Abstract
The circumscription and composition of the Hyaloscyphaceae are controversial and based on poorly sampled or unsupported phylogenies. The generic limits within the hyaloscyphoid fungi are also very poorly understood. To address this issue, a robust five-gene Bayesian phylogeny (LSU, RPB1, RPB2, TEF-1α, mtSSU; 5521 bp) with a focus on the core group of Hyaloscyphaceae and Arachnopezizaceae is presented here, with comparative morphological and histochemical characters. A wide representative sampling of Hyaloscypha supports it as monophyletic and shows H. aureliella (subgenus Eupezizella) to be a strongly supported sister taxon. Reinforced by distinguishing morphological features, Eupezizella is here recognised as a separate genus, comprising E. aureliella, E. britannica, E. roseoguttata and E. nipponica (previously treated in Hyaloscypha). In a sister group to the Hyaloscypha-Eupezizella clade a new genus, Mimicoscypha, is created for three seldom collected and poorly understood species, M. lacrimiformis, M. mimica (nom. nov.) and M. paludosa, previously treated in Phialina, Hyaloscypha and Eriopezia, respectively. The Arachnopezizaceae is polyphyletic, because Arachnoscypha forms a monophyletic group with Polydesmia pruinosa, distant to Arachnopeziza and Eriopezia; in addition, Arachnopeziza variepilosa represents an early diverging lineage in Hyaloscyphaceae s.str. The hyphae originating from the base of the apothecia in Arachnoscypha are considered anchoring hyphae (vs a subiculum) and Arachnoscypha is excluded from Arachnopezizaceae. A new genus, Resinoscypha, is established to accommodate Arachnopeziza variepilosa and A. monoseptata, originally described in Protounguicularia. Mimicoscypha and Resinoscypha are distinguished among hyaloscyphoid fungi by long tapering multiseptate hairs that are not dextrinoid or glassy, in combination with ectal excipulum cells with deep amyloid nodules. Unique to Resinoscypha is cyanophilous resinous content in the hairs concentrated at the apex and septa. Small intensely amyloid nodules in the hairs are furthermore characteristic for Resinoscypha and Eupezizella. To elucidate species limits and diversity in Arachnopeziza, mainly from Northern Europe, we applied genealogical concordance phylogenetic species recognition (GCPSR) using analyses of individual datasets (ITS, LSU, RPB1, RPB2, TEF-1α) and comparative morphology. Eight species were identified as highly supported and reciprocally monophyletic. Four of these are newly discovered species, with two formally described here, viz. A. estonica and A. ptilidiophila. In addition, Belonium sphagnisedum, which completely lacks prominent hairs, is here combined in Arachnopeziza, widening the concept of the genus. Numerous publicly available sequences named A. aurata represent A. delicatula and the confusion between these two species is clarified. An additional four singletons are considered to be distinct species, because they were genetically divergent from their sisters. A highly supported five-gene phylogeny of Arachnopezizaceae identified four major clades in Arachnopeziza, with Eriopezia as a sister group. Two of the clades include species with a strong connection to bryophytes; the third clade includes species growing on bulky woody substrates and with pigmented exudates on the hairs; and the fourth clade species with hyaline exudates growing on both bryophytes and hardwood. A morphological account is given of the composition of Hyaloscyphaceae and Arachnopezizaceae, including new observations on vital and histochemical characters.
Citation: Kosonen T, Huhtinen S, Hansen K. 2021. Taxonomy and systematics of Hyaloscyphaceae and Arachnopezizaceae. Persoonia 46: 26–62. https://doi.org/10.3767/persoonia.2021.46.02.
Keywords: Arachnoscypha, epibryophytic, genealogical species, Helotiales, subiculum, type studies
INTRODUCTION
Helotiales is one of the remaining unsolved pieces in the great puzzle of Leotiomycetes systematics. Depending on the taxonomic concept, the number of recognised species ranges from c. 2 360 (Baral 2016) to 3 881 (Kirk et al. 2008). There are no recent speculations on the total number of existing species, but e.g., Hawksworth (2001) gives an estimate of c. 70 000 species. Members of Helotiales are morphologically very variable and exhibit saprobic, parasitic as well as mycorrhizal life strategies. Hyaloscyphaceae in the traditional wide sense (Nannfeldt 1932) is the largest and most diverse family in the order (Kirk et al. 2008, Baral 2016 as Lineage D/Hyaloscyphaceae s.lat. with 68 genera and 673 species).
When Nannfeldt (1932) introduced the concept of Hyaloscyphaceae, it included three tribes, i.e., Arachnopezizeae, Hyaloscypheae and Lachneae, and altogether 13 genera. Nannfeldt emphasized especially the shape and structure of the excipular cells as a delimiting character at the family-level. Lachneae included species with lanceolate paraphyses and multiseptate granulate hairs and Arachnopezizeae species with a subiculum surrounding the apothecia. The species of Hyaloscypheae were not united by any unique combination of characters, but the majority of genera had cylindrical paraphyses and hairs of very diverse shapes and size.
Hyaloscyphaceae received growing attention during the following years and the systematics of the whole group was treated in studies by Dennis (1949, 1962, 1981), Raitviir (1970, 1987), Spooner (1987) and Svrček (1987). Although only a few monographs on hyaloscyphaceous genera have been published so far (Korf 1951b, Zhuang 1988, Huhtinen 1989), various authors contributed in documenting the diversity of the hairy Helotiales, e.g., Korf (e.g., 1951a, 1978, 1981, 2007), Haines (1974, 1989), Raschle (1977), Svrček (e.g., 1977, 1984, 1985), Korf & Kohn (1980), Raitviir & Sharma (1984), Baral & Krieglsteiner (1985), Spooner & Dennis (1985), Raitviir & Galán (1986), Baral (e.g., 1987, 1993), Huhtinen (e.g., 1987a, b, 1993a), Galán & Raitviir (1994, 2004), Cantrell & Hanlin (1997), Hosoya & Otani (1997), Raitviir & Huhtinen (1997), Leenurm et al. (2000), Zhuang (2000), Raitviir (2001), Quijada et al. (2014), and Baral & Rämä (2015). The number of recognised genera today is c. 70 and the number of species c. 1 000 (Kirk et al. 2008, Baral 2016).
There were no major revisions to the higher-level systematics until the 21st century. In his revised synopsis of the Hyaloscyphaceae s.str., Raitviir (2004) concluded that the morphological differences between the core members of the two largest tribes, Hyaloscypheae and Lachneae were ‘strictly contrasting’. With evidence on the ultra-structural differences in the hair wall layers (Leenurm et al. 2000) and the first phylogenetic analyses of the ITS region available (Cantrell & Hanlin 1997), Raitviir (2004) emended Hyaloscyphaceae to conform to the tribe Hyaloscypheae, and elevated Lachneae to the rank of family.
Recent multi-gene phylogenetic analyses have clearly shown that Hyaloscyphaceae sensu Raitviir (2004) is polyphyletic (Han et al. 2014, Johnston et al. 2019). The species occur in six or more clades, spread among other clades of Helotiales. These hyaloscyphoid clades correspond to emended tribes/sub-families/families, and to two newly described families (Arachnopezizaceae, Hyaloscyphaceae s.str., Lachnaceae, Pezizellaceae, Vandijckellaceae), as well as to unnamed groups (Raitviir 2004, Han et al. 2014, Crous et al. 2017, Johnston et al. 2019). The sampling of taxa or molecular characters is, however, very limited in the molecular studies (Han et al. 2014, Johnston et al. 2019), and the evolutionary history of these fungi is still poorly understood. In this study, we focus specifically on taxa closely related to Arachnopezizaceae, and the core group of Hyaloscyphaceae.
Arachnopezizaceae
Of the three original tribes, Arachnopezizeae comprised only a small number of species. Nannfeldt (1932) was unsure about the status of the tribe and gave only a provisional description. Korf (1951b) validated the tribe and stressed the significance of subiculum together with septate spores as the key morphological characters defining a natural group. Subiculum refers to the protruding hyphal elements forming an interconnected web-like structure on the substratum surrounding the apothecia (Kirk et al. 2008, see also Fig. 1). Korf (1951b) defined the tribe further as having septate hairs and partially thick-walled excipular cells. He included the genera Arachnopeziza, Eriopezia and Tapesina in Arachnopezizeae and placed Arachnoscypha in synonymy with Arachnopeziza. Dennis (1949, 1981) still recognised Arachnoscypha (for A. aranea), but did not comment on the close relationship between A. aranea and Arachnopeziza eriobasis shown by Korf (1951b). Raitviir (1970) removed Arachnopezizeae from Hyaloscyphaceae, emphasizing the similarities in the excipular structure to Durelloideae and Phialeoideae (Helotiaceae); he thought these three tribes should be placed in a separate family. Doing this, Raitviir gave less weight to the presence or absence of hairs in the overall systematics, suggesting that the presence of ‘true hairs’ is a polyphyletic character within Helotiales. Korf (1978) recognised Arachnopezizeae as a subfamily within Hyaloscyphaceae. At the same time he divided Arachnopezizoideae into two tribes: Arachnopezizeae including Arachnopeziza and provisionally Velutaria; and Polydesmiaeae, including Eriopezia, Parachnopeziza and Polydesmia. Later, a newly established genus Proliferodiscus was placed in Polydesmiaeae (Haines & Dumont 1983).
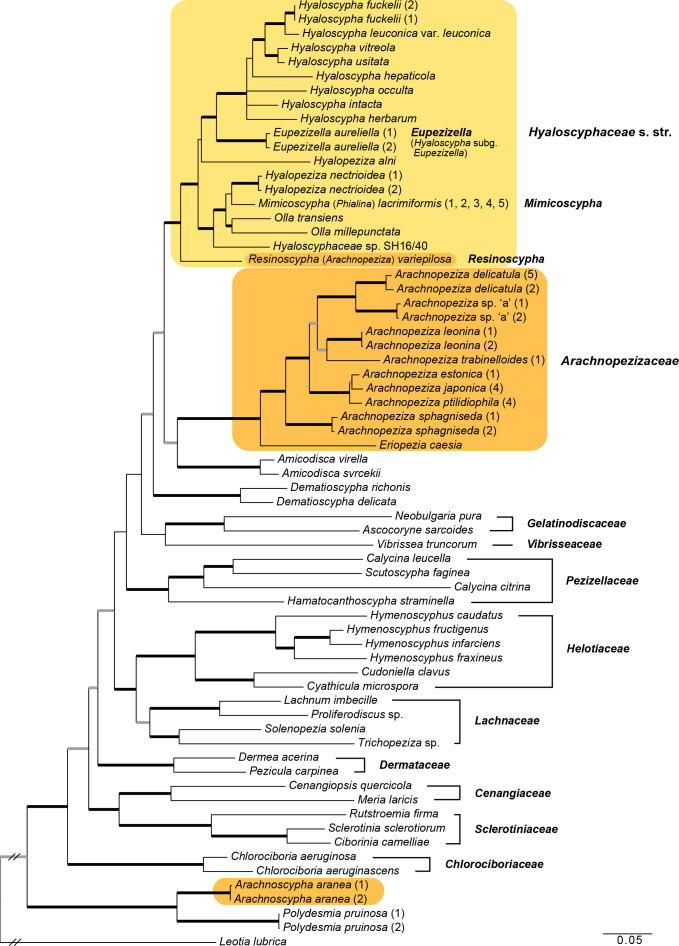
Fig. 1.

Examples of apothecia and habitats in Arachnopezizaceae, Arachnoscypha and Hyaloscyphaceae. a–c. Arachnopeziza leonina, showing hairs with crystals and subiculum; d. Arachnopeziza ptilidiophila; e. Eriopezia caesia; f. Arachnoscypha aranea†; g. Eupezizella aureliella; h. Olla transiens (a: T. Kosonen 7294; b: S. Huhtinen 16/42; c: S. Huhtinen 16/58; d: T. Kosonen 7289; e: T. Kosonen 7005; f: DPP-11788; g: DMS-9189563; h: T. Kosonen 7017). — Scale bars: a, e–f, h = 0.5 mm; b–d, g = 0.25 mm; † dried material. — Photos: a, d, f, h. N. Llerena; b–c. K. Hansen; e. T. Kosonen; g. J.H. Petersen.
Han et al. (2014) addressed the delimitation and relationship between Hyaloscyphaeae and Arachnopezizeae using phylogenetic analyses of the ITS, partial LSU rDNA, mtSSU and RPB2 sequences. Polydesmia and Proliferodiscus were suggested not to be close relatives of Arachnopeziza. Eventually, the rank of Arachnopezizeae was raised to its present status as a family including the genera Arachnopeziza, Arachnoscypha, Austropezia, Eriopezia and Parachnopeziza (Baral 2015). Although many species in Arachnopeziza are relatively conspicuous and new species have continued to be described, the number of Arachnopeziza sequences available is still relatively low as is the number of species sampled. A phylogenetic approach to clarify the family limits of Arachnopezizaceae, and the generic limits of the largest genus Arachnopeziza, is lacking.
Two different species have been interpreted as the type species of Arachnopeziza: A. aurata and A. aurelia. We follow here Korf (1951b: 132–133, 152) who concluded that A. aurata is the type species, as indicated by Saccardo (1884) when he combined Arachnopeziza as a subgenus in Belonidium. In his synopsis of the discomycete genera, Saccardo (1884) listed in general only one species per genus or subgenus that appear to have been selected as typical for them. Korf (1951b) also found it unfortunate if A. aurelia was to be considered the type species, because Fuckel (1872) excluded it from Arachnopeziza shortly after he described the genus. Several databases (e.g., MycoBank, Index Fungorum and Index Nominum Genericorum) list A. aurelia as the type species. Either they follow Clements & Shear (1931), who applied the ‘first species rule’ (A. aurelia is the first species listed by Fuckel (1870) in his description of Arachnopeziza), or Cannon et al. (1985) who first cite Korf (1951b) and then (erroneously!) list A. aurelia as the type species. The likeness of the epithets ‘aurelia’ and ‘aurata’ possibly contributed to that error.
Towards Hyaloscyphaceae sensu stricto
The disciplined use of histochemical methods and the reintroduction of vital taxonomy resulted in novel insights in inoperculate discomycetes systematics throughout the later part of the 20th century. The variation in iodine reactions and the significance of KOH pretreatment and its implications to taxonomy was observed and reviewed by Kohn & Korf (1975) and further explored by Baral (1987) resulting in the concept of hemiamyloidity. Nevertheless, the reactions in the cells produced by Melzer’s reagent were often reported inaccurately and an effort was made to distinguish between amyloid and dextrinoid reactions (Huhtinen 1987b, 1989, 1993b). The importance of studying living material was underlined by the works of Baral & Krieglsteiner (1985) and Baral (1992), showing the cells contained taxonomically valuable characters, changing or disappearing upon drying and/or using various reagents.
The type genus Hyaloscypha, as well as Phialina and Hamatocanthoscypha, were treated in detail by Huhtinen (1989). Many hyaloscyphaceous genera are characterised by a substance in the hair wall filling the space inside the hair partially or completely, giving it a glassy appearance. Several authors paid special attention to this character and proposed generic limits based on the morphology and chemical properties of the hairs (Raschle 1977, Korf & Kohn 1980, Huhtinen 1987b, 1989). Despite many advances, Hyaloscyphaceae remained essentially a combination of genera without a clear concept on their evolutionary relationship.
Combining the first phylogenetic results (Han et al. 2014) and vital taxonomy, Baral (2016) resurrected the family Pezizellaceae for 18 hyaloscyphaceous genera. The unifying morphological character is the frequent presence of vacuolar bodies (VB) in the cells of fresh specimens and a Chalara or similar asexual morph (Baral 2016). The family is supported as a distinct lineage in a recent study employing genome scale data (Johnston et al. 2019). In the systematic arrangement of the helotialean genera (Baral 2016), 26 genera representing c. 220 species were left in a restricted Hyaloscyphaceae, although the available phylogenetic evidence suggested it may still be polyphyletic (Han et al. 2014). As such, the family is delimited without any unique combination of characters, becoming distinguished rather by the lack of specific characters (i.e., the absence of Chalara asexual morph and VB’s). As a result, several taxa are included in Hyaloscyphaceae merely because no better placement is known.
Recent evidence shows, contrary to the traditional view, that at least some species of Urceolella and Cistella belong to a mono-phyletic clade Vandijckellaceae, distantly related to Hyaloscypha (Crous et al. 2017, Johnston et al. 2019). Han et al. (2014) proposed a very narrow concept of Hyaloscyphaceae, including only the genus Hyaloscypha. In their analyses of a dataset (the ‘inter-set’) including representatives from other families of Helotiales, Hyaloscypha formed a well-supported monophyletic group, resolved as a sister group to a clade including species of Olla, Hyalopeziza, Vibrisseaceae, Dermateaceae and Loramycetaceae. Johnston et al. (2019) showed that Amicodisca castanea and Dematioscypha are likely sisters to Hyaloscyphaceae s.str. However, many hyaloscyphoid genera have not been included in molecular phylogenetic studies and several might be non-monophyletic. Fehrer et al. (2018) provided some new samples, but most importantly broadened the view on the ecology of Hyaloscyphaceae by showing the connection between several well-known or new mycorrhizal-forming sterile, asexual or sexual morphs (including the Rhizoscyphus (Hymenoscyphus) ericae aggregate) and sexual Hyaloscypha species. The need to investigate the composition and circumscription of Hyaloscyphaceae is evident.
In the present study, we considerably expand the number of molecular characters, through the addition of sequence data from three protein-coding genes (RPB1, RPB2, TEF-1α), mitochondrial SSU and LSU rDNA, 5 521 bp in total. The ITS region was sequenced and used to aid in species identification or delimitation. With our observations of less known and novel species, together with phylogenetic analyses of sequences, we address the circumscription and relationships of Hyaloscyphaceae and Arachnopezizaceae. In Hyaloscyphaceae, we examine the delimitation of Hyaloscypha and the phylogenetic relationships of closely related taxa. In Arachnopezizaceae we investigate the phylogenetic placement and status of Arachnoscypha, and clarify species boundaries within Arachnopeziza, with a focus on Northern Europe, using genealogical concordance and comparative morphological studies, including the description of two new species and several new combinations.
MATERIALS AND METHODS
Material studied
This study is mainly based on samples collected by the authors in Estonia, Finland and Sweden during 2014–2018. These specimens are deposited in S and TUR. Several samples were collected as part of the project ‘Epibryophytic and lichenicolous fungi in Finland’ in 2003–2008 (Stenroos et al. 2010). In addition, we received both fresh and dry material from mycologists from Europe and North America (USA). From all of these, 69 samples were chosen for molecular phylogenetic study (Table 1). Fungarium material from CUP, H, K, NMNS, PRM, S, TAAM, TNS and TUR were studied.
Table 1.
Collections used in the molecular phylogenetic analyses, with voucher information and GenBank accession numbers. The number in parenthesis following a species name indicates the specific collection of a species (used in Fig. 2, 3, 4). Some GenBank sequences are re-identified by us and the names originally used in GenBank are listed after the taxon names (‘as’). Accession numbers in bold are sequences generated in this study. Sequences marked with ‘not used’ are available in GenBank but they were not used in the analyses. Unavailable sequence marked with an n-dash (–).
