Background:
Vascular smooth muscle cells (SMCs) undergo complex phenotypic modulation with atherosclerotic plaque formation in hyperlipidemic mice, which is characterized by de-differentiation and heterogeneous increases in the expression of macrophage, fibroblast, osteogenic, and stem cell markers. An increase of cellular cholesterol in SMCs triggers similar phenotypic changes in vitro with exposure to free cholesterol due to cholesterol entering the endoplasmic reticulum, triggering endoplasmic reticulum stress and activating Perk (protein kinase RNA-like endoplasmic reticulum kinase) signaling.
Methods:
We generated an SMC-specific Perk knockout mouse model, induced hyperlipidemia in the mice by AAV-PCSK9DY injection, and subjected them to a high-fat diet. We then assessed atherosclerotic plaque formation and performed single-cell transcriptomic studies using aortic tissue from these mice.
Results:
SMC-specific deletion of Perk reduces atherosclerotic plaque formation in male hyperlipidemic mice by 80%. Single-cell transcriptomic data identify 2 clusters of modulated SMCs in hyperlipidemic mice, one of which is absent when Perk is deleted in SMCs. The 2 modulated SMC clusters have significant overlap of transcriptional changes, but the Perk-dependent cluster uniquely shows a global decrease in the number of transcripts. SMC-specific Perk deletion also prevents migration of both contractile and modulated SMCs from the medial layer of the aorta.
Conclusions:
Our results indicate that hypercholesterolemia drives both Perk-dependent and Perk-independent SMC modulation and that deficiency of Perk significantly blocks atherosclerotic plaque formation.
Keywords: diet, high-fat; hypercholesterolemia; muscle, smooth, vascular; plaque, atherosclerotic
Highlights.
Vascular smooth muscle cells (SMCs) undergo complex phenotypic modulation with atherosclerotic plaque formation in hyperlipidemic mice, characterized by downregulation of their characteristic contractile markers and upregulation of macrophage-, fibroblast-, osteogenic cell-, and stem cell-specific markers. Our previous in vitro studies have shown that this transition is regulated by cholesterol-induced endoplasmic reticulum stress-induced unfolded protein response, particularly via the perk arm of the unfolded protein response.
When placed on a high-fat diet, hyperlipidemic male mice with SMC-specific deletion of Perk (PerkSMC−/−) demonstrated ~80% reduction in atherosclerotic plaque formation, despite having proatherogenic increases in circulating serum lipid levels.
Single-cell transcriptomic studies detected 2 modulated SMC clusters, one of which disappeared when Perk was specifically deleted from SMCs. These 2 clusters had significant overlap in their transcriptomic profiles, but the Perk-dependent cluster showed a unique phenotype of significantly decreased global transcript levels, a marker of an integrated stress response.
Pathway analysis identified cellular migration as one of the top downregulated pathways in PerkSMC−/− aortas, and our histopathologic and in vitro studies confirmed that Perk deletion blocked migration of contractile as well as modulated SMCs from the medial layer of the aorta into the intima.
Atherosclerosis is a complex process driven primarily by hypercholesterolemia and involving multiple cell types, including vascular smooth muscle cells (SMCs), endothelial cells, and inflammatory cells. Lineage tracing studies of atherosclerotic lesions in hyperlipidemic mice revealed a more prominent role of SMCs in plaque formation than originally proposed.1,2 Single-cell genomic technologies subsequently identified complex phenotypic modulation of SMCs associated with atherosclerotic plaque formation in hyperlipidemic mice.1,3,4 These modulated SMCs are de-differentiated with decreased expression of genes encoding contractile proteins (eg, Myh11, Acta2, Tagln, Cnn1) and have variable increased expression of genes that predict heterogeneous phenotypic changes, including increased expression of markers for macrophages (eg, Lgals3), fibroblasts (Lum and Fn1), osteogenic cells (Spp1), and stem cells (Ly6a). Importantly, this phenotypic modulation in both mouse and human atherosclerosis is heterogeneous in terms of the degree of de-differentiation and marker gene expression.
A critical question is how hypercholesterolemia drives atherosclerosis-associated phenotypic modulation of SMCs and atherosclerotic plaque formation. SMC-specific deletion of Klf4 in hypercholesterolemic mice decreases plaque size and the number and extent of phenotypic modulated SMCs, specifically increasing the expression of SMC differentiation markers and decreasing the number of Lgals3-positive and chondrocyte-like modulated SMCs.3,5 In addition, SMC-specific deficiency of other transcription factors, Tcf21 and aryl hydrocarbon receptor (Ahr), and activation of retinoic acid signaling in hyperlipidemic mice have reduced the number of modulated SMCs in atherosclerotic plaques.1,4,6
Hypercholesterolemia is the major risk factor for atherosclerotic plaque formation. SMC transcriptome changes similar to the molecular signature of modulated SMCs that appear with plaque formation in hypercholesterolemic mice can be modeled in vitro by exposing SMCs to cholesterol.7 Exposure to either free cholesterol or oxidized LDL (low-density lipoprotein) leads to de-differentiation of the SMCs and increases the expression of SMC modulation markers, including macrophage markers (eg, Lgals3), fibroblast markers (eg, Fn1), and osteogenic cell markers (eg, Spp1).7 Using this model system of cholesterol-driven SMC phenotypic modulation, we determined that the movement of free cholesterol into the endoplasmic reticulum (ER) induces ER stress and activates the 3 signaling pathways of an unfolded protein response (UPR): (1) Perk (protein kinase RNA-like ER kinase), which activates eIF2α (α-subunit of the eukaryotic initiation factor 2), increases the expression of Atf4 (activating transcription factor 4) and represses translation; (2) Ire-1 (inositol-requiring enzyme 1), which has dual kinase and ribonuclease activity and generates the activated transcription factor Xbp1 (X-box binding protein) by alternatively splicing its mRNA; and (3) Atf6 (activating transcription factor 6), which is activated through cleavage to a smaller form in the Golgi complex.8–10 Atf4 drives increased Klf4 expression and prevents the proteasomal degradation of the translated protein, and we found that SMCs exposed to free cholesterol increase Klf4 expression, protein levels, and transcriptional activity.7,11 Either knockdown of Perk expression or exposure to Perk inhibitors (ie, integrated stress response inhibitor) prevents both Klf4 activation and the majority of cholesterol-driven phenotypic modulation of SMCs. Based on these data, we sought to determine if cholesterol-induced Perk signaling in SMCs plays a similar role in SMC phenotypic modulation associated with plaque formation in hypercholesterolemic mice. SMC-specific Perk-deficient (PerkSMC−/−), and control mice were injected with a single dose of AAV-PCSK9DY and fed a high-fat diet (HFD) for 12 weeks. We found that Perk deletion in SMCs prevented up to 80% of plaque formation in the hyperlipidemic mice and inhibited both SMC phenotypic modulation and migration.
Methods
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The raw single-cell RNA sequencing data have been deposited to SRA (Sequence Read Archive) under accession number PRJNA850125.
Code Availability Statement
All bioinformatics analyses were performed using freely available software, and no custom codes were generated for this article.
Generation of SMC-Specific Perk Knockout Mice
Perkfloxed/floxed (Perkfl/fl, strain No. 023066; also designated as Eif2ak3tm.1.2Drc/J) and SM22αCre+/− mice [B6.Cg-Tg(Tagln-Cre)1Her/J, strain #017491] were obtained from The Jackson Laboratory (Bar Harbor, ME). The SM22αCre+/−Perkfl/+ obtained from crossing the above 2 strains were backcrossed into Perkfl/fl mice to obtain SM22αCre+/−Perkfl/fl mice (hereafter referred to as PerkSMC−/−). SM22αCre−/−Perkfl/fl littermates were used as controls (referred to as PerkSMC+/+). All animal experiments were performed in accordance with policies of the Animal Welfare Committee and the Center for Laboratory Animal Medicine and Care (CLAMC) of The University of Texas Health Science Center at Houston and guidelines from the National Institutes of Health.
AAV-PCSK9 DY Injection and High-Fat Diet
At 6 weeks of age, both male and female PerkSMC−/− and PerkSMC+/+ mice were injected with a single dose of AAV-PCSK9DY containing 1.1×1011 viral particles into a retro-orbital vein. At 7 weeks of age, the mice (12–15 mice per sex per genotype) were placed on HFD and maintained for 12 weeks.
Lipid Profile Analysis
Total cholesterol and triglycerides were analyzed in the sera of PerkSMC−/− and PerkSMC+/+mice 12 weeks after they were placed on the HFD, using Fast Performance Liquid Chromatography at the Baylor College of Medicine Mouse Metabolism and Phenotypic Core, as described earlier.12 (N=9–12 mice per sex per genotype)
En Face Oil Red O Staining of Aortas
Aortas were opened longitudinally to expose the lumen, washed with 60% isopropanol for 30 seconds, and stained with Oil Red O solution (0.3% in 60% isopropanol) using standard protocols. Ten aortas were analyzed per genotype (5 males and 5 females per group), according to the guidelines recommended by the American Heart Association.13 Plaques were quantified using ImageJ software and expressed as percentage area covered in plaque relative to the area of the entire aorta. Analysis was performed by 2 blinded individuals.
Histopathology
Paraffin-embedded cross-sections (5 μm) from formalin-fixed tissues (root and ascending aorta) were stained with hematoxylin and eosin (H&E) or used for immunofluorescent staining according to established protocols. Please see the Supplemental Materials and Methods for details. Images were acquired using a Zeiss LSM800 microscope. Five to 10 randomly chosen fields were imaged per sample for hematoxylin and eosin (H&E) stain. Lesion areas were quantified using ImageJ. For immunofluorescence, tissue sections were stained with antibodies against α-SMA, Vcam1, Pai1, and F4/80 along with anti-goat IgG with either anti-rabbit or anti-rat IgG, to ensure the specificity of the antibodies used for immunofluorescence staining (Figures S1F and S6A). Please refer to the Major Resources Table for detailed information on the antibodies used.
Single-Cell RNA Sequencing
Aortic tissue from the aortic root to the distal aortic arch was isolated from age-matched male mice of each genotype at baseline and following AAV-PCSK9DY injection and HFD. Tissues from 4 mice per genotype, per treatment were pooled together and digested to obtain single-cell suspensions, as previously described.14 Please see the Supplemental Methods for details.
