Abstract
Haemophilus influenzae, Streptococcus pneumoniae and Moraxella catarrhalis are bacterial species which frequently co-colonise the nasopharynx, but can also transit to the middle ear to cause otitis media. Chronic otitis media is often associated with a polymicrobial infection by these bacteria. However, despite being present in polymicrobial infections, the molecular interactions between these bacterial species remain poorly understood. We have previously reported competitive interactions driven by pH and growth phase between H. influenzae and S. pneumoniae. In this study, we have revealed competitive interactions between the three otopathogens, which resulted in reduction of H. influenzae viability in co-culture with S. pneumoniae and in triple-species culture. Transcriptomic analysis by mRNA sequencing identified a central role of arginine in mediating these interactions. Arginine supplementation was able to increase H. influenzae survival in a dual-species environment with S. pneumoniae, and in a triple-species environment. Arginine was used by H. influenzae for ATP production, and levels of ATP generated in dual- and triple-species co-culture at early stages of growth were significantly higher than the combined ATP levels of single-species cultures. These results indicate a central role for arginine-mediated ATP production by H. influenzae in the polymicrobial community.
Introduction
Otitis media (OM) is one of the most prevalent paediatric diseases, affecting 80% of children before the age of 3 [1]. OM frequently manifests as a chronic or recurrent infection [2], where conventional treatments such as antibiotic therapies [3], and tympanostomy tube placement [4] are often ineffective. The most common aetiologic agents of OM are the bacterial species Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis [5]. It is common that these species are present in the middle ear as a polymicrobial infection or in a polymicrobial biofilm [5,6]. They also regularly co-colonise the nasopharynx asymptomatically [7]. Concurrent carriage of all three pathogens has also been associated with clinical pneumonia [8].
We have previously identified the potential for both synergistic and competitive interactions between S. pneumoniae and H. influenzae in a dual-species polymicrobial environment, with the outcome of the interactions governed by environmental conditions, and characterised by a specific transcriptomic response [9,10]. However, the interactions between all three species, and the molecular aspects governing these interactions, have not been well defined [11], with current data showing both positive and negative interaction outcomes.
M. catarrhalis colonisation in children has been positively correlated to colonisation with H. influenzae [12], and another study has displayed positive associations of S. pneumoniae colonisation with co-colonisation by M. catarrhalis or H. influenzae [13]. However, a negative association between H. influenzae and S. pneumoniae has been reported during clinical studies of upper respiratory tract infection [14,15]. In addition, clinical studies have shown strain specificity to affect inter-species interactions [16], as well as significant variation in inter-species associations and species abundance between anatomical sites [5]. In a mouse colonisation model, co-infection of S. pneumoniae with M. catarrhalis was found to influence OM more significantly in comparison to co-infection of S. pneumoniae with H. influenzae [17]. In the context of antibiotic resistance of OM, M. catarrhalis has been shown to protect amoxicillin-susceptible H. influenzae and S. pneumoniae [18]. However, S. pneumoniae has also been shown to inhibit M. catarrhalis growth in a manner dependent on H2O2 and bacteriocin production [19].
The current study aims to elucidate the interactions between H. influenzae, S. pneumoniae and M. catarrhalis, and to assess the transcriptomic response of H. influenzae to growth in a triple-species co-culture with S. pneumoniae and M. catarrhalis.
Materials and methods
Bacterial growth and storage
All bacterial strains were stored at -80°C in 30% glycerol. For all assays the H. influenzae isolate 86-028NP from the nasopharynx of a pediatric patient with OM, or laboratory strain Rd KW20, S. pneumoniae serotype 3 OM middle ear isolate strain 11, and M. catarrhalis reference strain QC (23240) obtained from SA Pathology laboratory (originating from a lung infection) were used for analyses. H. influenzae was grown on heart infusion (HI) agar plates supplemented with 10% levinthals, S. pneumoniae was pre-cultured on blood agar plates and M. catarrhalis was pre-cultured on HI agar supplemented with 5% defibrinated horse blood [9]. For pre-culture, all three bacterial species were grown until mid-log phase. H. influenzae was cultured in 10 mL of HI medium supplemented with 10 μg/mL hemin and 2 μg/mL β-NAD, with shaking at 100 rpm. M. catarrhalis was pre-cultured in 5 mL HI media supplemented with hemin and β-NAD, as above. S. pneumoniae was pre-cultured HI media supplemented with hemin and β-NAD statically [10].
