ABSTRACT
Flowering at an appropriate time is crucial for plant development and reproduction. In Arabidopsis, the flowering process is managed by a regulatory network composed of at least 6 independent pathways. As a core protein in flowering regulation, FLOWERING LOCUS T (FT) participates in almost all these pathways. ANKYRIN REPEAT-CONTAINING PROTEIN 2A (AKR2A) was initially discovered as a 14-3-3-interacting protein. It was then found to be involved in the transportation of chloroplast outer membrane proteins and the resistance to low-temperature stress. Here, we identified an akr2a null mutant with a delayed flowering phenotype. Through the quantitative real-time PCR (qRT-PCR) and bimolecular fluorescence complementation (BiFC) assays, we demonstrated that AKR2A modulates the flowering process through its interaction with FT.
KEYWORDS: Arabidopsis thaliana, ANKYRIN REPEAT-CONTAINING PROTEIN 2A (AKR2A), FLOWERING LOCUS T (FT), flowering time
Introduction
Flowering is not only the transition from vegetative growth to reproductive growth but also the most significant change in the plant growth cycle.1 In the process of plant development, the flowering time is determined by a complex and elaborate molecular mechanism.2 Up to now, more than 300 genes have been identified in the flowering process of Arabidopsis thaliana.3 Six pathways co-regulate flower induction, i.e., the photoperiod pathway, vernalization pathway, ambient temperature pathway, gibberellin pathway, autonomous pathway, and age pathway.4 These pathways are independent and cooperate to form a complex and precise regulatory network.
FLOWERING LOCUS T (FT) is one of the most critical genes in the flowering regulation pathway.5–7 FT encodes a florigen protein that moves from leaves to plant growth points and activates the expression of downstream genes, thus promoting the transformation from vegetative growth to reproductive growth.8,9 Usually, CONSTANS (CO), induced by the long-day signal, binds to the promoter of FT, and activates its downstream genes.10 On the other hand, the transcription inhibitor Flowing LOCUS C (FLC) inhibits the expression of FT and negatively regulates Arabidopsis flowering.11
Arabidopsis thaliana ANKYRIN REPEAT-CONTAINING PROTEIN 2A (AKR2A) was recognized to function in disease resistance, antioxidation metabolism, and chloroplast outer membrane protein targeting.12–15 It has been reported to interact with peroxisomal membrane-bound ASCORBATE PEROXIDASE3 (APX3), 3-ketoacyl-CoA synthase 1 (KCS1), and chloroplast protein OEP7.12–15 Its mutants constructed using the antisense technique resulted in small necrotic areas in leaves accompanied by enhanced production of H2O2 under normal growth conditions.16 Under chilling temperature conditions, its point mutant T6 (Glu-to-Lys change at residue 150) seedlings were shorter and had smaller and curled rosette leaves compared with its wild-type (WT) parental line Columbia (Col) er105 (BM).15 A slightly delayed-flowering phenotype of the T6 mutant was also observed.15 However, these studies were not based on its null mutant, and some other functions of AKR2 might be missed.
In this study, we confirmed that the T-DNA insertion null mutant akr2a has a delayed-flowering phenotype, which might result from the protein-protein interaction between AKR2A and FT.
Results and discussion
AKR2A expression affects Arabidopsis flowering
To confirm the delayed-flowering phenotype of the AKR2A point-mutant T6, we acquired the T-DNA insertion line akr2a with a Col-0 background from ABRC. With the T-DNA insertion at the 7th exon, the transcript of AKR2A was not detected (Figure 1c, 1e). We then generated its homozygous complementation lines using its own promoter region to drive its full-length cDNA. All these lines were confirmed at the genomic and transcript levels (Figure 1c and 1e).
Figure 1.