Quality Control, Dimensionality Reduction, and Cell-Type Annotation
The CellRanger Software (10X genomics) was used to align the sequenced fragments to the reference genome. Successfully mapped regions were counted to generate the expression level of genes in each cell. Then, the count matrix was analyzed by the R package Seurat (V4.03).15 Cells with high levels of mitochondrial genes (>10%), indicating dead or dying cells during sequencing were removed, along with cells with extremely low levels (<0.3%) of mitochondrial genes. Cells with a limited number of expressed genes (≤800) were also removed. Details about the fraction reads in cells per group, number of genes detected and individual cells analyzed per annotated cell type are provided in Figure S2D and S2E. Next, the gene expression values underwent normalization to correct for sequence depth variation. Highly variable genes (Top 2000) were selected to perform canonical correlation analysis to identify “anchors”, which were used to integrate data from different conditions. After merging datasets, the normalized reads were scaled, and the mitochondrial expression ratio and number of reads counted were regressed out. We performed principal component analysis for dimensionality reduction. The selected principal components (Top 52) were used to create a shared nearest neighbor graph. On the graph, Louvain algorithm with multilevel refinement was applied to identify clusters. Uniform Manifold Approximation and Projection was used to visualize the clustering outcomes. Cell types were annotated manually by the expression pattern of marker genes. The marker genes of each cell type were identified by nonparametric Wilcoxon Rank-sum test with exclusion of genes that were detected in <30% of a given cell type.
Identification of Differentially Expressed Genes and Pathway Analyses
To detect the influence of the interaction between SMC-specific deletion of Perk and hypercholesterolemia, we applied generalized linear model implemented by R package MAST (V 1.18).16 The model is specified as follows:
is the expression level of genes in the cell i; indicates the genotype group, the hypercholesterolemia group of the cell i; indicate the proportion of genes detected in the cell i. We used the likelihood ratio test, testing for differences when we drop the interaction term to get P values.
For 2 group comparisons (SMC versus modulated smooth muscle cell [mSMC], mSMC1 versus mSMC2), similar generalized linear model was used. We estimated the coefficient for the main effect and tested the significance of it. The detection rate was used as a covariate.17 For tests, Benjamini and Hochberg’s method was used to control the false discovery rate.18 Corrected P values ≤0.05 were considered statistically significant.
For the differentially expressed genes, we ranked them by Z-scores of the coefficients: when comparing among 4 groups, it is that indicates the interaction effect on the tested gene; between 2 groups, it is that indicates main effects. Next, we performed preranked gene set analysis to identify whether the effect/change tends to occur toward the top (or bottom) of a pathway gene list from gene ontology (GO) annotations.19
RNAscope
RNAscope experiments were performed using materials from Advanced Cell Diagnostics, Inc. (Newark, CA), according to manufacturer’s instructions.
Trajectory Analyses and Transcription Factor Enrichment Analyses
To understand the effect of Perk during the modulation process, we conducted trajectory analyses. We extracted all SMCs and mSMCs, and then used potential of heat diffusion for affinity-based transition embedding implemented in phateR (V1.0.7) to generate a 2-dimensional embedding, which is highly suitable to identify developmental branches.20 Then, we performed trajectory inference with slingshot (V2.0.0) to identify the lineage structure and principal curves.21 TradeSeq (V1.6.0) was used to identify genes significantly associated within a lineage, that is, whether gene expression is constant across pseudotime.22 Next, we assessed differential expression patterns between conditions of those genes within a lineage, especially focusing on significantly different genes between PerkSMC−/− and PerkSMC+/+ at baseline and after HFD. Hierarchical clustering was used to identify the gene modules based on the dynamic gene expression pattern. To unveil the biological function of each module, we used hypergeometric test to determine whether a pathway from GO annotations is enriched.
We conducted transcription factor enrichment analyses to identify transcription factors responsible for the changes in gene expression of each lineage among different conditions.23 Chip-seq database was queried to determine which transcription factors may be most closely associated with genes, which had differential expression patterns between our various conditions. We prioritized transcription factors that are differentially expressed among the groups and whose targets are also enriched in the differentially expressed genes. The analysis was performed on R (V4.1.0). Ggplot2 (V3.3.5) and ComplexHeatmap (V2.8.0) were used for data visualization.24,25
SMC Explants and Cholesterol Treatment
Aortic SMCs were explanted from the ascending aortas of PerkSMC−/− and PerkSMC+/+ mice as described earlier.26 SMCs were treated with indicated amounts of free cholesterol complexed to methyl-β-cyclodextrin (MBD-Chol, Millipore Sigma) in DMEM containing high glucose (Cellgro), 10% FBS (Gibco), 1% antibiotic/anti-mycotic (Millipore Sigma), and 0.2% BSA (Fisher Scientific) for 72 hours at 37 °C and 5% CO2.
RNA Extraction, Quantitative Real-Time PCR, and Immunoblot Analyses
RNA extraction, quantitative real-time PCR, and immunoblot analyses were performed using standard protocols, which are described in the Supplemental Methods section. Please refer to Table S1 in the Major Resources Table for details of qPCR primers and antibodies used.
Transwell Migration Assay
PerkSMC−/− and PerkSMC+/+cells were plated on the upper layer of a cell culture insert containing a permeable membrane overnight and then treated with MBD-Chol for 72 hours. The cells were then washed with PBS, methanol and distilled water, followed by staining with NucBlue (Thermo Fisher Scientific). The permeable membranes containing the migrated cells were excised, mounted on glass slides using Permount (Thermo Fisher Scientific), covered with cover slips, and sealed with clear nail polish. The cells were then imaged using filters for DAPI on a Zoe Fluorescent Cell Imager (Bio-Rad Laboratories). The experiment was performed in triplicate using independent samples and 4 randomly chosen fields were imaged per sample each time. The migrated cells were counted using ImageJ software.
Klf4 Transcriptional Activity Assay
SMCs explanted from PerkSMC−/− and PerkSMC+/+ mice were transfected with Klf4 Cignal Reporter Plasmid (Qiagen), followed by treatment with MBD-Chol. Luciferase activity was measured using a Dual Luciferase Reporter Assay Kit (Promega), according to manufacturer’s instructions. Klf4 activity was expressed as the ratio of firefly to Renilla chemiluminescence.
Statistical Analysis
All data shown are expressed as ±SD. Data were tested for normality using Graph Pad Prism software version 9.2.0. (Graph Pad Software, Inc, San Diego, CA). The following data passed normality and were analyzed using unpaired 2-tailed t test with Welch correction: atherosclerotic lesion size using Oil Red O (genotypes were not separated by sex), LDL levels when the mice were not separated by sex, quantification of aortic root lesions by histology and quantification of cell density in the medial layers of the ascending aortas. The following data passed normality and were analyzed using 2-way ANOVA, followed by Tukey multiple comparisons test: LDL levels when separated by sex, qPCR for Lgals3, Klf4 luciferase activity and cholesterol-induced SMC migration studies. The following data did not pass normality and were analyzed by Kruskal-Wallis test followed by Dunn’s multiple comparisons test: atherosclerotic lesion size using Oil Red O when genotypes were separated by sex, total cholesterol, VLDL (very low-density lipoprotein), HDL, triglyceride levels (when genotypes were separated by sex), and all qPCR data except for Lgals3. The following data did not pass normality and were analyzed using Mann-Whitney U test: total cholesterol, VLDL, HDL, and triglyceride levels (when the mice were not separated by sex), lesion quantification in the ascending aorta and quantification of foam cells. P<0.05 was considered statistically significant.
Results
SMC-Specific Deletion of Perk in Hypercholesterolemic Mice Substantially Reduces Atherosclerotic Plaque Burden Despite Proatherogenic Increases in Serum Lipid Levels
The absence of Perk protein was confirmed in the aortas of PerkSMC−/−mice through immunoblot analyses (Figure S1A). Hyperlipidemia was induced in these mice by injection of a single dose of AAV-PCSK9DY (1.1×1011 viral particles) at 6 weeks of age and initiation of a HFD a week later. After 12 weeks on the HFD, male PerkSMC+/+ mice demonstrated higher weights than female PerkSMC+/+ mice, but there were no significant differences among weights of PerkSMC+/+ mice and PerkSMC−/− mice in either sex, or between those of male and female PerkSMC−/− mice (Figure S1B).
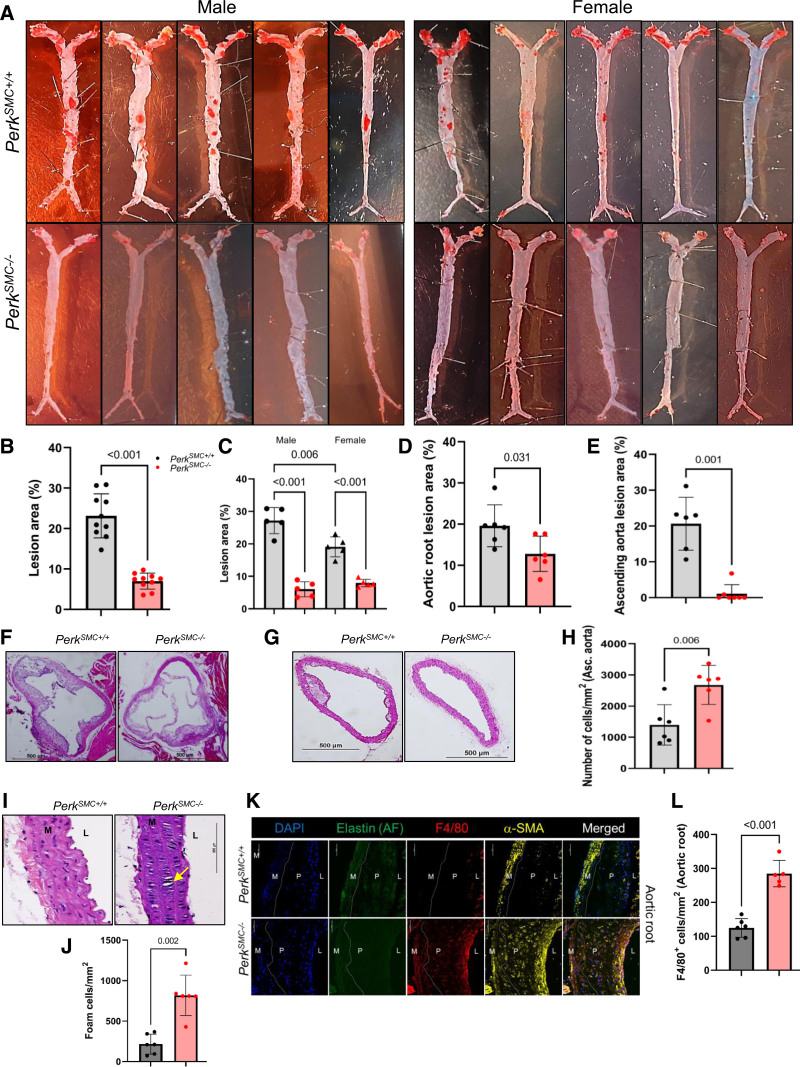
The mice were sacrificed after 12 weeks on a HFD and whole aortas harvested and stained with Oil Red O to assess plaque formation. In PerkSMC+/+ mice, extensive atherosclerotic plaques were observed in the root, ascending aorta, and aortic arch, with fewer plaques in the descending aorta (Figure 1A). In contrast, the PerkSMC−/−mice had greatly diminished plaque formation in the ascending, arch, and descending aorta and a few plaques in the aortic root (23.1±5.4% of the aorta showed plaques in controls versus 7.0±2.0% in PerkSMC−/−, Figure 1A and 1B; Figure S1C). The loss of Perk in SMCs prevented plaque formation to a greater degree in male than female mice: 28.4±2.1% versus 5.3±2.3%, and 19.1±3.1% versus 8.0±1.1 (Figure 1C). Histological assessment of plaque area determined that SMC-specific deficiency of Perk both significantly reduced the size of the plaques in the aortic root and lead to a lack of plaque in the ascending aorta (Figure 1D through 1G). The reduction of plaque occurred in the PerkSMC−/−mice despite significantly higher serum levels of total cholesterol, VLDL, LDL and triglycerides after 12 weeks on the HFD in the PerkSMC−/−mice when compared with PerkSMC+/+mice (Figure S1D). Total cholesterol was significantly elevated after hyperlipidemia was induced only in male PerkSMC−/− mice and VLDL only in female PerkSMC−/− mice compared with their corresponding PerkSMC+/+mice, while triglyceride levels went up in both sexes of PerkSMC−/− mice when compared with similarly treated PerkSMC+/+mice (Figure S1E). Thus, SMC-specific deletion of Perk prevents the majority of atherosclerotic plaque formation despite proatherogenic changes in serum lipid levels.
