Abstract
A survey of Penicillium in the fynbos biome from South Africa resulted in the isolation of 61 species of which 29 were found to be new. In this study we focus on Penicillium section Canescentia, providing a phylogenetic re-evaluation based on the analysis of partial beta-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) sequence data. Based on phylogenies we show that five fynbos species are new and several previously assigned synonyms of P. canescens and P. janczewskii should be considered as distinct species. As such, we provide descriptions for the five new species and introduce the new name P. elizabethiae for the illegitimate P. echinatum. We also update the accepted species list and synonymies of section Canescentia species and provide a review of extrolites produced by these species.
Citation: Visagie CM, Frisvad JC, Houbraken J, et al. 2021. A re-evaluation of Penicillium section Canescentia, including the description of five new species. Persoonia 46: 163–187. https://doi.org/10.3767/persoonia.2021.46.06.
Keywords: DNA barcodes, Genealogical Concordance, Phylogenetic Species, Recognition (GCPSR), new taxa, secondary metabolites, series Atroveneta, series Canescentia
INTRODUCTION
Penicillium section Canescentia species are mostly reported from soil and leaf litter (Raper & Thom 1949, Domsch et al. 1980, Pitt 1980, Ramírez 1982) and typically have terminal biverticillate conidiophores with subterminal branching and broad, short swollen phialides (Pitt 1980, Houbraken & Samson 2011). The section is mainly based on P. canescens and P. janczewskii but includes common species such as P. antarcticum, P. atrovenetum and P. novae-zeelandiae. Past classifications of section Canescentia highlight the difficulty of using morphology and more specifically conidiophore branching patterns to define groups in Penicillium. Penicillium canescens and P. janczewskii produce a high proportion of divaricate conidiophores and were therefore respectively placed in sections Asymmetrica (Raper & Thom 1949) and Divaricatum (Pitt 1980). On the other hand, species such as P. novae-zeelandiae and P. coralligerum produce symmetrical, biverticillate conidiophores and were classified in sections Biverticillata-Symmetrica (Raper & Thom 1949) and Furcatum (Pitt 1980), respectively. These past classifications have, however, long been shown to be relatively superficial and are not reflected in phylogenetic classifications (Peterson 2000, Houbraken & Samson 2011). As a result, a phylogenetic approach to subgeneric classifications has become the standard for Penicillium (Houbraken & Samson 2011, Visagie et al. 2014b, Houbraken et al. 2020).
Difficulties in using morphological characters to identify strains of either P. canescens and P. janczewskii were noted by Pitt (1980). Colonies of these two were found to be similar and were described on Czapek yeast autolysate agar (CYA) as having white to yellow coloured mycelia and reaching diameters between 25–32 mm after 7 d incubation. On malt extract agar (MEA) (Blakeslee 1915), colonies were found to typically be 15–25 mm wide, floccose and producing bluish to greenish grey conidia. As such, Pitt (1980) distinguished between the two based on the rough-walled stipes and smooth-walled conidia of P. canescens, in contrast to P. janczewskii that was characterised by smooth-walled stipes and rough-walled conidia. However, Pitt (1980) also reported on the existence of strains, such as IMI 149218, that bridge these characters forming roughened stipes and conidia, which he placed in P. canescens. As a result of this rather broad concept of both species, Pitt (1980) synonymised several species with P. canescens (= P. raciborskii, P. kapuscinksii, P. novae-caledoniae and P. yarmokense) and P. janczewskii (= P. echinatum (nom. illegit.), P. swiecickii, P. nigricans and P. nigricans var. sulphuratum). However, identification in this group remained problematic. In recent years, phylogenies helped to resolve some of these species. Houbraken & Samson (2011) reclassified Penicillium and divided the genus into 25 sections, classifying eight species (P. antarcticum, P. atrovenetum, P. canescens, P. coralligerum, P. janczewskii, P. jensenii, P. novae-zeelandiae and P. yarmokense) in section Canescentia. Since this classification, several new species were introduced in the section while ex-type strains of old names were also sequenced (Visagie et al. 2014a, 2016b, Grijseels et al. 2016, Kirichuk et al. 2016). In a recent Eurotiales review, a new series classification was introduced for Aspergillus and Penicillium and section Canescentia was divided into series Atroveneta and Canescentia, the latter containing species with biverticillate to terverticillate conidiophores and divergent branching, while the former have symmetrically biverticillate conidiophores (Houbraken et al. 2020).
A survey of Penicillium occurring in the diverse fynbos biome situated in South Africa was initiated in 2009 to characterise Penicillium isolated from soil, air, and mites associated with Protea repens infructescences. The ± 1 700 strains were found to represent 10 Penicillium sections and 61 species, including 29 described as new. The current study follows on from previous papers describing these new species in sections Aspergilloides (Houbraken et al. 2014), Citrina (Visagie et al. 2014c), Exilicaulis (Visagie et al. 2016c), Lanata-Divaricata (Visagie et al. 2015a), Sclerotiora (Visagie et al. 2013) and Torulomyces (Visagie et al. 2016a). The aim of this paper was to provide descriptions for five new Penicillium species and compare them with others from section Canescentia using phylogenetic inference from three gene regions, morphology and extrolite data. In the process, we review the classification of previously described species in the section, provide reference sequences for these and a review of extrolites produced by this group.
MATERIALS AND METHODS
Isolates
Isolates were obtained from soil, air and Protea repens infructescences collected from the fynbos biome as previously described by Visagie et al. (2014c). These strains were accessioned in the collections of the DAOMC (Canadian Collection of Fungal Cultures, Agriculture & Agri-Food Canada, Ottawa, Canada), IBT (the culture collection at the Department of Systems Biology, DTU, Lyngby, Denmark) and CBS (Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands). Reference strains were obtained from IBT, CBS and NRRL (United States Department of Agriculture, Agricultural Research Service, USDA-ARS, Peoria, United States). Table 1 summarises strains, their origin and GenBank accession numbers used for this study.
Table 1.
Strains used for phylogenetic analyses.
| Species name | Collection numbersa | Source, location | GenBank accession numbers |
|||
|---|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB2 | |||
| Penicillium allsoppiae | CBS:138943, DAOMC:241348, DTO:182-D5, CV:931 (ex-type) | Soil sample, Malmesbury, South Africa | JX140830 | JX140992 | JX157384 | KP016895 |
| CBS:138945, IBT:31952, DAOMC:241349, DTO:183-C8, CV:1704 | Soil sample, Struisbaai, South Africa | JX140822 | JX141004 | JX157399 | KP016910 | |
| CN086C6, S4C2 | Soil, Hopefield, South Africa | MW364385 | MW357820 | MW357831 | MW357840 | |
| CN086C7, S4C3 | Soil, Hopefield, South Africa | MW364386 | MW357821 | MW357832 | MW357841 | |
| CN086C8, S2E3 | Soil, Hopefield, South Africa | MW364387 | MW357822 | – | MW357842 | |
| Penicillium antarcticum | CBS:100492, FRR:4989 (ex-type) | Soil scraping, Ardery Island, Antarctica | KJ834503 | MN969371 | MN969236 | JN406653 |
| CBS:116938, IBT:3405, IBT:3742, IBT:4599, IBT:6834, DTO:187-D6 | Salami, Hillerod, Denmark | KP016845 | KP016925 | KP016827 | KP016848 | |
| CBS:116939, IBT:4017, DTO:187-B7 | Beach sand, Rorvig, Denmark | KP016829 | KP016921 | JX157255 | KP016849 | |
| KMM:4668 (ex-type of P. piltunense) | Subaqueous soil, Piltun Bay, Sakhalin island, Russia | KU358554 | KU358557 | KU358560 | – | |
| KMM:4670 (ex-type of P. ochotense) | Subaqueous soil, Piltun Bay, Sakhalin island, Russia | KU358553 | KU358556 | KU358559 | – | |
| KMM:4671 (ex-type of P. attenuatum) | Subaqueous soil, Sakhalin Bay, Sakhalin island, Russia | KU358555 | KU358558 | KU358561 | – | |
| Penicillium arizonense | DTO:216-H4 | Root tissue of Artemisia tridentata, USA | – | MF974900 | – | – |
| IBT:12289, CBS:141311 (ex-type) | Dry red soil, South rim of Grand Canyon, Arizona, USA | MH492021 | MH492019 | MH492020 | MH492022 | |
| Penicillium atrovenetum | CBS:241.56, ATCC:13352, FRR:2571, IFO:8138, IMI:061837 (ex-type) | Soil, Sussex Downs, England | AF033492 | JX140944 | KJ867004 | JN121467 |
| CBS:243.56, FRR:1666, IMI:061835 | Soil from spinach field, Norfolk, England | KP016835 | JX140945 | MN969241 | KP016854 | |
| Penicillium canescens | CBS:300.48, ATCC:10419, FRR:910, IMI:28260, MUCL:29169, NRRL:910 (ex-type) | Soil, England | AF033493 | JX140946 | KJ867009 | JN121485 |
| EN:1377 | Unknown source, Iran | – | KT285862 | – | – | |
| FMR:15028 | Dung, Spain | – | LT898237 | – | – | |
| IMI:149218, IBT:5978, DTO:189-A2 | Soil sample, Gharyan, Libya | KP016841 | JX140951 | KP027409 | KP016917 | |
| MUT<ITA>:1764 | Oil polluted water from the Mediterranean Sea, Italy | – | KU935636 | – | – | |
| MUT<ITA>:1815 | Oil polluted water from the Mediterranean Sea, Italy | – | KU935637 | – | – | |
| SQU14069 | Unknown | – | MH000349 | – | – | |
| Penicillium cf. murcianum | CBS:414.68 | Unknown source, Helsinki, Finland | KP016842 | KP016922 | KP016821 | KP016859 |
| CN086C9, S2C3 | Soil, Hopefield, South Africa | MW364388 | MW357823 | MW357833 | MW357843 | |
| CN086D1, S1C3 | Soil, Hopefield, South Africa | MW364389 | MW357824 | MW357834 | MW357844 | |
| CN086D2, S2B8 | Soil, Hopefield, South Africa | MW364390 | MW357825 | MW357835 | MW357845 | |
| CN086D7, S1C1 | Soil, Hopefield, South Africa | MW364395 | MW357830 | MW357839 | MW357850 | |
| IBT:31963, DAOMC:241111, DTO:182-A9, CV:816 | Air sample, Malmesbury, South Africa | JX140834 | JX140949 | JX157369 | KP016880 | |
| NRRL:35656 | Cheek pouch of kangaroo rat, USA | – | DQ658166 | – | – | |
| Penicillium coralligerum | CBS:114.69, FRR:1964, IMI:130675 | Soil under Hordeum sp., Canada | KP016836 | KJ866970 | KJ866991 | KP016847 |
| CBS:123.65, ATCC:16968, FRR:3465, IFO:9578, IMI:099159, NRRL:3465 (ex-type) | Seed of Hordeum vulgate, France | JN617667 | MN969378 | MN969248 | JN406632 | |
| Penicillium corvianum | CN086D3, S1G8 | Soil, Hopefield, South Africa | MW364391 | MW357826 | MW357836 | MW357846 |
| CN086D5, S1G1 | Soil, Hopefield, South Africa | MW364393 | MW357828 | MW357838 | MW357848 | |
| CN086D6, S1H8 | Soil, Hopefield, South Africa | MW364394 | MW357829 | – | MW357849 | |
| DAOMC:250517, CBS141000 (ex-type) | Soil, Natural Monument Corviano, Tuscany, Italy | KT887875 | KT887836 | KT887797 | MN969170 | |
| DAOMC:250518, CBS141001 | Soil, Natural Monument Corviano, Tuscany, Italy | KT887876 | KT887837 | KT887798 | – | |
| DTO:216-F4 | Root tissue of Psedotsuga menziesii, USA | – | MF974905 | – | – | |
| DTO:216-F7 | Root tissue of Psedotsuga menziesii, USA | – | MF974906 | – | – | |
| Penicillium doidgeae | CBS:138947, IBT:31950, DAOMC:241107, DTO:183-G7, CV:2189 (ex-type) | Mite from Protea repens infructescence, Struisbaai, South Africa | JX140804 | JX141006 | JX157413 | KP016915 |
| CBS:138948, IBT:31951, DAOMC:241108, DTO:183-G8, CV:2191 | Mite from Protea repens infructescence, Struisbaai, South Africa | JX140805 | JX141007 | JX157414 | KP016916 | |
| Penicillium dunedinense | CBS:138218, DTO:244-G1 (ex-type) | House dust, Dunedin, New Zealand | KJ775678 | KJ775171 | KJ775405 | MN969116 |
| Penicillium eickeri | CBS:138939, IBT:31921, DAOMC:241352, DTO:181-G3, CV:475 (ex-type) | Mite from Protea repens infructescence, Stellenbosch, South Africa | JX140824 | JX140979 | JX157365 | KP016876 |
| CBS:138940, IBT:31956, DAOMC:241350, DTO:181-G5, CV:487 | Bract from Protea repens infructescence, Stellenbosch, South Africa | JX140825 | JX140980 | JX157366 | KP016877 | |
| Penicillium elizabethiae | NRRL:917, MUCL:29170, IBT:21955, DTO:189-B8 (ex-type) | Soil, Scotland | KP016840 | KJ866964 | KJ867021 | KP016918 |
| Penicillium griseoazureum | CBS:162.42, FRR:1361 (ex-type) | Dune sand, France | KC411679 | KP016919 | KP016823 | KP016852 |
| Penicillium janczewskii | CBS:166.81, IMI:253795 (ex-type of P. granatense) | Air sample, Madrid, Spain | KC411682 | KJ866967 | KJ866998 | KP016853 |
| CBS:221.28, FRR:919, IMI:191499, NRRL:919 (ex-type) | Soil under Pinus sp., Poland | AY157487 | MN969386 | MN969267 | JN406612 | |
| CBS:279.47, ATCC:9439 | Soil, England | KP016837 | KJ866968 | KJ867008 | KP016855 | |
| CBS:413.69, FRR:518, IMI:140344 | Unknown source, Helsinki, Finland | KP016838 | KJ866969 | KJ867014 | KP016858 | |
| CBS:744.70, ATCC:18380 (ex-type of P. nigricans var. sulphureum) | Unknown source, Japan | KP016839 | KJ866966 | KJ867018 | KP016862 | |
| MUT<ITA>:2710 | Dysidea fragilis from Atlantic Ocean, Ireland | – | MG832199 | – | – | |
| MUT<ITA>:6182 | Contaminated soil, Italy | – | MK521578 | – | – | |
| Penicillium jensenii | CBS:216.28, ATCC:10456, IFO:5747, IMI:068233, NRRL:3431 | Unknown | JN617693 | KJ866963 | KJ867000 | JN606629 |
| CBS:327.59, ATCC:18317, FRR:909, IFO:5764, IMI:039768, NRRL:909 (ex-type) | Forest soil, Japan | AY443470 | JX140954 | AY443490 | JN406614 | |
| Penicillium linzhiense | CCTCC-M2019870 | Soil, Linzhi Town, Tibet Autonomous Region, China | MT461156 | MT461157 | MT461162 | – |
| Penicillium murcianum | CBS:161.81, ATCC:42239, IMI:253800 (ex-type) | Sandy soil, Madrid, Spain | KP016844 | MN969419 | MN969341 | MN969202 |
| CBS:458.69 | Soil, Turkey | KP016843 | KP016923 | KP016822 | KP016860 | |
| CN086D4, S1O1 | Soil, Hopefield, South Africa | MW364392 | MW357827 | MW357837 | MW357847 | |
| Penicillium nigricans | CBS:354.48, ATCC:10115, IFO:6103, IMI:039767, NRRL:915 (ex-type) | Unknown source, France | KC411755 | KJ866965 | KJ867012 | KP016857 |
| Penicillium novaezeelandiae | CBS:137.41, ATCC:10473, IFO:31748, IMI:040584ii, NRRL:2128 (ex-type) | Apothecium of Sclerotinia sp., Palmerston North, New Zealand | JN617688 | MN969390 | MN969279 | JN406628 |
| CBS:138936, IBT:31940, DAOMC:241112, DTO:180-G9, CV:42 | Air sample, Stellenbosch, South Africa | JX140853 | JX140956 | JX157352 | KP016864 | |
| CBS:138938, IBT:31937, DAOMC:241113, DTO:181-A6, CV:117 | Bract from Protea repens infructescence, Stellenbosch, South Africa | JX140846 | JX140958 | JX157356 | KP016868 | |
| CBS:138942, IBT:31947, DAOMC:241354, DTO:182-C8, CV:909 | Soil sample, Malmesbury, South Africa | JX140854 | JX140969 | JX157380 | KP016891 | |
