Abstract
The emergence of ceftazidime/avibactam (CZA) resistance among Guiana extended-spectrum β-lactamase (GES)-producing Pseudomonas aeruginosa isolates has rarely been described. Herein, we analyze the phenotypic and genomic characterization of CZA resistance in different GES-producing P. aeruginosa isolates that emerged in our institution. A subset of nine CZA-resistant P. aeruginosa isolates was analyzed and compared with thirteen CZA-susceptible isolates by whole-genome sequencing (WGS). All CZA-resistant isolates belonged to the ST235 clone and O11 serotype. A variety of GES enzymes were detected: GES-20 (55.6%, 5/9), GES-5 (22.2%, 2/9), GES-1 (11.1%, 1/9), and GES-7 (11.1%, 1/9). WGS revealed the presence of two mutations within the blaGES-20 gene comprising two single-nucleotide substitutions, which caused aspartic acid/serine and leucine/premature stop codon amino acid changes at positions 165 (D165S) and 237 (L237X), respectively. No major differences in the mutational resistome (AmpC, OprD porin, and MexAB-OprM efflux pump-encoding genes) were found among CZA-resistant and CZA-susceptible isolates. None of the mutations that have been previously demonstrated to cause CZA resistance were observed. Different mutations within the blaGES-20 gene were documented in CZA-resistant GES-producing P. aeruginosa isolates belonging to the ST235 clone in our institution. Although further analysis should be performed, according to our results, other resistance mechanisms might be involved in CZA resistance.
Keywords: Pseudomonas aeruginosa, GES β-lactamases, ceftazidime/avibactam resistance, whole-genome sequencing
1. Introduction
A great concern for Pseudomonas aeruginosa infections is the global emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) isolates, which can cause treatment failure and increased mortality [1]. The successful selection of chromosomal mutations and the growing acquisition of transferable resistance determinants, particularly those encoding carbapenemases (e.g., GES, KPC, VIM, and IMP enzymes) or extended spectrum β-lactamases (ESBLs), frequently co-transferred with aminoglycoside-modifying enzyme determinants (e.g., AAC (3′), AAC (6′), and ANT (2′)-I) are responsible for this increasing threat [2,3]. Furthermore, some MDR/XDR P. aeruginosa strains, denominated high-risk clones, have a clonal epidemic population structure with limited sequence types (e.g., ST235, ST175, ST111, and ST244) and a well-described ability to disseminate and cause severe infections [4,5,6].
The ceftazidime/avibactam (CZA) association, combining a cephalosporin with a novel established β-lactamase inhibitor, was recently introduced in clinical practice. Although unaffected by AmpC enzymes, ESBLs, and class-A carbapenemases (such as GES enzymes), CZA was shown to be ineffective against metallo-β lactamase (MBL)-producing isolates [7]. Several studies have demonstrated that CZA displays good in vitro activity against carbapenem-resistant P. aeruginosa isolates [8,9]. This novel combination has also been used for treating complicated MDR/XDR P. aeruginosa infections [10,11]. Nevertheless, other studies have reported limited CZA activity against carbapenemase-producing P. aeruginosa isolates [12,13].
In our institution, the production of GES enzymes is one of the most common causes of multidrug resistance in P. aeruginosa isolates [14,15]; thus, CZA is a valuable potential alternative against MDR/XDR P. aeruginosa infections. However, various GES variants resistant to CZA began to emerge last year. In general, GES enzymes confer resistance to penicillins, including ureidopenicillins, and oxyimino-cephalosporins but show less activity against carbapenems [2]. Nonetheless, specific substitutions can significantly alter this susceptibility profile, including G170S, which improves the hydrolyzing activity against carbapenems [3]. Whole-genome sequencing (WGS) techniques are promising tools for providing sufficient and reliable data for the surveillance and monitoring of antimicrobial resistance [16]. This study aimed to analyze the molecular epidemiology and resistome of a subset of CZA-resistant GES-producing XDR P. aeruginosa isolates by WGS, focusing on the mechanisms involved in CZA resistance, as a part of an institutional surveillance study.
2. Methods
2.1. Bacterial Sample Collection
All non-duplicated XDR P. aeruginosa isolates were collected from the Hospital Universitario 12 de Octubre, a 1300-bed tertiary-care hospital in Madrid, Spain, during 2020. The isolates were recorded at the Clinical Microbiology Laboratory from inpatients admitted to medical or surgical wards of the hospital. Only XDR isolates that were confirmed to be CZA-resistant GES-producing P. aeruginosa were further characterized. For a precise comparative analysis, a subset of well-characterized GES-producing XDR P. aeruginosa isolates with a CZA-susceptible phenotype were selected for WGS analysis [15].
2.2. Clinical Data Collection
Patient data were collected via chart review and included the following: age; gender; comorbidities; ward of admission (intensive care, medical, or surgical); sample type (respiratory, urinary, bloodstream, skin and soft tissue, or colonization); antimicrobial treatment received in the previous month; prior known MDR/XDR P. aeruginosa colonization; intensive care admission in the previous month; primary reason for hospital admission; antimicrobial therapy; and outcome (hospital discharge or death).