Growth analysis
HI media supplemented with hemin and β-NAD and, where specified, adjusted to pH 8, or unadjusted (pH 7) was used for growth analyses. Cultures were grown until mid-log phase for each species (OD600 = 0.2–0.25 for H. influenzae, 0.25 for S. pneumoniae and 0.2 for M. catarrhalis). Mono-culture, dual-culture and triple-culture inoculums contained 1/20 dilution of each species [20]. Cultures were grown statically in a 96-well microtitre plate. After 18 h of growth, mono- and co-cultures were collected, and plated onto HI agar or selective blood agar plates containing gentamicin (selecting for S. pneumoniae) at a concentration of 5 μg/mL, bacitracin (selecting for H. influenzae) at a concentration of 300 μg/mL, and HI agar supplemented with blood and vancomycin at a concentration of 3 μg/mL (selecting for M. catarrhalis). All growth assays were performed in triplicate.
Growth analysis with arginine
To assess H. influenzae growth with arginine, H. influenzae was inoculated into 10 mL of HI media containing no supplemented arginine or supplemented with an additional 2 or 4 g/L L-arginine (Sigma). The pH of the media with and without arginine was adjusted to pH 8 or 7 as specified, and cultures were grown statically or with shaking at 100 rpm for 18 h. For growth analysis OD600 was measured every 30 min for 18h on a SPECTRAmax spectrophotometer (Molecular Devices).
ATP production assay
ATP production was measured using the BacTiter-Glo Microbial Cell Viability Assay (Promega, Australia). To measure H. influenzae ATP production in the absence and presence of arginine, H. influenzae 86-028NP was pre-cultured to mid-log phase. Subsequently, the pre-culture was inoculated in a 1/20 dilution into 5 mL of HI media with 0, 2, 4 or 6 g/L arginine. Every 15 min, 50 μL of the culture was added to 50 μL of the BacTiter Glo reagent in a white 96-well plate, and luminescence was measured on the Synergy HTX Spectrophotometer (Biotek). OD600 of the culture was concurrently measured to ensure ATP production was not related to growth.
ATP production in single-species, dual-species and triple-species cultures was also measured using BacTiter-Glo Microbial Cell Viability Assay on the PHERAstar Spectrophotometer. Cultures were inoculated for 2h, following which ATP production was measured. Cultures were concurrently plated on selective media to ensure ATP production was unrelated to growth.
Intracellular pH assay
Intracellular pH was determined using the pH sensitive dye BCECF-AM (Invitrogen) using a protocol established for measuring intracellular pH in bacteria [21]. Cells were resuspended in PBS containing 1 μM BCECF-AM, followed by 30 min incubation at 37°C [21], washed in PBS to remove extracellular dye, and subsequently resuspended in 5 mL PBS. Fluorescence was measured using an excitation wavelength of 490nm and an emission wavelength of 530 nm on the (PHERAstar spectrophotometer, BMG LabTech). An intracellular pH standard curve was generated by using the intracellular pH calibration buffer kit (Invitrogen), according to the manufacturer’s guidelines.
RNA preparation
RNA sequencing was performed on cultures following 2 h of growth. Following pre-culture, 4 mL aliquots of mono-species, dual-species and triple-species cultures were prepared, in triplicate, as biological replicates of cell growth. Cultures were grown at 37°C for 2 h.
Following this, cultures were transferred directly to an equal volume of RNA Protect Bacterial Reagent (Qiagen), vortexed, incubated for 5 min at room temperature, and centrifuged (4000 × g for 10 min) [9,10]. The resulting cell pellet was stored at −80°C. For the RNA extraction, a combination of the hot phenol extraction method and a commercial RNA extraction kit (Qiagen, RNeasy Mini Kit) were utilised, as we have previously performed [9,10].
Transcriptomics analysis
RNA from each biological replicate was supplied to the Adelaide Cancer Genomic Research Facility (Adelaide, Australia) for library preparation and sequencing (RNAseq) using the Illumina NextSeq platform (Illumina). The fastq files were aligned to the reference genome of 86-028NP (for H. influenzae Genbank: NC_007146), using bowtie2 in the local mode. The resulting sam files were converted to bam file format, and then to the bed file format. Subsequently, the bed files were aligned to the reference genomes in gff format. The statistical analysis was performed with the DESeq package in R, to identify H. influenzae genes with the largest shift in expression in co-culture conditions compared to mono-culture conditions [9,10]. Differentially expressed genes presented in S1–S4 Tables display statistically significantly differentially expressed genes (p<0.01), which display a fold change in expression >2. The raw transcriptomic data has been made publicly available on the NCBI SRA database with accession number PRJNA820503 and submission ID SUB11224062.
Gene expression confirmation by qRT-PCR
Gene expression of artM was confirmed by a one-step relative qRT-PCR in a Roche LC480 real-time cycler, as we have performed previously [22]. RNA extracted as previously described from bacterial single-species, dual-species and triple species cultures following 2 h of growth was used to confirm gene expression by qualitative real-time PCR.