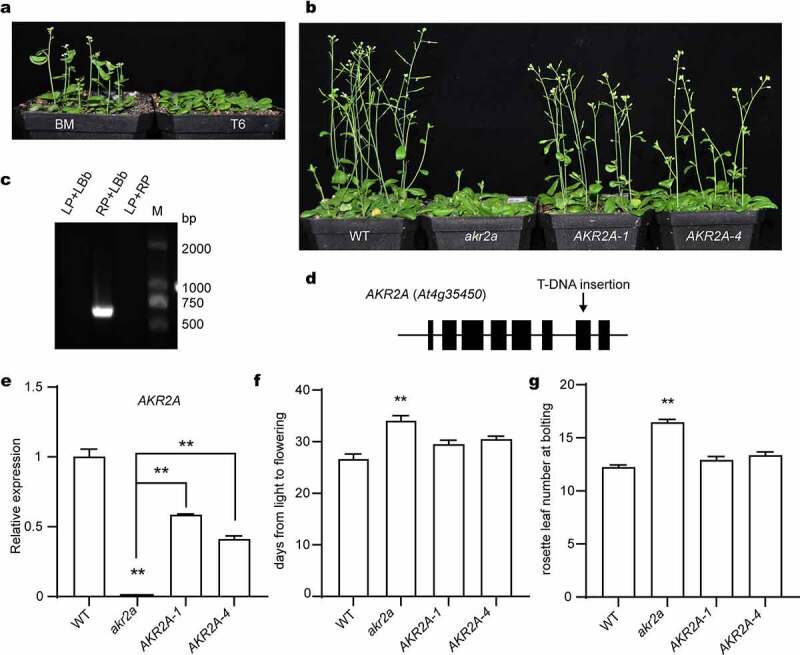
The silencing of AKR2A accounts for the delayed-flowering phenotype. (a) Flowering phenotype of the AKR2A point mutant T6 and its wild-type parent BM seedlings. (b) Flowering phenotype of the Col-0 WT, the T-DNA insertion akr2a mutant, and two complementation line (AKR2A-2, AKR2A-4) seedlings. For (a) and (b), seedlings were grown under a long-day (16 h/8 h light/dark) condition. Representative images were taken at 35 d after germination. (c) PCR confirmation of the T-DNA insertion akr2a mutant. (d) Scheme of the AKR2A gene structure. Black boxes indicate the exons. The arrow indicates the position of T-DNA insertion. (e) Relative expression of AKR2A in different lines at 28 d after germination. Transcript abundance was determined by qRT-PCR. ACTIN2 served as a reference. Data are means ± SEM (n = 3, Student’s t-test, **P < .01). (f) and (g) The days from light to flowering (f) and the rosette leaf number at bolting (g) of different lines. Data are means ± SEM (n = 9, Student’s t-test, **P < .01).
Under a 16 h/8 h light/dark long-day growth condition, the akr2a mutant seedlings flowered 7–8 d later than Col-0 WT, while the complementation seedlings flowered 4–5 d earlier than akr2a seedlings (Figure 1f). In the meantime, the rosette leaf number of akr2a was 4–6 more than Col-0 WT, but those of the two complementation lines were similar to the WT level (Figure 1g). Therefore, our results demonstrated the delayed flowering phenotype of the AKR2A null mutant.
Deletion of AKR2A affects the expression of flowering-related genes
Flowering is the transition from vegetative to reproductive growth in response to developmental and environmental cues.17 In the photoperiodic pathway, FT and TWIN SISTERS OF FT (TSF) are activated by CO and positively regulate Arabidopsis flowering.18,19 At the earlier stages, GIGANTEA (GI) forms a complex on the CO promoter to regulate CO expression.20 In the akr2a mutant, transcript abundances of FT and TSF were significantly lower than that in WT, but thelevels of CO or GI were similar to their corresponding WT levels (Figure 2). DELLA proteins (RGL1, RGL2, RGL3) and GA4 were negative regulators of GA responses to flowering,21–23 and the transcript levels of all their coding genes significantly increased in akr2a (Figure 2). Previous studies have proved that FRIGIDA (FRI), FLOWERING LOCUS C (FLC), SHORT VEGETATIVE PHASE (SVP), and TERMINAL FLOWER 1 (TFL1) inhibit the flowering of Arabidopsis by the vernalization pathway, ambient temperature pathway, and autonomous pathway.24–27 We also tested the transcript levels of these genes, and found enhanced transcript abundances of these genes in akr2a seedlings (Figure 2). Taken together, the silencing of AKR2A affects the expression of a bouquet of flowering-related genes.
Figure 2.
