Abstract
Some methane-oxidizing bacteria (methanotrophs) are known to be capable of expressing nitrogenase and utilizing N2 as a nitrogen source. However, no sequences are available for nif genes in these strains, and the known nitrogen-fixing methanotrophs are confined mainly to a few genera. The purpose of this work was to assess the nitrogen-fixing capabilities of a variety of methanotroph strains. nifH gene fragments from four type I methanotrophs and seven type II methanotrophs were PCR amplified and sequenced. Nitrogenase activity was confirmed in selected type I and type II strains by acetylene reduction. Activities ranged from 0.4 to 3.3 nmol/min/mg of protein. Sequence analysis shows that the nifH sequences from the type I and type II strains cluster with nifH sequences from other gamma proteobacteria and alpha proteobacteria, respectively. The translated nifH sequences from three Methylomonas strains show high identity (95 to 99%) to several published translated environmental nifH sequences PCR amplified from rice roots and a freshwater lake. The translated nifH sequences from the type II strains show high identity (94 to 99%) to published translated nifH sequences from a variety of environments, including rice roots, a freshwater lake, an oligotrophic ocean, and forest soil. These results provide evidence for nitrogen fixation in a broad range of methanotrophs and suggest that nitrogen-fixing methanotrophs may be widespread and important in the nitrogen cycling of many environments.
Methanotrophs, or methane oxidizers, are a group of bacteria capable of growth on methane as their sole source of carbon and energy. These bacteria can be divided into two major phylogenetic groups, the type I methanotrophs (gamma proteobacteria) and the type II methanotrophs (alpha proteobacteria) (15). These two groups are thought to differ in several ways, foremost among which is their carbon assimilation pathway. The type I methanotrophs use the ribulose monophosphate pathway, while the type II methanotrophs utilize the serine cycle (1).
Groups of methanotrophs have also been classified based on the types of methane monooxygenase (MMO) that they produce. Until recently, most type I methanotrophs were thought capable of producing only the membrane bound or particulate MMO (pMMO), whereas type II methanotrophs and the type I Methylococcus strains were known to also produce a different, cytoplasmic enzyme, or soluble MMO (sMMO) (15). However, recent work has shown that several type I strains, including members of the genera Methylomonas and Methylomicrobium, can also produce sMMO (2, 13, 18, 26, 27). The type of MMO expressed is of environmental significance because sMMO shows rates of oxidation of halogenated solvents such as trichloroethylene (TCE) that are 100- to 1,000-fold higher than those of pMMO (10, 22).
Nitrogen fixation capabilities in methanotrophs have also been thought to distinguish these two groups (20). Type II methanotrophs and members of the type I genus Methylococcus have been shown to be capable of nitrogen fixation, while other type I methanotrophs are not (9, 20, 21). However, some evidence from DNA hybridization studies and acetylene reduction assays has suggested that some members of the type I genus Methylomonas and the type I strain Methylobacter marinus A45 (formerly known as Methylomonas methanica A4) may also be capable of nitrogen fixation (4, 21, 24). However, acetylene reduction by Methylomonas and Methylobacter strains was not detected in the second study, and in the last study, the only acetylene reduction rate measured in whole cells for a type I strain (Methylomonas rubra) was very low (3.1 nmol/h/mg of cells, recalculated to be approximately 0.11 nmol/min/mg of protein). Thus, the significance of nitrogen fixation in type I methanotrophs other than Methylococcus has been unclear.
In order to efficiently degrade TCE in contaminated environments, methanotrophs require a sufficient nitrogen source in addition to their substrates of methane and oxygen. However, in some vadose zone and aquifer environments, fixed nitrogen may be limiting (5), and methanotrophs capable of nitrogen fixation would have an advantage. Evidence also exists that nitrogen-fixing methanotrophs have an increased capacity for TCE oxidation (5, 6, 7). Because both type I and type II methanotrophs are now known to possess sMMO, both groups may be important in the bioremediation of TCE. For these reasons, we were interested in assessing the nitrogen fixation capabilities of both type I and type II methanotroph strains. This work provides genetic and biochemical evidence for the presence of nitrogenase, the key enzyme involved in nitrogen fixation, in both type I and type II methanotrophs. In addition, sequence analysis of nifH, the gene that encodes the highly conserved Fe protein of nitrogenase, suggests that nitrogenase genes from type I and type II methanotrophs may be present in a variety of environments, indicating that nitrogen-fixing methanotrophs may be widespread.
MATERIALS AND METHODS
Strains and media.
The methane-oxidizing strains used in this study were described previously. They included strains isolated from Lake Washington, Seattle, Wash., (2), and the type strains Methylosinus trichosporium OB3b, Methylobacter marinus A45, Methylomonas methanica S1, Methylomonas rubra, and Methylomicrobium albus BG8 (4). For chromosomal DNA extraction, strains were grown on nitrate mineral salts medium (NMS) (31) with a vitamin solution (12) and 10 μM CuSO4 · 5H2O and incubated at 30°C under a 50% methane–50% air atmosphere (vol/vol). To promote nitrogen fixation, strains were grown on nitrate-free mineral salts (NFMS) medium with vitamins and 10 μM CuSO4 · 5H2O and incubated at 30°C under an 80% methane–20% air atmosphere (vol/vol). Cycloheximide and nystatin were dissolved in dimethyl sulfoxide (DMSO) and added to plates to final concentrations of 20 and 10 μg/ml, respectively, to minimize mold contamination. For assays, strains were inoculated from NFMS plates and grown in 160-ml serum vials with liquid NFMS medium containing vitamins and 10 μM CuSO4 · 5H2O and incubated at 30°C shaking at 200 rpm under a 90% methane–10% air atmosphere (vol/vol).
Acetylene reduction assays.
