Abstract
At the outset of solid organ transplantation, genetic variation between donors and recipients was recognized as a major player in mechanisms such as allograft tolerance and rejection. Genome-wide association studies have been very successful in identifying novel variant-trait associations, but have been difficult to perform in the field of solid organ transplantation due to complex covariates, era effects, and poor statistical power for detecting donor-recipient interactions. To overcome a lack of statistical power, consortia such as the International Genetics and Translational Research in Transplantation Network have been established. Studies have focused on the consequences of genetic dissimilarities between donors and recipients and have reported associations between polymorphisms in candidate genes or their regulatory regions with transplantation outcomes. However, knowledge on the exact influence of genetic variation is limited due to a lack of comprehensive characterization and harmonization of recipients’ or donors’ phenotypes and validation using an experimental approach. Causal research in genetics has evolved from agnostic discovery in genome-wide association studies to functional annotation and clarification of underlying molecular mechanisms in translational studies. In this overview, we summarize how the recent advances and progresses in the field of genetics and genomics have improved the understanding of outcomes after solid organ transplantation.
INTRODUCTION
Genetic factors have been established as major players in mechanisms such as transplant tolerance and rejection after solid organ transplantation. Previously, hypothesis driven candidate-gene studies, focusing on genes of which prior knowledge regarding their function is present, have led to the identification of genetic variants that are associated with clinical traits. In the past decade, genome-wide association studies (GWAS), which use an agnostic approach, have shown that millions of disease-associated variants lie in noncoding regions of the genome.1,2 In the field of solid organ transplantation, complex outcomes such as rejection or allograft dysfunction are likely influenced by multiple genetic polymorphisms that individually only contribute to a small proportion of the overall risk.3 Therefore, a GWAS, which screens cohorts for millions of single-nucleotide polymorphisms (SNPs), is an appealing approach to identify genetic variants related to posttransplantation outcomes in an unbiased manner.1
In this scoping review, we will provide an overview of the recent advances in genetics which have led to improvements of outcomes after transplantation, with a special focus on GWAS. A glossary of important methodological terminology can be found in the Table 1. We foresee that in the future personalized treatment based on knowledge of the donor’s and recipient’s genetic background will increase long-term quality of life posttransplantation. This goal can be reached by integrating knowledge obtained by current GWASs in solid organ transplantation with translational steps in post-GWAS analyses that are currently being undertaken.
TABLE 1.
Glossary of important methodological terminology
| eQTL | Expression quantitative trait loci are genetic variants associated with changes in gene expression and are identified by linking variations in transcript abundance with variations in genotypes. An eQTL is a locus that explains a fraction of the genetic variance of a gene expression phenotype. eQTL analysis is conducted to identify functional effects of GWAS-identified variants. |
| Genotype | A genotype is an individual’s collection of genes. The genotype is expressed when the information encoded in the genes’ DNA is used to make protein and RNA molecules. The expression of the genotype contributes to the individual’s observable traits, called the phenotype. |
| GWAS | An approach used in genetic research to associate specific genetic variations with particular diseases. Identified genetic markers can be used to understand how genes contribute to the disease and develop better prevention and treatment strategies. |
| GTEx | A comprehensive public resource to study tissue-specific gene expression and regulation. Samples were collected from 54 nondiseased tissue sites across nearly 1000 individuals, primarily for molecular assays including whole genome sequencing, whole-exome sequencing, and RNA sequencing. |
| Imputation | Genotype imputation is the term used to describe the process of predicting or imputing genotypes that are not directly assayed in a sample of individuals. Imputation has been used widely in the analysis of genome-wide association studies to boost power, fine-map associations, and facilitate the combination of results across studies using meta-analysis. |
| LD | Refers to the nonrandom association of alleles at 2 or more loci in a general population. LD is the correlation between nearby variants such that the alleles at neighboring polymorphisms are associated within a population more often than if they were unlinked. |
| Manhattan plot | A specific type of scatter plot widely used in genomics to visualize the association of genetic variants with given trait or disease as statistical significance in terms of P on a genomic scale. Each point represents a genetic variant. |
| Meta-analysis | Meta-analysis is a statistical procedure for combining data from multiple studies. Meta-analysis of genome-wide association datasets can increase the power to detect association signals by increasing sample size and by examining more variants throughout the genome than each dataset alone. |
| Mendelian randomization | A method of using measured variation in genes of known function to examine the causal effect of a modifiable exposure on disease in observational studies, which relies on the natural, random assortment of genetic variants during meiosis yielding a random distribution of genetic variants in a population. |
| PRS | A score reflecting the sum of all known risk alleles, weighted by how much risk for an outcome each variant carrier. PRS provides an overall estimate of the genetic propensity to a trait at the individual level which was, developed using genome-wide association study data. |
| SNP | A type of genetic variant involving variation of a single base pair. The advantage of using SNPs in population genetic studies lies in their abundance in the genome—approximately 85% of the human genetic variation can be attributed to SNPs. |
| WES | A genomic technique for sequencing all protein-coding regions of genes in a genome-wide manner. WES strategy starts by narrowing down the details of variants to be studied by filtering against databases, consisting of approximately 3.5 million SNPs, which enables a simpler way for discovery and validation of causative genes and common and rare variants. |
eQTL, expression quantitative trait loci; GTEx, genotype-tissue expression; GWAS, genome-wide association study; LD, linkage disequilibrium; PRS, polygenic risk score; SNP, single nucleotide polymorphism; WES, whole-exome sequencing.
HLA GENOTYPING IN SOLID ORGAN TRANSPLANTATION—THE CLASSIC GENETICS STUDY
The HLA complex, located on the short arm of chromosome 6, is the most polymorphic region of the human genome and includes the most immunologically relevant regions in the field of transplantation.4 The HLA is fundamental to the adaptive immune response and plays a critical role in the cellular and humoral response.5 In the field of kidney transplantation, the prevention of graft rejection, and subsequent graft dysfunction and loss, largely depends on minimizing the immunological variation between donors and recipients, by matching the recipient and their donor’s HLAs.6,7 In a series of 30 564 patients receiving a first deceased kidney transplant between 1984 and 1990, Held et al8 identified the importance of matching specific HLA types in improving allograft survival. Williams et al9 found for pediatric recipients of a primary kidney transplantation that, compared to zero mismatches, 1 mismatched HLA increased the allograft failure risk with 30%, and 6 HLA mismatches with 92%. Recent studies by Wiebe et al7,10 have identified single molecule eplet HLA-DR/DQ mismatch between donors and recipients as a prognostic biomarker for primary alloimmunity in kidney transplantation recipients, and subsequently report a correlation between single eplet HLA-DR/DQ mismatch and an increased risk of T-cell mediated rejection. Both studies emphasize the potential of eplet HLA-DR/DQ mismatch as a prognostic biomarker in the development of more personalized immune suppression regimes. The impact of HLA eplet mismatch is, however, still debatable in liver transplantation.11-13
The major progress in genetic analysis methods in the past 15 years have made high throughput genetic analysis an attractive way to perform detailed HLA typing. HLA typing can for example be enriched in silico, using prediction methods based on reference panels, as shown in a study combining data from the Scientific Registry of Transplant Recipients and HaploStats frequency statistics. They generated partial predictions of kidney transplant outcomes, based on refined HLA typing.14 In addition, multiple methodologies using low ambiguity rates (with 2 genotyping digits) have been developed to perform efficient typing and direct high-throughput of HLA genes. These HLA genes can be obtained by second-generation sequencing (SGS) and achieve substantially increased efficiency.15,16 Over the last few years, SGS panels targeted at HLA loci have become routinely available in the clinic, which is partially due to a reduction in the cost of these technologies to a level comparable to that of Sanger sequencing, as well as to the availability of commercial SGS HLA typing kits.17,18 These developments stimulate the need for reevaluation of the relevance of HLA mismatch in transplantation of solid organs where classically HLA matching is not considered clinically relevant, such as in liver transplantation.