| CBS:138949, DTO:184-D5, CV:47 | Air sample, Stellenbosch, South Africa | JX140835 | JX140957 | JX157353 | KP016865 | |
| CBS:138950, IBT:31931, DTO:185-D6, CV:818 | Air sample, Malmesbury, South Africa | JX140843 | JX140968 | JX157370 | KP016881 | |
| CBS:138952, IBT:31949, DTO:185-H9, CV:992 | Mite from Protea repens infructescence, Malmesbury, South Africa | JX140845 | JX140971 | JX157393 | KP016904 | |
| CBS:138953, IBT:31944, DTO:186-A3, CV:1290 | Bract from Protea repens infructescence, Malmesbury, South Africa | JX140847 | JX140972 | JX157395 | KP016906 | |
| CBS:138954, IBT:31946, DTO:186-B7, CV:1560 | Bract from Protea repens infructescence, Malmesbury, South Africa | JX140848 | JX140973 | JX157398 | KP016909 | |
| CBS:138955, IBT:31941, DTO:186-H1, CV:2029 | Mite from Protea repens infructescence, Struisbaai, South Africa | JX140850 | JX140975 | JX157401 | KP016913 | |
| CBS:138956, IBT:31948, DTO:186-H2, CV:2051 | Mite from Protea repens infructescence, Struisbaai, South Africa | JX140851 | JX140976 | JX157402 | KP016914 | |
| CBS:546.77 | Stem of Vitis vinifera, Auckland, New Zealand | KP016846 | KP016926 | KP016828 | KP016861 | |
| CV:0200 | Bract from Protea repens infructescence, Stellenbosch, South Africa | JX140838 | JX140961 | JX157360 | – | |
| IBT:31930, DTO:184-I1, CV:406 | Mite from Protea repens infructescence, Stellenbosch, South Africa | JX140840 | JX140964 | JX157363 | KP016874 | |
| IBT:31932, DTO:186-D9, CV:1812 | Mite from Protea repens infructescence, Struisbaai, South Africa | JX140849 | JX140974 | JX157400 | KP016912 | |
| IBT:31933, DTO:185-B7, CV:616 | Mite from Protea repens infructescence, Stellenbosch, South Africa | JX140842 | JX140967 | JX157368 | KP016879 | |
| IBT:31934, DTO:184-F1, CV:129 | Mite from Protea repens infructescence, Stellenbosch, South Africa | JX140836 | JX140959 | JX157357 | KP016869 | |
| IBT:31935, DTO:185-F3, CV:910 | Soil sample, Malmesbury, South Africa | JX140844 | JX140970 | JX157381 | KP016892 | |
| IBT:31936, DTO:185-B4, CV:587 | Mite from Protea repens infructescence, Stellenbosch, South Africa | JX140841 | JX140966 | JX157367 | KP016878 | |
| IBT:31938, DTO:184-F5, CV:147 | Mite from Protea repens infructescence, Stellenbosch, South Africa | JX140837 | JX140960 | JX157358 | KP016870 | |
| IBT:31939, DTO:184-I3, CV:452 | Mite from Protea repens infructescence, Stellenbosch, South Africa | KP016830 | JX140965 | JX157364 | KP016875 | |
| IBT:31942, DTO:184-H7, CV:337 | Bract from Protea repens infructescence, Stellenbosch, South Africa | JX140839 | JX140962 | JX157361 | KP016872 | |
| IBT:31943, DTO:181-E8, CV:355 | Mite from Protea repens infructescence, Stellenbosch, South Africa | JX140852 | JX140963 | JX157362 | KP016873 | |
| Penicillium nucicola | DAOMC:250521, CBS:140973, KAS:2101, W:109 | Fallen nuts of Fagus grandifolia, Milton, Ontario, Canada | KT887846 | KT887807 | KT887768 | – |
| DAOMC:250522, CBS:140987, KAS:2203, W:59 (ex-type) | Fallen nuts of Carya ovata, Niagara Falls, Ontario, Canada | KT887860 | KT887821 | KT887782 | MN969171 | |
| Penicillium pole-evansii | CBS:138946, IBT:31929, DAOMC:241106, DTO:183-D5, CV:1758 (ex-type) | Bract from Protea repens infructescence, Struisbaai, South Africa | JX140831 | JX141005 | JX157412 | KP016911 |
| Penicillium radiatolobatum | CBS:340.79 (ex-type) | Soil, Romania | KC411745 | MN969413 | MT066183 | MN969168 |
| FMR:15304 | Dung, Spain | – | LT898293 | – | – | |
| FMR:15305 | Dung, Spain | – | LT898294 | – | – | |
| FMR:15308 | Dung, Spain | – | LT898295 | – | – | |
| FMR:15485 | Dung, Spain | – | LT898299 | – | – | |
| FMR:15491 | Dung, Spain | – | LT898296 | – | – | |
| FMR:15845 | Dung, Spain | – | LT898297 | – | – | |
| IBT:31958, DAOMC:241110, DTO:184-G3, CV:198 | Bract from Protea repens infructescence, Stellenbosch, South Africa | JX140832 | JX140948 | JX157359 | KP016871 | |
| IBT:31959, DAOMC:241109, DTO:180-H5, CV:68 | Soil sample, Stellenbosch, South Africa | JX140833 | JX140947 | JX157354 | KP016866 | |
| Penicillium sacculum | CBS:231.61, MUCL:51025, IMI:68319 | Soil, Canary Islands, Spain | KC411707 | KJ834488 | KU896849 | JN121462 |
| Penicillium scottii | CBS:138935, IBT:31955, DAOMC:241192, DTO:180-G5, CV:30 | Air sample, Stellenbosch, South Africa | JX140823 | JX140977 | JX157351 | KP016863 |
| CBS:138937, IBT:31954, DTO:180-H9, CV:75 | Soil sample, Stellenbosch, South Africa | JX140826 | JX140978 | JX157355 | KP016867 | |
| CBS:138941, IBT:31957, DAOMC:241163, DTO:182-B4, CV:864 | Air sample, Malmesbury, South Africa | JX140827 | JX140981 | JX157371 | KP016882 | |
| CBS:138944, IBT:31964, DAOMC:241162, DTO:183-B9, CV:1502 | Bract from Protea repens infructescence, Malmesbury, South Africa | JX140820 | JX141002 | JX157396 | KP016907 | |
| CBS:138951, IBT:31905, DTO:185-F8, CV:930 (ex-type) | Soil sample, Malmesbury, South Africa | JX140812 | JX140991 | JX157383 | KP016894 | |
| DTO:185-G2, CV:939 | Soil sample, Malmesbury, South Africa | JX140814 | JX140994 | JX157386 | KP016897 | |
| IBT:31903, DTO:186-B4, CV:1512 | Bract from Protea repens infructescence, Malmesbury, South Africa | JX140821 | JX141003 | JX157397 | KP016908 | |
| IBT:31904, DTO:185-G6, CV:953 | Soil sample, Malmesbury, South Africa | KP016833 | JX140995 | JX157387 | KP016898 | |
| IBT:31906, DTO:185-H1, CV:958 | Soil sample, Malmesbury, South Africa | JX140815 | JX140996 | JX157388 | KP016899 | |
| IBT:31907, DTO:185-E7, CV:898 | Soil sample, Malmesbury, South Africa | KP016832 | JX140988 | JX157378 | KP016889 | |
| IBT:31908, DTO:185-H3, CV:967 | Soil sample, Malmesbury, South Africa | JX140816 | JX140997 | JX157389 | KP016900 | |
| IBT:31909, DTO:185-H4, CV:969 | Soil sample, Malmesbury, South Africa | KP016834 | JX140998 | JX157390 | KP016901 | |
| IBT:31910, DTO:185-H7, CV:975 | Soil sample, Malmesbury, South Africa | JX140817 | JX140999 | JX157391 | KP016902 | |
| IBT:31911, DTO:185-H8, CV:978 | Soil sample, Malmesbury, South Africa | JX140818 | JX141000 | JX157392 | KP016903 | |
| IBT:31912, DTO:185-E3, CV:890 | Soil sample, Malmesbury, South Africa | JX140808 | JX140984 | JX157374 | KP016885 | |
| IBT:31913, DTO:185-F7, CV:929 | Soil sample, Malmesbury, South Africa | JX140811 | JX140990 | JX157382 | KP016893 | |
| IBT:31914, DTO:185-G1, CV:937 | Soil sample, Malmesbury, South Africa | JX140813 | JX140993 | JX157385 | KP016896 | |
| IBT:31915, DTO:185-D8, CV:874 | Soil sample, Malmesbury, South Africa | JX140807 | JX140982 | JX157372 | KP016883 | |
| IBT:31916, DTO:185-E4, CV:892 | Soil sample, Malmesbury, South Africa | JX140809 | JX140985 | JX157375 | KP016886 | |
| IBT:31917, DTO:185-E6, CV:897 | Soil sample, Malmesbury, South Africa | JX140810 | JX140987 | JX157377 | KP016888 | |
| IBT:31918, DAOMC:241164, DTO:182-C2, CV:893 | Soil sample, Malmesbury, South Africa | JX140828 | JX140986 | JX157376 | KP016887 | |
| IBT:31919, DTO:185-E1, CV:881 | Soil sample, Malmesbury, South Africa | KP016831 | JX140983 | JX157373 | KP016884 | |
| IBT:31922, DAOMC:241353, DTO:182-H4, CV:1163 | Mite from Protea repens infructescence, Malmesbury, South Africa | JX140819 | JX141001 | JX157394 | KP016905 | |
| IBT:31953, DAOMC:241351, DTO:182-C4, CV:899 | Soil sample, Malmesbury, South Africa | JX140829 | JX140989 | JX157379 | KP016890 | |
| Penicillium yarmokense | CBS:410.69, FRR:520, IMI:140346 (ex-type) | Soil, Syria | KC411757 | MN969407 | MN969314 | JN406553 |
| DTO:216-C9 | Root tissue of Pinus ponderosa, USA | – | MF974897 | – | – | |
| DTO:216-D8 | Root tissue of Pinus monticola, USA | – | MF974898 | – | – | |
| DTO:216-E4 | Root tissue of Psedotsuga menziesii, USA | – | MF974899 | – | – | |
a Culture collection designations: ATCC, American Type Culture Collection, Manassas, USA; CBS, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CV, internal culture collection at the Department of Microbiology, University of Stellenbosch, South Africa; DAOM, culture collection and herbarium of the National Mycological Collections, Agriculture & Agri-Food Canada, Ottawa; DTO, internal culture collection of CBS; FRR, Food Science Australia, Ryde, Australia; IBT, culture collection of Center for Microbial Biotechnology (CMB) at Department of Systems Biology, Technical University of Denmark; IFO, Institute for Fermentation, Osaka, Japan; IMI, CABI Genetic Resources Collection, Surrey, UK; MUCL, Mycotheque de l’Universite catholique de Louvain, Belgium; NRRL, Agricultural Research Service Culture Collection, Peoria, Illinois, USA.
Morphology
Colony morphologies were recorded from fynbos strains grown on Czapek yeast autolysate agar (CYA), Czapek yeast autolysate agar with 5 % NaCl (CYAS), malt extract agar (MEA; Oxoid), yeast extract sucrose agar (YES), dichloran 18 % glycerol agar (DG18) and creatine sucrose agar (CREA) incubated at 25 °C for 7 d. Additional CYA plates were incubated at 30 and 37 °C for 7 d. Media preparation, inoculation technique, incubation conditions and microscope preparations were standardised according to recommended methods (Visagie et al. 2014b). Colour names and codes used in descriptions follow Kornerup & Wanscher (1967). Microscopic observations and measurements were made using an Olympus SZX12 dissecting and Olympus BX50 compound microscopes. These were equipped with an Evolution MP digital microscope camera and ImagePro v. 6.0 software. Affinity Photo v. 1.7.3 (Serif (Europe) Ltd) was used for creating photographic plates.
Phylogenetic analyses
DNA extractions were made using the ZR Fungal/Bacterial DNA kit (ZymoResearch, California) from 7-d-old colonies grown on MEA. PCR of the internal transcribed spacer rDNA region (ITS), beta-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) was carried out using primer pairs and amplification protocols as described in Visagie et al. (2014b), while reagents and master mixes were prepared following Visagie et al. (2013). Sequence contigs were assembled in Geneious Prime v. 2019.2.3 (Biomatters, NZ). GenBank accession numbers for sequences used in this study are summarised in Table 1.
A sequence dataset was compiled containing both ex-type reference sequences and fynbos Penicillium of section Canescentia. An ITS phylogeny of only ex-type sequences was calculated to determine if the ITS of the rDNA gene cluster distinguish between the species. Subsequently, single-gene trees were calculated for partial BenA, CaM and RPB2 genes, as well as trees representing concatenated datasets. All alignments were done using MAFFT v. 1.764b (Katoh & Standley 2013) and selecting the G-INS-i option. Phylogenetic analyses were performed using both Maximum Likelihood (ML) and Bayesian tree inference (BI). The multigene datasets were concatenated in Geneious Prime, with the partitioning scheme and substitution models selected using Partitionfinder v. 2.1.1 (Lanfear et al. 2017) allowing for introns, exons and codon positions to be independent datasets.
Maximum Likelihood trees were calculated using IQtree v. 1.6.12 (Nguyen et al. 2015) and nonparametric bootstrapping done with 1 000 replicates (Felsenstein 1985). Bayesian inference trees were calculated in MrBayes v. 3.2.7a (Ronquist et al. 2012). Analyses were run using two sets of four chains (one cold and three heated) and the stoprule was applied to stop the analyses once the average standard of deviation for split frequencies reached 0.01. Sample frequency was set at 100 with 25 % of the trees removed as burn-in. ML phylograms were used to represent data with both bootstrap values ≥ 80 % and/or posterior probabilities ≥ 0.95 given on thickened branches. Trees were visualised on the Interactive tree of life (iTOL) v. 3 (Letunic & Bork 2016) and edited for publication in Affinity Publisher and Designer v. 1.8.6 (Serif (Europe) Ltd, Nottingham, UK).
Extrolite analysis
Strains were grown on CYA and YES at 25 °C for 7 d with five agar plugs taken from each medium and pooled into one sample. Extractions were made with 0.75 mL ethyl acetate/dichloromethane/methanol (3 : 2 : 1) (v/v/v) with 1 % (v/v) formic acid. Extracts were filtered and analysed by HPLC using alkylphenone retention indices and diode array UV-VIS detection (Frisvad & Thrane 1987, 1993, Nielsen et al. 2011), with minor modifications described in Smedsgaard (1997). A 50 × 2 mm Luna C-18 (II) reversed-phase column (Phenomenex, CA, USA) fitted with a 2 × 2 mm guard column was used for analyses (Nielsen et al. 2011).
RESULTS
Isolates
Isolations resulted in ± 1 700 Penicillium isolates obtained from collected fynbos samples. Of these, 155 belonged in section Canescentia, which were placed into eight distinct morpho-groups based on colony characters on CYA and MEA. These groupings were later confirmed by sequencing representative strains included here in the phylogenetic analyses. Most strains belonged to two groups, later identified as P. novae-zeelandiae (115 strains) and a new species described here as P. scottii (30 strains). The largest proportion of P. novae-zeelandiae strains were obtained from Protea repens infructescence bracts, while most P. scottii strains originate from fynbos soil. From each morpho-group, a subset of strains was included for phylogenetic analyses (Fig. 1 2, 3 and 4). Additional strains of P. allsoppiae, isolated in another fynbos survey from soil obtained from wheat farms, were also included in the phylogenies.
Fig. 1.
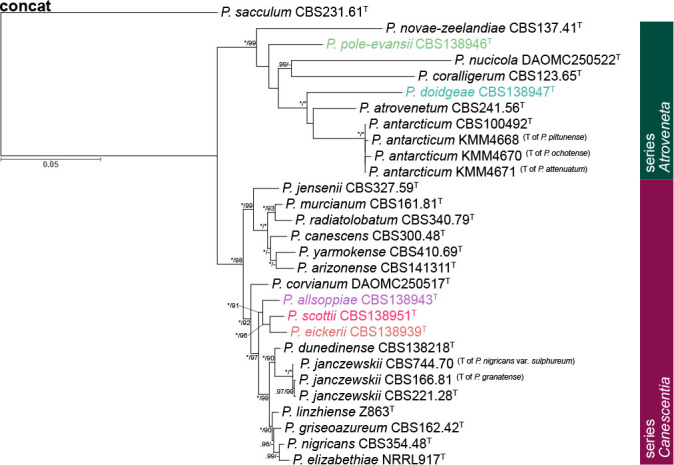
Phylogenetic tree of Penicillium section Canescentia ex-type strains using a concatenated dataset of BenA, CaM and RPB2. Penicillium sacculum was chosen as outgroup. Posterior probabilities (pp) and/or bootstrap values (bs) higher than 0.95 and 80, respectively, are given above thickened branches. Names in coloured text represent new species, T = ex-type strain.
Fig. 2.
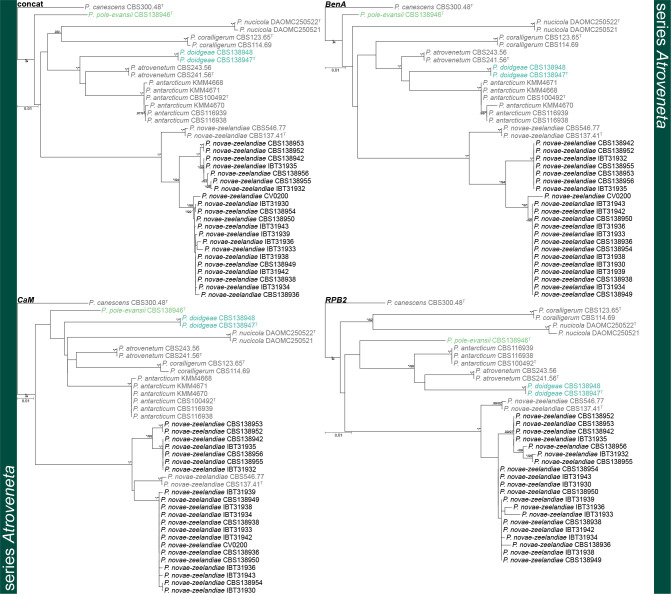
Phylogenetic trees of Penicillium section Canescentia series Atroveneta based on BenA, CaM, RPB2 and concatenated datasets. Penicillium canescens was chosen as outgroup. Posterior probabilities (pp) and/or bootstrap values (bs) higher than 0.95 and 80, respectively, are given above thickened branches. Names in grey text indicate reference strains, black text fynbos strains and coloured text new species, T = ex-type strain.
Fig. 3.
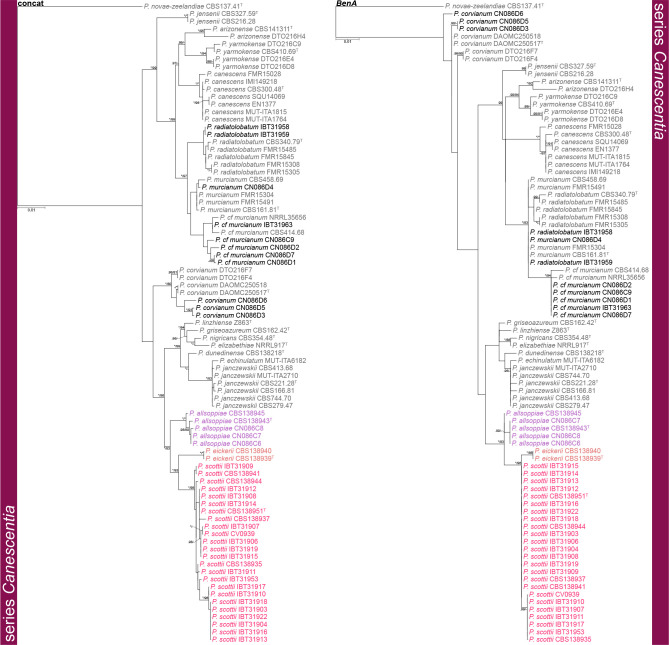
Phylogenetic trees of Penicillium section Canescentia series Canescentia based on BenA and concatenated datasets. Penicillium novae-zeelandiae was chosen as outgroup. Posterior probabilities (pp) and/or bootstrap values (bs) higher than 0.95 and 80, respectively, are given above thickened branches. Names in grey text indicate reference strains, black text fynbos strains and coloured text new species, T = ex-type strain.
