kb EcoRI fragments of PS-CTW31 containing IS1405d and 6.5-kb EcoRI fragments of PS95 containing IS1405f were eluted from agarose gel, respectively. The eluted fragments were ligated to EcoRI-digested pBluescript SK(+) vector and transformed into E. coli DH5α.
The analysis of flanking sequences of IS1405 in pCPP46-sacB1 (called IS1405a hereafter) and five endogenous IS1405s indicated that IS1405s insert preferentially into a CTAR (R = A or G) target site. When IS1405s transpose to a new site, they are flanked by a duplication of 4-bp target sequences of either CTAA or CTAG. The preferred target site of IS1405a and IS1405c is CTAA (the reverse sequence is TTAG), and that of other IS1405 insertions is CTAG. Comparison of 50-bp nucleotide sequences surrounding IS1405s revealed a consensus sequence upstream from the target sites (Fig. 3), but no sequence symmetry was found. The sequence alignment also indicated that insertions of IS1405 have two different orientations with respect to the target site. The G+C content of the flanking sequences ranges from 37 to 56%.
Strain characterization of R. solanacearum in Taiwan.
Southern blot hybridization of EcoRI-digested genomic DNAs of R. solanacearum race 1 strains was performed with an internal 175-bp PstI DNA fragment of IS1405 as a probe to examine genetic variability within the strains of R. solanacearum in Taiwan. Thirty-two strains of R. solanacearum isolated from eight different host plants were tested (Table 1). Complex hybridization banding patterns were obtained. The number of fragments containing IS1405 varied from 4 to more than 15. The representative result is shown in Fig. 1. Strains were first classified into two distinct groups based on number of hybridized DNA fragments. The number of fragments in group A varied from 4 to 10, and that in group B was more than 15. The strains in each group were subsequently classified into subgroups as to the similarity of hybridization patterns. Each subgroup represents strains with identical patterns with the exception of one or two bands.
Strains isolated from solanaceous hosts (called solanaceous strains hereafter) had extensive genetic variability and belonged to two groups and nine subgroups (A1 to A3 and B1 to B6). Solanaceous strains that were assigned to subgroups A1 and B1 had a broad host range and also occurred on nonsolanaceous hosts. Some subgroup strains isolated from nonsolanaceous hosts were limited to their original hosts and have not yet been found on other hosts. For example, A4 subgroup strains were found only on anthurium, A5 subgroup strains were found only on cat-tail willow, and B7 subgroup strains were found only on eustoma (Table 1).
Specific primers for detection of A1 and B1 subgroup strains of R. solanacearum by PCR.
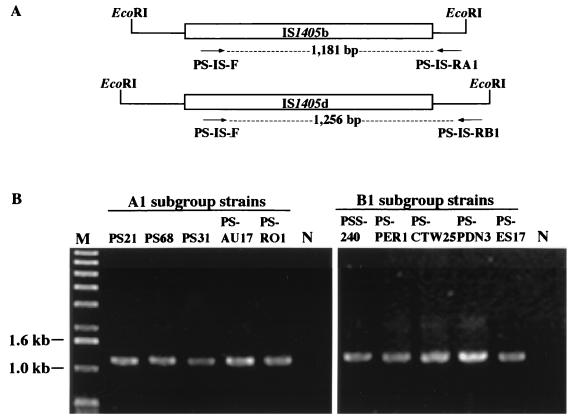
Strains in subgroups A1 and B1 appeared to be the most widespread in Taiwan. It was found that an IS1405 element located in a 1.4-kb EcoRI fragment (IS1405b) is common to all strains of the A1 subgroup and exists only in the A1 subgroup and that an IS1405 element in a 2.0-kb EcoRI fragment (IS1405d) was common to all strains of the B1 subgroup and only in the B1 subgroup. These unique locations were used as genetic markers for the identification of A1 and B1 strains, respectively. The flanking DNAs of these two IS1405s were amplified by an inverted PCR method and sequenced as described above. Two primers, PS-IS-RA1 and PS-IS-RB1, were designed according to the nucleotide sequence of one flanking region of IS1405b and IS1405d, respectively (Table 3; Fig. 4). The PS-IS-RA1 primer was paired with the PS-IS-F primer in PCR amplification, producing a DNA product of 1,181 bp, whereas the PS-IS-RB1 primer paired with the PS-IS-F primer producing a DNA product of 1,256 bp. The A1 and B1 primer sets amplified specific DNA fragments only from strains of A1 and B1 subgroups of R. solanacearum, respectively (Fig. 4). No amplification could be obtained when genomic DNAs from strains of other subgroups were tested as templates, confirming primer specificity for subgroups A1 and B1.