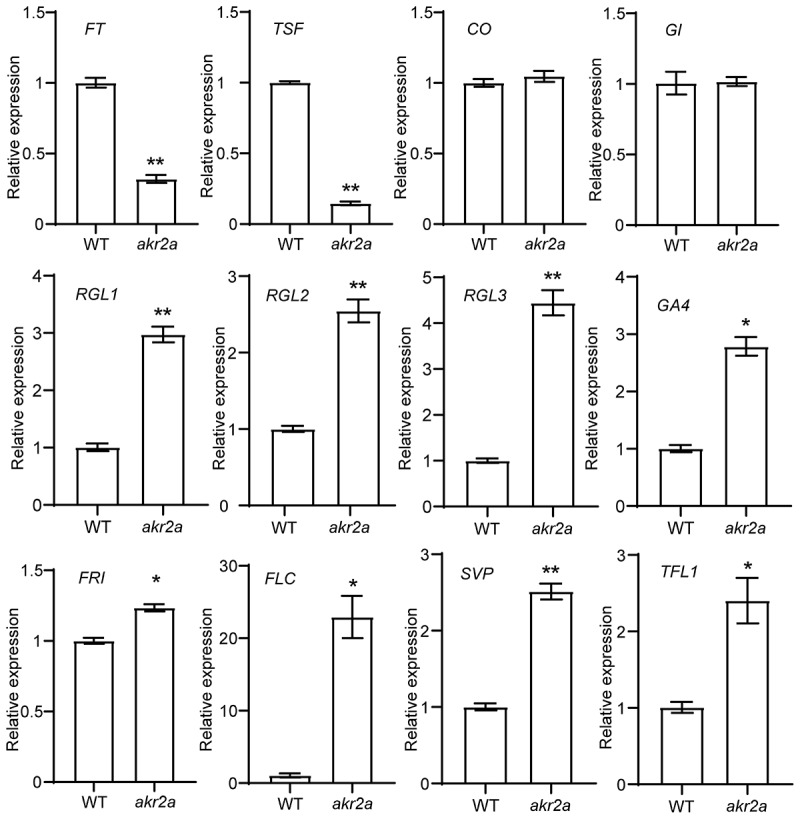
Transcript levels of flowering-related genes in akr2a mutant plants. Transcript abundances of flowering-related genes as indicated in akr2a mutant and the Col-0 WT seedlings at 28 d after germination were quantified by qRT-PCR. ACTIN2 served as a reference. Data are means ± SEM (n = 3, Student’s t-test, *P < .05, **P < .01).
In vivo interaction of AKR2A with FT
To further investigate how AKR2A regulates flowering time, we studied the subcellular localization of AKR2A. Full-length open-reading frame (ORF) of AKR2A was fused with enhanced yellow fluorescent protein (EYFP), and the fusion protein AKR2A-EYFP was transiently expressed in Nicotiana benthamiana mesophyll cells. As is shown in Figure 3a, AKR2A showed a cytosolic localization, resembling that of EYFP. We then tested whether AKR2A was able to interact with FT and/or CO, the two core components in flowering regulation. A BiFC assay was performed to test their in planta interactions. Our microscopic observation clearly revealed the positive interaction between AKR2A and FT in the cytosol, but not that between AKR2A and CO (Figure 3b).
Figure 3.
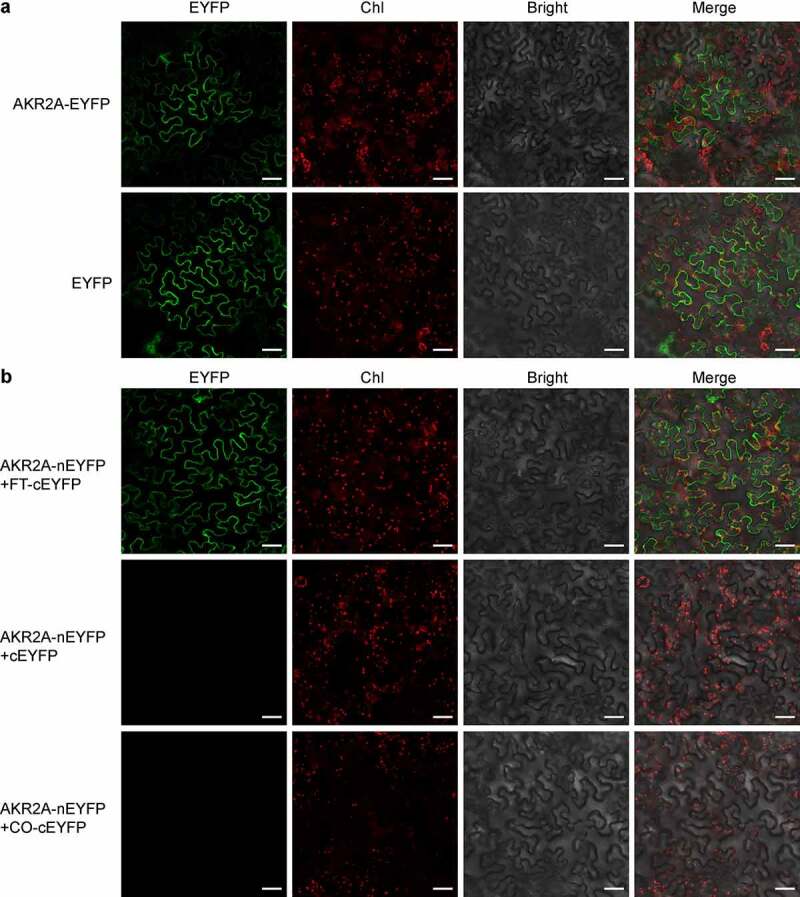
AKR2A interacts with FT in the cytosol. (a) Subcellular localization assay. Full-length AKR2A fused with EYFP or empty EYFP were transiently expressed in Nicotiana benthamiana mesophyll cells by infiltration. (b) BiFC detection. AKR2A was fused with the N-half of EYFP (nEYFP), and FT and CO were individually fused with the C-half of EYFP (cEYFP). Different combinations of fusion proteins as indicated were co-expressed in N. benthamiana mesophyll cells by infiltration. Images were collected 3 d after infiltration. Representative images under EYFP, chlorophyll (Chl), and bright field (Bright) channels and the merged signals are shown. Bar = 50 μm.
In this work, by utilizing the T-DNA insertion null mutant, we demonstrated the function of AKR2A in regulating the flowering process in A. thaliana. By comparing the transcript levels of flowering-related genes between akr2a and WT plants, we found that the silencing of AKR2A resulted in the down-regulation of the positive control