Fig. 4.
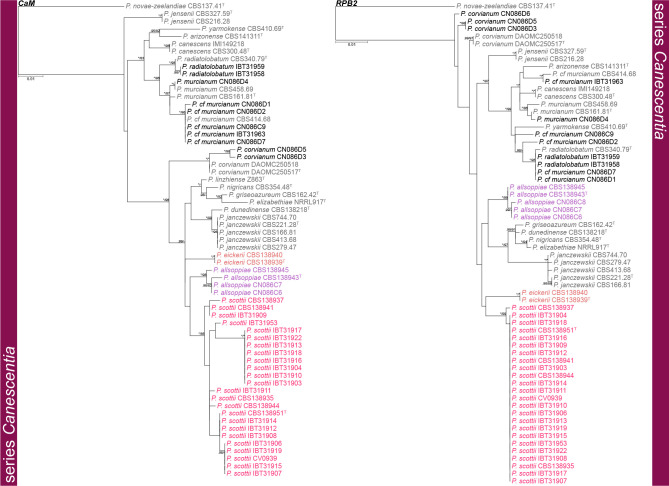
Phylogenetic trees of Penicillium section Canescentia series Canescentia based on CaM and RPB2. Penicillium novae-zeelandiae was chosen as outgroup. Posterior probabilities (pp) and/or bootstrap values (bs) higher than 0.95 and 80, respectively, are given above thickened branches. Names in grey text indicate reference strains, black text fynbos strains and coloured text new species, T = ex-type strain.
Phylogenetic analyses
Gene sequence alignments were uploaded to TreeBASE under submission ID 22924. Dataset characteristics, partitioning schemes and substitution models applied during data processing are summarized in Table 2. Phylogenies based on ex-type sequences resolved strains into two main clades: series Atroveneta and Canescentia. ITS has poor discriminating power between section Canescentia members and was omitted from further analyses (Fig. S1).
Table 2.
Partition-merging results and best substitution model for each partitiion.
| Description of dataset | Subset | Parition name | Best Model | # sites |
|---|---|---|---|---|
| sect. Canescentia ex-types (ITS) | 1 | ITS | K80 | 509 |
| sect. Canescentia ex-types (concat) | 1 | CaM_codon1, BenA_codon2, CaM_codon3, RPB2_codon1, BenA_codon1 | TRN+I | 658 |
| 2 | CaM_codon2, BenA_codon3 | TRN+G | 163 | |
| 3 | CaM_introns, BenA_introns | K80+G | 460 | |
| 4 | RPB2_codon2 | JC | 327 | |
| 5 | RPB2_codon3 | HKY+G | 326 | |
| ser. Atroveneta (BenA) | 1 | BenA_codon1, BenA_codon2, BenA_codon3, | TRNEF | 247 |
| 2 | BenA_introns | K80+G | 192 | |
| ser. Atroveneta (CaM) | 1 | CaM_codon1, CaM_codon3 | K80+I | 179 |
| 2 | CaM_codon2 | HKY+I | 88 | |
| 3 | CaM_introns | K80+I | 239 | |
| ser. Atroveneta (RPB2) | 1 | RPB2_codon1 | TRN+I | 284 |
| 2 | RPB2_codon2 | JC | 284 | |
| 3 | RPB2_codon3 | HKY+G | 283 | |
| ser. Atroveneta (concat) | 1 | CaM_codon1, BenA_codon2, CaM_codon3, BenA_codon1, RPB2_codon1 | TRN+G | 629 |
| 2 | CaM_codon2, BenA_codon3 | HKY+G | 169 | |
| 3 | BenA_introns, CaM_introns | K80+I | 431 | |
| 4 | CaM_codon2 | JC | 284 | |
| 5 | RPB2_codon3 | HKY+G | 283 | |
| ser. Canescentia (BenA) | 1 | BenA_codon1, BenA_codon3 | TRNEF | 143 |
| 2 | BenA_codon2 | JC | 71 | |
| 3 | BenA_introns | TRNEF | 201 | |
| ser. Canescentia (CaM) | 1 | CaM_codon1, CaM_codon3 | JC | 180 |
| 2 | CaM_codon2 | HKY | 89 | |
| 3 | CaM_introns | K80 | 231 | |
| ser. Canescentia (RPB2) | 1 | RPB2_codon1 | TRN | 284 |
| 2 | RPB2_codon2 | JC | 283 | |
| 3 | RPB2_codon3 | HKY+G | 283 | |
| ser. Canescentia (concat) | 1 | CaM_codon2, BenA_codon1 | TRN+G | 161 |
| 2 | CaM_codon1, BenA_codon2, RPB2_codon2 | JC | 445 | |
| 3 | BenA_codon3, BenA_introns, CaM_introns | K80+G | 503 | |
| 4 | CaM_codon3, RPB2_codon1 | TRN | 373 | |
| 5 | RPB2_codon3 | HKY+G | 28 |
Series Atroveneta — Fig. 2
Phylogenies confirmed two new species, described below as P. doidgeae and P. pole-evansii. We accept a high degree of infraspecies variation for P. novae-zeelandiae. Strains of this species resolved in three main clades with fynbos strains distinct from the ex-type strain (CBS 137.41T). However, as the morphological (Fig. 5) and extrolite data (Table 3, 4) were not consistent between these different clades, we opt to not introduce new species for them until more collections are made from different regions. Kirichuk et al. (2016) recently introduced three new species (P. attenuatum, P. ochotense and P. piltunense) and they considered P. antarcticum a close relative. However, alignments of published data (Fig. S2) revealed several frameshift mutations in coding regions at sequence ends, which may imply low-quality sequence reads. Phylogenetic trees (Fig. S3) obtained from untrimmed alignments place all three of these species on long branches within the P. antarcticum clade. Trimming of these sequences (Fig. S2) to remove suspected low-quality regions resulted in the long branches collapsing. Reference strains, unfortunately, were not available to study and re-sequence. For this reason, we reduce P. attenuatum, P. ochotense and P. piltunense to synonymy with P. antarcticum.
Fig. 5.
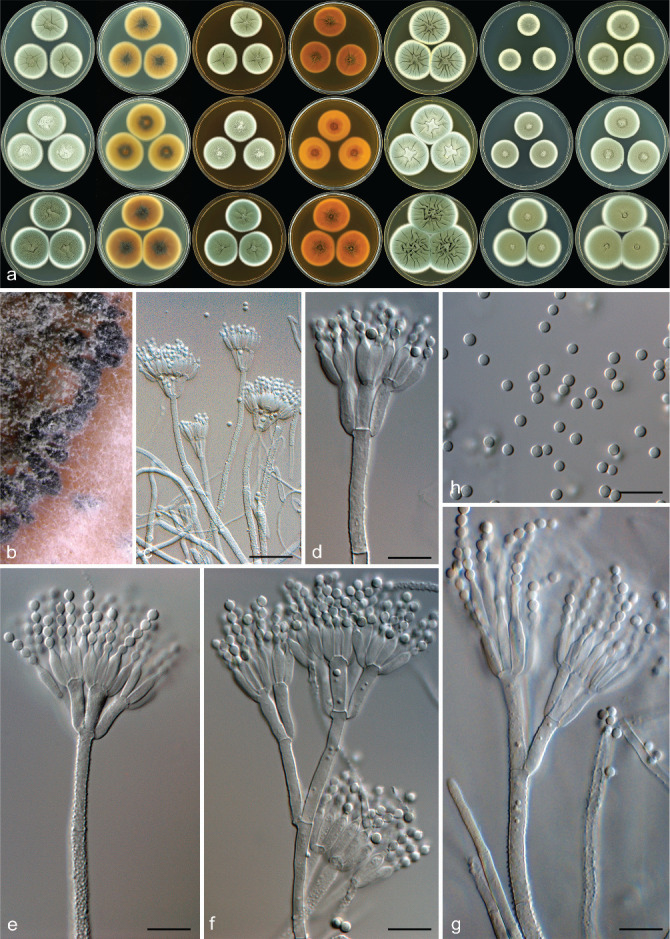
Penicillium novae-zeelandiae. a. Colonies left to right: CYA, CYA reverse, MEA, MEA reverse, YES, DG18, CYAS; top row: CBS 546.77; middle row CBS 138942; bottom row CBS 138949; b. black sclerotia produced on MEA; c–g. conidiophores; h. conidia. — Scale bars: b, c = 50 μm, d–h = 10 μm.
Table 3.
Extrolites produced by section Canescentia species (as detected in this study and reported in Visagie et al. 2016b).
| Species | Series | Extrolites |
|---|---|---|
| Penicillium allsoppiae | Canescentia | Penitrem A |
| Penicillium arizonense | Canescentia | Acetylaranotin, austalides, curvulinic acid, fumagillin, pseurotin A, pyripyropenes, tryptoquivalines, xanthoepocin |
| Penicillium canescens | Canescentia | Curvulinic acid, griseofulvin, patulodin, penicillic acid |
| Penicillium cf. murcianum | Canescentia | Not examined |
| Penicillium corvianum | Canescentia | Trichodermamide A (= penicillazine), trichodermamide C |
| Penicillium dunedinense | Canescentia | Not examined |
| Penicillium eickeri | Canescentia | Curvulinic acid, xanthoepocin |
| Penicillium elizabethiae | Canescentia | Chrysogine, communesin A & B, griseofulvin, patulodin, xanthoepocin |
| Penicillium griseoazureum | Canescentia | Curvulinic acid, a decaturin/ oxalicin, griseofulvin, pseurotin A, xanthoepocin |
| Penicillium janczewskii | Canescentia | Chrysogine, curvulinic acid, fumagillin, griseofulvin, penitrem A, penitremone A, perinadines, pseurotin A, tryptoquivalines, xanthoepocin |
| Penicillium jensenii | Canescentia | Asperpentyn?, curvulinic acid, griseofulvin, pseurotin A |
| Penicillium murcianum | Canescentia | Asperpentyn?, curvulinic acid, griseofulvin, tryptoquivalines, (apolar alkaloids / indoloterpenes) |
| Penicillium nigricans | Canescentia | Chrysogine, communesin A & B, curvulinic acid, griseofulvin, nigrifortine, patulodin, xanthoepocin |
| Penicillium radiatolobatum | Canescentia | Curvulinic acid, a decaturin / oxalicin, griseofulvin, penitrem A, penitremone A, xanthoepocin |
| Penicillium scottii | Canescentia | Curvulinic acid, dehydrogriseofulvin, griseofulvin, penitrem A, xanthoepocin |
| Penicillium yarmokense | Canescentia | Curvulinic acid, griseofulvin |
| Penicillium antarcticum | Atroveneta | Antarones, atlantinone A, asperentins, chrysogine, deoxyepifructigenine, fischerin, patulin, phyllostin, penitrem A, thomitrem A |
| Penicillium atrovenetum | Atroveneta | Atlantinone A, atrovenetin, communesin B, haenamindole, naphthalic anhydride, 3-nitropropionic acid |
| Penicillium coralligerum | Atroveneta | Austalides, chrysogine, coralligerin, naphthalic anhydride |
| Penicillium doidgeae | Atroveneta | Asperentins, atlantinone A, austalides, fischerin, patulin |
| Penicillium novae-zeelandiae | Atroveneta | Atlantinone A, benzomalvins, citreoviridin, cycloaspeptide A, cyclopiazonic acid in FRR 1905, decaturins / oxalicins, patulin, xanthoepocin in IMI 038496 and IBT 5831 |
| Penicillium nucicola | Atroveneta | Andrastin A-D, pulvilloric acid |
| Penicillium pole-evansii | Atroveneta | Atrovenetin, aurantiamine, communesin B, patulin |
Table 4.
Reported extrolites produced by members of Penicillium section Canescentia.
| Strain | Original identity | Identification method, ITS | Correct identity | Series | Extrolites |
|---|---|---|---|---|---|
| CBS 141311 | P. arizonense | MH492021, culture ex-type of P. arizonense | P. arizonense | Canescentia | Austalide B, J, K, L, curvulinic acid, dechlorogriseofulvin, 6-farnesyl-5,7-di-hydroxy-4-methylphthalide, fumagillin, griseofulvin, pseurotin A, pyripyropene A, E, F, O, tryptoquivaline C (or 27-epi-tryptoquivaline), tryptoquivaline G (or L), xanthoepocin1,2 |
| CGMCC 3.9958 | P. canescens | Reported to be 100% identical to ex-type of P. canescens | P. canescens | Canescentia | Penicanone, penicanescone A-C, integrastatin B3 |
| CGMCC 3.9958 | P. canescens | Reported to be 100% identical to ex-type of P. canescens | P. canescens | Canescentia | Canescone A-E4 (Canescone A-C, two stereoisomers each) |
| PL9A | P. canescens | Comparison to culture ex-type of P. canescens | P. canescens | Canescentia | Curvulic acid, dechlorogriseofulvin, griseofulvin, Sch 6423055 |
| ATCC 10419 | P. canescens | Culture ex-type | P. canescens | Canescentia | Griseofulvin6, Decaturin A, C, D, F, G, 15-deoxyoxalicine A & B, predecaturin E7 |
| 4.1.4.6a | P. canescens | MH820167 | P. arizonense, P. canescens, P. janczewskii, P. jensenii, P. murcianum, or P. radiatolobatum (all in series Canescentia). Presence of the bromphilone chromophore in the culture ex-type of P. canescens indicates that the fungus is indeed P. canescens | Canescentia | Bromophilone A & B, citreohybridinol, curvulinic acid, dechlorogriseofulvin, griseofulvin, griseophenone B, C, G, griseoxanthone C, methyl 3-chloro-2-(2,4-dimethoxy-6-methylphenoxy)-6-hydroxy-4-methoxybenzoate, methylcurvulinate, 3-O-methyl curvulinate, norlichexanthone, penicillic acid, piscarinine B, vulculic acid8 |
| VKM F-1148, VKM F-1287, VKM F-3108 | Unknown identity | Morphology | P. canescens | Canescentia | Fellutanine A, isorugulosuvine A & B9 |
| Sp. 4829 | Penicillium species | MH465534 | P. rubens but extrolite data suggest P. canescens | Canescentia | Citreohybriddione C, citreohybridinol, curvulinic acid, 2,4-dihydroxy-6-(oxopropyl) benzoic acid, (S)-2-(2-hydroxypropanamido) benzamide, griseofulvin, N-(2-hydroxypropanoyl)-2-amino benzoic acid, niacinamide, methyl 2-acetyl-3,5-dihydroxyphenylacetate, 6-O-methylnorlichexanthone, penicamide A & B, peni-isochroman A & B, 2-pyruvamidobenzamide, 2-pyruvoylaminobenzamide, pseurotin A, 3,6,8-trihydroxy-1-methylxanthone10,11 |
| MMS 194, MMS 460 | P. canescens | Morphology | P. cf. canescens | Canescentia | Dechlorogriseofulvin, griseofulvin, maculosin, oxaline, orselllinic acid, penicillic acid, penitremone A-C (tentative id.)12 |
| ILF-002 | P. canescens | FASTA files for ITS and β-tubulin sequences show that this strain is not P. canescens s.str., but a new species | New species sister to Penicillium canescens | Canescentia | Dechlorogriseofulvin, 6-desmethyldechlorogriseofulvin,6-desmethylgriseofulvin, 1,6-dihydroxy-3-methoxy-8-methylxanthone, griseofulvin, orsellinic acid, penicillic acid, pseurotin A, 1,2,3,5,6-pentahydroxy-8-methylxanthone, 1,3,5,6-tetra-hydroxy-8-methylxanthone, vulculic acid13 |
| 170A/28 | P. canescens | Morphology | Probably P. nigricans | Canescentia | Penitrem A14,15 |
| CBS 288.53 | P. canescens | Morphology | New species of Penicillium | Canescentia | Canescin16 |
| BAFC 3291 | P. canescens | Morphology | Probably P. aurantiogriseum | Canescentia | Aurantiamine, cyclo(L-Phe-L-Hyp), cyclo(L-Phe-L-Pro), cyclo(D-Phe-L-Val-D-Val-L-Tyr), pseurotin A17 |
| No number | P. canescens | No data | Unknown | Canescentia | Adamantanecarboxanilide, aphthosin, 9,10-anthracenedione, borane, decane, diethylphthalate, di-p-tolylacetylene, dodecane, eicosane, heptadecane, hexa-decane, hexadecanoic acid, 1,3,8-p-menthatriene, nicodicodine, orcinol, oxalic acid, phenanthrene, tetradecane, thioxanthene, tridecane, p-xylene18 (mostly volatiles) |
| No number | P. canescens | Morphology | Unknown | Canescentia | Citrinin19,20 |
| No number | P. canescens (two strains) | Morphology | P. cyclopium or P. canescens | Canescentia | Penicillic acid21,22 |
| ZJQY610 | P. canescens | GU556971 | P. canescens or other series Canescentia species | Canescentia | O-acetylbenzeneamidino-carboxylic acid, griseofulvin, 4-hydroxy-5-methoxy-2-methyl-naphtho[1,2-b]furan-3-carboxylic acid23 |
| No number | P. canescens | Morphology | Unknown | Canescentia | Citrinin, patulin, penicillic acid24 |
| SCSIOz053 | P. canescens | JN585930 | Talaromyces fusiformis? | Canescentia | Canescenin A & B25 |
| No number | P. canescens | Morphology | Talaromyces sp.? | Canescentia | Rugulosin26 |
| No number | P. canescens | Morphology | Probably P. verrucosum | Canescentia | Ochratoxin A27 |
| NRRL 2301 | P. janczewskii | Morphology | P. janczewskii | Canescentia | Bromogriseofulvin, dechlorogriofulvin, griseofulvin28,29,30, fungistatic and bacteriostatic red pigment31 |
| CBS 141000 | P. corvianum | KT887875, culture ex-type | P. corvianum | Canescentia | Compactin (= mevastatin), fiscalin C, trichodermamide C, C11H15O3N, C18H22O5, C29H30O7S232 |
| VKMF-312 | P. janczewskii | Morphology | P. janczewskii | Canescentia | Dechlorogriseofulvin, griseofulvin, patulin33 |
| VKMF-2191, VKMF-2378, VKMF-3023 | P. janczewskii | Morphology | P. janczewskii | Canescentia | Dechlorogriseofulvin, griseofulvin33 |
| VKMF-685 | P. janczewskii | Morphology | P. janczewskii or P. nigricans | Canescentia | Aurantioclavine, 6-N-ethylaurantioclavine33 |
| VKMF-2377 | P. janczewskii | Morphology | P. janczewskii? | Canescentia | Epicostaclavine33 |
| VKMF-2489 | P. janczewskii | Morphology | P. janczewskii? | Canescentia | 16-N-ethylroquefortine D, glandicolin B, (E)-3-(1H-imidazol-4-yl-methylene)-6-(1H-indol-3-yl methyl)-2,5-piperazinediol, meleagrin, roquefortine C33 |