FIG. 4.
(A) A scheme of the subgroup-correlated insertion site of IS1405 with the position of each primer. The locations of IS1405b and IS1405d in 1.4- and 2.0-kb EcoRI fragments are specific to the A1 and B1 subgroup strains, respectively. (B) Agarose gel electrophoresis of PCR products from genomic DNAs of five representative A1 or B1 subgroup strains of R. solanacearum race 1 with PS-IS-F and PS-IS-RA1 or PS-IS-F and PS-IS-RB1 primer pairs, respectively. M, molecular size marker (1-kb DNA ladder, Life Technologies); sizes (base pairs) are on the left. Lane N, a negative control.
DISCUSSION
The analysis of nucleotide and amino acid sequences assigned IS1405 to the IS5 family. The IS5 family can further be divided into subgroups IS1031, IS427, ISL2, ISH1, IS903, and IS5 (28). Based on overall nucleotide sequence identity, IS1405 was closely related to members of the IS5 subgroup, including two IS elements (IS1384 and ISPSMC) originating from Pseudomonas spp., three elements (IS1646, IS1479, and IS1051) from Xanthomonas spp. (1, 7), and IS5 from E. coli. The host organisms of IS1405-related IS5 subgroup members (Pseudomonas, Ralstonia, Xanthomonas, and Escherichia), which are all gram negative, non spore forming, and rod shaped, are also closely related. Therefore, the sequence similarity between members of IS5 subgroup corresponds to the relationship of their host organisms, suggesting that members of IS5 subgroup in these bacteria were inherited from a common ancestor and have been resident in these bacteria since their separation. Alternatively, the occurrence of horizontal transfer of IS5 elements between these related bacteria in the past could be an explanation for the high sequence similarity between members of the IS5 subgroups.
The diversity of the 5′ and 3′ noncoding regions makes IS1405 easily distinguishable from other elements of IS5 subgroups. The PCR primers designed based on the nucleotide sequences of these regions were shown to be specific to IS1405 and to all R. solanacearum race 1 strains tested. No amplification was observed with DNA from strains of other plant pathogenic bacteria. However, since other races of R. solanacearum have not yet been found in Taiwan, they were not tested in this study. It is unknown whether other races would also amplify with the PCR primers. Nevertheless, since the tested strains of R. solanacearum isolated from eight different host plants were considered to be representative samples of this species, the primers developed in this study will be useful for the identification and specific detection of strains of R. solanacearum race 1 in Taiwan. The PCR primers will further be used to detect the population of R. solanacearum in plant seeds and seedlings, irrigation water, or soil extracts. Since IS1405 was present as multiple copies in all strains of R. solanacearum tested, the PS-IS-F and PS-IS-R primer set should enable more sensitive detection. R. solanacearum cells could be detected with as few as 20 cells in suspension by this primer set, without the need for prior enrichment or cultivation. The strategy presented here may be applied to develop PCR-based diagnostics for other pathogenic bacteria. Because most plant pathogenic bacteria contain insertion sequences, it should be relatively easy to generate primer sets based on diversified 5′ and 3′ noncoding regions of ISs.
The Southern hybridization analysis indicated that a number of copies of IS1405 are inserted at different sites in strains of R. solanacearum. Although IS1405 can insert into many different sites, target site utilization may not be random. There have been few comprehensive analyses of the insertion preference of endogenous genomic insertion sequences, although a limited number of insertions have been examined using a plasmid as a target, which restricts both the availability of a preferred site and the size of the target (6, 24). Therefore, we decided to undertake target site analysis of endogenous genomic IS1405s. It was found that the six insertions of IS1405 were accompanied by duplication of a 4-bp target site sequence of CTAR (R = A or G) (often CTAG). Three members of the IS5 subgroups (IS5, IS427, and IS1479) and two members of the ISL2 group (IS112 and IS1373) of IS5 family also prefer a similar target site, YTAR (Y = C or T) (often CTAG) (6, 28). The degeneracy of the fourth base tolerated by these ISs will increase insertion sites in the bacterial genome.
