Abstract
A chitinase antigen has been identified in Pseudomonas aeruginosa strain 385 using sera from animals immunized with a whole-cell vaccine. The majority of the activity was shown to be in the cytoplasm, with some activity in the membrane fraction. The chitinase was not secreted into the culture medium. Purification of the enzyme was achieved by exploiting its binding to crab shell chitin. The purified enzyme had a molecular mass of 58 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and a pI of 5.2. NH2-terminal amino acid sequencing revealed two sequences of M(I/L)RID and (Q/M/V)AREDAAAAM that gave an exact match to sequences in a translated putative open reading frame from the P. aeruginosa genome. The chitinase was active against chitin azure, ethylene glycol chitin, and colloidal chitin. It did not display any lysozyme activity. Using synthetic 4-methylumbelliferyl chitin substrates, it was shown to be an endochitinase. The Km and kcat for 4-nitrophenyl-β-d-N,N′-diacetylchitobiose were 4.28 mM and 1.7 s−1 respectively, and for 4-nitrophenyl-β-d-N,N′,N′′-triacetylchitotriose, they were 0.48 mM and 0.16 s−1 respectively. The pH optimum was determined to be pH 6.75, and 90% activity was maintained over the pH range 6.5 to 7.1. The enzyme was stable over the pH range 5 to 10 for 3 h and to temperatures up to 50°C for 30 min. The chitinase bound strongly to chitin, chitin azure, colloidal chitin, lichenan, and cellulose but poorly to chitosan, xylan, and heparin. It is suggested that the chitinase functions primarily as a chitobiosidase, removing chitobiose from the nonreducing ends of chitin and chitin oligosaccharides.
Chitin is the second most abundant polysaccharide found in nature and consists of variable-length linear chains of β-1,4-linked polymers of N-acetylglucosamine hydrogen bonded into an ordered insoluble crystalline structure. The enormous amounts of chitin produced annually in the biosphere are degraded by chitinases. Chitinases are ubiquitous in nature, being found in eucaryotes, procaryotes, archaea, and viruses. They consist of a group of hydrolytic enzymes that are able to break down polymeric chitin to chitin oligosaccharides, diacetylchitobiose, and N-acetylglucosamine. Endochitinases catalyze the hydrolysis of chitin at random sites along the polymer, whereas exochitinases (β-1,4-N-acetylglucosaminidases) remove single N-acetylglucosamine residues from the nonreducing ends of chitin chains. Chitobiosidases that remove diacetylchitobiose from the nonreducing ends of chitin oligosaccharides are often considered exo- or endochitinases. They should be described as exochitinases only if it can be demonstrated that the specificity is restricted to the removal of diacetylchitobiose from the nonreducing ends of chitin and chitin oligosaccharides. Efficient breakdown of chitin to metabolizable monomers requires the action of both endochitinases and exochitinases to release monomeric N-acetylglucosamine, which can then be metabolized to generate energy, CO2, H2O, and NH3.
There is considerable interest in chitinases derived from various sources as potential biocontrol antifungal agents (5), for the processing of chitin waste (10), and as vaccine candidates to target parasitic diseases (14). Chitinases have been isolated from numerous bacterial sources including species from the genera Bacillus, Aeromonas, Vibrio, Enterobacter, Serratia, and Pseudomonas. Bacterial chitinases are classified on the basis of their amino acid sequence into family 18 of the glycosyl hydrolases (16). Structurally they consist of several different domains including the chitin-binding domain, fibronectin type III-like, and cadherin-like domains. The chitin-binding domain plays an important role in the degradation of insoluble chitin (27); the functions of the other two domains are yet to be elucidated.
Perhaps the most studied bacterial chitinases are those derived from various Serratia marcescens isolates, which produce numerous chitinase enzymes and isoenzymes that have been characterized at the gene level (3, 4, 12, 13, 28, 31). ChiA, ChiB, and ChiC have molecular masses of 50 to 52 kDa deduced from the genes sequenced from various strains. A 22-kDa chitinase has also been cloned from strain KCTC2172 (13). S. marcescens also secretes proteinases that can cleave the mature chitinases into active fragments with lower molecular weights (12). The crystal structure has been determined for ChiA from S. marcescens (28), and the enzyme has been shown to consist of three domains, an amino-terminal fibronectin III-like domain, a catalytic domain, and a small alpha- and beta-fold domain that is thought to involved in the binding to chitin.
Pseudomonas aeruginosa is a ubiquitous and opportunistic organism capable of inhabiting a large range of environments. There has been only one previous report on the purification and characterization of chitinases from P. aeruginosa (35). It can be a significant problem as a human pathogen in hospitals and accounts for more than 10% of hospital-acquired infections (2). It can colonize indwelling catheters, the lungs, eyes, and burn wounds and cause bacteremia. Mortality can be high with immunocompromised patients and in intensive care units due to the high antibiotic resistance of strains. For cystic fibrosis (CF) patients, it is a major problem, as colonization of the lungs causes a chronic infection that leads to the inflammation responsible for the majority of the morbidity and mortality in these patients.
Vaccines for P. aeruginosa have been under investigation for more than 30 years (9), and a number of clinical trials have used outer membrane proteins, lipolysaccharides, and alginate to vaccinate or prepare hyperimmune globulin for burns and CF patients. We have been studying the use of an inactivated whole-cell P. aeruginosa vaccine that protects against acute P. aeruginosa infections in rodents (8). As part of the characterization of the vaccine, Western blotting of whole-cell extracts identified a chitinase to be one of a restricted number of antigens detected.
Here we describe the purification and characterization of the chitinase produced by the vaccine strain.
MATERIALS AND METHODS
Bacterial strain and growth of cultures.
P. aeruginosa isolate 385 was obtained by culture on nutrient agar during routine microbiological analysis of sputum from a CF patient. Stocks of P. aeruginosa were stored at −80°C in tryptone soy broth (TSB) containing 20% (vol/vol) glycerol. Starter cultures were grown in 50-ml universal tubes by inoculating two loopfuls of stock into 25 ml of TSB and incubating for 16 to 20 h at 37°C in a shaking incubator at 250 rpm. Twenty-five milliliters of starter culture was used to inoculate each 1-liter shaking flask (four flasks, each containing 500 ml of TSB), and cultures were grown at 37°C in a shaking incubator at 250 rpm for 16 to 20 h. For culture on plates, medium was solidified by the incorporation of 1.5% (wt/vol) agar.
Subcellular fractionation.