Nitrogen fixation was estimated using the method of acetylene reduction described previously (8, 29) with a few modifications. Samples (1 ml) of liquid culture were removed from the growth vials and added to 21-ml serum vials. Methanol was added to a final concentration of 2% (vol/vol), and cell samples were incubated for 10 min with shaking at 30°C under a 90% argon–10% air atmosphere (vol/vol). Then 0.2 ml of acetylene was injected, and 0.5-ml samples of the headspace were removed at 0 min and approximately every 7 min up to 35 min postinjection. The rate of ethylene production was linear under these conditions. The amount of ethylene present in each sample was determined using a Carle analytical gas chromatograph (model 211) at 50°C equipped with a flame ionization detector, a 10-ft column (packed with a mixture of Porapak N and Porapak Q), and a Waters data module (model 740). To determine the protein concentration in the cultures, a sample of each culture was lysed by adding NaOH and sodium dodecyl sulfate to final concentrations of 1 N and 1%, respectively, and heating to 70°C for 15 min. The samples were then diluted fivefold with distilled water, and aliquots were used in the BCA protein assay (Pierce, Rockford, Ill.) as per the manufacturer's protocol. Bovine serum albumin samples were treated similarly and used as standards for the protein assay.
PCR amplification of nifH.
Chromosomal DNA was isolated from liquid cultures or from plates using methods described previously (25, 28). nifH gene fragments were amplified from chromosomal DNA samples using primers described previously (37). These degenerate primers, based on all known nifH genes at the time of design, were chosen from highly conserved amino acid sequences that required less than 200-fold degeneracy of the DNA coding sequences; the primers were synthesized with every possible combination of the base sequences, resulting in a mixture of 128 and 96 oligonucleotides for the upstream and downstream primers, respectively (37). For most strains, the reactions were carried out in an MJ Research PTC-200 thermocycler, with an initial denaturation step of 30 s at 94°C, followed by 30 cycles of 92°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final extension of 72°C for 5 min. For most strains, the PCRs contained final concentrations of 1×PCR buffer (Gibco-BRL, Rockville, Md), 1.5 mM MgCl2 (Gibco-BRL), 333 nM nifH-f, 333 nM nifH-r, 0.167 mM each deoxynucleoside triposphate (Boehringer Mannheim), and 2.5 U of Taq polymerase (Gibco-BRL) in a total volume of 30 μl. For LW5 and M. trichosporium OB3b, the reannealing temperature was lowered to 55°C, and DMSO was added to a final concentration of 5%. For PCR-positive strains, the nifH fragments were then cloned into pCR2.1 using the Topo-TA cloning kit (Invitrogen, San Diego, Calif.).
DNA sequencing and analysis.
DNA sequencing of the nifH PCR products was carried out on both strands using the ABI Prism BigDye terminator sequencing kit (PE Applied Biosystems, Foster City, Calif.). The sequencing reactions and analyses were performed by the University of Washington Department of Biochemistry Sequencing Facility using an Applied Biosystems automated sequencer. Analyses and translation of DNA sequences were performed using the Genetics Computer Group programs (Madison, Wis.). NifH sequences were aligned with translated nifH sequences obtained from the GenBank database using SeqPup (Indiana University) and GeneDoc (www.psc.edu/biomed/genedoc). Dendrograms were constructed using the programs PROTDIST, PROTPARS, NEIGHBOR, SEQBOOT, and CONSENSE from PHYLIP version 3.5c (11), and tree files were analyzed using Tree View (23). Related environmental nifH sequences were obtained using the tblastn program in BLAST version 2.1 (www.ncbi.nlm.nih.gov/BLAST).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nifH gene sequences described in this study are AF378714 to AF378724.
RESULTS
PCR amplification of nifH.
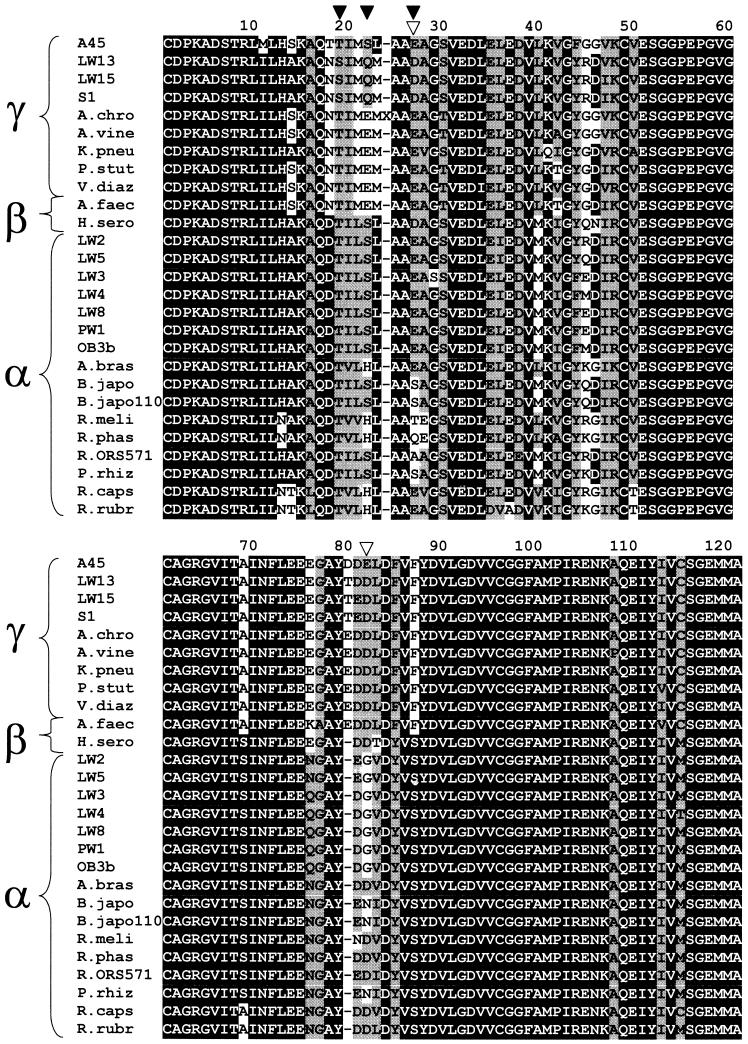
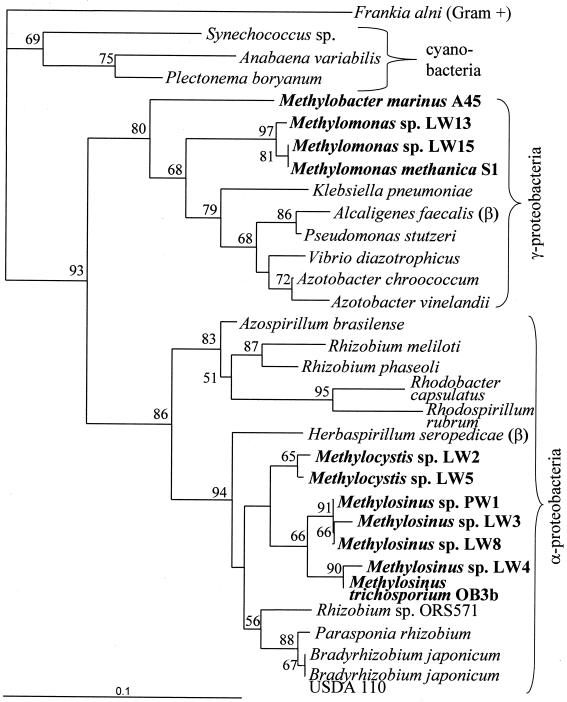