| Taxon | Voucher or culture1 | Origin, year, collector | GenBank accession number2 |
|||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | TEF-1α | RPB1 | RPB2 | mtSSU | |||
| Amicodisca svrcekii | TK7157 (S, TUR) | Sweden, 2016, T. Kosonen | MT231647 | MT231647 | MT434824 | MT216583 | MT228637 | MT231568 |
| A. virella | SBRH828 (S, TUR) | Netherlands, 2015, S. Helleman | MT231648 | MT231648 | MT254577 | MT216584 | MT228638 | MT231569 |
| Arachnopeziza araneosa (1) | ICMP 21731 | New Zealand, NA, NA | genome | genome | genome | genome | genome | genome |
| A. araneosa (2) | PDD 59117 /IMCP 22836 | New Zealand, 1991, P.R. Johnston | MH578555 | − | − | − | − | − |
| A. araneosa (3) | PDD 69076 /IMCP 22837 | New Zealand, 1994, P.R. Johnston | MH578556 | − | − | − | − | − |
| A. araneosa (4) | PDD 74085 /IMCP 22838 | New Zealand, 2001, P.R. Johnston & S. Whitton | MH578557 | − | − | − | − | − |
| A. aurata (1) | TUR 179456 | Denmark, 2007, J. Fournier | MT231649 | MT231649 | MT241676 | – | MT228639 | – |
| A. aurata (2) | CBS 116.54 | France, F. Mangenot | − | MH878241 | − | − | − | − |
| A. aurelia (1) | KUS-F51520 | Korea, 2007 | JN033409 | JN086712 | – | – | – | not used |
| A. aurelia (2) | CBS 117.54 | France, F. Mangenot | MH857261 | MH868796 | – | – | – | – |
| A. aurelia (3) | TNS-F-11211 / NBRC 102330 | Japan, 2003 | JN033435 | AB546937 | – | – | not used | not used |
| A. aurelia (4) | CBS 127675 | Japan, O. Izumi, single spore isolation | MH864618 | MH876056 | – | – | – | – |
| A. delicatula (1) | TNS-F-12770 | Japan, 2004, T. Hosoya | JN033433 | JN086736 | – | – | JN086877 | JN086802 |
| A. delicatula (2) | TK7076 (S, TUR) | Finland, 2015, T. Kosonen | MT231650 | MT231650 | MT254567 | MT216585 | MT228640 | MT231570 |
| A. delicatula (3) | JK14051801 (S, TUR) | USA, 2014, J. Karakehian | MT231651 | MT231651 | MT241687 | MT216586 | MT228641 | MT231571 |
| A. delicatula (4) | JP6655 (S, TUR) | Finland, 2013, J. Purhonen | MT231652 | MT231652 | MT241688 | MT216587 | MT228642 | – |
| A. delicatula (5) | TK7036 (S, TUR) | Finland, 2015, T. Kosonen | MT231653 | MT231653 | MT241689 | MT216588 | MT228643 | MT231572 |
| A. delicatula (6) as ‘A. aurata’ | TNS-F-11212 | Japan, 2003, T. Hosoya | JN033436 | AB546936 | – | – | JN086881 | JN086806 |
| A. delicatula (7) | TK7115 (S, TUR) | Finland, 2015, T. Kosonen | MT231654 | MT231654 | – | – | – | – |
| A. delicatula (8) | TK7127 (S, TUR) | Sweden, 2015, T. Kosonen | MT231655 | MT231655 | – | – | – | – |
| A. delicatula (9) | TK7152 (S, TUR) | Sweden, 2016, T. Kosonen | MT231656 | MT231656 | – | – | – | – |
| A. delicatula (10) as ‘A. aurata’ | TENN 074424 | USA, SC, 2018, D. Newman & K. Fleming | MH558278 | – | – | – | – | – |
| A. delicatula (11) as ‘A. aurata’ | Isolate CL0301_4_1 | South Africa | KY228700 | – | – | – | – | – |
| A. delicatula (12) as ‘A. aurata’ | JHH 2210 (NYS) | USA, MA | U57496 | – | – | – | – | – |
| A. delicatula (13) as ‘A. aurata’ | CBS 127674 | Japan | MH864617 | MH876055 | – | – | – | – |
| A. delicatula (14) as ‘A. aurata’ | KUS-F52038 | South Korea, 2008 | JN033393 | JN086696 | – | – | not used | not used |
| A. estonica (1) | SH15/38 (S, TUR) | Estonia, 2015, S. Huhtinen | MT231657 | MT231657 | MT241693 | MT216589 | MT228644 | MT231573 |
| A. estonica (2) | TL210 (TUR) | Finland, 2005, T. Laukka | MT231658 | MT231658 | MT241677 | MT216590 | MT228645 | – |
| A. estonica (3) | TL245 (TUR) | Finland, 2005, T. Laukka | MT231659 | MT231659 | MT241678 | MT216591 | MT228646 | – |
| A. estonica (4) | TL253 (TUR) | Finland, 2005, T. Laukka | MT231660 | MT231660 | MT241679 | MT216592 | MT228647 | – |
| A. japonica (1) | SH06/03 (TUR) | Sweden, 2006, S. Huhtinen | MT231661 | MT231661 | MT241683 | MT216593 | MT228648 | – |
| A. japonica (2) | RI194 (TUR) | Finland, 2006, R. Ilmanen | MT231662 | MT231662 | MT241684 | MT216594 | MT228649 | – |
| A. japonica (3) | RI239 (TUR) | Finland, 2006, R. Ilmanen | MT231663 | MT231663 | MT241685 | MT216595 | MT228650 | – |
| A. japonica (4) | TL267 (TUR) | Finland, 2006, T. Laukka | MT231664 | MT231664 | MT241686 | MT216596 | MT228651 | – |
| A. leonina (1) | KH.15.23 (S, TUR) | Estonia, 2015, K. Hansen | MT231665 | MT231665 | MT254566 | MT216597 | MT228652 | MT231574 |
| A. leonina (2) | TK7101 (S, TUR) | Estonia, 2015, T. Kosonen | MT231666 | MT231666 | MT241694 | MT216598 | MT228653 | MT231575 |
| A. leonina (3) | SH16/42 (S, TUR) | Sweden, 2016, T. Kosonen & S. Huhtinen | MT231667 | MT231667 | MT241695 | MT216599 | MT228654 | MT231576 |
| A. leonina (4) | JYV 11610 | Finland, 2013, J. Purhonen | MT277004 | – | – | – | – | – |
| A. obtusipila (1) | TNS-F-12768 | Japan, 2006, T. Hosoya | JN033445 | JN086746 | – | – | JN086890 | JN086815 |
| A. obtusipila (2) | TNS-F-12769 | Japan, 2006, T. Hosoya | JN033446 | JN086747 | – | – | not used | not used |
| A. ptilidiophila (1) | TK7287 (S, TUR) | Finland, 2018, T. Kosonen | MT231668 | MT231668 | – | MT216600 | MT228655 | MT231577 |
| A. ptilidiophila (2) | TK7289 (S, TUR) | Finland, 2018, T. Kosonen | MT231669 | MT231669 | – | MT216601 | MT228656 | MT231578 |
| A. ptilidiophila (3) | TK7290 (S, TUR) | Finland, 2018, T. Kosonen | MT231670 | MT231670 | – | – | MT228657 | MT231579 |
| A. ptilidiophila (4) | AL51 (TUR) | Finland, 2005, A. Lesonen | MT231671 | MT231671 | MT241680 | MT216602 | MT228658 | – |
| A. sp. ‘a’ (1) | RI199 (TUR) | Finland, 2006, R. Ilmanen | MT435517 | MT435517 | MT241681 | MT216603 | MT228659 | – |
| A. sp. ‘a’ (2) | TL260 (TUR) | Finland, 2006, T. Laukka | MT231672 | MT231672 | MT241682 | MT216604 | MT228660 | – |
| A. sp. ‘a’ (3) as ‘uncultured fungus’ | OTU19 (environ.) | n/a | EF521221 | – | – | – | – | – |
| A. sp. ‘a’ (4) as ‘uncultured fungus’ | SG031 E06 (environ.) | n/a | KP889888 | – | – | – | – | – |
| A. sp. ‘a’ (5) as ‘uncultured fungus’ | OTU18 (environ.) | n/a | EF521220 | – | – | – | – | – |
| A. sp. ‘b’ (1) | TK7223 (S, TUR) | Sweden, 2017, T. Kosonen | MT231673 | MT231673 | MT241696 | MT216605 | MT228661 | – |
| A. sp. ‘b’ (2) | TK7286 (S, TUR) | Finland, 2018, J. Purhonen | MT231674 | MT231674 | – | MT216606 | MT228662 | MT231580 |
| A. sp. (1) | L30_2013 /JCM 31955 | Japan, 2013, N. Nakamura | LC190976 | – | – | – | – | – |
| A. sp. (2) as ‘uncultured fungus’ | c19 (environ.) | USA, NC | HM030576 | – | – | – | – | – |
| A. sp. (3) as ‘Arachnopeziza sp.’ | PDD 112237 /ICMP 22830 | New Zealand, 2008, P.R. Johnston & P. White | MH578548 | – | – | – | – | – |
| A. sp. (4) as ‘Helotiales’ | EXP-0411F | n/a | DQ914728 | – | – | – | – | – |
| A. sp. (5) as ‘fungal sp.’ | Strain Mh859.5.8 | USA, NC | GQ996153 | – | – | – | – | – |
| A. sp. (6) | PDD 112226 | New Zealand, 2018, P.R. Johnston | MH578523 | – | – | – | – | – |
| A. sp. (7) | PDD 111524 /IMCP 22834 | Australia, 2009, P.R. Johnston | MH578552 | – | – | – | – | – |
| A. sp. (8) | PDD 105290 /IMCP 22833 | New Zealand, 1994, P.R. Johnston | MH578551 | – | – | – | – | – |
| A. sp. (9) | PDD 105289 /IMCP 22832 | New Zealand, 2008, P.R. Johnston & P. White | MH578550 | – | – | – | – | – |
| A. sphagniseda (1) | RI226 (TUR) | Finland, 2006, R. Ilmanen | MT231675 | MT231675 | – | MT216607 | MT228663 | – |
| A. sphagniseda (2) | RI267 (TUR) | Finland, 2006, R. Ilmanen | MT231676 | MT231676 | – | MT216608 | – | – |
| A. sphagniseda (3) | TUR 178046 | Finland, 2006, S. Huhtinen | MT231677 | MT231677 | – | – | – | – |
| A. sphagniseda (4) | TL268 (TUR) | Finland, 2006, T. Laukka | MT231678 | MT231678 | – | – | – | – |
| A. trabinelloides (1) | GJO 0071771 | Austria, 2014, I. Wendelin | MT231679 | MT231679 | MT241697 | MT216609 | MT228664 | MT231581 |
| A. trabinelloides (2) | JK14030203 (S, TUR) | USA, MA, 2014, J. Karakehian | MT252825 | – | – | – | – | – |
| Arachnoscypha aranea (1) | SH15/44 (S) | Finland, 2015, S. Huhtinen | MT231680 | MT231680 | MT241698 | MT216610 | MT228665 | MT231582 |
| A. aranea (2) | TK7129 (S) | Sweden, 2015, T. Kosonen | MT231681 | MT231681 | MT254568 | MT216611 | MT228666 | MT231583 |
| Ascocoryne sarcoides | NRRL 50072 | Chile, 2003 | not used | genome | genome | genome | genome | genome |
| Calycellina leucella | MP150937 (S, TUR) | Finland, 2015, M. Pennanen | MT231682 | MT231682 | MT241672 | MT216612 | MT228667 | – |
| Calycina citrina | CBS 139.62 | France | – | FJ176871 | – | – | FJ238354 | FJ176815 |
| Cenangiopsis quercicola | TAAM:178677 | Denmark | not used | KX090811 | KX090663 | KX090760 | KX090713 | – |
| Chlorociboria aeruginascens | TAAM:198512 | Germany | not used | LT158419 | KX090657 | KX090752 | KX090706 | – |
| C. aeruginosa | OSC 100056 | n/a | not used | AY544669 | DQ471053 | DQ471125 | DQ470886 | AY544734 |
| Ciborinia camelliae | ICMP 19812 | New Zealand, 2012, M. Denton-Giles | not used | genome | genome | genome | genome | genome |
| Cudoniella clavus | OSC 100054 | n/a | not used | DQ470944 | DQ471056 | DQ471128 | DQ470888 | DQ470992 |
| Cyathicula microspora | TF2006-BI (TUR) | Sweden, 2006 | EU940165 | EU940088 | – | – | EU940304 | EU940240 |
| Dematioscypha delicata | TK7123 (S, TUR) | Sweden, 2015, T. Kosonen | MT231683 | MT231683 | MT254569 | MT216613 | MT228668 | MT231584 |
| D. richonis | TK7082 (S, TUR) | Finland, 2015, T. Kosonen | MT231684 | MT231684 | MT254576 | MT216614 | MT228669 | MT231585 |
| Dermea acerina | CBS 161.38 | Canada, 1936 | – | DQ247801 | DQ471091 | DQ471164 | DQ247791 | DQ247809 |
| Eriopezia caesia | TK7005 (S, TUR) | Finland, 2014, S. Huhtinen | MT231685 | MT231685 | MT241673 | MT216615 | MT228670 | MT231586 |
| Eupezizella aureliella (1) | TK7300 (S, TUR) | Sweden, 2016, T. Kosonen & S. Huhtinen | MT231686 | MT231686 | MT241699 | MT216616 | MT228671 | MT231587 |
| E. aureliella (2) | s.n. (S, TUR) | Finland, 2016, O. Miettinen | MT231687 | MT231687 | MT241700 | MT216617 | MT228672 | MT231588 |
| Hamatocanthoscypha straminella | JHP-15.170 (S, TUR) | Estonia, 2015, J.H. Petersen | MT231688 | MT231688 | MT254565 | MT216618 | MT228673 | MT231589 |
| Hyalopeziza alni | TK7210 (S, TUR) | Sweden, 2016, T. Kosonen | MT231689 | MT231689 | MT241701 | MT216619 | MT228674 | MT231590 |
| H. nectrioidea (1) | CBS 597.77 | Switzerland, 1973, P. Raschle | JN033381 | JN086684 | – | – | JN086836 | JN086761 |
| H. nectrioidea (2) | ES-2016.35 (S, TUR) | Switzerland, 2016, E. Stöckli | MT231690 | MT231690 | – | MT216620 | – | MT231591 |
| Hyaloscypha fuckelii (1) | AMFB1780 (S, TUR) | Belgium, 2016, B. Clesse | MT231691 | MT231691 | MT254571 | MT216621 | MT228675 | MT231592 |
| H. fuckelii (2) | TK7053 (S, TUR) | Finland, Åland, 2015, T. Kosonen | MT231692 | MT231692 | MT254572 | MT216622 | MT228676 | MT231593 |
| H. hepaticola | MK37 (TUR) | Finland, 2006, M. Kukkonen | JN943614 | EU940150 | – | JN985233 | EU940359 | EU940290 |
| H. herbarum | MP150945 (S, TUR) | Finland, 2015, M. Pennanen | MT231693 | MT231693 | MT241674 | MT216623 | MT228677 | MT231594 |
| H. intacta | TK7111 (S, TUR) | Estonia, 2015, T. Kosonen | MT231694 | MT231694 | MT241675 | MT216624 | MT228678 | MT231595 |
| H. leuconica var. leuconica | TK7014 (S, TUR) | Finland, 2014, T. Kosonen | MT231695 | MT231695 | MT254573 | MT216625 | MT228679 | MT231596 |
| H. occulta as ‘Hyaloscypha sp.’ | TNS-F-31287 | Japan, 2007, T. Hosoya | JN033454 | JN086754 | – | – | JN086900 | JN086825 |
| H. usitata | TK7083 (S, TUR) | Finland, 2015, T. Kosonen | MT231696 | MT231696 | MT254574 | MT216626 | MT228680 | MT231597 |
| H. vitreola | SH14/10 (S, TUR) | Finland, 2014, S. Huhtinen | MT231697 | MT231697 | MT254575 | MT216627 | MT228681 | MT231598 |
| Hyaloscyphaceae sp. | SH16/40 (S, TUR) | Sweden, 2016, T. Kosonen & S. Huhtinen | MT231698 | MT231698 | MT241702 | MT216628 | MT228682 | MT231599 |
| Hymenoscyphus caudatus | KUS-F52291 | Korea, 2008 | not used | JN086705 | – | – | JN086856 | JN086778 |
| H. fraxineus | CBS 133217 (MNHNL) | Luxembourg, 2012, G. Marson | not used | genome | genome | genome | genome | genome |
| H. fructigenus | TNS-F-44644 | Japan, 2011, T. Hosoya | AB926057 | AB926144 | – | – | AB926189 | – |
| H. infarciens | CBS 122016 | France, 2001, G. Marson | not used | genome | genome | genome | genome | genome |
| Lachnum imbecille | TK7121 (S, TUR) | Finland, 2015, T. Kosonen | MT231699 | MT231699 | MT254570 | MT216629 | MT228683 | MT231600 |
| Leotia lubrica | OSC 100001 | n/a | – | AY544644 | DQ471041 | DQ471113 | DQ470876 | AY544746 |
| Meria laricis | CBS 298.52 | Switzerland, 1952, E.Müller | MH857046 | DQ470954 | DQ842026 | DQ471146 | DQ470904 | DQ471002 |
| Mimicoscypha lacrimiformis (1) | SH17/8 (S, TUR) | Sweden, 2017, S. Huhtinen | MT231700 | MT231700 | MT241703 | MT216630 | MT228684 | MT231601 |
| Mimicoscypha lacrimiformis (2) | TK7224 (S, TUR) | Sweden, 2017, T. Kosonen | MT231701 | MT231701 | – | – | – | – |
| Mimicoscypha lacrimiformis (3) | SH17/6 (S, TUR) | Sweden, 2017, S. Huhtinen | MT231702 | MT231702 | – | – | MT228685 | MT231602 |
| Mimicoscypha lacrimiformis (4) | SH17/7 (S, TUR) | Sweden, 2017, S. Huhtinen | MT231703 | MT231703 | MT434823 | MT216631 | MT228686 | MT231603 |
| Mimicoscypha lacrimiformis (5) | KH.17.02 (S) | Sweden, 2017, K. Hansen | MT231704 | MT231704 | – | – | MT228687 | – |
| Neobulgaria pura | CBS 477.97 | USA, 1996, K. Cameron | – | FJ176865 | FJ238397 | FJ238434 | FJ238350 | – |
| Olla millepunctata | TK7167 (S, TUR) | Sweden, 2016, T. Kosonen | MT231705 | MT231705 | MT241705 | MT216632 | MT228688 | MT231604 |
| O. transiens | TK7125 (S, TUR) | Sweden, 2015, T. Kosonen | MT231706 | MT231706 | MT241706 | MT216633 | MT228689 | MT231605 |
| Pezicula carpinea | CBS 282.39 | Canada, 1932, H.S. Jackson | – | DQ470967 | DQ479932 | DQ842032 | DQ479934 | DQ471016 |
| Polydesmia pruinosa (1) | TK7139 (S, TUR) | Sweden, 2015, T. Kosonen | MT231707 | MT231707 | MT241707 | MT216634 | MT228690 | MT231606 |
| P. pruinosa (2) | TK7236 (S, TUR) | Finland, 2017, T. Kosonen | MT231708 | MT231708 | MT241708 | – | MT228691 | MT231607 |
| Proliferodiscus sp. | KUS-F52660 | Korea, 2010 | not used | JN086730 | – | – | JN086871 | JN086796 |
| Resinoscypha variepilosa | SH16/41 (S, TUR) | Sweden, 2016, T. Kosonen & S. Huhtinen | MT231709 | MT231709 | MT241709 | MT216635 | MT228692 | MT231608 |
| Rustroemia firma | CBS 341.62 | France | – | DQ470963 | DQ471082 | DQ471155 | DQ470912 | DQ471010 |
| Sclerotinia sclerotiorum | 1980_UF70 | n/a | not used | genome | genome | genome | genome | genome |
| Scutoscypha fagina | TK7178 (S, TUR) | Sweden, 2016, T. Kosonen | MT231710 | MT231710 | MT241710 | MT216636 | MT228693 | MT231609 |
| Solenopezia solenia | SH17/18 (S, TUR) | Sweden, 2017, S. Huhtinen | MT231711 | MT231711 | MT241711 | MT216637 | MT228694 | MT231610 |
| Trichopeziza sp. | s.n. (S, TUR) | Spain, 2016, J. Castillo | MT427744 | MT427744 | MT241712 | MT216638 | – | MT231611 |
| Vibrissea truncorum | CBS 258.91 | Canada, 1987, R.G. Thorn | not used | FJ176874 | FJ238405 | FJ238438 | FJ238356 | FJ190635 |
1 In personal collection numbers names have been shortened and only the initials are given. Herbaria are cited according to acronyms in Index Herbariorum (http://sweetgum.nybg.org/ih/).
2 ITS: Internal transcribed spacers (ITS1 and ITS2) and the 5.8S gene of the nrDNA; LSU: 28S large subunit of the nrRNA gene; TEF-1α: Translation elongation factor 1-alpha; RPB1: RNA polymerase II largest subunit; RPB2: RNA polymerase II second largest subunit.
Fungal cultures
Fresh collections were cultured on malt-agar plates in order to ensure ample living material for DNA extraction. Depending on the size of the collection, 4–10 small pieces of substratum (c. 2 × 2 mm) with fresh apothecia were cut under a dissecting microscope and placed on moist paper tissue of a slightly larger size. In cases where apothecia from different species were growing intermixed, the apothecia of the untargeted species were removed. A 50 mm Petri dish, half filled with growth media, was placed upside down on top of each piece of apothecia assembly. The growth media consisted of sterile ion-exchanged water with 2 % (w/v) malt, glucose, agar, 0.1 % peptone and 0.01 % chloramphenicol. Spore release was triggered by altering the air pressure inside the Petri dish by carefully lifting it partially for 1–2 s. After 12–24 h, the Petri dish was closed and sealed. Similar hyphal growth pattern, on the 4–10 plates made from one collection, was used as an indication of the target species being successfully isolated. Cultures were grown for 3–6 mo at room temperature prior to DNA extraction. We observed no asexual reproductive structures during the first 3 mo when cultures were under consistent observation. Fungal cultures will be deposited in the Westerdijk Fungal Biodiversity Institute (CBS-KNAW collection).
Morphological methods
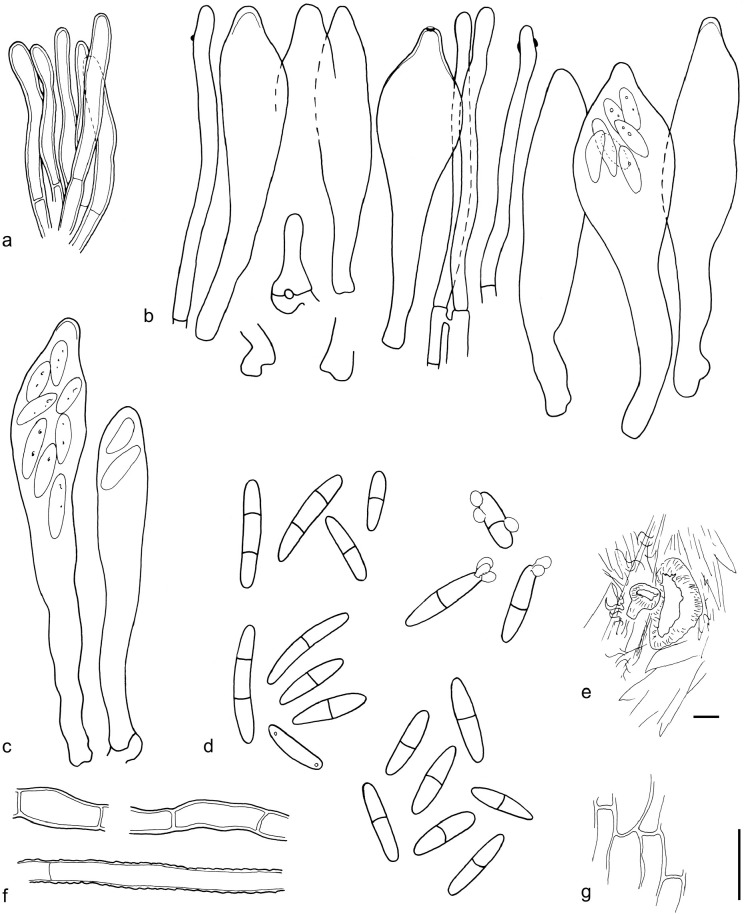
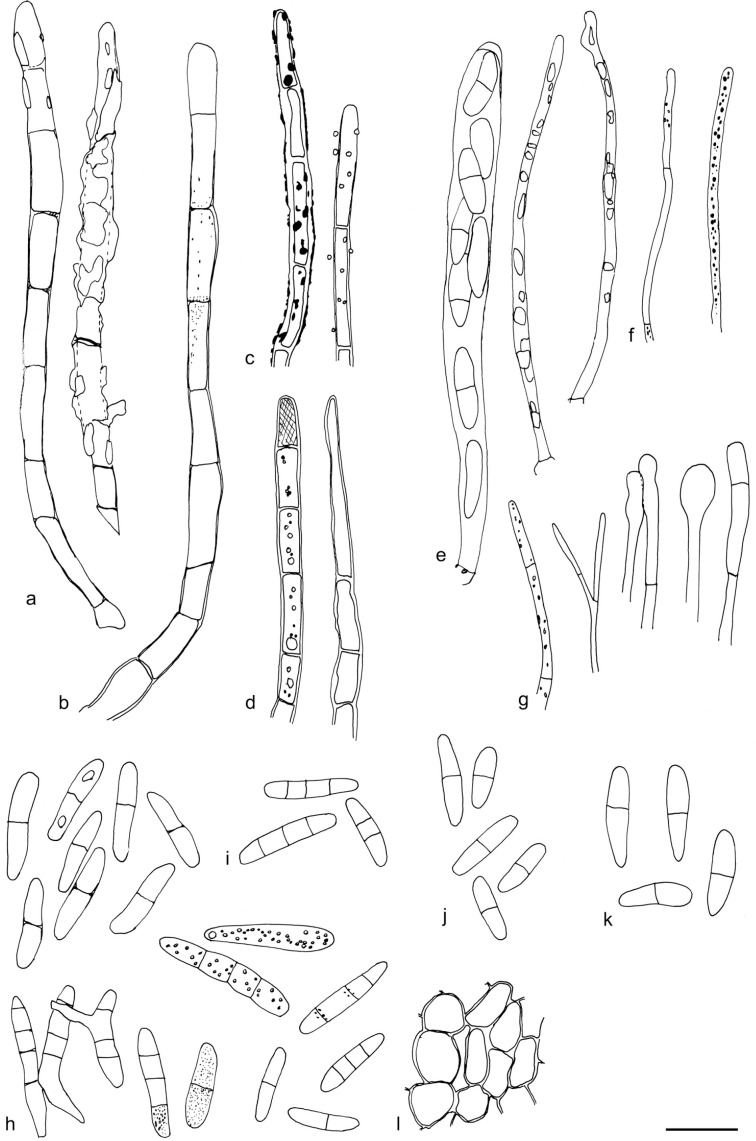
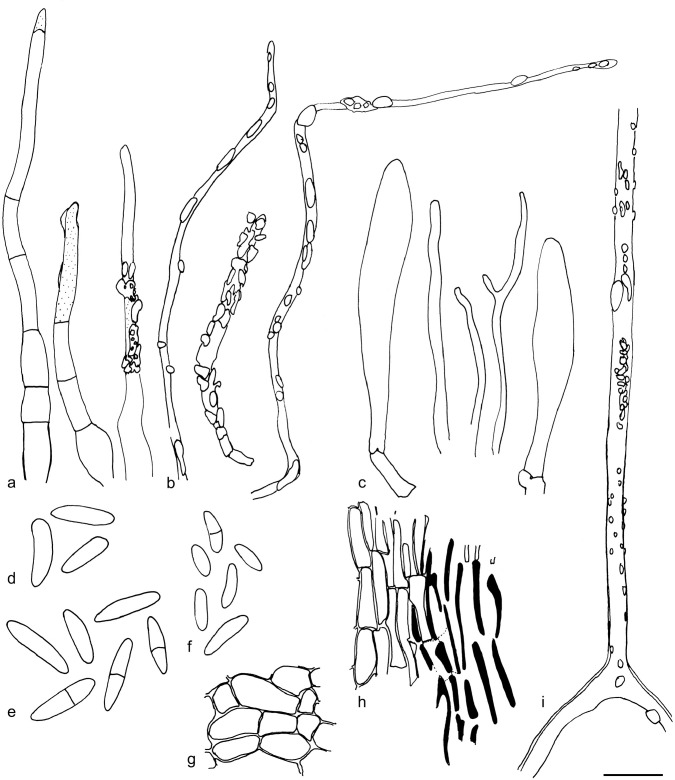
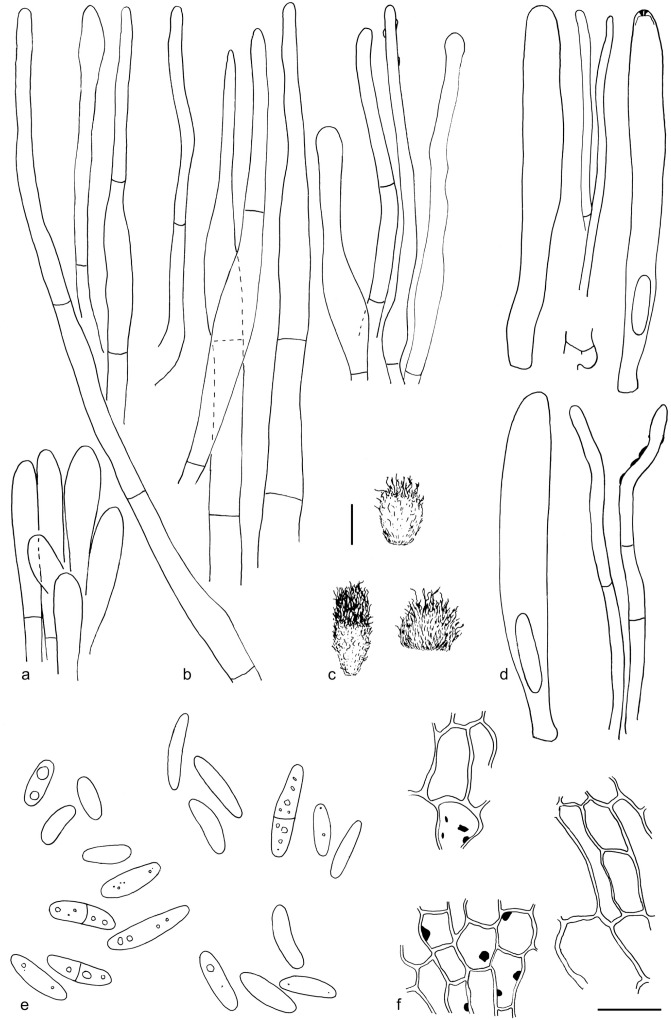
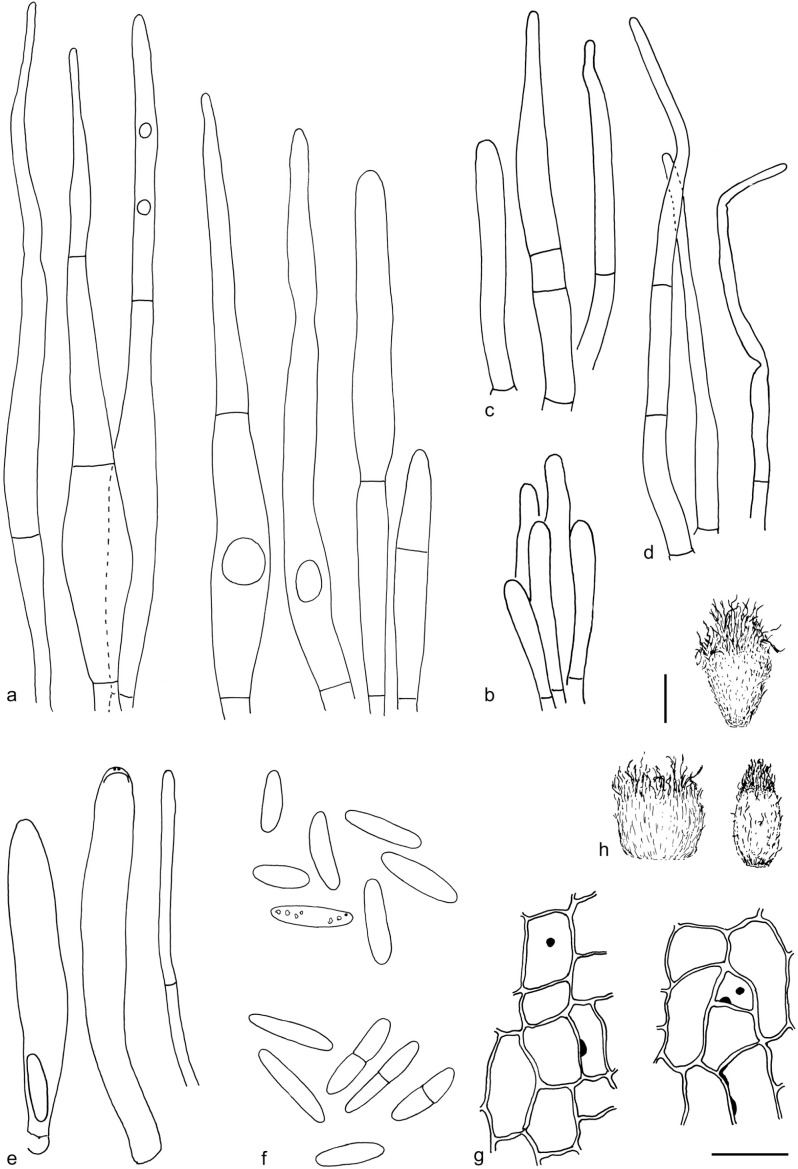
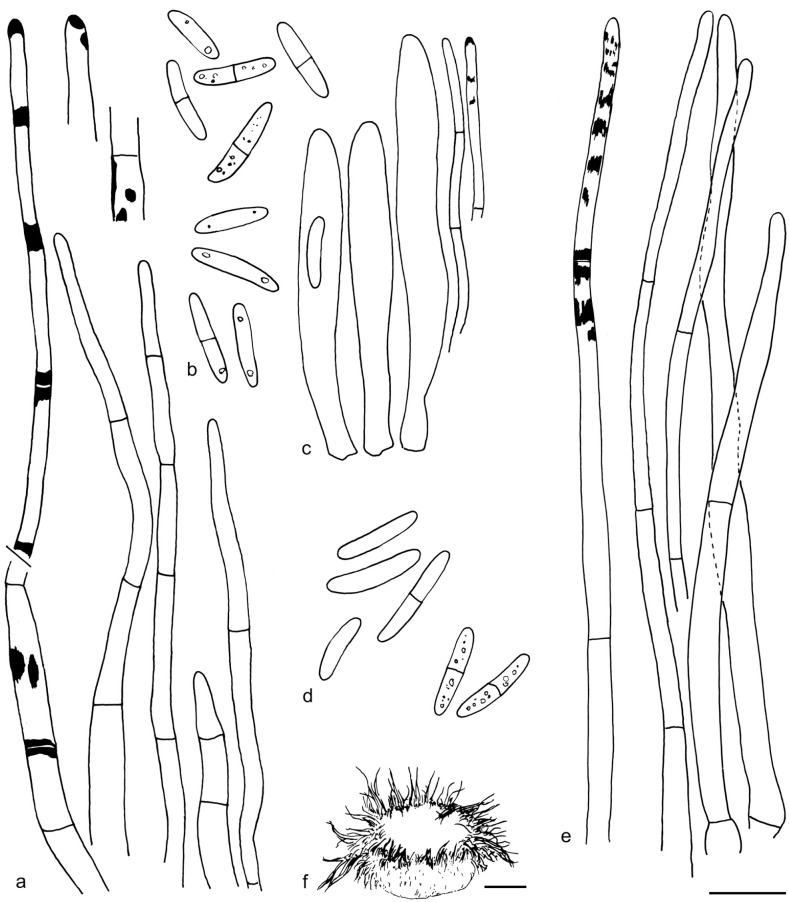
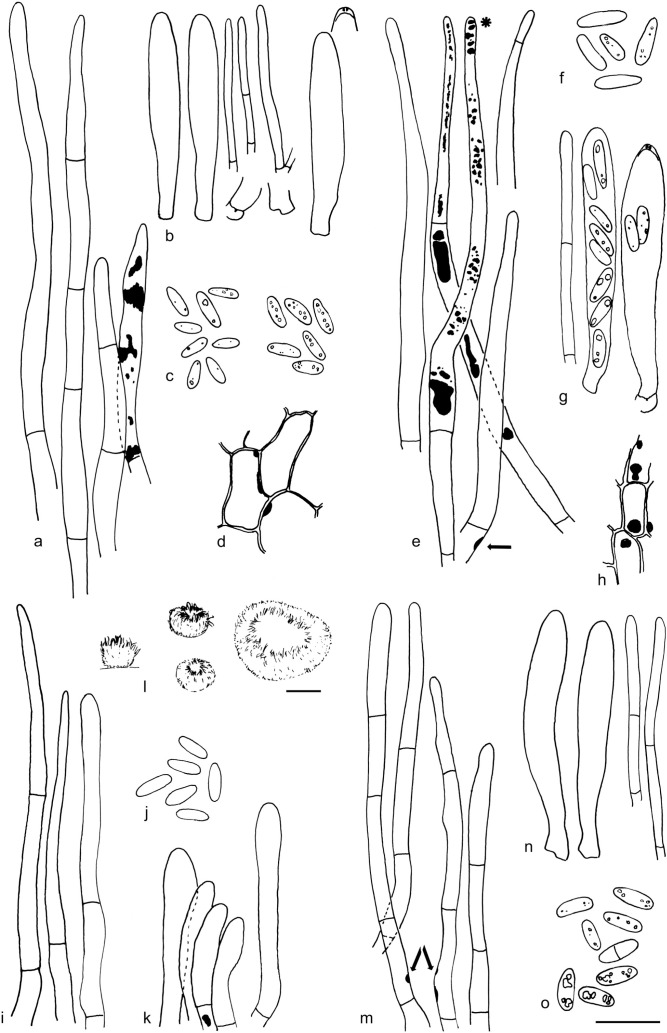
The material was studied with Olympus BX53 and Nikon i80 microscopes with bright field optics. Measurements were made using a 1 000× magnification. Fresh samples were first studied in tap water. Other mounting media used were: Melzer’s reagent (MLZ), IKI solution (LUG), Cotton blue (CB), ammoniacal Congo red (CR) and c. 5 % potassium hydroxide (KOH). The formulas for reagents follow Huhtinen (1989). Large apothecia were cut in half or sectioned for mounting, whereas minute and fragile apothecia were squash mounted. Thirty discharged spores were measured, when possible, from each population. Dimensions were recorded subjectively at the accuracy of 0.1 μm. The mean spore Q values were calculated separately from the individual Q values. The total number of spores measured is given (n), followed by the number of populations used as a source. Spore sizes include 90 % of the measured variation; the smallest and the largest 5 % of the values are excluded, although the largest measured value is given in parentheses. Colours, when accurately described, are given according to Cailleux (1981). Line drawings were made using a drawing tube and when present, photographs of vital characters were used as an aid. The exclamation mark (!) indicates that we examined type or other original.
Molecular techniques