A small portion of the culture of P. aeruginosa 385 was used for subcellular fractionation to determine the distribution of the chitinase. The remainder was processed to provide material for purification of the chitinase. Cells were harvested from cultures by centrifugation at 4,000 × g for 2.5 h and washed with 1 volume of 25 mM sodium phosphate buffer, pH 7.0. Periplasmic proteins were extracted from cells obtained from 20 ml of culture by cold shock treatment, essentially by the method of Hoshino (18). Following periplasmic extraction, the cells were resuspended in 20 ml of 25 mM sodium phosphate buffer (pH 7.0) containing 1 mM 1-(2-aminoethyl)benzenesulfonylfluoride-HCl (AEBSF protease inhibitor) and disrupted by sonication using a Sanyo Soniprep 150 (19-mm-wide probe) at an amplitude of 6 μm for 25 cycles (1 cycle consists of 30 s on and 60 s off). Cell debris and unbroken cells were removed by centrifugation at 11,000 × g for 30 min. The 11,000 × g supernatant was centrifuged further at 200,000 × g for 90 min to collect membranes. The resulting supernatant contains cytoplasmic proteins. The membrane pellet was suspended in 1.4 ml of 25 mM sodium phosphate buffer, pH 7.0, by passage through a 23-gauge hypodermic needle. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, protein content, and chitinase activity were determined for all fractions. The remaining cells from the culture (equivalent to 1,980 ml of culture) were resuspended in 60 ml of 25 mM sodium phosphate buffer (pH 7.0) containing 1 mM AEBSF and sonicated as described above in two 30-ml batches. The resulting sonicate was centrifuged at 11,000 × g for 1 h to remove cell debris and intact cells. The supernatant from this step was centrifuged further at 200,000 × g for 90 min. Chitinase was purified from the resulting supernatant soluble protein extract (periplasmic and cytoplasmic proteins).
Purification of chitinase by chitin binding.
Chitinase was purified by exploiting its strong binding to chitin. Five 100-mg aliquots of purified crab shell chitin (Sigma Chemical Co.) were mixed thoroughly in microcentrifuge tubes with 1 ml of soluble protein extract from P. aeruginosa 385. The mixtures were incubated overnight at room temperature on an end-over-end carousel mixer. Tubes were then centrifuged at 16,000 × g for 5 min, and the supernatants were removed. The chitin was washed three times with 1 ml of 200 mM sodium phosphate buffer (pH 7.0) for 30 min, with centrifugation performed as described above after each wash. The chitinase was then eluted from the chitin with 1 ml of 70% (vol/vol) ethylene glycol (in 200 mM sodium phosphate buffer [pH 7.0]) for 30 min at room temperature, and the chitin was removed by centrifugation as described above. The 70% (vol/vol) ethylene glycol supernatants were pooled and then buffer exchanged into 25 mM sodium phosphate buffer (pH 7.0) using PD10 desalting columns (Pharmacia). Recoveries and specific activity were determined by protein content and chitinase activity. The purity of the eluted chitinase was determined by SDS-PAGE, isoelectric focusing (IEF), and Western blotting.
Chitinase assays. (i) Chromogenic 4-nitrophenol chitinase assay.
4-Nitrophenyl-β-d-N,N′-diacetylchitobiose and 4-nitrophenyl-β-d-N,N′,N′′-triacetylchitotriose were prepared as stock solutions in dimethyl sulfoxide (DMSO). Two hundred microliters of 200 mM sodium phosphate buffer (pH 7.0) was dispensed into three microwells of a microtiter plate. Twenty-five microliters of sample was then added, briefly mixed, and preincubated for 15 min at 37°C in a Labsystems plate reader (model iEMS MF). The reaction was started by the addition of 25 μl of stock substrate. The absorbance at 410 nm was read every 15 s for a total of 15 min, and the maximum initial rate of reaction was calculated. The extinction coefficient for 4-nitrophenol at 410 nm was calculated by using the same assay buffer and volumes with standard 4-nitrophenol solutions in microtiter plates and used to convert rates to nanomoles of substrate per minute. For routine assays, to monitor purification, 1 mM 4-nitrophenyl-β-d-N,N′-diacetylchitobiose was used. One unit was defined as the conversion of 1 nmol/min. The assay was linear with respect to the initial rate and enzyme concentration. Km and kcat for 4-nitrophenyl-β-d-N,N′-diacetylchitobiose and 4-nitrophenyl-β-d-N,N′,N′′-triacetylchitotriose were determined for the purified enzyme using the microtiter plate assay described above but changing the substrate concentration. Stocks of 30 mM 4-nitrophenyl-β-d-N,N′-diacetylchitobiose and 4.45 mM 4-nitrophenyl-β-d-N,N′,N′′-triacetylchitotriose were prepared in DMSO and diluted to substrate concentrations in the ranges 3 to 24 mM and 0.45 to 3.6 mM, respectively, with DMSO. The Km and Vmax were calculated from direct linear plots (7) of the data.
(ii) Fluorogenic 4-methylumbelliferone chitinase assay.
4-Methylumbelliferyl N-acetyl-β-d-glucosaminide, 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside, 4-methylumbelliferyl-β-d-N,N′,N′′-triacetylchitotrioside, and 4-methylumbelliferyl-β-d-N,N′,N′′,N′′′-tetraacetylchitotetraoside were prepared as 1 mM stock solutions in DMSO. Chitinase activity was determined in black microtiter plates. Portions (150 μl) of 200 mM sodium phosphate buffer (pH 7.0) were dispensed into three microwells of a microtiter plate. Twenty microliters of substrate was then added and briefly mixed before incubating for 15 min at 37°C using a Fluoroskan plate reader (Labsystems). The reaction was started by the addition of 30 μl of chitinase. After 15 min of incubation, the reaction was stopped by the addition of 50 μl of 3 M sodium carbonate, and the fluorescence caused by the release of 4-methylumbelliferone at an excitation wavelength of 390 nm and emission wavelength of 485 nm was determined.
(iii) Ethylene glycol chitin chitinase assay.
The degradation of ethylene glycol chitin was measured on agar plates. Luria-Bertani medium (29) agar plates containing 0.05% (wt/vol) ethylene glycol chitin and 0.01% (wt/vol) trypan blue were prepared. One microliter of a bacterial culture (grown for 16 to 20 h) was pipetted onto the surface of the agar, and the plate was incubated at 37°C for 15.5 h. Degradation of the ethylene glycol chitin was shown by a clear halo around the bacteria against a blue background.
(iv) Chitin azure chitinase assay.
Ten micrograms of chitin azure was added to microcentrifuge tubes containing 950 μl of 200 mM sodium phosphate buffer, pH 7.0. Fifty microliters of chitinase was added, and the mixture was incubated at 37°C for 24 h on an end-over-end carousel mixer. The mixture was then centrifuged at 16,000 × g for 10 min, and the absorbance at 570 nm of the supernatant was determined. Samples were compared to blanks containing sample buffer without chitinase. One enzyme unit was defined as a change in the optical density at 570 nm (ΔOD570) of 1.0 in 24 h.
(v) Colloidal chitin chitinase assay.
