Abstract
Methanethiol (MT) and dimethyl sulfide (DMS) have been shown to be the dominant volatile organic sulfur compounds in freshwater sediments. Previous research demonstrated that in these habitats MT and DMS are derived mainly from the methylation of sulfide. In order to identify the microorganisms that are responsible for this type of MT and DMS formation, several sulfide-rich freshwater sediments were amended with two potential methyl group-donating compounds, syringate and 3,4,5-trimethoxybenzoate (0.5 mM). The addition of these methoxylated aromatic compounds resulted in excess accumulation of MT and DMS in all sediment slurries even though methanogenic consumption of MT and DMS occurred. From one of the sediment slurries tested, a novel anaerobic bacterium was isolated with syringate as the sole carbon source. The strain, designated Parasporobacterium paucivorans, produced MT and DMS from the methoxy groups of syringate. The hydroxylated aromatic residue (gallate) was converted to acetate and butyrate. Like Sporobacterium olearium, another methoxylated aromatic compound-degrading bacterium, the isolate is a member of the XIVa cluster of the low-GC-content Clostridiales group. However, the new isolate differs from all other known methoxylated aromatic compound-degrading bacteria because it was able to degrade syringate in significant amounts only in the presence of sulfide.
The production of dimethyl sulfide (DMS) and methanethiol (MT) has been intensively studied because of the roles that the oxidation products of these compounds (e.g., methanesulfonic acid and SO2) play in the processes of global warming and acid precipitation and in the global sulfur cycle (15). In marine and estuarine systems, DMS and MT are derived primarily from the degradation of dimethylsulfoniopropionate, a widespread osmolyte in marine macroalgae and phytoplankton (16). In freshwater habitats, formation of MT and DMS originates mainly from the methylation of sulfide (10, 17, 25) and to a lesser extent from the degradation of sulfur-containing amino acids (14). In many heterotrophic aerobic bacteria the methylation of sulfide is catalyzed by an S-adenosylmethionine-dependent thiol methyltransferase (7). Since this pathway is not observed in obligately anaerobic bacteria (21), it will be of minor importance in organic-rich freshwater sediments, which generally are oxygen limited (26, 27). One of the mechanisms for MT and DMS formation through sulfide methylation is the anaerobic O demethylation of methoxylated aromatic compounds (3, 10). Bacteria performing this kind of methylation use sulfide or MT as a methyl group acceptor instead of CO or CO2, which are used by several acetogenic bacteria (23, 24). However, in Holophaga foetida, Sporobacter termitidis (13), and Sporobacterium olearium (31), the O demethylation of methoxylated aromatic compounds did not strictly depend on sulfide (or MT) as methyl group acceptor. O demethylation in strain SA2 appeared to be strictly sulfide dependent (3), but unfortunately this strain was lost, which made a complete description of the isolate impossible.
The present study reports on the impact of methoxylated aromatic compounds on the production of volatile organic sulfur compounds (VOSC) in freshwater sediments. Further, the isolation of an obligately anaerobic bacterium which produces DMS and MT during growth on methoxylated aromatic compounds in sulfide-reduced mineral media is described. O demethylation of methoxylated aromatic compounds by our isolate appears to be strictly sulfide dependent.
MATERIALS AND METHODS
Source of inoculum.
Sediment samples were collected from a eutrophic freshwater pond (campus of the Dekkerswald Institute, Nijmegen, The Netherlands). The sediment samples were taken by suction in anaerobic bottles as described by Lomans et al. (25).
Slurry incubations.
Sediment slurries were prepared and dispensed in anaerobic bottles as described previously (25). Additions were made from neutral-pH stock solutions prepared in distilled water. These additions included bromoethanesulfonic acid (BES), syringate, and 3,4,5-trimethoxybenzoate (TMB). The sediment slurries (duplicates or triplicates) were incubated in the dark without shaking at 30°C. Sterilized sediment slurries (121°C, 20 min) served as abiotic controls.
Media and culture techniques.
The defined sulfide-reduced and bicarbonate-buffered medium described before (28) was supplemented with Na2SO4 to a final concentration of 28 mM. Instead of sulfide, other compounds were used as reducing agents: sodium thiosulfate (0.5 mM), cysteine-HCl (2 mM), sodium dithionite (0.5 mM), and titanium(III) nitrilotriacetic acid [Ti(III)NTA] (0.1 mM). Additions of carbon sources (syringate, TMB, gallate, pyrogallol, or pyroglucinol) (final concentration, 5 mM) were made 1 to 2 h before inoculation from anaerobic filter-sterilized stock solutions prepared in distilled water and neutralized with NaOH. Enrichment cultures were done in an anaerobic chemostat fed with syringate as the sole carbon source, continuously gassed with an H2S-containing gas stream (N2-CO2 [80:20, vol/vol]) (28). The chemostat was inoculated with 10% of a sediment slurry which had been pulsed with syringate. Batch cultivation was carried out in either 60-, 120-, or 500-ml serum bottles filled with 25, 50, or 350 ml of medium, respectively. The bottles were sealed with black butyl rubber stoppers, gassed with an O2-free mixture of N2 and CO2 (80:20, vol/vol), sterilized (121°C, 20 min), and incubated in the dark at 30°C. Purity was tested by growth experiments in the medium (with or without syringate) supplemented with yeast extract (0.5%) and Trypticase peptone (0.5%) and was checked by microscopy. Stock cultures were transferred into fresh medium once a month and stored in glass ampoules under N2-CO2 (80:20, vol/vol) after the addition of glycerol (final concentration, 5%) at −80°C.