2.3. Microbiological Methods
The identification of P. aeruginosa isolates was carried out using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics Inc., Bremen, Germany). Antimicrobial susceptibility testing was performed using a semi-automated microdilution system (MicroScan, Beckman Coulter Diagnostics, Indianapolis, IN, USA), including the following antimicrobial agents: ceftazidime, cefepime, aztreonam, piperacillin/tazobactam, imipenem, meropenem, gentamicin, tobramycin, amikacin, ciprofloxacin, and colistin. Additionally, ceftolozane/tazobactam and CZA minimum inhibitory concentrations (MICs) were determined by gradient strips (bioMérieux, Marcy l’Etoile, France). MIC50 and MIC90 values were determined, and percentages of susceptible (standard dose and incremented exposure) and resistant isolates were calculated using European Committee on Antimicrobial Susceptibility Testing (EUCAST) v. 10. 0 (2020) clinical breakpoints (www.eucast.org (accessed on 6 March 2021)). Isolates were considered XDR if they were non-susceptible to at least one agent in all but two or fewer antimicrobial categories [17]. For this study, fosfomycin was not considered. Carbapenemase genes (blaGES) were screened by polymerase chain reaction (PCR) using LightCycler 2.0 (Roche) and Sanger sequencing using an ABI prism 3100 DNA Sequencer (PE Applied Biosystems, Norkwalk, CT, USA). P. aeruginosa ATCC 27853 was used as a quality control for antimicrobial susceptibility testing.
2.4. Whole-Genome Sequencing (WGS) and Bioinformatics Analysis
WGS was performed in all P. aeruginosa isolates, prepared using indexed pair-end Nextera XT library according to manufacturer’s instructions and sequenced on the MiSeq platform (Illumina Inc., San Diego, CA, USA) with the MiSeq reagent kit v. 2 (Illumina Inc.), resulting in 250-bp paired-end reads. The generated raw reads were quality-trimmed with Trimmomatic tool v. 0. 32 [18] and de novo assembled using the SPAdes assembler v.3.11.1 [19]. Genome assembly was evaluated by QUAST v.5.0.2 [20]. Bacterial identification was confirmed using KmerFinder [21]. Assembled sequences were annotated using Prokka v.1.13.3 [22]. Antimicrobial resistance genes were identified using Antimicrobial Resistance Identification by Assembly (ARIBA) v.2.6.1 [23] with the Comprehensive Antibiotic Resistance Database (CARD) [24] and ResFinder [25] databases. A total of 65 chromosomal genes were selected, and the main relevant mutations known to be involved in antimicrobial resistance were investigated (Supplementary Materials Table S1) [2,3]. The in silico multilocus sequence type (MLST) was determined using a BLAST-based approach. The PAst program (https://cge.cbs.dtu.dk/services/PAst-1.0 (accessed on 20 April 2021)) was used for in silico O-serotyping [26]. To determine core genome single-nucleotide polymorphisms (SNPs), all genomes were aligned to the P. aeruginosa PAO1 reference genome (GenBank accession: NC_002516.2) using Snippy v.4.6.0 (https://github.com/tseemann/snippy (accessed on 24 April 2021)). Additionally, a non-recombinant core genome SNP was performed using ModelFinder and IQ-TREE v.1.6.3 with 1000 bootstrap replicates [27,28] and visualized using the iTOL tool (https://itol.embl.de/ (accessed on 4 May 2021)). Sequence files were deposited in GenBank under BioProject PRJNA697852 and PRJNA723160 and accession numbers JAFBXZ000000000-JAFCAB000000000 and JAGSOK000000000-JAGSOS000000000.
2.5. Ethical Consideration
This study was designed and performed in accordance with the ethical standards of the Helsinki Declaration. The study protocol was approved by the Clinical Research Ethics Committee of our institution (Instituto de Investigación Sanitaria Hospital 12 de Octubre imas12, Hospital Universitario 12 de Octubre, ref.: 19/441). The need for written informed consent was waived due to the retrospective and non-interventional study design.
3. Results
3.1. Bacterial Isolates and Clinical Data
During a one-year survey study, a total of 102 non-duplicated XDR P. aeruginosa isolates were recovered. Of them, 32 (31.4%) were GES producers. Nine (28.1%) isolates were CZA-resistant GES-producing P. aeruginosa and all were selected for further genomic analysis. This study also included 13 previously well-characterized CZA-susceptible GES-producing XDR P. aeruginosa isolates [15]. The demographics and clinical characteristics of the patients with CZA-resistant and CZA-susceptible isolates, respectively, are shown in Table 1. Overall, most patients colonized or infected by CZA-resistant GES-producing XDR P. aeruginosa isolates were older adults, with several underlying conditions and previous broad antibiotic exposure, mainly carbapenems and colistin. However, treatment with CZA prior to isolation was only documented in two (9.1%) patients.
Table 1.