Colloidal crab shell chitin was prepared essentially by the method of Jeuniaux (19) using 2 g of practical grade crab shell chitin (Sigma Chemical Co.). Hydrolysis of colloidal chitin was determined in microtiter plates by measuring the decrease in OD620 on a Labsystems plate reader (model iEMS MF) at 37°C. Briefly, 125 μl of 200 mM sodium phosphate buffer (pH 7.0) was dispensed into microwells, and 50 μl of a colloidal chitin suspension (40 mg/ml in 200 mM sodium phosphate buffer [pH 7.0]) was added and preincubated at 37°C for 15 min. The reaction was started by the addition of 25 μl of chitinase. The OD620 was measured every 5 min for 1 h. Prior to each measurement, the plate was shaken to ensure that the colloidal chitin was well dispersed. The maximum rate of the reaction was calculated (Labsystems Genesis software), after subtraction of a blank. One enzyme unit was defined as a ΔOD620 of 1.0 per min.
Lysozyme activity.
Lysozyme activity was determined by measuring the decrease in OD620 of a suspension of Micrococcus luteus cells. The assay was performed in a microtiter plate as described above for colloidal chitin, except that the substrate was a suspension of lyophilized M. luteus (called Micrococcus lysodeikticus in the Sigma Chemical Co. catalog) (40 mg/ml) cells in 200 mM sodium phosphate buffer (pH 7.0). Hen egg white lysozyme (Sigma Chemical Co.) was used as a positive control for the assay.
Effects of pH and temperature on activity and stability.
Chitinase activity was determined with 0.1 mM 4-nitrophenyl-β-d-N,N′-diacetylchitobiose over the pH range 4.4 to 9.0, using 100 mM sodium citrate buffers up to pH 5.35 and 200 mM sodium phosphate buffers from pH 5.35 to pH 9.0, as described above for the 4-nitrophenol assay.
The pH stability was determined over the range pH 3 to 10 by diluting purified chitinase 10-fold in either sodium citrate or sodium phosphate buffer, followed by incubation at 37°C for 3 h. After incubation, chitinase was then diluted 10-fold in 200 mM sodium phosphate buffer (pH 7.0) and assayed for activity using 0.1 mM 4-nitrophenyl-β-d-N,N′-diacetylchitobiose at pH 7.0 and 37°C, as described above.
The temperature stability was determined by incubating 100 μl of chitinase (31 μg/ml in phosphate buffer) for 30 min at temperatures ranging from 30 to 70°C using a water bath. After incubation, samples were stored on ice before assaying for chitinase activity.
Binding studies.
The binding of chitinase to crab shell chitin, chitin azure, colloidal chitin, chitosan, xylan, lichenan, heparin agarose, microgranular cellulose, and N,N′-diacetylchitobiose agarose was investigated by the addition of 1 ml of chitinase (230 μg/ml) to 100 mg of the insoluble polysaccharides. The suspension was mixed continuously on a rotating end-over-end mixer at room temperature for 3 h and then centrifuged at 16,000 × g for 10 min to remove the insoluble polysaccharides. The chitinase activity in the supernatant was determined and compared to that of the control to estimate the percentage bound.
Protein assay.
Protein concentrations were determined using a Pierce bicinchoninic acid protein assay kit according to the manufacturer's instructions, with bovine serum albumin (BSA) as a standard.
SDS-PAGE.
Chitinase samples (2 to 10 μg) were analyzed by SDS-PAGE under reducing conditions on precast Bio-Rad 4 to 20% T Tris-HCl gradient gels, using a Mini PROTEAN II electrophoresis system (Bio-Rad) and following the manufacturer's instructions. Bio-Rad SDS-PAGE broad-molecular-weight-range proteins were used as a standard. After electrophoresis, gels were stained either with silver nitrite (17) or with 0.1% (wt/vol) Coomassie brilliant blue R-250 in 30% (vol/vol) methanol and 10% (vol/vol) acetic acid. For Coomassie blue staining, proteins were visualized by destaining gels in 30% (vol/vol) methanol and 10% (vol/vol) acetic acid.
IEF.
IEF was performed on purified chitinase samples to determine the pI and purity of the protein using 3–9 Phast gels on a Pharmacia Phast system. Pharmacia broad-pI-range proteins were used as standards. Following electrophoresis, the gel was fixed in 20% (wt/vol) trichloroacetic acid for 30 min and then silver stained.
Western blotting and immunostaining.
Rabbit anti-P. aeruginosa 385 serum was prepared by subcutaneous administration with 1 ml of 1-mg/ml formalin-killed whole-cell P. aeruginosa 385 lyophilizate in Freund's complete adjuvant. Booster immunizations were given on a monthly basis for 6 months and thence at 3-month intervals with 1 ml of 1-mg/ml lyophilizate in incomplete Freund's adjuvant. After 16 months, the animals were killed and sera were prepared. Nonimmune rabbit serum was obtained from Sigma Chemical Co.
For Western blotting, SDS-PAGE was performed as described above, except Bio-Rad biotinylated broad-molecular-weight-range standards were used. Following SDS-PAGE, proteins were transferred onto a nitrocellulose membrane using a Bio-Rad TransBlot module (1 h at 15 V) following the manufacturer's instructions. Following Western blotting, the membrane was washed first with Tris-buffered saline (TBS) (20 mM Tris-HCl, 500 mM NaCl [pH 7.5]) and then with TBS containing 0.05% (vol/vol) Tween 20 (TTBS) and then incubated for 2 h with primary antibody (1:750 dilution of rabbit anti-P. aeruginosa 385 serum in TTBS containing 1% [wt/vol] BSA). After the membrane was washed with TTBS, it was incubated for 1 h with swine anti-rabbit immunoglobulin G horseradish peroxidase conjugate (1:500 dilution in TTBS containing 1% [wt/vol] BSA) and avidin peroxidase (1:500). Following incubation, the membrane was washed twice in TTBS and twice in TBS. The blot was developed by incubating with the horseradish peroxidase substrate 4-chloronaphthol (4-CN; 30 mg of 4-CN in 10 ml of methanol plus 50 ml of TBS and 30 μl of 30% [vol/vol] H2O2).
NH2-terminal amino acid analysis.
Following SDS-PAGE, the chitinase was Western blotted onto a polyvinylidene difluoride membrane for analysis by NH2-terminal sequencing. After the polyvinylidene difluoride membrane was blotted, it was stained with 0.025% (wt/vol) Coomassie brilliant blue R-250 dissolved in 40% (vol/vol) methanol plus 1% (vol/vol) acetic acid for 10 min, followed by destaining in 50% (vol/vol) methanol. The membrane was dried and the stained proteins were excised and subject to NH2-terminal amino acid analysis by Edman degradation using a model 471A sequencer (Applied Biosystems).
RESULTS
Identification of chitinase.
Western blotting of soluble protein extracts of P. aeruginosa 385 that were immunostained with sera isolated from rabbits immunized with killed whole-cell P. aeruginosa 385 identified a number of antigens. NH2-terminal amino acid sequencing of a protein band with an apparent molecular mass of approximately 60 kDa revealed the amino acid sequences M(I/L)RID and (Q/M/V)AREDAAAAM on two occasions. Search of the GenBank (National Center for Biotechnology Information) database with these sequences did not reveal any matching sequences. However, by searching the P. aeruginosa Genome Project Database (http://www.pseudomonas.com), an exact match was found to MIRID and QAREDAAAAM in a single putative open reading frame, suggesting that these sequences are derived from the same protein. Searching GenBank with the translated protein sequence from this open reading frame gave matches to several bacterial chitinases.