Determination of optimal growth conditions.
Specific growth rates were determined by measuring the sum of MT and DMS formed during growth on syringate, which correlated very well with the increase of optical density. Effects of various culture conditions (pH, temperature, and salt concentrations) were tested with preadapted cultures. Cultures were transferred under these conditions at least two times sequentially.
Syringate conversion by cell suspension.
Exponential cultures were centrifuged (30 min, 10,000 × g), and the pellet was washed in anaerobic medium without syringate and reduced with Ti(III)NTA (0.05 mM)-sodium dithionite (0.25 mM). Incubations were carried out under N2-CO2 (80:20, vol/vol) at 35°C. Incubation was started by addition of syringate or syringate plus sodium sulfide.
Cell extract incubations.
A 20-liter fermentor was inoculated with a pure culture. After depletion of the carbon source, the culture received another pulse of syringate (5 mM). After growth for 200 h at 30°C, cells were harvested by continuous centrifugation (Sharples; 20,000 × g, 17°C) under an N2 atmosphere. The cell pellet was resuspended in TES buffer {50 mM 2-[tris(hydroxymethyl)methylamino]-1-ethanesulfonic acid (TES), 5 mM MgCl2, 1 mM dithiothreitol, pH 7.0}. Cells were broken by passage through a French pressure cell under an N2 atmosphere at 120 MPa. The crude extract was centrifuged (40,000 × g, 30 min, 4°C), resulting in a clear supernatant which was stored under N2-CO2 (80:20, vol/vol) at −80°C. Cell extract incubations were done in 120-ml serum bottles with 10 to 100 μl of crude cell extract, 50 mM TES buffer, 1 to 2 mM ATP, 5 mM sodium sulfide, and 0.2 mM syringate adjusted to a total volume of 1 ml with MilliQ water. The bottles were prepared in an anaerobic cabinet and sealed with gray butyl rubber stoppers. Incubations (25°C) were started by addition of syringate, and demethylation activity was monitored by measuring the formation of MT.
Analytical techniques.
Gas samples were analyzed for H2S, MT, DMS, and methane as described before (6, 25). Acetate, butyrate, and methanol were measured as described by Cazemier et al. (4). Methoxylated and hydroxylated aromatic compounds were analyzed by isocratic separation on a high-pressure liquid chromatograph equipped with a Merck LiChrospher 100 RP-18 column and an HP1040 diode array detector. The flow rate of the eluent, 40% (vol/vol) methanol in ammonium phosphate buffer (100 mM, pH 2.6), was 0.5 ml/min. The column temperature was 25°C.
Microscopy and photography.
Transmission and scanning electron micrographs were made with a transmission electron microscope (Philips 201) and a Cryo-FEG scanning electron microscope (Jeol JSM 633OF) from cells of a late-exponential-phase culture. Negative staining was done with uranyl acetate (0.5 to 1%).
Phylogenetic analysis.
DNA from the pure culture was isolated by harvesting the cells in an Eppendorf centrifuge (10,000 × g for 5 min at room temperature). The cell pellet was washed two times in fresh nonreduced medium, suspended in 0.5 ml of lysis buffer (0.1 M sodium EDTA, 0.15 M NaCl, 0.02% lysozyme), and incubated for 2 h at 37°C. Sodium dodecyl sulfate (SDS) (1% wt/vol) was added, and incubation was continued for 30 min at 37°C. Proteinase K (50 μg/ml) was added, and the suspension was incubated for 30 min at 37°C. The solution was extracted with phenol-chloroform-isoamyl alcohol (25:24:1). The pellet obtained after ethanol precipitation of the water phase was dissolved in 10 μl of sterile distilled water and stored at −20°C. The DNA was used as a template for PCR amplification of an approximately 1,530-bp segment of the 16S rRNA gene with 2.5 mM MgCl2, an annealing temperature of 50°C, and 35 cycles. The PCR amplification primers used were pA (19EubFOR) (5′-GAGTTTGATCCTGGCTCAG-3′) and pH′ (1522REV) (5′-AAGGAGGTGATCCAGCCGCA-3′) (8). The amplification product was ligated in the pCR II vector and transformed into Escherichia coli cells from the TA cloning kit (Invitrogen). Plasmid DNA of clones was isolated by using the FlexiPrep kit (Pharmacia P-L Biochemicals Inc.). The sequence of the cloned PCR product, which represented the original 16S rRNA sequence, was analyzed on a DNA sequencer (model 373A; Applied Biosystems, Inc., Foster City, Calif.) using the Taq DyeDeoxy terminator cycle sequencing method (30; Taq DyeDeoxy terminator cycle sequencing kit user bulletin no. 901497, revision E [Applied Biosystems, Inc.]). Besides the primers of the TA cloning kit (M13FOR and M13REV) and primers mentioned in the literature (pA [19FOR], pE′ [908REV], and pH′ [1522REV] [8] and 517REV [1]), the following primers were used for sequencing: 500FOR (5′-TGTGCCAGCAGCCGCGGTAA-3′), 1050FOR (5′-GTGCATGGCTGTCGTCAGYTC-3′), and 1161REV (5′-TGACGTCATCCCCACCTT-3′). The primers 500FOR, 1050FOR, and 1162REV were designed on the basis of published eubacterial and archaeal 16S rRNA sequences. The resulting sequences were assembled to produce a 1,487-bp DNA sequence. The deduced 16S rRNA sequence of strain SYR1 was aligned with homologous 16S rRNA sequences of closely related members of the Eubacteria domain by using the Pileup method (Dutch CMBI Center Facility, Nijmegen, The Netherlands). Distance matrix trees were constructed by using the method of Fitch and Margoliash (12) and the neighbor-joining method of Saitou and Nei (33) in the FITCH and NEIGHBOR programs of the PHYLIP (version 3.4) program package (9). Parsimony and bootstrap parsimony analyses were performed using the DNAPARS and DNABOOT programs as implemented in the PHYLIP package.