NON-HLA GENOTYPING IN SOLID ORGAN TRANSPLANTATION—CANDIDATE-GENE STUDIES
Differences in genetic variants between donors and recipients can range between 3.5 million and 10 million, depending on ancestral and admixture differences.19 The first report on non-HLA genetic research in solid organ transplantation originates from 1998.20 To further explore the role of genetic polymorphisms and mismatched alleles in outcomes after solid organ transplantation, an increasing amount of research has been conducted over the years. Variants in non-HLA regions could impact outcomes depending on their presence in donors or recipients, or as mismatches between a donor-recipient pair. A prospective GWAS reported that genetic mismatches of non-HLA haplotypes, coding for transmembrane or secreted proteins, was associated with an increased risk of functional graft loss in kidney transplant recipients, independent of HLA incompatibility.21,22 Furthermore, allo-antibodies were shown to be directed against a number of these peptide differences in the sera of the same transplant recipients from this study.
Most studies investigating the effect of donor genetics are conducted for outcomes after kidney transplantation, and focus on specific candidate genes or gene variation for which an effect on transplantation outcome was expected.23 As a landmark discovery, a candidate-gene study identified the caveolin-1 rs4730751 SNP in kidney donors to be associated with an accelerated rate of fibrosis and risk of allograft failure.24 A candidate-gene approach focusing on deletion polymorphisms found that kidney recipients with 2 copies of a deletion near the LIMS1 gene had a significantly higher risk of developing allograft rejection than when the donor kidney had at least 1 full-sized version of the same gene.25 Another study focusing on transmembrane proteins estimated all possible cell surface antigen mismatches, independent of HLA matching, for kidney recipients and their living donor. These cell surface antigen mismatches were used to generate an allogeneic mismatch score, which was associated with long-term graft function.26 Finally, a study using the complement pathway as a starting point, found that donor polymorphisms in the promoter regions including CD46 (rs2796267) and CD59 (rs147788946) showed a protective effect on developing acute rejection.27
For solid organ transplantation other than kidney transplantation, research into the influence of genetic variation on outcomes is still in its infancy: study populations are small and replication cohorts are often not available. In studies focusing on liver- and lung transplantation, it was found that matching of donors and recipients could potentially help optimize allograft outcome. The presence of polymorphism mismatches in lectin pathway genes, such as MBL2, FCN2, and MASP2, between the donor and recipient conferred a 2-fold higher risk of infection after liver transplantation.28 A small liver transplantation cohort study showed that donor polymorphisms of 1-carbon metabolism play an important role in posttransplant hepatocellular carcinoma recurrence, as the enzymes involved in 1-carbon metabolism are considered to play a part in carcinogenesis.29 In the field of lung transplantation, a study on local complement3 (C3) production by donor lung cells found that the presence of a single C3 SNP in recipients affects postoperative short-term outcomes, while this SNP in donors has an opposite effect on long-term outcome.30
GENOME-WIDE ASSOCIATION STUDIES IN SOLID ORGAN TRANSPLANTATION
GWASs have been very successful in identifying novel variant-trait associations and have been performed in solid organ transplantation cohorts with increasing frequency over the past years. For this overview, we used MEDLINE (PubMed) and Embase databases to search for relevant literature including GWASs with clinical outcomes after solid organ transplantation (search strategy including the terms “transplantation,” “genome wide association study,” and their synonyms and related terms). An overview of all performed GWASs in the field of solid organ transplantation is shown in Table 2.
TABLE 2.
Summaries of genome-wide association studies in the solid organ transplantation.
| Study | Gene | Variants | Outcome | Sample size | Graft | Ethnicity |
|---|---|---|---|---|---|---|
| O’Brien et al54 | ZNF516, TRA | rs6565887, rs3811321 | 5-y creatinine variance and long-term allograft function | 326 transplant recipients | Kidney | Irish |
| McCaughan et al86 | ATP5F1P6, DNAJC16, CELA2B, AGMAT, CASP9, NOX4, NPPA, INPP5A | rs10484821, rs7533125, rs2861484, rs11580170, rs2020902, rs1836882, rs198372, rs4394754 | New-onset diabetes after transplantation | 529 individuals consisting 57 NODAT patients | Kidney | United Kingdom |
| Sanders et al40 | CACNA1D, CSMD1 | rs3774611, rs13270945 | Cutaneous squamous cell carcinoma developed after transplantation | 71 kidney- and 17 heart- recipients as discovery; 265 kidney- and 35 heart-recipients as controls | Kidney and heart | American |
| Oetting et al34 | CYP3A5*6, CYP3A5*7 | rs10264272, rs41303343 | Tacrolimus trough concentrations in blood | 197 adult transplant recipients and 160 recipients for validation | Kidney | African American |
| Ghisdal et al31 | PTPRO, CCDC67 | rs7976329, rs10765602 | Acute renal rejection | 275 cases and 503 controls as discovery, 313 cases and 531 controls as replication | Kidney | European |
| Hernandez-Fuentes et al33 | – | None donor or recipient genetic variant | Long- or short-term allograft survival | 2094 transplant pairs as discovery and 5866 pairs as replication | Kidney | United Kingdom |
| Oetting et al35 | CYP3A4*22 | rs35599367 | TAC concentrations | 1345 adult recipients | Kidney | European American |
| Pihlstrøm et al41 | 27-SNP genetic risk score | rs9818870, rs17609940, rs4977574, rs4773144 | Cardiovascular diseases | 1640 participants | Kidney | European |
| Liu et al87 | – | rs1927321, rs1057192 (donors) | Tacrolimus concentration in convalescence period | 115 donors and 115 matched recipients | Liver | Chinese |
| Liu et al87 | CYP3A5 | rs776746, rs2667662, rs7980521, rs4903096 (donors) and rs7828796, rs776746 (recipients) | Tacrolimus concentration in stabilizing period | 115 donors and 115 matched recipients | Liver | Chinese |
| Stapleton et al42 | Polygenic risk scores | eGFR at 1-y posttransplant | 10 844 donors and recipients from 5 cohorts | Kidney | European | |
| Stapleton et al88 | Polygenic risk scores | Squamous cell carcinoma, basal cell carcinoma and nonmelanoma skin cancer | 889 transplant recipients, 239 developed NMSC with 106 developed BCC and 150 developed SCC | Kidney | European | |
| Zhang et al89 | Proportion of genome-shared identity-by-descent | Death-censored allograft loss | 385 donor-recipient pair transplants | Kidney | United States | |
| Li et al44 | AK4, RGS5 | rs11208611-T, rs10917696-C | Thrombosis after transplantation | 1085 donors of 775 for adult recipients and 310 for paediatric recipients | Liver | European |
BCC, basal cell carcinoma; eGFR, estimated glomerular filtration rate; NMSC, nonmelanoma skin cancer; NODAT, New-onset diabetes after transplantation; SCC, squamous cell carcinoma; TAC, tacrolimus.