Figure 1.
Smooth muscle cell (SMC)-specific deletion of Perk in hypercholesterolemic mice substantially reduces atherosclerotic plaque burden despite proatherogenic increases in serum lipid levels. A, En face Oil Red O staining of aortas shows significantly reduced plaque formation in PerkSMC−/− mice compared with PerkSMC+/+ mice, which is quantified in B (N=10, P<0.001, by unpaired Student t test with Welch correction). C, Oil Red O staining of aortas shows that Perk-deficiency prevents plaque formation to a higher degree in males than in females (N=5 per sex per genotype, analyzed by Kruskal-Wallis test followed by Dunn multiple comparisons test). H&E staining demonstrates that PerkSMC−/− mice have smaller atherosclerotic lesion areas in both the aortic roots (D, F, N=6, P=0.031, by unpaired Student t test with Welch correction) and the ascending aortas (E, G, N=6, P=0.001, by Mann-Whitney U test). H. Medial layers of PerkSMC−/− ascending aortas demonstrate higher cellular density that those of PerkSMC+/+ascending aortas (N=6, P=0.006, by unpaired Student t test with Welch correction). I and J, The medial layers of the ascending aortas of PerkSMC−/− mice contain a significantly higher number of foam cells compared with PerkSMC+/+mice (N=6, P=0.002, by Mann-Whitney U test). K and L, Immunostaining against the macrophage-specific marker F4/80 shows significantly higher staining in the aortic roots of PerkSMC−/− mice (P<0.001, using unpaired Student t test with Welch correction). Medial layers of the aortic roots and ascending aortas of PerkSMC−/− mice demonstrated more intense staining for α-SMA (smooth muscle α-actin) when compared with those of PerkSMC+/+ mice. Data are represented as mean±SD. AF indicates autofluorescence; L, lumen; M, medial layer; and P, plaque.
Histological analyses of the root and ascending aorta were pursued, and a number of changes were detected in the medial layers of the mutant and wild-type (WT) mice after hyperlipidemia was induced. First, the cell density in the ascending aorta medial layer increased in the PerkSMC−/−ascending aorta (Figure 1H). Second, the PerkSMC−/−ascending aorta had a 4-fold increase in the number of cells with the appearance of foam cells when compared with PerkSMC+/+ mice (818±249 versus 216±122 cells/mm2, Figure 1I and 1J). Finally, the medial layers of the aortic roots and ascending aortas of PerkSMC−/− mice had more intense staining for α-SMA (smooth muscle α-actin) when compared with those of PerkSMC+/+ mice (Figure 1K). Atherosclerotic plaques in the aortic roots of PerkSMC−/− mice had >2-fold more cells positive for F4/80 (a marker specific for macrophages based on single-cell RNA sequencing data) compared with the plaques in the aortic roots of PerkSMC+/+ mice (Figure 1K and 1L; Figure S1F). It is notable that lesions in the ascending aortas and root of PerkSMC+/+ mice showed foci of osteogenic metaplasia, which were absent in the PerkSMC−/− aortas (Figure S1G). These data indicate that SMC deletion of Perk may prevent loss of medial SMCs but does not block cholesterol uptake based on the formation of SMC-derived foam cells in the medial layer.
Single-Cell Transcriptomics of Perk SMC−/− and Perk SMC+/+ Aortas at Baseline and After Hypercholesterolemia Show Loss of a Specific Subset of Modulated SMCs in the Perk SMC−/− Aortas
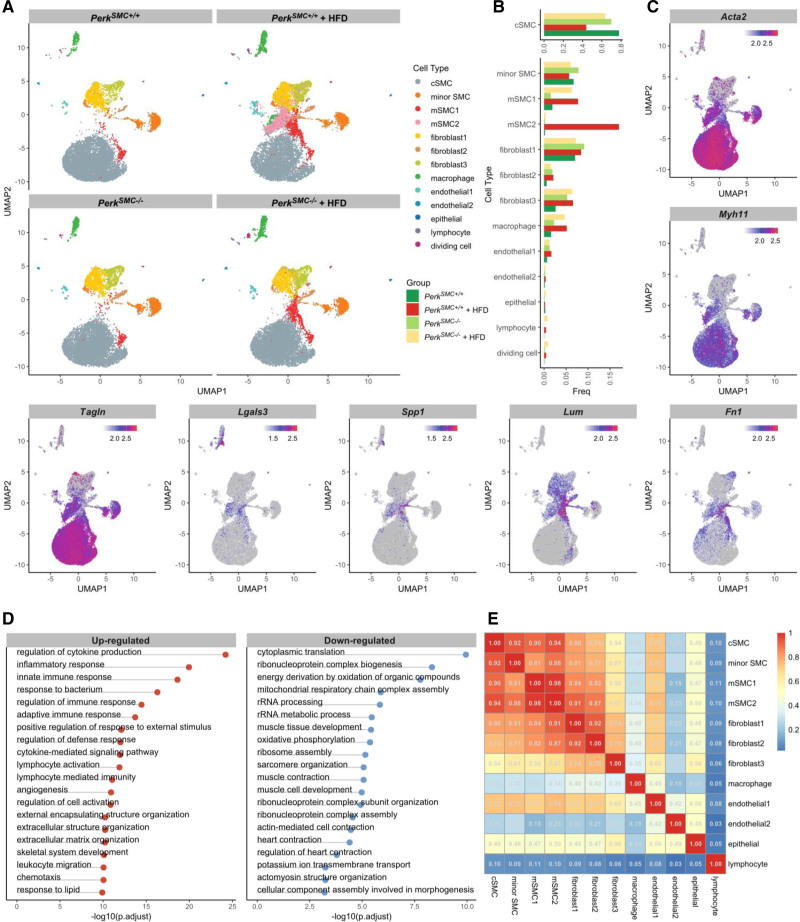
Single-cell transcriptomic data of cells from the aortic root to the distal aortic arch from PerkSMC+/+ and PerkSMC−/− at baseline and after hypercholesterolemia were used to identify cell clusters, which included contractile SMCs, 2 modulated SMC clusters (mSMC1 and mSMC2), fibroblasts, macrophages, and endothelial cells (Figure S2A through S2C). The mSMC1 cells were present at baseline and increased 4-fold in both PerkSMC+/+ and PerkSMC−/− aortas with hypercholesterolemia. In contrast, mSMC2 cells were rare at baseline for both genotypes, and increased 70-fold with hypercholesterolemia in PerkSMC+/+ aortas but only 2-fold in PerkSMC−/− aortas (Figure 2A and 2B). Notably, there was a small cluster of cells classified as macrophages grouped near the mSMC2 cells (green cluster; Figure 2A). The 2 separate macrophage clusters were compared, and the main difference between them is that the cells clustering near mSMC2s have decreased expression of macrophage markers and increased expression of contractile genes, suggesting they are SMC-derived. Due to the uncertainty of their origin, these cells were excluded from subsequent analyses (Figure S2F and S2G). Contractile SMC numbers were similar in PerkSMC+/+ and PerkSMC−/− aortas at baseline but decreased by half in the PerkSMC+/+ aortas after induction of hypercholesterolemia, and this decrease did not occur in the PerkSMC−/− aortas (Figure 2A).
Figure 2.
Single-cell transcriptomics of PerkSMC−/− and PerkSMC+/+ aortas at baseline and after hypercholesterolemia show loss of a specific subset of modulated smooth muscle cells (SMCs) in the PerkSMC−/− aortas. A, Modulated SMC clusters mSMC1 and mSMC2 expanded and appeared in the hypercholesterolemic PerkSMC+/+ mice with plaque formation, but mSMC2 did not appear in the PerkSMC−/− aortas with hypercholesterolemia. B, Perk deletion rescues the loss of contractile SMCs in PerkSMC+/+ aortas following HFD (top). mSMC1 cells are present at baseline in both genotypes and increase to a similar level following hypercholesterolemia. However, mSMC2 cells are rare at baseline in both mice but expand dramatically in PerkSMC+/+ aortas compared with PerkSMC−/− aortas. C, Expression patterns of contractile and SMC modulation markers in PerkSMC+/+ aortas following hypercholesterolemia. D, Top pathways in which DEGs of mSMC and cSMC clusters are enriched. E, Correlation plot indicates that mSMC1 and mSMC2 cells highly correlate with each other, as well as with contractile SMCs. Data are represented as mean±SD.
The established transcriptomic markers for atherosclerosis-associated modulation of SMCs were assessed in the modulated SMC clusters.1,3,4 When compared with contractile SMCs, mSMC1 and mSMC2 clusters expressed SMC contractile genes at decreased levels (Acta2, Myh11, and Tagln) and increased expression of previously described SMC modulation markers, including Lgals3, Spp1, Lum, and Fn1 (Figure 2C).1,3,4 To verify that mSMC1 and mSMC2 are SMC-derived cells, we assessed pathway changes based on the differentially expressed genes between contractile SMCs and all mSMCs and applied correlation analysis based on the top 2000 highly variable genes. Marker genes of mSMC1 and mSMC2 are enriched in pathways related to extracellular matrix organization, cell adhesion, and migration, which align with the previously identified pathways altered in atherosclerosis-associated modulated SMCs (Figure 2D).4 Based on previous studies, we calculated gene signatures specific to contractile and modulated SMCs and clearly identified downregulation of the contractile signature and a specific modulation signature in mSMCs when compared with other cell types (Figure S2H).1 Furthermore, we performed pairwise correlation analysis based on highly variable gene expression across all cell types, and mSMC1 and mSMC2 exhibited a strong degree of similarity with each other (r=0.98) and contractile SMCs (r=0.91 for mSMC1 and r=0.94 for mSMC2, Figure 2E). Thus, Perk deficiency in SMCs in hypercholesterolemic mice increased the number of contractile SMCs and blocked the formation of 1 of the 2 clusters of modulated SMCs when compared with wild-type hypercholesterolemic mice.