The preferred target sites of IS1405 include stop codons (TAG of CTAG and TAA of CTAA), and preferential insertion into these sites can possibly avoid insertion of coding regions. Sites remote from coding regions may be selectively neutral or subject to only very weak selection. Obviously, elements are more likely to be found in sites where insertions have little or no negative effects on fitness than in sites where insertions have deleterious effects. Another advantage of preferential insertion is to provide a mechanism whereby a transposon insertion into nonself DNA segments is encouraged (10). We searched the nucleotide sequence of IS1405 for preferred target sequences and neither CTAG nor CTAA was found within this IS. Thus, a preference for CTAR could tend to promote insertion of IS1405 into a target DNA other than the IS itself, thereby promoting dispersal of the element.
Various nucleotide sequence-based approaches, such as PCR-RFLP, Rep-PCR, and RAPD, have been developed to identify genetic diversity of R. solanacearum isolates for strain characterization (25, 27, 31). However, based on the sequence comparison between two strains of Helicobacter pylori, the genomic diversity seen by these approaches is primarily caused by nucleotide differences located at the third position of the triplet codons. The differences usually could not be translated into protein sequences. Furthermore, it was found that the divergence between strains of H. pylori could be mainly attributed to inversion and/or transposition of some chromosomal fragments and horizontal DNA transfer mediated by various IS elements (14). Therefore, IS1405 of R. solanacearum race 1 could have contributed to the strain diversity in Taiwan by its ability to insert at a number of sites and thereby bring about DNA rearrangements of the host chromosome. Strain characterization by molecular analysis using IS can reflect the overall genomic organization of a bacterial strain rather than nucleotide sequence difference.
Using IS1405 as a probe for RFLP analysis, extensive genetic variation was observed among strains of R. solanacearum race 1 in Taiwan, and RFLP patterns generated are readily amenable to analysis. Based on RFLP analysis, strains isolated from solanaceous hosts had extensive genetic variability. The genetic diversity of tomato strains in Taiwan has also been found by rep-PCR and random amplified polymorphic DNA techniques, respectively (25). Solanaceous hosts are widely grown in Taiwan, and bacterial wilt has been known for more than 50 years. Therefore, strains isolated from solanaceous hosts in Taiwan are considered to be endemic, such as strains of subgroups A1 and B1, which had a broad host range and also occurred on many nonsolanaceous hosts.
In addition to endemic strains (A1 and B1 subgroups), some other strains (A4, A5, and B7 subgroups) were isolated from nonsolanaceous hosts but have not yet been found on solanaceous hosts. For example, A4 subgroup strains were found only on anthurium, A5 subgroup strains were found only on cat-tail willow, and B7 subgroup strains were found only on eustoma (Tables 1 and 2). The nonsolanaceous hosts that were tested were introduced and grown in Taiwan for just a few years, and the disease was observed only recently (5, 37). The identities of the strains on these crops were confirmed in this study. Thus, A4, A5, and B7 subgroup strains may not be indigenous to Taiwan and may have been introduced through infected (or infested) tubers and seeds from foreign countries. Although the sample sizes used in this study were not adequate to draw definite conclusions about the genetic variability of R. solanacearum race 1 populations in Taiwan, the results reported here emphasized that the RFLP analysis using IS1405 as a probe was efficient in revealing genetic variability in the R. solanacearum population and allowed for strain identification to track strains of R. solanacearum through a host plant community.
The locations of insertion sequences within the genome can change, and strains with identical position of insertion elements should be closely related (16). A specific insertion of transposable element has been used to develop race-associated primers for detection of Fusarium oxysporum f. sp. dianthi (7). The same strategy was applied to identify A1 and B1 subgroup strains. It was found that one location of IS1405 was common and existed only in A1 or B1 subgroup strains. This specific location of IS1405 was used as a molecular marker to identify A1 or B1 strains and could be determined by PCR amplification by using the A1 or B1 primer set. Therefore, A1 and B1 subgroup strains can be easily identified through a rapid PCR assay rather than RFLP analysis.