Demographic and clinical characteristics of the study cohort.
| ID | Collection Date | Age | Gender | Ward | Sample Type | Prior Antipseudomonal Antibiotics | Prior CZA | Primary Reason for Admission | Patient Outcome |
|---|---|---|---|---|---|---|---|---|---|
| PACZA-01 | 2020-01-08 | 69 | Female | ICU | Urine | MEM, CST | None | Hepatic transplant | Death |
| PACZA-02 | 2020-08-03 | 60 | Male | Haematology | Respiratory | TZP, MEM, CST | None | Febrile neutropenia | Hospital discharge |
| PACZA-04 | 2020-01-09 | 34 | Female | Medical | Blood | MEM | None | Catheter-related bloodstream infection | Hospital discharge |
| PACZA-05 | 2020-04-10 | 64 | Female | ICU | Respiratory | TZP, MEM, CST, CZA, AMK | Yes | Ventilator-associated pneumonia | Hospital discharge |
| PACZA-06 | 2020-09-04 | 38 | Female | Surgical | Urine | CIP | None | Heart transplant | Death |
| PACZA-07 | 2020-09-09 | 80 | Male | Medical | Blood | MEM | None | Urinary tract infection | Hospital discharge |
| PACZA-08 | 2020-11-09 | 81 | Male | Medical | Urine | MEM, CST | None | Urinary tract infection | Hospital discharge |
| PACZA-09 | 2020-02-11 | 29 | Male | Surgical | Soft tissue | MEM, CZA | Yes | Wound infection | Hospital discharge |
| PACZA-10 | 2020-02-14 | 70 | Female | Medical | Colonization | CIP | None | Decompensate heart failure | Death |
| PA14-13 | 2014-09-26 | 82 | Male | Medical | Blood | CIP | None | Respiratory tract infection | Death |
| PA15-05 | 2015-05-25 | 68 | Male | Medical | Blood | IPM, CIP | None | Decompensation of liver cirrhosis | Death |
| PA15-18 | 2015-12-16 | 57 | Male | ICU | Blood | TZP | None | Respiratory tract infection | Death |
| PA16-05 | 2016-06-19 | 59 | Male | ICU | Blood | MEM, CST, ATM, AMK | None | Haematopoietic transplantation | Death |
| PA16-13 | 2016-09-16 | 86 | Male | ICU | Blood | CIP | None | Schönlein-Henoch purpura vasculitis | Death |
| PA16-16 | 2016-10-10 | 63 | Female | Haematology | Blood | MEM, AMK | None | Febrile neutropenia | Death |
| PA16-19 | 2016-11-16 | 51 | Male | Haematology | Blood | CIP | None | Febrile neutropenia | Death |
| PA16-22 | 2016-02-24 | 62 | Male | Haematology | Blood | MEM, AMK | None | Febrile neutropenia | Death |
| PA17-01 | 2017-10-26 | 76 | Male | Oncology | Blood | None | None | Late-stage lung carcinoma | Death |
| PA17-08 | 2017-02-11 | 39 | Female | ICU | Blood | None | None | Subdural haematoma | Death |
| PA17-11 | 2017-04-02 | 66 | Male | Haematology | Blood | MEM, CIP | None | Leukaemia treatment | Death |
| PA17-13 | 2017-05-05 | 42 | Male | ICU | Blood | TZP | None | Coronary acute syndrome | Death |
| PA17-15 | 2017-05-18 | 79 | Female | Medical | Blood | TZP, MEM, AMK | None | Paralytic ileus | Death |
ICU, intensive care unit; CST, colistin; TZP, piperacillin/tazobactam; ATM, aztreonam; MEM, meropenem; CIP, ciprofloxacin; AMK, amikacin; CZA, ceftazidime/avibactam; IPM, imipenem.
3.2. Antimicrobial Susceptibility Data
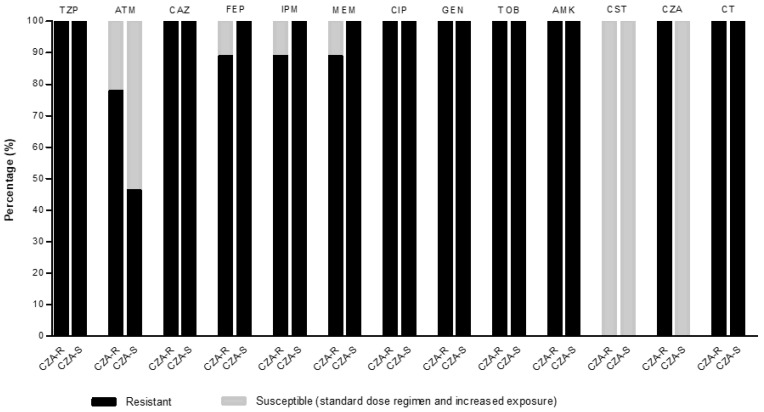
The antimicrobial susceptibility data are displayed in Figure 1. Increased MICs were observed for aztreonam (from 4/> 16 mg/L to 8/> 16 mg/L) and ceftolozane/tazobactam (from 6/24 mg/L to 24/> 256 mg/L) between CZA-resistant and CZA-susceptible isolates. The most active antipseudomonal agent in CZA-resistant isolates was colistin (100%, MIC50/90 = 1/2 mg/L). For the CZA-susceptible isolates, the most active antipseudomonal agents were colistin (100%, MIC50/90 = 1/2 mg/L) and CZA (100%, MIC50/90 = 2/6 mg/L). The activity of all other antibiotics was lower in both groups. All isolates were classified as XDR phenotypes. The carriage of the blaGES genes were confirmed in all isolates.