Primers used to amplify H. influenzae artM (CAAGAATATTTAAGCGTGATCG; AGTAAGGTATAAATTGACCGCA) and 16SrRNA (TGGCAACAAAGGATAAGGGTT; TCCTAAGAAGAGCTCAGAGAT) were used at a final concentration of 200 nM. H. influenzae artM and 16S rRNA primers were tested for absence of amplification of artM and 16S rRNA of mono-species S. pneumoniae and M. catarrhalis cultures. Amplification data was analysed using the comparative critical threshold method, as previously described and is presented as a percentage of total expression relative to 86-028NP [22]. Assays were performed in triplicate in two experiments. Statistical analyses were performed using the t-test, *p<0.05.
Results
There is strain-specific competition with H. influenzae in a dual-species environment with S. pneumoniae and in a triple-species environment with S. pneumoniae and M. catarrhalis
Our work has previously assessed the co-existence of H. influenzae strain Rd KW20 and S. pneumoniae in a polymicrobial environment and has identified both competitive and synergistic interaction outcomes [9,10]. In this study we aimed to assess the interactions of H. influenzae in a triple-species environment harbouring M. catarrhalis as well as S. pneumoniae. We have previously identified the interactions between H. influenzae and S. pneumoniae to result in either a synergistic or competitive outcome, as a consequence of specific parameters, including pH, growth phase and nutrient availability [9,10]. Hence, four analyses in this study, we used a pH of 8 for the growth media, conditions which we have previously shown resulted in reduced species competition [9]. For our current research we assessed both the H. influenzae laboratory strain Rd KW20 as was used in our previous analysis, as well as the genetically distinct, nontypeable, clinical OM isolate 86-028NP.
Determination of CFUs following 18 h growth in mono- or co-culture, showed that both strains were negatively affected by co-culture with S. pneumoniae and by co-culture in a triple-species environment, both in planktonic and biofilm form (Fig 1). However, strain 86-028NP was more severely affected than Rd KW20, reaching cell numbers below the detection limit in planktonic co-culture with S. pneumoniae (Fig 1A), and significantly lower cell numbers compared to Rd KW20 in triple-species co-culture. While Rd KW20 was affected to a similar level by co-culture in planktonic and biofilm state, 86-028NP was more affected by co-culture in the planktonic state, whereas in a biofilm, it had a better capacity to co-exist in dual- and triple-species culture (Fig 1).
Fig 1. Growth of H. influenzae in dual- and triple-species co-cultures with S. pneumoniae and M. catarrhalis after 18 h.
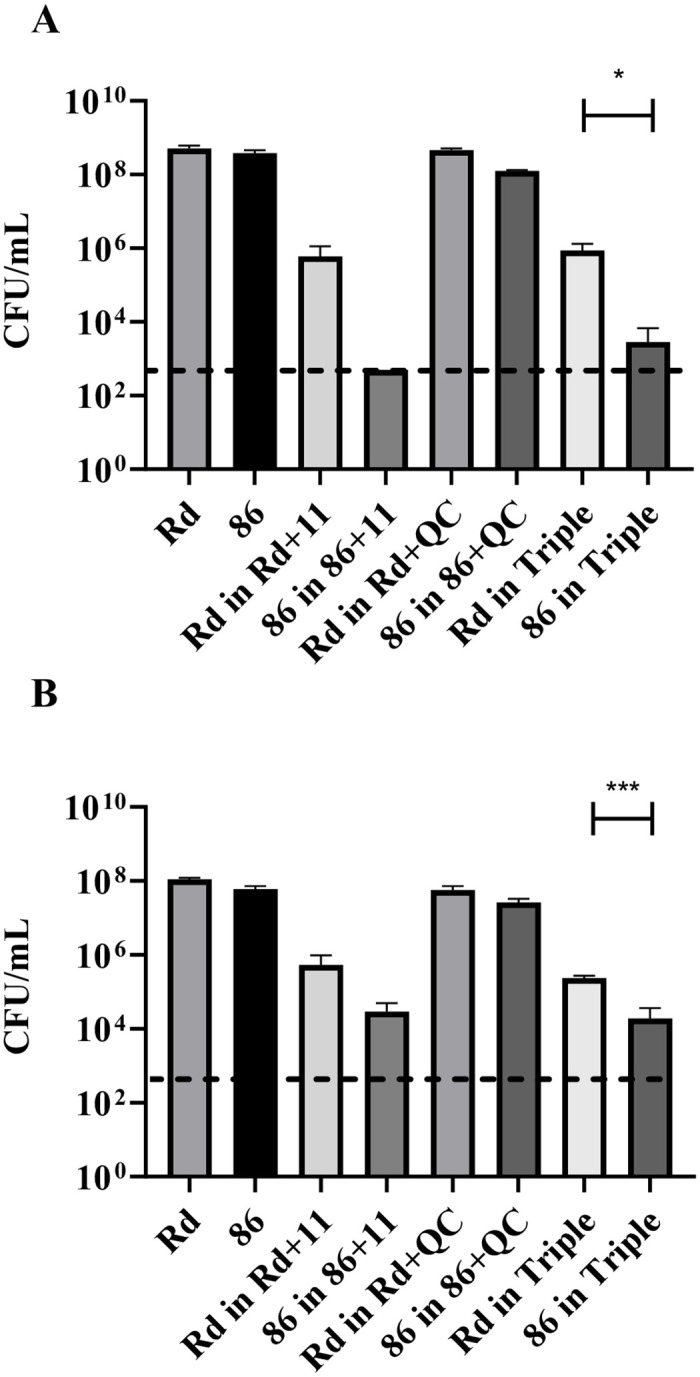
Growth in (A) planktonic and (B) biofilm state of H. influenzae middle ear isolate 86-028NP and laboratory strain Rd KW20 in mono-culture, in co-culture with S. pneumoniae strain 11, in co-culture with M. catarrhalis strain QC, and in triple-species co-culture with S. pneumoniae 11 and M. catarrhalis QC after 18h is represented as CFU/mL. * p<0.05, *** p<0.001.