| DSM 17433 = KMPB H-TW5/869 | P. janczewskii | Morphology | P. janczewskii? | Canescentia | 3R*,4R*- and 3S*,4R*-Dihydroxy-4-(4’-methoxyphenyl)-3,4-dihydro-2(1H)-quinolinone, 3-methoxy-4-hydroxy-4-(4’-methoxyphenyl)-3,4-dihydro-2(1H)-quinolinone, peniprequinolone34 |
| F 2757 & F 2641 | P. janczewskii | Morphology | P. janczewskii? | Canescentia | Fumagillin methyl ester, cis-fumagillin methyl ester35,36 |
| No number | P. janczewskii | Morphology | P. janczewskii? | Canescentia | Cycloaspeptide A, gliovictin, gliovictin acetate, peniprequinolone, pseurotin A37,38 |
| IFO 7745 = CBS 744.70 = ATCC 18380 | P. nigricans var. sulphuratum | Morphology | P. janczewskii | Canescentia | Griseofulvin39 |
| IMI 228669 and no number17 | P. janczewskii | Morphology | P. janczewskii | Canescentia | 19-Deoxypaxillin-16β-ol, nigrifortine (= amauromine), paxilline, penitrem A, penitrem E, pennigritrem15,40,41,42,43 |
| SIIA-F3933 | P. janczewskii | Morphology | Probably P. corvianum | Canescentia | Compactin (= mevastatin)44 |
| NRRL 909 | P. jensenii | AY443470, culture ex-type | P. jensenii | Canescentia | Griseofulvin6 |
| VKMF-292, VKMF-293 | P. jensenii | Morphology | P. jensenii | Canescentia | Meleagrin, roquefortine C & D9 |
| F-2813 | P. jensenii | Morphology | P. jensenii | Canescentia | Fumagillin, fumagillol45 |
| No number | P. nigricans | Morphology | P. nigricans | Canescentia | Griseofulvin46,47 |
| No number & LSHTM P38 = BRL 250 | P. nigricans | Morphology | P. nigricans | Canescentia | Griseofulvin48,49 |
| No number | P. nigricans | Morphology | P. nigricans? | Canescentia | MT8150 |
| F-5261 | P. nigricans | Morphology | P. nigricans | Canescentia | Fumagillin, fumagillol45 |
| IMI 228669 | P. nigricans | Morphology | P. janczewskii | Canescentia | 19-Deoxypaxillin-16β-ol, nigrifortine (= amauromine), paxilline, penitrem A, penitrem C, penitrem E, PC-M6, pennigritrem40,41,45,51 |
| LSHTM P38 | P. nigricans | Morphology | P. nigricans | Canescentia | Cyclo(L-Phe-L-Phe)52 |
| 170/D2 & 170/D4 | P. nigricans | Morphology | P. nigricans? or P. brasilianum | Canescentia | Verruculogen (as toxin X)53,54 |
| No number | P. nigricans | Morphology | P. nigricans? | Canescentia | Albidin48 |
| CBS 410.69 | P. yarmokense | Ex type, KC411757 | P. yarmokense | Canescentia | Griseofulvin6 |
| MMS 351 = LCP 99.43.43, MMS 747 | section Canes-centia species | JN676192 (and β-tubulin: JN794530) | P. yarmokense (first identified as P. waksmanii)56 | Canescentia | Agroclavine, dechlorogriseofulvin, festuclavine, griseofulvin, penicillic acid, ligerin, nortryptoquivaline, orcinol, orsellinic acid12,55 |
| FH-14 | P. antarcticum | Morphology, identified by Westerdijk Fungal Biodiversity Institute (WFBI) | P. antarcticum | Atroveneta | Antarone A & B56 |
| MMS 14, MMS 15 | P. antarcticum | Morphology, ITS | P. antarcticum | Atroveneta | Antarone A, aurantioclavine, chrysogine, cladosporin (= asperentin), 5-hydroxy-asperentin, patulin, terrestric acid?, violaceic acid12,57 |
| AF3-117C | Penicillium species | JX967116 | P. antarcticum | Atroveneta | Cladosporin (= asperentin), epiepoformin, patulin, phyllostin58 |
| YK-247 | P. coralligerum | LC2145672 | P. antarcticum | Atroveneta | 5′-hydroxyasperentin, cladomarine, cladosporine (= asperentin)59 |
| No number | P. antarcticum | Polyphasic identification, no data | P. antarcticum | Atroveneta | cis-Cyclo(4R-Hyp, L- Leu), trans-cyclo(4R-Hyp, L-Leu), cis-cyclo(4R-Hyp, L-Phe), cyclo-(L-Pro, Gly), ethyl 8-hydroxyhexylitaconate, ethyl-9-hydroxyitaconate, (-)-hydroxyhexylitaconate, cis-4-hydroxymellein, methyl-8-hydroxyhexylitaconate, methyl-9-hydroxyitaconate, methylitaconate, 2-phenetylalcohol60 |
| KMM 4668 | P. piltunense | Culture ex-type (but synonym of P. antarcticum) | P. antarcticum | Atroveneta | Penigrisacid D, piltunine A-F61 |
| No number | Marine-derived P. atrovenetum | Morphology | Probably P. antarcticum, as P. atrovenetum is a soil fungus | Atroveneta | Citreohybridinol62 |
| 9 strains, no numbers | P. antarcticum | No data | P. antarcticum (?) | Atroveneta | Butylphthalate, dibutylphthalate, di-2-ethylhexylphthalate, hydrocarbons, squalene, and fatty acids (the latter are general metabolites)63 |
| Strain 68 & 56 | P. atrovenetum & P. coralligerum | MH881491, MH881479 | P. antarcticum | Atroveneta | Aspinonene, atrovenetin, atrovenetinone, brevicompanine E, citreohydridonol, deoxyherquenone, griseofulvin, marcfortine C*, penitrem B, phytosphingosine64 |
| Strains 4-7 & 9-13 | P. atrovenetum & P. antarcticum | ITS and morphology | P. antarcticum (P. atrovenetum is regarded as a soil-borne species) | Atroveneta | 2-Acetyl-4(3H)-quinazolinone, aflatoxin M1*, andrastin A, andrastin “X” (wrongly identified as (wia) antibiotic UK 88051-9), antarone B (wia purpactin A), anti-biotic SMTP8*, arisugacin B*, asperentin (wia (S)-curvularin), atrovenetin, atro-venetinone (wia tetrahydrohalenaquinone A), bionectin B*, (S)-chrysogine, chaetoglobosin 510*, cladosporin-8-methylether (wia antibiotic PO1), cytosporone A*, cytosporone E*, deacetoxyfructigenine A (wia tubingensin A or B), deoxy-herquenone (= UP31), dictyosphaeric acid*, hamigeromycin*, hamigerone (wia austalide K), hongoquercin A*, (S)- 5-hydroxyasperentin (wia 11α-hydroxycurvularin), a hydroxyasperentin (wia 7,8-dihydro-7α-hydroxyresorcylide), 4-hydroxy-1-methoxy-5-phenyl-3-(tetrahydro-6-(3-hydroxy-1-methylpropyl)-3,5-dimethyl-2H-pyran-2-yl)-2(1H)-pyridinone*, libertellone B*, 3-O-methylterprenin*, patulin, penitrem A, penitrem E, podosporin A*, sporidesmin D*, stachybosin*, talaro-convolutin B*, territrem C*, 2’,2’’,3’,3’’-tetrahydrobisvertinolone*, tropolactone C*, UP10, 11, 12, 15, 9, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32** 65 |
| IMI 061837 = CBS 241.56 | P. atrovenetum | AF033492, culture ex-type, morphology | P. atrovenetum | Atroveneta | 3-nitropropionic acid66, atrovenetin67 |
| HO9-2 | P. atrovenetum | EMBL no. KC009765 | P. atrovenetum? | Atroveneta | Cyclopiazonic acid68 |
| CECT 2886 | P. novae-zeelandiae | Morphology | P. novae-zeelandiae | Atroveneta | Gentisylalcohol, 3-hydroxybenzylalcohol, patulin (in 1949 reported as expansine)6,32,69,70 |
| IBT 21392 (= IMI 204086), IBT 22457 | P. novae-zeelandiae | Morphology | P. novae-zeelandiae | Atroveneta | Atlantinone A71 |
| No number | P. novae-zeelandiae | Morphology | P. novae-zeelandiae? | Atroveneta | Penitrem A, verruculogen15 |
| No number | P. novae-zeelandiae | Morphology | P. novae-zeelandiae? | Atroveneta | Curvulinic acid72 |
| CBS 140987 | P. nucicola | KT887860, culture ex-type | P. nucicola | Atroveneta | Andrastin A-D, austin, C20H20O632 |
* The metabolites tentatively identified may also be other secondary metabolites with the same molecular mass. For example if aflatoxin M1 in P. antarcticum would have been correctly identified, one would also expect presence of aflatoxin B1 and B2, which are usually the main biosynthetic products in the aflatoxin secondary metabolite biosynthetic family.
** UP: Unknown secondary metabolites with a known molecular mass.
*** Penicillium linzhiense was recently described and thus not examined during this study.
1Grijseels et al. 2016; 2Prigent et al. 2018; 3Zang et al. 2020; 4Zang et al. 2019; 5Nicoletti et al. 2007; 6Frisvad & Filtenborg 1990; 7Yaegashi et al. 2015; 8Frank et al. 2019; 9Kozlovskii et al. 1997a; 10Chen et al. 2019; 11Niaz et al. 2019; 12Vansteelandt et al. 2012, Vansteelandt et al. 2013; 13Malik et al. 2020; 14Shreeve et al. 1978; 15Di Menna et al. 1986; 16Brian et al. 1953; 17Bertinetti et al. 2009; 18Ghanbari et al. 2014; 19Bilai 1963; 20Gedek 1977; 21Kharchenko 1970; 22Keromnes & Thouvenot 1985; 23Wang et al. 2010; 24Moslem et al. 2013; 25Dasanayaka et al. 2020; 26Leistner & Pitt 1977; 27Ueno et al. 1991; 28Brian et al. 1949; 29MacMillan 1951; 30Grove & Mcgowan 1947; 31Curtis & Grove 1947; 32Visagie et al. 2016b; 33Kozlovskii et al. 1997b; 34He et al. 2005; 35Kwon et al. 2000; 36Bae et al. 1999; 37Schmeda-Hirschmann et al. 2008; 38Schmeda-Hirschmann et al. 2005; 39Udagawa & Abe 1961; 40Laws & Mantle 1985; 41Mantle & Penn 1989; 42Penn et al. 1992; 43Penn & Mantle 1994; 44Chu et al. 1999; 45Goto et al. 1986; 46Wright 1955; 47Leistner 1979; 48Jefferys et al. 1953; 49Macmillan 1954; 50Gupta et al. 1984; 51Mantle et al. 1984; 52Birkinshaw & Mohammed 1962; 53Patterson et al. 1981; 54Patterson et al. 1979; 55Petit et al. 2004; 56Shiono et al. 2008; 57Geiger et al. 2013; 58Flewelling et al. 2013; 59Takahashi et al. 2017; 60Marchese et al. 2020; 61Afiyatullov et al. 2019; 62Özkaya et al. 2018; 63Oleinikova et al. 2018; 64Fan et al. 2019; 65Petersen et al. 2019; 66Raistrick & Stössl 1958; 67Neill & Raistrick 1957; 68Alapont et al. 2014; 69Burton 1949; 70Alfaro et al. 2003; 71Dalsgaard et al. 2012; 72Turner & Aldridge 1983.
Series Canescentia — Fig. 3, 4
Phylogenetic analyses revealed three new species in this clade, noting that the BenA phylogeny did not fully resolve the relationship between these clades in contrast to the CaM and RPB2 phylogenies that did. A similar observation was made for P. murcianum and P. radiatolobatum as discussed below. The largest proportion of strains belonged to the clade described as P. scottii, which showed a high degree of infraspecies variation in CaM, but less so for BenA and RPB2. The three new species are introduced in the P. janczewskii species complex that contains seven previously described species. The phylogenetic analyses resolved ex-type strains of P. granatense (CBS 166.81T) and P. nigricans var. sulphuratum (CBS 744.70T) with P. janczewskii confirming their status as synonyms (Pitt 1980). Also, P. echinatum nom. inval. (= P. janczewskii fide Pitt 1980), P. griseoazureum (= P. waksmanii fide Pitt 1980) and P. nigricans (= P. janczewskii fide Pitt 1980) represent distinct phylogenetic species. The novelty of the more recently described species P. dunedinense (Visagie et al. 2014b), P. corvianum, P. nucicola (Visagie et al. 2016b) and P. arizonense (Grijseels et al. 2016) is confirmed. One of Pitt’s (1980) morphologically intermediate strains (IMI 149218) is shown here to be P. canescens. The relationship between P. murcianum and P. radiatolobatum is problematic with no coherent clades produced, noting that ex-type strains for these two (CBS 340.79T, CBS 161.81T) are quite distinct. Strains were thus named based on the concatenated multigene phylogeny. Strains identified as Penicillium cf. murcianum probably represent a new species. However, more strains, sequencing of additional gene regions and extrolite data will be needed to resolve species boundaries in this complex.
Extrolite analysis
Series Canescentia is generally characterised by the production of curvulinic acid (11/14 species) and griseofulvin (10/14 species), but xanthoepocin (7/14 species), pseurotins (4/14 species), tryptoquivalines (4/14 species), fumagillin/ligerin (3/14 species) and penicillic acid (2/14 species) are also found in more than one species. In series Atroveneta, one or more species produce asperentins (2/7 species), atlantinones/andrastins (4/7 species), patulin (3/7 species), atrovenetins (2/7 species), antarones (1/7 species), aurantiamine (1/7 species), benzomalvins (1/7 species), fischerin (1/7 species) and haenamindole (1/7 species) (Table 3). Many species in section Canescentia produce other extrolites (Table 4), and in general this section contain species that produce a broad range of bioactive secondary metabolites. Both clades contain species that produce austalides, chrysogines, communesins, decaturins, and penitrems. The frequency of species producing the extrolites in each series may have been higher, if more media have been used. Likewise genomic analysis may show if the potential of producing these extrolites is larger than expected, and silent gene clusters may be activated by growing the fungi on different substrates and under different conditions.
TAXONOMY
Penicillium allsoppiae Visagie, A. Visagie, Frisvad & K. Jacobs, sp. nov. — MycoBank MB 834426; Fig. 6
Fig. 6.
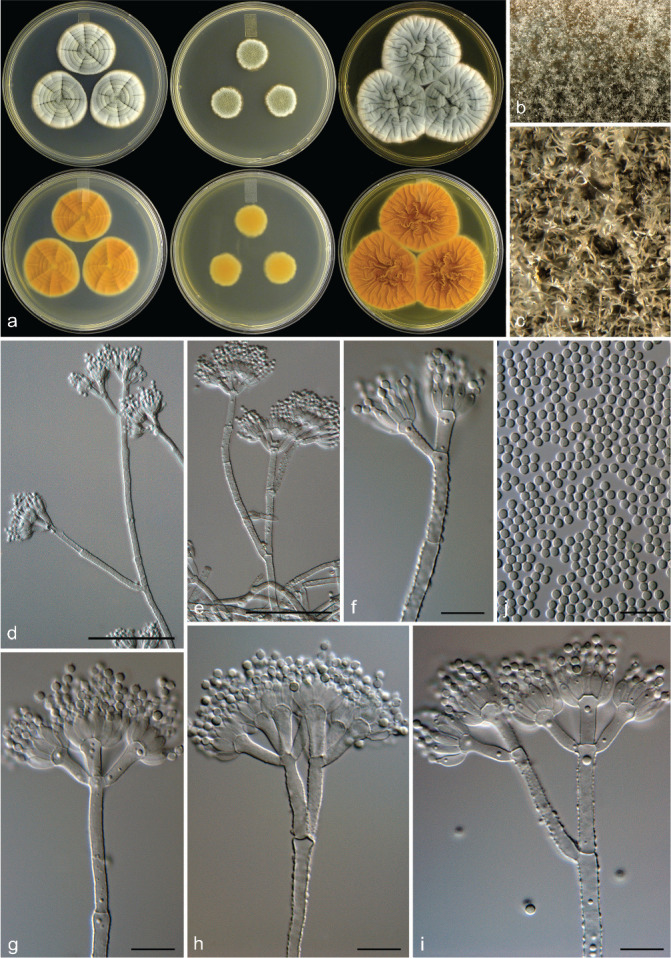
Penicillium allsoppiae. a. Colonies (top row, left to right: CYA, MEA, YES; bottom row, left to right: CYA reverse, MEA reverse, YES reverse); b, c. colony texture on MEA; d–i. conidiophores; j. conidia. — Scale bars: d = 25 μm, e–j = 10 μm.
Etymology. Latin, allsoppiae, named after Nicky Allsopp who reported several Penicillium species during a survey from the same Malmesbury sampling site we collected samples during our study.
Typus. SOUTH AFRICA, Malmesbury, soil sample, July 2009, coll. C.M. Visagie (CBS H-22036 holotype, culture ex-type CBS 138943 = DAOM 241348 = DTO 182-D5 = CV 931).
Subgeneric classification — subgenus Penicillium, section Canescentia, series Canescentia.
ITS barcode — JX140830. Alternative identification markers: BenA = JX140992, CaM = JX157384, RPB2 = KP016895.
Colony diam, 7 d (in mm) — CYA 25–30; CYA 30 °C 30–32; CYA 37 °C 4–6; MEA 20–30; YES 34–38; DG18 20–22; CYAS 25–30; CREA 11–12.