Figure 1.
Antimicrobial susceptibility of ceftazidime/avibactam (CZA)-resistant (n = 9) and CZA-susceptible (n = 13) P. aeruginosa isolates. Ceftolozane/tazobactam and CZA minimum inhibitory concentrations (MICs) were determined by gradient strips. Percentages of resistant and susceptible (standard dose and incremented exposure) isolates were calculated using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) v.10. 0 (2020) clinical breakpoints. TZP, piperacillin/tazobactam; ATM, aztreonam; CAZ, ceftazidime; FEP, cefepime; IPM, imipenem; MEM, meropenem; CIP, ciprofloxacin; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; CST, colistin; CZA, ceftazidime/avibactam; CT, ceftolozane/tazobactam.
3.3. Molecular Epidemiology
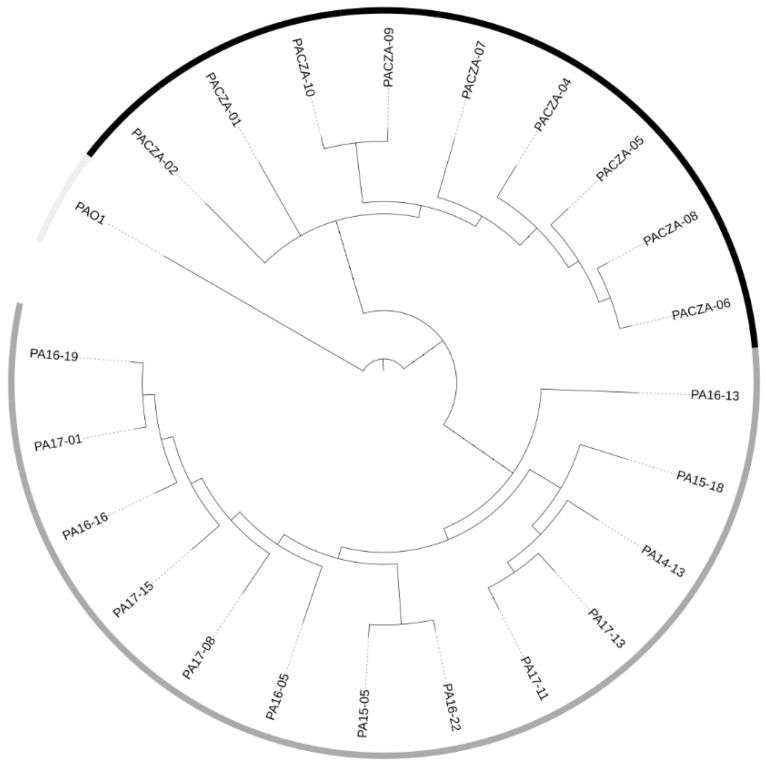
Genomic analysis showed that all isolates belonged to the widespread P. aeruginosa high-risk clone ST235. A core-genome phylogenetic tree reconstruction of all isolates and the PAO1 reference isolate is shown in Figure 2. According to the CZA phenotype, isolates were categorized as belonging to two different cluster types (CT-1 and CT-2). The isolates belonging to CT-1 included CZA-resistant phenotypes, and CT-2 comprised CZA-susceptible isolates. A core SNP analysis showed that the genetic diversity of the CZA-resistant and CZA-susceptible isolates ranged from 25 to 21 and from 19 to 83 SNPs, respectively (Tables S2 and S3). In silico O-antigen serotyping confirmed that all isolates belonged to the O11 serotype.
Figure 2.
Core-genome maximum-likelihood phylogenetic tree of all P. aeruginosa isolates and the P. aeruginosa PAO1 reference genome. The units of the scale are single-nucleotide polymorphisms (SNPs) by position. Ceftazidime/avibactam (CZA) phenotypes of P. aeruginosa isolates are highlighted by shaded squares: resistant (black), susceptible (dark grey), and not applicable (light grey). CZA, ceftazidime/avibactam.