Subcellular distribution of chitinase in P. aeruginosa 385.
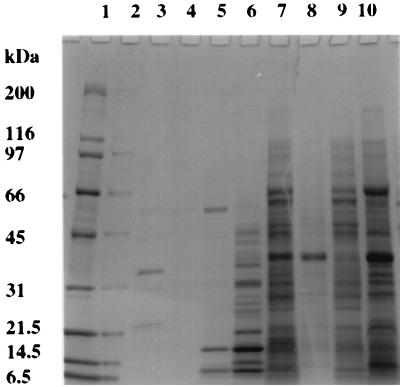
Cells of P. aeruginosa 385 were fractionated, and then the various cell fractions were analyzed by SDS-PAGE (Fig. 1) and for chitinase activity (Table 1). Inspection of the gel in Fig. 1 shows that the different fractions have distinct protein compositions. The growth medium, collected after harvesting of cells, clearly contains a number of proteins, and P. aeruginosa is known to secrete a wide variety of enzymes. The protein concentration of the medium could not be determined because one or more of the colored compounds secreted by the cells interfered in the protein assay.
FIG. 1.
SDS-PAGE analysis of subcellular fractions prepared from P. aeruginosa isolate 385. Proteins were separated on a 4 to 20% T polyacrylamide gel and stained with Coomassie brilliant blue R-250. Lanes: 1 and 2, molecular mass standards (with masses given in kilodaltons); 3, growth medium after harvesting of cells; 4, wash solution after cell wash; 5, TMK (10 mM Tris-HCl [pH 7.4], 1 mM MgCl2, 1 mM KCl) extract; 6, periplasmic proteins (cold shock extract); 7, supernatant from 11,000 × g centrifugation of sonicate; 8, pellet from 11,000 × g centrifugation of sonicate; 9, cytoplasmic proteins (supernatant from 200,000 × g centrifugation); 10, membrane proteins (pellet from 200,000 × g centrifugation).
TABLE 1.
Distribution of chitinase activity in subcellular fractions of P. aeruginosa strain 385a
| Sample | Chitinase activity (nmol/min/ ml) | Total chitinase activity (U) | % Chitinase activity | Total amt (mg) of protein | Sp act of chitinase (U/mg) |
|---|---|---|---|---|---|
| TMK extract | 0 | 0 | 0 | 0.4 | 0 |
| Cold shock extract | 0 | 0 | 0 | 1.5 | 0 |
| 11,000 × g supernatant | 10.4 | 182 | 97.8 | 36.2 | 5.0 |
| 11,000 × g pellet | 0.8 | 4 | 2.2 | 2.7 | 1.5 |
| Cytoplasmic fraction | 7.6 | 123 | 66.1 | 21.8 | 5.6 |
| Membrane fraction | 2.3 | 3 | 1.6 | 8.0 | 0.4 |
To calculate percent chitinase activity, the sum of the units in the 11,000 × g supernatant and 11,000 × g pellet samples (186 U) was defined as 100%.
No chitinase activity was detected in the medium, despite the expected requirement for an extracellular localization of this enzyme. In another series of experiments, we determined the SDS-PAGE profile of proteins bound to purified native crab shell chitin from culture supernatants. NH2-terminal sequencing of the major 21-kDa protein bound to chitin identified it as a truncated form of a 42-kDa chitin-binding protein (CBP) that has recently been described (11). These researchers stated that truncated CBP does not bind to colloidal chitin, whereas our studies show it can bind to native chitin.
Furthermore, no chitinase activity was located in the periplasmic extract (cold shock extract) that was prepared. The majority of the chitinase was detected in the cytoplasmic fraction (66%), with a small amount in the membrane fraction (2%). Approximately 30% of the total activity in the sonicate could not be accounted for after fractionation. This could be due to adsorption (e.g., onto plastic tubes), loss of activity, or an underestimate of the activity in the membrane fraction. The turbidity of the membrane fraction and/or components contained therein may interfere with the enzyme assay.
Purification of chitinase from P. aeruginosa 385.
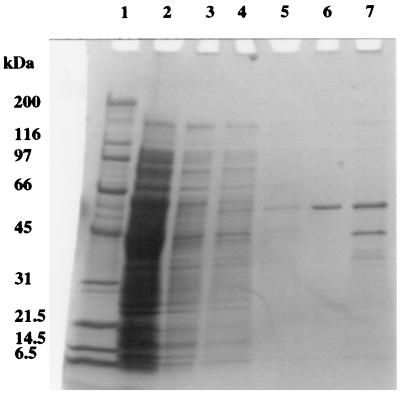
Chitinase was purified from the soluble protein extract (periplasmic and cytoplasmic) of P. aeruginosa 385 using a chitin-binding method. SDS-PAGE analysis (Fig. 2) shows that most of the proteins in the extract did not bind to chitin and were therefore washed from the substratum with the use of 200 mM sodium phosphate buffer, pH 7.0. After the third wash, no protein was recovered and the chitinase was then eluted from chitin with 70% (vol/vol) ethylene glycol. This gave an apparently pure protein of 58 kDa by SDS-PAGE analysis (Fig. 2). A sample of the chitin, after the 70% (vol/vol) ethylene glycol wash, was also analyzed by SDS-PAGE, which showed that some chitinase was still bound, along with a protein of 42 kDa. The 42-kDa protein was identified by NH2-terminal sequencing as the full-length CBP (the truncated version was identified in culture supernatants [see above]). A protein assay and chitinase assay were performed on the purified chitinase. Chitinase (1.6 mg) with a specific activity of 106 nmol/min/mg of protein was recovered from 5 ml of soluble protein extract (equivalent to 165 ml of culture) using 500 mg of purified crab shell chitin. The overall recovery was 35% of the chitinase activity measured in the soluble protein extract, with a purification factor of 29.5.
FIG. 2.
SDS-PAGE analysis of the purification of native chitinase by chitin binding. Proteins were separated on a 4 to 20% T polyacrylamide gel and silver stained. Lanes: 1, molecular mass standards (with masses given in kilodaltons); 2, unbound P. aeruginosa soluble protein extract; 3 to 5, 200 mM sodium phosphate buffer (pH 7.0) washes 1, 2, and 3, respectively; 6, 70% (vol/vol) ethylene glycol wash of chitin; 7, proteins remaining bound to chitin following elution with 70% (vol/vol) ethylene glycol.