nifH encodes the Fe protein of nitrogenase and has been used as a marker for nitrogenase (36). Phylogeny based on NifH sequences has been shown to parallel that based on 16S ribosomal DNA (rDNA) sequences (33, 34). In order to assess the nitrogen-fixing capabilities of both type I and type II methanotrophs, nifH was studied in several type strains as well as pure cultures recently isolated from Lake Washington. Existing degenerate nifH primers (37) were used to attempt amplification of an approximately 360-bp product from five type strains and 10 strains isolated from Lake Washington (2). Methylosinus trichosporium OB3b and Methylomicrobium albus BG8 were used as positive and negative controls, respectively, based on their known nitrogen fixation capabilities (21). PCR products were obtained for all type II strains tested, two Methylocystis strains and five Methylosinus strains, including M. trichosporium OB3b, the positive control (Table 1). In addition, PCR products were obtained for several type I strains, including three Methylomonas strains and one Methylobacter strain. PCR products could not be obtained for several type I strains, including two Methylomonas strains, a Methylobacter strain, and M. albus BG8, the negative control. These PCR products were sequenced and translated, and the amino acid alignments are highly similar, with 78% identity overall (Fig. 1). Translated PCR products were aligned with NifH sequences from other proteobacteria, revealing high conservation (67% identity overall). However, within the variable residues, some signature sequences were conserved only in specific groups of methanotrophs. For example, the combination of S, Q, and D at residues 19, 22, and 27 in the translated PCR products appears to be diagnostic of Methylomonas NifH sequences, while the combination of E and G at residues 27 and 82 appears to be a marker for type II methanotroph NifH sequences (Methylosinus and Methylocystis). The alignments were used to generate a phylogenetic tree (Fig. 2). The phylogeny of the translated PCR products corresponded to that predicted by 16S rDNA sequences, with the NifH sequences from type I strains clustering together within a group of sequences from other gamma proteobacteria and the NifH sequences from type II strains also clustering together within a group of sequences from other alpha proteobacteria.
TABLE 1.
Evidence for nitrogen fixation in methanotrophs
| Strain | PCR product obtained | Growth on N-free medium | Acetylene reduction activity (nmol/min/mg of protein) |
|---|---|---|---|
| Methylocystis sp. LW2 | + | + | 3.30 |
| Methylocystis sp. LW5 | + | + | 0.43 |
| Methylosinus sp. LW3 | + | + | 0.59 |
| Methylosinus sp. LW4 | + | + | NTa |
| Methylosinus sp. LW8 | + | + | 0.72 |
| Methylosinus sp. PW1 | + | + | 1.84 |
| Methylosinus trichosporium OB3b | + | + | NTa |
| Methylobacter sp. A45 | + | NTc | NTa |
| Methylomonas sp. LW13 | + | + | 2.76 |
| Methylomonas sp. LW15 | + | + | NTa |
| Methylomonas methanica S1 | + | + | NTa |
| Methylobacter sp. LW14 | − | − | NTb |
| Methylomonas sp. LW21 | − | − | NTb |
| Methylomonas rubra | − | − | NTb |
| Methylomicrobium albus BG8 | − | − | NTb |
These strains grew poorly in NFMS and were not tested for acetylene reduction.
No attempts to grow these strains in liquid NFMS and perform acetylene reduction assays were made because these strains did not grow on NFMS plates.
This strain was difficult to maintain and was subsequently lost.
FIG. 1.
Alignment of deduced amino acid sequences of the approximately 360-bp partial nifH genes from Methylobacter marinus A45, Methylomonas sp. strain LW13, Methylomonas sp. strain LW15, M. methanica S1, A. chroococcum (accession no. M73020), A. vinelandii (M11579), K. pneumoniae (J01740), P. stutzeri (AJ297529), V. diazotrophicus (AF111110), A. faecalis (X96609), H. seropedicae (Z54207), Methylocystis sp. strain LW2, Methylocystis sp. strain LW5, Methylosinus sp. strain LW3, Methylosinus sp. strain LW4, Methylosinus sp. strain LW8, Methylosinus sp. strain PW1, M. trichosporium OB3b, A. brasilense (M64344), B. japonicum (E00713), B. japonicum USDA 110 (K01620), R. meliloti (V01215), R. phaseoli (M10587), Rhizoboium sp. strain ORS571 (M16710), P. rhizobium (K00487), R. capsulatus (M15270), and R. rubrum (M33774). Identical residues are in black boxes, and similar residues are in gray boxes. Methanotroph strain names are in bold face. Solid triangles indicate signature amino acids for Methylomonas strains, while open triangles indicate signature amino acids for type II strains.
FIG. 2.
Phylogenetic analysis of the derived amino acid sequences of nifH genes. Bootstrap values of >50% are shown near the clades. The bar represents 10% sequence divergence, as determined by the lengths of the horizontal lines connecting any two species. The tree includes sequences shown in Fig. 1 as well as F. alni L41344, Synechococcus sp. strain U22146, A. variabilis U89346, and P. boryanum D00666.
Nitrogenase activity.