3.4. Acquired Resistome
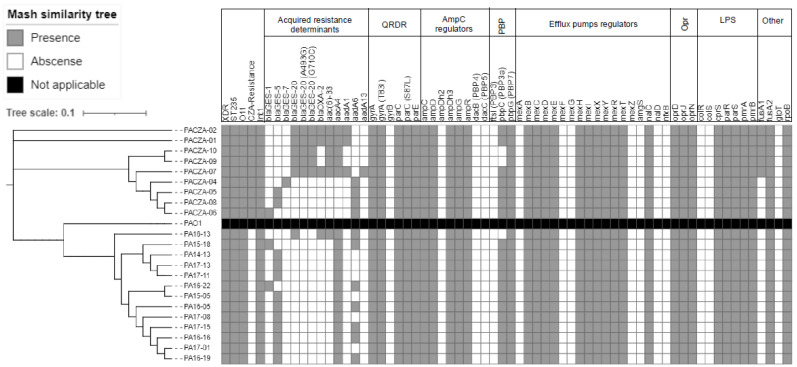
The most frequent horizontal acquired β-lactamases and aminoglycoside-modifying enzymes are summarized in Figure 3. Genomic analysis revealed a wide variety of GES enzymes among the CZA-resistant isolates. Among them, GES-20 (55.6%, 5/9) was the most frequent carbapenemase documented. The BLAST analysis of blaGES-20 revealed the presence of two mutations comprising two single-nucleotide substitutions (G to A and T to G), which caused aspartic acid/serine and leucine/premature stop codon amino acid changes at positions 165 (D165S) and 237 (L237X), respectively. GES-5 (22.2%, 2/9), GES-1 (11.1%, 1/9), and GES-7 (11.1%, 1/9) β-lactamases/carbapenemases were also detected. Additionally, OXA-2 β-lactamases were found to be mainly associated with GES-20 carbapenemases. Mutations in OXA-2 (D149, OXA-539) related to CZA resistance were not found. In CZA-susceptible isolates, GES-5 (76.9%, 10/13) was the most frequent carbapenemase, distantly followed by GES-1 (15.4%, 2/13) and GES-20 (7.7%, 1/3). A wide range of antimicrobial resistance determinants (aacA4, aac(6′)-33, aadA1, and aadA6) conferring co-resistance to amynoglycosides were also found in both CZA-resistant and CZA-susceptible isolates.
Figure 3.
Mash similarity tree of the P. aeruginosa isolates analyzed by whole-genome sequencing. Main acquired and mutational genes involved in antimicrobial resistance are also included. Brach length is indicative of the Mash distance. QRDR, quinolone-resistance-determining region; PBP, penicillin-binding protein; LPS, lipopolysaccharide; XDR, extensively drug-resistant; ST, sequence type; CZA, ceftazidime/avibactam.
3.5. Mutational Resistome
The complete list of chromosomal genes and mutations investigated is shown in Table S1. Up to 60.0% (39/65) of the analyzed genes showed non-synonymous or missense mutations. Figure 3 shows the main chromosomal genes involved in resistance to β-lactams, aminoglycosides, and fluoroquinolones. No major differences in the mutational resistome were found among CZA-resistant and CZA-susceptible isolates. All isolates contained non-synonymous mutations in the ampC gene (G27D, A97V, T105A, V205L, and G391A). Notably, none of the CZA-resistant isolates showed ampC substitutions (T96I, G183D, and E247G) known to be related to CZA resistance. Additionally, mutations in other well-known AmpC regulator genes, such as ampR and ampD, were also detected. However, previously described mutations (R504C and F533L) in ftsI (PBP3) related to CZA resistance were not found. Genes involved in the expression and regulation of efflux pumps were frequently mutated, including inactivating mutations in the MexAB-OprM-negative regulators mexR/nalB and nalC. Another frequently mutated gene was oprD, including mutations suggestive of OprD deficiency. Other mutations detected among all isolates included quinolone-resistance-determining region (QRDR) mutations gyrA (T83I) and parC (S87L), which are known to cause fluoroquinolone resistance. Finally, five CZA-resistant isolates showed a fusA1 (elongation factor G) mutation (F582I), linked to aminoglycoside resistance.
4. Discussion
Following the introduction of the novel CZA combination for the treatment of GES-producing XDR P. aeruginosa infections in our institution, the emergence of resistance to this antimicrobial agent was documented in vitro. Exposure to broad-spectrum antibiotics, including CZA, has been described as one of the main factors related to CZA resistance [1]. Despite this, the confirmation of CZA treatment prior to isolation was documented in a few patients in our cohort. In this study, we used a WGS approach to analyze the genomic characteristics of a subset of CZA-resistant GES-producing XDR P. aeruginosa isolates collected at a tertiary hospital as part of a surveillance study. We also focused on the main acquired and mutational antibiotic resistance determinants known to be involved in CZA resistance.
Depending on the underlying mechanisms of antimicrobial resistance, CZA could be an appropriate option for some MDR/XDR P. aeruginosa isolates, such as those harboring class-A carbapenemases (such as GES enzymes) or chromosomal combinations (such as OprD deficiency and AmpC hyperproduction) [8,9,10,11,12,13]. In a recent Spanish nationwide study, Del Barrio-Tofiño et al. demonstrated that the prevalence of MDR/XDR P. aeruginosa isolates due to transferable ESBLs or carbapenemases was 16.7%, with VIM being the most frequent carbapenemase detected (9.5%), distantly followed by GES enzymes (3.6%) [3]. In our institution, up to 59% of the MDR/XDR isolates demonstrated acquired GES or VIM carbapenemases. This prevalence is higher than that reported in other areas of Spain [2,3,4,6]. In 2019, the distribution of each of these two carbapenemases was 50%. However, in the last year, we experienced a dramatic increase in GES enzymes (81%). The cases reported here offer several important insights into the evolving landscape of GES-producing XDR P. aeruginosa isolates in a high-endemicity setting for high-risk clones [14,15].