genes, such as FT and TSF. On the other hand, the negative control genes, such as RGL, FRI, and FLC, were upregulated in the akr2a mutant (Figure 2). Our BiFC observation indicated the protein-protein interaction between AKR2A and FT (Figure 3). Such an interaction might explain the delayed-flowering phenotype of akr2a. It is interesting that FT was also reported to interact with the 14-3-3 protein.28 The protein-protein interaction between AKR2A and FT in the cytosol, as observed in our BiFC assay, might affect the translocation of FT from the cytosol to the nucleus (Figure 3b). Further analysis of the relationships among AKR2A, 14-3-3 proteins, and AKR2A-interacting proteins will help to decipher the plethora of functions of AKR2A.29
Materials and methods
Plant materials and growth condition
The akr2a mutant lines (SALK_204151C, with the T-DNA insertion at the exon) and wild-type ecotype Columbia-0 (Col-0 WT) were obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, USA). Seeds of Col er105 (BM) and its AKR2A TILLING line (T6) were kindly provided by Dr. Guoxin Shen (Zhejiang Academy of Agricultural Sciences, Hangzhou, China).13
All seeds were stratified at 4°C in the dark for 3 d and then allowed to germinate on Murashige-Skoog (MS) plates containing 2% sucrose and 0.8% agar at 22°C under 120 µmol photons m−2 s−1 irradiance with a 16 h/8 h light/dark regime (as the long-day condition) and relative humidity of 60% in a growth chamber. Two-week-old seedlings were moved to grow in soil (a mixture of peat moss, vermiculite, and perlite at 1:1:1) under the same conditions.
To rescue the akr2a null mutant, we fused a 2-kb DNA fragment upstream of the translation initiation codon (ATG) of AKR2A, the full-length ORF of AKR2A, and a 700-bp DNA fragment downstream of the stop codon of AKR2A in a tandem array. The cassette was cloned into pCAMBIA1300 (CAMBIA, Canberra, Australia). The akr2a mutant seedlings were transformed with this construct using the Agrobacterium-mediated floral-dipping method.30 Transgenic plants were screened on MS plates with 40 μg mL−1 hygromycin B and further confirmed by qRT-PCR for the expression of corresponding transgenes.