An identification system based on the specific insertion of an IS would be more reliable if the inserted copy was inactive or if the IS transposed in a replicative manner by which the inserted copy at the original site remained unchanged. IS102 and IS903 of the IS5 family can form cointegrates and are thus supposed to transpose in a replicative manner (17, 30). IS1405 may use the same mechanism for transposition and make a specific insertion as a permanent marker for identification. However, if IS1405s of different subgroup strains transpose into the locations of A1 or B1 subgroups used for identification, this possibility would make the diagnostic system unreliable. Indeed, the possibility cannot be excluded. One way to increase the reliability of strain identification based on specific insertions of an IS is to design PCR primer sets targeted at two different insertion sites. The presence of multiple copies of IS1405 in the genome of R. solanacearum race 1 could make this approach practical for these bacteria.
ACKNOWLEDGMENTS
We express sincere thanks to Shih-Tien Hsu, Kuo-Ching Tzeng (National Chung Hsing University, Taichung, Taiwan), Jaw-Fen Wang (Asian Vegetable Research and Development Center, Tainan, Taiwan), Chiu-Chu Su (Taiwan Agricultural Chemicals and Toxic Substances Research Institute, Taichung, Taiwan) for donating bacterial strains, and Clarence I. Kado (Department of Plant Pathology, University of California at Davis) for providing pUCD800. We also thank Ya-Chun Chang (National Taiwan University, Taipei, Taiwan) for helpful discussions and critical reading of the manuscript.
This research was supported by a grant from the National Science Council Project NSC 84-2321-B-030-006-B13.
REFERENCES
- 1.Berthier Y, Thierry D, Lemattre M, Guesdon J-L. Isolation of an insertion sequence (IS1051) from Xanthomonas campestris pv. dieffenbachiae with potential use for strain identification and characterization. Appl Environ Microbiol. 1994;60:377–384. doi: 10.1128/aem.60.1.377-384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher C A, Barberis P A, Trigalet A P, Demery D A. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J Gen Microbiol. 1985;131:2449–2457. [Google Scholar]
- 3.Buddenhagen I W. Bacterial wilt revisited. In: Persley G J, editor. Bacterial wilt disease in Asia and the South Pacific, PRO13. Proceedings of an International Workshop, PCARRD, Los Banos, Philippines. Canberra, Australia: Australia Centre for International Agricultural Research; 1986. pp. 126–143. [Google Scholar]
- 4.Buddenhagen I W, Kleman A. Biological and physiological aspects of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 1964;2:203–230. [Google Scholar]
- 5.Chao Y C, Liang W J, Huang L L, Ho W C, Tsai C H. Bacterial wilt of Texas blue bell (Eustoma grandiflorum L.) Plant Pathol Bull (Taiwan) 1995;4:193–195. [Google Scholar]
- 6.Chen J-H, Hsieh Y-Y, Hsiau S-L, Lo T-C, Shau C-C. Characterization of insertions of IS476 and two newly identified insertion sequences, IS1478 and IS1479, in Xanthomonas campestris pv. campestris. J Bacteriol. 1999;181:1220–1228. doi: 10.1128/jb.181.4.1220-1228.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiocchetti A, Bernardo I, Daboussi M-J, Garibaldi A, Gullino M L, Langin T, Migheli Q. Detection of Fusarium oxysporum f. sp. dianthi in carnation tissue by PCR amplification of transposon insertions. Phytopathology. 1999;89:1169–1175. doi: 10.1094/PHYTO.1999.89.12.1169. [DOI] [PubMed] [Google Scholar]
- 8.Collmer A, Bauer D W, He S Y, Lindeberg M, Kelemu S, Rodriguez-Palenzuela P, Burr T J, Chatterjee A K. Pectic enzyme production and bacterial plant pathogenicity. In: Hennecke H, Verma D P S, editors. Advances in molecular genetics of plant-microbe interactions. Vol. 1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 65–72. [Google Scholar]
- 9.Cook D, Barlow E, Sequeira L. Genetic diversity of Pseudomonas solanacearum: detection of restriction fragment length polymorphisms that specify virulence and the hypersensitive response. Mol Plant-Microbe Interact. 1989;2:113–121. [Google Scholar]
- 10.Craig N L. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 11.Daniels M J, Barber C E, Turner D C, Cleary W G, Sawzyc M K. Isolation of mutants of Xanthomonas campestris pv. campestris showing altered pathogenicity. J Gen Microbiol. 1984;130:2447–2455. [Google Scholar]