Nucleotide sequence accession numbers.
The deduced almost-complete primary structure (1,487 bp) of the 16S rRNA gene of strain SYR1 has been deposited in the EMBL sequence database under accession number AJ272036.
RESULTS
Slurry incubations.
Slurries from various origins amended with either syringate (0.5 mM) or TMB (0.5 mM) all showed transient accumulation of DMS and traces of MT. After prolonged incubation (>200 h), the accumulated DMS and MT disappeared. In contrast, in nonamended slurries MT and DMS concentrations were below the detection limit (<0.3 μM). Addition of BES to syringate- or TMB-amended slurries resulted in a dramatic increase of the rate of accumulation of MT and DMS, which reached concentrations of higher than 0.5 and 1.5 mM, respectively. In BES-syringate- or BES-TMB-amended slurries, DMS and MT did not disappear even after prolonged incubation. All of the sediments tested showed similar patterns, although the final concentrations of the MT and DMS accumulated differed. In all incubations DMS was the major VOSC produced.
Enrichment and isolation of strain SYR1.
The sediment of a eutrophic freshwater pond (Dekkerswald) was used to inoculate a chemostat in order to isolate anaerobic syringate-utilizing and DMS-producing microorganisms. Syringate was converted to acetate, MT, and DMS. The use of this chemostat (D = 0.10 h−1) resulted in an MT- and DMS-producing enrichment consisting of one dominant morphologically distinct bacterium. Deep-agar tube dilution series on syringate plus sulfide eventually led to a pure culture of an MT- and DMS-producing anaerobic bacterium, which was named strain SYR1. Purity was confirmed by microscopic analysis and absence of growth after addition of yeast extract (0.5%) and Trypticase peptone (0.5%) to the medium.
Morphology.
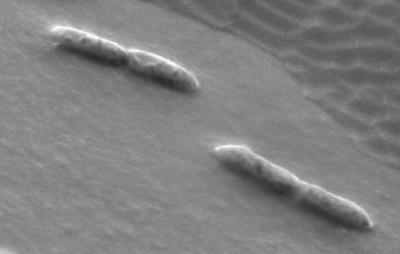
Deep-agar colonies of strain SYR1 were white circular disks and reached a diameter of 1 to 2 mm in 1 week while growing on syringate plus sulfide. Cells occur as double rods (Fig. 1). The average length and width of individual cells were 2.0 and 0.4 μm, respectively. Motility and spore formation were not observed. Cells did not lyse within 15 min after addition of SDS (0.1 to 1%). The Gram stain reaction was negative.
FIG. 1.
Scanning electron micrograph of a logarithmic-phase culture of strain SYR1 grown on syringate (5 mM). The photograph clearly shows the double-rod morphology of strain SYR1.
Catabolic substrates.
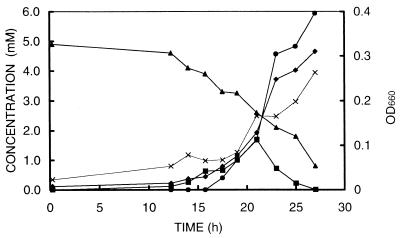
Growth of strain SYR1 was observed only on syringate (5 mM), TMB (5 mM), and gallate (5 mM). Growth on syringate and TMB was accompanied by concomitant formation of MT and DMS, whereas growth on gallate was not. Degradation of syringate was characterized by transient accumulation of MT (mostly during the logarithmic phase), directly followed by accumulation of DMS (Fig. 2). This increase of the MT and DMS correlated with an increase in optical density and acetate concentration and a decrease in syringate. Cultures normally had a lag phase of about 10 to 15 h followed by exponential growth. Transferring SYR1 from syringate to gallate did not result in a prolonged lag phase. The maximum specific growth rate estimated from MT and DMS formation as well as from optical density measurements was 0.2 h−1. No growth or MT or DMS formation was observed on 3,4-dimethoxybenzoate, 3,5-dimethoxybenzoate, vanillate, pyrogallol, phloroglucinol, benzoate, pyruvate, H2-CO2, methanol, malate, fructose, glucose, pectin, or glycine-betaine. Concentrations used were 5 mM, except for methanol (10 mM), H2-CO2 (80:20, vol/vol), phloroglucinol (2.5 mM), and pectin (1 g/liter).
FIG. 2.
Time courses of growth of strain SYR1 on syringate and sulfide. Symbols: ▴, syringate; ▪, MT; ♦, DMS; ●, acetate; ×, optical density at 660 nm (OD660).
Reducing agents.