Molecular manipulation
Genomic DNA was extracted from A. thaliana leaves using the CTAB method.31 The presence of the T-DNA insertion was detected by PCR according to the SIGnAL iSect tool (http://signal.salk.edu/tdnaprimers.2.html).
For RNA isolation, leaves were ground in liquid nitrogen into a fine powder and extracted using the RNAiso reagent (TaKaRa, Shiga, Japan). For each sample, 1 μg of total RNA was used for synthesizing the first-strand cDNA using the HiScript III RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China) following the manufacturer’s instruction. qRT-PCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme) in a Thermal Cycler Dice Real Time System TP800 (TaKaRa) following the manufacturers’ instructions. The cycling program consisted of 95°C for 5 min, followed by 40 cycles of 95°C for 10s, and then 60°C for 15s. Transcript abundance of ACTIN2 (At3g18780) was quantified as a reference. Relative expression levels were calculated by the comparative CT method.32 All primer sequences used in this study are listed in Table S1.
Subcellular localization and BiFC assays
For subcellular localization assay, the full-length ORF of AKR2A was cloned into the pCNHP-EYFP at the NcoI site as previously described.33 In this construct, the enhanced cauliflower mosaic virus (CaMV) 35S promoter, synthetic 5’ and 3’ untranslated regions of cowpea mosaic virus RNA-2, the coding region of AKR2A, enhanced yellow fluorescent protein (EYFP), and the Heat Shock Protein (HSP) terminator from A. thaliana were linked sequentially in the pCNHP-EYFP to express 35S:AKR2A-EYFP.33
For BiFC assay, the full-length ORF of AKR2A was cloned into pCNHP-nEYFP to generate 35S:AKR2A-nEYFP, and the full-length ORFs of FT and CO were individually cloned into pCNHP-cEYFP to generate 35S:FT-cEYFP and 35S:FT-cEYFP, respectively. The pCNHP-cEYFP and pCNHP-nEYFP vectors are almost the same as pCNHP-EYFP, except that they harbor the C- and N-halves of EYFP, respectively, instead of the intact EYFP of pCNHP-EYFP.
Each of the constructs was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation.34 Agrobacterium cells were collected by centrifugation at 5000 g for 5 min and then resuspended in infiltration media (10 mM MES, 10 mM MgCl2, and 200 μM acetosyringone) to an OD600 of 1. Before infiltration on Nicotiana benthamiana leaves, the mixture was activated for about 3 h at room temperature. Finally, the transfected plants were allowed to grow for 3 d under a 16 h light/8 h dark light cycle.
Fluorescence signals were observed using a confocal laser scanning microscope (FluoView FV1000, Olympus, Tokyo, Japan). The excitation wavelength and the emission filters for EYFP were 515 nm and 530–560 nm, respectively. Chlorophyll autofluorescence was monitored using 543 nm excitation wavelengths and 680–720 nm detection windows.
Statistical analysis
GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA) was used for statistical analysis. We employed Student’s t-test to determine statistical significance. Differences were considered significant at P < .05 and P < .01 levels.
Acknowledgments
We thank Zhong Zhuang for his help with the microscopic analysis.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Author contributions
Conceptualization, Q. T. and S. Lu; Investigation, Q. T., Y. Z., S. Luo; Writing, Q. T. and S. Lu. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary Materials
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592324.2022.2100685
References
- 1.Huijser P, Schmid M.. The control of developmental phase transitions in plants. Development. 2011;138(19):4117–6. doi: 10.1242/dev.063511. [DOI] [PubMed] [Google Scholar]
- 2.Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125(4):655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Bouche F, Lobet G, Tocquin P, Perilleux C. FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016;44(D1):1167–1171. doi: 10.1093/nar/gkv1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fornara F, de Montaigu A, and Coupland G. SnapShot: Control of flowering in Arabidopsis. Cell. 2010;141(3):550. doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 5.Corbesier L, Vincent C, Jang SH, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 6.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59(1):573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Li W, Liu Y, Li Y, Li Y, Yang W, Chen X, Pi L, and Yang H. Replication protein RPA2A regulates floral transition by cooperating with PRC2 in Arabidopsis. New Phytol. 2022. (in press 10.1111/nph.18279) [DOI] [PubMed] [Google Scholar]