To confirm the nitrogen-fixing capabilities of methanotroph strains that were PCR positive for nifH, these strains were grown on NFMS plates under low oxygen tension (see Materials and Methods). The PCR-negative strains were used as negative controls and showed no growth on these plates. All PCR-positive strains grew slowly under these conditions (Table 1), in some cases taking up to 10 days to form small isolated colonies. Acetylene reduction has been shown to be a suitable assay for nitrogen fixation in methanotrophs (8, 20, 29). Because acetylene is a potent MMO inhibitor, 2% methanol was provided as an alternate oxidizable substrate (8). Representative strains that grew best in nitrogen-free liquid medium were assayed for their ability to reduce acetylene (Table 1). Several type II strains as well as a type I Methylomonas strain showed activity, ranging between 0.43 and 3.30 nmol/min/mg of protein. These acetylene reduction activities fall into the range of previously reported activities for type II methanotrophs (20, 29).
Sequence comparison to environmental clones.
Environmental clone banks of nifH sequences have been generated from a number of different environments (19, 30, 32, 35, 38). In order to assess whether any of these environmental clones might have originated from nitrogen-fixing methanotrophs, translated BLAST nucleotide searches were performed with the methanotroph NifH sequences against the nonredundant nucleotide GenBank database. Translated environmental nifH sequences were found that were more closely related to Methylomonas NifH sequences than any others in the database, showing 95 to 99% identity at the amino acid level (Table 2). These environmental sequences were initially PCR amplified from rice roots and a freshwater lake (30, 35). Translated environmental nifH sequences were also found that were more closely related to type II methanotroph NifH sequences than any others in the database, showing 94 to 99% identity at the amino acid level (Table 3). These environmental sequences were PCR amplified from an oligotrophic ocean, a freshwater lake (by reverse transcription [RT]-PCR), rice roots, and a Douglas fir forest soil (30, 32, 35, 38). The signature amino acid combinations indicative of either Methylomonas or type II NifH sequences were found in the Methylomonas-like or type II-like environmental sequences, respectively, in all of these cases.
TABLE 2.
nifH PCR product identities for Methylomonas and related bacteriaa
| Clone or culture | M. methanica S1 | Methylomons sp. LW13 | Methylomonas sp. LW15 | H-RIC7 D26290 | H-RIC8 D26291 | H-RIC13 D26296 | H-RIC16 D26299 | H-RIC18 D26301 | H-RIC22 D26305 | H-RIC23 D26306 | LG1111 H AF212874 | LG1112 H AF212875 | K. pneumoniaeJ01740 | A. chroococcumM73020 | A. vinelandiiM11579 | V. diazotrophicusAF111110 | P. stutzeriAJ297529 | A. faecalisX96606 | M. purpuratumAF059648 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. methanica S1 | 98.2 (109) | 100 (109) | 97.6 (85) | 95.3 (85) | 96.5 (85) | 95.3 (85) | 97.6 (85) | 96.5 (85) | 96.5 (85) | 97.2 (109) | 97.2 (109) | 89.0 (109) | 90.0 (110) | 89.0 (109) | 89.9 (109) | 90.8 (109) | 89.0 (109) | 94.5 (109) | |
| Methylomons sp. LW13 | 93.6 (328) | 98.2 (109) | 95.3 (85) | 96.5 (85) | 98.8 (85) | 95.3 (85) | 97.6 (85) | 98.8 (85) | 98.8 (85) | 95.4 (109) | 95.4 (109) | 90.8 (109) | 91.8 (110) | 90.8 (109) | 91.7 (109) | 90.8 (109) | 89.0 (109) | 92.7 (109) | |
| Methylomonas sp. LW15 | 95.4 (328) | 92.7 (328) | 97.6 (85) | 95.3 (85) | 96.5 (85) | 95.3 (85) | 97.6 (85) | 96.5 (85) | 96.5 (85) | 97.2 (109) | 97.2 (109) | 89.0 (109) | 90.0 (110) | 89.0 (109) | 89.9 (109) | 90.8 (109) | 89.0 (109) | 94.5 (109) | |
| H-RIC7 D26290 | 88.6 (255) | 85.9 (255) | 88.2 (255) | 94.0 (116) | 94.0 (116) | 94.8 (116) | 95.7 (116) | 94.8 (116) | 93.1 (116) | 95.3 (85) | 95.3 (85) | 87.1 (116) | 87.2 (117) | 86.8 (91) | 86.2 (116) | 89.7 (116) | 86.8 (91) | 91.8 (85) | |
| H-RIC8 D26291 | 86.3 (255) | 84.3 (255) | 85.9 (255) | 87.4 (349) | 97.4 (116) | 97.4 (116) | 97.4 (116) | 98.3 (116) | 96.6 (116) | 91.8 (85) | 91.8 (85) | 88.8 (116) | 89.7 (117) | 89.0 (91) | 88.8 (116) | 90.5 (116) | 87.9 (91) | 90.6 (85) | |
