Abstract
The monomer composition of the exopolysaccharides (EPS) produced by Streptococcus thermophilus LY03 and S. thermophilus Sfi20 were evaluated by high-pressure liquid chromatography with amperometric detection and nuclear magnetic resonance spectroscopy. Both strains produced the same EPS composed of galactose, glucose, and N-acetylgalactosamine. Further, it was demonstrated that the activity of the precursor-producing enzyme UDP-N-acetylglucosamine 4-epimerase, converting UDP-N-acetylglucosamine into UDP-N-acetylgalactosamine, is responsible for the presence of N-acetylgalactosamine in the EPS repeating units of both strains. The activity of UDP-N-acetylglucosamine 4-epimerase was higher in both S. thermophilus strains than in a non-EPS-producing control strain. However, the level of this activity was not correlated with EPS yields, a result independent of the carbohydrate source applied in the fermentation process. On the other hand, both the amounts of EPS and the carbohydrate consumption rates were influenced by the type of carbohydrate source used during S. thermophilus Sfi20 fermentations. A correlation between activities of the enzymes α-phosphoglucomutase, UDP-glucose pyrophosphorylase, and UDP-galactose 4-epimerase and EPS yields was seen. These experiments confirm earlier observed results for S. thermophilus LY03, although S. thermophilus Sfi20 preferentially consumed glucose for EPS production instead of lactose in contrast to the former strain.
Exopolysaccharides (EPS) produced by lactic acid bacteria (LAB) have gained increasing attention over the last few years. LAB are food-grade microorganisms, and the EPS that they produce contribute to the rheology and texture of food products (4, 9a). Recently, important advances have been made concerning the chemical structure of EPS from lactococci, lactobacilli, and streptococci, as well as concerning the factors influencing their production and rheological properties (4, 9a, 29). Most EPS are composed of the neutral sugars glucose, galactose, and/or rhamnose (3, 12, 15, 20, 21, 23, 27, 30, 31, 34–36, 40–42, 44, 49). Some of them also consist of (acetylated) amino sugars (11, 32, 33, 37, 48), and only one EPS containing fucose has been described (25). Gene clusters directing EPS biosynthesis in mesophilic Lactococcus lactis subsp. cremoris and thermophilic Streptococcus thermophilus strains are organized in four functional regions: a central region with genes for glycosyltransferases specifically required for the assemblage of the EPS repeating unit, two regions flanking the central region that show homology to enzymes involved in polymerization and export, and a regulatory region located at the 5′ end of the EPS gene cluster (1, 2, 17, 37, 45). The chimeric structure of the eps loci suggests a very complex evolution, probably involving both horizontal transfer and exchanges within L. lactis and S. thermophilus species (1, 2).
Glycosyltransferase genes from both mesophilic and thermophilic EPS producers have been studied by homologous and heterologous expression and seem to be the determining factors for EPS biosynthesis, monomer composition, and linkages (38, 39, 46, 47). These enzymes form a repeating unit that is most probably connected to a lipid carrier anchored in the cytoplasmic membrane, which is most likely followed by transport of the repeating units across the membrane and polymerization of several hundred to several thousand repeating units to form the final EPS. However, not only the glycosyltransferases but also the enzymes involved in the biosynthesis of sugar nucleotides and sugar interconversions seem to play an important role in EPS production (6, 13, 18, 24). For instance, a correlation has been shown between the activities of the enzymes α-phosphoglucomutase, glucose pyrophosphorylase, and UDP-galactose 4-epimerase, on the one hand, and EPS yields for S. thermophilus LY03, a strain that produces EPS consisting of glucose and galactose (6), on the other. For Lactobacillus sakei 0-1 producing EPS composed of glucose and rhamnose, an additional correlation is seen between the activities of enzymes involved in dTDP-rhamnose synthesis and EPS yields (7). However, contradictory results are reported in the literature concerning the relationship between enzyme activities and EPS production (9a). At present, all of the genes encoding the enzymes that are putatively involved in the biosynthesis of the sugar nucleotides UDP-glucose (galU), UDP-galactose (galE), and dTDP-glucose and dTDP-rhamnose (rfbACBD) from glucose-1-phosphate have been cloned from L. lactis MG 1363 (19, 22). The mechanism of incorporation of the amino sugar N-acetylgalactosamine in the EPS repeating unit is still unclear. However, it was seen that this component was absent in the EPS from the recombinant L. lactis MG 1363 strain, when MG 1363 was transformed with a plasmid harboring the eps gene cluster from S. thermophilus Sfi6, although the native EPS was present in incorporation of the latter strain. The synthesis of an EPS with a similar size, but in which incorporation of galactose instead of N-acetylgalactosamine took place, is most probably due to the lack of UDP-N-acetylglucosamine 4-epimerase activity in the former strain (39).
In this study, the exact monomer composition and structure of the EPS produced by S. thermophilus LY03 and S. thermophilus Sfi20 was determined through high-pressure liquid chromatography (HPLC) and nuclear magnetic resonance (NMR), respectively. Both strains produced the same EPS consisting of galactose, glucose and N-acetylgalactosamine. Furthermore, this study explored the association of the activity of the precursor-producing enzyme UDP-N-acetylglucosamine 4-epimerase, converting UDP-N-acetylglucosamine into UDP-N-acetylgalactosamine, with the presence of the particular monomer N-acetylgalactosamine, by measuring its activity in both S. thermophilus strains compared to that in a non-EPS-producing control strain. In addition, this quantitative study was set up to confirm results, reported earlier for S. thermophilus LY03, on the correlation between activities of the enzymes α-phosphoglucomutase, UDP-glucose pyrophosphorylase, and UDP-galactose 4-epimerase, on the one hand, and EPS yields, on the other. In parallel, levels of carbohydrate utilization were compared between the two EPS-producing S. thermophilus strains.
MATERIALS AND METHODS
Bacterial strains and media.
S. thermophilus LY03 (kindly provided by V. Marshall, University of Huddersfield, Huddersfield, United Kingdom) and S. thermophilus Sfi20 (kindly provided by B. Mollet, Nestec, Ltd., Research Centre, Lausanne, Switzerland) were used as the EPS-producing strains throughout this study. The non-EPS-producing strain S. thermophilus NR (kindly provided by V. Marshall, University of Huddersfield) was used as a control for measuring UDP-N-acetylglucosamine 4-epimerase activities. The strains were stored at −80°C in de Man Rogosa Sharpe (MRS) broth (Oxoid, Basingstoke, United Kingdom), containing 25% (vol/vol) glycerol (9). To obtain fresh cultures, the bacteria were propagated twice (12 h at 42°C) in a medium identical to the one used for the fermentations later on (see below). The fermentor inoculum was always prepared in two steps. First, 10 ml of customized MRS or milk medium (see below) was inoculated with 100 μl of a freshly prepared culture. After 12 h of incubation at 42°C, it was used to inoculate 100 ml of customized MRS or milk medium. After another 12 h of growth at 42°C, this second preculture was used to inoculate the fermentor (10 liters).