Most GWASs in solid organ transplantation were performed in kidney transplantation, focusing on graft related outcomes (ie, acute rejection or graft loss), and patient related outcomes (ie, immunosuppressive concentrations or cutaneous squamous cell carcinoma). The first GWAS, which used pooled DNA data, that investigated acute rejection in a cohort of 778 kidney transplant patients was published in 2017. It identified 2 loci associated with T-cell mediated rejection, which were replicated in an independent cohort.31 One of these loci encompasses PTPRO (rs7976329), which encodes a receptor-type tyrosine kinase that is essential for B cell receptor signaling. This signal could not be replicated in a systematic meta-analysis, indicating the possibility of it being a false-positive association. On the other hand, this result was restricted to GWASs and meta-analyses, which could overlook the effect of gene-gene or gene-environment interactions on the risk of acute graft rejection.32 Another large-scale GWAS including 2094 kidney transplant pairs and 5866 replication pairs reported no convincing donor or recipient genetic factors contributing to short- or long-term graft survival. Importantly, the authors did not replicate any proposed findings from previous candidate-gene studies.33 Recently, other outcomes after kidney transplantation have been investigated using GWAS with varying sample sizes. Several studies reported that 3 allelic variants of CYP3A5 (rs776746 with CYP3A5*3; rs10264272 with CYP3A5*6 and rs41303343 with CYP3A5*7) could explain a great proportion of the variability in levels of tacrolimus blood concentrations. CYP3A4*22 and CYP3A5*3 were associated with tacrolimus metabolism in the European population.34,35 The CYP3A5*1 allele, preponderant in African Americans, was reported to be associated with rapid metabolism, subtherapeutic concentrations, and higher dose requirements for tacrolimus.34,36 The clearance of tacrolimus in patients with the CYP3A5*1 allele was 2.5-fold greater compared to those with the CYP3A5*3/*3 genotype.37 A recent study identified a novel association between CYP1A1 rare variants and Tacrolimus pharmacokinetic variability in CYP3A5 nonexpressers by targeted sequencing of 114 absorption, distribution, metabolism, and excretion or pharmacogenomics genes. These studies show the potential of pharmacogenetic research for the optimization of treatment with immunosuppressive medication.38,39 Another GWAS with a modest cohort size of 388 kidney and heart transplant recipients, focusing on cutaneous squamous cell carcinoma, a long-term complication, found no genetic variants that reached a genome-wide significant association probably due to the size of the study cohort.40
Studies using a polygenic risk score (PRS) have emerged the last few years, which estimate the genetic propensity to the trait at an individual level. A PRS is calculated by integrating an individual’s genotype profile and known relevant GWAS data and can be used as an estimate of the genetic liability to a disease phenotype. PRSs have been used as a determinant of the risk of certain outcomes in transplant cohorts, based on the current GWAS findings (Table 1). The first composite PRS was calculated based on 27 SNPs that were reported to predict the risk of cardiovascular events in the general population. This PRS was evaluated in kidney transplant recipients, confirming a high predictive value in recipients with an increased cardiovascular risk.41 A study conducted by the International Genetics and Translational Research in Transplantation Network (iGeneTRAiN) consortium analyzed the impact of eGFR associated PRSs on postoperative eGFR to examine the composite effect of risk variants on kidney function in the general population. They found that 32% of the variability in eGFR at 1-y posttransplant could be explained by the donor-recipient PRS, combined with clinical covariates including the weights for death/graft loss.42 There are several potential limitations to be overcome in the construction of PRS that could affect how they perform in transplantation populations, such as a lack of diversity in the transplant individuals recruited for genetic studies and a strong ascertainment bias coming from the commercial SNP genotyping arrays for GWAS.43
Unlike in kidney transplantation, only few studies have been conducted with relevant postoperative outcomes in liver transplantation. Our research team conducted 2 GWASs in adults and pediatrics, focusing on early postoperative thrombosis after liver transplantation and performed a meta-analysis with a total of 1085 liver transplant recipients. We identified 2 candidate risk loci (AK4 and RGS5) but also found that previously associated thrombophilia risk loci in donors did not increase the thrombosis risk in liver transplant recipients.44 Another study assessed 4 separate genetic matching scores, which were utilized to calculate differences between transplant donors and recipients, for liver transplant outcomes with robust statistical power. This study showed joint testing could help with detecting SNPs significantly associated with acute rejection in liver transplantation.45 Much still remains to be done to identify bona fide causal variants, particularly those with low allele frequency and small effect size in the current transplantation-GWAS results, which require large cohorts for potential replication. Performing further GWASs in additional solid organ transplantation cohorts is not only relevant for increasing power for variant replication; it can also help to gain insight into mechanisms underlying transplantation complications or drug metabolism. Such insights potentially benefit future patient care greatly. For instance, SNPs in the CYP3A4 and CYP3A5 genes, which encode the main tacrolimus-metabolizing enzymes, have been strongly linked to varying levels of drug metabolization, which helps determine the interindividual differences in tacrolimus pharmacokinetics after transplantation, so that dosing can be adjusted accordingly.46
ADVANCING POST-GWAS ANALYSIS: FROM CORRELATION TO CAUSALITY
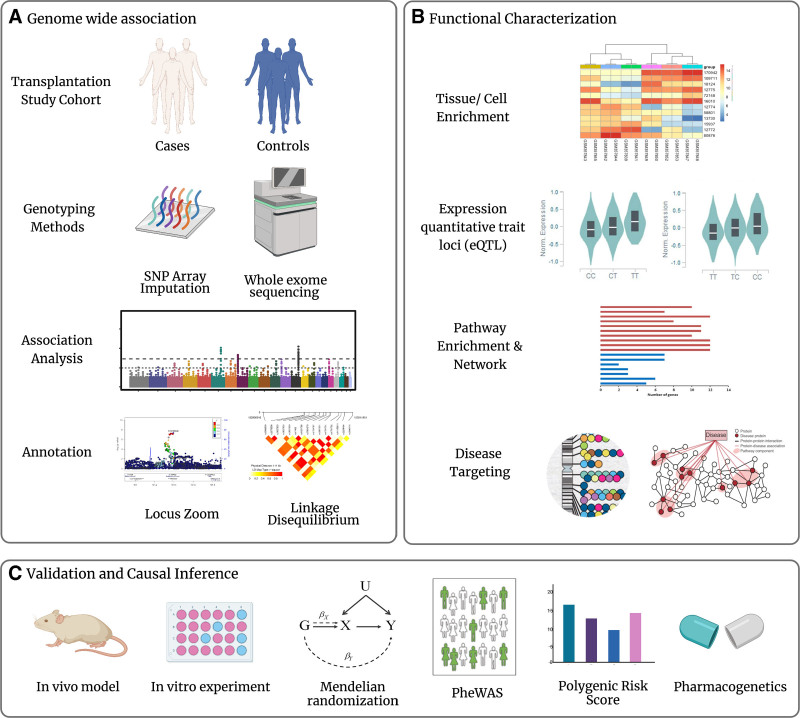
The first step of a proper GWAS study design is selecting an appropriate trait/disease and dividing the study cohort into case and control groups (Figure 1A). GWAS loci often implicate genes of unknown function or of previously unsuspected relevance. Functional characterization of identified genetic variants is often required to move from statistical association to functional investigation (Figure 1B). Experimental follow-up of such loci can lead to the discovery of novel biological mechanisms that underlie diseases (Figure 1C).1,47 Thousands of SNP associations throughout the genome that link common genetic variations to complex diseases have been identified. However, only a small fraction of these statistical associations has been thoroughly investigated to determine (1) which variant(s) are causal, (2) which genes or pathways are impacted by the causal variants, and (3) how changes in the regulation or function of the causal genes lead to a disease risk.48 Causal investigations in genetics have propelled the field from agnostic discovery in GWAS towards functional annotation and instrumental variable informed inference.