Colony characters — CYA 25 °C, 7 d: Colonies moderately deep, sulcate, raised at centre; margins low, narrow, entire; mycelia white, sometimes pinkish orange; texture floccose; sporulation sparse to moderately dense, conidia en masse dull green (25D4–26E4); soluble pigments absent; exudates minute clear droplets; reverse brownish orange to brown (6C6–D6), sometimes pale to light yellow (3A3–4). MEA 25 °C, 7 d: Colonies moderately deep, lightly sulcate; margins low, wide, entire; mycelia white; texture floccose; sporulation sparse to moderately dense, conidia en masse greyish to dull green (25C4–D4); soluble pigments absent; exudates clear minute droplets; reverse brownish orange to brown (6C7–E7). YES 25 °C, 7 d: Colonies moderately deep, sulcate, raised at centre; margins low, narrow, entire; mycelia white to pinkish orange; texture floccose; sporulation sparse, conidia en masse greenish grey (26B2–C2); soluble pigments absent; exudates absent; reverse brown (7E7), greyish orange (6B6), some light yellow (4A4) areas. DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, entire; mycelia white; texture floccose; sporulation moderately dense, conidia en masse dull to greyish green (25D4–5–E5); soluble pigments absent; exudates absent; reverse yellowish brown (5D6) to greyish beige (4C2) to yellowish white (4A2). CREA 25 °C, 7 d: Acid not produced.
Micromorphology — Conidiophores mostly biverticillate, terverticillate also present; stipes rough walled, smooth also present, 200–800 × 3.5–4 μm; branches/rami two when present, 13.5–34 × 3.5–4 μm; metulae divergent, 2–6 per stipe/branch, 10–18 × 2.5–4 μm, vesicle 4–5 μm; phialides ampulliform, 6–7 × 2.5–3.5 μm; average length phialide/metula 0.57; conidia finely rough to rough, globose, 2–2.5 × 2–2.5 μm (2.1 ± 0.1 × 2.1 ± 0.1), average width/length = 0.97, n = 40.
Extrolites — Penitrem A.
Distinguishing characters — Penicillium allsoppiae is morphologically most similar to P. dunedinense, P. eickeri and P. scottii, all showing relatively fast growth on MEA compared to other series Canescentia species. Penicillium allsoppiae lacks the dark brownish grey colony reverse on CYA observed in P. dunedinense, generally grows more restricted than P. scottii and P. eickeri, and consistently produces a higher proportion of roughened stipes. This distinguishes it from its closest relatives, but see also distinguishing characters for P. scottii.
Penicillium doidgeae Visagie, Frisvad & K. Jacobs, sp. nov. — MycoBank MB 834427; Fig. 7
Fig. 7.
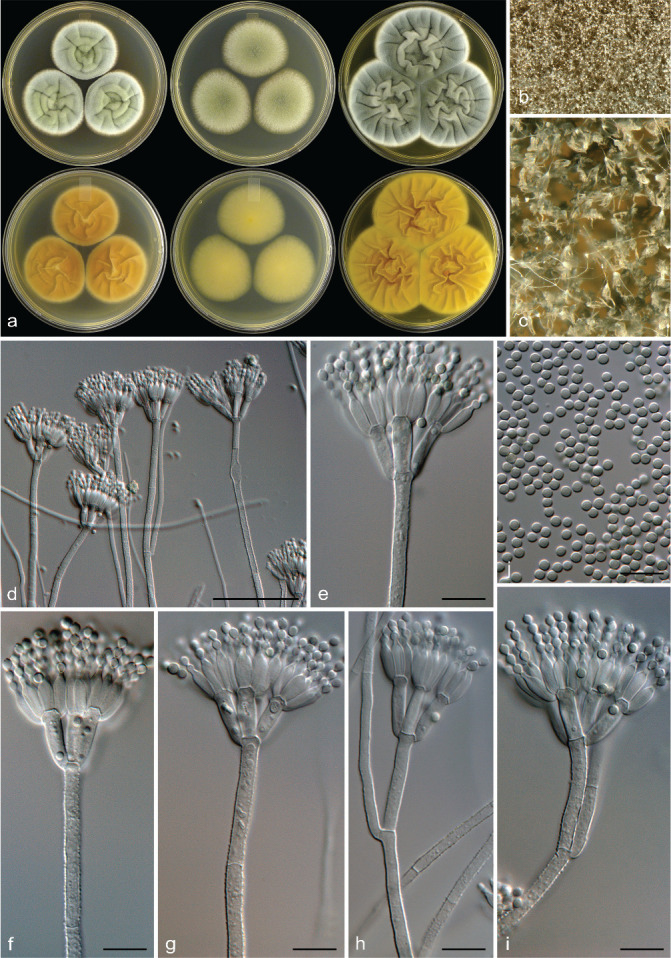
Penicillium doidgeae. a. Colonies (top row, left to right: CYA, MEA, YES; bottom row, left to right: CYA reverse, MEA reverse, YES reverse); b, c. colony texture on MEA; d–h. conidiophores; i. conidia. — Scale bars: d = 25 μm, e–i = 10 μm.
Etymology. Latin, doidgeae, named after Ethel Mary Doidge who had a long career as a mycologist/bacteriologist listing, amongst many others, Penicillium occurring in South Africa and became the first woman to receive a doctorate in South Africa.
Typus. SOUTH AFRICA, Struisbaai, bract from Protea repens infructescence, Aug. 2009, coll. C.M. Visagie (CBS H-22038 holotype, culture ex-type CBS 138947 = IBT 31950 = DAOM 241107 = DTO 183-G7 = CV 2189).
Subgeneric classification — subgenus Penicillium, section Canescentia, series Atroveneta.
ITS barcode — JX140804. Alternative identification markers: BenA = JX141006, CaM = JX157413, RPB2 = KP016915.
Colony diam, 7 d (in mm) — CYA 36–41; CYA 5 °C germination; CYA 30 °C 20–27; CYA 37 °C no growth; MEA 34–40; YES 48–52; DG18 40–42; CYAS 41–42; CREA 18–23.
Colony characters — CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, wide, entire; mycelia white, sometimes inconspicuously yellow near centre; texture floccose; sporulation moderately dense, conidia en masse greyish green (25C4–D6); soluble pigments absent; exudates absent; reverse brown (5E7–6E7), sometimes greyish yellow to greyish orange to brownish orange (4C5–5B5–C5). MEA 25 °C, 7 d: Colonies moderately deep, plane; margins low, wide, entire; mycelia white; texture floccose; sporulation moderately dense to dense, conidia en masse greyish to dull green (25E4–26E4); soluble pigments absent; exudates absent; reverse greenish white to pale yellow (30A2–1A3), sometimes greyish orange (5B4) centrally, brown (6E8) elsewhere. YES 25 °C, 7 d: Colonies moderately deep, sulcate, raised at centre; margins low, wide, entire; mycelia white; texture floccose; sporulation moderately dense to dense, conidia en masse greyish to dull green (25C4–E4–5); soluble pigments absent; exudates absent; reverse greyish brown (4B5). DG18 25 °C, 7 d: Colonies low to moderately deep, plane; margins low, wide, entire; mycelia white; texture velutinous and floccose areas; sporulation dense, conidia en masse dull green (26E4–27E4); soluble pigments absent; exudates absent; reverse orange (6B7) at centre, fading into dull green (29D4), sometimes centre white. CREA 25 °C, 7 d: Acid very weak only within colony periphery.
Micromorphology — Conidiophores mostly biverticillate, terverticillate also present, sometimes a long subterminal branch forms that extends up to 85 μm; stipes finely roughened to rough, minor proportion smooth, 85–600 × 3–4 μm; branches/rami two when present, 23–30(–85) × 3–4 μm; metulae appressed to slightly divergent, 4–6 per stipe/branch, 11–15 × 3–4.5 μm, vesicle 3.5–5 μm; phialides ampulliform, 8–10.5 ×2–4 μm; average length phialide/metula 0.74; conidia smooth, subglobose, 2–3 × 2–3 μm (2.5 ± 0.1 × 2.4 ± 0.1), average width/length = 0.95, n = 60.
Extrolites — Asperentins, atlantinone A, austalides, fischerin and patulin.
Distinguishing characters — Penicillium doidgeae produces fast-growing colonies on most agar media. Conidiophores sometimes are borne subterminally and have mostly roughened stipes and smooth conidia. The observed subterminal conidiophore branch is not borne at an angle to the main stipe, rather the branch leading to the terminal conidiophore is borne at an angle to the main stipe, a character similar to conidiophores of Penicillium section Thysanophora. The species is morphologically most similar to P. antarcticum. However, at cooler temperatures (5 °C) P. doidgeae conidia will at most only germinate on CYA, whereas P. antarcticum will produce microcolonies. Also, stipes of P. doidgeae are mostly finely roughened and generally longer (85–600 μm) compared to the smooth and generally shorter (100–250 μm) stipes of P. antarcticum (McRae et al. 1999).
Penicillium eickeri Visagie, Frisvad & K. Jacobs, sp. nov. — MycoBank MB 834428; Fig. 8
Fig. 8.
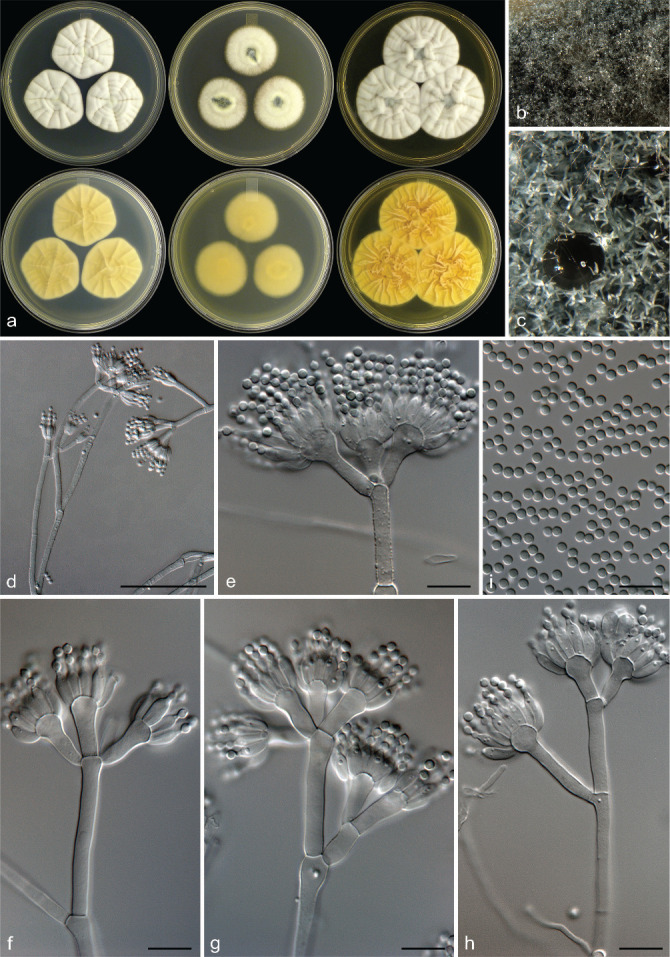
Penicillium eickeri. a. Colonies (top row, left to right: CYA, MEA, YES; bottom row, left to right: CYA reverse, MEA reverse, YES reverse); b, c. colony texture on MEA; d–h. conidiophores; i. conidia. — Scale bars: d = 25 μm, e–i = 10 μm.
Etymology. Latin, eickeri, named after Albert Eicker who reported several Penicillium species isolated from the old Transvaal region in South Africa.
Typus. SOUTH AFRICA, Stellenbosch, mite from Protea repens infructescence, Mar. 2009, coll. C.M. Visagie (CBS H-22034 holotype, culture ex-type CBS 138939 = IBT 31921 = DAOM 241352 = DTO 181-G3 = CV 475).
Subgeneric classification — subgenus Penicillium, section Canescentia, series Canescentia.
ITS barcode — JX140824. Alternative identification markers: BenA = JX140979, CaM = JX157365, RPB2 = KP016876.
Colony diam, 7 d (in mm) — CYA 38–39; CYA 30 °C 39–42; CYA 37 °C 1–5; MEA 34–38; YES 42–44; DG18 27–28; CYAS 30–31; CREA 15–17.
Colony characters — CYA 25 °C, 7 d: Colonies low, sulcate, raised at centre; margins low, narrow, entire; mycelia white; texture floccose; sporulation absent, conidia en masse not determined; soluble pigments absent; exudates clear; reverse greyish yellow (3C3) at centre, yellowish white (2A2–3A2) elsewhere. MEA 25 °C, 7 d: Colonies low to moderately deep, plane to sulcate, raised at centre; margins low, narrow, entire; mycelia white; texture floccose; sporulation sparse to moderately dense only at centre, conidia en masse greyish green (26C3); soluble pigments absent; exudates absent to minute clear droplets; reverse brownish orange yellowish brown (5C5–D8), sometimes a darker brown (6E7). YES 25 °C, 7 d: Colonies moderately deep, sulcate, raised at centre; margins low, narrow, entire; mycelia white; texture floccose; sporulation absent, conidia en masse not determined; soluble pigments absent; exudates absent; reverse light brown (6D6) at centre, light yellow (3A5) elsewhere. DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, entire; mycelia white; texture floccose; sporulation moderately dense centrally, conidia en masse greyish turquoise to greyish green (24E4–25E4); soluble pigments absent; exudates absent; reverse light brown (5D7) centrally, pale orange (5A3) elsewhere. CREA 25 °C, 7 d: Acid not produced.
Micromorphology — Conidiophores mostly biverticillate, terverticillate also present; stipes rough walled, smooth also present, 200–820 × 3–4.5 μm; branches/rami two when present, 11–37 × 3–4.5 μm; metulae divergent, 2–6 per stipe/branch, 11–14(–17) × 3–4 μm, vesicle 3–4.5 μm; phialides ampulliform, 7–9.5 × 2.5–3 μm; average length phialide/metula 0.60; conidia finely rough to rough, globose, 2–2.5 × 2–2.5 μm (2.3 ± 0.03 ×2.3 ± 0.1), average width/length = 0.97, n = 30.
Extrolites — Curvulinic acid and xanthoepocin.
Distinguishing characters — Penicillium eickeri is morphologically most similar to P. dunedinense, P. allsoppiae and P. scottii, all showing relatively fast growth on MEA compared to other series Canescentia species. Penicillium eickeri lacks the dark brownish grey colony reverse on CYA observed in P. dunedinense, generally grows faster than P. scottii and P. allsoppiae and does not sporulate on CYA after 7 d. This distinguishes it from its closest relatives, but see also distinguishing characters for P. scottii.
Penicillium elizabethiae Visagie & Frisvad, nom. nov. — MycoBank MB 834432
Etymology. Latin, elizabethiae, named after Elizabeth Dale who first described this new species but used an illegitimate name.
Basionym. Penicillium echinatum E. Dale, Ann. Mycol. 24: 137. 1926 (nom. illegit. Art. 53.1; non Rivolta 1873).
Typus. SCOTLAND, soil sample, 1914, coll. unknown (CBS H-22052 holotype, culture ex-type NRRL 917 = MUCL 29170 = IBT 21955 = DTO 189-B8).
Subgeneric classification — subgenus Penicillium, section Canescentia, series Canescentia.
ITS barcode — KP016840. Alternative identification markers: BenA = KJ866964, CaM = KJ867021, RPB2 = KP016918.
Notes — This species was described by Dale (1926) who used an already occupied P. echinatum Rivolta (Dei Parassiti Vegetali: 451, 1873; MB263715) currently classified as Memnoniella echinata (Riv.) Galloway, Trans. Brit. Mycol. Soc. 18: 165. 1933; MB263706). Penicillium elizabethiae is morphologically similar to P. janczewskii. However, it is phylogenetically distinct and closely related to P. nigricans and P. griseoazureum.
Penicillium pole-evansii Visagie, Frisvad & K. Jacobs, sp. nov. — MycoBank MB 834429; Fig. 9
Fig. 9.
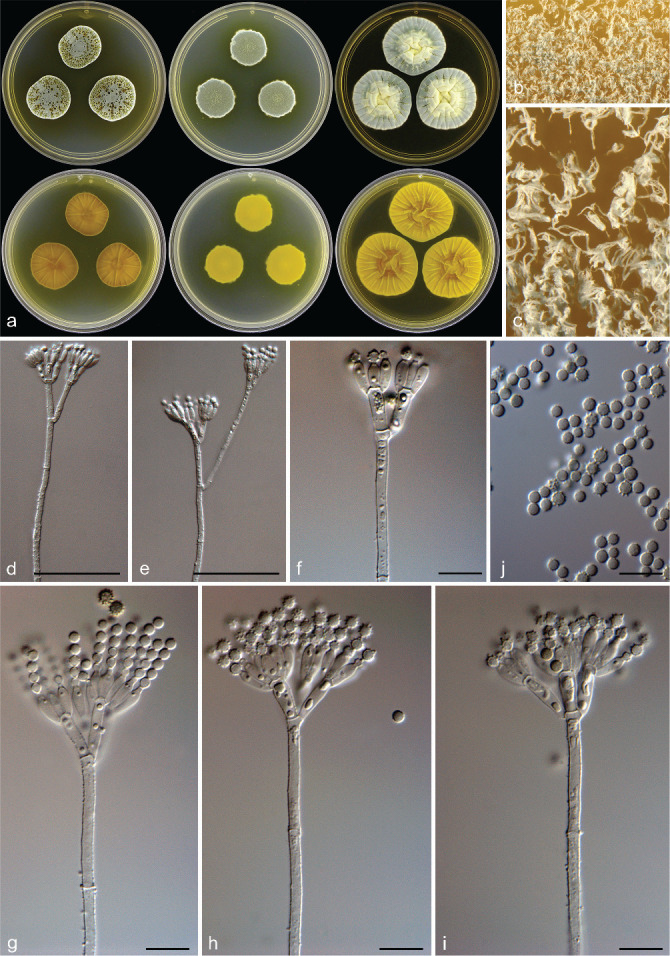
Penicillium pole-evansii. a. Colonies (top row, left to right: CYA, MEA, YES; bottom row, left to right: CYA reverse, MEA reverse, YES reverse); b, c. colony texture on MEA; d–i. conidiophores; j. conidia. — Scale bars: d, e = 25 μm, f–j = 10 μm.
Etymology. Latin, pole-evansii, named after Illtyd Buller Pole-Evans who recorded the first record of Penicillium in South Africa (P. digitatum from Citrus sp. collected in Kwazulu Natal, 1903).
Typus. SOUTH AFRICA, Struisbaai, bract from Protea repens infructescence, Aug. 2009, coll. C.M. Visagie (CBS H-22037 holotype, culture ex-type CBS 138946 = IBT 31929 = DAOM 241106 = DTO 183-D5 = CV 1758).
Subgeneric classification — subgenus Penicillium, section Canescentia, series Atroveneta.
ITS barcode — JX140831. Alternative identification markers: BenA = JX141005, CaM = JX157412, RPB2 = KP016911.