Milk medium (10.0% [wt/vol] skimmed milk powder) was used for fermentations to produce EPS for both monomer composition analysis by pulsed amperometric detection through HPLC and structure elucidation by NMR spectroscopy (see below). For all other fermentations, a customized MRS medium was used; it contained (in grams liter−1): peptone (Oxoid), 30; yeast extract (Merck, Darmstadt, Germany), 12; Lab Lemco (Oxoid), 8; K2HPO4, 2; sodium acetate, 5; triammonium citrate, 2; MgSO4 · 7H2O, 0.2; MnSO4 · H2O, 0.038; and Tween 80 (1 ml liter−1) (5).
Determination of the EPS monomer composition.
Routine monomer analysis of the EPS produced by S. thermophilus LY03 and S. thermophilus Sfi20 was done by HPLC (Waters Corp., Milford, Mass.), with refractive index detection (Waters 410 differential refractometer; Waters Corp.) and equipped with a Polyspher OA KC column (Merck). The detection range of all sugars was 0.5 to 10.0 g liter−1, and the standard deviation averaged 5.0%.
To determine the exact monomer composition of the EPS, both strains were inoculated (1%, vol/vol) from an overnight milk culture into 1-liter glass bottles of sterile milk medium (121°C, 20 min) and grown at 42°C for 12 h. The EPS were isolated as described before (10), dialyzed against ultrapure water for 6 days, and hydrolyzed (6 N trifluoroacetic acid at 100°C for 3 h). The monomer composition of the hydrolyzed EPS was determined using HPLC with a pulsed amperometric detector (Dionex, Sunnyvale, Calif.) and equipped with a CarboPac PA10 column (Dionex). A sodium hydroxide-sodium acetate gradient was used. Using this technique, the monomer composition could be determined accurately within a detection range of 0.001 to 0.010 g liter−1.
EPS structure elucidation by one-dimensional NMR spectroscopy.
Strains were inoculated (2%, vol/vol) from an overnight milk culture into 1-liter glass bottles of sterile milk medium (121°C, 20 min) and incubated at 42°C for 18 h. EPS were extracted by precipitating proteins with trichloroacetic acid (final concentration, 17%) and subsequently precipitating the EPS from the supernatant with an equal volume of chilled (4°C) ethanol. After the EPS was dialyzed against tap water for 72 h (the water was changed at least three times), it was freeze-dried. For NMR spectroscopy analysis, the lyophilized polysaccharides (10 mg ml−1) were dissolved directly in D2O (99.9% D; Goss Scientific Instruments, Ltd., Essex, United Kingdom). NMR spectra were recorded at a probe temperature of 70°C. The elevated temperature shifted the HOD signal to a higher field into a clear region of the spectrum. The higher temperature also increased the spectral resolution by reducing the sample viscosity. The NMR spectra were recorded on a Bruker Avance DPX400 MHz spectrometer operating with Z-field gradients and using Bruker's pulse programs. Chemical shifts (δ) were expressed in parts per million relative to internal acetone, δ 2.225. The one-dimensional 1H-NMR spectra were processed with 32,768 data points. The two-dimensional gs-DQF-COSY spectrum was recorded in magnitude mode at 70°C; the time domain data were multiplied by a squared-sine-bell function (SSB 0). After application of a linear prediction and after Fourier transformation, data sets of 1,024 by 1,024 points were obtained.
Fermentation conditions.
All fermentations were done in 15-liter laboratory fermentors (BiostatC; B. Braun Biotech International, Melsungen, Germany), with a working volume of 10 liters. The fermentors were computer controlled (MicroMFCS for WindowsNT; B. Braun Biotech International) and were sterilizable in situ. Sterilization was performed at 121°C for 20 min. For all fermentations, the optimal ratio of the initial carbohydrate-complex nitrogen concentration as determined earlier was applied (5). Lactose (0.22 M) and glucose (0.42 M), as well as additions of 0.14 M glucose, 0.14 M galactose, or 0.14 M fructose to 0.15 M lactose and additions of 0.07 M lactose, 0.14 M galactose, or 0.14 M fructose to 0.28 M glucose, were examined as the carbohydrate source(s). Carbohydrates were sterilized separately (20 min at 121°C) and aseptically pumped into the fermentor. The pH was controlled at 6.2 ± 0.1 by the automatic addition of 10 N NaOH. The pH level and the amount of base added were monitored on line. The temperature was kept constant at 42 ± 0.1°C. To keep the fermentation broth homogeneous, agitation was performed at 100 rpm with a stirrer composed of three standard impellers.
At regular time intervals, samples were aseptically withdrawn from the fermentor to determine the biomass (cell dry mass [CDM]), EPS production (polymer dry mass [PDM]), lactic acid concentration, galactose concentration, and residual carbohydrate concentrations (lactose, glucose, and fructose) as described elsewhere (10). EPS isolation, including both high-molecular-mass EPS (HMM-EPS) and low-molecular-mass EPS (LMM-EPS), and routine monomer analysis were done as described previously, with standard deviations of ca. 20.0 and 5.0%, respectively (5). The maximum specific growth rate (μmax, hour−1) was calculated as the maximum slope from the linearized values of the biomass (grams of CDM liter−1) as a function of fermentation time (in hours). The maximum carbohydrate consumption rates (rmax, hour−1) were calculated from the residual concentrations of the carbohydrate in the medium.
Correlations between enzyme activities and EPS yields.
Samples of 50 ml were taken at three time points to prepare cell extracts for measuring enzyme activities: once during the exponential growth phase, at the end of the exponential growth phase when EPS production reached its maximum, and during the stationary phase or beyond the EPS maximum. Cell extracts were prepared as described previously (6). The protein content of the cell extracts was determined with a commercial DC protein assay kit (Bio-Rad Laboratories, Hercules, Calif.) that is based on the method of Lowry et al. (26). Crude cell extracts of both S. thermophilus Sfi20 and S. thermophilus LY03 and of a non-EPS-producing S. thermophilus NR control strain were assayed for UDP-N-acetylglucosamine 4-epimerase activity. This assay was performed as described by Estrela et al. (14). The activities of nine enzymes either involved in the Embden-Meyerhof-Parnas (EMP) pathway (phosphoglucose isomerase, 6-phosphofructokinase, and fructose-1,6-bisphosphatase) or in the biosynthesis of glucose-1-phosphate (α- or β-phosphoglucomutase) and sugar nucleotides as EPS precursor molecules (UDP-glucose pyrophosphorylase, UDP-galactose 4-epimerase, and dTDP-glucose pyrophosphorylase) were determined for S. thermophilus Sfi20 only, as described elsewhere (6).
All enzyme activities were measured in triplicate and are expressed as mean values and standard deviations. In all of the assays, the reaction velocity was linearly proportional to the amount of cell extract. All reactions were carried out in a final volume of 0.5 ml. The statistical significance of associations between enzyme activities and amounts of EPS was determined based on a correlation test as outlined previously (6).
RESULTS
EPS production kinetics of S. thermophilus Sfi20.