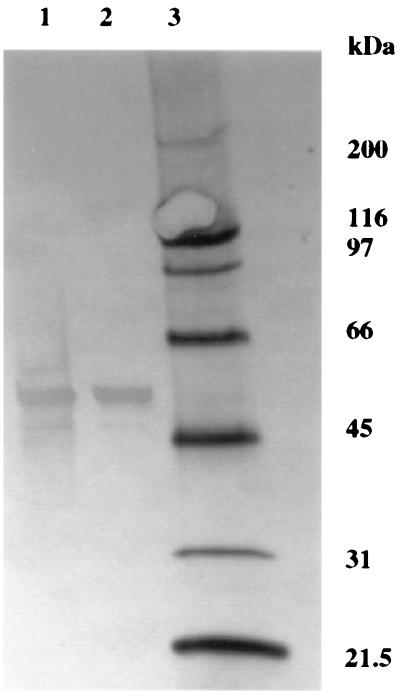
The purified chitinase was subjected to Western blotting, and the blot was immunostained with rabbit anti-P. aeruginosa 385 serum (Fig. 3). The chitinase was detected by the antiserum, as evidenced by a strong antigenic response at 58 kDa. There is also a small antigenic response at approximately 10 kDa, which is not detected by SDS-PAGE and silver staining (Fig. 2), indicating that it is a minor contaminant.
FIG. 3.
Immunostained Western blot of chitinase purified by chitin binding. Lanes: 1 and 2, chitinase purified by chitin binding and eluted with 70% (vol/vol) ethylene glycol, respectively; 3, biotinylated molecular mass standards (with masses given in kilodaltons).
The pI was estimated as 5.2, which is in good agreement with the pI of 5.1 calculated from the chitinase sequence of strain PAO1 of P. aeruginosa (www.Pseudomonas.com). The silver-stained IEF gel showed an apparently pure chitinase band (results not shown).
The NH2-terminal amino acid sequence of the purified chitinase was determined to be AREDA. This is identical to the sequence of amino acid residues 12 to 16 (inclusive) of the chitinase from P. aeruginosa strain PAO1 described above, suggesting that some proteolytic processing had occurred to remove the 11 NH2-terminal residues.
pH Activity profile and stability.
The activity of purified chitinase as a function of pH was studied using 4-nitrophenyl-β-d-N,N′-diacetylchitobiose as a substrate. Maximum enzyme activity was attained at pH 6.75, with more than 90% of activity retained in the pH range 6.5 to 7.1 and 50% in the pH range 6.0 to 7.5. Below pH 6.0 and above pH 8.0, activity fell rapidly with less than 10% activity detected below pH 5.0 and above pH 8.6.
The pH stability of chitinase was determined in the pH range 3 to 10 after incubation at 37°C for 3 h. Residual activity was determined using 4-nitrophenyl-β-d-N,N′-diacetylchitobiose. The enzyme was inactivated at pH 3.0. At pH 4.0, the enzyme was unstable with a 46% loss of activity. Throughout the pH range 5 to 10, the enzyme was stable under the conditions employed.
Maximum temperature stability of chitinase after 30 min of exposure was observed at 45°C, and 90% of the activity was retained up to 50°C. Above 50°C, the stability fell rapidly, with no activity detected after incubation for 30 min at 60°C or above.
Substrate specificity.
The chitinase showed activity against ethylene glycol chitin, chitin azure, colloidal chitin, and a range of synthetic substrates. The chitinase was active against chitin azure (1.12 U/mg), colloidal chitin (870 U/mg), and ethylene glycol chitin. Table 2 shows the activity against a range of methylumbelliferyl β-1,4-linked glucosamine oligosaccharides. There was no activity against 4-methylumbelliferyl N-acetyl-β-d-glucosaminide, indicating a lack of exochitinase (β-1,4-hexosaminidase) activity. Activity was measured against 4-methylumbellifer- yl-β-d-N,N′-diacetylchitobioside, 4-methylumbelliferyl-β-d- N,N′,N′′-triacetylchitotrioside, and 4-methylumbelliferyl-β-d-N,N′,N′′,N′′′-tetraacetylchitotetraoside. These all represent oligosccharide analogues of two or more sugars and demonstrate the endo-splitting nature of the chitinase activity. The highest activity was against 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside, which suggests that the primary function of the enzyme is the removal of diacetylchitobiose from the nonreducing end of chitin or chitodextrins.
TABLE 2.
Substrate specificity of P. aeruginosa strain 385 chitinase against a range of fluorogenic N-acetylglucosamine derivatives
| Substrate | Substrate specificitya |
|---|---|
| 4-Methylumbelliferyl N-acetyl-β-d-glucosaminide | 0 |
| 4-Methylumbelliferyl-β-d-N,N′-diacetylchitobioside | 7.24 × 105 |
| 4-Methylumbelliferyl-β-d-N,N′,N′′-triacetylchitotrioside | 4.93 × 105 |
| 4-Methylumbelliferyl-β-d-N,N′,N′′,N′′′-tetraacetylchitotetraoside | 1.86 × 105 |
Substrate specificity measured in fluorescence units per milligram of chitinase. Values shown are the means of three samples.
Many chitinases from eucaryotes display lysozyme activity and are able to depolymerize the peptidoglycan of bacterial cell walls by the hydrolysis of β-1,4 linkages between N-acetylmuramic acid and N-acetyl-d-glucosamine. However, the chitinase from P. aeruginosa 385 did not display any lysozyme activity, as determined by the lack of lysis of M. luteus cells.
Kinetic studies.
The Km, kcat, and specificity constant (kcat/Km) were calculated for the substrates 4-nitrophenyl-β-d-N,N′-diacetylchitobiose and 4-nitrophenyl-β-d-N,N′,N′′-triacetylchitotriose and are shown in Table 3. The specificity constants for both substrates are similar, but the Kms and kcats differ by an order of magnitude. The lowest Km value found, 0.48 mM, is that for 4-nitrophenyl-β-d-N,N′,N′′-triacetylchitotriose, and the highest Vmax value, 1.96 μmol/min/mg, giving a turnover number of 1.7 s−1, is for 4-nitrophenyl-β-d-N,N′-diacetylchitobiose. Although the specificity constants are comparable, indicating similar substrate preferences, the kinetic data suggest that the enzyme may have evolved to maximize rates against the removal of diacetylchitobiose from the nonreducing end of chitin or chitodextrins by increasing both the Km and kcat.
TABLE 3.
Kinetic parameters for P. aeruginosa strain 385 chitinase using two substrates
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (s−1 M−1) |
|---|---|---|---|
| 4-Nitrophenyl-β-d-N,N′-diacetylchitobiose | 4.28 | 1.7 | 404 |
| 4-Nitrophenyl-β-d-N,N′,N′′-triacetylchitotriose | 0.48 | 0.16 | 341 |
Binding studies.
Table 4 shows the results of investigations into the binding of chitinase to a number of different insoluble carbohydrate substrates. One hundred percent of the chitinase was bound to three of the chitin substrates; however, only 25% bound to chitosan. More than 80% of the chitinase bound to lichenan and microgranular cellulose, but less than 45% bound to xylan, heparin agarose, and N,N′-diacetylchitobiose agarose.
TABLE 4.