DNA was isolated from fresh mycelia scraped from the agar plates, or in cases of unsuccessful culturing from fresh or very recently dried apothecia. In the latter case, 10–30 apothecia were handpicked to a sterile Eppendorf tube. The material was shaken in a cell disrupter Mini-BeadBeaterTM (BioSpec Products, Bartlesville, Oklahoma) with 1.2 mm disposable steel beads in a 2 mL screw-cap microvial, at 4 200 rpm for 20 s. If necessary, shaking was repeated once after allowing the sample to cool down for 2–3 min. Alternatively, fresh mycelia were ground in an Eppendorf tube with a pestle together with a tiny amount of sterilized sea sand. DNA was extracted using the DNeasy® Plant Mini Kit (Qiagen), following the standard protocol for fresh plant material. In the final step, the DNA extract was eluted with the provided elution buffer in a volume of 200 μL. A 1 : 10 dilution of the DNA with sterile DNase-free water was used as template for the polymerase chain reaction (PCR). The DNA extract from scanty specimens was (in the final step) eluted in only a volume of 100 μL or less, and without further dilution used for PCR. For a small (random) part of the material, DNA was extracted using the Promega Wizard® Genomic DNA purification kit, following the provided protocol for filamentous fungi. The brightness of ITS-LSU PCR bands was used as an initial indication of the concentration of the DNA that was diluted even further, if necessary, for subsequent reactions.
Six different gene regions were amplified: rDNA ITS1–5.8S–ITS2 and the D1–D2 regions of LSU, c. 1300–1400 bp; mitochondrial small subunit (mtSSU), regions U2–U6, c. 800–1000 bp; RNA polymerase I (RPB1), A–C region, c. 700 bp; RNA polymerase II (RPB2), 5–11 regions, c. 1800 bp; and translation elongation factor 1-alpha (TEF-1α), c. 1000–1300 bp. The respective primers used were: ITS1, ITS4, LR0R, LR3 and LR5 (White et al. 1990); mrSSU1 and mrSSU3R (Zoller et al. 1999); RPB1A, RPB1C (Matheny et al. 2002); 5F, 6F, 7R, 7F, 11aR (Liu et al. 1999) and both RPB2-9R (Taşkin et al. 2010, referred as ‘RPB2-9f’ in the article) and RPB2-9mR (ATY AAA TGD GCA ATN GTC ATR CG), a modification of the RPB2-9R primer; 526F, 2F, 1567R and 2218R (S. Rehner unpubl., Rehner & Buckley 2005) and EF–3AR (Taşkin et al. 2010). Generally, the gene regions were PCR amplified in one piece. If unsuccessful, the genes were PCR amplified in two or more overlapping pieces. A standard approach for the RPB2 and TEF-1α genes was to use primers 5F and 9mR (RPB2) and 526F and EF-3AR (TEF-1α). This resulted in strong target product without multiple bands in 80–90 % of the sampled species. PCR amplifications were done using IllustraTM Hot Start Mix RTG PCR beads (GE Healthcare, UK) in a volume of 25 μL. All PCR programs included an initial hot start at 95 °C for 5 min and a final incubation at 72 °C for 7 min. The actual cycles were as follows: ITS and LSU regions of rDNA: 35 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 45 s and extension at 72 °C for 1–1.5 min; for mtSSU the program was identical, except that the annealing was at 56 °C for 1 min; for RPB1: 35 cycles at 95 °C for 1 min, annealing at 52 °C with 1 °C increase every 5 s until 72 °C and extension at 72 °C for 1.5 min; for RPB2: 34–40 cycles of denaturation at 95 °C for 1 min, annealing at 55–58 °C for 1 min and extension at 72 °C 1 min with an increase of 1 s each cycle; for TEF-1α a touchdown program: 9 cycles of denaturation at 95 °C for 30 s, annealing at 66 °C for 30 s with temperature reduced 1 °C every cycle and extension at 72 °C for 1 min followed by an additional 30–33 cycles with denaturation at 95 °C, annealing at 56 °C for 30 s and extension at 72 °C for 1 min. PCR products were examined on 1 % agarose gel and either purified directly using ExoSap–IT (Thermo Fisher Scientific) to remove excess primers and nucleotides. In the case of multiple bands, the total volume of PCR was run on a 1.5 % agarose gel and the band of correct size was excised and retained using the Gel Extraction Kit (Qiagen). Purified PCR products were sequenced by Macrogen Inc. (the Netherlands). The primers used for PCR (listed above) were also used for sequencing. Two internal primers were in addition used for sequencing long PCR products (> 1 000 bp), to produce multiple overlapping sequences.
Alignment, data-partitioning and phylogenetic analyses
Sequences were assembled and edited using Sequencher v. 4.10 (Gene Codes Corporation, Ann Arbor, MI, USA). Nucleotide sequences were aligned manually using Se-Al v. 2.0a11 (Rambaut 2002). Each alignment of the protein-coding genes was translated to amino acids using MacClade v. 4.08 (Maddisson & Maddisson 2000) to verify the alignment and determine the intron positions. The introns were too variable to align unambiguously and were therefore excluded from the analyses. All gene regions were analysed using the nucleotides. Two multi-gene datasets were assembled:
the Helotiales dataset, to resolve the boundaries and relationships of Arachnopezizaceae and Hyaloscyphaceae (including also representatives of other Helotiales); and
the Arachnopezizaceae dataset, to delimit species and elucidate the relationships in Arachnopeziza (including Arachnopezizaceae s.str. as delimited from analyses of the Helotiales dataset).
Leotia lubrica was used as an outgroup for the Helotiales dataset based on its placement outside Helotiales in various studies (Spatafora et al. 2006, Johnston et al. 2019). Amicodisca virella was used as an outgroup for the Arachnopezizaceae dataset, based on our results from analyses of the Helotiales dataset. Arachnopeziza aurelia (TNS-F11211) was not included in the final multi-gene Arachnopezizaceae dataset, because of missing data (no RPB1 and TEF-1α sequences are available and the RPB2 sequence is short (716 bp) compared to our sequences) and because it in preliminary analyses was found to be divergent from the other species sampled by us. The combination of lack of characters and closely related species (occurring on a very long branch) resulted in loss of resolution in the backbone of the multi-gene Bayesian and ML phylogenies (in the relationships among the Arachnopeziza clades). Arachnopeziza obtusipila was included, despite missing similar data, because it is closely related to another taxon (A. leonina) and the impediment of the missing data appeared to have no (or less) effect on the analyses. For each dataset the single gene regions were analysed separately and combined. Each of the three protein coding gene regions (RPB1, RPB2 and TEF-1α) were analysed with two distinct partitions:
first and second codon positions; and
third codon positions.
The LSU rDNA and mtSSU were each analysed as one distinct partition. Thus, the combined five-gene datasets were analysed with eight partitions.
The ITS was too variable to align across Helotiales and it was therefore not included in the Helotiales dataset. For the Arachnopezizaceae dataset, the ITS sequences were alignable. Analyses of the six-gene dataset, including the ITS region, improved the support values for the Arachnopeziza leonina clade, but weakened the support values for some of the backbone nodes. Also, we observed that analyses including the ITS were very sensitive to minor, but equally justified alterations in the ITS alignment. As a result, we did not include the ITS in the combined Arachnopezizaceae dataset. Nevertheless, to provide further insight into the species and ecological diversity and geographical distributions of Arachnopeziza, a third dataset was assembled that included additional Arachnopeziza species and collections with only ITS and, if available, LSU sequences (from GenBank and our own data). For an improved alignment, no outgroup was included in the analyses of this dataset and the phylogenies were rooted along the branch leading to A. sphagniseda. The beginning and tail of the mtSSU sequences were highly variable and not possible to align, and were not included in the Helotiales or Arachnopezizaceae data-sets. Alignments of the combined five-gene Helotiales and Arachnopezizaceae datasets, and the more inclusive ITS-LSU alignment of Arachnopeziza, are available at TreeBASE under accession number S25441.
Individual and combined analyses of the six different gene regions were performed using Metropolis-coupled Markov chain Monte Carlo (MCMCMC) in MrBayes v. 3.2.6 (Ronquist & Huelsenbeck 2003) and Maximum Likelihood-based inference (ML) in RAxML-HPC2 v. 8.2.10 (Stamatakis 2014). All analyses were run on CIPRES science Gateway (Miller et al. 2010). The Bayesian analyses were run in parallel using model jumping (/mixed models) (Ronquist et al. 2012), and with all parameter values, except branch length and tree topologies, unlinked. The analyses consisted of four parallel searches, with four chains each, initiated with random trees. For the single gene datasets the analyses were run for 5 M generations and the combined five-gene datasets for 10 M generations. The chains were sampled every 500 generations in the 5 M generation runs and every 1 K generations in the 10 M runs. A majority rule consensus tree was assembled and the posterior probabilities (PP) were calculated from the last 75 % of the posterior tree samples. The ML analyses used a GTRGAMMA model for the rate heterogeneity with all free model parameters estimated by the program. Maximum likelihood bootstrap analyses (ML-BP) were performed using 1 000 rapid bootstrap replicates from random starting trees, followed by a thorough ML search similarly using 1 000 replicates to find the best tree.
We applied the concept of Genealogical Concordance Phylogenetic Species Recognition (GCPSR) (Taylor et al. 2000) to delimit species in Arachnopeziza. To identify independent evolutionary lineages using genealogical concordance we employed two criteria based on Dettman et al. (2003):
the clade had to be present in the majority (3/4) of the single-gene phylogenies; and
the clade was well supported in at least one single-gene phylogeny (as judged by both PP ≥ 0.95 and ML-BP ≥ 75 %), and if its existence was not contradicted by any of the other single-gene phylogenies at the same level of support.
The ITS, LSU, RPB1, RPB2 and TEF-1α genealogies were visually compared to find concordance.
RESULTS
Sequences produced, congruence and data partitions
Altogether 346 sequences from 69 samples were produced in this study. GenBank accession numbers for the specific gene regions are listed in Table 1. In addition, 126 sequences of 28 samples were retrieved from GenBank. The concatenated datasets for Helotiales and Arachnopezizaceae contained 5 521 and 5 124 characters, respectively. The Arachnopeziza ITS-LSU dataset had 1 045 characters. All Bayesian analyses converged: the average standard deviation of split frequencies reached values below 0.01, except in the individual analysis of RPB2 in the Helotiales dataset, which reached a value of c. 0.013 after 5 M generations. In all analyses, the Potential Scale Reduction Factor values stabilized at 1.000. In the ML analyses of the combined Helotiales and Arachnopezizaceae datasets, the single best scoring trees were recovered with –InL = 73800.332097 and –InL = 18838.975670, respectively. In the ML analyses of the Arachnopeziza ITS-LSU dataset, the single best scoring tree was recovered with –InL = 3423.787414. Individual trees for each gene marker for both concatenated datasets were studied for conflicts. There were no supported (PP ≥ 0.95 and ML-BP ≥ 75 %) conflicts between the individual gene trees of the Helotiales dataset.
The major lineages and relationships of Arachnopezizaceae, Hyaloscyphaceae s.str. and related genera
Bayesian and ML analyses of the five-gene Helotiales dataset produced topologies with identical deeper branching patterns. The back-bone nodes are supported by Bayesian analysis (PP values ≥ 0.95, Fig. 2), except for two nodes that have low support (both PP 0.73) i.e., in the placement of Pezizellaceae, Vibrissea(ceae) and Gelatinodiscaceae. The Arachnopezizaceae is polyphyletic, because Arachnoscypha forms a highly supported monophyletic group with Polydesmia pruinosa as a sister group to the rest of the Helotiales (PP 1.00, ML-BP 91 %), very distant to Arachnopeziza and Eriopezia. Eight species of Arachnopeziza and Eriopezia caesia form a highly supported monophyletic group (PP 1.00, ML-BP 100 %). Arachnopeziza variepilosa is supported as a separate distinct lineage. Hyaloscyphaceae (Hyaloscypha and closely related taxa) is supported as a sister group to a clade of Arachnopezizaceae and Amicodisca in Bayesian analyses (PP 0.96), with Dematioscypha as a sister group to all of those (PP 0.96) (Fig. 2). The ML analysis, however, resolves Dematioscypha and Amicodisca as successive sister taxa to Hyaloscyphaceae, but without support (ML-BP below 50 %). The placement of Gelatinodiscaceae and Vibrissea differs likewise in the ML phylogeny, but also without support. The Hyaloscyphaceae s.str. here includes novel sequences of poorly understood hyaloscyphoid taxa. All other families, as represented here, are supported as monophyletic in both Bayesian and ML analyses, except Lachnaceae that is resolved as two successive sister lineages to members of Helotiaceae in the ML-analysis but without support (ML-BP 59 %). Members of Lachnaceae and Helotiaceae form a highly supported monophyletic group (PP 1.00, ML-BP 94 %) as do members of Cenangiaceae and Sclerotiniaceae (PP 1.00, ML-BP 94 %).
Fig. 2.
Phylogenetic relationships of Arachnopezizaceae (orange square) and Hyaloscyphaceae (yellow square) among members of Helotiales based on Bayesian analyses of combined LSU, RPB1, RPB2, TEF-1α and mtSSU loci. Arachnoscypha aranea and Resinoscypha variepilosa (orange squares) were previously treated in Arachnopezizaceae. Thick black branches received both Bayesian posterior probabilities (PP) ≥ 0.95 and maximum likelihood bootstrap value (ML-BP) ≥ 75 %. Thick grey branches received support by either PP ≥ 0.95 or ML-BP ≥ 75 %. The number in parenthesis after a species name refers to an exact collection (see Table 1).
Generic relationships in Hyaloscyphaceae s.str.
The nine selected species of Hyaloscypha form a highly supported monophyletic clade (Fig. 2). Hyaloscypha aureliella is strongly supported as a sister lineage to all other Hyaloscypha species (PP 1.00, ML-BP 99 %), confirming that it is distinct; Huhtinen (1989) recognised it within Hyaloscypha in subgenus Eupezizella. It is here accepted in the separate genus Eupezizella (see Taxonomy). The relationships within Hyaloscypha s.str. are not fully resolved (Fig. 2). Two species of Olla, O. transiens and O. millepunctata (type species of Olla), form a monophyletic group (PP 1.00, ML-BP 81 %). Two species of Hyalopeziza, H. alni and H. nectrioidea, do not form a monophyletic group. Our preliminary results on the type species of Hyalopeziza (H. ciliata) indicate that it belongs in Pezizellaceae (not shown) and that it is not closely related to any of the other sequenced Hyalopeziza species. Hyalopeziza alni forms a separate distinct lineage within Hyaloscyphaceae s.str., but its placement is without support. Hyalopeziza nectrioidea forms a monophyletic group with Olla and Mimicoscypha (Phialina) lacrimiformis. The relationships of M. lacrimiformis are resolved differently between the two analyses and with only low support from Bayesian PP and ML bootstrap analyses, but it is clearly distinct morphologically from Olla and from the two Hyalopeziza species (see Taxonomy) and therefore we erect a new genus Mimicoscypha for this taxon. Based on morphology this is not a species of Phialina. Other species of Phialina or Calycellina are placed in Pezizellaceae (Han et al. 2014, Baral 2016, Johnston et al. 2019).
A possible new species or genus constitutes a distinct lineage (Hyaloscyphaceae sp., SH 16/40), sister to the Mimicoscypha-Olla clade. Additional material is needed to fully understand and describe this taxon. Arachnopeziza variepilosa is highly supported as a sister group to all other Hyaloscyphaceae s.str. representatives. It is clearly distant from other species of Arachnopeziza and a new genus, Resinoscypha, is therefore created (Fig. 2, see Taxonomy).
Phylogenetic species recognition and diversity in Arachnopeziza
Eight terminal independent evolutionary lineages were identified in Arachnopeziza, using the two grouping criteria for GCPSR (see Materials and Methods), and these are inferred as phylogenetic species (marked by a triangle on the node in Fig. 3). All of the species were strongly supported as monophyletic by Bayesian PP (≥ 0.95) and ML-BP (≥ 75 %) in at least two of the single gene trees (Table 2). Four of them present newly discovered species of which two are formally described in the present paper, A. estonica and A. ptilidiophila. Analyses of the LSU did not resolve A. delicatula and A. estonica as monophyletic, but their monophyly was not significantly contradicted in the LSU genealogies. Bayesian and ML analyses of the combined LSU, RPB1, RPB2, and TEF-1α data supported all eight species as monophyletic (PP 1.00, ML-BP 100 %). The monophyly of four putative species (A. araneosa, A. aurata, A. obtusipila and A. trabinelloides), represented by only single collections, could not be tested, but they were considered to be distinct because they were genetically divergent from their sisters (Fig. 3). ITS and LSU sequences were available in GenBank from one to three additional collections of these four species and our analyses of the taxon-expanded ITS-LSU dataset support them as monophyletic groups (Fig. 4). The two collections each of A. aurata (from Denmark and France), A. obtusipila (Japan) and A. trabinelloides (Austria and USA, MA) showed identical ITS and/or LSU sequences. Arachnopeziza delicatula showed internal phylogenetic structure in analyses of the five-gene dataset, with two subgroups: A. delicatula (4) and (5) (PP 1.00) and A. delicatula (2) and (6) (ML-BP 88 %) (Fig. 3), but these groupings were strongly contradicted among the single genealogies suggesting recent recombination within a single lineage/species. The A. delicatula populations sampled for multiple gene analyses were from three different continents, Japan, Northern Europe and USA, and did not show any geographical pattern.
Fig. 3.
Phylogenetic tree based on Bayesian analysis of the Arachnopezizaceae dataset (combined LSU, RPB1, RPB2, TEF-1α and mtSSU loci). Amicodisca virella was used as an outgroup in the analysis and for rooting the tree. Thick black branches received both Bayesian posterior probabilities (PP) ≥ 0.95 and maximum likelihood bootstrap value (ML-BP) ≥ 75 %. Thick grey branches received support by either PP ≥ 0.95 or ML-BP ≥ 75 %. The triangles at the nodes indicate eight species recognised by genealogical concordance phylogenetic species recognition. The number in parenthesis after a species name refers to an exact collection (see Table 1). Four clades are named for discussion.