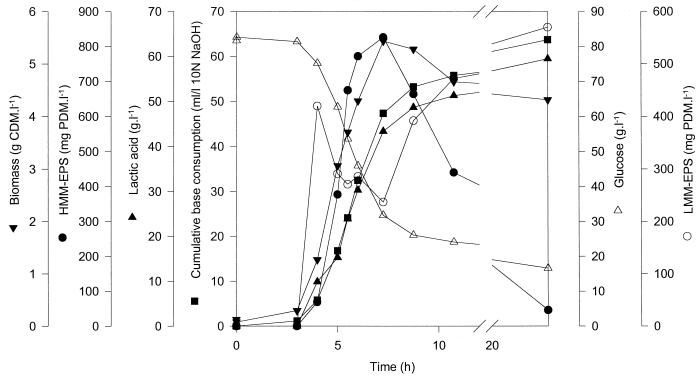
Figure 1 represents a typical fermentation profile of the EPS-producing S. thermophilus Sfi20 strain grown in customized MRS medium with 0.42 M glucose as the sole carbohydrate source under optimal conditions of controlled temperature (42°C) and pH (6.2). Exponential growth took place during ca. 5 h. The stationary phase began after about 8 h of fermentation. Glucose was homofermentatively converted into lactic acid (by glycolytic degradation), and after 24 h, 16 g of glucose liter−1 remained unused in the fermentation broth. EPS production displayed primary metabolite kinetics (10). S. thermophilus Sfi20 produced an HMM-EPS and an LMM-EPS, as was the case for S. thermophilus LY03 (5). HMM-EPS was produced mainly during the exponential growth phase. The production started after about 4 h of fermentation and reached a maximum of 826 mg of PDM liter−1 after 7 h of fermentation. LMM-EPS production started at the beginning of the exponential growth phase, decreased at the beginning of the stationary phase, and reached a maximum at the end of the fermentation. The decrease of HMM-EPS and the concomitant increase of LMM-EPS, after 10 h of fermentation, might possibly be due to enzymatic degradation. In contrast, at the beginning of the fermentation, both fractions were produced simultaneously. A comparable fermentation profile was observed for all fermentations carried out.
FIG. 1.
Batch fermentation profile of S. thermophilus Sfi20 growth and EPS production at 42°C and at a constant pH of 6.2. Cells were grown in a BiostatC fermentor containing 10 liters of customized MRS medium with 0.42 M glucose as the sole carbohydrate source. Symbols: ▪, cumulative base consumption; ▾, biomass; ●, HMM-EPS; ○, LMM-EPS; ▵, residual glucose concentration; ▴, lactic acid.
Monomer composition of the EPS produced by S. thermophilus Sfi20 and S. thermophilus LY03.
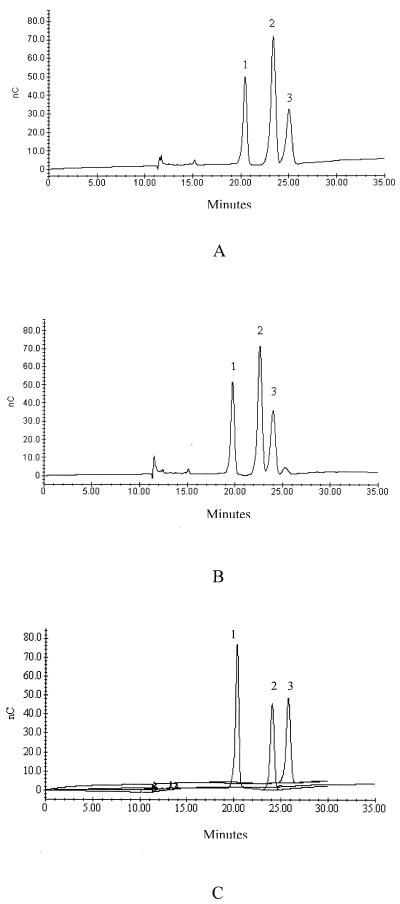
Routine HPLC analysis with refractive index detection of both HMM-EPS and LMM-EPS of both S. thermophilus Sfi20 and S. thermophilus LY03, grown in customized MRS medium, resulted in the following galactose-glucose monomer compositions: 3.0:1.0 for S. thermophilus Sfi20 and 4.0:1.0 for S. thermophilus LY03 (10). Determination of the monomer composition of the EPS from both strains grown in milk medium by HPLC with pulsed amperometric detection resulted in a more precise composition, namely, galactose–glucose–N-acetylgalactosamine in 3.1:1.5:1.0 and 3.2:1.4:1.0 ratios for S. thermophilus Sfi20 and S. thermophilus LY03, respectively (Fig. 2). HPLC with refractive index detection could not detect galactosamine and could not distinguish between galactose and mannose. During acid hydrolysis, N-acetylgalactosamine is converted into galactosamine. When we used HPLC with amperometric detection, we found that glucose, galactose, and galactosamine could be separated efficiently. When the strains are grown and subcultured in MRS medium, mannose is also detected, derived from the glucomannans present in yeast extract. This mannose contributes to the higher galactose peak for routine HPLC analysis with refractive index detection. At present, it is not known why more mannose is isolated from the medium in the case of S. thermophilus LY03 than in the case of S. thermophilus Sfi20. For the remainder of this study, all EPS monomer compositions were determined using the routine HPLC method, since the only objective was to check whether the EPS monomer composition changed during fermentation. Unless stated otherwise, reference is made to HMM-EPS. However, both EPS possess the same monomeric composition and are produced throughout the fermentation cycle.
FIG. 2.
Determination of the monomer composition of the EPS from S. thermophilus LY03 (A) and S. thermophilus Sfi20 (B) by HPLC with pulsed amperometric detection. (C) The standards (10 ppm) are visualized to reflect their differences in sensitivity. Peaks are designated as follows: 1, galactosamine; 2, galactose; and 3, glucose.
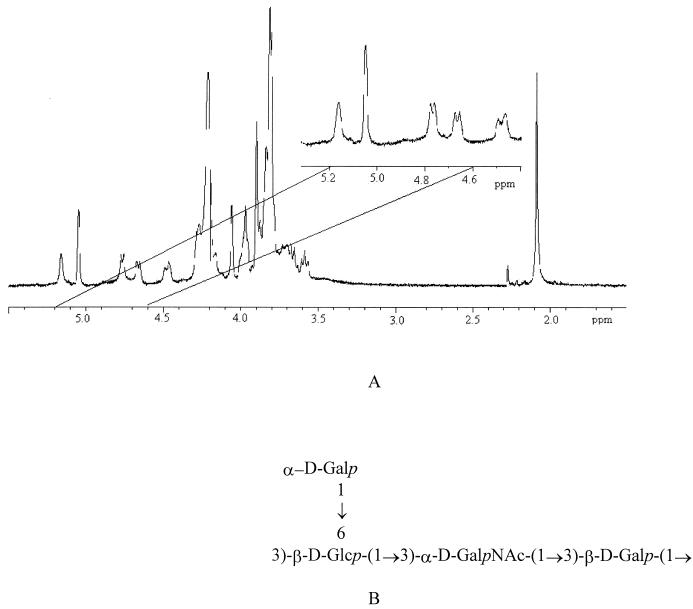
Structure elucidation of the EPS produced by S. thermophilus LY03 and S. thermophilus Sfi20 by NMR spectroscopy.