In cultures of strain SYR1 on syringate, TMB, or gallate, no growth was observed when sulfide was replaced by cysteine, thiosulfate, or Ti(III)NTA. Cysteine, thiosulfate, and Ti(III)NTA were not toxic, since they did not inhibit growth of strain SYR1 when added to sulfide-reduced cultures. Moreover, if sulfide was added to cysteine- or Ti(III)NTA-reduced cultures after prolonged incubation, growth was observed (see also Fig. 3B). The combined addition of sodium sulfide, cysteine, sodium thiosulfate, and Ti(III)NTA usually resulted in higher final MT and DMS concentrations in fully grown cultures. Addition of cysteine also resulted in a slightly higher optical density (at 660 nm) of the final culture (0.184, versus 0.155 on 5 mM syringate). Sulfide toxicity was tested by growing strain SYR1 at various sulfide concentrations (0.9 to 17 mM). The growth rate calculated from MT and DMS production was highest (0.18 h−1) in cultures with 2.5 mM sulfide and decreased dramatically in cultures with higher sulfide concentrations.
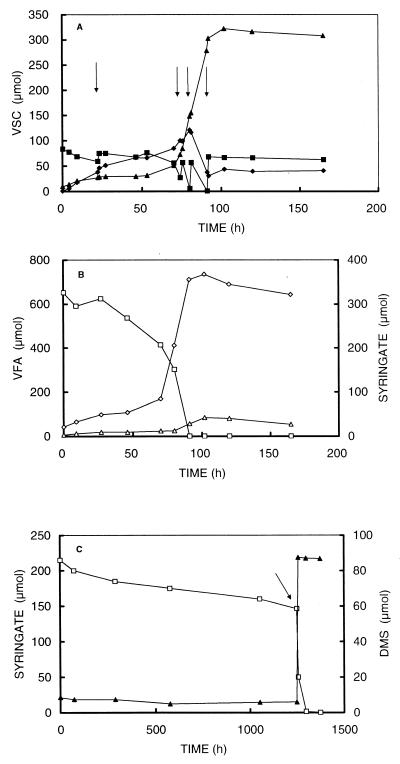
FIG. 3.
Time courses of volatile sulfur compounds (VSC) (A) and syringate and volatile fatty acids (VFA) (B) of a sulfide-rich culture of strain SYR1 and time courses of syringate and VOSC of a sulfide-free culture of strain SYR1 (C). Arrows indicate pulsewise additions of sodium sulfide. Symbols: ▪, sulfide; ♦, MT; ▴, DMS; □, syringate; ⋄, acetate; ▵, butyrate.
Optimal growth conditions.
Strain SYR1 grew optimally between pH 6.5 and 7.0 and between 34 and 37°C. No growth was observed above 40°C. Strain SYR1 showed optimal growth at NaCl concentrations of 0 to 30 mM (note that the standard medium already contained 28 mM sodium sulfate). MT and DMS formation occurred up to 135 mM NaCl. The tolerance of strain SYR1 to high concentrations of syringate and acetate was tested in order to be able to predict the behavior of strain SYR1 during cultivation of high quantities of cell biomass. In cultures with higher concentrations of syringate, sodium sulfide was added in pulses (2.5 mM) to avoid both toxicity and depletion of sulfide. Strain SYR1 showed optimal growth at syringate concentrations of 2.5 to 7.5 mM. At higher concentrations (tested up to 20 mM), growth was observed but appeared to be lower and less predictable. Moreover, in cultures grown with increasing concentrations of syringate, the concentrations of acetate produced did not increase proportionally to the amount of syringate added. In addition, increasing amounts of butyrate were formed (Table 1). Addition of 0.05% (wt/vol) yeast extract stimulated growth but did not appear to be essential for growth.
TABLE 1.
Formation of acetate and butyrate in cultures amended with various concentrations of syringatea
| Syringate added (mM) | Acetate formed (mM)b | Butyrate formed (mM)b |
|---|---|---|
| 2.5 | 5.3 ± 0.4 | <0.1 |
| 5 | 8.4 ± 0.6 | <0.1 |
| 7.5 | 12.6 ± 0.9 | 0.8 ± 0.1 |
| 10 | 15.9 ± 1.1 | 3.7 ± 0.3 |
| 15 | 22.7 ± 1.5 | 7.1 ± 0.5 |
| 20 | 26.0 ± 1.2 | 9.4 ± 0.6 |
Culture bottles were incubated for 14 days at 30°C without shaking. When sulfide was depleted, extra additions were made.
Values are means ± standard deviations (n = 3).
Impact of sulfide on syringate degradation.
The sulfide dependency of the syringate degradation was investigated by cultivation of strain SYR1 under sulfide-rich and sulfide-free conditions.
(i) Cultivation in an excess of sulfide.
During the incubation, cultures were pulsewise amended with sodium sulfide to prevent both sulfide-limited conditions and sulfide toxicity. Under sulfide-rich conditions, syringate degradation resulted in the formation of large amounts of MT, DMS, and acetate (Fig. 3A). Formation of MT, DMS, acetate, and butyrate stopped when syringate was depleted (90 h). The amount of butyrate was very low compared to amount of acetate produced (Fig. 3B). After complete degradation of syringate, in the presence of residual sulfide, MT remained in the medium.
(ii) Cultivation in sulfide-free media.
Cultures without sodium sulfide and with Ti(III)NTA (0.6 mM) or cysteine (3.4 mM) as a reducing agent did not show any DMS formation and showed only very limited syringate degradation (Fig. 3C). Furthermore, no increase in optical density was observed. Cultures amended with Ti(III)NTA turned yellowish. When after an incubation of more than 40 days sulfide was added to the Ti(III)NTA-reduced cultures, syringate was degraded with concomitant formation of DMS (Fig. 3C) and the yellow color disappeared. The cells in this culture apparently were still viable.
(iii) Cell suspension experiments.