To assess if the mSMCs are pericyte-derived or derived from cells undergoing endothelial-mesenchymal transition (EMT), we defined a pericyte signature and an EMT signature based on previous studies.1,27 Considering the significant overlap between EMT and modulation signatures, we also investigated an endothelial signature in these clusters. We found that the pericyte signature was expressed at similar levels in mSMCs, contractile SMCs and macrophages. We did detect a higher EMT signature but the endothelial signature was not present, so we concluded that the EMT signature was due to overlap with the modulated SMC signature (Figure S2H). Taken together, these data exclude the possibility that the majority of mSMCs are pericyte or endothelial cell derived.
Contractile SMC Clusters in the Hypercholesterolemic Perk SMC+/+ Mice Activate Perk Signaling
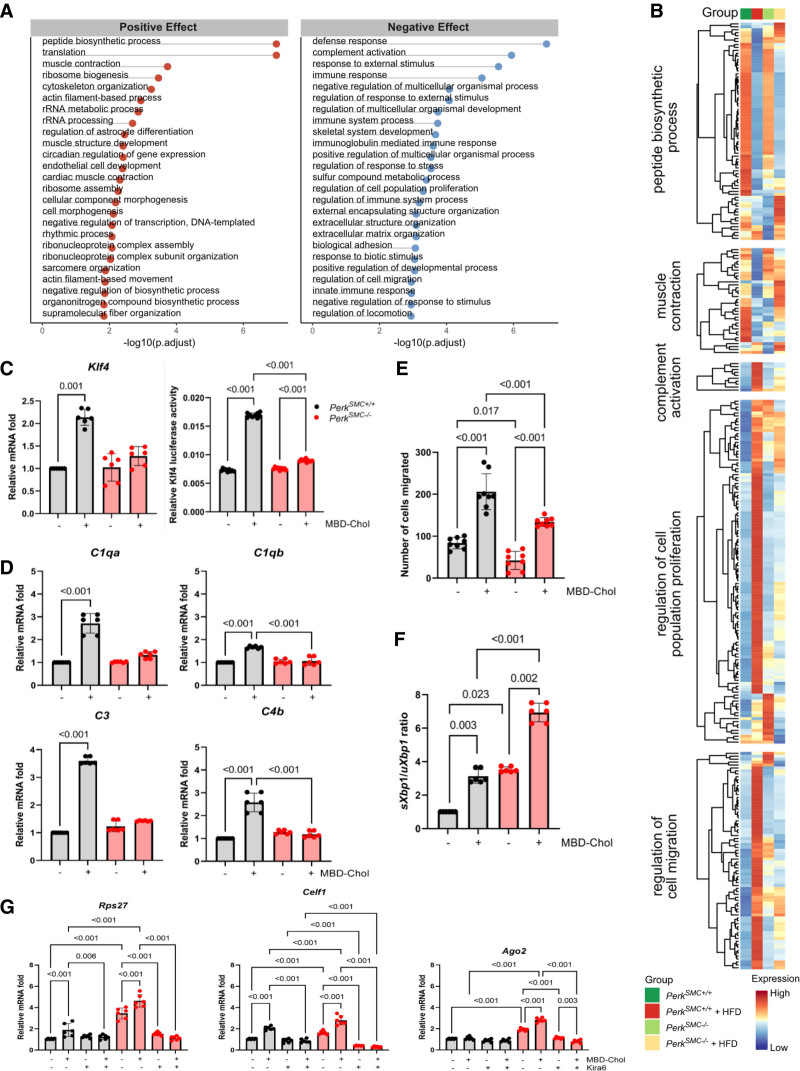
Since exposure to free cholesterol increases Perk signaling in SMCs in vitro, we initially sought to determine if Perk signaling was increased in contractile SMCs in hypercholesterolemic PerkSMC+/+ mice.7 Transcriptomic changes in contractile SMC clusters in the aortas of the PerkSMC+/+ and PerkSMC−/− mice at baseline and with hypercholesterolemia were assessed by MAST to identify differentially expressed genes, and gene set enrichment analysis based on GO annotations was used to identify pathways altered with loss of Perk signaling in the hypercholesterolemic PerkSMC−/− SMCs (Figure 3A).16 Two major pathways activated in these analyses, peptide biosynthetic process and translation, suggest hypercholesterolemia induces Perk signaling in PerkSMC+/+ contractile SMCs, and loss of Perk re-activated these pathways. In fact, 1 activated pathway is negative regulation of the biosynthetic pathways, which is specifically processes (in this case, loss of Perk) that reduce the disruption of translation driven by ER stress. Another activated pathway in the PerkSMC−/− contractile SMCs from the hypercholesterolemic mice is muscle contraction, and we have previously shown that blocking Perk signaling prevents cholesterol-driven de-differentiation of SMCs (Figure 3A and 3B).7 Suppressed pathways in the hypercholesterolemic PerkSMC−/− SMCs include complement activation and regulation of migration (Figure 3A and 3B).
Figure 3.
Contractile smooth muscle cell (SMC) clusters in the hypercholesterolemic PerkSMC+/+ mice activate Perk signaling. A, Gene ontology (GO) enrichment analysis of the DEGs identified the Top 25 up- and downregulated pathways altered specifically with loss of Perk signaling in contractile SMCs. B, Heatmap of the genes up- and downregulated in select pathways in contractile SMCs that are specifically altered due to deletion of Perk. C, Cholesterol-induced increases in expression and transcriptional activation of Klf4 are abrogated by Perk deletion in explanted PerkSMC−/− SMCs. D, Cholesterol-induced increased expression of the complement pathway components C1qa, C1qb, C3, and C4b in SMCs is abolished by the deletion of Perk. E, Perk deletion reduces migration of SMCs. F, The Ire1 arm of unfolded protein response is upregulated in PerkSMC−/− SMCs and is further activated by cholesterol treatment. G, Ribonuclear protein assembly associated genes are upregulated at baseline in PerkSMC−/− SMCs and show further activation with cholesterol exposure, and this increased expression is abrogated by the chemical inhibition of Ire by Kira6. All qPCR data were analyzed by Kruskal-Wallis test followed by Dunn multiple comparisons test. Data are represented as mean±SD. HFD indicates hypercholesterolemia followed by high-fat diet.
To further assess the role of Perk in these cholesterol-driven signaling pathways, ascending aortic SMCs were explanted from PerkSMC+/+ and PerkSMC−/− mice and exposed to free cholesterol (MBD-Chol). Similar to our previous findings, the PerkSMC+/+ aortic SMCs de-differentiate and increase the expression of UPR markers downstream of Perk activation (Atf4, Ddit3, Hspa5, Hsp90b1, and Edem) with exposure to MBD-Chol, and these transcriptional changes are blocked in the PerkSMC−/− SMCs (Figures S3A through S3C and S4A).7 PerkSMC+/+ SMCs increased both Klf4 expression and activity with MBD-Chol exposure, whereas PerkSMC−/− SMCs did not, further supporting that Klf4 activation with hypercholesterolemia is Perk-dependent (Figure 3C). MBD-Chol exposure upregulated expression of genes in the complement activation pathway (C1qa, C1qb, C3, and C4b) in PerkSMC+/+ SMCs, and this upregulation was prevented in PerkSMC−/− SMCs, which potentially explains the suppression of the complement pathway with loss of Perk (Figure 3D; Figure S3D).
Since our GO analysis indicated cellular migration was suppressed in Perk-deficient aortas, we sought to determine whether Perk activation with cholesterol exposure increases migration of SMCs. PerkSMC+/+ SMCs indeed increased migration with cholesterol exposure, and PerkSMC−/− SMCs migrated significantly slower both at baseline and with cholesterol exposure (Figure 3E; Figure S3E). Assessment of the effect of Perk deficiency on MBD-Chol-induced proliferation and apoptosis of SMCs did not identify any significant differences between mutant and WT SMCs (Figure S4B and S4C).
Another pathway activated in the hypercholesterolemic PerkSMC−/− aortic contractile SMCs is ribonucleoprotein complex assembly. Activation of another arm of UPR, Ire1, is established to drive RNP (ribonucleoprotein) assembly, so we assessed Ire1 activity in SMCs based on altered splicing of its target, Xbp1.28 PerkSMC−/− aortic SMCs exhibited significantly higher levels of spliced Xbp1 (sXbp1) compared with PerkSMC+/+ both at baseline and with MBD-Chol treatment (Figure 3F). To determine whether increased Ire1 activity is responsible for augmented expression of ribonuclear proteins in the hypercholesteremic PerkSMC−/− aortas, the expression of ribosomal genes increased in the pathway were assessed in the explanted SMCs. PerkSMC−/− SMCs had increased expression of RpII7, Ago2, and Celf1 at baseline and with exposure to MBD-Chol when compared with similarly treated PerkSMC+/+ SMCs; the Ire1 inhibitor Kira6 blocked this increased gene expression in the PerkSMC−/− SMCs (Figure 3G; Figure S4D).