Binding P. aeruginosa strain 385 chitinase to insoluble polysaccharidesa
| Substrate | % Chitinase activity bound |
|---|---|
| Purified crab shell chitin | 100 |
| Chitin azure | 100 |
| Colloidal chitin | 100 |
| Chitosan | 25 |
| Xylan | 42 |
| Lichenan | 88 |
| Heparin agarose | 35 |
| Microgranular cellulose | 81 |
| N,N′-Diacetylchitobiose agarose | 36 |
Chitinase activity in the supernatant was measured using 4-nitrophenyl-β-d-N,N′-diacetylchitobiose.
DISCUSSION
We have identified and characterized a novel chitinase from a clinical isolate of P. aeruginosa. The enzyme was identified by Western blotting and NH2-terminal amino acid sequencing. The only other report for P. aeruginosa is of two extracellular enzymes with bifunctional chitinase and lysozyme activity that were purified from the cell-free culture supernatant of the soil isolate K-187 (35), when grown in the presence of crab shell chitin. Lee et al. (25) also describe the characterization of a chitobiosidase from a marine Pseudomonas species that is secreted into culture media when grown on colloidal chitin.
The chitinase from P. aeruginosa 385 appears to be produced constitutively, and not under catabolite repression, when the bacteria is grown in the absence of chitin. Many bacteria have been shown to produce chitinases, and although most appear to be under tight control of expression, the mechanisms involved in the control of expression are poorly understood. The chitinases of S. marcescens have been extensively studied, and their expression is induced by the presence of chitin or chitobiose (26) and catabolite repressed by glucose or N-acetylglucosamine in the culture medium. However, little is known about the control mechanism(s) governing expression of these enzymes. Studies of three strains of Enterobacter agglomerans revealed only one isolate that produced chitinases constitutively when grown on glucose or sucrose in the absence of chitin (5). A chitinase promoter has been cloned from a Pseudoalteromonas sp. (32) and shown that expression was induced with 10% CO2 and in stationary-phase cultures. Also, catabolite repression was not observed when glucose was used as a carbon source. The metabolism of chitin by the marine bacterium Vibrio furnissii is a complex process that involves a regulatory system in which N,N′-diacetylchitobiose is probably the inducer of several chitinases (1). There are also two reports of quorum sensing regulating production of chitinases in gram-negative bacteria. Winson et al. (37) working with P. aeruginosa PAO1 were able to demonstrate that quorum sensing with N-acyl-l-homoserine (AHL) regulates the production of a number of virulence determinants including the production of chitinase activity in the culture supernatant. Using a mutant derived from PAO1 that is defective in AHL production, they were able to demonstrate that the mutant was unable to produce chitinase activity in the culture supernatant. Addition of AHL restored activity in the supernatant. Chernin et al. (6) studying the chitinolytic system of Chromobacterium violaceum showed that the production of six chitinases was induced in the presence of chitin and regulated by quorum sensing with N-hexanonyl-l-homoserine (HHL). Mutants defective in HHL production were completely deficient in chitinase production.
Another unusual feature of the chitinase from P. aeruginosa 385 is that we were unable to detect it in the culture medium. Since the chitinase appeared to be produced constitutively in the cytoplasm, this opens up the intriguing possibility that AHL regulates secretion, rather than production, of the chitinase. Lack of production in the culture supernatant may be because AHL failed to reach the critical concentration. However, it is also possible that our strain is defective in the transport mechanism or quorum-sensing system that exists in P. aeruginosa PAO1 for chitinase. Although there have been relatively few detailed investigations of the subcellular location of bacterial chitinases, most described thus far are secreted, including those for other P. aeruginosa strains (35, 37) and Pseudomonas sp. (25). Notable exceptions can be found in the marine bacterium V. furnissii, which has two periplasmic chitinases (22), and ChiB from S. marcescens isolate BJL200 which was found mostly in the periplasm (less than 1% was found extracellularly) (3): however, this enzyme is secreted in other strains (36). Keyhani and Roseman (21) argue that for marine bacteria at least, chitinases would ideally be cell associated to yield products that can be immediately assimilated. Perhaps as P. aeruginosa can inhabit a large range of freshwater ecological niches and can exist as biofilms, it could be expected to adopt a similar strategy.
Although we were unable to detect secretion of chitinase into culture supernatants, we were able to detect a truncated CBP (21 kDa) that has recently been described (11). Unlike folders et al. (11), we were unable to detect the full-length version (43 kDa) in culture supernatants but were able to isolate the full-length version from the cytoplasmic fraction. We found that both species bound to native chitin, whereas they found that the truncated version did not bind to colloidal chitin. We can only assume that this difference is due to the poor binding of the truncated version to colloidal chitin. The role of this protein is unknown, but it may be involved in the pathogenicity and in attachment to chitin or N-acetylglucosamine-containing substrates.
Subcellular fractionation of P. aeruginosa 385 cells was undertaken to determine the localization of the chitinase within the cell. No chitinase activity could be detected in the periplasmic extract, and the majority of the activity was detected in the cytoplasmic extract, with a small proportion in the membrane fraction. Other researchers have detected chitinase activity in the cytoplasmic fraction at levels that exceed those in the extracellular medium (5). In addition, Brurberg et al. (3) fractionated Escherichia coli cells expressing chiB and S. marcescens overexpressing chiB using two methods and probed for chitinase using immunocytochemistry. The majority of the activity was located in the periplasm for S. marcescens and in the cytoplasm for E. coli harboring the chiB gene. Less than 1% and 5% activity, respectively, was found in the cell-free culture medium. ChiB was characterized as a chitobiosidase, and the researchers suggested that the enzyme functions in digestion of soluble chitin oligosaccharides (possibly GlcNac trimers) capable of entering the periplasmic space.
For P. aeruginosa 385, the large proportion of chitinase found in the cytoplasm is difficult to reconcile in the absence of any evidence for transport of chitin oligosaccharides or secretion mechanism for the enzyme. However, it should be noted that P. aeruginosa has a large capacity for the transport of small molecules, as evidenced by identification of 408 putative transporter genes in its genome; a cytoplasmic membrane phosphotransferase transporter for N-acetylglucosamine has also been identified (30). The chitinolytic system of the marine bacterium V. furnissii has been studied in some detail. An outer membrane chitoporin (20) and N,N′-diacetylchitobiose permease have been characterized (24). The chitoporin transports chitin oligosaccharides (n = 2 to 6) into the periplasm, and the permease transports N,N′-diacetylchitobiose into the cytoplasm. Such a transport mechanism may exist in P. aeruginosa for the transport of chitin oligosaccharides into the periplasmic space and cytoplasm. However, it is also possible that the cytoplasm is a reserve that supplies the enzyme for secretion (see above) or to the cell membrane in which we have found chitinase activity. For an opportunist such as P. aeruginosa, the controlled secretion or location in the outer membrane of a chitinase would serve a scavenger function in microbial communities that contain efficient chitin degraders (secreting a whole battery of chitinolytic enzymes). A membrane-bound chitinase would be ideally poised to take advantage of soluble oligosaccharides released by the digestion of chitin. By releasing N,N′-diacetylchitobiose (see below) from chitin oligosaccharides, these disaccharides could be transported to the cytoplasm for digestion by a β-N-acetylglucosaminidase. Searching the Pseudomonas Genome Database identified a β-N-acetylglucosaminidase; no other chitinolytic enzymes could be identified.