Competitive interactions with H. influenzae in dual- and triple-species environments are time-point specific
As we have previously shown that there were time-dependent competitive interactions between H. influenzae Rd KW20 and S. pneumoniae [9], to assess whether the inhibitory effects of S. pneumoniae or a triple-species environment on strain 86-028NP were also time-dependent, growth of each species was assessed in planktonic culture at early log phase, following 2 h of growth (Fig 2). The results showed that all three species displayed an equivalent growth in mono-, dual- and triple-species co-culture (Fig 2).
Fig 2. Growth dynamics of dual- and triple-species co-cultures of H. influenzae, S. pneumoniae and M. catarrhalis after 2 h.
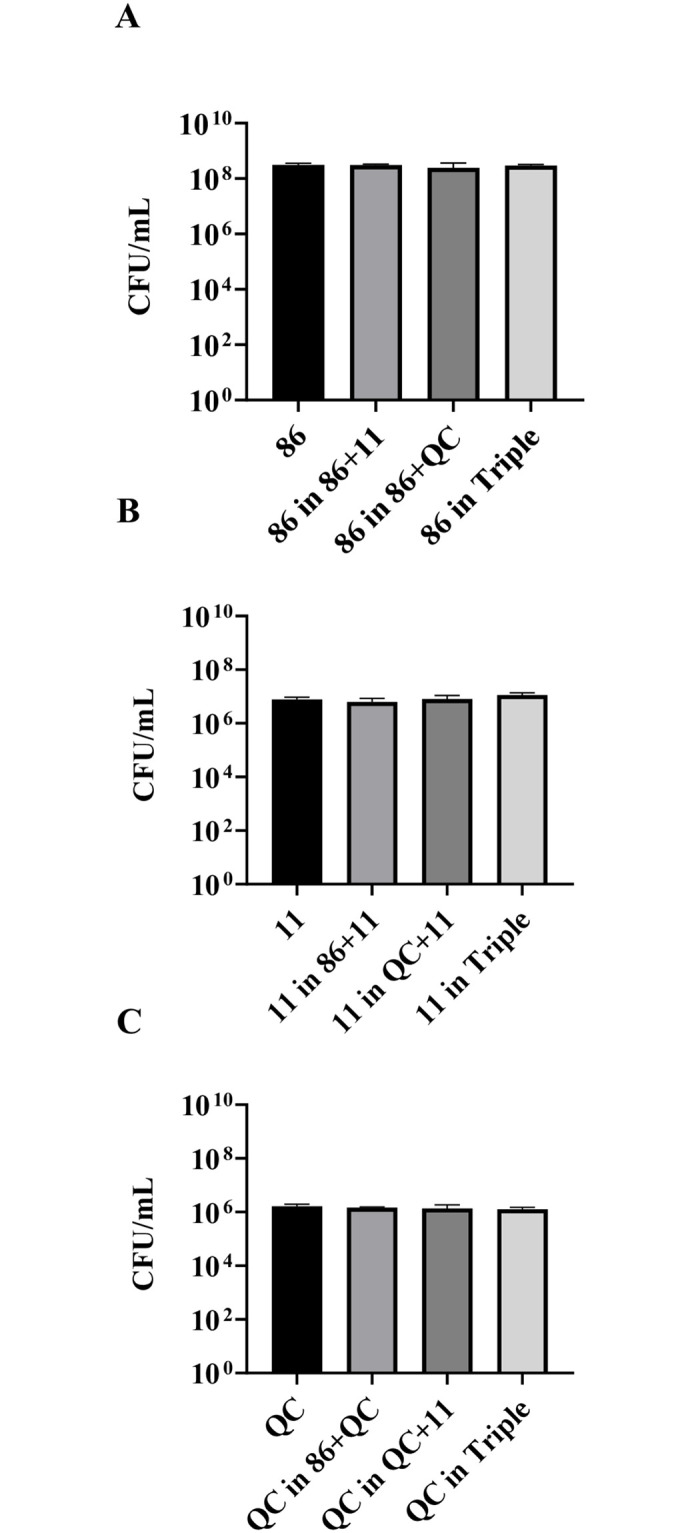
Growth in mono-, dual- and triple-species co-culture of (A), H. influenzae 86-028NP, (B), S. pneumoniae 11, and (C), M. catarrhalis QC in the planktonic state after 2h of growth are demonstrated as CFU/mL.