Table 2.
Support values for Arachnopeziza species recognised by genealogical concordance in the analysis of individual gene regions and in the combined dataset (LSU, RPB1, RPB2, EF-1α and mtSSU3): Maximum Likelihood values (RAxML) and Bayesian posterior probabilities (PP). NA, only one or no sequences available. Limits of these species correspond to the nodes with triangles in Fig. 3.
| Species1 | ITS ML-BP / PP | LSU ML-BP / PP | RPB1 ML-BP / PP | RPB2 ML-BP / PP | TEF-1α ML-BP / PP | Combined five-gene data |
|---|---|---|---|---|---|---|
| A. delicatula | 87 / 0.82 | –2 / – | 96 / 1.00 | 100 / 1.00 | 100 / 1.00 | 100 / 1.00 |
| A. estonica | 100 / 1.00 | – / – | 100 / 1.00 | 100 / 1.00 | 99 / 1.00 | 100 / 1.00 |
| A. japonica | 100 / 1.00 | 95 / 1.00 | 100 / 1.00 | 100 / 1.00 | 100 / 1.00 | 100 / 1.00 |
| A. leonina | 100 / 1.00 | 97 / 0.96 | 100 / 1.00 | 98 / 0.99 | 100 / 1.00 | 100 / 1.00 |
| A. ptilidiophila | 97 / 0.68 | 64 / 0.84 | 100 / 1.00 | 100 / 1.00 | NA | 100 / 1.00 |
| A. sphagniseda | 100 / 1.00 | 100 / 1.00 | 99 / 1.00 | NA | NA | 100 / 1.00 |
| A. sp. ‘a’ | 99 / 1.00 | 77 / 0.84 | 97 / 1.00 | 92 / 1.00 | NA | 100 / 1.00 |
| A. sp. ‘b’ | 98 / 0.97 | 100 / 1.00 | 100 / 1.00 | 100 / 1.00 | 99 / 1.00 | 100 / 1.00 |
1 Support values not available for the following species represented by a single collection: A. aurata, A. araneosa, A. obtusipila and A. trabinelloides.
2 –, the clade was not resolved as monophyletic.
3 Support values based on mtSSU alone are not given due to the limited taxon sampling.
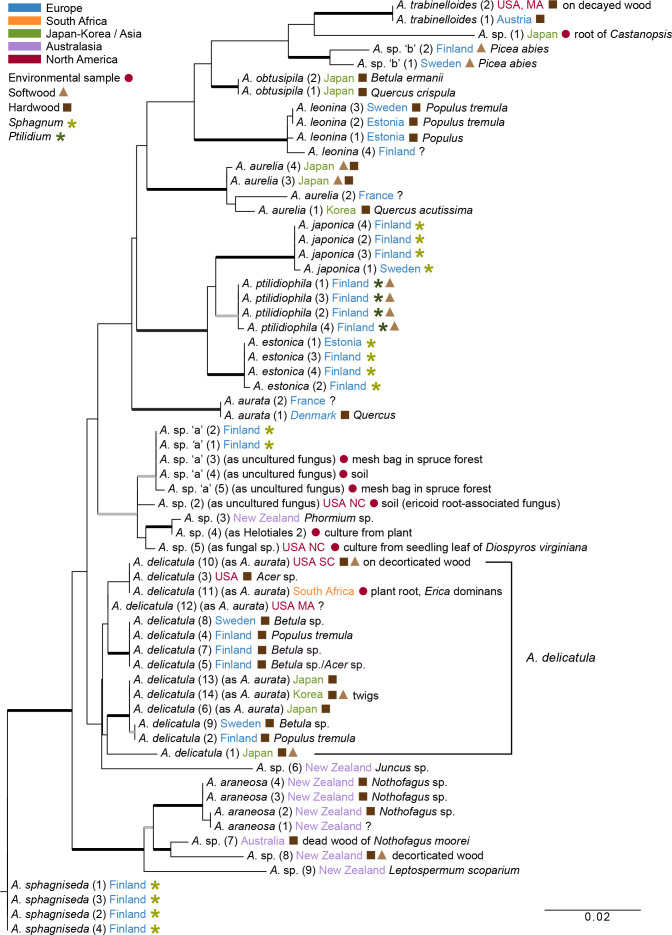
Fig. 4.
The best scoring maximum likelihood phylogeny of Arachnopeziza based on ITS-LSU. Thick black branches received both Bayesian posterior probabilities (PP) ≥ 0.95 and maximum likelihood bootstrap value (ML-BP) ≥ 75 %. Thick grey branches received support by either PP ≥ 0.95 or ML-BP ≥ 75 %. The analyses were run without an outgroup. The tree was rooted on the branch leading to A. sphagniseda. All sequences are derived from ascomata or cultures derived from ascomata, unless marked as environmental samples. The number in parenthesis after a species name refers to an exact collection (see Table 1).
Bayesian and/or ML analyses of the taxon-expanded ITS-LSU dataset supported all of the species recognised using GCPSR or genetic divergence, except for A. delicatula (Fig. 4). In addition to the six collections included in our multi-gene analyses (Fig. 3), nine other A. delicatula collections were included in the ITS-LSU dataset. Six of these were retrieved from GenBank as A. aurata, but are for the time being referred to the A. delicatula lineage, because they had ITS (± LSU) sequences that were identical to ITS and LSU sequences of A. delicatula studied by us (Fig. 4). They include populations from four different continents, i.e., Northern Europe, Eastern North America, South Africa and East Asia. Arachnopeziza aurata, the type species of Arachnopeziza, forms a separate distinct lineage (Fig. 3, 4). Included in the ITS-LSU phylogeny is one species not represented in our five-gene phylogeny, A. aurelia, which is supported as a distinct lineage (PP 1.00, ML-BP 99 %). Three undetermined sequences of Arachnopeziza are closely related or conspecific with A. araneosa, originating from apothecia on hardwood or softwood from Australia and New Zealand (A. sp. 7, 8, 9). Three environmental ITS sequences from clones originating from mesh bag in spruce forest and soil, i.e., A. sp. ‘a’ (3, 4, 5) (Fig. 4), were 100 % identical or nearly so to our two ITS sequences from A. sp. ‘a’ (1, 2) collections from Finland on Sphagnum, and are considered conspecific. Another four undetermined ITS sequences are likely closely related or conspecific with A. sp. ‘a’ based on ML analyses (ML-BP 75 %): three are environmental sequences from plant leaves (A. sp. 4, 5) and from soil (A. sp. 2), and one from apothecia on Phormium (A. sp. 3) (Fig. 4).
Relationships among phylogenetic species in Arachnopeziza
No supported conflict (PP ≥ 0.95, ML-BP ≥ 75 %) was detected between the single gene Arachnopezizaceae phylogenies in terms of relationships among the twelve species recognised. The five-gene phylogeny of Arachnopezizaceae is fully resolved and highly supported in all branches as inferred from Bayesian PP and/or ML-BP, except for the node joining A. aurata and the A. leonina clade, which has no support (Fig. 3). Eriopezia caesia is placed as the earliest diverging lineage in Arachnopezizaceae (as also found in the five-gene Helotiales phylogeny, Fig. 2). Four major clades are identified in Arachnopeziza and to facilitate results and discussion we have named these as indicated on Fig. 3. These clades receive high support from Bayesian PP (1.00) and ML-BP (100 %) except for the A. leonina clade that is supported only by Bayesian PP 0.99 (ML-BP 37 %).
The A. japonica clade consists of three closely related species with a lifestyle connected to bryophytes. Arachnopeziza estonica and A. japonica are morphologically very alike and they both form apothecia on stems and branches of Sphagnum. They are strongly supported as sister species in the five-gene phylogeny (Fig. 3); although Bayesian and ML analysis of RPB1 resolve A. ptilidiophila and A. japonica as sister species (PP 0.92, ML-BP 70 %), analysis of RPB2 and TEF-1α strongly supports A. japonica and A. estonica as sisters (PP 1.00, ML-BP 98 % and PP 0.53, ML-BP 87 %). Arachnopeziza ptilidiophila is associated with Ptilidium spp. and Pinus sylvestris, forming apothecia both on wood and Ptilidium shoots. Its connection to Ptilidium is unclear, not least because Ptilidium is omnipresent on Pinus sylvestris trunks. The A. japonica clade forms a highly supported monophyletic group with the A. delicatula clade as inferred from ML-BP 78 % (PP 0.86). The A. delicatula clade is composed of A. delicatula, A. araneosa and a newly discovered species A. sp. ‘a’ (to be described later when more material has been collected, including observations on vital morphological features). Arachnopeziza delicatula and A. sp. ‘a’ form a monophyletic group (PP 1.00, ML-BP 83 %). The A. leonina clade includes two highly supported subclades: A. leonina and A. obtusipila; and A. trabinelloides and a newly discovered species A. sp. ‘b’. Arachnopeziza sp. ‘b’ is morphologically difficult to distinguish from A. leonina, but appears to be restricted to Picea abies. The two records from Sweden and Finland show marked variation in all the studied gene regions and additional material is needed to explore this further. The A. leonina clade (and A. aurata) is placed as sister to the A. japonica and A. delicatula clades (PP 1.00, ML-BP 88 %). Arachnopeziza sphagniseda (previously Belonium sphagnisedum) constitutes a distinct separate clade, strongly supported as sister to the rest of Arachnopeziza in the five-gene phylogeny (PP = 1.00, ML-BP = 98 %, Fig. 3).
TAXONOMY
Based on GCPSR or genetic divergence using five loci (Fig. 3, Table 2), we delimited 12 species within Arachnopeziza, and with newly collected material from Northern Europe, nine of these are treated and discussed below. Based on the five-gene Bayesian phylogeny (Fig. 2) and morphological and histochemical characters, the two new genera within Hyaloscyphaceae, Mimicoscypha and Resinoscypha, are described with two and three species, respectively, that are combined in the genera. Updated descriptions or notes are given for all of these. Eupezizella is recognised as a separate genus for four species that are combined in the genus. Hyaloscypha usitata is reported for the first time from Europe. An account is given for Arachnoscypha that is removed from Arachnopezizaceae and placed in the Arachnoscypha-Polydesmia clade.
Arachnopeziza aurata Fuckel, Jahrb. Nassauischen Vereins Naturk. 23–24: 304. 1870 — Fig. 5
Fig. 5.
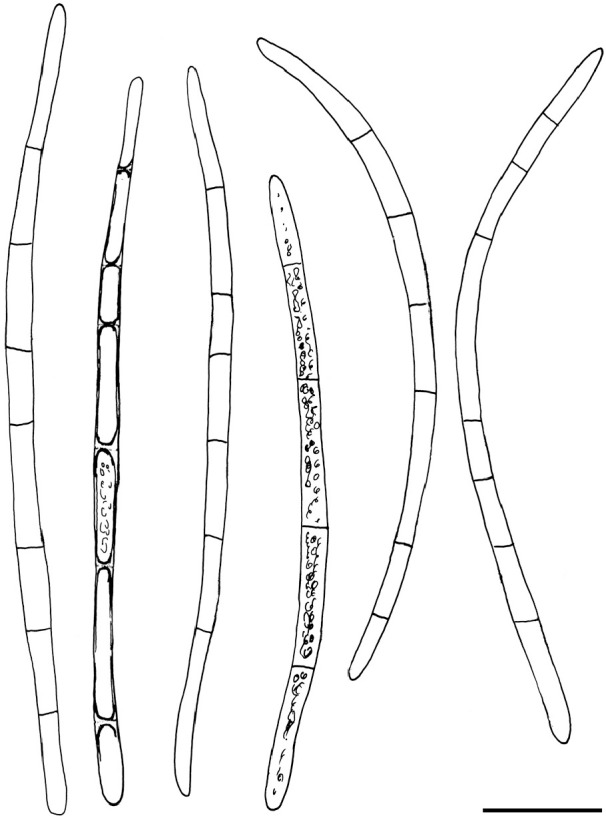
Arachnopeziza aurata. Spores in water (TUR 179456). — Scale bar = 10 μm. — Drawings: T. Kosonen.
Original material. GERMANY, Oestrich, on very moist inner bark of Populus pyramidalis, in the autumn, Fuckel (no date) (S-F92643, ex Herb. Fuckel 1894, ex Herb. Barbey-Boissier 1266) ! (duplicate S-F92641).
Specimen examined. DENMARK, Sjælland, Allindelille Fredskov, on hardwood, 26 May 2007, J. Fournier (TUR 179456).
Notes — We apply the name A. aurata to a collection from Denmark and, based on identical LSU sequences, to CBS 116.54 from France (Fig. 4). It is a distinct, well-delimited species based on our multi-gene phylogenetic analyses and morphology (Fig. 3, 5). In his monograph, Korf (1951b) distinguished A. aurata and A. delicatula primarily on spore measurements and septation: A. aurata with longer, (43–)48–73(–80) × 1.4–2.7(–3.4) μm, 7-septate spores; and A. delicatula with shorter, (24–)28–40(–48) × 2–2.7(–3.4) μm, 3–5 septate spores. Based on our molecular results using GCPSR, A. delicatula populations have often longer and up to 6–7-septate spores thus overlapping with A. aurata (see further under A. delicatula). We suggest that A. aurata is distinguished from A. delicatula by narrower, pointed and straight or curved spores, with regularly 6–7 septa. The spore measurements for the studied material are: 51.8–70.6(–78.3) × 1.7–2.7(–2.9) μm, mean 61.1 × 2.2 μm (n = 30), Q = 21.4–34.0, mean Q = 28.7. The apothecia of the Danish collection are intensively yellow orange. Resin is present on the hairs, among the excipulum cells as large droplets and on the subicular hyphae. No resin was observed on the paraphyses, but the exact placement of the pigment should be studied from fresh material.
Arachnopeziza aurata is resolved as a sister species to the A. leonina clade (Fig. 3), characterized by ample resin depositions on the hairs and in the excipulum. Arachnopeziza delicatula, although with overlapping spore morphology, belongs to a separate clade. Since the large material studied and referred to A. aurata by Korf (1951b) may include collections representing A. delicatula, the reported variation in morphology is possibly variation between A. delicatula and A. aurata populations. The distribution and ecological range of A. aurata is thus also unclear, and will need further study. It is interesting that the collections of A. aurata sensu Korf were not only from various decaying hardwood trees, but also from herbaceous plants, i.e., Typha and Andromeda (Korf 1951b), a substrate not commonly reported for Arachnopeziza species. We made no observations of A. aurata from Estonia, Finland and Sweden during forays in a wide range of habitats, whereas A. delicatula was observed continuously.
Arachnopeziza delicatula Fuckel, Jahrb. Nassauischen Vereins Naturk. 23–24: 304. 1870 — Fig. 6
Fig. 6.
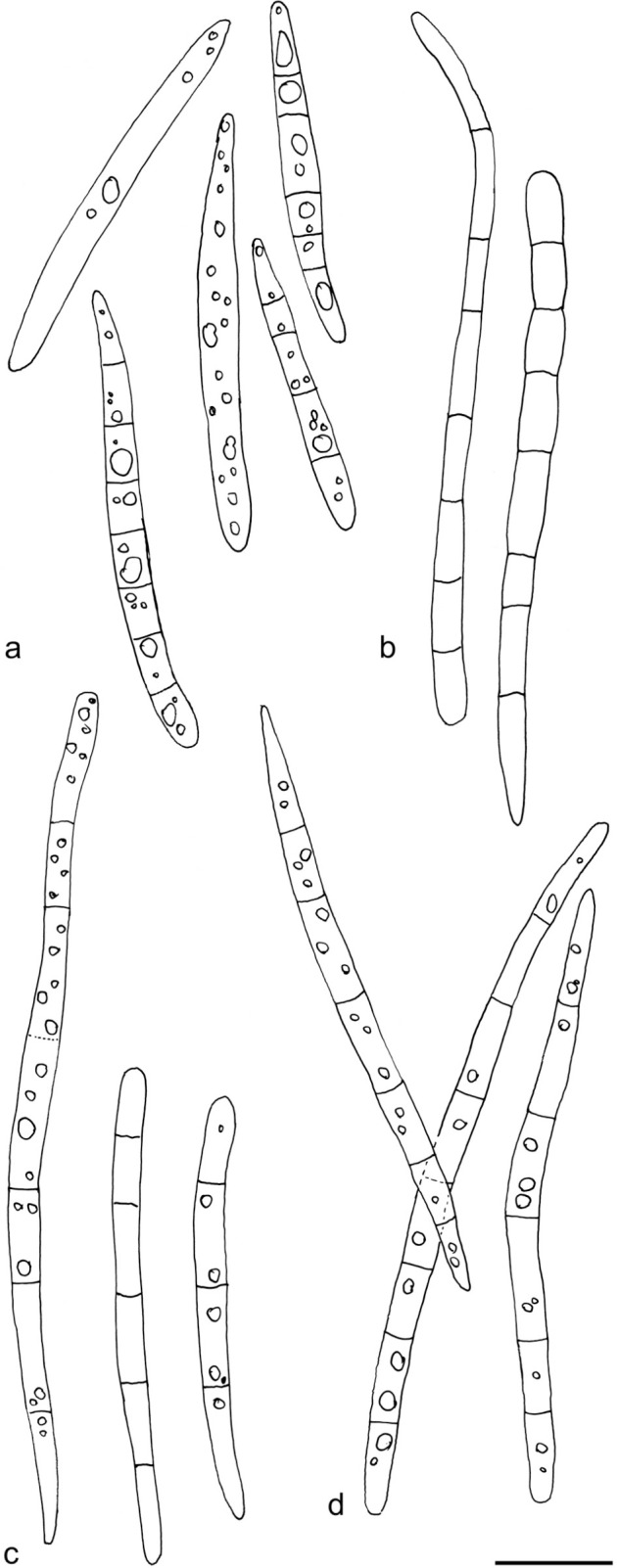
Arachnopeziza delicatula. Spores in CR (a: TNS-F12770; b: T. Kosonen 7036; c: T. Kosonen 7076; d: T. Kosonen 7127). — Scale bar = 10 μm. — Drawings: T. Kosonen.
Original material. GERMANY, Eberbach, Eichberg forest, on Quercus sp., Fuckel, no date (S-F153832, Fuckel Fungi Rhen. Exs. 2384) !
Specimens examined. CANADA, Yukon, Kluane Lake, NW of Sulphur Lake, on Populus tremuloides, 22 Sept. 1987, S. Huhtinen 87/145 (TUR). – FINLAND, Varsinais-Suomi, Turku, Piipanoja, 14 May 2015, T. Kosonen 7036 (S, TUR); Etelä-Häme, Urjala, Raikonkulma, Rantakaski, on a fallen trunk of Betula, 25 Sept. 2015, T. Kosonen 7115 (S, TUR); same date and location, T. Kosonen 7116 (S, TUR); Pohjois-Savo, Suonenjoki, Viippero, Ilmakkamäki, on a trunk of P. tremula, 23 Sept. 2013, J. Purhonen 6655 (JYV 11610); Muurame, Kuusimäki, 25 Aug. 2015, T. Kosonen 7076 (S, TUR). – JAPAN, Hokkaido, Iwamizawa-shi, Tonebetsu, 25 July 2004, T. Hosoya (TNS-F12770). – SWEDEN, Dalsland, Håverud, Forsbo Nature Reserve, on a trunk of Betula, 23 Sept. 2016, S. Huhtinen 16/54 (S, TUR); Västergötland, Falköping, Forentorpa ängars Nature Reserve, on a trunk of Betula, 15 Oct. 2015, T. Kosonen 7127 (S, TUR); Kinnekulle, Munkängerna, on Ulmus, 17 Oct. 2015, K. Hansen & T. Kosonen 7132 (S, TUR); Öland, Byxelkrok, Trollskogen, on a trunk of Betula, 29 Mar. 2016, T. Kosonen 7152 (S, TUR). – USA, Massachusetts, Bristol County, North Easton, Borderland State Park, on the underside of a decayed hardwood log, 19 May 2012, J. Karakehian 12051901 (S, TUR).
Notes — Arachnopeziza delicatula appears to be a phylogenetically and morphologically diverse species. Both Fuckel (1870) in the original description and Korf (1951b) in his monograph describe A. delicatula spores being on average less than 50 μm long and with up to 5 septa. Such populations exist in the material reported here, but using GCPSR they do not represent a distinct species from those populations with on average longer spores (above 50 μm). Instead, our results suggest that A. delicatula have spores with a wide range in length: 30–61.0(–63.5) × 2.4–3.8(–4.0) μm in CR, mean 47.6 × 2.9 μm, Q = 9.7–23.8(–25.7), mean Q = 16.5 (n = 35, from three populations). It can be distinguished from A. aurata by the width of the spores that are on average wider in A. delicatula (3–3.5 μm wide in the longest spores) and by the Q-value that is in average less than 20 in A. delicatula and above 20 in A. aurata. Most likely this is the main reason for the numerous ITS and LSU sequences deposited in GenBank under the name A. aurata, which we consider to represent A. delicatula (see Fig. 4). Despite the wide range in spore size and high divergence in ITS sequences we found no support for delimiting further species based on GCPSR or on geographical origin (Fig. 3, Table 2). All the samples studied by us were from hardwood and most of the ITS and LSU sequences retrieved from GenBank originated from apothecia on wood (if specified, on hardwood). One GenBank ITS sequence is from a plant root (CL0301_4_1 from South Africa) (Fig. 4). It is identical to an ITS sequence of a North American collection (J. Karakehian 12051901), which is also represented in our multi-gene analyses (A. delicatula 3, Fig. 3). There are earlier reports of A. delicatula from softwood (e.g., Korf 1951b), as well as some more recent collections in TAAM. Our preliminary morphological study of the TAAM A. delicatula collection (TAAM062455) from softwood, found it conforms to the other A. delicatula material examined here. This could well be a demonstration of a broad ecology, but there are no sequences available from material on softwood.
Arachnopeziza estonica T. Kosonen, Huhtinen, K. Hansen, sp. nov. — MycoBank MB835728, Fig. 7
Fig. 7.
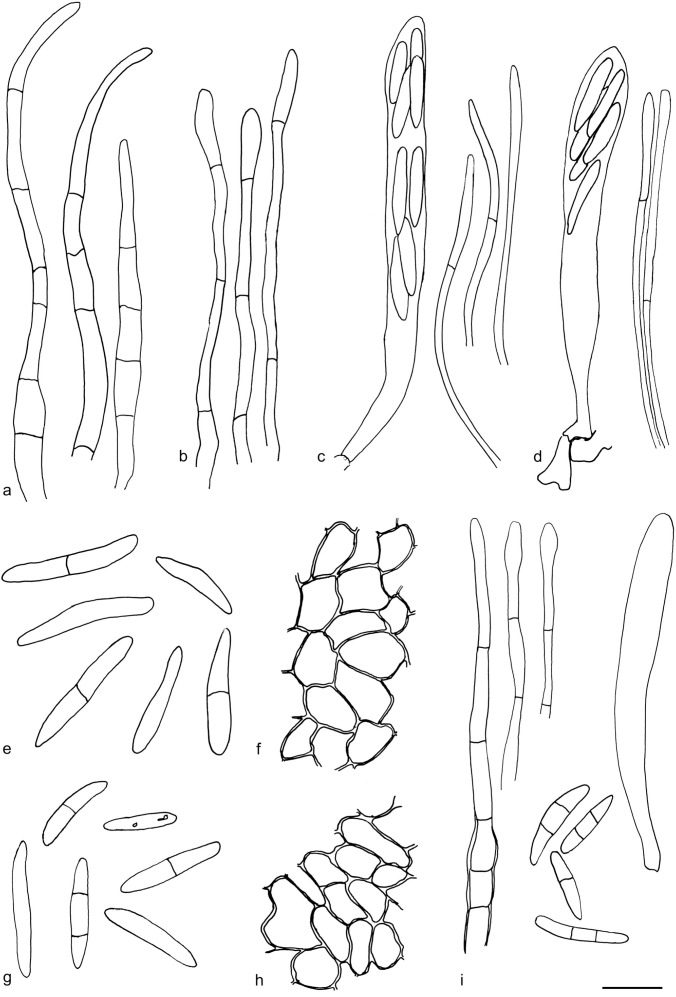
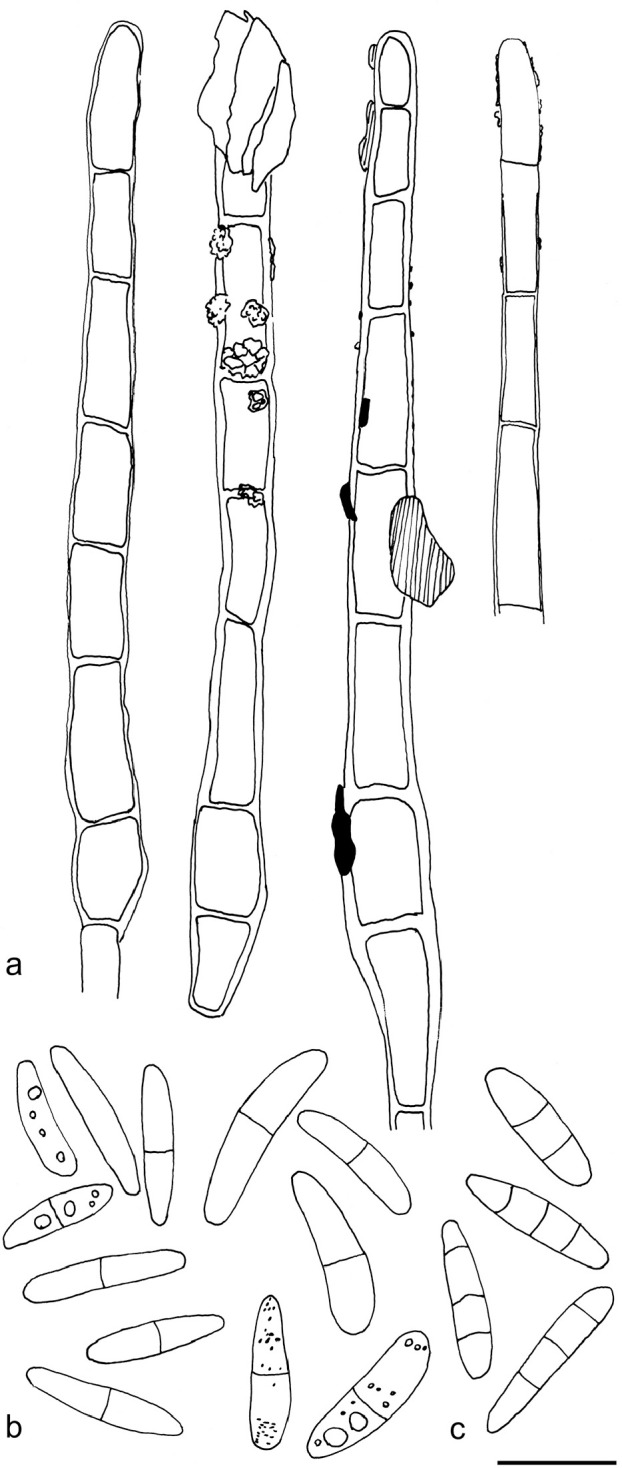
a, c, e–f. Arachnopeziza estonica (holotype) and b, d, g–i. A. japonica. All in CR. a–b. Hairs; c–d. asci and paraphyses; e–f. spores; g–h. excipulum; i. hairs, asci and spores (a, c, e–f: S. Huhtinen 15/38; b, d, g–h: T. Laukka 267; i: holotype). — Scale bar = 10 μm. — Drawings: T. Kosonen.