FIGURE 1.
Genome-wide association studies (GWAS) design and post-GWAS analysis. A, The first step is selecting an appropriate trait/disease and dividing the study cohort into a case and control group. Genotyping can be performed using single nucleotide polymorphism (SNP) arrays combined with imputation or by using whole-exome sequencing (WES). Association analysis is used to identify candidate loci associated with the phenotype of interest at genome-wide significance. Next, a common step is visualizing the statistics of the tested SNPs using a Manhattan plot and Locuszoom. Causal variants are often not directly genotyped but in linkage disequilibrium with the genotyped SNPs. B, Functional characterization of identified genetic variants is often required to move from statistical association to functional investigation. Several advances have aided prioritizing variants for a functional follow-up; for example, databases of gene expression enable to assess tissue or cell enrichment of candidate genes, databases of genetic variation influencing gene expression aid in deciding whether candidate risk variants are expression quantitative trait loci, and databases of genetic variation enriched in function pathways enable to evaluate the enrichment pathway of candidate genes and databases of targeting loci in complex diseases or traits (ie, GWAS Catalog). C, The validation, causal inference, and determination of clinical significance of GWAS results can be done in a number of ways. An experimental approach in vivo or in vitro is available to determine the molecular mechanisms. Mendelian randomization (MR), in coupling with phenome-wide association studies (PheWAS) present a potential way forward by providing necessary and sufficient conditions to isolate a particular causal effect. Polygenic risk scores generated by GWAS estimate the genetic propensity of the trait at an individual level. GWAS can aid in the development of new drugs based upon the genetic make-up of patients.
Although GWASs are not usually directly informative with respect to disease mechanisms, post-GWAS functional experiments reveal novel targets for therapeutic intervention (Figure 1B and 1C). To understand the specific biological effects of genetic variants in cells, the field of human genetics has extensively studied expression quantitative trait loci. An expression quantitative trait is an amount of a mRNA transcript or a protein. Expression quantitative trait loci (eQTL) are genomic loci that explain the variance of expression of mRNA. Detection of eQTLs is helpful when trying to connect the genetic trait with the disease pathogenesis. With available public resources like GTEx, eQTL analysis could be applied to hundreds of individuals with dozens of tissues retrieved by the expression levels of given SNPs.49 Another methodological approach to identify the genetic variant that is most likely causal to a disease association identified in GWAS analysis is Mendelian randomization. Mendelian randomization uses genetic variation as a natural experiment to estimate the causal relations between potentially modifiable risk factors and outcomes in observational data.50 This method makes it possible to estimate the causality using a putative causal SNP that is linked to an intermediate biomarker (instrument) for transplantation outcomes. However, because of the multiple phenotypic effects associated with transplant outcomes and lack of suitable polymorphisms for studying modifiable exposures of interest, functional follow-ups are needed to validate genetic results from earlier studies.51
One of the pressing challenges in the field of solid organ transplantation is to explicate the biological context through which identified and validated risk alleles impact function.3,48 Such mechanistic studies will be essential to translate findings from genetic association studies into actual changes in clinical practice and, finally, to truly personalized medicine. On the other hand, advanced techniques provide more powerful approaches to investigate genetic impact on transplantation outcomes. A recent study performed whole-exome sequencing and single-cell RNA sequencing of kidney transplant biopsies and defined precise immune cell chimerism and transcriptional profiles in donor derived grafts.52
BIOBANKS AND CONSORTIA: ESSENTIAL TO OVERCOME FALSE-POSITIVE ASSOCIATIONS IN GWAS
Many GWASs have the limitation of small cohorts with subsequently insufficient statistical power when performing multiple variable statistical analyses. There is a need for large cohort sizes to get sufficient power for robust identification of genetic predictors of posttransplant outcomes.53 For example, in kidney transplant recipients treated with calcineurin inhibitors, 2 genetic variants were reported to be associated with impaired kidney function, using 5-y creatinine levels as a measure with a genome-wide approach.54 The impact of these 2 SNPs on death-censored graft survival or all-cause mortality was, however, not confirmed in the Assessment of LEscol in Renal Transplant study with a larger cohort.55 A similar observation was made for 2 SNPs (rs7976329 and rs10765602 harbored in PTPRO and SLC36A4) that were reported to be associated with acute kidney graft rejection identified by a GWAS, however, could not be confirmed in a large meta-analysis.32 Out of 75 common genetic variants, only 1 variant (rs2910164) was reported to exhibit a significant association with acute rejection within the African American replication cohort.56 These examples illustrate the risk of false-positive associations in genetic analyses, resulting from having a small sample size. Cohort sizes and cohort diversity need to be adjusted to the targeted disease, and the outcome of interest as the inheritance models and effect sizes will vary regarding the disease prevalence.57,58
More research into the identified genetic factors associated with outcomes posttransplantation may lead to the discovery of novel pathways or biological processes that are involved and will lead to a better understanding of previously known biological pathways. Collaborative initiatives such as the Clinical Trials in Organ Transplantation59 consortium are built up to bring together investigators and facilitate the collective additions of transplantation samples from enrolling in clinical trials to biobanks. TransplantLines, a Dutch initiative, was designed as a biobank that includes all different types of solid organ transplant recipients as well as living organ donors.60 In addition, the iGeneTRAiN consortium (2014, www.igenetrain.org) aims to combine genomic data of multiple international solid organ transplant recipient cohorts with their corresponding phenotypic data, to discover and validate genetic contributors to transplant outcomes.61,62
CURRENT LIMITATIONS FOR GWAS IN TRANSPLANTATION: REPRODUCIBILITY AND DISEASE HETEROGENEITY
Current issues with transplantation are the many layers of potential confounders that impact statistical power to detect potential effects, including: (1) harmonization of the effect from covariates or transplant phenotypes; (2) transplant era effects; and (3) interactions between donor and recipient.