Transcriptomic Differences Between the mSMC1 and mSMC2 Clusters Identify Increased Perk Signaling in mSMC2 Cells and Changes in Global Transcript Levels
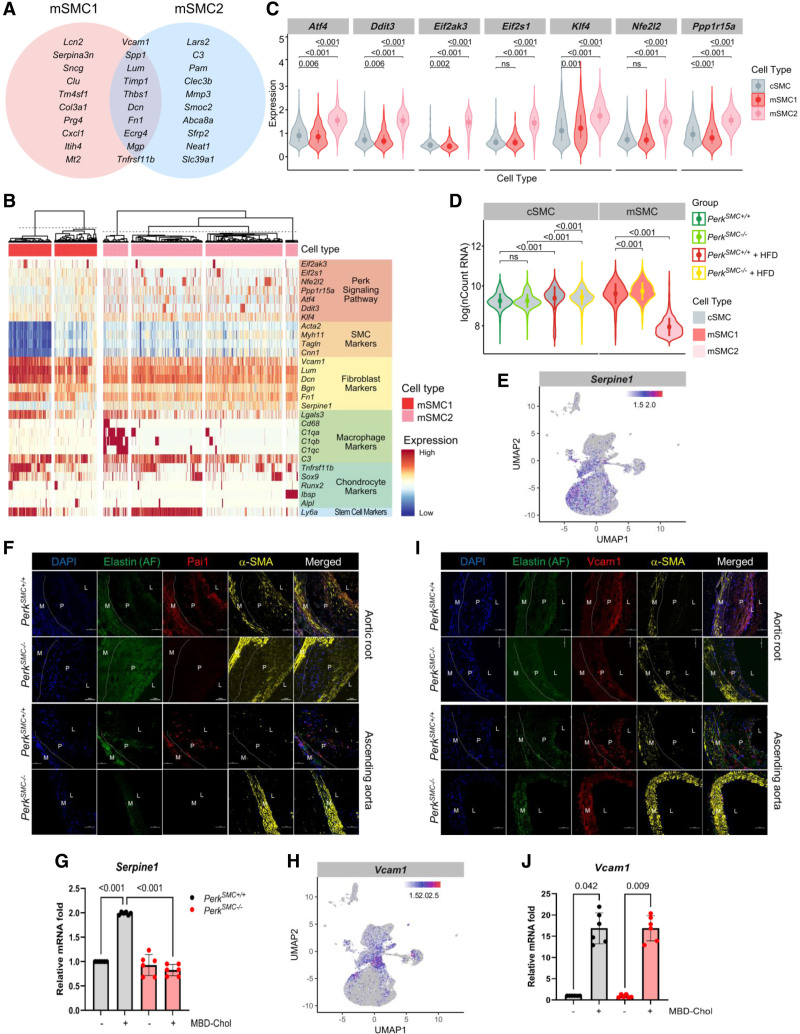
We sought to characterize transcriptomic differences between mSMC1 and mSMC2 cells and assess Perk signaling in the mSMC2 cells. The top 20 genes upregulated in the mSMC1 and mSMC2 clusters were identified and half of these markers overlapped between the clusters (Figure 4A; Figure S5A). Heatmaps were generated with the expression of genes that are upregulated with Perk signaling, along with markers of SMC differentiation and modulated SMC genes. These maps clearly illustrate the heterogeneity and overlap of expression of modulated SMC markers of both mSMC clusters (Figure 4B). This map, along with analysis of expression levels, illustrates the expression of Perk targets is greater in the mSMC2 than mSMC1 cells (Figure 4B and 4C). The heatmap also illustrates that mSMC1 are less differentiated and have increased expression of Vcam1 and other “fibromyocyte” markers (Lum, Dcn, Fn1) than mSMC2.1 Importantly, the mSMC2 cluster uniquely contains a subset of cells that have upregulated macrophage markers. Both clusters contain cells that have increased expression of osteogenic markers and Sca1 (Ly6a). Finally, the 2 clusters differ in global transcript levels with mSMC2 cells having decreased transcript levels when compared with mSMC1 and contractile SMCs (Figure 4D). Assessment of transcript levels in all SMC clusters found that the levels in contractile SMCs do not differ at baseline based on genotype and increase with hypercholesterolemia in both genotypes. For both hypercholesterolemic contractile SMCs and mSMC1 cells, PerkSMC+/+ have significantly lower transcript levels than PerkSMC−/−, likely due to a subset of cells with very low transcript levels. Importantly, this subset of cells does not form to the same extent in the PerkSMC−/− aortas.
Figure 4.
Transcriptomic differences between the mSMC1 and mSMC2 clusters identify changes in global transcript levels and increased Perk signaling in mSMC2 cells. A, The top 20 genes upregulated in the mSMC1 and mSMC2 clusters show substantial overlap. B, Heatmap compares expression of Perk targets, contractile genes, fibromyocyte markers, macrophage and complement pathway markers, and osteogenic markers between mSMC1 and mSMC2. C, Compared with cSMCs and mSMC1 cells, mSMC2 cells show significantly increased expression of Perk targets (only cells that had detectable expression of the concerned genes were included in the analysis). D, mSMC2 cells have significantly decreased mRNA transcripts when compared with cSMCs in both genotypes with and without hyperlipidemia and HFD as well as mSMC1 cells in both genotypes. E, Serpine1 shows higher expression in mSMC2 than mSMC1. F, Pai1+ mSMC2 cells were abundant in the media and the lesions in both the aortic root and the ascending aortas of control mice and quite rare in those of PerkSMC−/− mice. G, Expression of Serpine1 is dependent on Perk. H, Vcam1 expression is significantly higher in mSMC1 compared with mSMC2. I, Vcam1 immunostaining in the aortic root in the PerkSMC+/+ and PerkSMC−/− aortas identified staining in the plaques, but more intense staining was present in cells in the fibrous cap of the PerkSMC+/+ aortas. In the ascending aortas, Vcam1+ staining cells were in the plaque in the PerkSMC+/+ mice, but stronger Vcam1 staining was observed in the cells the medial layer of PerkSMC−/− mice. J, Cholesterol-induced upregulation of Vcam1 is independent of Perk. All qPCR data were analyzed by Kruskal-Wallis test followed by Dunn multiple comparisons test. Data are represented as mean±SD. AF indicates autofluorescence; L, lumen; M, medial layer; and P, plaque.
To assess global changes between mSMC1 and mSMC2 cells from the hypercholesterolemic PerkSMC+/+ aortas, we performed gene set enrichment analysis based on GO annotations. mSMC2 cells showed significant downregulation of genes involved in translation and peptide biosynthesis, further supporting that Perk signaling is increased in these cells. Ribosomal assembly and rRNA processing genes are downregulated in mSMC2, which potentially contributes to lower RNA transcripts in these cells. Genes involved in SMC migration, cell adhesion, and actin filament bundle assembly pathways are activated in mSMC2 cells, likely due to signaling pathways distinct from Perk increasing the expression of SMC differentiation markers in these cells (Figure S5B). The UPR activates regulated Ire1α-dependent decay of specific mRNA transcripts, and the levels of some regulated Ire1α-dependent decay-specific transcripts are present in mSMC1 cells but absent or decreased in the majority of mSMC2 cells (Figure S5C).29
Finally, we sought to localize these modulated SMCs in atherosclerotic plaques. Serpine1 is primarily expressed in mSMC2 and contractile SMCs in hypercholesterolemic PerkSMC+/+ aortas when compared with mSMC1 (Figure 4E). We used immunostaining for its corresponding protein, Pai1, to assess the localization of mSMC2 cells in the aortic root and ascending aortas of hypercholesterolemic PerkSMC+/+ and PerkSMC−/− mice. In both the aortic roots and ascending aortas of WT mice, Pai1+ cells were primarily localized to the lesions and medial layer, while the PerkSMC−/− aortas had little to no staining (Figure 4F, Figure S6A). Similarly, RNAscope analyses identified positive staining for Serpine1 mRNA in the media of PerkSMC+/+ aortic roots and ascending aortas, with less intense staining throughout the plaque and the fibrous cap, while the corresponding PerkSMC−/− tissues demonstrated little to no signal (Figure S6B). Correlated with these findings, cholesterol-driven expression of Serpine1 was found to be Perk-dependent in the explanted SMCs (Figure 4G). Vcam1 expression was significantly higher in the mSMC1 cluster than mSMC2 in hypercholesterolemic PerkSMC+/+ aortas (Figure 4H). Vcam1 immunostaining in the aortic root in the PerkSMC+/+ and PerkSMC−/− aortas identified staining in the plaques, but more intense staining was present in the fibrous cap of the PerkSMC+/+ aortas (Figure 4I; Figure S6A). In the ascending aortas, Vcam1+ staining cells were in the plaque in the PerkSMC+/+ aortas, while strong Vcam1 staining was observed in the cells in the medial layer of PerkSMC−/− mice. Staining for Vcam1 mRNA was observed in the fibrous cap region of the aortic root lesions of both PerkSMC+/+ and PerkSMC−/− mice, and in the medial layer of PerkSMC−/− roots. Vcam1 transcripts were present in the fibrous caps of the lesions in the ascending aortas of PerkSMC+/+mice and in the medial layer of PerkSMC−/− ascending aortas (Figure S6C). Importantly, Vcam1 expression increased to a similar extent in both the PerkSMC+/+ and PerkSMC−/− SMCs with exposure to MBD-Chol, further confirming that cholesterol-driven Vcam1 expression is not Perk-dependent (Figure 4J).
SMC Modulation Driven by Hypercholesterolemia in Mice Occurs Through 2 Lineages, One of Which Is Dependent on Perk Signaling
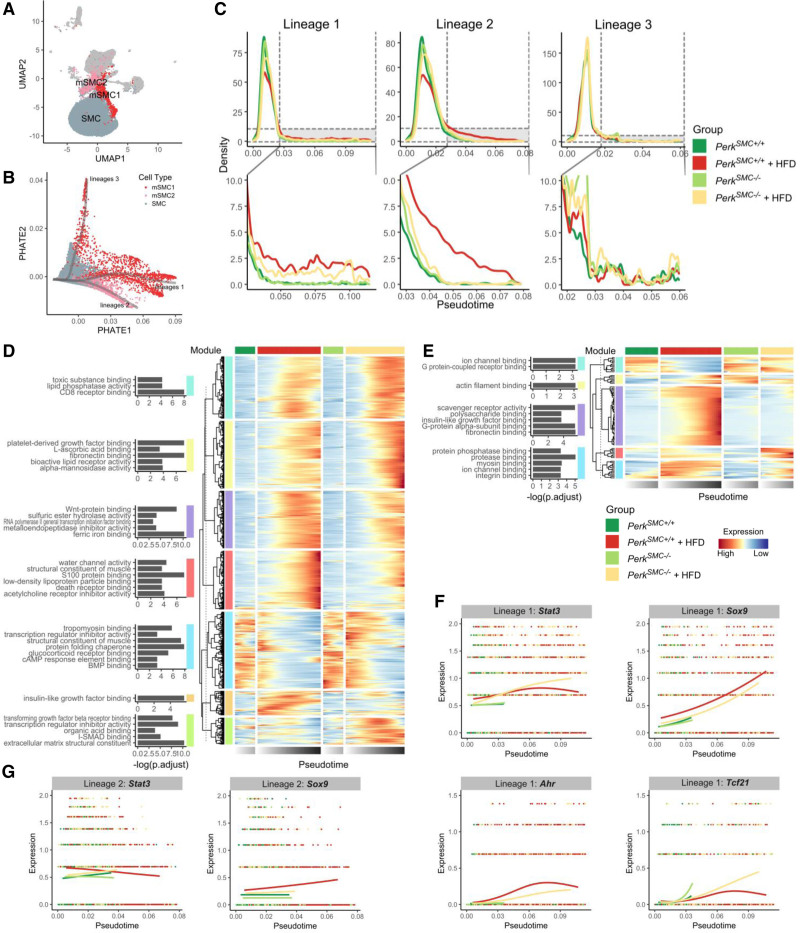
To study the influence of Perk deletion in SMCs on the dynamic changes of SMC modulation with hypercholesterolemia, the trajectories of transcriptomic changes were analyzed. Potential of heat diffusion for affinity-based trajectory embedding representation of transcriptomic data from the SMC clusters was used to capture the modulation process; in this representation, contractile SMCs remained closely grouped together (gray cells), while mSMC1 (red) and mSMC2 (pink) cells align briefly, then distribute radially (Figure 5A and 5B).20 The mSMC1 cells further disperse along multiple lineages, while mSMC2 form a single lineage. We then performed trajectory inference on this embedding. Contractile SMCs were used as reference and 3 major lineages were identified by Slingshot.21 The distribution of the pseudotime trajectories (ie, dynamic changes in gene expression) within each lineage showed differences among the 3 groups (Figure 5C). At the late stage of modulation, lineage 1 showed the greatest increase in the number of cells in both the hypercholesterolemic PerkSMC+/+ and PerkSMC−/− aortas, while the aortas not exposed to hypercholesterolemia had few cells in this lineage. The late stage of lineage 2 is dominated by cells from hypercholesterolemic PerkSMC+/+, whereas PerkSMC−/− cells track with the contractile SMCs from mice not exposed to hypercholesterolemia. The density curves in lineage 3 completely overlap in all 4 groups and most like represent minor modulation of contractile SMCs.