H. influenzae up-regulates arginine uptake in dual- species co-culture with S. pneumoniae, and in triple-species co-culture
Our previous analysis of the H. influenzae Rd KW20 transcriptome in co-culture with S. pneumoniae has identified molecular factors central to polymicrobial co-existence in distinct environmental conditions [9,10]. As 86-028NP was more affected by dual- and triple- polymicrobial co-culture than Rd KW20, in this study we endeavoured to characterise the strain-specific H. influenzae 86-028NP transcriptome, but within a triple-species context which is relevant to the OM scenario. For the transcriptomic assessment of molecular events facilitating H. influenzae survival in the dual- and triple-species conditions, we performed a transcriptomic analysis following 2 h of growth, a timepoint in which all the bacterial species were viable and prior to competition affecting inter-species interactions.
Transcriptomic analysis identified H. influenzae genes up- and down-regulated in dual- and triple-species co-culture compared to monoculture (S1–S4 Tables), and importantly, showed a consistent up-regulation of genes of the arginine uptake system artM-artP (artM, artQ, artI and artP) in H. influenzae in dual-species culture with S. pneumoniae, as well as in the triple-species co-culture (S1–S3 Tables). No genes were significantly differentially expressed in H. influenzae when in dual-species co-culture with M. catarrhalis.
The expression of the first gene of the artM-artP operon, artM, following H. influenzae growth for 2 h in dual-species and triple-species conditions was confirmed by qRT-PCR (S1 Fig), supporting H. influenzae having a requirement for arginine in a dual- and triple species environment.
Arginine supplementation does not impact H. influenzae growth, but restores its survival in co-culture with S. pneumoniae, and in the triple-species culture
Assessment of arginine supplementation on growth phenotype of the mono-, dual- and triple-species cultures showed that the presence of exogenous supplemented arginine did not affect growth of H. influenzae or M. catarrhalis, but did produce a biphasic growth for S. pneumoniae, which was also reflected in the dual- and triple-species growth curves containing S. pneumoniae (S2 Fig).
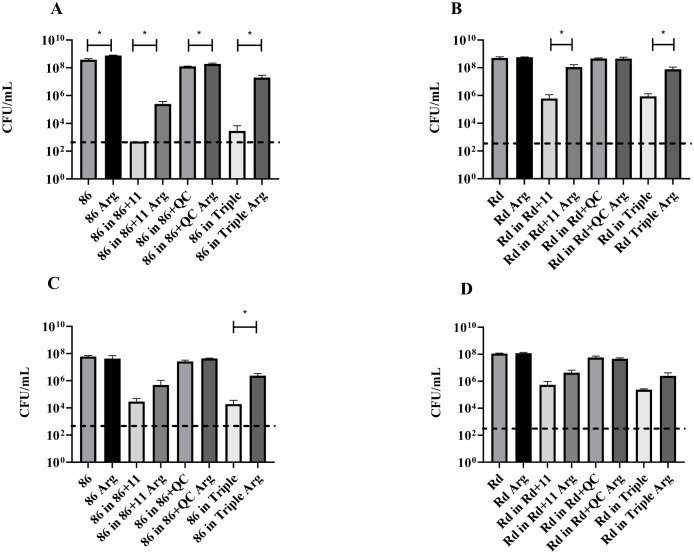
To assess a necessity for arginine in conditions of polymicrobial culture, H. influenzae 86-028NP and Rd KW20 were separately grown in dual- and triple-species co-cultures in the absence and presence of exogenous arginine supplemented at 4 g/L (Fig 3), and viable cell counts were determined following 18 h growth. In both strains the presence of exogenous arginine enabled the recovery of H. influenzae survival in co-culture with S. pneumoniae, and in triple-species culture. However, the recovery effect of arginine was significantly more pronounced for strain 86-028NP than for Rd KW20 (Fig 3), and was greater in the planktonic lifestyle (Fig 3A and 3B) than in the biofilm lifestyle (Fig 3C and 3D).
Fig 3. Arginine restores H. influenzae survival in dual- and triple-species co-culture.