In common with many chitinases, P. aeruginosa 385 chitinase was purified to apparent homogeneity in a simple one-step adsorption to purified chitin. The enzyme can be eluted with high concentrations of ethylene glycol, which suggests a strong hydrophobic interaction with chitin. Some chitinase and a CBP could be removed only by boiling chitin in SDS-PAGE buffer, which may be due to stronger interactions with a different binding site(s) on the chitin polymer. By SDS-PAGE analysis, the enzyme was shown to be pure and had an estimated molecular mass of 58 kDa. Analysis by IEF and immunostaining of Western blots confirmed the high purity of the preparation. The acidic isoelectric point of 5.2 is common with many bacterial chitinases. NH2-terminal analysis from blots of crude and purified preparations gave a variety of amino acid sequences which suggests proteolytic clipping at the N terminus. P. aeruginosa contains a number of proteinases, including elastase and aminopeptidase, that could be responsible for this action.
The activity against synthetic analogues of N-acetylglucosamine was maximal at neutral pH, but >90% activity was maintained over the pH range 6 to 7.5. Many chitinases derived from bacteria, fungi, actinomycetes, and plants have acidic pH optima; neutral pH optimum are relatively uncommon. Interestingly, it has been shown for a periplasmic β-N-acetylglucosaminidase from the marine bacterium V. furnissii (23) that the pH optimum changed with the chain length of chitin oligosaccharides. Thus, it is perhaps difficult to make generalizations when comparing pH optima across bacterial species when a large number of different substrates are employed to assay for chitinolytic activity.
The enzyme was stable to changes in pH for 3 h over the pH range 5 to 10 but lost activity rapidly below pH 5. This is similar to chitinases I and chitinase II isolated from P. aeruginosa K-187, which were stable in the pH ranges 6 to 9 and 5 to 10, respectively (35), although these were tested for 30 min. The enzyme was stable up to a maximum of 50°C, after which there was a rapid decline in activity.
In binding experiments, the chitinase was shown to bind strongly to native chitin, chitin azure, and colloidal chitin. It also bound strongly to lichenan, a β-1,3-1,4 glucan and cellulose. The enzyme bound only poorly to chitosan, but a significant amount bound to xylan (β-1,4 heteropolymer) and heparin (α-1,3, β-1,4 heteropolymer). A number of chitinases have been shown to contain a conserved domain that shows homology to the cellulose-binding domains of cellulases (33) and are thought to function to bind chitin. Chitinases from S. marscescens (31), Alteromonas sp. (34), Clostridium paraputrificum (27), and Pseudomonas sp. (25) were shown to have affinity for cellulose and avicel. In view of these data, it is perhaps not surprising that the chitinase from P. aeruginosa can also bind to cellulose. However, it should be noted that the chitin-binding domain of chitinase A1 from Bacillus circulans was shown to be specific for insoluble chitin only (15).
The chitinase from P. aeruginosa 385 was active against a range of chitin derivatives and synthetic analogues of chitin but showed a preference for diacetylchitobiose derivatives. The determination of kinetic constants with two substrates showed that Km decreased by 1 order of magnitude by increasing the chain length by one sugar unit, but this was also accompanied by a 10-fold decrease in kcat. The specificity constant was the same for both substrates. The enzyme appears to have evolved as an endochitinase with a preference to generate diacetylchitobiose units. The enzyme has lower activity against 4-nitrophenyl-β-d-N,N′-diacetylchitobiose compared to the chitin dextrinase of V. furnissii (22), which has both a lower Km (0.17 mM) and a higher Vmax (34 μmol/min/mg). However, a secreted chitobiosidase from a Pseudomonas sp. (25) displayed both a lower Km (1.06 mM) and lower Vmax (0.74 μmol/min/mg) for the same substrate. Other workers have determined kinetic constants using methylumbelliferyl derivatives for chitinases from C. paraputrificum (27) and S. marcescens (12) both of which have a preference for diacetylchitobiose substrates. It should be noted that although the chitinase from P. aeruginosa 385 does not appear to be of the highest catalytic efficiency for either of the substrates tested compared to other chitobiosidases, others have noted that the activity of chitinases are often better against natural substrates. The chitinase did show activity against a range of substrates, although the activity was low against chitin azure and colloidal chitin. Some workers have shown that pH and temperature optima can differ for synthetic versus natural substrates and specificity can change with pH. Thus, the specificity and physiological role for P. aeruginosa 385 chitinase cannot at this stage be specified unambiguously, although the enzyme may function as a chitobiosidase to scavenge or utilize chitin oligosaccharides and degrade these oligosaccharides to chitobiose units. The fact that the bacterium does not contain the full complement of enzymes required for efficient chitin degradation and the low activity of the chitinase against colloidal chitin and chitin azure supports this notion.