In silico MLST analysis demonstrated that all isolates belonged to the epidemic P. aeruginosa high-risk clone ST235. In addition, core SNP-based phylogenetic analysis confirmed the high diversity among these isolates, suggesting that person-to-person transmission is scarce. The available evidence indicates that ST235, the founder of the CC235 clonal complex, is the most relevant P. aeruginosa high-risk clone [4,5,6]. It shows a worldwide dissemination and is associated with MDR/XDR phenotypes by the acquisition of different ESBLs (e.g., OXA and CTX-M) and class-A and -B carbapenemases (e.g., GES, KPC, VIM, IMP, and NDM) [4]. Indeed, the association of ST235 with horizontally acquired resistance determinants, including integrons, transposons, and plasmids, is overwhelming [6]. In a recent genomic analysis, Treepong et al. suggest that the specific presence of DrpA, a determinant involved in homologous recombination, likely increases the ability of this high-risk clone to acquire and maintain foreign resistance elements at a higher rate than other P. aeruginosa high-risk clones [5].
A variety of blaGES genes were observed among these isolates, and blaGES-20 was the most frequent carbapenemase. In general, the GES enzymes were class-A ESBLs, although certain variants (GES-4, -5, -6, -14, -15, -16, -18, -20, and -24) exhibited carbapenemase activity due to the presence of a single missense mutation at nucleotide position 493 (G493A), which changed glycine to serine at amino acid position 170 (G170S) [2,3]. Recently, GES mutations related to CZA resistance have been reported [29]. In this regard, Fraile-Ribot et al. demonstrated that P162S substitution reverted the carbapenemase phenotype determined by the G170S change of GES-5 (into GES-1), significantly increasing ceftolozane/tazobactam and CZA MICs. Of note, our GES-20-producing isolates exhibited D165S and L237X substitutions, and we hypothesize that both amino acid changes and gene expression may influence CZA resistance phenotypes. The functional impact of SNPs on the protein sequence was predicted using Protein Variation Effect Analyzer (PROVEAN) V.1.1.3 [30]. Although functional validation is necessary for the D165S variant, a PROVEAN analysis predicted that it had a deleterious effect on function (score −4.000). However, L237X had a neutral effect on function (score 0). This is a concern for clinical sites where GES enzymes seem to be an important contributor to MDR/XDR phenotypes, such as our institution [14,15]. Further studies of the differential expression and mechanisms underlying blaGES-conferred CZA resistance in other clonal backgrounds are needed.
Additional mechanisms of resistance to CZA have been proposed [31,32,33]. In a previous study, specific mutations leading to the modification of the AmpC catalytic center were found to be the first step in developing resistance to CZA [32]. The changes in the Ω-loop conferred resistance to ceftolozane/tazobactam and cross-resistance to CZA. Moreover, the presence of an OXA-2 mutant (OXA-539, harboring the duplication of the key residue D149) contributed to resistance to CZA [33]. Likewise, mutations R504C and F533L in ftsI (PBP3), MexAB-OprM overexpression, OprD inactivation, and AmpC hyperproduction are well-known to be involved in CZA resistance [31]. Unfortunately, none of the aforementioned mutations that have been previously demonstrated to cause CZA resistance were observed in our isolates, suggesting the involvement of other resistance mechanisms. The potential contribution of these determinants to CZA resistance will be assessed in future clinical and experimental studies.
Our study has some limitations that should be acknowledged. First, it was a retrospective study with a small sample size due to the number of available cases, and the results should be interpreted with caution. Second, our study reflects the experience of a single medical center, and the results may not be applicable to other locations with a different molecular epidemiology. Third, we used the consensus definitions of MDR/XDR P. aeruginosa phenotypes offered by Magiorakos et al. [17]. While this proposal was certainly useful for harmonizing the definitions of P. aeruginosa resistance profiles, the following issues remain: (1) the result varies depending on whether CLSI or EUCAST clinical breakpoints are considered, and (2) the comprehensive application of the proposed definitions is limited by the lack of EUCAST or CLSI clinical breakpoints for fosfomycin. We used EUCAST (2020) clinical breakpoints, and therefore fosfomycin was not considered. According to previous national surveillance studies [2,3], the prevalence of XDR phenotypes may be slightly underestimated when fosfomycin is not considered. Finally, we did not evaluate gene expression, which could have explained the CZA-resistant phenotypes observed in these GES-producing P. aeruginosa isolates. Additional transcriptomic analyses are needed to achieve a better understanding of the influence of resistance genes on the evolution of CZA resistance.