Viable cell numbers of H. influenzae Rd KW20 or H. influenzae 86-028NP in planktonic (A,B) and biofilm (C,D) growth in mono-culture, dual-culture with S. pneumoniae 11, dual-culture with M. catarrhalis QC, and in triple-species culture for 18h, with and without supplementation with exogenous arginine at a concentration of 4g/L. Dotted line indicates the limit of detection. *, p<0.05.
Arginine enhances H. influenzae ATP production
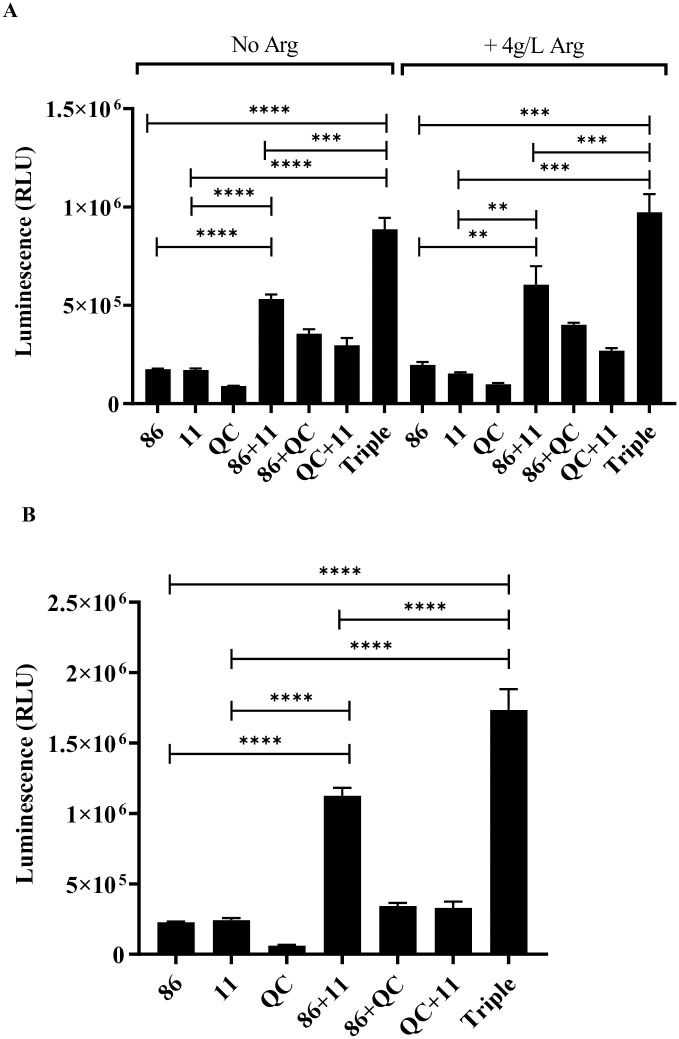
Uptake of arginine by H. influenzae could serve to enhance ATP production at a particular time-point during polymicrobial co-culture. We therefore measured ATP production (BacTiter Glo kit, Promega, Australia) at 15 min intervals with varying concentrations of exogenously supplemented arginine. Arginine significantly increased ATP production in 86-028NP during early lag phase of growth. Addition of higher arginine concentrations showed a trend for higher ATP production, although this was not statistically significant (Fig 4).
Fig 4. Arginine supplementation enhances H. influenzae ATP production.
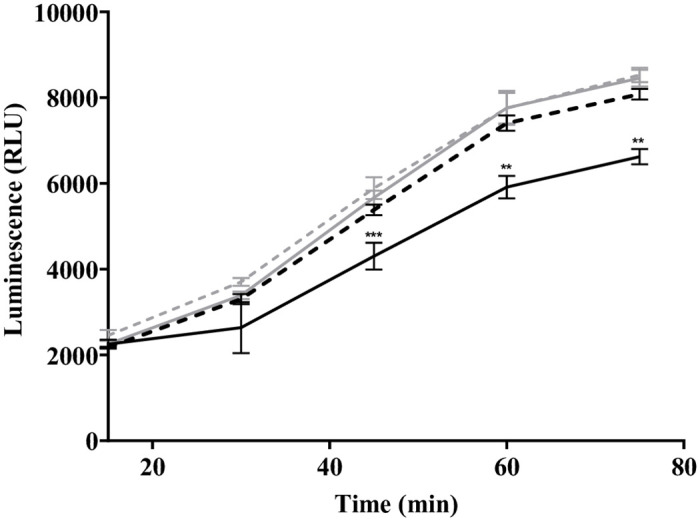
Relative ATP production measured by luminescence (relative light units, RLU), of H. influenzae without supplementation of arginine (black), and with supplementation of 2 g/L (black dotted line), 4 g/L arginine (grey filled line) and 6 g/L arginine (grey dotted line). Luminescence (ATP production) is significantly higher in all concentrations of arginine, compared to no arginine supplementation. Significant differences were determined with a student t-test (*** p ≤ 0.001, ** p ≤ 0.01).