Figure 5.
Smooth muscle cell (SMC) modulation driven by hypercholesterolemia in mice occurs through 2 lineages, one of which is dependent on Perk signaling. A, Relative location of contractile SMC (cSMC), mSMC1, and mSMC2 cells among the various cell clusters. B, Distribution of contractile SMCs (gray), mSMC1 (red), and mSMC2 (pink) along different trajectories. C, Distribution of the pseudotime trajectories within each lineage highlights the differences among the 3 groups of SMCs. D and E, Modules of genes that have a similar pattern of expression through the lineage trajectory 1 (D) and trajectory 2 (E). F and G, Transcription factors affected by Perk enriched in lineages 1 (F) and 2 (G). HFD indicates hypercholesterolemia followed by high-fat diet.
To investigate signaling that drives the changes identified by the trajectories, we first determined significant gene expression changes associated with the trajectories. Then, we tested the differences among the groups, identified modules based on patterns of dynamic gene expression, and then performed overrepresentation analysis based on GO annotations to identify the pathways associated with the modules. For the Perk-independent trajectory (lineage 1), we identified 7 modules based on gene expression patterns through the lineage trajectory (Figure 5D). For example, the light blue bar designates a module that shows decreasing expression of SMC differentiation genes with hypercholesterolemia, including Myh11, and Cnn1, with this decreased expression slowed but not prevented in mSMC1 cells in hypercholesterolemic PerkSMC−/− aortas. The red module of genes encodes many of the markers of SMC modulation, including Lgals3, Spp1, Anxa2, Dcn, and Smoc1 and shows less robust increases in expression in PerkSMC−/− aortas when compared with PerkSMC+/+ aortas. Overall, most of the modules show modest changes between the PerkSMC−/− and PerkSMC+/+ aortas, except the last 2 modules. For the Perk-dependent trajectory (lineage 2), the gene expression changes associated with the trajectory are limited but more dramatically altered (Figure 5E). The purple module contains genes that are increased in mSMC2 cells in the hypercholesterolemic PerkSMC+/+ aortas and includes scavenger receptors.
The ChIP-seq database was queried to identify transcription factors responsible for the changes in gene expression among groups in the modulated SMC clusters. We focused on transcription factors differentially expressed among the groups that had their target genes similarly altered. We identified seventeen transcription factors linked with lineage 1‚ and 2 transcription factors for lineage 2 (Figure S7A and S7B). Both Perk-dependent and Perk-independent lineages involved Stat3 and Sox9 transcription factors (Figure 5F and 5G). Specific for mSMC1 lineage were Tcf21 and Ahr (Figure 5F), transcription factors already identified to be involved in SMC modulation with hypercholesterolemia.1,6
Finally, in vitro assessment of PerkSMC+/+ SMCs found Sox9 expression increased with exposure to free cholesterol, and loss of Perk prevented this increase in the PerkSMC−/− SMCs and in lineage 2 cells (Figure S7C; Figure 5G). In contrast, the increase in Sox9 expression in mSMC1 cells is primarily Perk-independent but is influenced by Perk loss (Figure 5F). Interestingly, Tcf21 expression is not increased with cholesterol exposure in PerkSMC+/+ SMCs but is significantly upregulated in PerkSMC−/− SMCs and this increased expression of Tcf21 is also evident in the PerkSMC−/− trajectory of lineage 1 cells (Figure 5F; Figure S7D).
Discussion
Despite decades of research, the molecular and cellular links between hypercholesterolemia and atherosclerotic plaque formation have not been fully delineated. We previously found that cholesterol activates the Perk arm of the UPR and leads to atherosclerotic plaque-associated phenotypic modulation of SMCs in vitro, characterized by de-differentiation and increased expression of macrophage, osteogenic, and fibroblast markers.1,3,4,7 Previous studies found UPR-initiated apoptosis in macrophages contributed to atherosclerotic plaque formation.30,31 Additionally, treatment with a general chemical inhibitor of ER stress rescued loss of medial SMCs and reduced plaque burden in a mouse model of Hutchinson-Gilford Progeria Syndrome.32 Here, we investigated the contribution of UPR-driven Perk signaling in SMCs and found that Perk deletion in SMCs prevents up to 80% of plaque formation that occurs in hypercholesterolemic mice and blocks the formation of one of the 2 modulated SMC clusters that form in the aortas of hypercholesterolemic wild-type mice. The mSMC2 cells in the hypercholesterolemic PerkSMC+/+ aortas express genes upregulated by Perk signaling (eg, Atf4 and Klf4) at higher levels than either the contractile SMCs or mSMC1 clusters, implicating Perk signaling in the formation of these cells. Furthermore, transcriptomic changes in the contractile SMC clusters with hypercholesterolemia show ER stress is activated in PerkSMC+/+ contractile SMCs and specifically blocked with Perk-deficiency. Thus, our findings support the conclusion that hypercholesterolemia-driven Perk signaling alters contractile SMCs and contributes to phenotypic modulation to mSMC2 cells, and both of these Perk-dependent changes in SMCs contribute to atherosclerotic plaque formation. Our data also indicate that another major modulated SMC population, mSMC1 cells, which are characterized by Vcam1 expression, is formed through Perk-independent pathways, and we show here that Vcam1 expression is cholesterol-driven in SMCs in vitro but not Perk-dependent. Performing these studies in SMC lineage-traced mice would conclusively establish that mSMC1 and mSMC2 cells are derived from SMCs, and the lack of those experiments remains a limitation of this study; however, based on our correlation analyses (Figure 2E), these cells are very likely derived from SMCs.
We previously found that blocking Perk signaling suppresses SMC de-differentiation associated with hypercholesterolemia, and here we show that loss of Perk also decreases SMC migration, both at baseline and with cholesterol treatment. These observations, along with the single-cell RNA sequencing data that Perk deficiency prevents the hypercholesterolemia-induced decreases in the number of contractile SMCs in PerkSMC+/+ aortas, suggest that Perk deficiency blocks SMC migration from the media to the intima in hypercholesterolemic PerkSMC−/− mice. These findings align with data showing that chemical inhibition of Perk signaling decreases neointimal formation and increases differentiation of medial SMCs in a rat arterial injury model.33 With hypercholesterolemia, SMCs in the medial layer of PerkSMC−/− aortas do take up cholesterol and assume a foam cell appearance, but Perk deficiency prevents these cholesterol-rich SMCs from undergoing phenotypic modulation to mSMC2. Importantly, the increased number of SMC-derived foam cells in the aortic media of the PerkSMC−/− mice does not lead to cholesterol crystal deposits and necrotic areas, or to an influx of macrophages into the medial layer. Finally, Perk deficiency does not prevent modulation to mSMC1 cells with hypercholesterolemia but does decrease the migration of these mSMC1 to the intima. While our single-cell RNA sequencing analysis did not identify proliferation or apoptosis pathways as being altered, we cannot completely rule out that blocking Perk-driven SMC apoptosis also contributed to the maintenance of contractile SMCs in the medial layer.
Trajectory analyses of SMC modulation indicate that contractile SMCs initially undergo phenotypic modulation in a uniform manner, then diverge into mSMC1 and mSMC2 clusters. mSMC2 cells do not form in the hypercholesterolemic PerkSMC−/− aortas, while the mSMC1 trajectory is populated in both the PerkSMC+/+ and PerkSMC−/− hypercholesterolemic aortas. These cells potentially contribute to the fibrous cap of the plaques.4 How hypercholesterolemia drives the formation of mSMC1 cells is unknown, but other potential pathways have been identified and were present in our lineage data analyses.1,34 Tcf21 is a transcription factor identified to be involved in mSMC1 modulation in our study, and Tcf21 has an established role in SMC differentiation and phenotypic modulation with plaque formation in mice, and in the pathogenesis of atherosclerosis in humans.1,35–37 Another transcription factor, Ahr, was found to be active in cells in the fibrous cap of plaques and is involved in mSMC1 cell modulation in our study.6 Our data indicate that Perk deficiency prevents the migration of the majority of Vcam1+ mSMC1 cells into the intima, so the role of mSMC1 in driving atherosclerotic plaque burden is not defined in this study.
Similar to previous studies, the transcriptomic changes in mSMC1 and mSMC2 clusters are heterogeneous with substantial overlap between the 2 clusters.1,3,4 The expression of Perk-regulated genes and pathway analyses indicate Perk activation in mSMC2 cells. The presence of increased levels of oxidized LDL in the neointima, along with increased expression of scavenger receptors identified with lineage tracing, may be responsible for augmented UPR and Perk signaling in these cells. Cholesterol loading of SMCs has been shown to activate ER stress, disrupt energy utilization, and lead to SMC death.38 Increased cholesterol uptake by macrophages drives an UPR and subsequent CHOP-driven apoptosis; a subset of mSMC2 cells highly express the gene encoding CHOP, Ddit3.11,39 A subset of mSMC2 cells also express macrophage markers, along with elevated expression of complement pathway genes, suggesting these mSMC2 cells may escape immune surveillance.40 Furthermore, C3 is upregulated in SMCs with cholesterol in a Perk-dependent manner, similar to data that other cells downregulate C3 expression with loss of Perk.41 It is important to note that cells with high expression of macrophage markers clustering close to the mSMC2 group (see Figure 2A, green next to the pink cluster) were not included as SMCs in our analyses to avoid any contamination of macrophages in the analyses of SMC modulation but may have led to an underestimation of the phenotypic modulation to a macrophage-like cell. Modulated SMCs in atherosclerotic plaques drive inflammation in part through activation of complement, and C3 can trigger macrophage migration and clonal expansion of SMCs.40 Macrophage and other inflammatory cells secrete and activate TGF-β in plaques, which could lead to re-expression of SMC differentiation markers in mSMC2 cells.3,42 All these cellular stresses, along with UPR-driven regulated Ire1α-dependent decay, may ultimately decrease global transcript levels in the mSMC2.29,43,44 Importantly, our data support that Perk signaling is activated in the mSMC2 cells, but we cannot determine how much of the modulation of these cells is due to Perk signaling versus factors in the environment of the plaque.