Most solid organ transplant genetic studies used individuals of European descent (Table 1),63-65 and thus the applicability of their findings to a more diverse patient population remains to be determined. One source of variation that strengthens differences in transplant outcomes between population groups are unique, non-HLA population-specific variants from ethnically diverse donor-recipient pairings. The relative risk of kidney graft loss was reportedly higher between African American donors and Caucasian recipients (21.3%) than in Caucasian recipients who received from a Caucasian donor.66 Over the past several years, APOL1 risk variants have emerged as important predictors of kidney allograft failure in individuals of African descent. However, there is no adequate evidence for the use of APOL1 risk variants in deceased or living donor transplantation because of inherent limitations.67,68
Various clinical variables and differences in study groups may result in inaccurate conclusions and statistical heterogeneity due to limited statistical power.65 An organ specific manner of effect of a genetic variation on transplantation outcomes between different allograft types needs to be investigated further. The APOL1 variants, for example, predict graft failure and have been implicated in liver necrosis in mouse studies, but were found to have only a small effect on outcomes after liver transplantation.69,70 Moving to other types of organ transplantation and other ancestral groups will be essential in gaining a full comprehension of transplantation outcomes and their molecular pathways.
Studies have started to address the “genomic collision” hypothesis which states that the risk of rejection may be increased in recipients homozygous for loss-of-function variants with grafts from nonhomozygous donors. An example is the reported genomic collision at the LIMS1 locus, which was associated with an increased risk of rejection in kidney transplantation.25,64 Outcomes that are determined by composite effects of genetic interactions between a donor and recipient still need further investigation. GWASs have established a basic set of genes and variants that are associated with independent transplant outcomes, but not enough follow-up studies to determine functional effects of polymorphisms have been performed to date. In conclusion, more diverse cohorts than those in previous studies will be required in the field of transplantation, and initiatives such as iGeneTRAiN with GWAS performed for >50 000 transplant genomes will be essential for future variant replication.
FUTURE PERSPECTIVES: FROM TRANSPLANT-OMICS TO PERSONALIZED MEDICINE
To make genetic studies in the field of solid organ transplantation clinically useful, the diagnostic assays will need to discriminate between recipients with relatively good outcomes and those with unfavorable outcomes. In contrast to a variety of clinical prediction models which use donor-, recipient-, and even operative factors to estimate posttransplant survival, only few cost-effective diagnostic assays are available for commercial use in donor screening.71-73 The transcriptional kidney solid organ response test and the IFN-γ enzyme-linked immunosorbent spot assay were assessed as predictive probabilities for subclinical kidney acute rejection.74 However, the availability of more robustly validated assays such as laboratory developed tests is warranted for more precise patient and graft monitoring.
Recently, efforts have been made to identify protein panels for rapid and noninvasive differentiation of different causes of unfavorable transplant outcomes, without requiring invasive diagnostic procedures like biopsies. These unfavorable outcomes include acute rejection, interstitial fibrosis, tubular atrophy, and BK virus nephropathy.75,76 While none are yet ready to replace the standard of care, there are several promising, minimally invasive, blood-derived biomarkers, for example cell free DNA, that are cost-effective and under intensive research for the diagnosis of transplantation outcomes, especially for detection of acute rejection.77
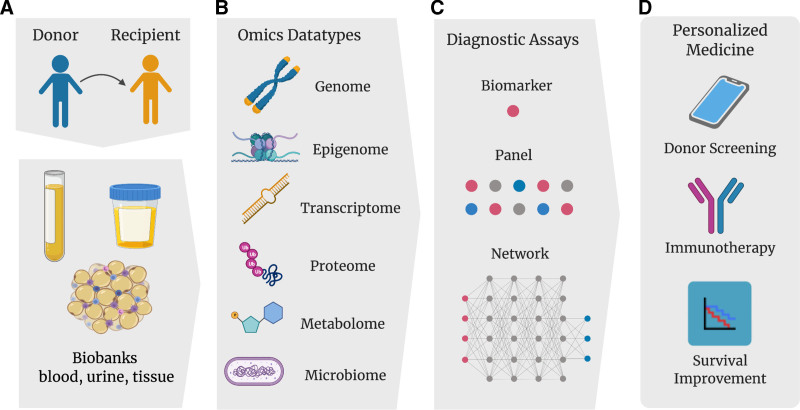
With GWAS, although thousands of SNPs have been identified as associated to complex traits, the functional implications and mechanisms of the associated loci are largely unknown.78 In order to attain a deeper understanding of transplant outcomes, an approach that synthesizes knowledge from genomics, transcriptomics, proteomics, metabolomics and other -omics approaches are essential (Figure 2). A study in nontransplant settings (ie, atrial fibrillation) showed that a risk variant identified by a multi-omics approach explained a greater disease variance than those identified by GWAS alone.79 The application of multi-omics in fields other than transplantation has steadily increased over the years. Numerous reviews describing multi-omics approaches to improve diagnostics or treatment for diseases ranging from ovarian cancer to inflammatory bowel disease have been published.80-82 Such a multi-omics approach in the field of transplantation has been coined ‘transplant-omics’. As an example, a study combining proteomics and metabolomics delineates which key cellular processes are perturbed in the kidney after brain death.83 We propose that such a broad approach is essential for achieving the goals of repositioning existing drugs and developing novel interventional treatments.84,85 Key components to biomarker discovery include associating a specific signature with a phenotype (ie, rejection, tolerance, graft failure, or response to therapy), and subsequently deciding which biospecimens are the most relevant (ie, blood, urine, or tissue). Importantly, for adequate biomarker discovery, independent replication and validation of an exploratory dataset must be performed.77
FIGURE 2.