In conclusion, our study illustrates the potential molecular complexity of CZA resistance that can emerge in ST235 MDR/XDR P. aeruginosa isolates carrying different GES variants. Notably, two single amino acid substitutions within the blaGES-20 gene were found in CZA-resistant isolates. In addition, although further analysis should be performed, our results indicated that other resistance mechanisms might be involved in CZA resistance. This emerging scenario highlights the need to optimize the use of current antimicrobial agents to minimize the emergence of resistance and track the evolution of resistance with novel genome-based approaches, as well as the urgent need for novel treatments against MDR/XDR P. aeruginosa infections.
Acknowledgments
The authors thank the staff of the clinical services from the Hospital Universitario 12 de Octubre for their dedication to exceptional clinical service. The preliminary results of this study were delivered as a poster presentation at the XXIV Congress of the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC), online format from 5–11 June 2021.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11070871/s1, Table S1: Phenotypic and genotypic characteristics of the P. aeruginosa isolates, Table S2: Distances of ceftazidime/avibactam-resistant P. aeruginosa isolates, Table S3: Distances of ceftazidime/avibactam-susceptible P. aeruginosa isolates.
Author Contributions
All authors (R.R., J.V., S.G.-B., P.B., M.Á.O., M.M.-L., J.L.-T., F.C. and E.V.) made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; provided final approval of the version to be published; and agreed to be accountable for all aspects of the work. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Clinical Research Ethics Committee of our institution (Instituto de Investigación Sanitaria imas12, Hospital Universitario 12 de Octubre, ref.:19/441).
Informed Consent Statement
The informed written consent requirement was waived because of the retrospective and observational nature of this study.
Data Availability Statement
The data presented in this study are available in the article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Plan Nacional de I + D + i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016), co-financed by the European Regional Development Fund ERDF “A way to achieve Europe”, Operative Program Intelligent Growth 2014–2020. This study was supported by the Fondo de Investigación Sanitaria (grant PI19/00571 to E.V.). R.R. and M.M-L. are supported by the Subprograma Río Hortega, Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Ciencia, Innovación y Universidades, Spain (CM19/00229 and CM19/00226). E.V. is supported by the Subprograma Juan Rodés, Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Ciencia, Innovación y Universidades, Spain (JR18/00048).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Horcajada J.P., Montero M., Oliver A., Sorlí L., Luque S., Gómez-Zorrilla S., Benito N., Grau S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019;32:e00031-19. doi: 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Barrio-Tofiño E., López-Causapé C., Cabot G., Rivera A., Benito N., Segura C., Montero M.M., Sorlí L., Tubau F., Gómez-Zorrilla S., et al. Genomics and Susceptibility Profiles of Extensively Drug-Resistant Pseudomonas aeruginosa Isolates from Spain. Antimicrob. Agents Chemother. 2017;61:e01589-17. doi: 10.1128/AAC.01589-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Barrio-Tofiño E., Zamorano L., Cortes-Lara S., López-Causapé C., Sánchez-Diener I., Cabot G., Bou G., Martínez-Martínez L., Oliver A., Galán F., et al. Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J. Antimicrob. Chemother. 2019;74:1825–1835. doi: 10.1093/jac/dkz147. [DOI] [PubMed] [Google Scholar]
- 4.Oliver A., Mulet X., López-Causapé C., Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updates. 2015;21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Treepong P., Kos V., Guyeux C., Blanc D., Bertrand X., Valot B., Hocquet D. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 2018;24:258–266. doi: 10.1016/j.cmi.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Del Barrio-Tofiño E., López-Causapé C., Oliver A. Pseudomonas aeruginosa Epidemic High-Risk Clones and Their Association with Horizontally-Acquired β-Lactamases: 2020 Update. Int. J. Antimicrob. Agents. 2020;56:106196. doi: 10.1016/j.ijantimicag.2020.106196. [DOI] [PubMed] [Google Scholar]