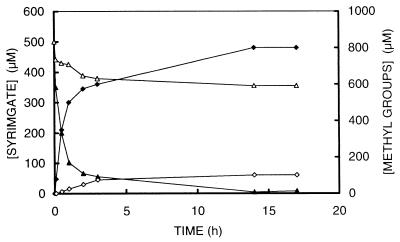
The growth experiments with sulfide limitation mentioned above are likely to be affected by sulfur source limitation, since strain SYR1 is strictly dependent on sulfide as a sulfur source. Therefore, cell suspension experiments were performed under sulfide-rich and sulfide-free [Ti(III)NTA-reduced] conditions in which no growth and thus no sulfur source is needed. Indeed, syringate degradation was limited by the absence of sulfide. In cell suspensions with sulfide, syringate was demethylated very rapidly within 60 min with concomitant production of MT and DMS (Fig. 4). Although no sulfide was added (or detected) in the sulfide-free washed-cell suspensions, a limited degradation of syringate with concomitant MT and DMS formation was observed (Fig. 4). The potential for demethylation of the cell suspensions without sulfide was not irreversibly affected, since addition of sulfide after prolonged incubation of these cell suspensions resulted in an immediate production of MT, DMS, and demethylated aromatic residues (data not shown). These aromatic residues disappeared after prolonged incubation. The Ks for sulfide of strain SYR1 was determined by measuring the initial MT formation rate of cell suspensions incubated with various concentrations of sulfide (added as sodium sulfide). The Ks of strain SYR1 for sulfide was on the order of 200 to 400 μM.
FIG. 4.
Time courses of cell suspensions incubated under sulfide-rich (closed symbols) and sulfide-free (open symbols) conditions. Triangles, syringate; diamonds, methyl groups (MT plus DMS).
Demethylation of methoxylated aromatic compounds by strain SYR1.
The influence of temperature on the demethylation activity was tested by incubation of cell extracts at various temperatures (20 to 40°C). Maximum demethylation activity, measured as the MT formed, was found at 25°C. The oxygen sensitivity of demethylation was tested by preincubating the cell extract under an air atmosphere in the absence and presence of ATP. Subsequently, MT formation capacity was measured under an anaerobic atmosphere. The MT formation capacity of the cell extracts preincubated under air appeared to be dramatically lower (38 pmol · min−1 · μl of cell extract−1) than that of the control (258 pmol · min−1 · μl of cell extract−1) and was not restored by the addition of ATP either before of after the exposure to oxygen (15 pmol · min−1 · μl of cell extract−1). The enzymatic formation of MT was strictly ATP dependent (optimum at an ATP concentration of 2 mM). Addition of both glucose and hexokinase during the incubation significantly inhibited the MT formation. Addition of propyliodide, a known inhibitor of corronoid-dependent methyltransferases, did not affect the MT formation by the cell extract. Doubling the amount of cell extract in the assay mixture resulted in proportionally increased MT formation rates. Activity screening of fractions obtained by fast performance liquid chromatography gel filtration demonstrated that all of the activity resided in only one fraction with a molecular mass of about 200 kDa. MT formation was found after addition of the following substrates: 3,4-dimethoxybenzoate, TMB, 4-hydroxy-3-methoxybenzoate (vanillate), and 4-hydroxy-3,5-dimethoxybenzoate (syringate). No MT formation was found after addition of 3,5-dimethoxybenzoate.
Phylogenetic and taxonomic analysis.
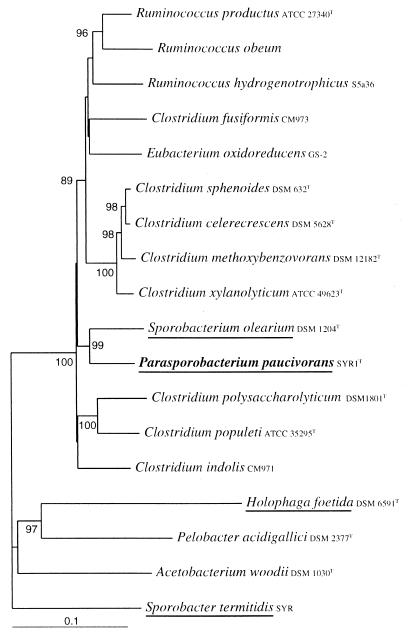
A 1,530-bp DNA fragment homologous to the rRNA gene of strain SYR1 was amplified in vitro, cloned in the pCR II vector, transformed in E. coli, and sequenced. Use of the Chimera Check analysis of the Ribosomal Database Project (29) showed that the sequence was indeed derived from a single target DNA sequence. Database searches revealed that the highest similarity value (91.8%) was found for the sequences of strain SYR1 and Sporobacterium olearium. A phylogenetic tree based on a matrix of distances of a 1,487-bp fragment of the 16S rRNA gene clearly shows that strain SYR1 clusters within subclass XIVa of the very polymorphic Clostridiales (Fig. 5). Besides 16S rRNA sequences of closely related microorganisms, those of several other methoxylated aromatic compound-degrading bacteria, including H. foetida and Sporobacter termitidis, are incorporated in this tree.
FIG. 5.