The 1H-NMR spectra recorded were identical for the EPS samples isolated during the stationary phase from several fermentations of S. thermophilus LY03 and S. thermophilus Sfi20. There are four low field H-1 signals (δ 5.16 H-1, 4.67 H-1, 4.76 H-1, and 5.05 H-1) and a high field N-acetyl methyl resonance (δ 2.06) (Fig. 3A). The locations of the related H-2 resonances were available from the COSY spectrum (δ 4.49 H-2, 3.58 H-2, 3.76 H-2, and 3.91 H-2). The spectra and the H-1 and H-2 chemical shifts are in remarkable agreement with those reported for the EPS isolated from S. thermophilus CNCMI 733 (11). The structure for the EPS from both strains is the same as that reported for S. thermophilus CNCMI 733 (11) and S. thermophilus Sfi6 (37), i.e., a branched tetrasaccharide repeating unit consisting of galactose, glucose, and N-acetylgalactosamine (Fig. 3B).
FIG. 3.
The structure of the exopolysaccharides produced by Streptococcus thermophilus LY03 and S. thermophilus Sfi20 determined by 400-MHz 1H-NMR spectroscopy (A) are identical to the one reported by Doco et al. (11) for S. thermophilus CNCMI 733 (B). Only the 1H-NMR spectrum of the S. thermophilus LY03 exopolysaccharide is shown.
Influence of the carbohydrate source on EPS yields and monomer composition.
As found with S. thermophilus LY03 (6), S. thermophilus Sfi20 did not consume galactose, fructose, rhamnose, maltose, and sucrose as the sole carbohydrate source (data not shown). The highest amounts of EPS were obtained on glucose as the sole carbohydrate and with a combination of lactose and glucose (Table 1). The rmax values confirm that glucose seemed to be used preferentially compared to lactose, in particular when both carbohydrate sources were present. For both strains, fructose was fermented only when applied in combination with glucose or lactose (data not shown; see also reference 6). For all fermentations carried out with two carbohydrate sources, except for the ones where galactose was used as additional carbohydrate source, both sugars were consumed simultaneously and converted into lactic acid. Galactose as the sole carbohydrate source was not consumed; when galactose was added to glucose or lactose, it was consumed only upon prolonged fermentation (data not shown). The amount of EPS (total EPS, HMM-EPS and LMM-EPS) was influenced by the nature of the carbohydrate source used. However, the EPS monomer composition and galactose/glucose ratio remained unchanged for all carbohydrates and carbohydrate combinations tested; an average value of 3:1 was always reported.
TABLE 1.
Influence of carbohydrate source(s) on growth and EPS production during S. thermophilus Sfi20 fermentationsa
| Carbohydrate source(s) (concn [M]) | μmax (h−1) (r2) | rmax (h−1)b (r2) | Biomassmax (g of CDM liter−1) (h) | (HMM-EPS)max (mg of PDM liter−1) (h) | (LMM-EPS)max (mg of PDM liter−1) (h) | (Total EPS)max (mg of PDM liter−1) (h) |
|---|---|---|---|---|---|---|
| Lactose (0.22) | 1.1 (0.997) | 0.5 (0.996) | 5.5 (11.25) | 685 (5.50) | 547 (3.75) | 880 (5.50) |
| Lactose (0.15) + fructose (0.14) | 1.2 (0.999) | 0.3 (0.999) | 5.0 (8.00) | 272 (12.00) | 435 (10.00) | 651 (12.00) |
| 0.3 (0.990) | ||||||
| Lactose (0.15) + galactose (0.14) | 0.9 (0.971) | 0.5 (0.999) | 3.3 (7.00) | 187 (7.00) | 494 (24.00) | 597 (7.00) |
| Lactose (0.15) + glucose (0.14) | 1.1 (0.954) | 0.4 (0.995) | 3.9 (5.75) | 1,162 (5.75) | 456 (23.50) | 1,238 (5.75) |
| 0.7 (0.880) | ||||||
| Glucose (0.42) | 1.2 (0.980) | 0.3 (0.999) | 5.4 (7.25) | 826 (7.25) | 570 (24.50) | 1,096 (8.75) |
| Glucose (0.28) + fructose (0.14) | 1.2 (0.993) | 0.4 (0.995) | 4.8 (7.00) | 401 (10.50) | 537 (7.00) | 771 (7.00) |
| 0.1 (0.999) | ||||||
| Glucose (0.28) + galactose (0.14) | 1.1 (0.977) | 0.7 (0.999) | 4.4 (14.75) | 326 (12.75) | 433 (13.25) | 691 (12.00) |
| Glucose (0.28) + lactose (0.07) | 1.2 (0.966) | 0.7 (0.962) | 5.4 (26.50) | 793 (16.50) | 390 (13.50) | 1,018 (16.50) |
| 0.1 (0.993) |
Fermentations were performed in MRS broth at 42°C and at a controlled pH of 6.2. The biokinetic parameters μmax and rmax were estimated through modeling of the experimental data.
Dual values refer to the respective carbohydrate sources.
Correlations between enzyme activities and EPS yields.
Based on the observation that N-acetylgalactosamine was present in the EPS from S. thermophilus Sfi20 and S. thermophilus LY03, the UDP-N-acetylglucosamine 4-epimerase activity was measured in cell extracts of both strains at three different time points of fermentations carried out with glucose or glucose combinations and with lactose or lactose combinations as the carbohydrate source(s) (Tables 2 and 3). UDP-N-acetylglucosamine 4-epimerase converts the cell wall biosynthesis precursor UDP-N-acetylglucosamine into UDP-N-acetylgalactosamine. Both strains displayed UDP-N-acetylglucosamine 4-epimerase activity in all phases of the fermentation for all fermentable carbohydrates and carbohydrate combinations tested. No activity was observed in the non-EPS-producing S. thermophilus NR strain. However, it was not possible to correlate UDP-N-acetylglucosamine 4-epimerase activity with the total amount of EPS produced. Indeed, correlations (r) of 0.19 and 0.47 were observed for the S. thermophilus Sfi20 strain and the S. thermophilus LY03 strain, respectively (P > 0.10). Interestingly, the highest UDP-N-acetylglucosamine 4-epimerase activity was observed for all fermentations at the end of the exponential growth phase, the time point at which the maximum amount of EPS was produced as well.
TABLE 2.
Amounts of total EPS measured in fermented medium of the EPS-producing strain S. thermophilus Sfi20a grown on glucose or on combinations of glucose and fructose, galactose, or lactose on the one hand and on lactose or on combinations of lactose and fructose, galactose, or glucose on the other hand
| Carbohydrate source(s) (concn [M]) | Amt (mg of PDM liter−1) of EPS produced at time point:
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| Glucose (0.42) | 668 | 1,063 | 912 |
| Glucose (0.28) + fructose (0.14) | 386 | 771 | 482 |
| Glucose (0.28) + galactose (0.14) | 437 | 691 | 552 |
| Glucose (0.28) + lactose (0.07) | 595 | 1,018 | 590 |
| Lactose (0.22) | 644 | 880 | 736 |
| Lactose (0.15) + fructose (0.14) | 549 | 506 | 651 |
| Lactose (0.15) + galactose (0.14) | 390 | 597 | 480 |
| Lactose (0.15) + glucose (0.14) | 961 | 1,283 | 1,057 |
S. thermophilus Sfi20 was grown in batch cultures in MRS broth on glucose or lactose or on combinations of glucose or lactose with other carbohydrates at 42°C and at a constant pH of 6.2. Samples were taken at three time points of the fermentation course, representing the exponential growth phase (point 1), the end of the exponential growth phase (point 2), and the stationary phase (point 3). Each value is always the average of two measurements; the standard deviation is approximately 20%. For the results of S. thermophilus LY03, see reference 6.