The mSMC2 phenotype characterized by low global transcript levels has not been described in previous single-cell transcriptomic studies in hypercholesterolemic mice. We identified these mSMC2 cells based on clustering using both genotypes, with and without hypercholesterolemia, potentially amplifying the mSMC2 cluster based on the importance of Perk signaling in the modulation. Importantly, our quality control (QC) protocols of the transcriptomic data removed both cells with ≤800 genes detected and cells in which mitochondrial gene reads are >10%, similar to protocols used in other studies. Our mSMC1 and mSMC2 clusters both share significant overlap of gene expression with modulated SMC clusters identified in other publications. However, we do not know if these previously described modulated SMC clusters contained cells with low transcript levels, ie, similar to the subset of low transcript level cells identified in our hypercholesterolemic mSMC1 and contractile SMC clusters.
Pathway analyses on transcriptomic data from aortic contractile SMC clusters indicated that RNP assembly was activated in hypercholesterolemic PerkSMC−/− aortas, and Ire1 activation was found to be significantly higher in explanted PerkSMC−/− SMCs than PerkSMC+/+ SMCs. We also found evidence of augmented regulated Ire1α-dependent decay due to Ire1 activation in mSMC2 in hypercholesterolemic PerkSMC+/+ aortas. Previous studies found Ire1 inhibition suppresses plaque progression in hyperlipidemic mice, but we cannot exclude that Ire1 activation in PerkSMC−/− SMCs contributed to reduced plaque formation.45
Breakthrough atherosclerotic plaques did form in the aortic root in the hypercholesterolemic PerkSMC−/− aortas, though these plaques were significantly smaller than the plaques in the PerkSMC+/+ mice. Increased sheer stress from the turbulent blood flow in the root may augment plaque formation by alternative Perk-independent pathways in the aortic root. Macrophage numbers increased with hypercholesterolemia to similar levels in both the PerkSMC+/+ and PerkSMC−/− aortas despite the significant decrease in plaque burden in the PerkSMC−/− aortas. Immunofluorescent staining demonstrated a significantly higher number of cells that stained positive for the macrophage-specific marker F4/80 in the aortic root lesions of PerkSMC−/− mice, raising the possibility that the plaques in the aortic root of the hypercholesterolemic PerkSMC−/− are enriched in macrophages (Figure 1K and 1L).
Single-cell transcriptomic data and histology of the atherosclerotic plaques indicate similar changes in modulated SMCs when either Perk or Klf4 are deleted from SMCs.3 SMC deficiency of either gene in hypercholesterolemic mice decreases the number of cells in modulated SMC clusters and increases the number of contractile SMCs. Brachiocephalic artery atherosclerotic plaques in SMC Klf4-deficient Apoe−/− mice are 50% smaller than similarly treated Apoe−/− mice, but aortic plaque formation has not been assessed.5 Thus, additional studies are required to determine if Perk deficiency in SMCs prevents plaque formation to a greater extent than Klf4 SMC deletion, and if so, what are the additional cellular pathways blocked with Perk deficiency. Additionally, we did not use Apoe−/− mice but rather induced hypercholesterolemia in wild-type mice through injection of AAV-PCSK9DY, and therefore, direct comparison of our data with others’ data may be influenced by our use of wild-type mice.
A surprising observation was that the PerkSMC−/− mice have higher total cholesterol, VLDL, LDL, and triglycerides and lower HDL compared with PerkSMC+/+ mice. The expression of Lipa (lysosomal acid lipase, Lal), which hydrolyzes cholesteryl esters in SMCs and macrophages, was reduced in the hypercholesterolemic PerkSMC−/− aortas when compared with PerkSMC+/+ aortas, which may contribute to reduced uptake of esterified cholesterol (data not shown). Exogenous Lal administration enhances uptake of cholesterol from the circulation into the tissues.46,47 Adipocyte-specific Lipa overexpression also reduces blood cholesterol levels in HFD-fed mice and Lipa deficiency leads to impaired formation of HDL in LDL-loaded human dermal fibroblasts.48,49 Decreased Lipa expression in SMCs could also decrease free cholesterol uptake in the PerkSMC−/− SMCs but did not prevent formation of SMC foam cells in the PerkSMC−/− aortic walls.
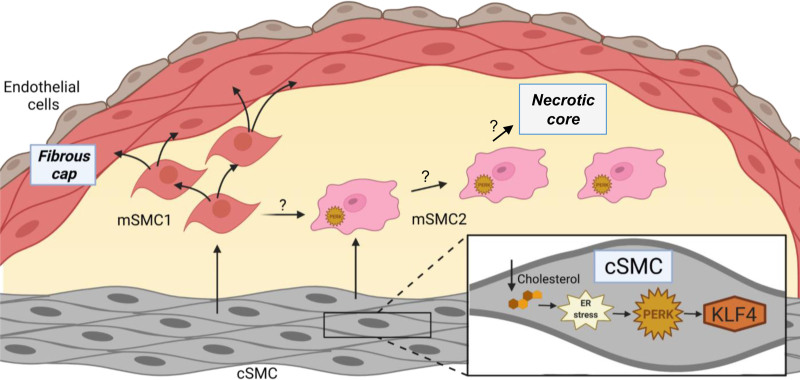
In summary, hypercholesterolemia drives modulation of contractile SMCs to both mSMC2 by a Perk-dependent pathway and mSMC1 in a Perk-independent manner. Perk deletion in SMCs also blocks a subset of cells in the contractile SMC and mSMC1 clusters from decreasing their transcript levels with hypercholesterolemia, and Perk-dependent mSMC2 cells are characterized by decreased global transcript levels. Based on the overall data, we propose a model of plaque formation in which hypercholesterolemia in mice drives Perk-dependent de-differentiation and migration of SMCs into the intima, and with that migration, the SMCs are exposed to additional environmental factors that lead to modulation into 2 distinct fates (Figure 6). Specifically, mSMC1 cells remain de-differentiated, thus primed to proliferate and migrate, and contribute to the fibrous cap. In contrast, mSMC2 cells experience further cholesterol-driven ER stress, leading to decreased global transcript levels and cell death, thus likely contributing to the necrotic core. Perk loss in SMCs prevents both the migration and the phenotypic modulation into mSMC2 and, therefore, we cannot be certain whether mSMC2 cell formation is dependent on Perk signaling or migration into the intima, or both. Regardless, our findings establish a clear mechanistic link from hypercholesterolemia-driven Perk activation to Perk-dependent migration and phenotypic modulation of SMCs to atherosclerotic plaque formation.
Figure 6.
Overall model. With hypercholesterolemia, contractile smooth muscle cells (cSMCs) in the medial layers of the aorta activate ER stress and Perk signaling, which leads to Klf4 activation, de-differentiation of cSMCs and migration into the intima. In the intima, the migrated cells are exposed to additional environmental insults, which lead to modulation into 2 distinct fates: the mSMC1 cells proliferate and contribute to the fibrous cap and may also give rise to mSMC2 cells by an unknown mechanism. mSMC2 cells experience further ER stress in the intima, leading to decreased transcript levels and cell death, ultimately contributing to the necrotic core.
Article Information
Acknowledgments
Confocal microscopy was performed at the Center for Advanced Microscopy, a Nikon Center of Excellence, Department of Integrative Biology & Pharmacology at McGovern Medical School, UTHealth Houston.
Sources of Funding
This work was supported by the National Heart, Lung and Blood Institute (RO1 HL146583 to Dr Milewicz), an American Heart Association Merit Award (Dr Milewicz), Marfan Foundation McKusick Fellowship Award (A. Chattopadhyay), NIH T32GM120011 (K. Kaw) and American Heart Association Grant 20CDA35310689 (C.S. Kwartler). Lipid profile analysis was performed at the Mouse Metabolism and Phenotypic Core at the Baylor College of Medicine (BCM), funded by NIH RO1DK114356 & UM1HG006348. The scRNA-seq was performed at the Single Cell Genomics Core at BCM partially supported by NIH shared instrument grants (S10OD023469, S10OD025240) and P30EY002520.
Disclosures
None.
Supplemental Material
Expanded Materials and Methods
Figures S1–S7
Data Set S1 and S2
Supplementary Material
Nonstandard Abbreviations and Acronyms
- α-SMA
- smooth muscle α-actin
- Atf4
- activating transcription factor 4
- DEG
- differentially expressed genes
- eIF2α
- α-subunit of the eukaryotic initiation factor 2
- EMT
- endothelial-mesenchymal transition
- GO
- gene ontology
- HFD
- high-fat diet
- Ire-1
- inositol-requiring enzyme 1
- LDL
- low-density lipoprotein
- MBD-Chol
- methyl-β-cyclodextrin cholesterol
- mSMC
- modulated smooth muscle cell
- Perk
- protein kinase RNA-like ER kinase
- RNP
- ribonucleoprotein
- SMC
- smooth muscle cell
- TC
- total cholesterol
- UPR
- unfolded protein response
- Xbp1
- X-box-binding protein
A. Chattopadhyay and P. Guan contributed equally.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.121.317451.
For Sources of Funding and Disclosures, see page 1020 & 1021.