Phylogenetic tree based on a distance matrix prepared from an alignment of a partial 16S rRNA sequences (1,478 bp) of P. paucivorans and other selected closely related bacteria. Bootstrap values indicate the percentage of occurrence of 100 bootstrap trees. Only values above 80 are given. Reference sequences were from the GenBank and EMBL databases: X95624, Ruminococcus hydrogenotrophicus; X94966, Ruminococcus productus; X85101, Ruminococcus obeum; AF116854, Sporobacterium olearium; Z49863, Sporobacter termitidis; X77215, Holophaga foetida; X77216, Pelobacter acidigallici; X96954, Acetobacterium woodii; X71858, Clostridium polysaccharolyticum; X71853, Clostridium populeti; AF028351, Clostridium indolis; X71855, Clostridium xylanolyticum; AF067965, Clostridium methoxybenzovorans; X73449, Clostridium sphenoides; X71848, Clostridium celerecrescens; AF028349, Clostridium fusiformis; AF202259, Eubacterium oxidoreducens. Sporobacter termitidis was used as the outgroup. Bar, 10 base substitutions per 100 bases. Names of MT- or DMS-producing microorganisms are underlined.
DISCUSSION
Isolation and physiology of strain SYR1.
The in situ concentrations of the most dominant VOSC, MT and DMS, showed strong correlation with the rate of methane formation and the sulfide concentration in the sediment, the dominant compartment in VOSC formation of freshwater ecosystems (25). Methylation of sulfide with methoxylated aromatic compounds as methyl group donors is the major mechanism for VOSC formation in freshwater sediments (10, 17, 25). Addition of syringate or TMB to slurries prepared from various freshwater sediments immediately resulted in elevated levels of DMS, indicating an active bacterial population. From one of the sediments an anaerobic bacterium was isolated with syringate as the sole carbon source. The isolated strain, SYR1, has a very limited substrate spectrum, being capable of anaerobic growth only on syringate, TMB, and gallate as sole carbon and energy sources. The fact that the production of MT and DMS was restricted to growth on syringate or TMB with sulfide (it was absent with gallate plus sulfide) demonstrates that the carbon atoms in the produced MT and DMS are derived from the methoxy groups of syringate or TMB, as is the case for the previously described isolates strain SA2, H. foetida, Sporobacter termitidis, and Sporobacterium olearium (3, 13, 31). Similar to the case for H. foetida and SA2, strain SYR1 is likely to degrade syringate via O demethylation of the methoxy groups , since the strain does not grow on methanol. In contrast to H. foetida (3), however, the demethylation by strain SYR1 appeared to be strictly sulfide dependent. Sulfide could not be replaced by cysteine, thiosulfate, or sulfate. A similar dependency was reported for strain SA2 (3). However, this organism could grow on gallate without addition of sulfide.
The transient accumulation of MT in cultures of strain SYR1 demonstrates that DMS is formed via MT as an intermediate. In this respect strain SYR1 differs from the closely related Sporobacterium olearium, which only produces MT from the methoxy groups of syringate (31). The use of MT or sulfide as a methyl group acceptor is likely to depend on the actual concentration of either of the two compounds, since MT formation was higher in sulfide-rich cultures, whereas DMS was the dominant product under sulfide-limited conditions. The Ks for sulfide of strain SYR1 (200 to 400 μM) was on the same order of magnitude as that found for the freshwater sediment from which it had been isolated (25).
The different isolation procedures for H. foetida and strain SA2 (most-probable-number series) and strain SYR1 (chemostat) is reflected in the fact that strain SYR1 has a higher maximum specific growth rate (0.2 h−1) than H. foetida (0.06 h−1), strain SA2 (0.10 h−1), and Sporobacter termitidis (0.05 h−1) (3, 13). The maximum specific growth rate of Sporobacterium olearium was not reported (31). On the basis of the experiments outlined in Fig. 2 and Table 1, we assume the following stoichiometries for acetate and butyrate formation, respectively:
C9H10O5 (syringic acid) + H2S + 3H2O → 3CH3COOH + CH3SCH3 + CO2
5C9H10O5 (syringic acid) + 5H2S + 9H2O → 6CH3CH2CH2COOH + 5CH3SCH3 + 11CO2
Biochemical analysis of a cell extract of strain SYR1 revealed that several methoxylated aromatic compounds, which do not support growth, were also demethylated. This lack of growth might be caused by the absence of the appropriate transport mechanism for the demethylated products. Moreover, it was demonstrated that the demethylating enzyme system was ATP dependent and oxygen sensitive, making MT and DMS synthesis a strictly anaerobic process, as was the case for H. foetida (18, 19). The absence of inhibition by propyliodide revealed that the enzyme systems of strain SYR1 are likely to be corronoid independent.
Phylogenetic and taxonomic analysis.
Strain SYR1 appeared to be a member of cluster XIVa of the genus Clostridiales (4, 5). Phylogenetically, strain SYR1 appeared to be most closely related to Sporobacterium olearium; however, there are numerous physiological differences between the two isolates. Strain SYR1 has a very limited substrate range, whereas Sporobacterium olearium could use a wide variety of substrates. Moreover, strain SYR1 did not produce spores, whereas Sporobacterium olearium did. Gram staining of Sporobacterium olearium was positive, whereas that of strain SYR1 was negative. Finally, our isolate produces both MT and DMS from methoxy groups of syringate, whereas Sporobacterium olearium produced MT only. Also, the syringate metabolism of strain SYR1 did not appear to be influenced by the presence of H2 in the culture like that of Sporobacterium olearium (31).
In contrast to most of the other closely related organisms of the genera Clostridium, Eubacterium, and Ruminococcus, both strain SYR1 and Sporobacterium olearium could not utilize common carbohydrates (e.g., glucose and fructose) as carbon and energy sources. Remarkably, the feature of (methoxylated) aromatic compound metabolism is distributed among various genera (2, 3, 13, 20, 22, 23, 24, 31, 32, 34, 35).