TABLE 3.
Activity of UDP-N-acetylglucosamine 4-epimerase in cell extracts of S. thermophilus LY03 and S. thermophilus Sfi20a grown either on glucose or combinations of glucose and fructose, galactose, or lactose or on lactose or combinations of lactose and fructose, galactose, or glucosea
| Carbohydrate source (concn [M]) | Time point | Mean activity of UDP- N-acetylglucosamine 4-epimerase (nmol mg of cell protein−1 min−1) ± SD in S. thermophilus:
|
|
|---|---|---|---|
| LY03 | Sfi20 | ||
| Glucose (0.42) | 1 | 165 ± 23 | 215 ± 65 |
| 2 | 209 ± 22 | 230 ± 55 | |
| 3 | 145 ± 34 | 201 ± 30 | |
| Glucose (0.28) + fructose (0.14) | 1 | 175 ± 37 | 210 ± 29 |
| 2 | 210 ± 11 | 240 ± 16 | |
| 3 | 136 ± 19 | 209 ± 40 | |
| Glucose (0.28) + galactose (0.14) | 1 | 169 ± 10 | 178 ± 9 |
| 2 | 189 ± 11 | 219 ± 21 | |
| 3 | 123 ± 42 | 188 ± 10 | |
| Glucose (0.28) + lactose (0.07) | 1 | 201 ± 43 | 187 ± 23 |
| 2 | 209 ± 13 | 211 ± 13 | |
| 3 | 132 ± 12 | 187 ± 4 | |
| Lactose (0.22) | 1 | 180 ± 33 | 144 ± 12 |
| 2 | 203 ± 34 | 174 ± 14 | |
| 3 | 155 ± 12 | 163 ± 12 | |
| Lactose (0.15) + fructose (0.14) | 1 | 199 ± 45 | 189 ± 18 |
| 2 | 210 ± 27 | 197 ± 43 | |
| 3 | 145 ± 23 | 182 ± 24 | |
| Lactose (0.15) + galactose (0.14) | 1 | 179 ± 23 | 175 ± 15 |
| 2 | 189 ± 12 | 201 ± 5 | |
| 3 | 166 ± 14 | 200 ± 10 | |
| Lactose (0.15) + glucose (0.14) | 1 | 187 ± 11 | 189 ± 12 |
| 2 | 174 ± 12 | 212 ± 38 | |
| 3 | 168 ± 11 | 190 ± 14 | |
S. thermophilus LY03 and S. thermophilus Sfi20 were grown in batch cultures in MRS broth on lactose or glucose or combinations of lactose or glucose with other carbohydrates at 42°C and at a constant pH of 6.2. Samples were taken at three time points of the fermentation course, representing the exponential growth phase (point 1), the end of the exponential growth phase (point 2), and the stationary phase (point 3). Each value is always the average of two measurements; the standard deviation is approximately 12%.
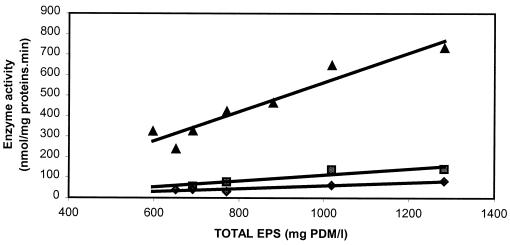
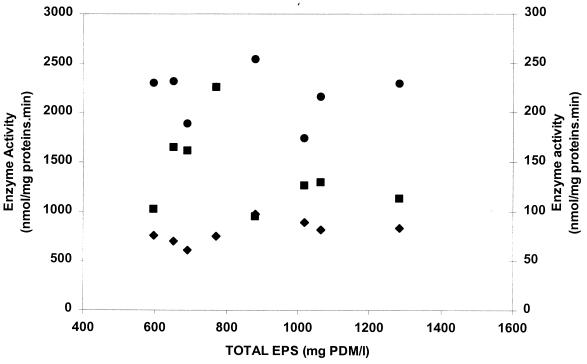
Among the other enzyme activities measured in cell extracts of S. thermophilus Sfi20, phosphoglucose isomerase and 6-phosphofructokinase involved in the EMP pathway, α-phosphoglucomutase involved in the biosynthesis of glucose-1-phosphate, and both UDP-glucose pyrophosphorylase and UDP-galactose 4-epimerase involved in the biosynthesis of the sugar nucleotides UDP-glucose and UDP-galactose were found to be highly active during growth and EPS biosynthesis. The activity of fructose-1,6-bisphosphatase was very low. The activities of β-phosphoglucomutase and dTDP-glucose pyrophosphorylase (involved in dTDP-rhamnose biosynthesis) were almost zero (data not shown). A significant correlation was found between the total amounts of EPS produced and the enzyme activities of α-phosphoglucomutase, UDP-galactose 4-epimerase, and UDP-glucose pyrophosphorylase (Fig. 4). Despite the fact that the activities of the last two enzymes were lower than for S. thermophilus LY03, all three enzyme activities were significantly correlated with EPS yield (P < 0.05). No correlation at all was observed between the total amount of EPS produced and the activities of phosphoglucose isomerase, 6-phosphofructokinase, and fructose-1,6-bisphosphatase (Fig. 5). The correlation coefficient (r) was always less than 0.60 (P > 0.05).
FIG. 4.
Relationship between the activities of the enzymes α-phosphoglucomutase (▴; r = 0.96), UDP-galactose 4-epimerase (⧫; r = 0.93), and UDP-glucose pyrophosphorylase (▪; r = 0.93) and the total amount of EPS produced (n = 8).
FIG. 5.
Relationship between the activities of the enzymes phosphoglucose isomerase (primary axis) (⧫; r = 0.55; n = 8), 6-phosphofructokinase (primary axis) (▪; r = −0.32; n = 8), and fructose-1,6-bisphosphatase (secondary axis) (●; r = −0.09; n = 7) and the total amount of EPS produced.
DISCUSSION
The yogurt strains S. thermophilus LY03 and S. thermophilus Sfi20 produce at the same time two heterotype EPS with different molecular sizes, an HMM-EPS and an LMM-EPS. All EPS produced by both strains display the same monomer composition, namely, galactose, glucose, and (N-acetyl)galactosamine in the average ratio 2:1:1. The same primary structure was observed as for S. thermophilus CNCMI 733 (11) and S. thermophilus Sfi6 (37).