Etymology. Referring to the geographical origin of the holotype.
Holotype. ESTONIA, Otepää, Kääriku, NE of lake Kääriku, on Sphagnum squarrosum, in a boggy mixed forest, 13 Sept. 2015, S. Huhtinen 15/38 (S-F399746); Isotype (TUR 212780) !
Apothecia 0.2–0.5 mm diam, hyaline to white when fresh, white with a yellow hue when dry, on leaves of Sphagnum, narrowly attached, often only with a few visible long hairs originating from the margin and upper flanks, subiculum present but indistinct to naked eye. Ectal excipulum in the upper flanks of textura prismatica, cells c. 15–20 × 5 μm, below and towards the base of textura angularis to globulosa, cells c. 5–10 μm diam. Hairs 50–90 × 3–4 μm, with 3–5 septa, cylindrical, thin-walled, but moderately to clearly thick-walled in the basal parts, uppermost cell tapering distinctly, smooth, hyaline resin present on the outside. Asci 70–90 × 6–10(–14) μm, cylindrical to clavate, apical pore MLZ+, arising from croziers. Ascopores 15.6–23.2(–24.2) × 2.8–4.4(–5.2) μm, mean 18.8–3.7 μm, Q = 3.9–8.3, mean Q = 5.2 (n = 55, from two populations), cylindrical, often narrowing more prominently towards basal end, with characteristic angular appearance, usually with 0–1 septa, or more rarely with two. Paraphyses cylindrical, regularly branched at the basal cell, smooth and without content in CR. Subicular hyphae 3–5 μm wide, thick-walled, finely warted.
Specimens examined. FINLAND, Varsinais-Suomi, Turku, Kuhankuono, on Sphagnum, 13 Nov. 2005, T. Laukka 253 (TUR); Etelä-Häme, Tammela, Liesjärvi, on Sphagnum, 4 July 2005, T. Laukka 210 (TUR); Pohjois-Häme, Ruovesi, Siikaneva National Park, on Sphagnum squarrosum, 7 Sept. 2005, T. Laukka 245 (TUR).
Notes — This species is very closely related to A. japonica and morphologically very similar. It is supported as a separate species using GCPSR (Fig. 3, Table 2) and it can be distinguished by the on average larger spores and wider hairs as compared to A. japonica. The hair apices are slightly inflated in A. japonica, whereas in A. estonica the hairs taper evenly and no apical widening is observed. All observations are from Sphagnum and often from Sphagnum squarrosum. The two close species, A. japonica and A. estonica, appear to have very similar ecology at least in Scandinavia.
Arachnopeziza japonica Korf, Bull. Natl. Sci. Mus., Tokyo 44: 392. 1959 — Fig. 7
Holotype. JAPAN, Kyushu, Miyazaki prefecture, Yabitsu valley, Kijomura village, on leaves, leaf-galls and debris, 6 Nov. 1957, S. Imai et al. (CUP-JA-422) !
Apothecia 0.2–0.6 mm diam, hyaline to whitish when fresh, white with a yellow hue when dry, at margin distinctly hairy, broadly attached, subiculum present, scanty. Ectal excipulum of firm walled textura prismatica, cells c. 18 × 5 μm in the upper flanks, somewhat thick-walled textura angularis – textura globulosa towards the base, cells 3.5–10 μm wide. Hairs 50–80 × 2–3 μm, cylindrical, with 3–5 septa, thin-walled, in the basal parts with moderately to clearly thickened walls, apices typically widened, smooth, hyaline resin present, only partly soluble in MLZ or lactic acid, hairs often tightly glued together in dry material. Asci 70–90 × 6–10(–14) μm, cylindrical to clavate, apical pore clearly MLZ+, arising from croziers. Ascospores 12.5–21.6(–23.2) × 2.7–4.1(–4.4) μm, mean 16.3 × 3.3 μm (n = 60, from two populations), Q = 3.9–7.9, mean Q = 5.1, cylindrical, but usually narrowing more prominently towards the basal end, slightly angular, usually with 1–2 septa, the 2-septate spores are relatively common, rarely with 3 septa. Paraphyses regularly branched at the basal cell, smooth and without content in CR, with a round apex. Subicular hyphae 3–5 μm wide, thick-walled, warted.
Specimens examined. All specimens on Sphagnum. FINLAND, Varsinais-Suomi, Nousiainen, Pukkipalo, 26 May 2006, R. Ilmanen 194 (TUR); Parainen, Kirjalansaari, 22 Sept. 2006, R. Ilmanen 239 (TUR); same location, 22 Sept. 2006, T. Laukka 267 (TUR). – SWEDEN, Söderåsen Nature Park, 5 June 2006, S. Huhtinen 06/3 (TUR).
Notes — Arachnopeziza japonica is recorded for the first time from Europe. The four collections from Finland and Sweden agree well with the original description and our study of the holotype. In spore size, the holotype (CUP-JA-422, spores: 11.5–16.0 × 2.7–3.0, n = 4) represents the smaller-spored part of the variability, but the observed range is relatively wide and the number of measured spores from the scanty specimen is very small. New observations include a population (R. Ilmanen 194) with a roughly similar average spore size and variation as the holotype. Spores with 3 septa are rare and usually then clearly overmature. Spores with 2 septa are relatively common and have the same size and shape as spores with one or no septa. The original description was based on only the holotype from forest litter without any mention of bryophytes or mires. The four recent collections are from Sphagnum from paludified forest habitats. Arachnopeziza japonica belongs to a clade of three closely related species sharing a connection to bryophytes. Often apothecia of these Arachnopeziza species were found by studying randomly picked Sphagnum tuffs under a dissecting microscope. Based on the frequently found (single) apothecia on Sphagnum, we conclude that the species are relatively common (on Sphagnum), but the limits of their ecological niche are poorly known. Larger groups of apothecia of these species were found less often.
Arachnopeziza leonina (Schwein.) Dennis, Kew Bull. 17: 351. 1963 — Fig. 1a–c, 8
Fig. 8.
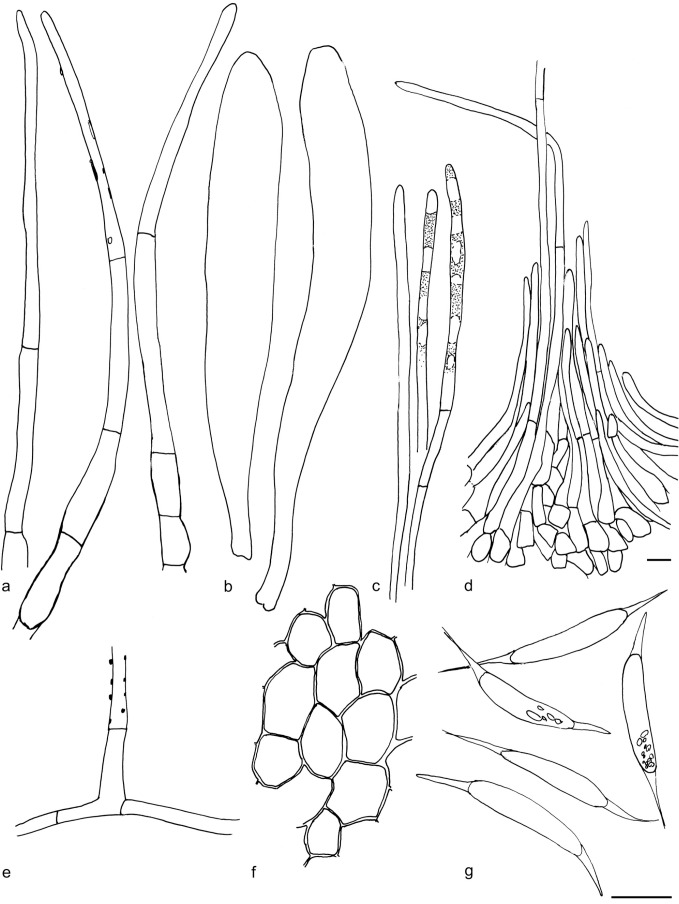
Arachnopeziza leonina. a. Hairs in water; b. spores in water; c. four spores with multiple septa in CR (a–b: GJO 0071770; c: KH.15.23). — Scale bar = 10 μm. — Drawings: T. Kosonen.
Basionym. Peziza leonina Schwein., Schr. Naturf. Ges. Leipzig 1: 93. 1822.
Synonym. Arachnopeziza candido-fulva (Schwein.) Korf, Lloydia 14: 163. 1951.
Specimens examined. AUSTRIA, Steiermark, Graz, Andritz, near the church of St. Ulrich, on hardwood, 1 Mar. 2014, I. Wendelin (GJO 0071770). – ESTONIA, Valga, Sangaste, Lauküla, Keesliku forest, on a trunk of Populus tremula, 12 Sept. 2015, T. Kosonen 7101 (S, TUR); same date and location, K. Hansen, KH.15.23 (S, TUR). – FINLAND, Pohjois-Häme, Muurame, Kuusimäki, on a trunk of Betula sp., 25 Aug. 2015, T. Kosonen 7078 (S, TUR); same date and location, on a trunk of P. tremula, T. Kosonen 7091 (S, TUR); on a trunk of Betula sp., T. Kosonen 7092 (S, TUR); on trunk of Betula sp., 14 May 2018, J. Purhonen & T. Kosonen 7294 (S, TUR). – SLOVAKIA, Chorvátsky Grob, Čierna voda, National Nature Reserve Šúr, on a decayed hardwood trunk, 22 Nov. 2017, A. Polhorský 18/26 (S, TUR). – SWEDEN, Stockholm, Haninge, Tyresta National Park, on a trunk of P. tremula, 1 June 2016, T. Kosonen & S. Huhtinen 16/42 (S, TUR); Västra Götaland, Falköping, Forentorpa ängars Nature Reserve, 15 Oct. 2015, T. Kosonen 7128 (S, TUR).
Notes — Arachnopeziza leonina is a phylogenetically distinct species. It is best characterised by the less than 18 μm long, relatively wide spores with three or less septa and by the yellow resin on the hairs and between the excipular cells. It appears to be restricted to decayed hardwood trunks (Populus, Betula and Quercus) or to very coarse debris. No observations were made from recently (< 5 yr) fallen trunks. Morphologically, it overlaps with Arachnopeziza sp. ‘b’ found on softwoods (Picea abies). Arachnopeziza trabinelloides can often be distinguished from A. leonina by the abundant resin on the hairs and the practically non-existent subiculum, but herbarium specimens of A. trabinelloides with little resin can be morphologically inseparable from A. leonina samples with (for that species) ample resin. We found very little help, if any, in the number of septa in the spores since mature spores seem to have 1–3 septa in both A. leonina and A. trabinelloides. Fresh samples of A. leonina do not react in MLZ as A. trabinelloides (see notes under A. trabinelloides). For herbarium material we found the best delimiting character to be the lower part of the hairs: in A. trabinelloides hairs are fairly cylindrical and of even width for the whole length, whereas in A. leonina the hairs are slightly, but clearly, basally widened. In Scandinavia A. leonina has an ecology similar to A. delicatula; both are regularly present on fallen trunks of hardwood (Betula or Populus tremula) already in the early stages of decomposition. In his monograph, Korf (1951b) missed the older description by Schweinitz (1822) and applied the name A. candido-fulva instead. This was pointed out by Dennis (1963) and later accepted by Korf (Korf & Gruff 1981). Arachnopeziza leonina is a relatively common species in the boreal and temperate zones, with observations from Europe and North America. Observations from the southern hemisphere are lacking.
Arachnopeziza ptilidiophila T. Kosonen, Huhtinen, K. Hansen, sp. nov. — MycoBank MB835729, Fig. 1d, 9
Fig. 9.
Arachnopeziza ptilidiophila (T. Kosonen 7291). All in water. a. Hairs; b. asci; c. paraphyses; d. multiple hairs glued together; e. subicular hypha; f. excipulum; g. spores. — Scale bars: a–c, e–g = 10 μm, d = 10 μm. — Drawings: T. Kosonen.
Etymology. Referring to co-occurrence with Ptilidium.
Holotype. FINLAND, Pohjois-Häme, Muurame, Kuusimäki, on decayed, still quite hard Pinus sylvestris trunk, beside or directly on Ptilidium shoots, 14 May 2018, J. Purhonen & T. Kosonen 7287 (S-F399747) !
Apothecia 0.2–0.7 mm diam, hyaline when fresh, white to yellow when dry, narrowly attached, hymenium flat, hairs concolorous with the receptacle surface, protruding c. 50–70 μm above the hymenium, hairs often slightly bent at upper-third and attached to each other giving a shaggy appearance, subiculum distinct but sparse. Ectal excipulum of textura prismatica, cells slightly rounded, c. 12–20 × 5 μm in the upper flanks, excipulum below of textura angularis – textura globulosa, cells c. 10–13 × 6–10. Hairs 80–130 × 3–5 μm, cylindrical, thin-walled, basal cells more thick-walled, slightly widened, usually 1–3-septate, apical cell c. one third of the whole length, resin present as small hyaline droplets on the hairs. Asci 70–100 × 6–12 μm, clavate, apical pore MLZ+, arising from croziers. Ascospores 15.7–23.4(–25.6) × 3.9–5.3(–5.5) μm, mean = 20.1 × 4.6 μm, Q = 3.3–6.1, mean Q = 4.4 (n = 60, from two populations), inequilateral, freshly released spores with 0–1 septa, aseptate spores dominating, most spores with c. 6–10 μm long, hyaline, thin, slender, gel-like polar appendages, tapering to a very sharp end. Paraphyses filiform, 1.5–2.5 μm wide. Subicular hyphae c. 1.5–3 μm wide, thick-walled, smooth or minutely warted.
Specimens examined. FINLAND, Satakunta, Yläne, Vaskijärvi Nature Protection Area, Kuusela, on Ptilidium pulcherrimum, 15 Apr. 2005, T. Laukka & A. Lesonen 51 (TUR); Pohjois-Häme, Muurame, Kuusimäki, on Pinus sylvestris trunk and on Ptilidium shoots, 14 May 2018, T. Kosonen 7287 (S, TUR); same date and location, T. Kosonen 7289 (S, TUR); same date and location, T. Kosonen 7291 (S, TUR).
Notes — The most striking feature of A. ptilidiophila is the long and slender polar spore appendages (Fig. 9g). The character is only visible in living material; already more than 1-mo-old dried specimens failed to show the appendages in various rehydration trials with different reagents. Although gelatinous appendages are rare in hyaloscyphoid fungi, they seem to be less so in Arachnopeziza. Spore appendages have also been observed in A. aurelia, A. cornuta and A. floriphila. Boudier (1910: Plate 520, n°. 530) illustrated A. aurelia with similar spore appendages to those in A. ptilidiophila, but in A. aurelia the appendages are often shorter and more blunt. Arachnopeziza floriphila has also similar spore appendages to A. ptilidiophila, but it differs in spore size and by the thick-walled multiseptate hairs (Baral 1989). In his monograph, Korf (1951b) did not observe the appendages in (dried) specimens of A. cornuta, but according to the original description (Ellis 1882) spores have ‘short, blunt polar appendages’. Spore size (less than 15 μm long) and hair morphology (distinctly multiseptate and thick-walled) distinguish A. cornuta from A. ptilidiophila. Another noteworthy feature are the thin-walled hairs attached to each other forming ‘teeth’. This is rare in Arachnopeziza with only A. fitzpatrickii sharing the character. We observed A. ptilidiophila growing directly on liverwort shoots as well as on barkless softwood, but always with liverwort shoots or protonema in the immediate vicinity. Many of the observed populations were often relatively small, comprising less than five apothecia in a small notch on the Pinus bark. The specimens examined by us are of populations with more (> 10) apothecia. Ptilidium ciliare and P. pulcherrimum are ubiquitous liverworts growing as epiphytes on a range of softwood and hardwood. Further sampling of Arachnopeziza species on Ptilidium growing on non-coniferous or non-woody substrates should elucidate the more specific ecological niche of this species. All the observations are from sites with relatively undisturbed continuity of dead wood and with large dead Pinus sylvestris trunks.
Arachnopeziza sphagniseda (Velen.) T. Kosonen, Huhtinen & K. Hansen, comb. nov. — MycoBank MB835730; Fig. 10
Fig. 10.
Arachnopeziza sphagniseda (PRM611379). a. Detail from margin; b. asci and paraphyses in CR; c. asci in CR; d. spores, including rare spores producing conidia; e. dry apothecia; f. subicular hyphae; g. ectal excipulum. — Scale bars: a–d, f–g = 10 μm, e = 100 μm. — Drawings: S. Huhtinen.
Basionym. Belonium sphagnisedum Velen., Monogr. Discom. Bohemiae Pars. 1: 179, pl. iv, f. 16. 1934.
Synonym. Hymenoscyphus sphagnisedus (Velen.) Svrček, Česká Mykol. 33: 200. 1979.
Holotype. CZECH REPUBLIC, Krkonoše Mountains, on Sphagnum, Aug. 1927, K. Cejp (PRM 149855) ! (the convolute is empty).
Lectotype designated here: pl. IV, f. 16 in Velenovsky 1934: Monogr. Discom. Bohemiae Pars. 1. MycoBank MBT392710.
Superseded neotype. Designated by Svrček 1979 in Česká Mykol. 33: 200. CZECH REPUBLIC, Southern Bohemia, Třeboň, ‘Hrádeček’, in a ditch on living Sphagnum sp., 18 May 1964, M. Svrček (PRM 611379) !
Apothecia 0.2–2 mm diam, discoid to cupulate, smooth, white, apricot when dry, sessile to subsessile with scanty subicular hyphae around the base. Ectal excipulum of textura oblita to textura prismatica, more globose below, cells relatively thick-walled. Margin with cylindrical-clavate 2–3 μm wide hyphal ends forming a smooth rim, thin-walled to somewhat thick-walled, varying between the populations, no true hairs observed. Asci 49–67 × 5.6–8.0 μm in CR, mean 52.2 × 7.0 μm (n = 10, from six populations), cylindrical, in neotype often clearly widened, maximally to 14 μm, characterized by an abruptly narrowed apex, apical pore blue both in MLZ and LUG, arising from croziers. Ascospores 8.6–13.2(–14.0) × 2.0–3.0 μm in CR, mean 10.6 × 2.4 μm, Q = 3.0–5.8(–6.0), mean Q = 4.4 (n = 38, from six populations), in neotype 9.3–15.2(–16.6) × 2.0–2.8 μm in CR, mean 12.3 × 2.5 μm, Q = 4.0–6.0(–7.5), mean Q = 5.1, ellipsoid to narrowly ellipsoid to subfusoid, septation varying from aseptate to sparsely 1-septate populations, to populations where 1-septate free spores are a majority, 2–3-septate spores rare. Paraphyses cylindrical, obtuse, 2–3 um wide, unbranched, not exceeding the asci. Subicular hyphae hardly observable under dissecting microscope but always present in a mount, 3–4 μm wide, smooth to minutely warty, thick-walled.
Specimens examined. All on Sphagnum. FINLAND, Varsinais-Suomi, Kaarina, Kuusisto, 10 June 2003, T. Laukka 06 (TUR 165732); Lieto, Mellilä, Liedonperäntie 294, 5 Sept. 2007, R. Ilmanen 338 (TUR 179358); Nousiainen, Kurjenrahka National Park, 11 July 2006, T. Laukka et al. (TUR 174929); Parainen, Kirjalansaari, 22 Sept. 2006, R. Ilmanen 246 (TUR); same date and location, T. Laukka 268 (TUR); Turku, Ruissalo, Choraeuksen lähde, 26 May 2004, M. Paajanen 804 (TUR 173706); same date and location, M. Paajanen 805 (TUR 173700); Satakunta, Luvia, Rekojärvi, 13 Aug. 2006, R. Ilmanen 224 (TUR 174912); same date and location, R. Ilmanen 226 (TUR 174911); Rekojärvi, 28 Oct. 2006, R. Ilmanen 267 (TUR 178007); same date and location, R. Ilmanen 269 (TUR 178009); Pohjois-Karjala, Kitee, 15 Sept. 2006, S. Huhtinen 06/18 (TUR); same date and location, S. Huhtinen 06/20 (TUR).
Notes — The presence of a subiculum and relatively big, septate spores are typical characters of an Arachnopeziza. Most likely it was the presence of a subiculum that prompted Velenovský to name the single collection initially as ‘Arachnopeziza sphagnicola’ in his fungarium notes on the original convolute. In 1934 the collection was described as Belonium, most likely owing to the lack of hairs. Due to the missing holotype collection (thereafter found by us in PRM, but only as an empty convolute), Svrček (1979) designated a neotype. This was, however, superfluous since there is a plate to choose as lectotype (Velenovský 1934). As Svrček stated, this ‘neotype’ is a perfect match to Velenovský’s description, and therefore we use it as a reference collection. We, nevertheless, refrain from selecting it as an epitype, because it is a very scanty specimen and it is not suitable for DNA extraction and sequencing. Our material is from a totally different biogeographical area, although also from Sphagnum. This species seems to be relatively easily found, so a new collection from the Czech Republic (with photographs and DNA sequences) would make the most suitable epitype. Our material fits well both the original diagnosis and Svrček’s collection. Although there is substantial morphological variation among our material, there is no variation in the sequence data and the growth form in culture was also identical. We did not obtain TEF-1α or mtSSU sequences for A. sphagniseda, but the four ITS and LSU rDNA sequences were 100 % identical, and the two RPB1 sequences had identical amino acid translation and differed only by six base pairs. In the superseded neotype, especially the subtruncate apex and width of the asci are prominent (Fig. 10b), and represent the extreme end of the observed overall variation.
Only one species of Arachnopeziza known, A. nuda, resembles A. sphagniseda in lacking distinct apothecial hairs. It is also described as having asci with an abruptly narrowed and thickened apex. Arachnopeziza nuda has, however, short clavately enlarged cells, i.e., hairs, 9–18 μm long, arising from the outermost cells of the ectal excipulum (Korf 1959). It also has longer and broader spores than A. sphagniseda and occurs on decorticated wood. We were not able to sample A. nuda for molecular phylogenetic study, but our five-gene phylogeny suggests A. sphagniseda is sister to all other Arachnopeziza species sampled (Fig. 3).
Arachnopeziza trabinelloides (Rehm) Korf, Lloydia 14: 169. 1952 — Fig. 11
Fig. 11.
Arachnopeziza trabinelloides. a. Hairs in water, hair on the right almost totally covered by resin; b. hair in CR; c. upper part of hairs in water, loose resin around the hair on the right; d. upper part of hairs in CR; e. asci and paraphyses with pigmented resin in water; f. paraphyses in CR; g. paraphyses apex variation in CR; h. spores of different maturity in water; i. spores in MLZ; j–k. spores in CR; l. ectal excipulum in water (a, c, e, h–i, l: GJO 0071771; b, g, k: holotype; d, f, j: DHP-02493). — Scale bar = 10 μm. — Drawings: T. Kosonen.
Basionym. Helotium trabinelloides Rehm, Hedwigia 26 (3): 82. 1887.
Holotype. ROMANIA [FORMER AUSTRIA-HUNGARY], Caraş-Severin, Băile Herculane, on a fallen decorticated trunk of Fagus, Apr. 1885, Lojka (S F7139) !