- 12.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 13.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Z, Tayor D E. Contributions of genome sequencing to understanding the biology of Helicobacter pylori. Annu Rev Microbiol. 1999;53:353–387. doi: 10.1146/annurev.micro.53.1.353. [DOI] [PubMed] [Google Scholar]
- 15.Gillings M, Fahy P. Genetic diversity of Pseudomonas solanacearum biovars 2 and N2 assessed using restriction endonuclease analysis of total genomic DNA. Plant Pathol. 1993;42:744–753. [Google Scholar]
- 16.Green L, Miller R D, Dykhuizen D E, Hartl D L. Distribution of DNA insertion element IS5 in natural isolates of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:4500–4504. doi: 10.1073/pnas.81.14.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grindley N D F, Joyce C M. Genetic DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci USA. 1981;77:7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter sequences. Nucleic Acids Res. 1983;8:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayward A C. Characteristics of Pseudomonas solanacearum. J Appl Bacteriol. 1964;27:265–277. [Google Scholar]
- 20.He L Y, Sequeira L, Kelman A. Characteristics of strains of Pseudomonas solanacearum from China. Plant Dis. 1983;67:1357–1361. [Google Scholar]
- 21.Hermans P W M, Van Soolingen D, Dale J W, Schuitema A R J, McAdam R A, Catty D, Van Embden J D A. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990;28:2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu S-T, Hong W-F, Tzeng K-C, Chen C-C. Bacterial wilt of perilla caused by Pseudomonas solanacearum and its transmission. Plant Dis. 1993;77:674–677. [Google Scholar]
- 23.Hsu S-T, Tsai T-T, Tzeng K-C. Pathovars of Pseudomonas solanacearum in Taiwan and their interaction on tobacco plants. Natl Sci Counc Mon. 1979;7:609–620. . (In Chinese.) [Google Scholar]
- 24.Hu W-Y, Derbyshire K M. Target choice and orientation preference of the insertion sequence IS903. J Bacteriol. 1998;180:3039–3048. doi: 10.1128/jb.180.12.3039-3048.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaunet T X, Wang J-F. Variation in genotype and aggressiveness of Ralstonia solanacearum race 1 isolated from tomato in Taiwan. Phytopathology. 1999;89:320–327. doi: 10.1094/PHYTO.1999.89.4.320. [DOI] [PubMed] [Google Scholar]
- 26.Lin C H, Hsu S T, Tzeng K C. Radish (Raphanus sativus L.), a new host of Pseudomonas solanacearum in Taiwan. Plant Pathol Bull. 1994;3:147–155. [Google Scholar]
- 27.Louws F J, Fulbright D W, Stephens E R, deBruijn F J. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol. 1994;60:2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLafferty M A, Harcus D R, Hewlett E L. Nucleotide sequence and characterization of a repetitive DNA element from the genome of Bordetella pertussis with characteristics of an insertion sequence. J Gen Microbiol. 1988;134:2297–2306. doi: 10.1099/00221287-134-8-2297. [DOI] [PubMed] [Google Scholar]
- 30.Ohtsubo H, Zenilman M, Ohtsubo E. Insertion element IS102 resides in plasmid pSC101. J Bacteriol. 1980;144:131–140. doi: 10.1128/jb.144.1.131-140.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poussier S, Vandewalle P, Luisetti J. Genetic diversity of African and worldwide strains of Ralstonia solanacearum as determined by PCR-restriction fragment length polymorphism analysis of the hrp gene region. Appl Environ Microbiol. 1999;65:2184–2194. doi: 10.1128/aem.65.5.2184-2194.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rezsohazy R, Hallet B, Delcour J, Mahillon J. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 36.Smith J J, Offord L C, Holderness M, Saddler G S. Pulsed-field gel electrophoresis analysis of Pseudomonas solanacearum. EPPO Bull. 1995;25:163–167. [Google Scholar]
- 37.Su C C, Leu L S. Bacterial wilt of anthurium caused by Pseudomonas solanacearum. Plant Pathol Bull (Taiwan) 1995;4:34–38. [Google Scholar]
- 38.Triglia T, Peterson M G, Kemp D J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 40.Yang T H, Hsu S T, Tzeng K C. Studies on bird-of-paradise strains of Pseudomonas solanacearum-physiological characteristics, pathogenicity and lectin induction. J Agric For (Taiwan) 1980;29:119–133. [Google Scholar]