On the basis of its low 16S rRNA sequence similarity (91.8%) with the closest related bacterium, Sporobacterium olearium, as well as its physiological properties, we propose that strain SYR1 is a representative of a novel genus. The isolate was named Parasporobacterium paucivorans.
Ecological niche of P. paucivorans.
The formation and degradation of VOSC in situ are well balanced (25). Production of MT and DMS occurs mainly anaerobically because of the steep oxygen gradient at the water column-sediment interface (26, 27). The major part of the endogenous produced MT and DMS is formed by means of the methylation of sulfide and subsequently MT (17, 25). According to its physiological properties (VOSC production and apparent Ks for sulfide), P. paucivorans strain SYR1 is likely to be responsible for the production of MT and DMS in freshwater sediments in situ. The degradation of methoxylated aromatic compounds in sulfide-poor sediments is likely to be dominated by acetate-producing bacteria such as Acetobacterium woodii, whereas in sulfide-rich sediment MT- and DMS-producing organisms like P. paucivorans will dominate. In sediments with alternating conditions (sulfide rich and sulfide poor), bacteria like H. foetida and Sporobacter termitidis will have an advantage compared to the other organisms. The transient accumulation of MT and DMS in slurry experiments confirms the interaction between syringate-degrading and methanogenic bacteria (10, 11). A representative methanogen, Methanomethylovorans hollandica, was previously isolated (28).
Description of Parasporobacterium paucivorans gen. nov., sp. nov.
Parasporobacterium paucivorans (Pa.ra.spo.ro.bac.te.′ri.um. Gr. prefix para, besides, next to; Gr. n. sporos, seed, spore; Gr. n. bacterion, small rod; N.L. neut. n. Parasporobacterium, a genus similar to Sporobacterium; pau.ci.vo′rans. L. plur. pron. pauci, a few; L. adj. part. vorans, devouring; L. adj. paucivorans, degrading a limited number of substrates), double rod-shaped cells (length, 1.5 to 2 μm; width, 0.3 to 0.5 μm) which exhibit a negative Gram reaction. Cells normally occur as double rods. Cells were not sensitive to lysis by 0.1 g of SDS per liter. Syringate, TMB, and gallate are catabolic substrates. Sulfide is essential for growth and demethylation of methoxylated aromatic compounds. Growth was most rapid at 0 to 1.8 g of NaCl; some MT and DMS formation occurred at up to 8 g of NaCl. Growth is most rapid at pH 6.5 to 7.0. Growth is most rapid at 34 to 37°C, and no growth was obtained at temperatures of above 40°C. The type strain SYR1 was isolated from a slurry of a eutrophic lake sediment (campus of the Dekkerswald Institute, Nijmegen, The Netherlands). Strain SYR1T, named P. paucivorans, has been deposited in the DSMZ culture collection (Braunschweig, Germany) (accession number not yet available) and in The Netherlands Culture Collection of Bacteria under accession number NCCB100009.
ACKNOWLEDGMENTS
We thank Hans Hippe of the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH for his advice on the nomenclature for our isolate.
This work was supported by The Netherlands Organization for the Advancement of Pure Research (NWO) as part of the program “Verstoring van Aardsystemen.”
REFERENCES
- 1.Achenbach L, Woese C. 16S and 23S rRNA-like primers. In: Robb F T, Place A R, Sowers K R, Scheier H J, DasSarma S, Fleischmann E M, editors. Archaea, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 521–523. [Google Scholar]
- 2.Bache R, Pfennig N. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol. 1981;130:255–261. [Google Scholar]
- 3.Bak F, Finster K, Rothfuss F. Formation of dimethyl sulfide and methanethiol from methoxylated aromatic compounds and inorganic sulfide by newly isolated anaerobic bacteria. Arch Microbiol. 1992;157:529–534. [Google Scholar]
- 4.Cazemier A E, Op den Camp H J M, Hackstein J H P, Vogels G D. Fibre digestion in arthropods. Comp Biochem Physiol. 1997;118A:101–109. [Google Scholar]
- 5.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 6.Derikx P J L, Op den Camp H J M, van der Drift C, Van Griensven L J L D, Vogels G D. Odorous sulfur compounds emitted during production of compost used as a substrate in mushroom cultivation. Appl Environ Microbiol. 1990;56:176–180. doi: 10.1128/aem.56.1.176-180.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drotar A, Burton G A, Jr, Tavernier J E, Fall R. Widespread occurrence of bacterial thiol methyltransferases and the biogenic emission of methylated sulfur gases. Appl Environ Microbiol. 1987;53:1626–1631. doi: 10.1128/aem.53.7.1626-1631.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards U. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein J. Numerical methods of inferring evolutionary trees. Q Rev Biol. 1982;57:379–404. [Google Scholar]
- 10.Finster K, King G M, Bak F. Formation of methyl mercaptan and dimethyl sulfide from methoxylated aromatic compounds in anoxic marine and freshwater sediments. FEMS Microbiol Ecol. 1990;74:295–302. [Google Scholar]
- 11.Finster K, Tanimoto Y, Bak F. Fermentation of methanethiol and dimethylsulfide by a newly isolated methanogenic bacterium. Arch Microbiol. 1992;157:425–430. [Google Scholar]