| H-RIC13 D26296 | 86.7 (255) | 85.9 (255) | 85.9 (255) | 86.2 (348) | 90.2 (348) | 96.6 (116) | 98.3 (116) | 99.1 (116) | 98.3 (116) | 92.9 (85) | 92.9 (85) | 90.5 (116) | 91.5 (117) | 91.2 (91) | 90.5 (116) | 91.4 (116) | 89.0 (91) | 91.8 (85) | |
| H-RIC16 D26299 | 86.3 (255) | 84.7 (255) | 85.9 (255) | 87.4 (348) | 98.9 (349) | 90.2 (348) | 96.6 (116) | 97.4 (116) | 95.7 (116) | 92.9 (85) | 92.9 (85) | 88.8 (116) | 89.7 (117) | 89.0 (91) | 88.8 (116) | 90.5 (116) | 89.0 (91) | 91.8 (85) | |
| H-RIC18 D26301 | 91.8 (255) | 91.0 (255) | 91.0 (255) | 88.2 (348) | 92.0 (348) | 91.1 (348) | 92.0 (348) | 99.1 (116) | 97.4 (116) | 94.1 (85) | 94.1 (85) | 89.7 (116) | 90.6 (117) | 90.1 (91) | 89.7 (116) | 93.1 (116) | 90.1 (91) | 92.9 (85) | |
| H-RIC22 D26305 | 89.8 (255) | 89.8 (255) | 89.8 (255) | 84.5 (348) | 86.8 (348) | 89.1 (348) | 87.4 (348) | 89.1 (348) | 98.3 (116) | 92.9 (85) | 92.9 (85) | 90.5 (16) | 91.5 (117) | 91.2 (91) | 90.5 (116) | 92.2 (116) | 89.0 (91) | 91.8 (85) | |
| H-RIC23 D26306 | 89.8 (255) | 88.6 (255) | 88.2 (255) | 85.9 (348) | 87.9 (348) | 90.5 (348) | 87.9 (348) | 89.4 (348) | 87.6 (348) | 92.9 (85) | 92.9 (85) | 91.4 (116) | 92.3 (117) | 91.2 (91) | 92.2 (116) | 92.2 (116) | 89.0 (91) | 91.8 (85) | |
| LG1111H AF212874 | 85.3 (326) | 83.7 (326) | 84.0 (326) | 84.3 (255) | 81.6 (255) | 83.1 (255) | 82.4 (255) | 85.5 (255) | 83.1 (255) | 82.4 (255) | 97.2 (109) | 88.1 (109) | 89.1 (110) | 88.1 (109) | 89.0 (109) | 89.0 (109) | 88.1 (109) | 93.6 (109) | |
| LG1112H AF212875 | 86.2 (326) | 84.7 (326) | 85.6 (326) | 85.5 (255) | 84.3 (255) | 85.1 (255) | 85.1 (255) | 83.5 (255) | 83.1 (255) | 85.1 (255) | 89.3 (326) | 88.1 (109) | 89.1 (110) | 88.1 (109) | 89.0 (109) | 89.9 (109) | 88.1 (109) | 93.6 (109) | |
| K. pneumoniaeJ01740 | 74.8 (326) | 74.2 (326) | 74.5 (326) | 76.7 (347) | 75.8 (347) | 75.5 (347) | 76.1 (347) | 74.6 (347) | 74.6 (347) | 76.1 (347) | 76.1 (326) | 76.1 (327) | 88.7 (291) | 88.6 (289) | 91.3 (229) | 93.0 (158) | 87.7 (292) | 89.0 (109) | |
| A. chroococcumM73020 | 80.5 (329) | 80.2 (329) | 81.5 (329) | 81.1 (350) | 78.1 (352) | 79.4 (350) | 77.7 (350) | 78.6 (350) | 82.3 (350) | 78.0 (350) | 79.9 (329) | 81.2 (330) | 79.8 (875) | 95.5 (288) | 93.5 (230) | 96.2 (159) | 91.8 (291) | 91.8 (110) | |
| A. vinelandiiM11579 | 78.5 (326) | 79.4 (326) | 79.1 (326) | 80.4 (347) | 77.4 (349) | 79.5 (347) | 76.9 (347) | 77.8 (347) | 81.6 (347) | 78.4 (347) | 78.5 (326) | 80.7 (327) | 80.2 (868) | 88.0 (1286) | 93.0 (229) | 96.2 (158) | 91.3 (289) | 90.8 (109) | |
| V. diazotrophicusAF111110 | 81.0 (326) | 82.5 (326) | 81.0 (326) | 79.0 (348) | 77.6 (348) | 79.9 (348) | 78.2 (348) | 79.6 (348) | 77.6 (348) | 79.3 (348) | 79.5 (327) | 77.9 (326) | 78.5 (688) | 78.0 (691) | 77.8 (688) | 94.9 (158) | 92.6 (229) | 91.7 (109) | |
| P. stutzeriAJ297529 | 76.1 (326) | 76.4 (326) | 77.0 (326) | 78.7 (347) | 75.8 (347) | 77.8 (347) | 76.1 (347) | 77.8 (347) | 78.4 (347) | 76.1 (347) | 77.6 (326) | 78.3 (327) | 83.8 (475) | 80.3 (676) | 83.2 (612) | 77.5 (475) | 98.7 (158) | 92.7 (109) | |
| A. faecalisX96609 | 75.2 (326) | 75.5 (326) | 76.1 (326) | 77.8 (347) | 74.9 (347) | 77.2 (347) | 75.5 (347) | 76.7 (347) | 77.5 (347) | 76.1 (347) | 76.7 (326) | 78.0 (327) | 81.6 (870) | 81.9 (1071) | 83.5 (1005) | 75.3 (688) | 98.3 (1081) | 90.8 (109) | |
| M. purpuratumAF059648 | 75.5 (326) | 75.2 (326) | 75.5 (326) | 75.6 (254) | 72.0 (254) | 75.2 (254) | 72.4 (254) | 75.2 (254) | 74.4 (254) | 74.4 (254) | 78.2 (326) | 78.6 (327) | 84.4 (327) | 84.5 (330) | 85.0 (327) | 74.8 (326) | 88.4 (327) | 88.1 (327) |
Identities for the nifH PCR product obtained for pure cultures of Methylomonas strains, environmental clones, and related bacteria. Values above the diagonal are percent amino acid identity for the translated nifH products. Values below the diagonal are percent nucleotide identity. The number in parentheses is the number of the residues compared. Environmental amino acid identities with NifH from Methylomonas pure cultures greater than identities with NifH from other bacteria are shown in bold.
TABLE 3.