- 7.Abboud M.I., Damblon C., Brem J., Smargiasso N., Mercuri P., Gilbert B., Rydzik A.M., Claridge T.D.W., Schofield C.J., Frère J.-M. Interaction of Avibactam with Class B Metallo-β-Lactamases. Antimicrob. Agents Chemother. 2016;60:5655–5662. doi: 10.1128/AAC.00897-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sader H.S., Castanheira M., Mendes R.E., Flamm R.K., Farrell D.J., Jones R.N. Ceftazidime-Avibactam Activity against Multidrug-Resistant Pseudomonas aeruginosa Isolated in U.S. Medical Centers in 2012 and 2013. Antimicrob. Agents Chemother. 2015;59:3656–3659. doi: 10.1128/AAC.05024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sader H.S., Castanheira M., Shortridge D., Mendes R.E., Flamm R.K. Antimicrobial Activity of Ceftazidime-Avibactam Tested against Multidrug-Resistant Enterobacteriaceae and Pseudomonas aeruginosa Isolates from U.S. Medical Centers, 2013 to 2016. Antimicrob. Agents Chemother. 2017;61:e01045-17. doi: 10.1128/AAC.01045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sader H.S., Castanheira M., Flamm R.K., Mendes R.E., Farrell D.J., Jones R.N. Ceftazidime/avibactam tested against Gramnegative bacteria from intensive care unit (ICU) and non-ICU patients, including those with ventilator-associated pneumonia. Int. J. Antimicrob. Agents. 2015;46:53–59. doi: 10.1016/j.ijantimicag.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Sader H.S., Castanheira M., Flamm R.K. Antimicrobial Activity of Ceftazidime-Avibactam against Gram-Negative Bacteria Isolated from Patients Hospitalized with Pneumonia in U.S. Medical Centers, 2011 to 2015. Antimicrob. Agents Chemother. 2017;61:e02083-16. doi: 10.1128/AAC.02083-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazmierczak K.M., De Jonge B.L.M., Stone G.G., Sahm D.F. In vitro activity of ceftazidime/avibactam against isolates of Pseudomonas aeruginosa collected in European countries: INFORM global surveillance 2012–15. J. Antimicrob. Chemother. 2018;73:2777–2781. doi: 10.1093/jac/dky267. [DOI] [PubMed] [Google Scholar]
- 13.Karlowsky J.A., Kazmierczak K.M., Bouchillon S.K., de Jonge B.L.M., Stone G.G., Sahm D.F. In Vitro Activity of Ceftazidime-Avibactam against Clinical Isolates of Enterobacteriaceae and Pseudomonas aeruginosa Collected in Asia-Pacific Countries: Results from the INFORM Global Surveillance Program, 2012 to 2015. Antimicrob. Agents Chemother. 2018;62:e02569-17. doi: 10.1128/AAC.02569-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Recio R., Mancheño M., Viedma E., Villa J., Orellana M., Lora-Tamayo J., Chaves F. Predictors of Mortality in Bloodstream Infections Caused by Pseudomonas aeruginosa and Impact of Antimicrobial Resistance and Bacterial Virulence. Antimicrob. Agents Chemother. 2020;64:e01759-19. doi: 10.1128/AAC.01759-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Recio R., Viedma E., González-Bodí S., Villa J., Orellana M., Mancheño-Losa M., Lora-Tamayo J., Chaves F. Clinical and bacterial characteristics of Pseudomonas aeruginosa affecting the outcome of patients with bacteraemic pneumonia. Int. J. Antimicrob. Agents. 2021;58:106450. doi: 10.1016/j.ijantimicag.2021.106450. [DOI] [PubMed] [Google Scholar]
- 16.Didelot X., Bowden R., Wilson D.J., Peto T.E.A., Crook D.W. Transforming clinical microbiology with bacterial genome sequencing. Nat. Rev. Genet. 2012;13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 18.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasman H., Saputra D., Sicheritz-Ponten T., Lund O., Svendsen C.A., Frimodt-Møller N., Aarestrup F.M. Rapid Whole-Genome Sequencing for Detection and Characterization of Microorganisms Directly from Clinical Samples. J. Clin. Microbiol. 2014;52:139–146. doi: 10.1128/JCM.02452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 23.Hunt M., Mather A.E., Sánchez-Busó L., Page A., Parkhill J., Keane J.A., Harris S.R. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J., Bhullar K., Canova M.J., De Pascale G., Ejim L., et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thrane S.W., Taylor V.L., Lund O., Lam J.S., Jelsbak L. Application of Whole-Genome Sequencing Data for O-Specific Antigen Analysis and In Silico Serotyping of Pseudomonas aeruginosa Isolates. J. Clin. Microbiol. 2016;54:1782–1788. doi: 10.1128/JCM.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen L.-T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraile-Ribot P.A., Fernández J., Gomis-Font M.A., Forcelledo L., Mulet X., López-Causapé C., Oliver A. In Vivo Evolution of GES β-Lactamases Driven by Ceftazidime/Avibactam Treatment of Pseudomonas aeruginosa Infections. Antimicrob. Agents Chemother. 2021;65:AAC0098621. doi: 10.1128/AAC.00986-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi Y., Chan A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Wang J., Wang R., Cai Y. Resistance to ceftazidime–avibactam and underlying mechanisms. J. Glob. Antimicrob. Resist. 2019;22:18–27. doi: 10.1016/j.jgar.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Lahiri S.D., Walkup G.K., Whiteaker J.D., Palmer T., McCormack K., Tanudra M.A., Nash T.J., Thresher J., Johnstone M.R., Hajec L., et al. Selection and molecular characterization of ceftazidime/avibactam-resistant mutants in Pseudomonas aeruginosa strains containing derepressed AmpC. J. Antimicrob. Chemother. 2015;70:1650–1658. doi: 10.1093/jac/dkv004. [DOI] [PubMed] [Google Scholar]
- 33.Fraile-Ribot P.A., Mulet X., Cabot G., del Barrio-Tofiño E., Juan C., Pérez J.L., Oliver A. In Vivo Emergence of Resistance to Novel Cephalosporin–β-Lactamase Inhibitor Combinations through the Duplication of Amino Acid D149 from OXA-2 β-Lactamase (OXA-539) in Sequence Type 235 Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2017;61:e01117-17. doi: 10.1128/AAC.01117-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the article.