Overview of transplant-omics and personalized medicine in solid organ transplantation. A, As the cost of high-throughput sequencing decreases and computational methods to analyze data improve, various types of omics datasets can now be combined to gain more in-depth insight into transplantation outcomes based on the biobanks of donor-recipient pairs. B, These omics data can be analyzed for biomarkers (a single gene, transcript, epigenetic modification, protein, metabolite, or microbiome that is associated with a transplantation outcome). C, The panels (a combination of multiple biomarkers) or networks (a complex mapping of many signatures accounting for the relationship between biomarkers within the panel) could be facilitated as diagnostic assays. D, The types of analyses will be able to guide diagnostic approaches and facilitate the development of personalized immunosuppressive therapies, thereby providing personalized medicine to help improve patient and graft survival and distinguish high-risk cases from low-risk cases in living donor screening.
The ultimate goal is to use knowledge of genetic architecture obtained by biobanks from large consortia such as iGeneTRAiN to improve outcomes for transplant recipients. Examples include the use of individualized dosing and selection of immunosuppressive medication and possibly even risk stratification for adverse outcomes after transplantation. The ability to link individual genetic differences to subphenotypes and transplant outcomes will be vital in gaining a complete picture of transplant phenomena.
Footnotes
Y.L. and L.M.N. contributed equally.
The work was supported by a grant from Stichting Louise Vehmeijer. This organization had no influence on the design, conduct, or data analysis of the study. Y.L. was supported by the China Scholarship Council (No. 201706940045).
The authors declare no conflicts of interest.
Y.L. and L.M.N. shared first authorship. E.A.M.F. and V.E.d.M. conceived the idea and were responsible for daily supervision. Y.L. performed the review process and wrote the initial paper draft, together with L.M.N. B.J.K. helped with the revision of the initial manuscript draft. All co-authorship places were awarded in accordance with international (ICMJE) guidelines. All authors have critically read the final paper draft and provided feedback before final submission.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Visscher PM, Wray NR, Zhang Q, et al. 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dehghan A. Genome-wide association studies. Methods Mol Biol. 2018;1793:37–49. [DOI] [PubMed] [Google Scholar]
- 3.Yang JY, Sarwal MM. Transplant genetics and genomics. Nat Rev Genet. 2017;18:309–326. [DOI] [PubMed] [Google Scholar]
- 4.Edgerly CH, Weimer ET. The past, present, and future of HLA typing in transplantation. Methods Mol Biol. 2018;1802:1–10. [DOI] [PubMed] [Google Scholar]
- 5.Kransdorf EP, Pando MJ, Gragert L, et al. HLA population genetics in solid organ transplantation. Transplantation. 2017;101:1971–1976. [DOI] [PubMed] [Google Scholar]
- 6.Jackson AM, Pinelli DF. Understanding the impact of HLA molecular mismatch in solid organ transplantation: are we there yet? Am J Transplant. 2021;21:9–10. [DOI] [PubMed] [Google Scholar]
- 7.Wiebe C, Kosmoliaptsis V, Pochinco D, et al. HLA-DR/DQ molecular mismatch: a prognostic biomarker for primary alloimmunity. Am J Transplant. 2019;19:1708–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Held PJ, Kahan BD, Hunsicker LG, et al. The impact of HLA mismatches on the survival of first cadaveric kidney transplants. N Engl J Med. 1994;331:765–770. [DOI] [PubMed] [Google Scholar]
- 9.Williams RC, West LJ, Opelz G. The risk of failure with HLA mismatch and recipient age in first pediatric (<18 years) kidney transplants. Transplant Direct. 2018;4:e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiebe C, Rush DN, Gibson IW, et al. Evidence for the alloimmune basis and prognostic significance of Borderline T cell-mediated rejection. Am J Transplant. 2020;20:2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Senev A, Coemans M, Lerut E, et al. Eplet mismatch load and De Novo occurrence of donor-specific anti-HLA antibodies, rejection, and graft failure after kidney transplantation: an observational cohort study. J Am Soc Nephrol. 2020;31:2193–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badawy A, Kaido T, Yoshizawa A, et al. Human leukocyte antigen compatibility and lymphocyte cross-matching play no significant role in the current adult-to-adult living donor liver transplantation. Clin Transplant. 2018;32:e13234. [DOI] [PubMed] [Google Scholar]
- 13.Kubal CA, Mangus R, Ekser B, et al. Class II Human Leukocyte antigen epitope mismatch predicts De Novo donor-specific antibody formation after liver transplantation. Liver Transpl. 2018;24:1101–1108. [DOI] [PubMed] [Google Scholar]
- 14.Manski CF, Tambur AR, Gmeiner M. Predicting kidney transplant outcomes with partial knowledge of HLA mismatch. Proc Natl Acad Sci USA. 2019;116:20339–20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou L, Enriquez E, Persaud M, et al. Next generation sequencing characterizes HLA diversity in a registry population from the Netherlands. HLA. 2019;93:474–483. [DOI] [PubMed] [Google Scholar]
- 16.Klasberg S, Surendranath V, Lange V, et al. Bioinformatics strategies, challenges, and opportunities for next generation sequencing-based HLA genotyping. Transfus Med Hemother. 2019;46:312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weimer ET, Montgomery M, Petraroia R, et al. Performance characteristics and validation of next-generation sequencing for human Leucocyte Antigen Typing. J Mol Diagn. 2016;18:668–675. [DOI] [PubMed] [Google Scholar]
- 18.Kuffel A, Gray A, Nic Daeid N. Human Leukocyte Antigen alleles as an aid to STR in complex forensic DNA samples. Sci Justice. 2020;60:1–8. [DOI] [PubMed] [Google Scholar]
- 19.Carja O, MacIsaac JL, Mah SM, et al. Worldwide patterns of human epigenetic variation. Nat Ecol Evol. 2017;1:1577–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo YMD, Tein MSC, Pang CCP, et al. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. 1998;351:1329–1330. [DOI] [PubMed] [Google Scholar]
- 21.Reindl-Schwaighofer R, Heinzel A, Kainz A, et al. ; iGeneTRAiN consortium. Contribution of non-HLA incompatibility between donor and recipient to kidney allograft survival: genome-wide analysis in a prospective cohort. Lancet. 2019;393:910–917. [DOI] [PubMed] [Google Scholar]
- 22.Reindl-Schwaighofer R, Heinzel A, Gualdoni GA, et al. Novel insights into non-HLA alloimmunity in kidney transplantation. Transpl Int. 2020;33:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meneghini M, Bestard O. Genotypic variants influencing acute allograft rejection: inherited susceptibility also matters. Transplantation. 2019;103:2466–2467. [DOI] [PubMed] [Google Scholar]