Specimens examined. AUSTRIA, Steiermark, Graz, Andritz, Reinerkogel N-side, near the church of St. Ulrich, on deciduous wood, 1 Mar. 2014, I. Wendelin (GJO 0071771). – UNITED STATES, Massachusetts, Princeton, Fay Lane, a trunk of Acer rubrum, 1 Apr. 2002, J. Choiniere, DHP-02493 (TUR); Wachusett Meadow Wildlife Sanctuary, West Trail, on decayed wood (white-rot), 2 Mar. 2014, J. Karakehian 14030203 (S, TUR).
Notes — A well annotated and illustrated collection of this relatively rarely collected and documented Arachnopeziza species was recently reported from Austria (Friebes & Wendelin 2014). Its morphological similarities to A. leonina are commented on under that species. A unique character of A. trabinelloides is the rapidly disappearing colour reaction, where MLZ turns the resinous exudates and oily pigments in the hymenia blue. The reaction, that is only visible in fresh samples, was already observed by Rehm (1887) and part of the original description. As noted by Korf (1951b), the variation in the shape of the paraphyses apices is high in A. trabinelloides compared to other species of Arachnopeziza (Fig. 11f, g). Typically, the hairs of A. trabinelloides have abundant resin expanding more or less the whole length of the hair, much more than in any other Arachnopeziza species known to us (Fig. 11a, c). In the field it is easily recognised by the large (often > 1 mm diam), bright yellow-orange apothecia and the scanty or nearly non-existing subiculum. Arachnopeziza trabinelloides belongs to the A. leonina clade (Fig. 3) of wood inhabiting Arachnopeziza species with substantial pigmented resin. We made no observations of A. trabinelloides during numerous forays in Estonia and Fennoscandia in this project. It appears to be restricted to Quercus, Fagus, Castanea and other hardwoods in the temperate region. It is strongly supported as a sister species to Arachnopeziza sp. ‘b’, an undescribed Arachnopeziza species on softwood (Fig. 3).
Arachnopeziza sp. ‘a’ — Fig. 12
Fig. 12.
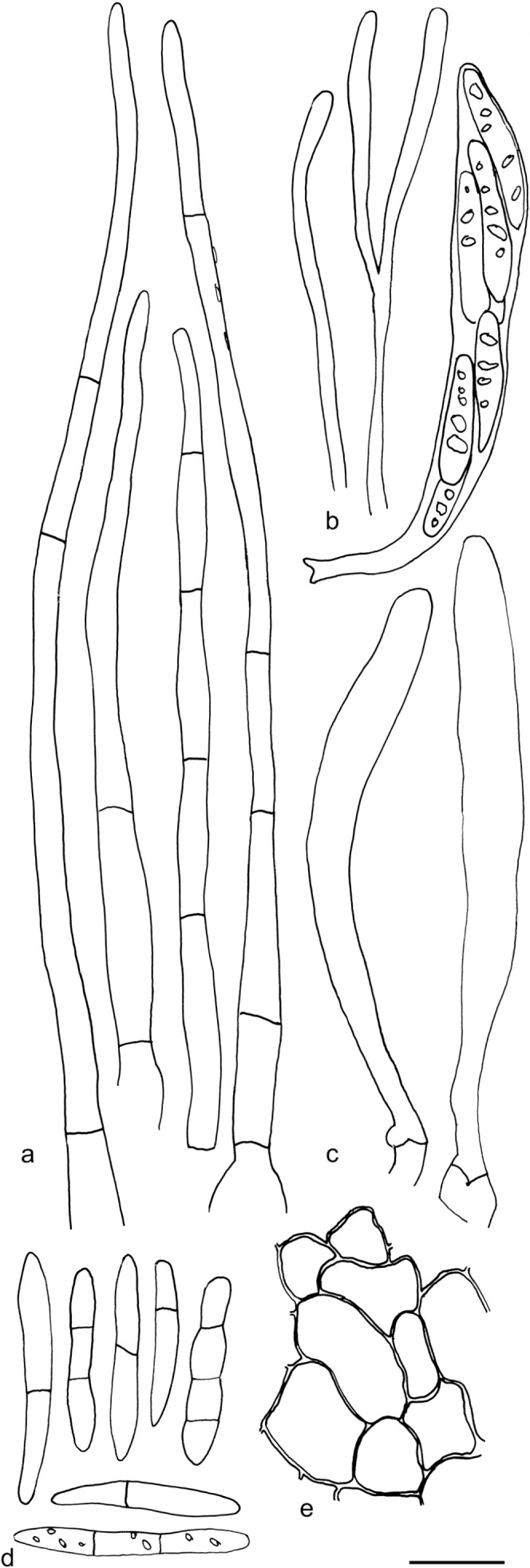
Arachnopeziza sp. ‘a’ (T. Laukka 260). All in CR. a. Hairs; b. paraphyses and ascus; c. asci; d. spores; e. excipulum. — Scale bar = 10 μm. — Drawings: T. Kosonen.
Apothecia on Sphagnum shoots, c. 0.3–0.5 mm diam, sessile, narrowly attached, margin hairy, white to hyaline when fresh, yellow when dry, subiculum not observed. Ectal excipulum of textura prismatica to textura angularis. Hairs 80–140 × 3–5 μm, cylindrical, thin-walled, with 2–5 septa, tapering, apex obtuse, c. 2 μm wide, fragile appearance, occasionally with small hyaline resinous exudates. Asci 50–65 × 5–8 μm, thin-walled, cylindrical, apical pore MLZ+, arising from croziers. Ascospores 18.3–23.7(–25.4) × 2.6–3.0 μm in CR, mean = 20.6 × 2.8 μm, Q = 6.0–8.5, mean Q = 7.4 (n = 17, from two populations), ellipsoid, unevenly cylindrical, tapering towards the somewhat angular ends, usually with one septum. Paraphyses filiform, occasionally bifurcate, no septa observed, 1–2 μm wide, smooth.
Specimens examined. FINLAND, Varsinais-Suomi, Turku, Kohmo, 20 June 2006, R. Ilmanen 199 (TUR); Pääskyvuori, on Sphagnum, 6 July 2006, T. Laukka 260 (TUR).
Notes — We have found no published name for this species that is easily distinguished by the large spores, long slender multi-septate hairs and occurrence on Sphagnum shoots. The two samples were collected less than 2 km apart, although on different forays, and it may be rare. The collections are relatively small and documentation of characters from fresh apothecia is incomprehensive and therefore we refrain from formally describing this novel species. The ecology appears similar to that of the species in the A. japonica clade (e.g., A. japonica and A. estonica) (Fig. 3). Based on our phylogenetic analysis, however, it is a sister species to A. delicatula and not a member of the A. japonica clade. There are three identical or nearly identical ITS sequences in GenBank (EF521221, KP889888, EF521220) obtained from environmental samples from soil and mesh bag in spruce forest. In addition, four other samples form a group with A. sp. ‘a’, as supported by ML-BP 75 % (Fig. 4), but it is uncertain if these are conspecific, differing in 5–10 base pairs.
Arachnoscypha Boud., Bull. Soc. Mycol. France 1: 118. 1885
Type species. Arachnoscypha aranea (De Not.) Boud. ex Dennis.
Notes — The genus Arachnoscypha was erected with a single species listed (‘L’espèce typique’), Peziza aranea, by Boudier (1885). Later, however, Boudier (1907) treated and combined P. aranea in Arachnopeziza. Korf (1951b) agreed on placing Arachnoscypha in synonymy with Arachnopeziza, based on morphological similarities, i.e., the multiseptate hairs, septate spores and the existence of a subiculum. Korf (1951b) also noted that since 1-septate spores are not uncommon in Arachnopeziza, Arachnoscypha (aranea) should not be separated based on this. Our molecular phylogenetic results, placing Arachnoscypha aranea as a sister taxon to Polydesmia, are somewhat surprising. When compared, Arachnoscypha and Polydesmia show some similarities in morphological characters. Both the type species of Polydesmia (P. pruinosa) and Arachnoscypha have apothecia with hairs on the flanks. However, in P. pruinosa the hairs are regarded as undifferentiated (Korf 1978, Verkley 2005) and undulating. In Arachnoscypha, especially on the upper flanks, there are relatively straight multiseptate hairs (Fig. 13a). Also, P. pruinosa and A. aranea both have long hyphal structures arising from the base of the apothecia. In A. aranea these hyphae are often bent, giving them a characteristic angular appearance (Fig. 13b). We consider the hyphae originating at the base of the apothecia in Arachnoscypha to be anchoring hyphae (as opposed to subicular hyphae, see further under Discussion). The branched and twisted paraphysis apices protruding above the asci in Polydesmia, giving the apothecia the characteristic pruinose appearance is a unique feature. The paraphyses in Arachnoscypha are bifurcate and undulating as in Polydesmia, but in Arachnoscypha they undulate more irregularly. This is also observed from fresh material. In Polydesmia the paraphyses have exudates somewhat like the exudates on Arachnoscypha hairs and hyphae at the apothecial base.
Fig. 13.
Arachnoscypha aranea. All in CR. a–b. Hairs with crystals; c. asci and paraphyses; d–f: spores; g. basal ectal excipulum; h. excipulum with gelatinized textura prismatica; i. anchoring hypha showing the thick-walled base not present in the hairs (a, c, e: T. Kosonen 7129; b, f–i: J. Purhonen 6063-B1; d: DPP-563). — Scale bar = 10 μm. — Drawings: T. Kosonen.
Arachnoscypha aranea (De Not.) Boud. ex Dennis, Mycol. Pap. 32: 87. 1949. — Fig. 1f, 13
Basionym. Peziza aranea De Not., Mem. Reale Accad. Sci. Torino, Ser. 2, 3: 57. 1841.
Synonym. Arachnopeziza aranea (De Not.) Boud. Icon. Mycol. livr. 14: n°. 236, pl. 521. 1907.
Apothecia up to 0.4–0.6 mm diam, white to pale yellow, narrowly attached, no visible stipe, surrounded by a ‘mesh’ of undulating hairs originating from the flanks and by superficially similar anchoring hyphae originating from the base. Hymenium clearly yellow when fresh. Ectal excipulum of textura prismatica with variable wall thickness, towards inner parts of gelatinized textura prismatica, cells c. 8–12 μm long. Hairs 40–100 × 4–6 μm with 2–4 septa, with clearly widened, bulbous, relatively short basal cell, thin-walled and often covered with crystalline exudates; numerous elongated hairs, reaching several hundred μm in length, slightly varying in width, but usually less than 5 μm wide, sparsely septate, with crystalline exudates, tapering towards a 1–2 μm wide apex, often bent at irregular intervals. Asci 40–60 × 5–7 μm, cylindrical to clavate, apical pore MLZ+, arising from distinct croziers. Ascospores 6.8–12.7(–12.9) × 2.0–3.2(–3.3) μm, mean = 10.4 × 2.7 μm, Q = 3.2–4.8(–5.2), mean Q = 3.8 (n = 38, from three populations), cylindrical, sub-fusoid to slightly allantoid, occasionally with one septum. Paraphyses simple or bifurcate near the apice, c. 1–2 μm wide, irregularly undulating. Anchoring hyphae 3–6 μm wide, slightly to clearly thick-walled, with crystalline exudates similar to hairs, sparsely branched.
Specimens examined. FINLAND, Varsinais-Suomi, Salo, Perniö, Lampyöli, on a leaf of a deciduous tree in a ditch, 28 Sept. 2015, S. Huhtinen 15/44 (TUR); Etelä-Häme, Jämsä, Kalmavirta, Hallinmäki, Sammalsuo Nature Reserve, on a large trunk of Populus tremula, 18 Sept. 2013, J. Purhonen 6063 (JYV). – SWEDEN, Västergötland, Skövde, Silverfallet-Karlsfors Nature Reserve, on fallen Acer platanoides leaves, 16 Oct. 2015, T. Kosonen 7129 (S, TUR); same location, 6 Oct. 2017, S. Huhtinen 17/52 (S, TUR). – USA, New York, Ithaca, Cascadilla Creek woods, on wood, 28 Sept. 1947, R. Korf 913 (CUP-K-0237) (sub Arachnopeziza eriobasis); Coy Glen, 3 Oct. 1902, H. Whetzel (DPP 563); Lower Six Miles Creek, on hull of Carya, 2 Oct. 1949, C. Rogerson & R. Korf (CUP-K-0239) (sub A. eriobasis); Ithaca, old burrs of Castanea dentata, c. 1929, H. Whetzel (DPP 11788).
Notes — In the monograph of Arachnopeziza, including both North American and European material, Korf (1951b) considered A. aranea to be a rare species restricted to Castanea. This strict ecology was the main character delimiting A. aranea from Arachnopeziza eriobasis, a species combined into Arachnopeziza in the same study. Korf (1951b) considered these as two closely related species, but A. eriobasis having a less specific ecology. Dennis (1981) recognised Arachnoscypha for A. aranea, but refrained from combining Arachnopeziza eriobasis into Arachnoscypha, although he considered it appropriate. He also considered A. eriobasis as ‘not uncommon’ occurring on various decaying substrates and A. aranea as ‘uncommon’ and restricted to Castanea. His descriptions of the two species are overlapping, as are Korf’s. We were not able to distinguish two species among the material studied here using morphology and therefore we choose to refer our material to A. aranea. None of our recent collections from Finland and Sweden, from which we obtained 100 % identical sequences, were from Castanea. Such a specific ecology is uncommon, but of course not impossible among helotialean fungi. If recent material from Castanea becomes available in future studies it should be further explored if two species exist.
Eupezizella Höhn., Mitt. Bot. Inst. Techn. Hochsch. Wien 3: 61. 1926
Type species. Eupezizella candida (Starbäck) Höhn., Mitt. Bot. Inst. Techn. Hochsch. Wien 3: 61. 1926.
Basionym. Pezizella candida Starbäck, Bih. Kongl. Svenska Vetensk.-Akad. Handl. 21: 30, pl. 1: 16a–c. 1895; non Pezizella candida Velen., Opera Bot. Cech. 4: 117. 1947, nom. illeg. (Art. 53.1).
Synonym. Hyaloscypha candida (Starbäck) Boud., Hist. Classific. Discomyc. Europe: 127. 1907.
Included species: Eupezizella aureliella, E. britannica, E. candida, E. nipponica, E. roseoguttata.
Notes — The genus Eupezizella conforms here to Hyaloscypha subgenus Eupezizella (Huhtinen 1989). It is distinguished from Hyaloscypha by the abundant resinous exudates (Fig. 1g), the predominantly blunt and aseptate hairs and by the lack of overall dextrinoid reactions. Some species and populations show deep amyloid nodules in the hairs and/or in the excipular cells. In very rare cases these nodules may be dextrinoid (Fig. 14b). Contrary to Hyaloscypha, Eupezizella is almost exclusively known from softwoods. Two Japanese collections of E. nipponica Huhtinen (1989; as Hyaloscypha nipponica) originate from hardwood. In the same study, the third cited specimen under H. nipponica seems to represent an unknown species of the new genus Resinoscypha.
Fig. 14.

Examples of hair shapes, inclusions and reactions, and of amyloid nodules in excipulum cells, in Hyaloscyphaceae. a. Thin-walled aseptate hairs, Hyaloscypha spiralis (in water); b. hairs showing amyloid and dextrinoid nodules, Eupezizella aureliella (in MLZ); c–d. dextrinoid glassy hairs in Olla (in MLZ): c. Olla transiens; d. Olla millepunctata; e. septate hairs showing resinous matter inside the hairs (arrows), Resinoscypha variepilosa (in water); f–h. Mimicoscypha lacrimiformis: f. septate hairs with refractive globules; g. purple nodules in ectal excipulum cells (in MLZ); h. bulbous basal cells of hairs with exudates (in water) (a: KH.16.02; b: T. Kosonen 7296; c: U. Söderholm 4829; d: T. Kosonen 7155; e: S. Huhtinen 16/41; f–h. KH.17.02). — Scale bars = 10 μm. — All from fresh material. — Photos: a, e–h. K. Hansen; b–d. T. Kosonen.
Starbäck (1895) described Eupezizella as a subgenus in Pezizella, which makes the name invalid (Art. 21.3). He included three species: Pezizella candida, P. atomaria (= Hyaloscypha aureliella) and P. minor (= Phialina lachnobrachya). When Hyaloscypha was monographed, Eupezizella was combined at subgeneric level under Hyaloscypha, as well as lectotypified (Huhtinen 1989). The taxa included in the subgenus were: Hyaloscypha aureliella, H. britannica, H. britannica var. roseoguttata, H. candida, H. nipponica and H. strobilicola. They are here all, with the exception of H. strobilicola, combined into the genus Eupezizella. Our phylogenetic analyses strongly support the placement of H. aureliella as a sister group to Hyaloscypha s.str. (Fig. 2), confirming the results of Huhtinen (1989). Some of the morphological features of Eupezizella are also characteristic for the new genera Resinoscypha and Mimicoscypha, but the aseptate hairs are a consistent feature distinguishing Eupezizella.
In previous molecular phylogenetic studies H. aureliella has been considered to be nested within Hyaloscypha, because two collections (TNS-F17137 and TNS-F11213) published as H. albohyalina var. albohyalina (Han et al. 2014), formed a sister group to Hyaloscypha including H. aureliella (see also Fehrer et al. 2018). Our morphological studies of TNS-F17137 have, however, convinced us that these do not represent H. albohyalina sensu Huhtinen (1989). Instead they represent a distinct species and genus to be described later when more material becomes available. We refer to Huhtinen (1989) for full descriptions and illustrations of all species here combined in Eupezizella, including their type specimens, because very little additional data has emerged.
Recently, the generic name Cheiromycella was proposed to be used for E. aureliella and closely related species through the synonymy of C. microscopica and E. aureliella (Fehrer et al. 2018). Since there is no sequence material available from either of the generic types (C. speiroidea and E. candida) we find it most appropriate, for the time being, to keep the name Eupezizella for this group of species.
Eupezizella aureliella (Nyl.) T. Kosonen, Huhtinen & K. Hansen, comb. nov. — MycoBank MB835731
Basionym. Peziza aureliella Nyl., Not. Sallsk. Fauna Fl. Fenn. Forh. 10: 49. 1869.
Eupezizella britannica (Huhtinen) T. Kosonen, Huhtinen & K. Hansen, comb. nov. — MycoBank MB835732
Basionym. Hyaloscypha britannica Huhtinen, Karstenia 29: 113. 1990.
Eupezizella nipponica (Huhtinen) T. Kosonen, Huhtinen & K. Hansen, comb. nov. — MycoBank MB835733
Basionym. Hyaloscypha nipponica Huhtinen, Karstenia 29: 155. 1990.
Eupezizella roseoguttata (Huhtinen) T. Kosonen, Huhtinen & K. Hansen, comb. & stat. nov. — MycoBank MB835734
Basionym. Hyaloscypha britannica var. roseoguttata Huhtinen, Karstenia 29: 116. 1990.
Notes — We recognise E. roseoguttata as a distinct species based on morphology. The type collection of H. britannica var. roseoguttata can be distinguished by the combination of two independent morphological characters: the rosy-coloured exudates at the margin and the MLZ-ascal pore. The culture (strain CBS 251.90) of the type is reddish orange, a colour not present in other cultures of Eupezizella (only obtained from E. aureliella so far) (Huhtinen 1989). Also, the single available sequence in GenBank of E. roseoguttata (ITS, MH862208) differs in 20 bp from the ITS sequences of E. aureliella, supporting it as a distinct species. A direct comparison between the ITS and LSU sequences of E. roseoguttata and E. britannica is not possible, because so far only an LSU sequence is available for E. britannica. The LSU region of E. britannica and E. aureliella differs in 10 base pairs. The available sequences strongly suggest E. aureliella, E. britannica and E. roseoguttata to be congeneric.
Hyaloscypha usitata Huhtinen, Karstenia 29: 173. 1990
Holotype. COLOMBIA, Dpto, Antipquia, Medellin, Forest research station Piedras Blancas, on wood, 19 July 1974, Dumont, Haines & Velasquez, Dumont-Co 1591 (NY).
Specimens examined. FINLAND, Pohjois-Häme, Toivakka, Vuorilampi, on a trunk of Betula, 27 Aug. 2015, T. Kosonen 7083 (TUR). – SWEDEN, Härjedalen, Hamrafjällen, a trunk of Betula, 4 Aug. 2015, S. Huhtinen 15/25 (S); Södermanland, Haninge, Tyresta National Park, on a small hardwood trunk, 1 June 2016, S. Huhtinen 16/47 (S); Västra Götland, Falköping, Tidaholm, Forentorpa nature reserve, on a trunk of Betula, 15 Oct. 2015, S. Huhtinen 15/54 (S); Skara, Vadet, on a trunk of Betula or Alnus, 14 Oct. 2015, T. Kosonen 7122 (TUR).
Notes — Hyaloscypha usitata was described from fungarium material from Colombia and Venezuela (Huhtinen 1989). It is here reported for the first time from Europe. The material of H. usitata and H. vitreola studied so far indicate that these two are phylogenetically and morphologically clearly defined, but closely related sister species. The vital characters of H. usitata differ from most other Hyaloscypha species in that the hairs and especially the paraphyses have usually one distinct VB in each cell. This feature was observed in all material studied fresh.
Mimicoscypha T. Kosonen, Huhtinen & K. Hansen, gen. nov. — MycoBank MB835735; Fig. 14f–h, 15, 16, 17, 18
Fig. 15.
Mimicoscypha lacrimiformis (holotype). a. Marginal end cells; b. marginal hairs in CR, the four on right in KOH; c. dry apothecia; d. asci and paraphyses; e. spores; f. ectal excipulum in MLZ showing the amyloid nodules. — Scale bars: a–b, d–f = 10 μm, c = 100 μm. — Drawings: S. Huhtinen.
Fig. 16.
Mimicoscypha lacrimiformis. a. Marginal hairs in CR showing several months old refractive globules; b. marginal end cells; c–d. marginal hairs in CR; e. asci and paraphysis; f. ascospores in CR; g. ectal excipulum in MLZ showing the amyloid nodules; h. dry apothecia (a, f: T. Kosonen 7224; b, d, g–h: S. Huhtinen 17/6; c, e: Huhtinen 17/8). — Scale bars: a–g = 10 μm, h = 100 μm. — Drawings: S. Huhtinen.
Fig. 17.
Mimicoscypha mimica (holotype). a. Marginal hairs, detail showing maximal wall thickness in CR; b. asci and paraphyses; c. ascospores; d. dry apothecia; e. ectal excipulum in CB. — Scale bars: a–c, e = 10 μm, d = 100 μm. — Drawings: S. Huhtinen.
Fig. 18.
Mimicoscypha paludosa. a. Marginal hairs in CR; b. ascospores; c. dry apothecia; d. asci and paraphyses; e. ascospores; f. marginal hairs; g. ectal excipulum in CR (a, g: PRM 817288; b–c, f (right): Clark 11 Sept. 1983; d–f (left): lectotype). — Scale bars: a–b, d–g = 10 μm, c = 100 μm. — Drawings: S. Huhtinen.
Etymology. For mimicking two other genera, Eupezizella and Resinoscypha.
Type species. Mimicoscypha lacrimiformis (Hosoya) T. Kosonen, Huhtinen & K. Hansen.
Included species: M. lacrimiformis, M. mimica, M. paludosa.
A genus with light-coloured, hairy apothecia on wood, bryophytes and plant debris. Hairs with 1–3 septate, thin- to somewhat thick-walled, hyaline, mostly smooth, sometimes with hyaline to yellowish, superficial resin; inside the hairs hyaline, refractive globules variably present in living, occasionally in dried material, amyloid nodules not present. Ectal excipulum of textura angularis – prismatica, typically without dextrinoid reactions, but may show strongly amyloid nodules inside the cells. Asci arising from croziers or simple septa, apical pore MLZ+, 8-spored. Ascospores ellipsoid to oblong-ellipsoid, aseptate to 1-septate. Paraphyses cylindrical.
Notes — Mimicoscypha earns its name by sharing morphological features with both Eupezizella and Resinoscypha, despite it is clearly distinct from these genera based on our multi-gene data (Fig. 2). Delimitation may prove difficult if only morphological features are used. It differs from Eupezizella in the septate hairs, and from both Eupezizella and Resinoscypha in not showing amyloid nodules in the hairs (see further in Notes under Resinoscypha below). These blunt-haired, hyaloscyphaceous taxa have been vaguely placed for decades and a strongly supported placement has not been offered until now. Our solution is based on a combination of morphological characters and molecular phylogenetic results. Mimicoscypha is closely related to the genus Olla and Hyalopeziza nectrioidea (Fig. 2) although species of Olla and H. nectrioidea have very different excipular hairs (see Fig. 14c–d, f). The long slender, thin- to somewhat thick-walled, hyaline hairs, without glassy elements (typical for Olla) or a dextrinoid reaction (typical for both Olla and H. nectrioidea), supports the creation of this new genus.
Mimicoscypha lacrimiformis (Hosoya) T. Kosonen, Huhtinen & K. Hansen, comb. nov. — MycoBank MB835736; Fig. 14f–h, 15, 16
Basionym. Phialina lacrimiformis Hosoya, Mycoscience 38: 181. 1997.
Holotype. JAPAN, Nagano Pref., Sanada-cho, Daimyojin water fall, on decaying wood, 24 May 1993, T. Hosoya (TNS-F-181594) !