Colony diam, 7 d (in mm) — CYA 21–26(–36); CYA 30 °C 18–21; CYA 37 °C no growth; MEA 21–28; YES 36–40; DG18 34–35; CYAS 33–34; CREA 15–21.
Colony characters — CYA 25 °C, 7 d: Colonies moderately deep, sulcate, sometimes shaped as irregular pentagon; margins low, narrow, entire; mycelia white; texture floccose; sporulation moderately dense, conidia en masse dull green (25D4–D5–26D4); soluble pigments yellowish brown; exudates yellow and brown droplets; reverse olive brown to brown (4F4–5F4) at centre, fading into olive to yellowish brown (4D6–5D6), with age becoming dark green (25F5–26F5) where colonies meet. MEA 25 °C, 7 d: Colonies moderately deep, lightly sulcate; margins low, narrow, entire; mycelia white; texture floccose; sporulation dense, conidia en masse greyish turquoise to dull green (24E4–26E4); soluble pigments absent; exudates absent, sometimes yellowish orange; reverse brown (6E8). YES 25 °C, 7 d: Colonies low to moderately deep, sulcate, raised at centre; margins low, narrow, entire; mycelia white; texture floccose; sporulation sparse to moderately dense, conidia en masse greyish turquoise (24B3–C5); soluble pigments absent; exudates minute yellow droplets; reverse dull to greyish yellow (4B4–4C6). DG18 25 °C, 7 d: Colonies low to moderately deep, plane; margins low, narrow, entire; mycelia white; texture velutinous and floccose areas; sporulation dense, conidia en masse dull green (25E4); soluble pigments absent; exudates absent; reverse light yellow (2A5) a centre, fading into dull green (29D4). CREA 25 °C, 7 d: Acid not produced.
Micromorphology — Conidiophores mostly biverticillate, terverticillate also present, sometimes subterminal branch very long extending up to 65 μm; stipes finely rough to rough, 120–475 × 2.5–3.5 μm; branches/rami two when present, 20–65 × 2.5–3.5 μm; metulae divergent, 3–6 per stipe/branch, 9.5–16 × 2.5–4 μm, vesicle 3–4.5 μm; phialides ampulliform, 6.5–9 × 2.5–3.5 μm; average length phialide/metula 0.65; conidia rough to spiny, globose, 2–3 × 2–3 μm (2.8 ± 0.2 × 2.8 ± 0.2), average width/length = 0.97, n = 64.
Extrolites — Atrovenetin, aurantiamine, communesin B and patulin.
Distinguishing characters — Penicillium pole-evansii is distinguished by the copious amounts of yellow exudates and soluble pigments produced on CYA. With age, colony reverses on CYA becomes dark green. Penicillium pole-evansii is morphologically similar to P. atrovenetum. However, P. atrovenetum do not produce the dark green reverses observed after prolonged incubation in P. pole-evansii.
Penicillium scottii Visagie, Frisvad & K. Jacobs, sp. nov. — MycoBank MB 834430; Fig. 10
Fig. 10.
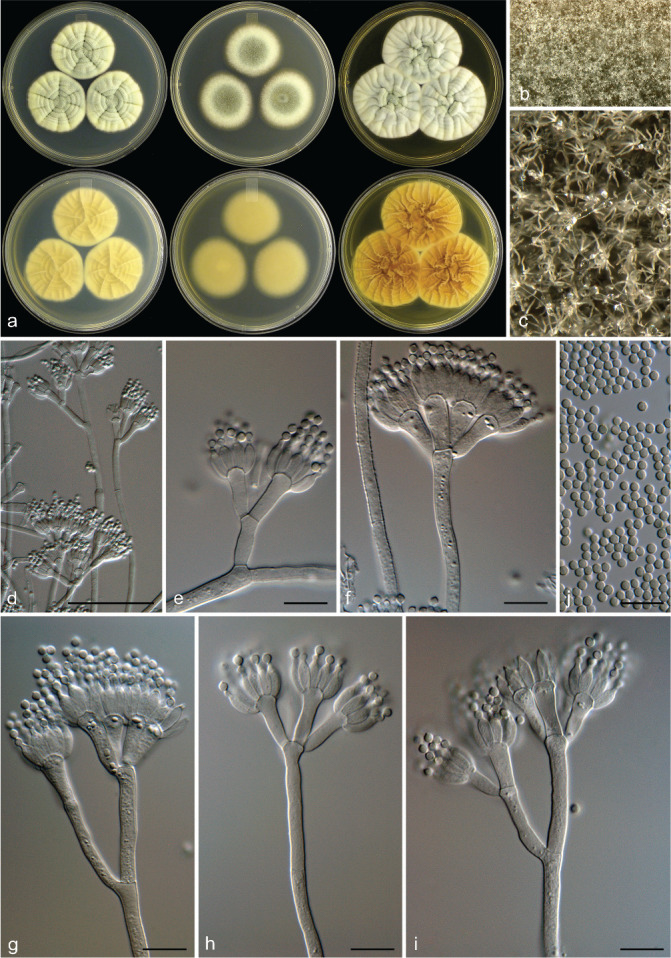
Penicillium scottii. a. Colonies (top row, left to right: CYA, MEA, YES; bottom row, left to right: CYA reverse, MEA reverse, YES reverse); b, c. colony texture on MEA; d–i. conidiophores; j. conidia. — Scale bars: d, e = 25 μm, f–j = 10 μm.
Etymology. Latin, scottii, named after De Buys Scott who described many new Penicillium species isolated from South Africa, all still considered distinct species today (Visagie et al. 2014b).
Typus. SOUTH AFRICA, Malmesbury, soil sample, July 2009, coll. C.M. Visagie (CBS H-22040 holotype, culture ex-type CBS 138951 = IBT 31905 = DTO 185-F8 = CV 930).
Subgeneric classification — subgenus Penicillium, section Canescentia, series Canescentia.
ITS barcode — JX140812. Alternative identification markers: BenA = JX140991, CaM = JX157383, RPB2 = KP016894.
Colony diam, 7 d (in mm) — CYA (28–)30–40; CYA 30 °C (15–)30–38; CYA 37 °C 0–11; MEA (25–)32–40; YES 38–42; DG18 18–26; CYAS 28–32; CREA 15–20.
Colony characters — CYA 25 °C, 7 d: Colonies moderately deep, sulcate, raised at centre; margins low, narrow, entire; mycelia white to yellow, sometimes pinkish orange; texture floccose; sporulation sparse, sometimes moderately dense, conidia en masse greyish green (28B4), dull green (25D4) in denser areas; soluble pigments absent; exudates clear; reverse greyish to brownish orange (5B5–5D6), sometimes yellowish grey to greyish yellow (4B2–3). MEA 25 °C, 7 d: Colonies moderately deep, plane to lightly sulcate, raised at centre; margins low, narrow, entire; mycelia white to yellow to sometimes pinkish orange; texture floccose; sporulation moderately dense, sometimes sparse, conidia en masse greyish to dull green (25D4–26D4); soluble pigments absent; exudates absent, sometimes clear; reverse brownish orange to brown (6C8–E8), sometimes olive brown to brown (4D5–5D5). YES 25 °C, 7 d: Colonies moderately deep, sulcate, raised at centre; margins low, narrow, entire; mycelia white to yellow to pinkish orange; texture floccose; sporulation absent, sometimes sparse, conidia en masse pale to greyish green (28A3–B3); soluble pigments absent; exudates absent; reverse brown (6E8), greyish orange (6B6), greyish yellow (4B5). DG18 25 °C, 7 d: Colonies moderately deep, sulcate, raised at centre; margins low, narrow, entire; mycelia white to pinkish orange; texture floccose; sporulation moderately dense, conidia en masse greyish to dull green (25B4–D4–26E4); soluble pigments absent; exudates absent; reverse light to brownish orange (5A5–6A5–C6), sometimes brown (6E7) at centre, sometimes white. CREA 25 °C, 7 d: Acid not produced.
Micromorphology — Conidiophores mostly biverticillate, terverticillate also present; stipes smooth to sometimes rough walled, 200–800 × 3–4 μm; branches/rami two when present, 10–38 × 3–4 μm; metulae divergent, 2–6 per stipe/branch, 10–19 × 2–4 μm, vesicle 3–8.5 μm; phialides ampulliform, 6.5–9.5 × 2.5–3.5 μm; average length phialide/metula 0.57; conidia finely rough to rough, globose, 2–2.5 × 2–2.5 μm (2.2 ± 0.1 × 2.2 ± 0.1), average width/length = 0.98, n = 100.
Extrolites — Curvulinic acid, dehydrogriseofulvin, griseofulvin, penitrem A and xanthoepocin.
Distinguishing characters — Penicillium scottii produces biverticillate to terverticillate conidiophores with smooth to rough stipes and roughened conidia. Micromorphologically, it thus resembles other species previously considered to represent P. janczewskii. However, colony growth rates on MEA is faster than other species from this group, which generally never reach 20 mm growth in 7 d. The exception is the recently described P. dunedinense and P. arizonense that phylogenetically is a close relative of P. janczewskii but produces colonies up to 36 and 28 mm diam on MEA, respectively (Visagie et al. 2014a, Grijseels et al. 2016). Penicillium dunedinense produces an intense and very dark brownish grey reverse, which is not observed in P. scottii. Penicillium eickeri and P. allsoppiae, described in this study, morphologically is most similar to P. scottii. However, P. eickeri generally grows faster than both these species, while P. allsoppiae generally grows more restricted than P. scottii. Penicillium allsoppiae also consistently produces conidiophores with roughened stipes more regularly than the other two species and P. eickeri fails to sporulate on CYA after 7 d incubation.
PENICILLIUM SECTION CANESCENTIA SERIES ATROVENETA ACCEPTED SPECIES AND THEIR SYNONYMS
Penicillium antarcticum A.D. Hocking & C.F. McRae, Polar Biol. 21: 103. 1999 (MB 482749). — Type: DAR 72813 (holotype). Ex-type: CBS 100492 = FRR 4989. Subgenus Penicillium section Canescentia series Atroveneta. ITS barcode: KJ834503 (alternative markers: BenA = MN969371; CaM = MN969236; RPB2 = JN406653).
= Penicillium attenuatum Kirichuk & Pivkin, Mycol. Progr. 16: 21. 2016 (2017) (MB 818673). — Type: LE 312279 (holotype). Ex-type: KMM 4671 (PIBOC). ITS barcode: KU358555 (alternative markers: BenA = KU358558; CaM = KU358561; RPB2 = n.a.).
= Penicillium ochotense Kirichuk & Pivkin, Mycol. Progr. 16: 21. 2016 (2017) (MB 818672). — Type: LE 312278 (holotype). Ex-type: KMM 4670 (PIBOC). ITS barcode: KU358553 (alternative markers: BenA = KU358556; CaM = KU358559; RPB2 = n.a.).
= Penicillium piltunense Kirichuk & Pivkin, Mycol. Progr. 16: 19. 2016 (2017) (MB 818671). — Type: LE 312276 (holotype). Ex-type: KMM 4668 (PIBOC). ITS barcode: KU358554 (alternative markers: BenA = KU358557; CaM = KU358560; RPB2 = n.a.).
Penicillium atrovenetum G. Sm., Trans. Brit. Mycol. Soc. 39: 112. 1956 (MB 302377). — Type: IMI 061837 (neotype, Pitt et al. 2000). Ex-type: CBS 241.56 = ATCC 13352 = FRR 2571 = IFO 8138 = IMI 061837 = LSHBSm683 = QM 6963. Subgenus Penicillium section Canescentia series Atroveneta. ITS barcode: AF033492 (alternative markers: BenA = JX140944; CaM = KJ867004; RPB2 = JN121467).
Penicillium attenuatum: see under Penicillium antarcticum.
Penicillium doidgeae Visagie, Frisvad & K. Jacobs, published here (MB 834427). — Type: CBS H-22038 (holotype). Ex-type: CBS 138947 = IBT 31950 = DAOM 241107 = DTO 183-G7 = CV 2189. Subgenus Penicillium section Canescentia series Atroveneta. ITS barcode: JX140804 (alternative markers: BenA = JX141006; CaM = JX157413; RPB2 = KP016915).
Penicillium novae-zeelandiae J.F.H. Beyma, Antonie van Leeuwenhoek 6: 275. 1940 (MB 522253). — Type: IMI 40584ii (neotype, Pitt 1980). Ex-type: CBS 137.41 = ATCC 10473 = IFO 31748 = IMI 040584ii = NRRL 2128 = QM 1934 = VKMF-2886. Subgenus Penicillium section Canescentia series Atroveneta. ITS barcode: JN617688 (alternative markers: BenA = MN969390; CaM = MN969279; RPB2 = JN406628).
Penicillium nucicola Visagie, Malloch & Seifert, Persoonia 36: 259. 2016 (MB 815771). — Type: DAOM 695770 (holotype). Ex-type: DAOMC 250522 = CBS 140987 = W 59 = KAS 2203. Subgenus Penicillium section Canescentia series Atroveneta. ITS barcode: KT887860 (alternative markers: BenA = KT887821; CaM = KT887782; RPB2 = MN969171).
Penicillium ochotense: see under Penicillium antarcticum.
Penicillium piltunense: see under Penicillium antarcticum.
Penicillium pole-evansii Visagie, Frisvad & K. Jacobs, published here (MB 834429). — Type: CBS H-22037 (holotype). Ex-type: CBS 138946 = IBT 31929 = DAOM 241106 = DTO 183-D5 = CV 1758. Subgenus Penicillium section Canescentia series Atroveneta. ITS barcode: JX140831 (alternative markers: BenA = JX141005; CaM = JX157412; RPB2 = KP016911).
PENICILLIUM SECTION CANESCENTIA SERIES CANESCENTIA ACCEPTED SPECIES AND THEIR SYNONYMS
Penicillium allsoppiae Visagie, A. Visagie, Frisvad & K. Jacobs, published here (MB 834426). — Type: CBS H-22036 (holotype). Ex-type: CBS 138943 = DAOM 241348 = DTO 182-D5 = CV 931. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: JX140830 (alternative markers: BenA = JX140992; CaM = JX157384; RPB2 = KP016895).
Penicillium arizonense Frisvad, Grijseels & J.C. Nielsen, Sci. Rep. 6: srep35112 p. 8. 2016 (MB 817128). — Type: Herb. C-F-101845 (holotype). Ex-type: IBT 12289 = CBS 141311. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: MH492021 (alternative markers: BenA = MH492019; CaM = MH492020; RPB2 = MH492022).
Penicillium canescens Sopp, Skr. Vidensk.-Selsk. Christiana, Math.-Naturvidensk. Kl. 11: 181. 1912 (MB 153765). — Type: IMI 28260 (neotype, Pitt 1980). Ex-type: CBS 300.48 = ATCC 10419 = DSM1215 = FRR 910 = IMI 028260 = MUCL 29169 = NCTC 6607 = NRRL 910 = QM 7550 = VKMF-1148. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: AF033493 (alternative markers: BenA = JX140946; CaM = MN969241; RPB2 = JN121485).
Penicillium coralligerum Nicot & Pionnat, Bull. Soc. Mycol. France 78: 245. 1963 (1962) (MB 335721). — Type: IMI 99159 (neotype, Pitt et al. 2000). Ex-type: CBS 123.65 = ATCC 16968 = FRR 3465 = IFO 9578 = IHEM 4511 = IMI 099159 = LCP 58.1674 = NRRL 3465. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: JN617667 (alternative markers: BenA = MN969378; CaM = MN969248; RPB2 = JN406632).
Penicillium corvianum Visagie & Seifert, Persoonia 36: 259. 2016 (MB 815770). — Type: DAOM 695764 (holotype). Ex-type: DAOMC 250517 = CBS 141000 = KAS 3618 = IT-2008-4-D. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: KT887875 (alternative markers: BenA = KT887836; CaM = KT887797; RPB2 = MN969170).
Penicillium dunedinense Visagie, Seifert & Samson, Stud. Mycol. 78: 121. 2014 (MB 809183). — Type: CBS H-21803 (holotype). Ex-type: CBS 138218. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: KJ775678 (alternative markers: BenA = KJ775171; CaM = KJ775405; RPB2 = MN969116).
Penicillium echinatum: see under Penicillium elizabethiae.
Penicillium eickeri Visagie, Frisvad & K. Jacobs, published here (MB 834428). — Type: CBS H-22034 (holotype). Ex-type: CBS 138939 = IBT 31921 = DAOM 241352 = DTO 181-G3 = CV 475. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: JX140824 (alternative markers: BenA = JX140979; CaM = JX157365; RPB2 = KP016876).
Penicillium elizabethiae Visagie, Frisvad & K. Jacobs, published here (MB 834432). — Type: CBS H-22052 (holotype). Ex-type: NRRL 917 = MUCL 29170 = IBT 21955 = DTO 189-B8. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: KP016840 (alternative markers: BenA = KJ866964; CaM = KJ867021; RPB2 = KP016918).
≡ Penicillium echinatum E. Dale, Ann. Mycol. 24: 137. 1926 (nom. illegit. Art. 53.1; non Rivolta 1873) (MB 505484).
Penicillium granatense: see under Penicillium janczewskii.
Penicillium griseoazureum C. Moreau & M. Moreau ex C. Ramírez, Manual and Atlas of the Penicillia: 61. 1982 (MB 115800). — Type: CBS 162.42 (holotype). Ex-type: CBS 162.42 = FRR 1361. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: KC411679 (alternative markers: BenA = KP016919; CaM = KP016823; RPB2 = KP016852).
≡ Penicillium griseo-azureum C. Moreau & M. Moreau, Rev. Mycol. 6: 59. 1941 (nom. inval., Art. 36.1) (MB 289084). — Herb.: CBS 162.42. Ex-type: CBS 162.42 = FRR 1361.
Penicillium janczewskii K.M. Zaleski, Bull. Int. Acad. Polon. Sci., Cl. Sci. Math., Ser. B., Sci. Nat. 1927: 488. 1927 (MB 120703). — Type: IMI 191499 (neotype, Pitt 1980). Ex-type: CBS 221.28 = FRR 919 = IMI 191499 = NRRL 919. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: AY157487 (alternative markers: BenA = MN969386; CaM = MN969267; RPB2 = JN406612).
= Penicillium granatense C. Ramírez et al., Mycopathologia 72: 31. 1980 (MB 113023). — Type: IJFM 5965. Ex-type: CBS 166.81 = IJFM 5965 = IMI 253795 = VKMF-2191. ITS barcode: KC411682 (alternative markers: BenA = KJ866967; CaM = KJ866998; RPB2 = KP016853).