nifH PCR product identities for type II methanotrophs and related bacteriaa
| Clone or culture | Methylocystis sp. LW2 | Methylocystis sp. LW5 | Methylosinus sp. LW3 | Methylosinus sp. LW4 | Methylosinus sp. LW8 | Methylosinus sp. PW1 | M. trichosporium OB3b | PO3120 AF059644 | PO3133 AF059645 | LG1120 AF212882 | LG1122 AF212884 | LG2369 AF212891 | H-RIC20 D26303 | AO12 AF016615 | B13 AF099789 | B. japonicum USDA 110 K01620 | P. rhizobiumK00487 | Rhizobium sp. ORS571 M16710 | H. seropedicaeZ54207 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methylocystis sp. LW2 | 99.1 (108) | 94.4 (108) | 94.4 (108) | 95.4 (108) | 95.4 (108) | 95.4 (108) | 99.1 (108) | 95.4 (108) | 98.1 (108) | 98.1 (108) | 97.2 (108) | 95.2 (84) | 95.4 (108) | 93.5 (108) | 95.4 (108) | 95.4 (108) | 94.4 (108) | 91.7 (108) | |
| Methylocystis sp. LW5 | 94.5 (325) | 94.4 (108) | 94.4 (108) | 95.4 (108) | 95.4 (108) | 95.4 (108) | 98.1 (108) | 94.4 (108) | 97.2 (108) | 97.2 (108) | 96.3 (108) | 95.2 (84) | 95.4 (108) | 92.6 (108) | 96.3 (108) | 95.4 (108) | 93.5 (108) | 92.6 (108) | |
| Methylosinus sp. LW3 | 89.5 (325) | 88.6 (325) | 95.4 (108) | 99.1 (108) | 99.1 (108) | 96.3 (108) | 95.4 (108) | 91.7 (108) | 94.4 (108) | 94.4 (108) | 93.5 (108) | 94.0 (84) | 93.5 (108) | 89.8 (108) | 92.6 (108) | 92.6 (108) | 90.7 (108) | 91.7 (108) | |
| Methylosinus sp. LW4 | 91.4 (325) | 91.1 (325) | 91.7 (325) | 96.3 (108) | 96.3 (108) | 99.1 (108) | 93.5 (108) | 89.8 (108) | 92.6 (108) | 92.6 (108) | 91.7 (108) | 95.2 (84) | 94.4 (108) | 89.8 (108) | 90.7 (108) | 90.7 (108) | 90.7 (108) | 91.7 (108) | |
| Methylosinus sp. LW8 | 90.2 (325) | 89.2 (325) | 98.5 (325) | 92.3 (325) | 100 (108) | 97.2 (108) | 96.3 (108) | 92.6 (108) | 95.4 (108) | 95.4 (108) | 94.4 (108) | 95.2 (84) | 94.4 (108) | 90.7 (108) | 93.5 (108) | 93.5 (108) | 91.7 (108) | 92.6 (108) | |
| Methylosinus sp. PW1 | 90.4 (356) | 89.8 (325) | 98.2 (325) | 93.2 (325) | 98.5 (325) | 97.2 (108) | 96.3 (108) | 92.6 (108) | 95.4 (108) | 95.4 (108) | 94.4 (108) | 95.2 (84) | 94.4 (108) | 90.7 (108) | 93.5 (108) | 93.5 (108) | 91.7 (108) | 92.6 (108) | |
| M. trichosporium OB3b | 92.6 (325) | 92.6 (325) | 91.1 (325) | 95.7 (325) | 91.7 (325) | 91.7 (325) | 94.4 (108) | 90.7 (108) | 93.5 (108) | 93.5 (108) | 92.6 (108) | 95.2 (84) | 95.4 (108) | 90.7 (108) | 91.7 (108) | 91.7 (108) | 91.7 (108) | 92.6 (108) | |
| PO3120 AF059644 | 88.0 (324) | 85.2 (324) | 84.9 (324) | 84.6 (324) | 85.5 (324) | 84.6 (324) | 85.2 (324) | 96.3 (108) | 99.1 (108) | 99.1 (108) | 98.1 (108) | 96.4 (84) | 94.4 (108) | 94.4 (108) | 96.3 (108) | 96.3 (108) | 95.4 (108) | 92.6 (108) | |
| PO3133 AF059645 | 85.8 (324) | 83.3 (324) | 82.7 (324) | 83.4 (325) | 83.3 (324) | 82.4 (324) | 83.0 (324) | 97.8 (324) | 95.4 (108) | 95.4 (108) | 94.4 (108) | 91.7 (84) | 90.7 (108) | 90.7 (108) | 92.6 (108) | 92.6 (108) | 93.5 (108) | 88.9 (108) | |
| LG1120 AF212882 | 87.7 (324) | 84.9 (324) | 84.6 (324) | 84.3 (324) | 85.2 (324) | 84.3 (324) | 84.9 (324) | 99.7 (324) | 97.5 (324) | 98.1 (108) | 97.2 (108) | 96.4 (84) | 93.5 (108) | 93.5 (108) | 95.4 (108) | 95.4 (108) | 94.4 (108) | 91.7 (108) | |
| LG1122 AF212884 | 87.7 (324) | 84.9 (324) | 84.6 (324) | 84.3 (324) | 85.2 (324) | 84.3 (324) | 84.9 (324) | 99.7 (324) | 97.5 (324) | 99.4 (324) | 97.2 (108) | 95.2 (84) | 93.5 (108) | 93.5 (108) | 95.4 (108) | 95.4 (108) | 94.4 (108) | 91.7 (108) | |
| LG2369 AF212891 | 87.0 (324) | 84.3 (324) | 84.0 (324) | 83.6 (324) | 84.6 (324) | 83.6 (324) | 84.3 (324) | 99.1 (324) | 96.9 (324) | 96.8 (324) | 98.8 (324) | 95.2 (84) | 92.6 (108) | 92.6 (108) | 94.4 (108) | 94.4 (108) | 93.5 (108) | 90.7 (108) | |
| H-RIC20 D26303 | 84.5 (252) | 85.7 (252) | 86.1 (252) | 86.5 (252) | 86.9 (252) | 86.5 (252) | 86.9 (252) | 86.9 (251) | 84.5 (251) | 86.9 (251) | 86.5 (251) | 86.5 (251) | 94.0 (84) | 92.9 (84) | 92.2 (115) | 93.0 (115) | 90.4 (115) | 91.3 (115) | |
| AO12 AF016615 | 85.8 (323) | 86.7 (323) | 85.8 (323) | 86.7 (323) | 86.7 (323) | 86.7 (323) | 87.9 (323) | 82.0 (323) | 81.1 (323) | 81.7 (323) | 81.7 (323) | 81.1 (323) | 85.7 (251) | 90.7 (108) | 91.7 (108) | 91.7 (108) | 91.7 (108) | 89.8 (108) | |
| B13 AF099789 | 84.9 (325) | 83.7 (325) | 82.2 (325) | 82.5 (326) | 83.4 (325) | 83.4 (326) | 82.8 (326) | 81.8 (324) | 80.9 (324) | 81.5 (324) | 81.5 (324) | 80.9 (324) | 78.0 (273) | 84.9 (324) | 91.7 (109) | 91.7 (109) | 92.7 (109) | 90.8 (109) | |
| B. japonicum USDA 110 K01620 | 79.6 (324) | 80.6 (324) | 79.9 (324) | 78.1 (324) | 79.6 (324) | 79.3 (324) | 79.0 (324) | 80.2 (323) | 79.3 (323) | 79.9 (323) | 79.9 (323) | 79.3 (323) | 77.9 (344) | 79.6 (324) | 79.1 (368) | 96.9 (294) | 92.5 (293) | 87.0 (293) | |
| P. rhizobiumK00487 | 81.8 (324) | 82.7 (324) | 80.6 (324) | 80.9 (324) | 80.9 (324) | 80.9 (324) | 82.1 (324) | 81.7 (323) | 80.8 (323) | 81.4 (323) | 81.4 (323) | 80.8 (323) | 79.9 (344) | 82.7 (324) | 77.2 (368) | 85.9 (1274) | 93.5 (293) | 86.3 (293) | |
| Rhizobium sp. ORS571 M16710 | 87.4 (325) | 88.9 (325) | 84.0 (324) | 87.1 (325) | 84.9 (325) | 84.6 (325) | 88.0 (325) | 85.5 (324) | 85.2 (324) | 85.2 (324) | 85.2 (324) | 84.6 (324) | 82.3 (344) | 83.6 (323) | 82.6 (368) | 78.5 (933) | 82.1 (914) | 86.3 (293) | |
| H. seropedicaeZ54207 | 85.8 (325) | 86.8 (325) | 84.6 (325) | 87.4 (325) | 85.5 (325) | 85.2 (325) | 86.8 (325) | 86.7 (324) | 84.6 (324) | 86.4 (324) | 86.4 (324) | 85.8 (324) | 82.0 (344) | 81.1 (323) | 81.8 (368) | 78.7 (890) | 77.8 (901) | 80.0 (935) |
Identities for the nifH PCR product obtained for pure culture type II methanotrophs, environmental clones, and related bacteria. Environmental amino acid identities with NifH from type II methanotroph pure cultures greater than identities with NifH from other bacteria are shown in bold. Also see Table 2, footnote a.