The biosynthesis of bacterial EPS is initiated by the synthesis of the repeating units of sugar nucleotides (9a). The incorporation of the constituting activated monosaccharides into these EPS repeating units will depend on the activity of the corresponding specific glycosyltransferases, their encoding genes being part of the eps gene clusters (37, 45). However, the supply of the activated sugars for EPS biosynthesis is dependent on the intracellular sugar nucleotide levels that are in turn influenced by the activities of the intracellular enzymes involved in their biosynthesis and interconversion. These enzymes have to be generated from housekeeping genes in the bacteria because they are involved in other indispensable cell processes as well. Common sugar nucleotides such as UDP-glucose and UDP-galactose are readily available, since they are required for cell wall biosynthesis (8). In addition, a positive correlation has been observed for S. thermophilus LY03 and S. thermophilus Sfi20 between the activities of enzymes UDP-glucose pyrophosphorylase and UDP-galactose 4-epimerase and the EPS yields, a correlation independent of the fermentable carbohydrate source(s) used (6, 13, 16). A higher activity of these enzymes will be responsible for an additional supply of UDP-glucose and UDP-galactose to be incorporated in the EPS repeating units. UDP-galactose 4-epimerase is also involved in galactose breakdown by the Leloir pathway, fueling UDP-galactose through galactose-1-phosphate from galactose. Since most S. thermophilus strains are galactose negative, UDP-galactose 4-epimerase is believed to be especially active in EPS-producing S. thermophilus strains (6, 19, 28). With EPS production by L. lactis subsp. cremoris NIZO B40, no relationship between activities of precursor-forming enzymes and the amounts of EPS produced on glucose and fructose was found. This may be explained by the difference in EPS production kinetics between mesophilic and thermophilic LAB strains (5). EPS production in mesophilic LAB is not necessarily growth associated, so that the pool of enzymes involved in the biosynthesis of EPS precursors is fully available for EPS production either in the stationary growth phase or under nonoptimal growth conditions. The growth-associated character of EPS production in thermophilic LAB requires increased activities of EPS precursor-forming enzymes. Hence, the higher the EPS yields, the higher are the activities of the key enzymes.
The sugar nucleotides UDP-N-acetylgalactosamine and dTDP-rhamnose are not necessary for the normal cell functions and have to be synthesized from glucose-1-phosphate and from the cell wall precursor UDP-N-acetylglucosamine (in turn derived from fructose-6-phosphate), respectively, by UDP-N-acetylglucosamine 4-epimerase and dTDP-rhamnose synthesizing enzymes, respectively. The presence of these enzymes will thus determine whether N-acetylgalactosamine or rhamnose may be present in the EPS repeating unit (7, 39). Indeed, the EPS repeating units from the strains S. thermophilus LY03 and S. thermophilus Sfi20 contain N-acetylgalactosamine but no rhamnose. Hence, both strains displayed high UDP-N-acetylglucosamine 4-epimerase activity and no dTDP-glucose pyrophosphorylase activity, while the activity of both enzymes was zero for the non-EPS-producing S. thermophilus NR strain. However, it is remarkable that UDP-N-acetylglucosamine 4-epimerase activities of the EPS-producing strains were not correlated with the amounts of EPS produced, indicating that the enzyme might possibly be involved in other, unknown metabolic processes.
With such data the identification of the rate-limiting enzymes in EPS production is thus difficult. On the one hand, some enzymes, in particular those involved in sugar nucleotide biosynthesis, take part in several cell processes. On the other hand, precursor molecules of the sugar nucleotides, such as glucose-6-phosphate and hence glucose-1-phosphate, serve as precursors or intermediates in many other pathways. As an example, the fructose-specific effect of lower EPS production levels on fructose than on glucose or lactose, seen in Lactobacillus delbrueckii subsp. bulgaricus (18) and L. lactis (24), was supposed to be a result of the (very) low activity of fructose 1,6-bisphosphatase converting fructose-1,6-bisphosphate to fructose-6-phosphate, which is essential for growth on fructose but not on the other sugars. This was also supposed to be an essential step in the generation of the central sugar nucleotide precursor glucose-1-phosphate from fructose-6-phosphate (via phosphoglucose isomerase and phosphoglucomutase). However, in S. thermophilus strains, there seems to be no “backward” flow from fructose-1,6-bisphosphate to glucose-6-phosphate, a finding corresponding with a very low fructose-1,6-bisphosphatase activity (6). Therefore, the breakdown of glucose or the glucose moiety of lactose is fueling EPS biosynthesis in LAB.
The monomer composition of the EPS produced by both strains S. thermophilus LY03 and S. thermophilus Sfi20 remained unchanged when they were grown on different carbohydrates, indicating no influence of the nature of the carbohydrate source on EPS composition (5, 13). On the other hand, the highest amounts of EPS were produced on lactose and glucose for S. thermophilus LY03 and S. thermophilus Sfi20, respectively, and on a combination of lactose and glucose for both strains. The differences in EPS yields could be explained by the differences of activities of the enzymes involved in the sugar nucleotide biosynthesis, probably resulting in changed EPS precursor levels. However, the EPS composition of galactose and glucose in a 4:1 or 3:1 ratio for S. thermophilus LY03 or S. thermophilus Sfi20, respectively, does not seem to be influenced by differences of activities of the enzymes involved in the biosynthesis of sugar nucleotides when the strains are grown on different carbohydrate source(s). Finally, it has been postulated that the glucose moiety of lactose is preferentially consumed by S. thermophilus instead of glucose that is internalized from the growth medium (43). While the glucose moiety of lactose is metabolized via glycolysis, in LAB two distinct pathways, the tagatose-6-phosphate pathway and the Leloir pathway, can be used for degradation of the galactose moiety. The genes of the tagatose-6-phosphate enzymes are absent in S. thermophilus; however, the gal genes for the Leloir enzymes are present but are not fully expressed due to a defect in the induction mechanism of the rate-limiting key enzyme galactokinase GalK. Apparently, during growth on lactose, either the pool of EPS precursor-forming enzymes or the metabolic flux in the direction of sugar nucleotides is less than during growth on glucose in S. thermophilus Sfi20. The situation with the two carbohydrates is reversed for S. thermophilus LY03 (6).
To conclude, the relatively low EPS production levels displayed by (thermophilic) LAB could be increased by enhancing the metabolic flux toward the sugar nucleotides. Knowledge of the enzymes investigated in this and previous studies combined with measurements of intracellular concentrations of sugar nucleotides could lead to the development of a metabolic flux model. This strategy would generate a rationale for improvement of the EPS production levels that result from a complex biosynthesis pathway.
ACKNOWLEDGMENTS
We acknowledge financing from the European Commission (grants FAIR-CT-98-4267 and INCO Copernicus IC15-CT98-0905). L.D.V. further acknowledges financial support from the Flemish Institute for the Encouragement of Scientific and Technological Research in the Industry (IWT), the Fund for Scientific Research (FWO—Flanders), the LINK 2000 Action of the Brussels Capital Region, and the Research Council of the Vrije Universiteit Brussel. B.D. and F.V. are recipients of an IWT fellowship.