- 12.Fitch W M, Margoliash E. Construction of phylogenetic trees: a method based on mutation distances as estimated by cytochrome c sequences is of general applicability. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 13.Grech-Mora I, Fardeau M-L, Patel B K C, Ollivier B, Rimbault A, Prensier G, Garcia J-L, Garnier-Sillam E. Isolation and characterization of Sporobacter termitidis gen. nov., sp. nov., from the digestive tract of the wood-feeding termite Nasutitermes lujae. Int J Syst Bacteriol. 1996;46:512–518. [Google Scholar]
- 14.Kadota H, Ishida Y. Production of volatile sulfur compounds by microorganisms. Annu Rev Microbiol. 1972;26:127–138. doi: 10.1146/annurev.mi.26.100172.001015. [DOI] [PubMed] [Google Scholar]
- 15.Kelly D P, Smith N A. Organic sulfur compounds in the environment: biochemistry, microbiology and ecological aspects. Adv Microb Ecol. 1990;11:345–385. [Google Scholar]
- 16.Kiene R P, Capone D G. Microbial transformations of methylated sulfur compounds in anoxic salt marsh sediments. Microb Ecol. 1988;15:275–291. doi: 10.1007/BF02012642. [DOI] [PubMed] [Google Scholar]
- 17.Kiene R P, Hines M E. Microbial formation of dimethyl sulfide in anoxic Sphagnum peat. Appl Environ Microbiol. 1995;61:2720–2726. doi: 10.1128/aem.61.7.2720-2726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreft J-U. The methyl ester cleaving enzyme system of the anaerobic bacterium Holophaga foetida. PhD thesis. Konstanz, Germany: Universität Konstanz; 1995. [Google Scholar]
- 19.Kreft J-U, Schink B. Demethylation and degradation of phenylmethylethers by the sulfide-methylating homoacotogenic bacterium strain TMBS4. Arch Microbiol. 1993;159:308–315. [Google Scholar]
- 20.Krumholz L R, Bryant M P. Eubacterium oxidoreducens sp. nov. requiring H2 or formate to degrade gallate, pyrogallol, phloroglucinol and quercetin. Arch Microbiol. 1986;144:8–14. [Google Scholar]
- 21.Larsen G L. Distribution of cysteine β-lyase in gastro-intestinal bacteria and in the environment. Xenobiotica. 1985;15:199–209. doi: 10.3109/00498258509045350. [DOI] [PubMed] [Google Scholar]
- 22.Liesack W, Bak F, Kreft J U, Stackebrandt E. Holophaga foetida gen. nov., sp. nov., a new homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch Microbiol. 1994;162:85–90. doi: 10.1007/BF00264378. [DOI] [PubMed] [Google Scholar]
- 23.Liu S, Suflita J M. H2-CO2-dependent anaerobic O-demethylation activity in subsurface sediments by an isolated bacterium. Appl Environ Microbiol. 1993;59:1325–1331. doi: 10.1128/aem.59.5.1325-1331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, Suflita J M. H2 as an energy source for mixotrophic acetogenesis from the reduction of CO2 and syringate by Acetobacterium woodii and Eubacterium limosum. Curr Microbiol. 1995;31:245–250. [Google Scholar]
- 25.Lomans B P, Smolders A J P, Intven L, Pol A, Op den Camp H J M, van der Drift C. Formation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl Environ Microbiol. 1997;63:4741–4747. doi: 10.1128/aem.63.12.4741-4747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomans B P, Op den Camp H J M, Pol A, Vogels G D. Anaerobic versus aerobic degradation of dimethyl sulfide and methanethiol in anoxic freshwater sediments Appl. Environ Microbiol. 1999;65:438–443. doi: 10.1128/aem.65.2.438-443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomans B P, Op den Camp H J M, Pol A, van der Drift C, Vogels G D. Role of methanogens and other bacteria in degradation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl Environ Microbiol. 1999;65:2116–2121. doi: 10.1128/aem.65.5.2116-2121.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomans B P, Maas R, Luderer R, Op den Camp H J M, Pol A, van der Drift C, Vogels G D. Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl Environ Microbiol. 1999;65:3641–3650. doi: 10.1128/aem.65.8.3641-3650.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride L J, Koepf S M, Gibbs R A, Salser W, Mayrand P E, Hunkapiller M W, Kronick M N. Automated DNA sequencing methods involving polymerase chain reaction. Clin Chem. 1989;35:2196–2201. [PubMed] [Google Scholar]
- 31.Mechichi T, Labat M, Garcia J-L, Thomas P, Patel B K C. Sporobacterium olearium gen. nov., sp. nov., a new methanethiol-producing bacterium that degrades aromatic compounds, isolated from an olive mill wastewater treatment digester. Int J Syst Bacteriol. 1999;49:1741–1748. doi: 10.1099/00207713-49-4-1741. [DOI] [PubMed] [Google Scholar]
- 32.Mechichi T, Labat M, Patel B K C, Woo T H S, Thomas P, Garcia J-L. Clostridium methoxybenzovorans sp. nov., a new aromatic O-demethylating homoacetogen from an olive mill waste treatment digester. Int J Syst Bacteriol. 1999;49:1201–1209. doi: 10.1099/00207713-49-3-1201. [DOI] [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. The neighbor-joining method: a new method for reconstruction of phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Schink B, Pfennig N. Fermentation of trihydroxybenzenes by Pelobacter acidigallici gen. nov., sp. nov., a new strictly anaerobic non-spore-forming bacterium. Arch Microbiol. 1982;133:195–201. [Google Scholar]
- 35.Stupperich E, Konle R. Corronoid-dependent methyl transfer reactions are involved in methanol and 3,4-dimethoxybenzoate metabolism by Sporomusa ovata. Appl Environ Microbiol. 1993;59:3110–3116. doi: 10.1128/aem.59.9.3110-3116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]