- 8.Mouradov A, Cremer F, Coupland G. Control of flowering time: interacting pathways as a basis for diversity. Plant Cell. 2002;14(suppl 1):S111–S130. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Putterill J, Varkonyi-Gasic E. FT and florigen long-distance flowering control in plants. Curr Opin Plant Biol. 2016;33:77–82. doi: 10.1016/j.pbi.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell. 2013;25(3):820–833. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20(7):898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae W, Lee YJ, Kim DH, Lee J, Kim S, Sohn EJ, and Hwang I. AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat Cell Biol. 2008;10(2):220–227. doi: 10.1038/ncb1683. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Hu WJ, Mishra N, Wei J, Lu HL, Hou YQ, Qiu X, Yu S, Wang C, and Zhang H. AKR2A interacts with KCS1 to improve VLCFAs contents and chilling tolerance of Arabidopsis thaliana. Plant J. 2020;4(4):1575–1589. doi: 10.1111/tpj.14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu WJ, Chen L, Qiu XY, Wei J, Lu HL, Sun GC, Ma X, Yang Z, Zhu C, Hou Y. AKR2A participates in the regulation of cotton fibre development by modulating biosynthesis of very-long-chain fatty acids. Plant Biotechnol J. 2020;18(2):526–539. doi: 10.1111/pbi.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen GX, Kuppu S, Venkataramani S, Wang J, Yan JQ, Qiu XY, Zhang H. ANKYRIN REPEAT-CONTAINING PROTEIN 2A is an essential molecular chaperone for peroxisomal membrane-bound ASCORBATE PEROXIDASE3 in Arabidopsis. Plant Cell. 2010;22(3):811–831. doi: 10.1105/tpc.109.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan JQ, Wang J, and Zhang H. An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J. 2002;29(2):193–202. doi: 10.1046/j.0960-7412.2001.01205.x. [DOI] [PubMed] [Google Scholar]
- 17.Blumel M, Dally N, Jung C. Flowering time regulation in crops - what did we learn from Arabidopsis? Curr Opin Biotechnol. 2015;32:121–129. doi: 10.1016/j.copbio.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288(5471):1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46(8):1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 20.Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410(6832):1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 21.Galvao VC, Horrer D, Kuttner F, Schmid M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development. 2012;139(21):4072–4082. doi: 10.1242/dev.080879. [DOI] [PubMed] [Google Scholar]
- 22.Khan SA, Hamayun M, Yoon H, Kim HY, Suh SJ, Hwang SK, Kim JM, Lee IJ, Choo YS, Yoon UH, et al. Plant growth promotion and Penicillium citrinum. BMC Microbiol. 2008;8(1):231–240. doi: 10.1186/1471-2180-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porri A, Torti S, Romera-Branchat M, Coupland G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development. 2012;139(12):2198–2209. doi: 10.1242/dev.077164. [DOI] [PubMed] [Google Scholar]
- 24.Johanson U, West J, Lister C, Michaels S, Amasino R, and Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290(5490):344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- 25.Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13(4):935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pose D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RGH, Schmid M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature. 2013;503(7476):414–417. doi: 10.1038/nature12633. [DOI] [PubMed] [Google Scholar]
- 27.Serrano-Mislata A, Fernandez-Nohales P, Domenech MJ, Hanzawa Y, Bradley D, Madueno F. Separate elements of the TERMINAL FLOWER 1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity. Development. 2016;143(18):3315–3327. doi: 10.1242/dev.135269. [DOI] [PubMed] [Google Scholar]
- 28.Purwestri YA, Ogaki Y, Tamaki S, Tsuji H, Shimamoto K. The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 2009;50(3):429–438. doi: 10.1093/pcp/pcp012. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Li X, Zhang Y, Kuppu S, Shen G. Is AKR2A an essential molecular chaperone for a class of membrane-bound proteins in plants? Plant Signal Behav. 2010;5(11):1520–1522. doi: 10.4161/psb.5.11.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clough SJ, and Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 31.Green MR, and Sambrook J. Molecular cloning: A laboratory manual. New York, USA: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 32.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q, Cao TJ, Zheng H, Zhou CF, Wang Z, Wang R, and Lu S. Manipulation of carotenoid metabolic flux by lycopene cyclization in ripening red pepper (Capsicum annuum var. conoides) fruits. J Agric Food Chem. 2019;67(15):4300–4310. doi: 10.1021/acs.jafc.9b00756. [DOI] [PubMed] [Google Scholar]
- 34.Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1(4):2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]