DISCUSSION
The ability to utilize N2 as a sole nitrogen source is an important trait for the use of methanotrophs for in situ bioremediation as well as for understanding the role of methanotrophs in nitrogen cycling in different environments. However, previous results had suggested that only type II and the type I moderately thermophilic Methylococcus strains were capable of N2 fixation. Therefore, type I strains have been assumed to be unable to fix N2 in mesophilic environments. The presence of both nifH gene fragments and acetylene reduction activity in a variety of type I and type II strains provides genetic and biochemical evidence that nitrogen fixation capabilities are broadly distributed among methanotrophs (Table 1). So far the only major group of mesophilic methanotrophs for which N2-fixing strains have not been identified are the Methylomicrobium strains, and it is possible that this is due to the small number of strains tested.
Comparison of the translated nifH sequences obtained in this study with translated nifH sequences in environmental clone banks suggests that nitrogen-fixing methanotrophs may be more widespread than was previously thought. Methylomonas-like nifH fragments were amplified from rice roots and a freshwater lake (30, 35). Although we cannot be certain these nifH fragments are from methanotrophs, they are more similar to methanotrophic nifH sequences than any others in the database, and they do contain the signature amino acids that are so far specific to methanotrophs. In addition, these are environments in which type I methanotrophs are generally present (15, 16, 17).
Type II-like nifH fragments were amplified from a greater variety of environments. The translated type II nifH fragments showed high identity to translated nifH fragments from rice roots, a freshwater lake, a Douglas fir soil site, and an oligotrophic ocean (30, 32, 35, 38). Type II strains are generally thought to be present in rice roots, freshwater environments, and soils (15). However, their role in nitrogen fixation in these environments has not been studied in detail. The fact that the type II-like nifH fragment was amplified via RT-PCR from the freshwater lake environment suggests that type II methanotrophs may play a significant role in nitrogen cycling in this environment (35). Further work will be necessary to address this question.
Comparison of type II translated nifH sequences with environmental nifH sequences also showed the presence of type II-like sequences in both Atlantic and Pacific ocean samples (38). This is surprising because marine environments are generally thought to be dominated by type I strains (15). However, one study has suggested that type II methanotrophs are present in such habitats (14), so it is possible that these nifH sequences originated in methanotrophs.
Traditionally, it has been assumed that in natural populations, type II strains will be the dominant sMMO-containing population, and most in situ bioremediation protocols involving methanotrophs focus on type II strains (3). However, both type I (LW13, LW15, and the thermophilic Methylococcus capsulatus Bath) and type II strains (LW3, LW4, LW8, PW1, and Methylosinus trichosporium OB3b) have been found that can both express sMMO and fix nitrogen (2, 4, 13, 18, 21, 24, 26, 27). The correlation between increased capacity for TCE oxidation and nitrogen fixation in methanotrophs suggests that both type I and type II strains may be suitable for the bioremediation of TCE (5, 6, 7).
ACKNOWLEDGMENTS
This work was supported by a grant from the NSF (DEB9707383).
We thank John Leigh (University of Washington) for his assistance during this study.