- 24.Moore J, McKnight AJ, Simmonds MJ, et al. Association of caveolin-1 gene polymorphism with kidney transplant fibrosis and allograft failure. JAMA. 2010;303:1282–1287. [DOI] [PubMed] [Google Scholar]
- 25.Steers NJ, Li Y, Drace Z, et al. Genomic mismatch at LIMS1 locus and kidney allograft rejection. N Engl J Med. 2019;380:1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mesnard L, Muthukumar T, Burbach M, et al. Exome sequencing and prediction of long-term kidney allograft function. Plos Comput Biol. 2016;12:e1005088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michielsen LA, van Zuilen AD, Kardol-Hoefnagel T, et al. Association between promoter polymorphisms in CD46 and CD59 in kidney donors and transplant outcome. Front Immunol. 2018;9:972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Rooij BJ, van Hoek B, ten Hove WR, et al. Lectin complement pathway gene profile of donor and recipient determine the risk of bacterial infections after orthotopic liver transplantation. Hepatology. 2010;52:1100–1110. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Lu D, Ling Q, et al. Donor one-carbon metabolism gene single nucleotide polymorphisms predict the susceptibility of cancer recurrence after liver transplantation. Gene. 2019;689:97–101. [DOI] [PubMed] [Google Scholar]
- 30.Kardol-Hoefnagel T, Budding K, van de Graaf EA, et al. A single Nucleotide C3 polymorphism associates with clinical outcome after lung transplantation. Front Immunol. 2019;10:2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghisdal L, Baron C, Lebranchu Y, et al. Genome-wide association study of acute renal graft rejection. Am J Transplant. 2017;17:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cargnin S, Galli U, Lee KS, et al. Gene polymorphisms and risk of acute renal graft rejection: a field synopsis of meta-analyses and genome-wide association studies. Transplant Rev (Orlando). 2020;34:100548. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez-Fuentes MP, Franklin C, Rebollo-Mesa I, et al. ; United Kingdom and Ireland Renal Transplant Consortium (UKIRTC) and the Wellcome Trust Case Control Consortium (WTCCC)-3. Long- and short-term outcomes in renal allografts with deceased donors: a large recipient and donor genome-wide association study. Am J Transplant. 2018;18:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oetting WS, Schladt DP, Guan W, et al. ; DeKAF Investigators. Genomewide association study of tacrolimus concentrations in African American kidney transplant recipients identifies multiple CYP3A5 alleles. Am J Transplant. 2016;16:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oetting WS, Wu B, Schladt DP, et al. Genome-wide association study identifies the common variants in CYP3A4 and CYP3A5 responsible for variation in tacrolimus trough concentration in Caucasian kidney transplant recipients. Pharmacogenomics J. 2018;18:501–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobson PA, Oetting WS, Brearley AM, et al. ; DeKAF Investigators. Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation. 2011;91:300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vannaprasaht S, Reungjui S, Supanya D, et al. Personalized tacrolimus doses determined by CYP3A5 genotype for induction and maintenance phases of kidney transplantation. Clin Ther. 2013;35:1762–1769. [DOI] [PubMed] [Google Scholar]
- 38.Yoon JG, Song SH, Choi S, et al. ; the Korean Organ Transplantation Registry (KOTRY) study group. Unraveling the genomic architecture of the CYP3A locus and ADME genes for personalized tacrolimus dosing. Transplantation. 2021;105:2213–2225. [DOI] [PubMed] [Google Scholar]
- 39.Gabardi S. Unraveling the genomic architecture of the CYP3A locus and ADME genes for personalized tacrolimus dosing. Transplantation. 2021;105:2135–2136. [DOI] [PubMed] [Google Scholar]
- 40.Sanders ML, Karnes JH, Denny JC, et al. Clinical and genetic factors associated with cutaneous squamous cell carcinoma in kidney and heart transplant recipients. Transplant Direct. 2015;1:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pihlstrøm HK, Mjøen G, Mucha S, et al. Genetic markers associated with long-term cardiovascular outcome in kidney transplant recipients. Am J Transplant. 2019;19:1444–1451. [DOI] [PubMed] [Google Scholar]
- 42.Stapleton CP, Heinzel A, Guan W, et al. ; UK Ireland Renal Transplant Consortium; DeKAF Genomics and GEN03 Studies; International Genetics and Translational Research in Transplantation Network. The impact of donor and recipient common clinical and genetic variation on estimated glomerular filtration rate in a European renal transplant population. Am J Transplant. 2019;19:2262–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De La Vega FM, Bustamante CD. Polygenic risk scores: a biased prediction? Genome Med. 2018;10:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Nieuwenhuis LM, Voskuil MD, et al. Donor genetic variants as risk factors for thrombosis after liver transplantation: a genome-wide association study. Am J Transplant. 2021;21:3133–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arthur VL, Guan W, Loza BL, et al. Joint testing of donor and recipient genetic matching scores and recipient genotype has robust power for finding genes associated with transplant outcomes. Genet Epidemiol. 2020;44:893–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Y, Xu Q, Li R, et al. CYP3A7, CYP3A4, and CYP3A5 genetic polymorphisms in recipients rather than donors influence tacrolimus concentrations in the early stages after liver transplantation. Gene. 2022;809:146007. [DOI] [PubMed] [Google Scholar]
- 47.Tam V, Patel N, Turcotte M, et al. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. [DOI] [PubMed] [Google Scholar]
- 48.Gallagher MD, Chen-Plotkin AS. The Post-GWAS era: from association to function. Am J Hum Genet. 2018;102:717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aguet F, Brown AA, Castel SE, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318:1925–1926. [DOI] [PubMed] [Google Scholar]
- 51.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 52.Malone AF, Wu H, Fronick C, et al. Harnessing expressed single nucleotide variation and single cell RNA sequencing to define immune cell chimerism in the rejecting kidney transplant. J Am Soc Nephrol. 2020;31:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sham PC, Purcell SM. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet. 2014;15:335–346. [DOI] [PubMed] [Google Scholar]
- 54.O’Brien RP, Phelan PJ, Conroy J, et al. A genome-wide association study of recipient genotype and medium-term kidney allograft function. Clin Transplant. 2013;27:379–387. [DOI] [PubMed] [Google Scholar]
- 55.Pihlstrøm HK, Mjøen G, Mucha S, et al. Single nucleotide polymorphisms and long-term clinical outcome in renal transplant patients: a validation study. Am J Transplant. 2017;17:528–533. [DOI] [PubMed] [Google Scholar]