ATP production is increased in dual- and triple-species cultures
As H. influenzae ATP production was related to the presence of arginine, we then assessed the ATP production in the dual- and triple-species polymicrobial culture after 2 h of growth, correlating to the time-point arginine uptake was up-regulated, in comparison to the single-species cultures. Comparisons were performed in a growth medium at pH 8, as well as a growth medium at pH 7, conditions we have previously reported to result in higher levels of species competition [9]. ATP production was significantly higher for the H. influenzae and S. pneumoniae dual-species culture (86-028NP+11), and in the triple-species culture, compared to the H. influenzae single-species culture (86-028NP) (Fig 5). Importantly, the 86-028NP+11 dual-species culture, produced a significantly higher amount of ATP than the combined ATP for 86-028NP and 11 in single-species culture, and this was more pronounced at pH 7 (Fig 5B). The ATP production in triple-species culture was also significantly higher than the combined ATP production of the 86-028NP+11 in co-culture and QC in mono-culture or 86-028NP+QC dual-species culture and 11 in mono-culture (Fig 5). The presence of exogenously supplemented arginine did not significantly affect ATP production.
Fig 5. ATP production is enhanced in dual- and triple-species cultures.
ATP production measured by luminescence (relative light units, RLU), of H. influenzae 86-028NP (86), S. pneumoniae 11 and M. catarrhalis QC following 2 h of incubation in single-species culture, dual-species culture, and triple species culture at A) pH 8, with and without arginine supplementation, and B) in pH 7, without arginine supplementation. Significant differences were observed between H. influenzae alone (86), in dual-species culture with S. pneumoniae (86+11), and in triple-species culture (Triple). Statistical significance was evaluated with a student’s t-test (** p<0.01, *** p<0.001, **** p≤0.0001).
Arginine does not impact H. influenzae intracellular pH
In other bacterial species, arginine has been shown to serve different cellular roles such as regulating the intracellular pH. Using the BCECF-AM intracellular pH dye, we performed an analysis of intracellular pH in H. influenzae following incubation with exogenously supplemented arginine. Supplementation with arginine did not result in a significant alteration of intracellular pH (S3 Fig).
Discussion
In this study, we have utilized a model for the polymicrobial culture of H. influenzae, S. pneumoniae and M. catarrhalis, which has enabled the analysis of planktonic and biofilm cells within their mono-, dual- and triple-species communities. This expands on our previous publications on the co-culture of H. influenzae and S. pneumoniae [9,10].
For each species within dual- and triple-species conditions, growth was equivalent after 2 h of culture compared to mono-species culture. However, inter-species competition was observed at 18 h of growth in both the planktonic and biofilm state, where H. influenzae was reduced in co-culture with S. pneumoniae and in the triple-species culture (Fig 1). This competitive effect was strain-specific; the H. influenzae clinical isolate 86-028NP was significantly more affected by dual-species culture with S. pneumoniae, and by triple-species culture, in comparison to the laboratory strain Rd KW20. This result suggests a specific importance of competition for clinical H. influenzae isolates within the polymicrobial community.
We had previously established the H. influenzae Rd KW20 transcriptional response to co-culture with S. pneumoniae. In the current study we assessed the transcriptional response of H. influenzae 86-028NP in the presence of S. pneumoniae and M. catarrhalis in dual- and triple-species co-culture [9,10].
Genes of the arginine uptake operon were consistently up-regulated in H. influenzae both in dual-species co-culture with S. pneumoniae, and in the triple-species co-culture (S1 and S2 Tables). While H. influenzae is known to have a requirement for arginine and lacks the initial 5 steps of the arginine biosynthesis pathway, the role of arginine for H. influenzae survival in a polymicrobial environment has not been established [23]. Addition of exogenous arginine was able to partially reverse the competitive effect of co-culture on H. influenzae. Interestingly, arginine had a greater effect in recovering the survival of 86-028NP in co-culture compared to Rd KW20, and a more pronounced effect was observed in planktonic co-culture, compared to the biofilm state (Fig 3). These results highlight a critical role for arginine in enabling the co-existence of H. influenzae, S. pneumoniae and M. catarrhalis, and strain-specificity of this role.