REFERENCES
- 1.Anthony C. The biochemistry of methylotrophs. London, England: Academic Press; 1982. [Google Scholar]
- 2.Auman A J, Stolyar S, Costello A M, Lidstrom M E. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol. 2000;66:5259–5266. doi: 10.1128/aem.66.12.5259-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman J P, Jimenez L, Rosario I, Hazen T C, Sayler G S. Characterization of the methanotrophic bacterial community present in a trichloroethylene-contaminated subsurface groundwater site. Appl Environ Microbiol. 1993;59:2380–2387. doi: 10.1128/aem.59.8.2380-2387.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman J P, Sly L I, Nichols P D, Hayward A C. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methantrophs. Int J Syst Evol Bacteriol. 1993;43:735–753. [Google Scholar]
- 5.Chu K-H, Alvarez-Cohen L. Trichloroethylene degradation by methane-oxidizing cultures grown with various nitrogen sources. Water Environ Res. 1996;68:76–82. [Google Scholar]
- 6.Chu K-H, Alvarez-Cohen L. Evaluation of toxic effects of aeration and trichloroethylene oxidation on methanotrophic bacteria grown with different nitrogen sources. Appl Environ Microbiol. 1999;65:766–772. doi: 10.1128/aem.65.2.766-772.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu K-H, Alvarez-Cohen L. Treatment of chlorinated solvents by nitrogen-fixing and nitrate-supplied methane oxidizers in columns packed with unsaturated porous media. Environ Sci Technol. 2000;34:1784–1793. [Google Scholar]
- 8.Dalton H, Whittenbury R. The acetylene reduction technique as an assay for nitrogenase activity in the methane oxidizing bacterium Methylococcus capsulatus strain Bath. Arch Microbiol. 1976;109:147–151. [Google Scholar]
- 9.Dedysh S N, Liesack W, Khmelinina V N, Suzina N E, Trotsenko Y A, Semrau J D, Bares A M, Panikov N S, Tiedje J M. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol. 2000;50:955–969. doi: 10.1099/00207713-50-3-955. [DOI] [PubMed] [Google Scholar]
- 10.Dispirito A A, Gulledge J, Murrell J C, Shiemke A K, Lidstrom M E, Krema C L. Trichloroethylene oxidation by the membrane associated methane monooxygenase in type I, type II and type X methanotrophs. Biodegradation. 1992;2:151–164. [Google Scholar]
- 11.Feselstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 12.Fulton G F, Nunn D N, Lidstrom M E. Molecular cloning of a malyl CoA lyase gene (Mcl) from Pseudomonas AM1, a facultative methylotroph. J Bacteriol. 1984;160:718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuse H, Ohta M, Takimura O, Murakami K, Inoue H, Yamaoka Y, Oclarit J M, Omori T. Oxidation of trichloroethylene and dimethyl sulfide by a marine Methylomicrobium strain containing soluble methane monooxygenase. Biosci Biotechnol Biochem. 1998;62:1925–1931. doi: 10.1271/bbb.62.1925. [DOI] [PubMed] [Google Scholar]
- 14.Guezennec J, Fiala-Medioni A. Bacterial abundance and diversity in the Barbado Trench determined by phospholipid analysis. FEMS Microbiol Ecol. 1996;19:83–93. [Google Scholar]
- 15.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King G M. Associations of methanotrophs with the roots and rhizomes of aquatic vegetation. Appl Environ Microbiol. 1994;60:3220–3227. doi: 10.1128/aem.60.9.3220-3227.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh S-C, Bowman J P, Sayler G S. Soluble methane monooxygenase production and trichloroethylene degradation by a type I methanotroph, Methylomonas methanica 68-1. Appl Environ Microbiol. 1993;59:960–967. doi: 10.1128/aem.59.4.960-967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lovell C R, Piceno Y M, Quattro J M, Bagwell C E. Molecular analysis of diazotroph diversity in the rhizosphere of the smooth cordgrass Spartina alterniflora. Appl Environ Microbiol. 2000;66:3814–3822. doi: 10.1128/aem.66.9.3814-3822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murrell J C, Dalton H. Nitrogen fixation in obligate methanotrophs. J Gen Microbiol. 1983;129:3481–3486. [Google Scholar]
- 21.Oakley C J, Murrell J C. nifH genes in the obligate methane oxidizing bacteria. FEMS Micriobiol Lett. 1988;49:53–57. [Google Scholar]
- 22.Oldenhuis R, Oedzes J Y, Van der Waarde J J, Janssen D B. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl Environ Microbiol. 1991;57:7–14. doi: 10.1128/aem.57.1.7-14.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 24.Romanovskaya V A, Ludvichenko E S, Sokolov E G, Malashenko Y R. Fixation of molecular nitrogen by methanotrophs. Mikrobiol Zh. 1980;42:683–688. [PubMed] [Google Scholar]
- 25.Saito H, Miura K-I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 26.Shen R-N, Yu C-L, Ma Q-Q, Li S-B. Direct evidence for a soluble methane monooxygenase from type I methanotrophic bacteria: purification and properties of a soluble methane monooxygenase from Methylomonas sp. GYJ3. Arch Biochem Biophys. 1997;345:223–229. doi: 10.1006/abbi.1997.0239. [DOI] [PubMed] [Google Scholar]
- 27.Shigematsu T, Hanada S, Eguchi M, Kamagata Y, Kanagawa T, Kurane R. Soluble methane monooxygenase gene clusters from trichloroethylene-degrading Methylomonas sp. strains and detection of methanotrophs during in situ bioremediation. Appl Environ Microbiol. 1999;65:5198–5206. doi: 10.1128/aem.65.12.5198-5206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stolyar S, Costello A M, Peeples T L, Lidstrom M E. Role of multiple gene copies in particulate methane monooxygenase activity in the methane-oxidizing bacterium Methylococcus capsulatus Bath. Microbiol. 1999;145:1235–1244. doi: 10.1099/13500872-145-5-1235. [DOI] [PubMed] [Google Scholar]
- 29.Toukdarian A E, Lidstrom M E. Nitrogen metabolism in a new obligate methanotroph, Methylosinus strain 6. J Gen Microbiol. 1984;130:1827–1837. doi: 10.1099/00221287-130-7-1827. [DOI] [PubMed] [Google Scholar]
- 30.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whittenbury R, Philips K, Wilkinson J F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 32.Widmer F, Shaffer B T, Porteous L A, Seidler R J. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade mountain range. Appl Environ Microbiol. 1999;65:374–380. doi: 10.1128/aem.65.2.374-380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young J P W. Phylogenetic classification of nitrogen-fixing organisms. In: Stacy G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 43–86. [Google Scholar]
- 34.Young J P W. Molecular phylogeny of rhizobia and their relatives. In: Palacios R, Mora J, Newton W E, editors. New horizons in nitrogen fixation. London, United Kingdom: Kluwer Academic Publications; 1993. pp. 587–592. [Google Scholar]
- 35.Zani S, Mellon M T, Collier J L, Zehr J P. Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl Environ Microbiol. 2000;66:3119–3124. doi: 10.1128/aem.66.7.3119-3124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zehr J P, Capone D G. Problems and promises of assaying the genetic potential for nitrogen fixation in the marine environment. Microb Ecol. 1996;32:263–281. doi: 10.1007/BF00183062. [DOI] [PubMed] [Google Scholar]
- 37.Zehr J P, McReynolds L A. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1989;55:2522–2526. doi: 10.1128/aem.55.10.2522-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zehr J P, Mellon M T, Zani S. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl Environ Microbiol. 1998;64:3444–3450. doi: 10.1128/aem.64.9.3444-3450.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]