Apothecia 0.2–0.5 mm diam, white, disc-shaped and broadly attached, to shallowly cupulate, to shortly stipitate, prominently hairy at the margin. Ectal excipulum of textura prismatica to textura angularis, cell walls thickened, with abundant amyloid nodules in the basal excipulum cells, sometimes present also in cells closer to the margin, basal excipulum covered with brown, crustose resin, not changing in MLZ. Hairs up to 150 μm long, narrowly to broadly conical, straight, thin-walled and hence tardily reviving to their original shape, apex blunt or tapering to 1–2 μm wide, close to base 4–8 μm wide, smooth, without apical solidifications, regularly with 1–2 septa, rarely up to 6–7 septa, occasionally with solitary refractive vacuoles inside, the amount varying between populations from prominent to lacking, external resinous matter gluing hairs together, present in water mounts or totally lacking, on lower flanks hairs occasionally completely covered in similar brown crustose resin as basal excipular cells. Asci 40–55 × 4–8 μm, cylindrical, 8-spored, apical pore MLZ+, arising from croziers. Ascospores 7.2–12.6(–14) × 2.0–3.0 μm, mean 9.9 × 2.5 μm, Q = 3.2–4.8, mean Q = 4.0 (n = 28, from 3 populations), ellipsoid to oblong-ellipsoid to cylindrical, often slightly allantoid, aseptate, more rarely 1-septate, with or without refractive vacuoles when fresh in water. Paraphyses cylindrical, 1–2 μm wide.
Specimens examined. SWEDEN, Östergötland, Ödeshög, Mörkahålkärrets Nature Reserve, on man-made coarse, coniferous wood chips, 26 Apr. 2017, S. Huhtinen 17/6 (S, TUR); same date, ecology and location, S. Huhtinen 17/7 (S, TUR); same date, ecology and location, S. Huhtinen 17/8 (S, TUR); same date, ecology and location, K. Hansen KH.17.02 (S, TUR); on a decayed trunk of Picea abies, T. Kosonen 7224 (S, TUR).
Notes — The absence of bright yellow pigment, typical for the genus Phialina (Huhtinen 1989), excludes its original placement by Hosoya (Hosoya & Otani 1997). In some populations from Sweden there were clear refractive globules in the hairs when studied in fresh condition (e.g., Fig. 14f, 16a), whereas other populations, collected simultaneously from the same locality, lacked the character completely. The four collections sequenced by us, had all identical ITS and LSU sequences. The RPB1, RPB2 and TEF-1α regions were in addition sequenced from two of these populations (S. Huhtinen 17/8, with refractive vacuoles and S. Huhtinen 17/7, without the vacuoles). All the sequences were identical between the two populations. The presence of amyloid nodules in the ectal excipulum (Fig. 14g, 15f, 16g) is a character common with Eupezizella and Resinoscypha. Swedish material of M. lacrimiformis shows marked variation in the amount of crustose, brown resin present on basal excipulum cells and basal hairs (Fig. 14h). Even in the same collection the apothecia vary from faintly coloured (as in the holotype) to basally dark brown. The species is new to Europe.
Mimicoscypha mimica T. Kosonen, Huhtinen & K. Hansen, nom. nov. — MycoBank MB835737; Fig. 17
Etymology. For mimicking Mimicoscypha paludosa.
Synonym. Hyaloscypha paludosa Dennis, Kew Bull. 16: 325. 1962, non Hyaloscypha paludosa Velen. 1934, Monogr. Discom. Bohemiae Pars. 2: pl. xiv, f. 25, nom. illeg. (Art. 33.1, lapsus calami pro Eriopeziza paludosa).
Holotype. UK, Derbyshire, Kinder Scout, Ashop Clough, on dead culms of Cyperaceae, 9 July 1960, J.T. Palmer (K) !
Apothecia 0.2–0.5 mm diam, 0.2–0.4 mm high, cupulate, narrowing towards base, with amber yellow (Cailleux K87) flanks and disc when dry, hair cover prominent, white. Ectal excipulum of textura prismatica, with small, deep amyloid nodules in the cells, walls thickened, at places up to 2 μm, walls CB-, MLZ-, CR-. Hairs 80–150 × 3–4 μm, cylindrical-conical with a blunt apex, 1–5-septate, somewhat firm-walled, smooth or bearing very small amount of yellowish brown, resinous substance seen in a water mount, lost in MLZ and CR, walls CB-, MLZ-, CR+. Asci cylindrical-clavate, 8-spored, 70–83 × 5.0–5.7 μm, apical pore deep MLZ+ even without KOH pretreatment, arising from croziers. Ascospores 8.5–14.0 × 2.0–3.0 μm in MLZ, mean 10.6 × 2.4 μm (n = 14), Q = 3.5–5.4, mean Q = 4.5, oblong-ellipsoid, slightly bent, narrowly tapering towards the ends, multiguttulate, often with one septum, sometimes already in asci. Paraphyses cylindrical, 2 μm wide.
Notes — Describing Hyaloscypha paludosa, Dennis (1962) missed the frequent septa in the hairs (Fig. 17a). As he also overemphasized the narrowness of the hairs, it is understandable that he placed the new taxon in the genus Hyaloscypha. This placement was later followed by Raitviir (1970), but criticized by Huhtinen (1989). This taxon has seldom been collected. Dennis (1962) mentioned another collection from UK. One additional collection from the Netherlands (leg. Rommelaars in herb. S. Helleman) may belong here, but it shows dextrinoid nodules (or wall thickenings) in the excipulum cells. This variation between dextrinoid and amyloid has proved to be very rare. For example, in E. aureliella c. 29 % of the populations studied showed the deep amyloid nodules and only 3 % dextrinoid (Huhtinen 1989).
Mimicoscypha paludosa (Velen.) T. Kosonen, Huhtinen & K. Hansen, comb. nov. — MycoBank MB835738; Fig. 18
Basionym. Eriopezia paludosa (as Eriopeziza paludosa) Velen., Monogr. Discom. Bohemiae Pars. 1: 266. 1934.
Synonym. Hyaloscypha paludosa Velen., Monogr. Discom. Bohemiae Pars. 2: pl. xiv, f. 25. 1934. nom. illeg. (Art. 33.1., lapsus calami pro Eriopezia paludosa).
Lectotype designated here: CZECH REPUBLIC, Mnihovice, Svojětice, 11 July 1927, J. Velenovský (PRM 147457) ! MycoBank MBT392711.
Apothecia 0.2–0.4 mm diam, shortly stipitate to seemingly sessile, stipe hidden below the disc, ochraceous yellow when dry (between L80 and L87), hair cover white. Ectal excipulum of textura prismatica-angularis, lacking MLZ+ nodules. Hairs 60–130 × 2–3.5 μm, regularly reaching 100 μm in length, cylindrical, tapering to a blunt apex, thin-walled, ranging from aseptate to more often 1–3-septate, rarely up to 5-septate, basally widened up to 5–6 μm, in water often tightly glued together, hyaline resin sometimes present, hard to notice even in a water mount where hairs mainly smooth and empty, as well as in MLZ, CB and CR, rarely some hair contents and resin cover CB+ and CR+. Asci cylindrical, 40–60 × 4–6 μm, 8-spored, apical pore clearly MLZ+, arising from simple septa. Ascospores (6.8–)7.7–9.7(–10.0) × 1.9–2.2 μm, mean 9.1 × 2.2 μm, Q = 3.3–5.0, mean Q = 4.1 (n = 50, from three populations), ellipsoid to oblong-ellipsoid, aseptate, rarely 1-septate, eguttulate to guttulate. Paraphyses mainly cylindrical, 1–2 μm wide, but in one specimen also moniliform and subcapitate paraphyses are common.
Specimens examined. CZECH REPUBLIC, Bohemia, Revnice, amongst pine needles buried in Sphagnum, 10 Oct. 1948, M. Svrček (PRM 817288). – FINLAND, Varsinais-Suomi, Nauvo, Seili, seashore thicket with Phragmites, on debris, 17 Aug. 1980, J. Vauras & S. Huhtinen s.n. (TUR). – UK, Yorkshire, Skipwith, on Juncus stem, 11 Sept. 1983, M.C. Clark s.n. (K).
Notes — Mimicoscypha paludosa is distinguished from M. mimica by asci that arise from simple septa and on average smaller spores. The wall of the hairs also differs, being somewhat thick-walled and CR+ in M. mimica. All observations are from wet environments, from e.g., Sphagnum, Juncus and dead bryophytes.
Apparently, this species has been overlooked in the literature and the few reports have been hiding under the later homonym Hyaloscypha paludosa Dennis (1962) (= M. mimica). The cited, abundant type in PRM has markings by Prof. Svrček, indicating it as a syntype and hence valid for lectotypification, although the exact collection locality, mentioned in the protoloque, seems to be different. But as many collection sites were included in the protoloque by Velenovský and all with ‘pr.’ (prope = near) and Svojětice is a neighbouring municipality to, e.g., Struharov and Ondřejov (mentioned in the protoloque) we feel it safe enough to validate Svrček’s selection.
The name Hyaloscypha paludosa was not explicitly accepted by Velenovský (1934) (Art. 33.1.). The proof of this is rather obvious. Under Eriopeziza paludosa, Velenovský refers to his plate 14: 25, where the legend erroneously bears the name Hyaloscypha paludosa. But only E. paludosa is listed in the index. Furthermore, H. paludosa is not treated in the texts on Hyaloscypha (Velenovský 1934). Looking at the lectotype cover markings of E. paludosa in PRM, made by Svrček, one can see that he was of the same opinion. In this case he had added to the lectotype: ‘Hyaloscypha paludosa Vel. in herb. = Eriopeziza paludosa’, so apparently he also thought that Velenovský had changed his mind of the placement at some point, but forgot to change it in the legend of plate 14.
Resinoscypha T. Kosonen, Huhtinen & K. Hansen, gen. nov. — MycoBank MB835739; Fig. 14e, 19, 20
Fig. 19.
Resinoscypha monoseptata. a. Marginal hairs in lactic acid, leftmost in CB showing the CB+ resinous substance, the two details in MLZ showing deep amyloid nodules; b. spores, upper five in CB, lower four in lactic acid; c. asci and paraphyses in lactic acid, paraphyse on right in CB showing the scanty resinous substance; d. spores, upper four in CB, lower two in MLZ; e. marginal hairs in lactic acid, leftmost in CB showing the CB+ resin; f. dry apothecium (a–c: isotype, Herb. Galán 6154; d–f: CUP 59279). — Scale bars: a–e = 10 μm, f = 100 μm. — Drawings: S. Huhtinen.
Fig. 20.
Resinoscypha variepilosa. a. Marginal hairs in CR, rightmost in CB showing the CB+ resinous contents; b. asci and paraphyses; c. spores in CB (left) and CR (right); d. ectal excipulum showing the amyloid nodules; e. marginal hairs showing the brown resinous substance inside, in MLZ, hair with asterisk (*) in water, one hair with basal amyloid nodule (arrow); f. spores in MLZ; g. asci and paraphysis; h. ectal excipulum showing the amyloid nodules; i. marginal hairs in CR, rightmost hair in water; j. spores in CR; k. short marginal hairs in MLZ showing one rare amyloid nodule; l. dry apothecia; m. marginal hairs in MLZ, some showing the amyloid nodules (arrows); n. asci and paraphyses; o. spores in CB, three lower in KOH (a–d: holotype; e–h: S. Huhtinen 87/131; i–k: S. Huhtinen 16/41; l–o: S. Huhtinen 86/60). — Scale bars: a–k, m–o = 10 μm, l = 100 μm. — Drawings: S. Huhtinen.
Etymology. Referring to the cyanophilous resinous substance inside the hairs.
Type species. Resinoscypha variepilosa (R. Galán & Raitv.) T. Kosonen, Huhtinen & K. Hansen.
Included species. R. monoseptata, R. variepilosa.
A light-coloured, hairy, lignicolous genus characterised by long, smooth, hyaline, thin-walled, regularly somewhat flexuous, cylindrical to narrowly conical hairs with 1–4 septa, tapering towards a blunt apex and showing cyanophilous, non-glassy resinous substance especially inside, shorter marginal hairs very variable in shape, refractive globules lacking. Excipulum of textura angularis-prismatica, without dextrinoid reactions, but both hairs and excipular cells may contain strongly amyloid nodules. Asci arising from croziers, apical pore MLZ+, 8-spored. Ascospores ellipsoid to oblong-ellipsoid, aseptate to 1-septate. Paraphyses cylindrical.
Notes — Emended descriptions of both included species were provided by Huhtinen (1993b) and also earlier by Huhtinen (1987b) and are therefore not reproduced here, but new additional illustrations are provided (Fig. 14e, 19, 20). Resinoscypha is phylogenetically a distinct genus, sister to all other taxa in the Hyaloscyphaceae s.str. clade (Fig. 2). The two species, originally described in Protounguicularia by Raitviir & Galán (1986), were separated from the type species, P. brevicapitata (now Olla transiens), mainly based on the notable differences in hair inclusions (Huhtinen 1987a). In O. transiens the material inside the hairs is glassy and strongly dextrinoid (Fig. 14c). The true glassiness is verified by observations in CB, where the colour does not penetrate the glassy substance. The two Resinoscypha species have resinous material concentrated at the apex of the hairs and at septal areas (Fig. 14e, 19a, e, 20a, e). These inclusions are MLZ- and strongly CB+. The taxonomic value of these differences, as well as the differences in ascal development (arising from simple septa in the type species versus from clear croziers in the others), passed largely unnoticed by the original authors. The characteristic CB+ resin inside the hairs, combined with the variably shaped and septate hairs, delimit this new genus from Eupezizella and Mimicoscypha, which both may have a variable number of amyloid nodules in the excipula.
When combining the two species to Arachnopeziza, Huhtinen (1987a) emphasized the cylindrical septate hairs. It was suggested, however, that these species occupy ‘a transitional position’ between Arachnopezizoideae and Hyaloscyphaceae and the author contemplated on alternative solutions (e.g., a separate genus).
Resinoscypha monoseptata (R. Galán & Raitv.) T. Kosonen, Huhtinen & K. Hansen, comb. nov. — MycoBank MB835741; Fig. 19
Basionym. Protounguicularia monoseptata R. Galán & Raitv., Int. J. Mycol. Lichenol. 2: 224. 1986.
Synonym. Arachnopeziza monoseptata (R. Galán & Raitv.) Huhtinen, Mycotaxon 30: 18. 1987.
Holotype. SPAIN, Malaga, Ronda, Sierra de las Nieves, 1500 m a.s.l., decorticated wood of Abies pinsapo, 2 Apr. 1984, R. Galán (TAA; Isotype in Herb. Galán 6154) !
Specimens examined. FINLAND, Inarin Lappi, Utsjoki, Kevo, on an old coniferous board lying in a collapsed cellar, 8 Aug. 1991, S. Huhtinen 91/29 (TUR 105080, sub A. cf. monoseptata). – NORWAY, Finnmark, Karasjåkka, near old customs house, on a coniferous board with Tomentella sp., 22 Aug. 1978, S. Sivertsen et al. s.n. (CUP 59279); Spitsbergen, Ny-Ålesund, old mining area near the village, on a coniferous board, 11 Aug. 1988, S. Huhtinen 88/10 (TUR 107784, sub. A. cf. monoseptata).
Notes — We have no sequences of this species, but it is clearly congeneric with R. variepilosa. The very obvious morphological features of the type specimens (the CB+ inclusions in the hairs and the variable hair morphology) linking the two taxa have been discussed before (e.g., Huhtinen 1987a, b). Some variability has since been observed, as summarized by Huhtinen (1993b). Resinoscypha monoseptata differs from the generic type species by longer (8.6–13.6 × 2–3 μm) and occasionally 1-septate spores (Raitviir & Galán 1986, see also our Fig. 19b, d, 20c, j, o). Interestingly, all four collections of R. monoseptata were collected from arctic/alpine to subarctic areas and always on decorticated softwood, especially construction timber. Resinoscypha variepilosa shows more variability, growing both on hardwood and softwood.
Resinoscypha variepilosa (R. Galán & Raitv.) T. Kosonen, Huhtinen & K. Hansen, comb. nov. — MycoBank MB835740; Fig. 14e, 20
Basionym. Protounguicularia variepilosa R. Galán & Raitv., Int. J. Mycol. Lichenol. 2: 223. 1986.
Synonym. Arachnopeziza variepilosa (R. Galán & Raitv.) Huhtinen, Mycotaxon 30: 14. 1987.
Holotype. SPAIN, Cádiz, Grazalema, Sierra del Pinar de Grazalema, 1200 m a.s.l., on dead wood of Abies pinsapo, 27 Nov. 1982, R. Galán (Herb. Galán 6117) !
Specimens examined. CANADA, Yukon, Kluane Lake, Outpost Mountain, on fallen, decayed trunk of Picea glauca, 19 Aug. 1987, S. Huhtinen 87/131 (TUR 99569). – DENMARK, Zealand, Sorø, Suserup forest reserve, on decorticated trunk of Fagus, 8 May 1994, J. Heilmann-Clausen 04-035 (TUR 124469). – SLOVAKIA, Bratislava Forest Park, along river Vydrica, on decorticated hardwood, 4 Aug. 1986, P. Lizon & S. Huhtinen 86/60 (TUR 92119). – SWEDEN, Upland, Haninge, Tyresta National Park, Bylsjöbäcken, on coniferous trunk, 1 June 2016, T. Kosonen & S. Huhtinen 16/41 (S, TUR). – UK, England, Hertfordshire, Wadesmill, on decayed wood of Ulmus, 24 Nov. 2010, K. Robinson (TUR 193603); Gloucestershire, Forest of Dean, Coalpit Hill, on a trunk of Betula pendula, 18 Apr. 2012, K. Robinson (TUR 196799).
Notes — This is a distinct but rarely collected species with collections from two continents. Based on morphology alone it is difficult to assign a suitable genus to this species. However, in addition to the CB+ resin (Fig. 20a), also the very variable shape of (especially the shorter marginal) hairs is a characteristic feature, already shown by Raitviir & Galán (1986: f. 8–11 of the holotype). The two collections, S. Huhtinen 87/131 and S. Huhtinen 16/41, from Canada and Sweden (Fig. 20e–k), have identical ITS and LSU sequences. The amount of resin on and in the hairs is highly variable even within one population, ranging from hairs practically without resin to hairs densely covered in resin (Fig. 20a, e, i). An ITS-LSU sequence of the Canadian sample has been previously deposited to GenBank (isolate M337 / collection S. Huhtinen 87/131: ITS, EU940163 and LSU, EU940086) (Stenroos et al. 2010). It was an erroneously annotated combination of two different species, the ITS presenting the correct species and the LSU apparently that of Pezoloma ciliifera of uncertain origin. The correct sequences were obtained by re-sequencing the original extraction of the isolate M337. The two sequenced samples were both from softwood.
DISCUSSION
The composition of the family Arachnopezizaceae
Our datasets included multiple sequences of Arachnopeziza species, the monotypic Eriopezia and Arachnoscypha aranea. Of these, Arachnoscypha is clearly shown to be distant to Arachnopezizaceae. Eriopezia forms a robust monophyletic group with species of Arachnopeziza and these two genera constitute, with the current sampling, the core Arachnopezizaceae. The included species produce a subiculum and usually apothecial hairs that are thin- to thick-walled and multiseptate. The ectal excipulum is mostly of textura angularis with somewhat thick to thick walls. Reinforcing the results by Han et al. (2014), our five-gene analyses suggest that the subiculum is a shared derived character for Arachnopezizaceae. It excludes species lacking a subiculum, such as A. variepilosa and A. monoseptata, now placed in Resinoscypha.
To our knowledge, a close relationship between Arachnoscypha and Polydesmia has not been proposed previously. Earlier authors have treated Polydesmia as a monogeneric unit without a well-established placement within Helotiales (Dennis 1960, 1968, Verkley 2005). At one point, Polydesmia was assigned to Arachnopezizoideae in the tribe Polydesmieae together with Eriopezia and Parachnopeziza (Korf 1978). The main point was, however, to include Polydesmia in Hyaloscyphaceae and no specific arguments were given for a close relationship of Polydesmia and Arachnopeziza, which, at that time, included Arachnoscypha (Korf 1978). Our results confirm, with high support, the recent multi-gene phylogenetic results by Johnston et al. (2019) that resolves Polydesmia as a sister taxon to Chlorociboria(ceae).
Two of the genera included in the recent description of Arachnopezizaceae (Baral 2015), Austropezia and Parachnopeziza, have not been included in robust molecular phylogenies. Based on analysis of the ITS-region across all of the Leotiomycetes, Johnston et al. (2019) suggested that A. samuelsii (the type species of Austropezia) does not belong in Arachnopezizaceae. There are some recent observations of other taxa that are, in preliminary analyses, early diverging in a monophyletic clade including Arachnopezizaceae. For example, Sokolski et al. (2006) have reported a black spruce (Picea mariana) needle endophyte with an aquatic stage, which, based on ITS/LSU sequences, appears to be related to Arachnopeziza. Moreover, the ITS/LSU sequences from Durella melanochlora and D. macrospora suggest that these are related to Arachnopezizaceae (Johnston et al. 2019). In addition to these, we have made some collections of herbicolous helotioid populations (unpubl. data) without distinctive morphological characters to assign them to known genera, but which likewise show strong phylogenetic affinity to Arachnopezizaceae.
Morphological characters and diversity in Arachnopeziza
The fact, that apothecia of A. sphagniseda have no true hairs, underlines the significance of the somewhat thick-walled subiculum as a unifying character among the members of Arachnopeziza. It is often less obvious among the species producing apothecia on bryophytes, but still regularly observed at least in mounts of apothecia. A subiculum is also produced in other groups of ascomycetes, for example, in Mollisia (Helotiales) as well as in Pyronema and Byssonectria (Pezizomycetes), but appears to be a result of convergent evolution. The subicular hyphae in Arachnopeziaceae are characteristic. The hyphae are thick-walled compared to the apothecial cells, but less than 2 μm wide and hyaline. The surface of the hyphae bears minute warts and is often partly covered with exudates similar to those on the apothecial hairs. In the literature, apothecia are sometimes described as being seated on the subiculum (Korf 1978). This is related to the idea of a ‘false’ and ‘true’ subiculum, where apothecia are borne on a ‘true’ subiculum, and a ‘false’ subiculum is a hyphal mat only surrounding the base of the apothecia. We find this kind of terminology problematic. The ontogeny of subicular hyphae in different genera has not been studied comprehensively and compared. There is large variation in the amount of subiculum produced among species of Arachnopeziza, but it is fairly constant for a given species. For example, A. trabinelloides has often very little subiculum, whereas A. aurelia or E. caesia always have copious subiculum.
Although Arachnoscypha is considered to have a subiculum there are some distinct differences between the aerial hyphae surrounding or connected to apothecia of Arachnoscypha and members of Arachnopezizaceae. Based on observations on fresh and dried material, we conclude that in Arachnoscypha the hyphae originate mainly from the lower part of the apothecia and there is no ‘hyphal web’ (i.e., subiculum) arising from the substrate surface even in the immediate vicinity of the apothecia as in Arachnopeziza. The subicular hypha in Arachnopeziza is of fairly even width and relatively thick-walled for the whole length and thus easily distinguishable from other apothecial elements. In Arachnoscypha, the basal parts of the hypha are thick-walled and branch as in Arachnopeziza, but towards the apices of the hypha are thin-walled, tapering and thoroughly covered in exudates and morphologically inseparable from the hairs. Based on these differences, we consider the thick-walled hyphae originating from the base of the apothecia in Arachnoscypha to be anchoring hyphae or simply basal mycelium.
All the species currently recognised in Arachnopeziza, including the species accepted in this study, have asci arising from croziers. The feature is mentioned in the description of the family (Baral 2015), but deserves to be noted, because in many species-rich helotialean genera there is variability in ascus development between species (e.g., in Hyaloscypha, Hymenoscyphus). The spore and excipulum characters show variation, but spores with at least one septum exist in most of the populations and the excipulum is at least partly thick-walled.
Using genealogical concordance (GCPSR), we recognise eight species within Arachnopeziza (indicated with triangles at the nodes in Fig. 3, Table 2). An additional four species are recognised based on single collections, because these are genetically divergent from their sisters (Fig. 3). Based on ITS (± LSU) sequences available in GenBank, we accept an additional species, A. aurelia (Fig. 4). However, the genetic exclusivity of this species still needs to be tested. To fully resolve the Arachnopeziza species limits and diversity requires further sampling. Several morphologically established species were lacking in our study and some were likely too narrowly sampled geographically, with our focus in Northwestern Europe. Species described from North America, such as A. cornuta, A. fitzpatrickii and A. major, treated in the Arachnopeziza monograph (Korf 1951b), should be recollected and investigated. Furthermore, three species of Arachnopeziza have been described fairly recently from subtropical Asia: A. colachna, A. hiemalis and A. subnuda (Korf & Zhuang 1985, Yu & Zhuang 2002, Wang 2009), and no sequences are available for these. Based on the descriptions and available literature, our estimate is that on top of the species treated in this study, there are c. 10–20 species described and assigned to Arachnopeziza that should be investigated and included in molecular studies to confirm their identity and phylogenetic relationships. Based on documents of Arachnopeziza species on softwood (Korf 1951b), together with our observations and some of Raitviir’s unpublished collections (in TAAM) from Siberia, we suggest that conifers are likely to harbour some undiscovered Arachnopeziza species that should be further searched for and studied.
Of the four clades in Arachnopeziza supported by our five-gene phylogeny (Fig. 3), two are composed solely of species with a strong association to bryophytes, i.e., the A. japonica clade and A. sphagniseda. The A. delicatula clade has also at least one species producing apothecia on Sphagnum. Our observations show that in Arachnopeziza a substantial proportion of the known species grow on or in bryophytes and sequences from environmental sampling indicates an unrecognised diversity (Fig. 4, around Arachnopeziza sp. ‘a’). The relationship of bryophytes and these fungi is not fully understood (e.g., Stenroos et al. 2010).