= Penicillium nigricans var. sulphureum S. Abe, J. Gen. Appl. Microbiol., Tokyo 2: 83. 1956 (nom. inval., Art. 36) (MB 347374). — Type: unknown. Ex-type: CBS 744.70 = ATCC 18380 = FAT536 = QM 7297. ITS barcode: KP016839 (alternative markers: BenA = KJ866966; CaM = KJ867018; RPB2 = KP016862).
Penicillium jensenii K.M. Zaleski, Bull. Int. Acad. Polon. Sci., Cl. Sci. Math., Ser. B., Sci. Nat. 1927: 494. 1927 (MB 120708). — Type: IMI 39768 (neotype, Pitt 1980). Ex-type: CBS 327.59 = ATCC 18317 = FRR 909 = IFO 5764 = IMI 039768 = LCP 89.1389 = NRRL 909 = QM 7587. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: AY443470 (alternative markers: BenA = JX140954; CaM = AY443490; RPB2 = JN406614).
= Penicillium siemaszkii K.M. Zaleski, Bull. Int. Acad. Polon. Sci., Cl. Sci. Math., Sér. B., Sci. Nat. 1927: 487. 1927 (Thom 1930, Raper & Thom 1949) (MB 278109). No culture available.
Penicillium linzhiense H.K. Wang & Jeewon, Front. Cell. Infect. Microbiol. 10: e-6045044. 2021 (MB 838576). — Type: CCTCC no: M2019870. Ex-type: CCTCC-M2019870. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: MT461156 (alternative markers: BenA = MT461157; CaM = MT461162; RPB2 = n.a.).
Penicillium murcianum C. Ramírez & A.T. Martínez, Mycopathologia 74: 37. 1981 (MB 112524). — Type: IJFM 7031 (holotype). Ex-type: CBS 161.81 = ATCC 42239 = IJFM 7031 = IMI 253800 = VKMF-2196. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: KC411678 (alternative markers: BenA = MN969419; CaM = MN969341; RPB2 = MN969202).
Penicillium nigricans Bainier in Thom, Penicillia: 351. 1930 (MB 119303). — Type: CBS H-22051 (holotype). Ex-type: CBS 354.48 = ATCC 10115 = IFO 6103 = IMI 039767 = NRRL 915 = QM 1933 = VKMF-313. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: KC411755 (alternative markers: BenA = KJ866965; CaM = KJ867012; RPB2 =KP016857).
Penicillium nigricans var. sulphureum: see under Penicillium janczewskii.
Penicillium radiatolobatum Lörinczi, Publ. Soc. Nat. Rom. Pent. Stiinta Sol. 10B: 435. 1972 (MB 114326). — Type: CBS H-7530 (isotype). Ex-type: CBS 340.79. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: KC411745 (alternative markers: BenA = MN969413; CaM = MT066183; RPB2 = MN969168).
Penicillium scottii Visagie, Frisvad & K. Jacobs, published here (MB 834430). — Type: CBS H-22040 (holotype). Ex-type: CBS 138951 = IBT 31905 = DTO 185-F8 = CV 930. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: JX140812 (alternative markers: BenA = JX140991; CaM = JX157383; RPB2 = KP016894).
Penicillium siemaszkii: see under Penicillium jensenii.
Penicillium yarmokense Baghd., Novosti Sist. Nizsh. Rast. 5: 99. 1968 (MB 335774). — Type: CBS H-7536 (isotype). Ex-type: CBS 410.69 = FRR 520 = IMI 140346 = VKMF-1076. Subgenus Penicillium section Canescentia series Canescentia. ITS barcode: KC411757 (alternative markers: BenA = MN969407; CaM = MN969314; RPB2 = JN406553).
DISCUSSION
Penicillium section Canescentia species are often reported in studies, but identifications in this group have remained problematic over the years. One major obstacle was species delineation based solely on morphological characters. Pitt (1980) mentions this difficulty because of, amongst others, the existence of intermediate strains (e.g., IMI 149218) that share characters between P. canescens and P. janczewskii. Here we show that this particular strain does belong to P. canescens which substantiates Pitt’s findings or indeed concerns with regards to the group’s variable morphologies. In this paper, we provide a sequence dataset for the 19 species previously considered to belong to section Canescentia (no culture is available for P. siemaszkii). We describe a further five new species P. allsoppiae, P. doidgeae, P. eickeri, P. pole-evansii and P. scottii. Furthermore, we synonymise P. granatense and P. nigricans var. sulphuratum with P. janczewskii, but show that P. griseoazureum, P. murcianum, P. nigricans, P. radiatolobatum and P. yarmokense should be considered as distinct species. We were unable to obtain a good representative set of strains for P. canescens and its close relatives P. murcianum, P. radiatolobatum and P. yarmokense. This particular clade will be subject to a future study to better resolve the group using many more strains and sequencing of additional gene regions. Similarly, P. novae-zeelandiae needs further revision with strains from a wider range of sources and locations needed. Even though more work is needed in section Canescentia, species identifications using only gene sequences should not present difficulties with the dataset provided in this study. The only concern is using BenA sequences to distinguish between P. murcianum and P. radiatolobatum; however, CaM sequences provides sufficient resolution. Penicillium chrysogenum, P. rubens and P. allii-sativii is an example of a similar situation where you may need more than BenA sequences for making an identification, both CaM and RPB2 sequences work well (Houbraken et al. 2011, 2012).
Correct species identifications of section Canescentia strains is important. Species in this section are often reported as good enzyme producers for biotechnology purposes (Brian et al. 1946, 1949, Pessoni et al. 1999, 2002, Weber et al. 2003, Chávez et al. 2006, Assamoi et al. 2008a, b, Terrasan et al. 2010). For example, JGI undertook whole genome sequencing of P. canescens ATCC 10419 with the aim to unravel mechanisms of phosphate solubilisation. In addition, some section Canescentia species are producers of antibiotics (Brian et al. 1953, Birch et al. 1965, Shiono et al. 2008), as well as mycotoxins (e.g. beta-nitropropionic acid from P. atrovenetum; patulin from P. antarticum, P. doidgeae, P. novae-zeelandiae and P. pole-evansii). The possible exploitation of these fungi for human purposes thus makes it very important to resolve the taxonomy of this section, resulting in well-defined species descriptions based on a polyphasic or consilient approach.
Most species from section Canescentia produce a diverse range of secondary metabolites that may provide a competitive advantage if produced in the soil environment (Table 3, 4). Fourteen out of 21 species produce known antibacterial compounds (patulin or penicillic acid and/or xanthoepocin) (Igarashi et al. 2000, Rasmussen et al. 2005, Frisvad 2018). Both patulin and penicillic acid are also quorum sensing inhibitors (Rasmussen et al. 2005) and could thus inhibit bacteria in soil and marine environments. Many species in series Canescentia also produce antifungal extrolites such as griseofulvin. Apart from the metabolites detected and reported from section Canescentia, several further secondary metabolite gene clusters appear to be silent (see for example data for P. arizonense (Grijseels et al. 2016)), at least under laboratory conditions. Thus one can anticipate to discover new antibiotics from this section.
Penicillium section Canescentia species have a worldwide distribution (Domsch et al. 1980, Houbraken & Samson 2011) and are commonly isolated from the fynbos biome of South Africa, especially from soil. Penicillium novae-zeelandiae and P. scottii were found to be common at sampling sites across the fynbos. One of our collection sites are situated in the Riverlands Nature Reserve near Malmesbury. This site also formed part of a previous survey that explored fungal communities associated with Protea rhizosphere soil and amongst other species, recovered P. novae-zeelandiae and P. janczewskii (Allsopp et al. 1987). Penicillium janczewskii has not been reported from any other South African surveys and it is our opinion that Allsopp’s strains may belong to P. scottii. Another survey also found P. novae-zeelandiae in Protea species (Schutte 1992). It is not often that one has the opportunity to collect samples from the same area as a previous survey, especially ones that identified Penicillium to species level. It is quite remarkable that the same species from 1987 were also recorded in our study. This was not only for the section Canescentia, but also for P. corylophilum classified in section Exilicaulis (Visagie et al. 2016c) and section Torulomyces (previously the genus Torulomyces) (Visagie et al. 2016a). Even though Penicillium species are often considered as degraders of dead organic waste, they arguably have additional roles in nature. The fact that the same Penicillium species were found almost 20 years later at the same site, might point to stable core fungal communities and suggest that these species either play an important role in this ecosystem or might be associated with specific plant species in the area and thus conidia also occur in soil or litter. Observations from this survey suggest that species might be subjected to vectored dispersal along with common conidia dispersal mechanisms such as air or water dispersal. This has opened a number of questions about the distribution of Penicillium species in the fynbos biome and is something that should be studied further. In terms of species diversity, this survey resulted in the recovery of 61 Penicillium (29 described as new) and 15 Talaromyces species (nine described as new) (Visagie et al. 2009, 2014a, 2015b, Visagie & Jacobs 2012, Yilmaz et al. 2016) with many new DNA reference sequences generated. The large number of species recovered and the high proportion of new species found was not unexpected considering the diversity of endemic plants in the region. However, considering the limited number of sites that were sampled and the diverse vegetation of the fynbos (Mucina & Rutherford 2006), we believe that there are many more new Penicillium and Talaromyces species waiting to be discovered from this diversity hotspot (Myers et al. 2000).
Acknowledgments
We are grateful to Keith A. Seifert for his support, guidance and insights provided while working on this manuscript, with whom CMV completed the morphological descriptions while working on the Alfred P. Sloan Foundation Program on the Microbiology of the Built Environment at the Ottawa Research and Development Centre (Agriculture and AgriFood Canada). We also acknowledge the South African Biosystematics Initiative (SABI; grant nr KFD2008052100003) and Foundational Biodiversity Information Programme (FBIP; grant nr 118607) of the National Research Foundation (NRF), and the Future Leaders - African Independent Research fellowship programme (FLAIR, FLR\R1\201831), for the financial support during this project. The FLAIR Fellowship Programme is a partnership between the African Academy of Sciences and the Royal Society funded by the UK Government’s Global Challenges Research Fund. The Western Cape Nature Conservation Board issued permits for collecting soil and Protea repens infructescences in the fynbos. JCF acknowledge the Danish National Research Foundation (DNRF137) for the Center for Microbial Secondary Metabolites (CeMiSt), the Novo Nordisk Foundation for the project Fungal quinone batteries (grant nr. NNF18OC0034952) and the project INTARACT (grant nr. NNF 19SA0059360).
Supplementary material
Phylogenetic tree of Penicillium section Canescentia ex-type strains using the ITS region. Penicillium sacculum was chosen as outgroup. Posterior probabilities (pp) and/or bootstrap values (bs) higher than 0.95 and 80, respectively, are given above thickened branches. Names in grey text indicate reference strains, black text fynbos strains and coloured text new species, T = ex-type strain.
Untrimmed ITS, BenA and CaM alignments of Penicillium section Canescentia. Annotations: blue = rRNA; green = internal transcribed spacer regions; orange = complete cds; yellow = cds as submitted; red = considered as suspicious base calls (regions trimmed from final alignment).
Phylogenetic trees of ITS, BenA and CaM calculated using untrimmed alignments as shown in Fig. S1. Penicillium brevicompactum was chosen as outgroup. Posterior probabilities and/or bootstrap values higher than 0.95 and 80, respectively, are given on supported branches. Names in green represent P. antarcticum, while red represents species described by Kirichuk et al. (2016).
REFERENCES
- Afiyatullov SS, Zhuravleva OI, Antonov AS, et al. 2019. Piltunines A–F from the marine-derived fungus Penicillium piltunense KMM 4668. Marine Drugs 17: 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alapont C, López-Mendoza MC, Gil JV, et al. 2014. Mycobiota and toxigenic Penicillium species on two Spanish dry-cured ham manufacturing plants. Food Additives & Contaminants: Part A 31: 93–104. [DOI] [PubMed] [Google Scholar]
- Alfaro C, Urios A, González MC, et al. 2003. Screening for metabolites from Penicillium novae-zeelandiae displaying radical-scavenging activity and oxidative mutagenicity: Isolation of gentisyl alcohol. Mutation Research - Genetic Toxicology and Environmental Mutagenesis 539: 187–194. [DOI] [PubMed] [Google Scholar]
- Allsopp N, Olivier DL, Mitchell DT. 1987. Fungal populations associated with root systems of proteaceous seedlings at a lowland fynbos site in South Africa. South African Journal of Botany 53: 365–369. [Google Scholar]
- Assamoi AA, Destain J, Delvigne F, et al. 2008a. Improvement of xylanase production by Penicillium canescens 10-10c in solid-state fermentation. Pentoses dérivés de la biomasse. Biotechnology, Agronomy, Society and Environment 12: 111–118. [Google Scholar]
- Assamoi AA, Destain J, Delvigne F, et al. 2008b. Solid-state fermentation of xylanase from Penicillium canescens 10-10c in a multi-layer-packed bed reactor. Applied Biochemistry and Biotechnology 145: 87–98. [DOI] [PubMed] [Google Scholar]
- Bae G-S, Jung H-W, Kim H-G-, et al. 1999. Novel compound, cis-fumagillin, inhibiting angiogenesis and angiogenesis-inhibiting composition containing the same. Patent KR2001011248–A pp. [Google Scholar]
- Bertinetti BV, Pena NI, Cabrera GM. 2009. An antifungal tetrapeptide from the culture of Penicillium canescens. Chemistry & Biodiversity 6: 1178–1184. [DOI] [PubMed] [Google Scholar]
- Bilai V. 1963. Antibiotic-producing microscopic fungi. edn. Elsevier, Amsterdam. [Google Scholar]
- Birch AJ, Loh L, Pelter A, et al. 1965. The structure of canescin. Tetrahedron Letters 6: 29–32. [DOI] [PubMed] [Google Scholar]
- Birkinshaw JH, Mohammed YS. 1962. Studies in the biochemistry of micro-organisms. 111. The production of l-phenylalanine anhydride (cis-l-3,6-dibenzyl-2,5-dioxopiperazine) by Penicillium nigricans (Bainier) Thom. Biochemical Journal 85: 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee AF. 1915. Lindner’s roll tube method of separation cultures. Phytopathology 5: 68–69. [Google Scholar]
- Brian PW, Curtis PJ, Hemming HG. 1946. A substance causing abnormal development of fungal hyphae produced by Penicillium janczewskii Zal.: I. Biological assay, production and isolation of ‘curling factor’. Transactions of the British Mycological Society 29: 173–187. [Google Scholar]
- Brian PW, Curtis PJ, Hemming HG. 1949. A substance causing abnormal development of fungal hyphae produced by Penicillium janczewskii Zal.: III. Identity of ‘curling factor’ with griseofulvin. Transactions of the British Mycological Society 32: 30–33. [Google Scholar]
- Brian PW, Hemming HG, Moffatt JS. 1953. Canescin, an antibiotic produced by Penicillium canescens. Transactions of the British Mycological Society 36: 243–247. [Google Scholar]
- Burton HS. 1949. Antibiotics from Penicillia. British Journal of Experimental Pathology 30: 151–158. [PMC free article] [PubMed] [Google Scholar]
- Chávez R, Bull P, Eyzaguirre J. 2006. The xylanolytic enzyme system from the genus Penicillium. Journal of Biotechnology 123: 413–433. [DOI] [PubMed] [Google Scholar]
- Chen S, Jiang M, Chen B, et al. 2019. Penicamide A, a unique N,N′-ketal quinazolinone alkaloid from ascidian-derived fungus Penicillium sp. 4829. Marine Drugs 17: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Yang X, Peng Y. 1999. A new producer of mevastatin. Chinese Journal of Antibiotics 24: 4–6. [Google Scholar]
- Curtis PJ, Grove JF. 1947. A fungistatic and bacteriostatic red pigment produced by a strain of the Penicillium nigricans-janczewskii series. Nature 160: 574–575. [DOI] [PubMed] [Google Scholar]
- Dale E. 1926. Note on three new species of Penicillium: P. echinatum, P. flexuosum and P. sacculum. Annales Mycologici 24: 137. [Google Scholar]
- Dalsgaard PW, Petersen BO, Duus JØ, et al. 2012. Atlantinone A, a meroterpenoid produced by Penicillium ribeum and several cheese associated Penicillium species. Metabolites 2: 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasanayaka SAHK, Nong X-H, Liang X, et al. 2020. New dibenzodioxocinone and pyran-3,5-dione derivatives from the deep-sea-derived fungus Penicillium canescens SCSIO z053. Journal of Asian Natural Products Research 22: 338–345. [DOI] [PubMed] [Google Scholar]
- Di Menna ME, Lauren DR, Wyatt PA. 1986. Effect of culture conditions on tremorgen production by some Penicillium species. Applied and Environmental Microbiology 51: 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsch KH, Gams W, Anderson T-H. 1980. Compendium of soil fungi. edn. IHW-Verlag, Eching. [Google Scholar]
- Fan B, Parrot D, Blümel M, et al. 2019. Influence of OSMAC-based cultivation in metabolome and anticancer activity of fungi associated with the brown alga Fucus vesiculosus. Marine Drugs 17: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Flewelling AJ, Johnson JA, Gray CA. 2013. Antimicrobials from the marine algal endophyte Penicillium sp. Natural Product Communications 8: 373–374. [PubMed] [Google Scholar]
- Frank M, Hartmann R, Plenker M, et al. 2019. Brominated azaphilones from the sponge-associated fungus Penicillium canescens strain 4.14.6a. Journal of Natural Products 82: 2159–2166. [DOI] [PubMed] [Google Scholar]
- Frisvad JC. 2018. A critical review of producers of small lactone mycotoxins: patulin, penicillic acid and moniliformin. World Mycotoxin Journal 11: 73–100. [Google Scholar]
- Frisvad JC, Filtenborg O. 1990. Revision of Penicillium subgenus Furcatum based on secondary metabolites and conventional characters. In: Samson RA, Pitt JI. (eds), Modern concepts in Penicillium and Aspergillus classification: 159–172. Plenum Press, New York. [Google Scholar]
- Frisvad JC, Thrane U. 1987. Standardized high performance liquid chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone indices and UV VIS spectra (diode array detection). Journal of Chromatography 404: 195–214. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Thrane U. 1993. Chapter 8 liquid column chromatography of mycotoxins. In: Betina V. (ed.), Journal of Chromatography library: 253–372 Elsevier, Amsterdam. [Google Scholar]