REFERENCES
- 1.Almirón-Roig E, Mulholland F, Gasson M J, Griffin A M. The complete cps gene cluster from Streptococcus thermophilus NCFB 2393 involved in the biosynthesis of a new exopolysaccharide. Microbiology. 2000;146:2793–2802. doi: 10.1099/00221287-146-11-2793. [DOI] [PubMed] [Google Scholar]
- 2.Bourgoin F, Pluvinet A, Gintz B, Decaris B, Guédon G. Are horizontal transfers involved in the evolution of the Streptococcus thermophilus exopolysaccharide synthesis loci? Gene. 1999;233:151–161. doi: 10.1016/s0378-1119(99)00144-4. [DOI] [PubMed] [Google Scholar]
- 3.Bubb W A, Urashima T, Fujiwara R, Shinnai T, Ariga H. Structural characterisation of the exocellular polysaccharide produced by Streptococcus thermophilus OR901. Carbohydr Res. 1997;301:41–50. doi: 10.1016/s0008-6215(97)00083-9. [DOI] [PubMed] [Google Scholar]
- 4.Cerning J, Marshall V M. Exopolysaccharides produced by the dairy lactic acid bacteria. Rec Res Dev Microbiol. 1999;3:195–209. [Google Scholar]
- 5.Degeest B, De Vuyst L. Indication that the nitrogen source influences both amount and size of exopolysaccharides produced by Streptococcus thermophilus LY03 and modeling of bacterial growth and exopolysaccharide production. Appl Environ Microbiol. 1999;65:2863–2870. doi: 10.1128/aem.65.7.2863-2870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Degeest B, De Vuyst L. Correlation of activities of the enzymes α-phosphoglucomutase, UDP-galactose 4-epimerase, and UDP-glucose pyrophosphorylase with exopolysaccharide biosynthesis by Streptococcus thermophilus LY03. Appl Environ Microbiol. 2000;66:3519–3527. doi: 10.1128/aem.66.8.3519-3527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degeest, B., B. Janssens, and L. De Vuyst. Exopolysaccharide (EPS) biosynthesis by Lactobacillus sakei 0-1: production kinetics, enzyme activities, and EPS yields. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 8.Delcour J, Ferain T, Deghorain M, Palulmbo E, Hols P. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:159–184. [PubMed] [Google Scholar]
- 9.de Man J C, Rogosa M, Sharpe M E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 9a.De Vuyst L, Degeest B. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol Rev. 1999;23:157–177. doi: 10.1111/j.1574-6976.1999.tb00395.x. [DOI] [PubMed] [Google Scholar]
- 10.De Vuyst L, Vanderveken F, Van de Ven S, Degeest B. Production by and isolation of exopolysaccharides from Streptococcus thermophilus grown in milk medium and evidence for their growth-associated biosynthesis. J Appl Microbiol. 1998;84:1059–1068. doi: 10.1046/j.1365-2672.1998.00445.x. [DOI] [PubMed] [Google Scholar]
- 11.Doco T, Wieruszeski J M, Fournet B. Structure of an exocellular polysaccharide produced by Streptococcus thermophilus. Carbohydr Res. 1990;198:313–321. doi: 10.1016/0008-6215(90)84301-a. [DOI] [PubMed] [Google Scholar]
- 12.Dueñas-Chasco M T, Rodriguez-Carvajal M A, Tejero-Mateo P, Franco-Rodriguez G, Espartero J L, Irastorza-Iribar A, Gil- Serrano A M. Structural analysis of the exopolysaccharides produced by Lactobacillus sp. G-77. Carbohydr Res. 1997;307:125–133. doi: 10.1016/s0008-6215(98)00034-2. [DOI] [PubMed] [Google Scholar]
- 13.Escalante A, Wacher-Rodarte C, Garcia-Garibay M, Farrés A. Enzymes involved in carbohydrate metabolism and their role on exopolysaccharide production in Streptococcus thermophilus. J Appl Microbiol. 1998;84:108–114. doi: 10.1046/j.1365-2672.1997.00330.x. [DOI] [PubMed] [Google Scholar]
- 14.Estrela A-I, Pooley H M, De Lencastre H, Karamata D. Genetic and biochemical characterization of Bacillus subtilis 168 mutants specifically blocked in the synthesis of the teichoic acid poly(3-O-β-d-glucopyranosyl-N-acetylgalactosamine 1-phosphate): gneA, a new locus, is associated with UDP-N-acetylglucosamine 4-epimerase activity. J Gen Microbiol. 1991;137:943–950. doi: 10.1099/00221287-137-4-943. [DOI] [PubMed] [Google Scholar]
- 15.Faber E J, Zoon P, Kamerling J P, Vliegenthart J F G. The exopolysaccharides produced by Streptococcus thermophilus Rs and Sts have the same repeating unit but differ in viscosity of their milk cultures. Carbohydr Res. 1998;310:269–276. doi: 10.1016/s0008-6215(98)00189-x. [DOI] [PubMed] [Google Scholar]
- 16.Forsén R, Häivä V M. Induction of stable slime-forming and mucoid states by p-fluorophenylalanine in lactic streptococci. FEMS Microbiol Lett. 1981;12:409–413. [Google Scholar]
- 17.Griffin A M, Morris V J, Gasson M J. The cpsABCDE genes involved in polysaccharide production in Streptococcus salivarius subsp. thermophilus strain NCBF 2393. Gene. 1996;183:23–27. doi: 10.1016/s0378-1119(96)00405-2. [DOI] [PubMed] [Google Scholar]
- 18.Grobben G J, Smith M R, Sikkema J, de Bont J A M. Influence of fructose and glucose on the production of exopolysaccharides and the activities of enzymes involved in the sugar metabolism and the synthesis of sugar nucleotides in Lactobacillus delbrueckii subsp. bulgaricus NCFB 2772. Appl Microbiol Biotechnol. 1996;46:279–284. [Google Scholar]
- 19.Grossiord B, Vaughan E E, Luesink E J, de Vos W M. Genetics of galactose-utilization via the Leloir pathway in lactic acid bacteria. Lait. 1998;78:77–84. [Google Scholar]
- 20.Gruter M, Leeflang B R, Kuiper J, Kamerling J P, Vliegenthart J F G. Structural characterisation of the exopolysaccharide produced by Lactobacillus delbrueckii subspecies bulgaricus rr grown in skimmed milk. Carbohydr Res. 1993;239:209–226. doi: 10.1016/0008-6215(93)84216-s. [DOI] [PubMed] [Google Scholar]
- 21.Gruter M, Leeflang B R, Kuiper J, Kamerling J P, Vliegenthart J F G. Structure of the exopolysaccharide produced by Lactococcus lactis subspecies cremoris H414 grown in a defined medium or skimmed milk. Carbohydr Res. 1992;231:273–291. doi: 10.1016/0008-6215(92)84025-n. [DOI] [PubMed] [Google Scholar]
- 22.Kleerebezem M, van Kranenburg R, Tuinier R, Boels I C, Zoon P, Looijesteijn E, Hugenholtz J, de Vos W M. Exopolysaccharides produced by Lactococcus lactis: from genetic engineering to improved rheological properties? Antonie Leeuwenhoek. 1999;76:357–365. [PubMed] [Google Scholar]