- 56.Oetting WS, Schladt DP, Dorr CR, et al. ; DeKAF Genomics and GEN03 Investigators. Analysis of 75 candidate SNPs associated with acute rejection in kidney transplant recipients: validation of rs2910164 in MicroRNA MIR146A. Transplantation. 2019;103:1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang M, Xu S. Statistical power in genome-wide association studies and quantitative trait locus mapping. Heredity (Edinb). 2019;123:287–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt SAJ, Lo S, Hollestein LM. Research techniques made simple: sample size estimation and power calculation. J Invest Dermatol. 2018;138:1678–1682. [DOI] [PubMed] [Google Scholar]
- 59.Hricik DE, Augustine J, Nickerson P, et al. ; CTOT-01 consortium. Interferon gamma ELISPOT testing as a risk-stratifying biomarker for kidney transplant injury: results from the CTOT-01 multicenter study. Am J Transplant. 2015;15:3166–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eisenga MF, Gomes-Neto AW, van Londen M, et al. Rationale and design of TransplantLines: a prospective cohort study and biobank of solid organ transplant recipients. BMJ Open. 2018;8:e024502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fishman CE, Mohebnasab M, van Setten J, et al. Genome-wide study updates in the international genetics and translational research in transplantation network (iGeneTRAiN). Front Genet. 2019;10:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keating BJ, Van Setten J, Jacobson PA, et al. Design and implementation of the international genetics and translational research in transplantation network. Transplantation. 2015;99:2401–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jindra PT, Cusick MF. Genetic polymorphism in cytokines and costimulatory molecules in stem cell and solid organ transplantation. Clin Lab Med. 2019;39:107–123. [DOI] [PubMed] [Google Scholar]
- 64.Zanoni F, Kiryluk K. Genetic background and transplantation outcomes: insights from genome-wide association studies. Curr Opin Organ Transplant. 2020;25:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Loon E, Naesens M. Single nucleotide polymorphisms in renal transplantation: cannot see the wood for the trees. Transplantation. 2019;103:2464–2465. [DOI] [PubMed] [Google Scholar]
- 66.Pisavadia B, Arshad A, Chappelow I, et al. Ethnicity matching and outcomes after kidney transplantation in the United Kingdom. PLoS One. 2018;13:e0195038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zwang NA, Shetty A, Sustento-Reodica N, et al. APOL1-associated end-stage renal disease in a living kidney transplant donor. Am J Transplant. 2016;16:3568–3572. [DOI] [PubMed] [Google Scholar]
- 68.Ojo A, Knoll GA. APOL1 genotyping of African American deceased organ donors: not just yet. Am J Transplant. 2015;15:1457–1458. [DOI] [PubMed] [Google Scholar]
- 69.Ma L, Divers J, Freedman BI. Mechanisms of Injury in APOL1-associated kidney disease. Transplantation. 2019;103:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomson R, Genovese G, Canon C, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci USA. 2014;111:E2130–E2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Flores A, Asrani SK. The donor risk index: a decade of experience. Liver Transpl. 2017;23:1216–1225. [DOI] [PubMed] [Google Scholar]
- 72.Miller CM, Quintini C, Dhawan A, et al. The international liver transplantation society living donor liver transplant recipient guideline. Transplantation. 2017;101:938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vorlat A, De Hous N, Vervaecke AJ, et al. Biomarkers and donor selection in heart transplantation. Transplant Proc. 2019;51:1673–1678. [DOI] [PubMed] [Google Scholar]
- 74.Crespo E, Roedder S, Sigdel T, et al. Molecular and functional noninvasive immune monitoring in the ESCAPE study for prediction of subclinical renal allograft rejection. Transplantation. 2017;101:1400–1409. [DOI] [PubMed] [Google Scholar]
- 75.Sigdel TK, Gao Y, He J, et al. Mining the human urine proteome for monitoring renal transplant injury. Kidney Int. 2016;89:1244–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marx D, Metzger J, Olagne J, et al. Proteomics in kidney allograft transplantation-application of molecular pathway analysis for kidney allograft disease phenotypic biomarker selection. Proteomics Clin Appl. 2019;13:e1800091. [DOI] [PubMed] [Google Scholar]
- 77.Kohut TJ, Barandiaran JF, Keating BJ. Genomics and liver transplantation: genomic biomarkers for the diagnosis of acute cellular rejection. Liver Transpl. 2020;26:1337–1350. [DOI] [PubMed] [Google Scholar]
- 78.Zhao S, Jiang H, Liang ZH, et al. Integrating multi-omics data to identify novel disease genes and single-neucleotide polymorphisms. Front Genet. 2019;10:1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang B, Lunetta KL, Dupuis J, et al. Integrative omics approach to identifying genes associated with atrial fibrillation. Circ Res. 2020;126:350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ye M, Lin Y, Pan S, et al. Applications of multi-omics approaches for exploring the molecular mechanism of ovarian carcinogenesis. Front Oncol. 2021;11:745808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jung HD, Sung YJ, Kim HU. Omics and computational modeling approaches for the effective treatment of drug-resistant cancer cells. Front Genet. 2021;12:742902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lacroix V, Cassard A, Mas E, et al. Multi-omics analysis of gut microbiota in inflammatory bowel diseases: what benefits for diagnostic, prognostic and therapeutic tools? Int J Mol Sci. 2021;22:11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akhtar MZ, Huang H, Kaisar M, et al. Using an integrated -omics approach to identify key cellular processes that are disturbed in the kidney after brain death. Am J Transplant. 2016;16:1421–1440. [DOI] [PubMed] [Google Scholar]
- 84.Sirota M, Sarwal MM. Transplantomics: toward precision medicine in transplantation research. Transplantation. 2017;101:1777–1782. [DOI] [PubMed] [Google Scholar]
- 85.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCaughan JA, McKnight AJ, Maxwell AP. Genetics of new-onset diabetes after transplantation. J Am Soc Nephrol. 2014;25:1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, Zhang C, Li L, et al. Genome-wide association study of tacrolimus pharmacokinetics identifies novel single nucleotide polymorphisms in the convalescence and stabilization periods of post-transplant liver function. Front Genet. 2019;10:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stapleton CP, Birdwell KA, McKnight AJ, et al. Polygenic risk score as a determinant of risk of non-melanoma skin cancer in a European-descent renal transplant cohort. Am J Transplant. 2019;19:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Z, Menon MC, Zhang W, et al. Genome-wide non-HLA donor-recipient genetic differences influence renal allograft survival via early allograft fibrosis. Kidney Int. 2020;98:758–768. [DOI] [PMC free article] [PubMed] [Google Scholar]