- Gedek B. 1977. Kind and frequency of toxin production by cereals contaminated with Aspergilli and Penicillia. Annals de Nutrition et Alimentation 31: 467–475. [PubMed] [Google Scholar]
- Geiger M, Guitton Y, Vansteelandt M, et al. 2013. Cytotoxicity and mycotoxin production of shellfish-derived Penicillium spp., a risk for shellfish consumers. Letters in Applied Microbiology 57: 385–392. 10.1111/lam.12143. [DOI] [PubMed] [Google Scholar]
- Ghanbari MAT, Mohammadkhani H, Babaeizad V. 2014. Identification of some secondary metabolites produced by four Penicillium species. Mycologia Iranica 1: 107–113. [Google Scholar]
- Goto T, Tanaka H, Okuhara M, et al. 1986. Immuno-regulator agents obtained by fermentation of Penicillium species. Japanese patent JP 61257921–A pp. [Google Scholar]
- Grijseels S, Nielsen JC, Randelovic M, et al. 2016. Penicillium arizonense, a new, genome sequenced fungal species, reveals a high chemical diversity in secreted metabolites. Scientific Reports 6: srep35112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove JF, Mcgowan JC. 1947. Identity of griseofulvin and ‘curling-factor’. Nature 160: 574. [DOI] [PubMed] [Google Scholar]
- Gupta M, Chatterjee T, Sengupta S, et al. 1984. Structure of a new mycotoxin (MT81). Indian Journal of Chemistry 23B: 393–394. [Google Scholar]
- He J, Lion U, Sattler I, et al. 2005. Diastereomeric quinolinone alkaloids from the marine-derived fungus Penicillium janczewskii. Journal of Natural Products 68: 1397–1399. [DOI] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Samson RA. 2011. Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus 2: 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Seifert KA, et al. 2012. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia 29: 78–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Kocsube S, Visagie CM, et al. 2020. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. 2011. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Visagie CM, Meijer M, et al. 2014. A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Studies in Mycology 78: 373–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Kuwamori Y, Takagi K, et al. 2000. Xanthoepocin, a new antibiotic from Penicillium simplicissimum IFO5762. The Journal of Antibiotics 53: 928–933. [DOI] [PubMed] [Google Scholar]
- Jefferys EG, Brian PW, Hemming HG, et al. 1953. Antibiotic production by the microfungi of acid heath soils. Journal of General Microbiology 9: 314–341. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keromnes J, Thouvenot D. 1985. Role of penicillic acid in the phytotoxicity of Penicillium cyclopium and Penicillium canescens to the germination of corn seeds. Applied and Environmental Microbiology 49: 660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko S. 1970. Identification of antibiotics from penicillia active against agents of mottled leaves and bunt fungi. Mikrobiologische Zhurnal (Kiev) 32: 115–119. [PubMed] [Google Scholar]
- Kirichuk NN, Pivkin MV, Matveeva TV. 2016. Three new Penicillium species from marine subaqueous soils. Mycological Progress 16: 15–26. [Google Scholar]
- Kornerup A, Wanscher JH. 1967. Methuen handbook of colour. 2nd edn. Methuen & Co Ltd, London, England. [Google Scholar]
- Kozlovskii AG, Vinokurova NG, Zhelifonova VP, et al. 1997a. Alkaloid formation by penicillia of the series Fellutana and Canescentia. Microbiology (Moscow) 66: 429–433. [Google Scholar]
- Kozlovskii AG, Vinokurova NG, Zhelifonova VP, et al. 1997b. Secondary metabolites of fungi belonging to the species Penicillium janczewskii. Applied Biochemistry and Microbiology 33: 61–64. [Google Scholar]
- Kwon JY, Jeong HW, Kim HK, et al. 2000. Cis-fumagillin, a new methionine aminopeptidase (type 2) inhibitor produced by Penicillium sp. F2757. The Journal of Antibiotics 53: 799–806. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. [DOI] [PubMed] [Google Scholar]
- Laws I, Mantle PG. 1985. Nigrifortine, a diketopiperazine metabolite of Penicillium nigricans. Phytochemistry 24: 1395–1397. [Google Scholar]
- Leistner L. 1979. Vorkommen toxinogener Penicillien bei Fleischerzeugnissen. Die Fleischwirtschaft 59: 1892–1896. [Google Scholar]
- Leistner L, Pitt J. 1977. Miscellaneous Penicillium toxins. In: Rodricks JV, Hesseltine CW, Mehlman MA. (eds), Mycotoxins in human and animal health: 639–653. Pathotox Publishers, Park Forest South. [Google Scholar]
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research 44: W242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan J. 1951. Dechlorogriseofulvin – a metabolic product of Penicillium griseofulvum Dierckx and Penicillium janczewskii Zalessky. Chemistry & Industry 1951: 719. [Google Scholar]
- MacMillan J. 1954. Griseofulvin. Part IX. Isolation of the bromo-analogue from Penicillium griseofulvum and Penicillium nigricans. Journal of the Chemical Society (Resumed): 2585. [Google Scholar]
- Malik A, Ardalani H, Anam S, et al. 2020. Antidiabetic xanthones with α-glucosidase inhibitory activities from an endophytic Penicillium canescens. Fitoterapia 142: 104522. [DOI] [PubMed] [Google Scholar]
- Mantle PG, Laws I, Tan MJ, et al. 1984. A novel process for the production of penitrem mycotoxins by submerged fermentation of Penicillium nigricans. Journal of General Microbiology 130: 1293–1298. [DOI] [PubMed] [Google Scholar]
- Mantle PG, Penn J. 1989. A role for paxilline in the biosynthesis of indole-diterpenoid penitrem mycotoxins. Journal of the Chemical Society, Perkin Transactions 1: 1539–1540. [Google Scholar]
- Marchese P, Mahajan N, O’Connell E, et al. 2020. A novel high-throughput screening platform identifies itaconate derivatives from marine Penicillium antarcticum as inhibitors of mesenchymal stem cell differentiation. Marine Drugs 18: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae CF, Hocking AD, Seppelt RD. 1999. Penicillium species from terrestrial habitats in the Windmill Islands, East Antarctica, including a new species, Penicillium antarcticum. Polar Biology 21: 97–111. [Google Scholar]
- Moslem MA, El-Samawaty AE-R, Yassin M. 2013. Unconventional control method of mycotoxigenic Penicillium spp. associated with apple blue mold. Fresenius Environmental Bulletin 22: 813–817. [Google Scholar]
- Mucina L, Rutherford MC. 2006. The vegetation of South Africa, Lesotho and Swaziland. edn. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, et al. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- Neill KG, Raistrick H. 1957. Studies in the biochemistry of microorganisms. 100. Metabolites of Penicillium atrovenetum Smith, George.1. Atrovenetin, a new crystalline colouring matter. Biochemical Journal 65: 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, et al. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaz S-I, Zhang P, Shen H, et al. 2019. Two new isochromane derivatives penisochromanes A and B from ascidian-derived fungus Penicillium sp. 4829. Natural Product Research 33: 1262–1268. [DOI] [PubMed] [Google Scholar]
- Nicoletti R, Lopez-Gresa MP, Manzo E, et al. 2007. Production and fungitoxic activity of Sch 642305, a secondary metabolite of Penicillium canescens. Mycopathologia 163: 295–301. [DOI] [PubMed] [Google Scholar]
- Nielsen KF, Mansson M, Rank C, et al. 2011. Dereplication of microbial natural products by LC-DAD-TOFMS. Journal of Natural Products 74: 2338–2348. [DOI] [PubMed] [Google Scholar]
- Oleinikova GK, Kirichuk NN, Afiyatullov SS. 2018. Nonpolar compounds and free fatty acids from several isolates of marine fungus Penicillium antarcticum. Chemistry of Natural Compounds 54: 535–537. [Google Scholar]
- Özkaya FC, Ebrahim W, Klopotowski M, et al. 2018. Isolation and X-ray structure analysis of citreohybridonol from marine-derived Penicillium atrovenetum. Natural Product Research 32: 840–843. [DOI] [PubMed] [Google Scholar]
- Patterson DS, Roberts BA, Shreeve BJ, et al. 1979. Tremorgenic toxins produced by soil fungi. Applied and Environmental Microbiology 37: 172–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson DS, Shreeve BJ, Roberts BA, et al. 1981. Verruculogen produced by soil fungi in England and Wales. Applied and Environmental Microbiology 42: 916–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn J, Biddle JR, Mantle PG, et al. 1992. Pennigritrem, a naturally-occurring penitrem A analogue with novel cyclisation in the diterpenoid moiet. Journal of the Chemical Society, Perkin Transactions 1, 1992: 23–26. [Google Scholar]
- Penn J, Mantle PG. 1994. Biosynthetic intermediates of indole-diterpenoid mycotoxins from selected transformations at C-10 of paxilline. Phytochemistry 35: 921–926. [Google Scholar]
- Pessoni RA, Figueiredo-Ribeiro RC, Braga MR. 1999. Extracellular inulinases from Penicillium janczewskii, a fungus isolated from the rhizosphere of Vernonia herbacea (Asteraceae). Journal of Applied Microbiology 87: 141–147. [DOI] [PubMed] [Google Scholar]
- Pessoni RAB, Freshour G, Figueiredo-Ribeiro RdCL, et al. 2002. Woronin bodies in Penicillium janczewskii Zaleski. Brazilian Journal of Microbiology 33: 127–130. [Google Scholar]
- Petersen L-E, Marner M, Labes A, et al. 2019. Rapid metabolome and bioactivity profiling of fungi associated with the leaf and rhizosphere of the baltic seagrass Zostera marina. Marine Drugs 17: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SW. 2000. Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences. In: Samson RA, Pitt JI. (eds), Integration of modern taxonomic methods for Penicillium and Aspergillus classification: 163–178. Harwood Academic Publishers, Amsterdam. [Google Scholar]
- Petit KE, Mondeguer F, Roquebert MF, et al. 2004. Detection of griseofulvin in a marine strain of Penicillium waksmanii by ion trap mass spectrometry. Journal of Microbiological Methods 58: 59–65. [DOI] [PubMed] [Google Scholar]
- Pitt JI. 1980. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. edn. Academic Press, London. [Google Scholar]
- Pitt JI, Samson RA, Frisvad JC. 2000. List of accepted species and their synonyms in the family Trichocomaceae. In: Samson RA, Pitt JI. (eds), Integration of modern taxonomic methods for Penicillium and Aspergillus classification: 9–79. Harwood Academic Publishers, Reading. [Google Scholar]
- Prigent S, Nielsen JC, Frisvad JC, et al. 2018. Reconstruction of 24 Penicillium genome-scale metabolic models shows diversity based on their secondary metabolism. Biotechnology and Bioengineering 115: 2604–2612. [DOI] [PubMed] [Google Scholar]
- Raistrick H, Stössl A. 1958. Studies in the biochemistry of micro-organisms. 104. Metabolites of Penicillium atrovenetum G. Smith: β-nitropropionic acid, a major metabolite. Biochemical Journal 68: 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez C. 1982. Manual and atlas of the Penicillia. edn. Elsevier Biomedical Press, Amsterdam. [Google Scholar]
- Raper KB, Thom C. 1949. A manual of the Penicillia. edn. The Williams & Wilkins company, Baltimore. [Google Scholar]
- Rasmussen TB, Skindersoe ME, Bjarnsholt T, et al. 2005. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151: 1325–1340. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeda-Hirschmann G, Hormazabal E, Astudillo L, et al. 2005. Secondary metabolites from endophytic fungi isolated from the Chilean gymnosperm Prumnopitys andina (Lleuque). World Journal of Microbiology and Biotechnology 21: 27–32. [Google Scholar]
- Schmeda-Hirschmann G, Hormazabal E, Rodriguez JA, et al. 2008. Cycloaspeptide A and pseurotin A from the endophytic fungus Penicillium janczewskii. Zeitschrift fur Naturforschung C 63: 383–388. [DOI] [PubMed] [Google Scholar]
- Schutte AL. 1992. An overview of Penicillium (Hyphomycetes) and associated teleomorphs in southern Africa. Bothalia 22: 77–91. [Google Scholar]
- Shiono Y, Seino Y, Koseky T, et al. 2008. Antarones A and B, two polyketides from an endophytic Penicillium antarcticum. Verlag der Zeitschrift für Naturforschung 63b: 909–914. [Google Scholar]
- Shreeve BJ, Patterson DS, Roberts BA, et al. 1978. Isolation of potentially tremorgenic fungi from pasture associated with a condition resembling ryegrass staggers. Veterinary Record 103: 209–210. [DOI] [PubMed] [Google Scholar]
- Smedsgaard J. 1997. Micro-scale extraction procedure for standardization screening of fungal metabolite production in cultures. Journal of Chromatography A 760: 264–270. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Sakai K, Nagano Y, et al. 2017. Cladomarine, a new anti-saprolegniasis compound isolated from the deep-sea fungus, Penicillium coralligerum YK-247. The Journal of Antibiotics 70: 911–914. [DOI] [PubMed] [Google Scholar]
- Terrasan CRF, Temer B, Duarte MCT, et al. 2010. Production of xylanolytic enzymes by Penicillium janczewskii. Bioresource Technology 101: 4139–4143. [DOI] [PubMed] [Google Scholar]
- Thom C. 1930. The Penicillia. edn. Williams & Wilkins, Baltimore. [Google Scholar]
- Turner WB, Aldridge DC. 1983. Fungal metabolites II. edn. Academic Press, New York. [Google Scholar]
- Udagawa K, Abe S. 1961. Production of griseofulvin by some strains of the genus Penicillum. The Journal of Antibiotics, Series A 14: 215–220. [Google Scholar]
- Ueno Y, Kawamura O, Sugiura Y, et al. 1991. Use of monoclonal antibodies, enzyme-linked immunosorbent assay and immunoaffinity column chromatography to determine ochratoxin A in porcine sera, coffee products and toxin-producing fungi. In: Castagnero M, Plestina R, Dirheimer G, et al. (eds), IARC Scientific Publications: 71–75. [PubMed] [Google Scholar]
- Vansteelandt M, Blanchet E, Egorov M, et al. 2013. Ligerin, an antiproliferative chlorinated sesquiterpenoid from a marine-derived Penicillium strain. Journal of Natural Products 76: 297–301. [DOI] [PubMed] [Google Scholar]
- Vansteelandt M, Kerzaon I, Blanchet E, et al. 2012. Patulin and secondary metabolite production by marine-derived Penicillium strains. Fungal Biology 116: 954–961. [DOI] [PubMed] [Google Scholar]
- Visagie CM, Hirooka Y, Tanney JB, et al. 2014a. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology 78: 63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Dijksterhuis J, et al. 2016a. A taxonomic review of Penicillium species producing conidiophores with solitary phialides, classified in section Torulomyces. Persoonia 36: 134–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC, et al. 2014b. Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Rodriques C, et al. 2013. Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia 31: 42–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Seifert KA, et al. 2015a. Four new Penicillium species isolated from the fynbos biome in South Africa, including a multigene phylogeny of section Lanata-Divaricata. Mycological Progress 14: 96. [Google Scholar]
- Visagie CM, Jacobs K. 2012. Three new additions to the genus Talaromyces isolated from Atlantis sandveld fynbos soils. Persoonia 28: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Renaud JB, Burgess KM, et al. 2016b. Fifteen new species of Penicillium. Persoonia 36: 247–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Roets F, Jacobs K. 2009. A new species of Penicillium, P. ramulosum sp. nov., from the natural environment. Mycologia 101: 888–895. [DOI] [PubMed] [Google Scholar]
- Visagie CM, Seifert KA, Houbraken J, et al. 2014c. Diversity of Penicillium section Citrina within the fynbos biome of South Africa, including a new species from a Protea repens infructescence. Mycologia 106: 537–552. [DOI] [PubMed] [Google Scholar]
- Visagie CM, Seifert KA, Houbraken J, et al. 2016c. A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. IMA Fungus 7: 75–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Yilmaz N, Frisvad JC, et al. 2015b. Five new Talaromyces species with ampulliform-like phialides and globose rough walled conidia resembling T. verruculosus. Mycoscience 56: 486–502. [Google Scholar]
- Wang Y, Wang G, Wang L, et al. 2010. Isolation and identification of an endophytic fungus of Polygonatum cyrtonema and its antifungal metabolites. Acta Microbiologica Sinica 50: 1036–1043. [PubMed] [Google Scholar]
- Weber RWS, Kuhn A, Anke H. 2003. Soil-borne Penicillium spp. and other microfungi as efficient degraders of the explosive RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine). Mycological Progress 2: 83–93. [Google Scholar]
- Wright JM. 1955. The production of antibiotics in soil. II. Production of griseofulvin by Penicillium nigricans. Annals of Applied Biology 43: 288–296. [Google Scholar]
- Yaegashi J, Romsdahl J, Chiang Y-M, et al. 2015. Genome mining and molecular characterization of the biosynthetic gene cluster of a diterpenic meroterpenoid, 15-deoxyoxalicine B, in Penicillium canescens. Chemical Science 6: 6537–6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz N, Visagie CM, Frisvad JC, et al. 2016. Taxonomic re-evaluation of species in Talaromyces section Islandici, using a polyphasic approach. Persoonia 36: 37–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y, Gong Y, Gong J, et al. 2020. Fungal polyketides with three distinctive ring skeletons from the fungus Penicillium canescens uncovered by OSMAC and Molecular Networking Strategies. The Journal of Organic Chemistry 85: 4973–4980. [DOI] [PubMed] [Google Scholar]
- Zang Y, Gong Y-H, Li X-W, et al. 2019. Canescones A–E: aromatic polyketide dimers with PTP1B inhibitory activity from Penicillium canescens. Organic Chemistry Frontiers 6: 3274–3281. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of Penicillium section Canescentia ex-type strains using the ITS region. Penicillium sacculum was chosen as outgroup. Posterior probabilities (pp) and/or bootstrap values (bs) higher than 0.95 and 80, respectively, are given above thickened branches. Names in grey text indicate reference strains, black text fynbos strains and coloured text new species, T = ex-type strain.
Untrimmed ITS, BenA and CaM alignments of Penicillium section Canescentia. Annotations: blue = rRNA; green = internal transcribed spacer regions; orange = complete cds; yellow = cds as submitted; red = considered as suspicious base calls (regions trimmed from final alignment).
Phylogenetic trees of ITS, BenA and CaM calculated using untrimmed alignments as shown in Fig. S1. Penicillium brevicompactum was chosen as outgroup. Posterior probabilities and/or bootstrap values higher than 0.95 and 80, respectively, are given on supported branches. Names in green represent P. antarcticum, while red represents species described by Kirichuk et al. (2016).