References
- 1.Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–1289. doi: 10.1038/s41591-019-0512-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146 [DOI] [PubMed] [Google Scholar]
- 3.Alencar GF, Owsiany KM, Karnewar S, Sukhavasi K, Mocci G, Nguyen AT, Williams CM, Shamsuzzaman S, Mokry M, Henderson CA, et al. Stem cell pluripotency genes Klf4 and Oct4 regulate complex SMC phenotypic changes critical in late-stage atherosclerotic lesion pathogenesis. Circulation. 2020;142:2045–2059. doi: 10.1161/CIRCULATIONAHA.120.046672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan H, Xue C, Auerbach BJ, Fan J, Bashore AC, Cui J, Yang DY, Trignano SB, Liu W, Shi J, et al. Single-cell genomics reveals a novel cell state during smooth muscle cell phenotypic switching and potential therapeutic targets for atherosclerosis in mouse and human. Circulation. 2020;142:2060–2075. doi: 10.1161/CIRCULATIONAHA.120.048378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015;21:628–637. doi: 10.1038/nm.3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JB, Zhao Q, Nguyen T, Pjanic M, Cheng P, Wirka R, Travisano S, Nagao M, Kundu R, Quertermous T. Environment-sensing aryl hydrocarbon receptor inhibits the chondrogenic fate of modulated smooth muscle cells in atherosclerotic lesions. Circulation. 2020;142:575–590. doi: 10.1161/CIRCULATIONAHA.120.045981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay A, Kwartler CS, Kaw K, Li Y, Kaw A, Chen J, LeMaire SA, Shen YH, Milewicz DM. Cholesterol-induced phenotypic modulation of smooth muscle cells to macrophage/fibroblast-like cells is driven by an unfolded protein response. Arterioscler Thromb Vasc Biol. 2021;41:302–316. doi: 10.1161/ATVBAHA.120.315164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- 9.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou AX, Wang X, Lin CS, Han J, Yong J, Nadolski MJ, Borén J, Kaufman RJ, Tabas I. C/EBP-Homologous Protein (CHOP) in vascular smooth muscle cells regulates their proliferation in aortic explants and atherosclerotic lesions. Circ Res. 2015;116:1736–1743. doi: 10.1161/CIRCRESAHA.116.305602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bissig-Choisat B, Wang L, Legras X, Saha PK, Chen L, Bell P, Pankowicz FP, Hill MC, Barzi M, Leyton CK, et al. Development and rescue of human familial hypercholesterolaemia in a xenograft mouse model. Nat Commun. 2015;6:7339. doi: 10.1038/ncomms8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daugherty A, Tall AR, Daemen MJAP, Falk E, Fisher EA, García-Cardeña G, Lusis AJ, Owens AP, 3rd, Rosenfeld ME, Virmani R; American Heart Association Council on Arteriosclerosis, Thrombosis and Vascular Biology; and Council on Basic Cardiovascular Sciences. Recommendation on design, execution, and reporting of animal atherosclerosis studies: a scientific statement from the american heart association. Arterioscler Thromb Vasc Biol. 2017;37:e131–e157. doi: 10.1161/ATV.0000000000000062 [DOI] [PubMed] [Google Scholar]
- 14.Luo W, Wang Y, Zhang L, Ren P, Zhang C, Li Y, Azares AR, Zhang M, Guo J, Ghaghada KB, et al. Critical role of cytosolic DNA and its sensing adaptor STING in aortic degeneration, dissection, and rupture. Circulation. 2020;141:42–66. doi: 10.1161/CIRCULATIONAHA.119.041460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x [Google Scholar]
- 18.Soneson C, Robinson MD. Bias, robustness and scalability in single-cell differential expression analysis. Nat Methods. 2018;15:255–261. doi: 10.1038/nmeth.4612 [DOI] [PubMed] [Google Scholar]
- 19.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. doi: 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB, Chen WS, Yim K, Elzen AVD, Hirn MJ, Coifman RR, et al. Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol. 2019;37:1482–1492. doi: 10.1038/s41587-019-0336-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van den Berge K, Roux de Bézieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S, Clement L. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun. 2020;11:1201. doi: 10.1038/s41467-020-14766-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, Jagodnik KM, Kropiwnicki E, Wang Z, Ma’ayan A. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47(W1):W212–W224. doi: 10.1093/nar/gkz446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; 2016. [Google Scholar]
- 25.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 26.Kwartler CS, Zhou P, Kuang SQ, Duan XY, Gong L, Milewicz DM. Vascular smooth muscle cell isolation and culture from mouse aorta. Bio-Protocol. 2016;6:e2045. doi: 10.21769/BioProtoc.2045 [Google Scholar]
- 27.Zhao G, Lu H, Liu Y, Zhao Y, Zhu T, Garcia-Barrio MT, Chen YE, Zhang J. Single-cell transcriptomics reveals endothelial plasticity during diabetic atherogenesis. Front Cell Dev Biol. 2021;9:689469. doi: 10.3389/fcell.2021.689469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acosta-Alvear D, Karagöz GE, Fröhlich F, Li H, Walther TC, Walter P. The unfolded protein response and endoplasmic reticulum protein targeting machineries converge on the stress sensor IRE1. Elife. 2018;7:e43036. doi: 10.7554/eLife.43036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, Sweeney M, Rong JX, Kuriakose G, Fisher EA, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035 [DOI] [PubMed] [Google Scholar]
- 31.Thorp E, Li G, Seimon TA, Kuriakose G, Ron D, Tabas I. Reduced apoptosis and plaque necrosis in advanced atherosclerotic lesions of Apoe-/- and Ldlr-/- mice lacking CHOP. Cell Metab. 2009;9:474–481. doi: 10.1016/j.cmet.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamczyk MR, Villa-Bellosta R, Quesada V, Gonzalo P, Vidak S, Nevado RM, Andrés-Manzano MJ, Misteli T, López-Otín C, Andrés V. Progerin accelerates atherosclerosis by inducing endoplasmic reticulum stress in vascular smooth muscle cells. EMBO Mol Med. 2019;11:e9736. doi: 10.15252/emmm.201809736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B, Zhang M, Urabe G, Huang Y, Chen G, Wheeler D, Dornbos DJ, 3rd, Huttinger A, Nimjee SM, Gong S, et al. PERK Inhibition Mitigates Restenosis and Thrombosis: A Potential Low-Thrombogenic Antirestenotic Paradigm. JACC Basic Transl Sci. 2020;5:245–263. doi: 10.1016/j.jacbts.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis-Knowlton J, Turner JE, Turner A, Damian-Loring S, Hagler N, Henderson T, Emery IF, Bond K, Duarte CW, Vary CPH, et al. Characterization of smooth muscle cells from human atherosclerotic lesions and their responses to Notch signaling. Lab Invest. 2019;99:290–304. doi: 10.1038/s41374-018-0072-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. ; Cardiogenics; CARDIoGRAM Consortium. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller CL, Anderson DR, Kundu RK, Raiesdana A, Nürnberg ST, Diaz R, Cheng K, Leeper NJ, Chen CH, Chang IS, et al. Disease-related growth factor and embryonic signaling pathways modulate an enhancer of TCF21 expression at the 6q23.2 coronary heart disease locus. PLoS Genet. 2013;9:e1003652. doi: 10.1371/journal.pgen.1003652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nurnberg ST, Cheng K, Raiesdana A, Kundu R, Miller CL, Kim JB, Arora K, Carcamo-Oribe I, Xiong Y, Tellakula N, et al. Coronary artery disease associated transcription factor TCF21 regulates smooth muscle precursor cells that contribute to the fibrous cap. PLoS Genet. 2015;11:e1005155. doi: 10.1371/journal.pgen.1005155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedi X, Ming Y, Yongping W, Yi Y, Xiaoxiang Z. Free cholesterol overloading induced smooth muscle cells death and activated both ER- and mitochondrial-dependent death pathway. Atherosclerosis. 2009;207:123–130. doi: 10.1016/j.atherosclerosis.2009.04.019 [DOI] [PubMed] [Google Scholar]
- 39.Devries-Seimon T, Li Y, Yao PM, Stone E, Wang Y, Davis RJ, Flavell R, Tabas I. Cholesterol-induced macrophage apoptosis requires ER stress pathways and engagement of the type A scavenger receptor. J Cell Biol. 2005;171:61–73. doi: 10.1083/jcb.200502078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Nanda V, Direnzo D, Ye J, Xiao S, Kojima Y, Howe KL, Jarr KU, Flores AM, Tsantilas P, et al. Clonally expanding smooth muscle cells promote atherosclerosis by escaping efferocytosis and activating the complement cascade. Proc Natl Acad Sci U S A. 2020;117:15818–15826. doi: 10.1073/pnas.2006348117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith HL, Freeman OJ, Butcher AJ, Holmqvist S, Humoud I, Schätzl T, Hughes DT, Verity NC, Swinden DP, Hayes J, et al. Astrocyte unfolded protein response induces a specific reactivity state that causes non-cell-autonomous neuronal degeneration. Neuron. 2020;105:855–866.e5. doi: 10.1016/j.neuron.2019.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toma I, McCaffrey TA. Transforming growth factor-β and atherosclerosis: interwoven atherogenic and atheroprotective aspects. Cell Tissue Res. 2012;347:155–175. doi: 10.1007/s00441-011-1189-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631 [DOI] [PubMed] [Google Scholar]
- 44.Dukler N, Booth GT, Huang YF, Tippens N, Waters CT, Danko CG, Lis JT, Siepel A. Nascent RNA sequencing reveals a dynamic global transcriptional response at genes and enhancers to the natural medicinal compound celastrol. Genome Res. 2017;27:1816–1829. doi: 10.1101/gr.222935.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tufanli O, Telkoparan Akillilar P, Acosta-Alvear D, Kocaturk B, Onat UI, Hamid SM, Çimen I, Walter P, Weber C, Erbay E. Targeting IRE1 with small molecules counteracts progression of atherosclerosis. Proc Natl Acad Sci U S A. 2017;114:E1395–E1404. doi: 10.1073/pnas.1621188114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowden KL, Dubland JA, Chan T, Xu YH, Grabowski GA, Du H, Francis GA. LAL (Lysosomal Acid Lipase) Promotes Reverse cholesterol transport in vitro and in vivo. Arterioscler Thromb Vasc Biol. 2018;38:1191–1201. doi: 10.1161/ATVBAHA.117.310507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dubland JA, Allahverdian S, Besler KJ, Ortega C, Wang Y, Pryma CS, Boukais K, Chan T, Seidman MA, Francis GA. Low LAL (Lysosomal Acid Lipase) expression by smooth muscle cells relative to macrophages as a mechanism for arterial foam cell formation. Arterioscler Thromb Vasc Biol. 2021;41:e354–e368. doi: 10.1161/ATVBAHA.120.316063 [DOI] [PubMed] [Google Scholar]
- 48.Bowden KL, Bilbey NJ, Bilawchuk LM, Boadu E, Sidhu R, Ory DS, Du H, Chan T, Francis GA. Lysosomal acid lipase deficiency impairs regulation of ABCA1 gene and formation of high density lipoproteins in cholesteryl ester storage disease. J Biol Chem. 2011;286:30624–30635. doi: 10.1074/jbc.M111.274381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gamblin C, Rouault C, Lacombe A, Langa-Vives F, Farabos D, Lamaziere A, Clément K, Gautier EL, Yvan-Charvet L, Dugail I. Lysosomal Acid Lipase Drives Adipocyte Cholesterol Homeostasis and Modulates Lipid Storage in Obesity, Independent of Autophagy. Diabetes. 2021;70:76–90. doi: 10.2337/db20-0578 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The raw single-cell RNA sequencing data have been deposited to SRA (Sequence Read Archive) under accession number PRJNA850125.