The A. leonina clade appears to be composed solely of species forming apothecia on wood (Fig. 3). Many of the wood-associated species produce excessive amounts of resin especially on the hairs, resulting in apothecia with a strong, yellow-orange overall appearance (Fig. 1b, c). The ecological spectrum of the studied species is relatively wide. Species such as, for example, A. floriphila (Baral 1989; on dead flowers of Fagus sylvaticus) or A. groenlandica (Raitviir 2003; dead branches of Salix glauca) expand the spectrum even further. Arachnopeziza is also one of the inoperculate genera that have unrecognised species present in the rhizobiome of woody plants (GenBank: LC190976, see also Fig. 4).
For barcoding purposes the ITS-region in Arachnopeziza has enough variation to provide a good estimate on the species identity. The LSU-region is conservative, and sequences differ often in only few single nucleotides between species and do not offer a reliable mean for species identification. RPB1 proved to have a strong species recognition power, resolving all species with high support values (Table 2). Due to missing data we were not able to fully judge the RPB2 and TEF-1α as barcoding regions, but both regions seem promising. With the high amplification success of the short RPB1 region, we suggest it may serve as the best barcoding locus for Arachnopeziza species.
Towards a natural Hyaloscyphaceae
Based on our results Hyaloscyphaceae s.str. embraces Hyaloscypha, Eupezizella, Olla, Mimicoscypha and Resinoscypha. In addition, several species currently accepted in Hyalopeziza belong to this clade. The genera included have hyaline or whitish to greyish apothecia, and they all have hairs, which are often dextrinoid. The hair morphology varies, from cylindrical to tapering, from aseptate to multiseptate, as well as ranging from thin-walled to thick-walled or to solidified (‘glassy’). The ectal excipulum is of predominantly thin-walled textura prismatica, which is occasionally dextrinoid or has small amyloid inclusions. Most species have been considered saprobic, but recent molecular phylogenetic studies and in vitro re-synthesis experiments (Fehrer et al. 2018) have shown some species to have the ability to form ericoid mycorrhizae, ectomycorrhizae, and/or to grow in rhizoids of liverworts. The majority of the species are lignicolous or herbicolous, and some occur on bryophytes. Strictly foliicolous species are rare.
Our strongest estimate of the phylogeny suggests Hyaloscyphaceae s.str. is sister to a clade of Arachnopezizaceae and Amicodisca (Fig. 2). Based on analyses of 3 156 single copy genes, but with a very limited taxon sampling, Johnston et al. (2019) recently suggested that the closest sister group to Arachnopezizaceae is Erysiphaceae. We did not include members of Erysiphaceae in our analyses, but otherwise our results are in general consistent with those of Johnston et al. (2019).
Hyaloscypha encompass c. 35 recognised species (Huhtinen 1989, Fehrer et al. 2018). It is the most species-rich genus in the family. Based on our analyses, including nine species selected from a larger (unpublished) sampling, Hyaloscypha species form a monophyletic clade. The genus is well defined morphologically: the hairs of Hyaloscypha species do not have septa in the protruding part, i.e., the hairs are aseptate. There are taxa, closely related to Hyaloscypha, that have caused confusion in the delimitation of the genus. For example, the undescribed taxon represented by TNS-F-17137 (as H. albohyalina in Han et al. 2014; see our Taxonomy section under Eupezizella) has thin-walled, cylindrical hairs as in Hyaloscypha, but they are multiseptate, unlike in Hyaloscypha or in Eupezizella.
Another example is Hyaloscypha leuconica var. leuconica and var. bulbopilosa that were combined into Hyalopeziza (Raitviir 2004) based on partially thick, glassy walls typical in many populations (Huhtinen 1989). Most likely, material matching Raitviir’s description is represented by for example KUS-F-52474 (Han et al. 2014). We have, however, sequences of populations that fit the morphological concept of Hyaloscypha leuconica var. leuconica (asci arising from simple septa) as well as H. leuconica var. bulbopilosa (asci arising from croziers). Based on our phylogenetic analyses, these are members of Hyaloscypha (results not shown for var. bulbopilosa). Our preliminary phylogenetic analyses also suggest that these varieties represent separate species. In Huhtinen (1989) the morphological concept of Hyaloscypha leuconica var. leuconica hairs was wide and included also collections with relatively thick glassy walls, perhaps morphologically close to KUS-F-52474. We have not been granted a study of the KUS-material, but it most likely represents an undescribed species related to species in the clade of Hyalopeziza nectrioidea, Mimicoscypha and Olla (Fig. 2).
The morphological delimitation of Eupezizella largely follows Huhtinen (1989; as a subgenus). Hyaloscypha and Eupezizella both have aseptate apothecial hairs, but in Eupezizella the hairs are always blunt and are never dextrinoid. The occasional amyloid nodules in the excipulum of Eupezizella have not been observed in Hyaloscypha. The latter character is shared with some of the other members of Hyaloscyphaceae s.str. (e.g., Mimicoscypha). Eupezizella is highly supported as a sister group to Hyaloscypha.
The erection of a new genus, Mimicoscypha, solves some long-lasting taxonomic questions among the hyaloscyphoid taxa. The species, Mimicoscypha paludosa and M. mimica were previously considered to belong to Hyaloscypha (Velenovský 1934, Dennis 1962). Although the multiseptate hairs indicate that these species do not belong to Hyaloscypha or Eupezizella, there has been no suitable genus for these species with long (sparsely) multiseptate thin-walled tapering hairs (Huhtinen 1989). The observed variation in vital taxonomy of M. lacrimiformis is interesting, but it should be interpreted with caution. Vacuolar bodies in paraphyses and often in hairs have been suggested as a character for several members of Pezizellaceae (Baral 2016) and there are only few observations of vacuolar bodies in strictly hyaloscyphoid genera. Mimicoscypha is forming a group with Olla and Hyalopeziza nectrioidea (Fig. 2). They all have multiseptate hairs, but Olla and H. nectrioidea differ from Mimicoscypha in having dextrinoid hairs. Since the type species of Hyalopeziza, H. ciliata, is not related to the species of Hyalopeziza nested within Hyaloscyphaceae s.str. (unpubl. data), eventually a new genus is needed for these. Hyalopeziza alni has similarly dextrinoid, multiseptate hairs, but it is resolved outside the Mimicoscypha-Olla clade (Fig. 2). Also, for example Han et al. (2014) have two species identified as ‘Hyalopeziza sp.’ in their analyses that are possibly related to this clade. Based on morphological characters, Hyalopeziza corticicola with dextrinoid hairs, is most likely another closely related species. In our five-gene phylogeny the type species of Olla (O. millepuctata) and Protounguicularia (P. brevicapitata = O. transiens) form a supported monophyletic group and therefore we follow here Baral (1993) and treat Protounguicularia as a synonym of Olla. Nevertheless, it should be noted that these two representatives of Olla are morphologically very different (see Fig. 14c, d for their apothecial hairs) and the long branches leading to these species suggest they diverged a long time ago (Fig. 2). ‘Hyaloscyphaceae sp.’ (SH 16/40) presents an undescribed lignicolous species. It has multiseptate, thin-walled hairs, no dextrinoid reactions and the spores are characteristically large, measuring close to 20 μm in length. We have no suggestion for a genus that could host such a species. It could be described as a monotypic genus, but with a single collection only, we refrain from doing so.
The dextrinoid reaction of hairs and excipulum cells appears to be a unique character for Hyaloscyphaceae s.str. As discussed above, some genera lack the reaction completely and there is also variation inside some genera, e.g., in Hyaloscypha, Huhtinen (1989) lists nine species with dextrinoid hairs, five without, and eight occasionally with dextrinoid hairs. Importantly, in Hyaloscyphaceae, species with solidified hair walls always show a dextrinoid reaction. Resinoscypha has some unique features within Hyaloscyphaceae, i.e., the CB+ resinous inclusions in the hairs (Fig. 19, 20) and our results suggest it is an early diverging lineage within the family.
Other genera and species associated with Hyaloscyphaceae s.str.
Our datasets did not include recognised members of Psilocistella. However, based on our preliminary analyses of the ITS/LSU region, P. quercina and P. vernalis belong to Hyaloscyphaceae s.str. As long as the generic type P. obsoleta is unavailable for phylogenetic analysis, it is difficult to treat the morphologically rather diverse genus with necessary certainty. Psilocistella obsoleta, with the relatively short, aseptate hairs, resembles morphologically P. quercina, whereas P. vernalis has relatively long multiseptate hairs (Svrček 1977, 1985, Huhtinen 1993a, Quijada et al. 2014). We have observations on taxa that based on phylogenetic analyses, nest in Hyaloscyphaceae s.str., close to Hyaloscypha and Eupezizella. They are morphologically similar to Cistella, with short, thin-walled lageniform hairs with small spines. The type of Cistella (C. dentata) has not been sequenced, but sequences available from other species of Cistella indicate these do not belong to Hyaloscyphaceae s.str., but are more closely related to Urceolella (Han et al. 2014, Johnston et al. 2019).
Acknowledgments
We are thankful to David Hawksworth and Alexander Sennikov for advice on nomenclatural issues; to the curators and other staff at CUP, H, K, NMS, S, TAAM, TNS and TUR for arranging loans; to Joseba Castillo, Bernard Clesse, Jason Karakehian, Otto Miettinen, Marja Pennanen, Jenna Purhonen, Kadri Pärtel, Elisabeth Stöckli, Unto Söderholm, Elina Varis and Ilse Wendelin and many others for providing specimens; to Nelly Llerena for macrophotography; to Gustavo Romero and Xianghua Wang for advice on scanning the line drawings; and to Kadri Pärtel, Luis Quijada and one anonymous reviewer for their valuable comments. The Swedish Taxonomy Initiative provided funding for this research by a grant to K.H. and S.H. (grant no. 2013-138).
REFERENCES
- Baral H-O. 1987. Der Apikalapparat der Helotiales. Eine lichtmikroskopische Studie über Arten mit Amyloidring. Zeitschrift für Mykologie 53: 119–136. [Google Scholar]
- Baral H-O. 1989. Beiträge zur Taxonomie der Discomyceten I. Zeitschrift für Mykologie 55: 119–130. [Google Scholar]
- Baral H-O. 1992. Vital versus herbarium taxonomy: morphological differences between living and dead cells of ascomycetes, and their taxonomic implications. Mycotaxon 44: 333–390. [Google Scholar]
- Baral H-O. 1993. Beiträge zur Taxonomie der Discomyceten III. Zeitschrift für Mykologie 59: 3–22. [Google Scholar]
- Baral H-O. 2015. Nomenclatural novelties. Index Fungorum 225: 1–3. [Google Scholar]
- Baral H-O. 2016. Leotiomycetes. In: Jaklitsch W, Baral H-O, Lücking R, et al. (eds), Syllabus of plant families – A. Engler’s Syllabus der Pflanzenfamilien Part 1/2: 157–205. 13th edn. Borntraeger Science Publishers, Germany. [Google Scholar]
- Baral H-O, Krieglsteiner GJ. 1985. Bausteine zu einer Askomyzeten-Flora der Bundesrepublik Deutschland: in Süddeutschland gefundene inoperculate Diskomyceten mit taxonomischen, ökologischen und chorologischen Hinweisen. Beihefte zur Zeitschrift für Mykologie 6: 1–226. [Google Scholar]
- Baral H-O, Rämä T. 2015. Morphological update on Calycina marina (Pezizellaceae, Helotiales, Leotiomycetes), a new combination for Laetinaevia marina. Botanica Marina 58: 523–534. [Google Scholar]
- Boudier JLÉ. 1885. Nouvelle classification naturelle des Discomycètes charnus. Bulletin de la Société Mycologique de France 1: 91–120. [Google Scholar]
- Boudier JLÉ. 1907. Histoire et classification des Discomycètes d’Europe. Paris, France. [Google Scholar]
- Boudier JLÉ. 1910. Icones Mycologicae, livr. 27. Klincksieck, Paris, France. [Google Scholar]
- Cailleux A. 1981. Code des couleurs des sols. Paris, France. [Google Scholar]
- Cannon PF, Hawksworth DL, Sherwood-Pike MA. 1985. The British Ascomycotina – An annotated checklist. Huddersfield, UK. [Google Scholar]
- Cantrell SA, Hanlin RT. 1997. Phylogenetic relationships in the family Hyaloscyphaceae inferred from sequences of ITS regions, 5.8S ribosomal DNA and morphological characters. Mycologia 89: 745–755. [Google Scholar]
- Clements FE, Shear CL. 1931. The genera of fungi. New York, USA. [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. 2017. Fungal planet description sheet: 625–715. Persoonia 39: 270–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis RWG. 1949. A revision of the British Hyaloscyphaceae with notes on related European species. Mycological papers 32: 1–97. [Google Scholar]
- Dennis RWG. 1960. British cup fungi and their allies. London, UK. [Google Scholar]
- Dennis RWG. 1962. New or interesting British Helotiales. Kew Bulletin 16: 317–327. [Google Scholar]
- Dennis RWG. 1963. A redisposition of some fungi ascribed to the Hyaloscyphaceae. Kew Bulletin 17: 319–379. [Google Scholar]
- Dennis RWG. 1968. British Ascomycetes. Lehre, Germany. [Google Scholar]
- Dennis RWG. 1981. British Ascomycetes. Vaduz, Germany. [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW. 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57: 2703–2720. [DOI] [PubMed] [Google Scholar]
- Ellis JB. 1882. New species of North American fungi. Bulletin of the Torrey Botanical Club 9: 73. [Google Scholar]
- Fehrer J, Réblová M, Bambasová V, et al. 2018. ‘2019’. The root-symbiotic Rhizoscyphus ericae aggregate and Hyaloscypha (Leotiomycetes) are congeneric: phylogenetic and experimental evidence. Studies in Mycology 92: 195–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebes G, Wendelin I. 2014. Über einige seltene und interessante Ascomyceten-Funde vom Reinerkogel (Graz, Steiermark, Österreich). Joannea Botanik 11: 5–33. [Google Scholar]
- Fuckel L. 1870 ‘1869–1870’. Symbolae mycologicae. Jahrbuücher des Nassauischen Vereins fuür Naturkunde 23–24: 1–459. [Google Scholar]
- Fuckel L. 1872 ‘1871–1872’. Symbolae mycologicae. Jahrbücher des Nassauischen Vereins für Naturkunde 25–26: 287–346. [Google Scholar]
- Galán R, Raitviir A. 1994. Some new or interesting species of the Hyaloscyphaceae from Spain. Nova Hedwigia 58: 453–473. [Google Scholar]
- Galán R, Raitviir A. 2004. Deux Hyaloscyphaceae jaunes (Ascomycetes, Helotiales). Bulletin de la Société Mycologique de France 120: 187–198. [Google Scholar]
- Haines JH. 1974. Notes on the genus Trichopezizella with descriptions of new taxa. Mycologia 66: 231–241. [Google Scholar]
- Haines JH. 1989. Studies in the Hyaloscyphaceae V: species described by C.H. Peck. Mycotaxon 35: 317–352. [Google Scholar]
- Haines JH, Dumont KP. 1983. Studies in the Hyaloscyphaceae II: Prolifero-discus, a new genus of Arachnopezizoideae. Mycologia 75: 535–543. [Google Scholar]
- Han J-G, Hosoya T, Sung G-H, et al. 2014. Phylogenetic reassessment of Hyaloscyphaceae sensu lato (Helotiales, Leotiomycetes) based on multigene analyses. Fungal Biology 118: 150–167. [DOI] [PubMed] [Google Scholar]
- Hawksworth DL. 2001. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycological Research 105: 1422–1432. [Google Scholar]
- Hosoya T, Otani Y. 1997. Hyaloscyphaceae in Japan (2): glassy-haired members of the tribe Hyaloscypheae. Mycoscience 38: 187–205. [Google Scholar]
- Huhtinen S. 1987a. The genus Protounguicularia in Europe. Beiträge Kenntnis Pilze Mitteleuropas 3: 457–463. [Google Scholar]
- Huhtinen S. 1987b. Taxonomic studies in the genera Protounguicularia, Arachnopeziza and Dematioscypha. Mycotaxon 30: 9–28. [Google Scholar]
- Huhtinen S. 1989 ‘1990’. A monograph of Hyaloscypha and allied genera. Karstenia 29: 1–252. [Google Scholar]
- Huhtinen S. 1993a. New or less known hyaloscyphaceous fungi from the Canadian timberline. Bibliotheca Mycologica 150: 93–103. [Google Scholar]
- Huhtinen S. 1993b. Some hyaloscyphaceous fungi from tundra and taiga. Sydowia 45: 188–198. [Google Scholar]
- Johnston PR, Quijada L, Smith CA, et al. 2019. A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, et al. 2008. Dictionary of the fungi. 10th edn. CABI International, United Kingdom. [Google Scholar]
- Kohn LM, Korf RP. 1975. Variation in ascomycete iodine reactions: KOH pretreatment explored. Mycotaxon 3: 165–172. [Google Scholar]
- Korf RP. 1951a. Arachnopeziza obtusipila Grelet description emended. Mycologia 43: 211–214. [Google Scholar]
- Korf RP. 1951b ‘1952’. A monograph of the Arachnopezizeae. Lloydia 14: 129–180. [Google Scholar]
- Korf RP. 1959. Japanese discomycete notes IX–XVI. Bulletin of the National Science Museum 4: 389–400. [Google Scholar]
- Korf RP. 1978. Revisionary studies in the Arachnopezizoideae: a monograph of the Polydesmieae. Mycotaxon 7: 457–492. [Google Scholar]
- Korf RP. 1981. A preliminary discomycete flora of Macaronesia: part 2, Hyaloscyphaceae subf. Arachnopezizoideae. Mycotaxon 13: 137–144. [Google Scholar]
- Korf RP. 2007. On the genus Solenopezia (Fungi: Lachnaceae): Peziza solenia and ICBN Art. 58 – A sleeping dog bites back. Boletin de la Sociedad Argentina de Botanica 42: 29–32. [Google Scholar]
- Korf RP, Gruff SC. 1981. Discomycetes exsiccati, Fasc. IV. Mycotaxon 13: 5–15. [Google Scholar]
- Korf RP, Kohn LM. 1980. Revisionary studies in the Hyaloscyphaceae. I. On genera with ‘glassy’ hairs. Mycotaxon 10: 503–512. [Google Scholar]
- Korf RP, Zhuang W-Y. 1985. Some new species and new records of discomycetes in China. Mycotaxon 22: 483–514. [Google Scholar]
- Leenurm K, Raitviir A, Raid R. 2000. Studies on the ultrastructure of Lachnum and related genera (Hyaloscyphaceae, Helotiales, Ascomycetes). Sydowia 52: 30–45. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Maddisson DR, Maddison WP. 2000. MacClade 4: Analysis of phylogeny and character evolution. Version 4.0. Sinauer Ass., Sunderland, MA, United States. [Google Scholar]
- Matheny PB, Liu YJ, Ammirati JF, et al. 2002. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). American Journal of Botany 89: 688–698. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA, United States: 1–8. [Google Scholar]
- Nannfeldt JA. 1932. Studien über die Morphologie und Systematik der nicht-lichenisierten inoperculaten Discomycete. Nova Acta Regiae Societatis Scientiarum Upsaliensis Ser. 4, 8: 1–368. [Google Scholar]
- Quijada L, Huhtinen S, Hairaud M, et al. 2014. Is Psilocistella quercina (Velen.) Svrček a good taxon? Ascomycete.Org 6: 143–146. [Google Scholar]
- Raitviir A. 1970. Synopsis of the Hyaloscyphaceae. Scripta Mycologica 1: 1–115. [Google Scholar]
-
Raitviir A. 1987.
 . (System of hyaloscyphic fungi.)
Mikologija i Fitopatologija
21: 200–206. [Google Scholar]
. (System of hyaloscyphic fungi.)
Mikologija i Fitopatologija
21: 200–206. [Google Scholar] - Raitviir A. 2001. Taxonomic notes on Dematioscypha and Amicodisca. Czech Mycology 52: 289–294. [Google Scholar]
- Raitviir A. 2003. New or forgotten Helotiales from Greenland 1. Dermateaceae and Hyaloscyphaceae. Mycotaxon 87: 359–378. [Google Scholar]
- Raitviir A. 2004. Revised synopsis of the Hyaloscyphaceae. Scripta Mycologica 20: 1–133. [Google Scholar]
- Raitviir A, Gálan R. 1986. A new genus of the Hyaloscyphaceae. International Journal of Mycology and Lichenology 2: 221–234. [Google Scholar]
- Raitviir A, Huhtinen S. 1997. Glassy-haired Hyaloscyphaceae: new taxa and new synonymies. Mycotaxon 62: 445–460. [Google Scholar]
- Raitviir A, Sharma MP. 1984. Some new species of Helotiales from the North-West Himalayas (India). Proceedings of the Estonian Academy of Science, Biology and Ecology 33: 194–197. [Google Scholar]
- Rambaut A. 2002. Se-Al. Sequence Alignment Editor. Version 2.0 alpha 11. University of Oxford, Oxford, United Kingdom. [Google Scholar]
- Raschle P. 1977. Taxonomische Untersuchungen an Ascomyceten aus der Familie der Hyaloscyphaceae Nannfeldt. Sydowia 29: 170–236. [Google Scholar]
- Rehm H. 1887. Ascomyceten XVIII. Hedwigia 26: 81–98. [Google Scholar]
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Mark P, et al. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo PA. 1884. Conspectus generum Discomycetum hucusque cognitorum. Botanisches Centralblatt 18: 1–16. [Google Scholar]
- Schweinitz LD. 1822. Synopsis fungorum Carolinae superioris. Schriften der naturforschenden Gesellschaft zu Leipzig 1: 2–131. [Google Scholar]
- Sokolski S, Piché Y, Chauvet É, et al. 2006. A fungal endophyte of black spruce (Picea mariana) needles is also an aquatic hyphomycete. Molecular Ecology 15: 1955–1962. [DOI] [PubMed] [Google Scholar]
- Spatafora JW, Sung G-H, Johnson D, et al. 2006. A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028. [DOI] [PubMed] [Google Scholar]
- Spooner BM. 1987. Helotiales of Australasia: Geoglossaceae, Orbiliaceae, Sclerotiniaceae, Hyaloscyphaceae. Bibliotheca Mycologica 116: 1–711. [Google Scholar]
- Spooner BM, Dennis RWG. 1985. New or interesting ascomycetes from the Highlands and Islands. Sydowia 38: 294–316. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starbäck K. 1895. Discomyceten-Studien. Bihang till Svensk Vetenskapsakademiens handlingar 21: 1–42. [Google Scholar]
- Stenroos S, Laukka T, Huhtinen S, et al. 2010. Multiple origins of symbioses between ascomycetes and bryophytes suggested by a five-gene phylogeny. Cladistics 26: 281–300. [DOI] [PubMed] [Google Scholar]
- Svrček M. 1977. New or less known Discomycetes . Nové nebo méněznámé diskomycety 6. Česka Mykologia 31: 193–200. [Google Scholar]
- Svrček M. 1979. New or less known Discomycetes 1. Nové nebo méněznámé diskomycety 10. Česka Mykologia 33: 193–206. [Google Scholar]
- Svrček M. 1984. A taxonomic revision of inoperculate discomycetes described by J. Velenovský in the genus Helotium, preserved in National Museum, Prague. Acta Musei Nationalis Pragae 15B: 129–215. [Google Scholar]
- Svrček M. 1985. Notes on genus Hyaloscypha (Helotiales). Poznámky o rodu Hyaloscypha (Helotiales). Česka Mykologia 39: 205–219. [Google Scholar]
- Svrček M. 1987. The European genera of the family Hyaloscyphaceae (Helotiales). Evropské rody diskomycetů čeledi Hyaloscyphaceae (Helotiales). Česka Mykologia 41: 193–206. [Google Scholar]
- Taşkin H, Büyükalaca S, Doğan HH, et al. 2010. A multigene molecular phylogenetic assessment of true morels (Morchella) in Turkey. Fungal Genetics and Biology 47: 672–682. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, et al. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Velenovský J. 1934. Monographia Discomycetum Bohemiae. Prague, Czech Republic. [Google Scholar]
- Verkley GJM. 2005. Brefeldochium pruinosum, gen. et sp. nov., the anamorph of Polydesmia pruinosa (Hyaloscyphaceae, Helotiales, Ascomycota). Nova Hedwigia 80: 503–510. [Google Scholar]
- Wang Y-Z. 2009. A new species of Arachnopeziza from Taiwan. Mycotaxon 108: 485–489. [Google Scholar]
- White TJ, Bruns TD, Lee SB, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR – Protocols and Applications – A Laboratory Manual. Academic Press, San Diego, CA, United States. [Google Scholar]
- Yu Z-H, Zhuang W-Y. 2002. New taxa and new records of Lachnum and Arachnopeziza (Helotiales, Hyaloscyphaceae) from tropical China. Nova Hedwigia 74: 415–428. [Google Scholar]
- Zhuang W-Y. 1988. A monograph of the genus Unguiculariopsis (Leotiaceae, Encoelioideae). Mycotaxon 32: 1–83. [Google Scholar]
- Zhuang W-Y. 2000. Hyaloscyphaceous discomycetes from Ningxia Province, China. Mycologia 92: 593–597. [Google Scholar]
- Zoller S, Scheidegger C, Sperisen C. 1999. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming Ascomycetes. Lichenologist 31: 511–516. [Google Scholar]