- 23.Lemoine J, Chirat F, Wieruszeski J-M, Strecker G, Favre N, Neeser J-N. Structural characterization of the exocellular polysaccharides produced by Streptococcus thermophilus SFi39 and SFi12. Appl Environ Microbiol. 1997;63:3512–3518. doi: 10.1128/aem.63.9.3512-3518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Looijesteijn P J, Boels I C, Kleerebezem M, Hugenholtz J. Regulation of exopolysaccharide production by Lactococcus lactis subsp. cremoris by the sugar source. Appl Environ Microbiol. 1999;65:5003–5008. doi: 10.1128/aem.65.11.5003-5008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Low D, Ahlgren J A, Horne D, McMahon D J, Oberg C J, Broadbent J R. Role of Streptococcus thermophilus MR-1C capsular exopolysaccharide in cheese moisture retention. Appl Environ Microbiol. 1998;64:2147–2151. doi: 10.1128/aem.64.6.2147-2151.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 27.Nakajima H, Hirota T, Toba T, Adachi S. Structure of the extracellular polysaccharide from slime-forming Lactococcus lactis subsp. cremoris SBT 0495. Carbohydr Res. 1992;224:245–253. doi: 10.1016/0008-6215(92)84110-e. [DOI] [PubMed] [Google Scholar]
- 28.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDP-glucose 4-epimerase. J Bacteriol. 1990;171:244–253. doi: 10.1128/jb.172.7.4037-4047.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricciardi A, Clementi F. Exopolysaccharides from lactic acid bacteria: structure, production, and technological applications. It J Food Sci. 2000;12:23–45. [Google Scholar]
- 30.Robijn G W, Thomas J R, Haas H, van den Berg D J C, Kamerling J P, Vliegenthart J F G. The structure of the exopolysaccharide produced by Lactobacillus helveticus 766. Carbohydr Res. 1995;276:137–154. doi: 10.1016/0008-6215(95)00171-o. [DOI] [PubMed] [Google Scholar]
- 31.Robijn G W, van den Berg D J C, Haas H, Kamerling J P, Vliegenthart J F G. Determination of the structure of the exopolysaccharide produced by Lactobacillus sake 0-1. Carbohydr Res. 1995;276:117–136. doi: 10.1016/0008-6215(95)00172-p. [DOI] [PubMed] [Google Scholar]
- 32.Robijn G W, Wienk H L J, van den Berg D J C, Haas H, Kamerling J P, Vliegenthart J F G. Structural studies of the exopolysaccharide produced by Lactobacillus paracasei 34-1. Carbohydr Res. 1996;285:129–139. doi: 10.1016/s0008-6215(96)90178-0. [DOI] [PubMed] [Google Scholar]
- 33.Robijn W G, Gallego G R, van den Berg D J C, Haas H, Kamerling J P, Vliegenthart J F G. Structural characterization of the exopolysaccharide produced by Lactobacillus acidophilus LMG9433. Carbohydr Res. 1996;288:203–218. doi: 10.1016/s0008-6215(96)90799-5. [DOI] [PubMed] [Google Scholar]
- 34.Staaf M, Widmalm G, Yang Z, Huttunen E. Structural elucidation of an extracellular polysaccharide produced by Lactobacillus helveticus. Carbohydr Res. 1996;291:155–164. doi: 10.1016/s0008-6215(96)00166-8. [DOI] [PubMed] [Google Scholar]
- 35.Staaf M, Yang Z, Huttunen E, Widmalm G. Structural elucidation of the viscous exopolysaccharide produced by Lactobacillus helveticus Lb161. Carbohydr Res. 2000;326:113–119. doi: 10.1016/s0008-6215(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 36.Stingele F, Lemoine J, Neeser J-R. Lactobacillus helveticus Lh59 secretes an exopolysaccharide that is identical to the one produced by Lactobacillus helveticus TN-4, a presumed spontaneous mutant of Lactobacillus helveticus TY1-2. Carbohydr Res. 1997;302:197–202. doi: 10.1016/s0008-6215(97)00119-5. [DOI] [PubMed] [Google Scholar]
- 37.Stingele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stingele F, Newell J W, Neeser J-R. Unraveling the function of glycosyltransferases in Streptococcus thermophilus Sfi6. J Bacteriol. 1999;181:6354–6360. doi: 10.1128/jb.181.20.6354-6360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stingele F, Vincent S J F, Faber E J, Newell J W, Kamerling J P, Neeser J-R. Introduction of the exopolysaccharide gene cluster from Streptococcus thermophilus Sfi6 into Lactococcus lactis MG1363: production and characterization of an altered polysaccharide. Mol Microbiol. 1999;32:1287–1295. doi: 10.1046/j.1365-2958.1999.01441.x. [DOI] [PubMed] [Google Scholar]
- 40.van Casteren W H M, de Waard P, Dijkema C, Schols H A, Voragen A G J. Structural characterisation and enzymic modification of the exopolysaccharide produced by Lactococcus lactis subsp. cremoris B891. Carbohydr Res. 2000;327:411–422. doi: 10.1016/s0008-6215(00)00065-3. [DOI] [PubMed] [Google Scholar]
- 41.van Casteren W H M, Dijkema C, Schols H A, Beldman G, Voragen A G J. Structural characterisation and enzymic modification of the exopolysaccharide produced by Lactococcus lactis subsp. cremoris B39. Carbohydr Res. 2000;324:170–181. doi: 10.1016/s0008-6215(99)00292-x. [DOI] [PubMed] [Google Scholar]
- 42.van Casteren W H M, Dijkema C, Schols H A, Beldman G, Voragen A G J. Characterisation and modification of the exopolysaccharide produced by Lactococcus lactis subsp. cremoris B40. Carbohydr Polym. 1998;37:123–130. doi: 10.1016/s0008-6215(99)00292-x. [DOI] [PubMed] [Google Scholar]
- 43.Van Den Boogaard P, Kleerebezem M, Kuipers O P, de Vos W M. Control of lactose transport, β-galactosidase activity, and glycolysis by CcpA in Streptococcus thermophilus: evidence for carbon catabolite repression by a non-phosphoenolpyruvate-dependent phosphotransferase system sugar. J Bacteriol. 2000;182:5982–5989. doi: 10.1128/jb.182.21.5982-5989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanhaverbeke C, Bosso C, Colin-Morel P, Gey C, Gamar-Nourani L, Blondeau K, Simonet J-M, Heyraud A. Structure of an extracellular polysaccharide produced by Lactobacillus rhamnosus strain C83. Carbohydr Res. 1998;314:211–220. doi: 10.1016/s0008-6215(98)00297-3. [DOI] [PubMed] [Google Scholar]
- 45.van Kranenburg R, Marugg J D, van Swam I I, Willem N, de Vos W M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 46.van Kranenburg R, van Swam I I, Marugg J D, Kleerebezem M, de Vos W M. Exopolysaccharide biosynthesis in Lactococcus lactis NIZO B40: functional analysis of the glycosyltransferase genes involved in synthesis of the polysaccharide backbone. J Bacteriol. 1999;181:338–340. doi: 10.1128/jb.181.1.338-340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Kranenburg R, Vos H R, van Swam I I, Kleerebezem M, de Vos W M. Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: complementation, expression, and diversity. J Bacteriol. 1999;181:6347–6353. doi: 10.1128/jb.181.20.6347-6353.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto Y, Murosaki S, Yamauchi R, Kato K, Sone Y. Structural study on an exocellular polysaccharide produced by Lactobacillus helveticus TY1-2. Carbohydr Res. 1994;261:67–78. doi: 10.1016/0008-6215(94)80006-5. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto Y, Nunome T, Yamauchi R, Kato K, Sone Y. Structure of an exocellular polysaccharide of Lactobacillus helveticus TN-4, a spontaneous mutant strain of Lactobacillus helveticus TY1-2. Carbohydr Res. 1995;275:319–332. doi: 10.1016/0008-6215(95)00077-7. [DOI] [PubMed] [Google Scholar]