REFERENCES
- 1.Bassler B L, Yu C, Lee Y C, Roseman S. Chitin utilisation by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24276–24286. [PubMed] [Google Scholar]
- 2.Beck-Sague C M, Banerjee S N, Williams J R. Epidemiology and control of Pseudomonas aeruginosa in U.S. hospitals. In: Baltch A L, Smith R P, editors. Pseudomonas aeruginosa infections and treatment. New York, N.Y: Marcel Dekker, Inc; 1994. p. 51. [Google Scholar]
- 3.Brurberg M B, Eijsink V G H, Haandrikman A J, Venema G, Nes I F. Chitinase B from Serratia marcescens BJL200 is exported to the periplasm without processing. Microbiology. 1995;141:123–131. doi: 10.1099/00221287-141-1-123. [DOI] [PubMed] [Google Scholar]
- 4.Brurberg M B, Haandrikman A J, Leenhouts K J, Venema G, Nes I F. Expression of a chitinase gene from Serratia marcescens in Lactococcus lactis and Lactobacillus plantarum. Microbiol Biotechnol. 1994;42:108–115. doi: 10.1007/BF00170232. [DOI] [PubMed] [Google Scholar]
- 5.Chernin L S, Ismailov Z, Haran S, Chet I. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl Environ Microbiol. 1995;61:1720–1726. doi: 10.1128/aem.61.5.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernin L S, Winson M K, Thompson J M, Haran S, Bycroft B W, Chet I, Williams P, Stewart G S. Chitinolytic activity in Chromobacterium violaceum: substrate analysis and regulation by quorum sensing. J Bacteriol. 1998;180:4435–4441. doi: 10.1128/jb.180.17.4435-4441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornish-Bowden A, Eisenthal R. Statistical considerations in the estimation of enzyme kinetic parameters by the direct linear plot and other methods. Biochem J. 1974;139:721–730. doi: 10.1042/bj1390721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cripps A, Dunkley M L, Clancy R L. Mucosal and systemic immunizations with killed Pseudomonas aeruginosa protect against acute respiratory infection in rats. Infect Immun. 1994;62:1427–1436. doi: 10.1128/iai.62.4.1427-1436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cryz S J. Vaccines, immunoglobulins and monoclonal antibodies for the prevention and treatment of Pseudomonas aeruginosa infections. In: Baltch A L, Smith R P, editors. Pseudomonas aeruginosa infections and treatment. New York, N.Y: Marcel Dekker, Inc; 1994. p. 519. [Google Scholar]
- 10.Deshpande M V. Enzymatic degradation of chitin and its biological applications. J Sci Ind Res. 1986;45:273–281. [Google Scholar]
- 11.Folders J, Tommassen J, van Loon L C, Bitter W. Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J Bacteriol. 2000;182:1257–1263. doi: 10.1128/jb.182.5.1257-1263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gal S W, Choi J Y, Kim C Y, Cheong Y H, Choi Y J, Lee S Y, Bahk J D, Cho M J. Cloning of the 52-kDa chitinase gene from Serratia marcescens KCTC2172 and its proteolytic cleavage into an active 35-kDa enzyme. FEMS Microbiol Lett. 1998;160:151–158. doi: 10.1111/j.1574-6968.1998.tb12905.x. [DOI] [PubMed] [Google Scholar]
- 13.Gal S W, Choi J Y, Kim C Y, Cheong Y H, Choi Y J, Bahk J D, Lee S Y, Cho M J. Isolation and characterisation of the 54-kDa and 22-kDa chitinase genes of Serratia marcescens KCTC2172. FEMS Microbiol Lett. 1997;151:197–204. doi: 10.1111/j.1574-6968.1997.tb12570.x. [DOI] [PubMed] [Google Scholar]
- 14.Harrison R A, Yu Y, Egerton G, Bianco A E. DNA immunisation with Onchocerca volvulus chitinase induces partial protection against challenge infection with L3 larvae in mice. Vaccine. 2000;18:647–655. doi: 10.1016/s0264-410x(99)00274-1. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto M, Ikegami T, Seino S, Ohuchi N, Fukada H, Sugiyama J, Shirakawa M, Watanabe T. Expression and characterization of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12. J Bacteriol. 2000;182:3045–3054. doi: 10.1128/jb.182.11.3045-3054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heukeshoven J, Dernick R. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis. 1985;6:103–112. [Google Scholar]
- 18.Hoshino T. Transport systems for branched chain amino acids in Pseudomonas aeruginosa. J Bacteriol. 1979;139:705–712. doi: 10.1128/jb.139.3.705-712.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeniaux C. Chitinases. Methods Enzymol. 1996;8:644–650. [Google Scholar]
- 20.Keyhani N O, Li X B, Roseman S. Chitin catabolism in the marine bacterium Vibrio furnissii: identification and molecular cloning of a chitoporin. J Biol Chem. 2000;275:33068–33076. doi: 10.1074/jbc.M001041200. [DOI] [PubMed] [Google Scholar]
- 21.Keyhani N O, Roseman S. Physiological aspects of chitin catabolism in marine bacteria. Biochim Biophys Acta. 1999;1473:108–122. doi: 10.1016/s0304-4165(99)00172-5. [DOI] [PubMed] [Google Scholar]
- 22.Keyhani N O, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Molecular cloning, isolation, and characterization of a periplasmic chitodextrinase. J Biol Chem. 1996;271:33414–33424. doi: 10.1074/jbc.271.52.33414. [DOI] [PubMed] [Google Scholar]
- 23.Keyhani N O, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Molecular cloning, isolation and characterization of a periplasmic beta-N-acetylglucosaminidase. J Biol Chem. 1996;271:33425–33432. doi: 10.1074/jbc.271.52.33425. [DOI] [PubMed] [Google Scholar]
- 24.Keyhani N O, Wang L X, Lee Y C, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Characterization of an N,N′-diacetyl-chitobiose transport system. J Biol Chem. 1996;271:33409–33413. doi: 10.1074/jbc.271.52.33409. [DOI] [PubMed] [Google Scholar]
- 25.Lee H-S, Han D-S, Choi S-J, Choi S-W, Kim D-S, Bai D-H, Yu J-H. Purification, characterization, and primary structure of a chitinase from Pseudomonas sp. YHS-A2. Appl Microbiol Biotechnol. 2000;54:397–405. doi: 10.1007/s002530000408. [DOI] [PubMed] [Google Scholar]
- 26.Monreal J, Reese E. The chitinase of Serratia marcescens. Can J Microbiol. 1969;15:689–696. doi: 10.1139/m69-122. [DOI] [PubMed] [Google Scholar]
- 27.Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J Bacteriol. 1997;179:7306–7314. doi: 10.1128/jb.179.23.7306-7314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrakis A, Tews I, Dauter Z, Oppenheim A B, Chet I, Wilson K S, Vorgias C E. Crystal structure of a bacterial chitinase at 2.3 A resolution. Structure. 1994;2:1169–1180. doi: 10.1016/s0969-2126(94)00119-7. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrook-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K-S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olsen M V. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki K, Taiyoji M, Sugawara N, Nikaidou N, Henrissat B, Watanabe T. The third chitinase gene (chi C) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem J. 1999;343:587–596. [PMC free article] [PubMed] [Google Scholar]
- 32.Techkarnjanaruk S, Pongpattanakitshote S, Goodman A E. Use of a promoterless lacZ gene insertion to investigate chitinase gene expression in the marine bacterium Pseudoalteromonas sp. strain S9. Appl Environ Microbiol. 1997;63:2989–2996. doi: 10.1128/aem.63.8.2989-2996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomme P, Warren R A J, Gilkes N R. Cellulase degradation by bacteria and fungi. Adv Microb Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 34.Tsujibo H, Orikoshi H, Shiotani K, Hayashi M, Umeda J, Miyamoto K, Imada C, Okami Y, Inomori Y. Characterization of chitinase C from the marine bacterium Alteromonas sp. strain O-7 and its corresponding gene and domain structure. Appl Environ Microbiol. 1998;64:472–478. doi: 10.1128/aem.64.2.472-478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S-L, Chang W-T. Purification and characterization of two bifunctional chitinases/lysozymes extracellularly produced by Pseudomonas aeruginosa K-187 in a shrimp and crab shell powder medium. Appl Environ Microbiol. 1997;63:380–386. doi: 10.1128/aem.63.2.380-386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe T, Kimura K, Sumiya T, Nikaidou N, Suziki K, Suziki M, Taiyoji M, Ferrer S, Regue M. Genetic analysis of the chitinase system of Serratia marcescens 2170. J Bacteriol. 1997;179:7111–7117. doi: 10.1128/jb.179.22.7111-7117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P C, Bycroft B W, Lazdunski A, Stewart G S A B, Williams P. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]