These results imply that arginine may have a previously unrecognised role in nutrient sharing between the bacterial species. M. catarrhalis is unable to utilise extracellular carbohydrates [24], and is reliant on alternative carbon sources, including amino acids. While M. catarrhalis possesses genes for the synthesis of most amino acids, it does not possess pathways for the biosynthesis of arginine [24]. S. pneumoniae also lacks a complete pathway of arginine metabolism [25], and H. influenzae likewise lacks the initial 5 steps of the arginine biosynthesis pathway [23]. Therefore, it seems likely that competition driven by the ability to uptake arginine may result in this polymicrobial environment. Despite potential for such competition, growth phenotype analysis showed that only S. pneumoniae growth was different in the presence of exogenous arginine, resulting in a biphasic growth phenotype. This suggested arginine utilisation in S. pneumoniae following utilisation of a preferred nutrient source, but showed that arginine did not increase mono-species growth of H. influenzae or M. catarrhalis (S2 Fig).
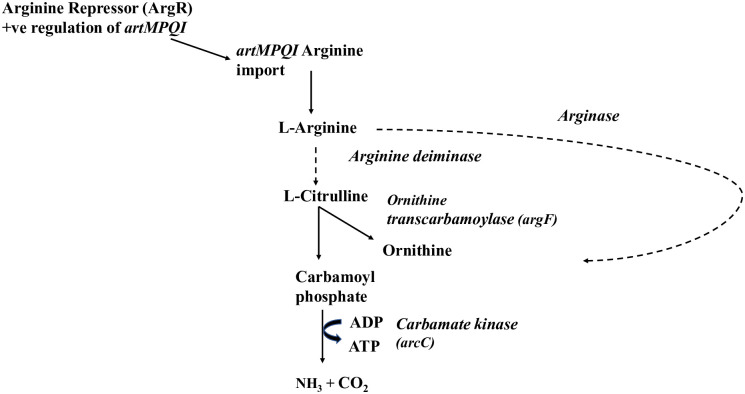
Conceivably, arginine could be important for additional reasons, including ATP generation, or acidity regulation. In other bacterial species, arginine was found to enhance acid tolerance and/or generate ATP via the arginine deiminase pathway [26]. The arginine deiminase pathway comprises a three-step reaction, catalysed by arginine deiminase, ornithine transcarbamoylase and carbamate kinase, respectively [27] (Fig 6). The reaction converts arginine to ornithine, ammonia and carbon dioxide, generating 1 mol of ATP per 1 mol of arginine. In this context, ammonia acts to buffer the environmental pH (25). However, while H. influenzae possesses both the ornithine transcarbamoylase and carbamate kinase, it does not possess the arginine deiminase gene [27], therefore in the absence of an orthologous enzyme which could catalyse the reaction, H. influenzae would not be expected to produce ammonia or ATP through this pathway (Fig 6).
Fig 6. Arginine import and catabolic pathways in H. influenzae 86-028NP as determined from KEGG pathway analysis.
Pathways for which genes have been identified in 86-028NP are indicated with black line, and pathways for which genes are missing in 86-028NP are indicated with dashed line.
Despite the lack of a complete arginine deiminase pathway, the presence of supplemented arginine did result in enhanced ATP production in H. influenzae (Fig 4). In the context of nutrient limitation and competition in a polymicrobial environment, ATP production in H. influenzae may be critical to continue to survive in both a dual-species and triple-species environment. Indeed, ATP production was shown to increase in the dual-species and triple-species environment of H. influenzae, S. pneumoniae and M. catarrhalis (Fig 5). This indicates that the condition of polymicrobial co-culture stimulates ATP production in H. influenzae, and potentially also stimulates ATP production in S. pneumoniae and M. catarrhalis. Supplementation with exogenous arginine did not enhance ATP production, suggesting that the polymicrobial community and not arginine levels drives the ATP production in co-culture. The increase in ATP generation in S. pneumoniae and H. influenzae dual-species culture and triple-species culture was greater at pH 7, where competitive interactions have previously been observed to be greater, suggesting that early ATP generation may be necessary for H. influenzae viability in the polymicrobial environment [9].
In other bacterial species, ATP production from arginine utilisation is achieved primarily through the arginine deiminase pathway. The arginine deiminase pathway is also important in the pH regulation of bacteria, as ammonia is produced in the last step, resulting in alkalisation of the intracellular pH (25). As arginine utilisation was induced in H. influenzae in conditions of co-culture with S. pneumoniae, where we have previously shown acid stress to play a role [9], arginine, through a modified arginine deiminase pathway, could potentially result in alkalization of intracellular pH. However, our results have indicated that the presence of supplemented arginine did not affect the intracellular pH of H. influenzae (S3 Fig). Importantly, our transcriptomic data showed a concurrent increase in expression of an aspartate ammonia lyase in co-culture with S. pneumoniae, and in triple species co-culture (S3 Table), as well as artM. Aspartate ammonia lyase can neutralise the produced ammonia, by catalysing the conversion of ammonia and fumaric acid to aspartate [28], therefore utilisation of arginine through a similar pathway may not necessarily result in detectable increases in intracellular pH. In addition, in a dual-species or triple species environment harbouring H. influenzae, the potential release of ammonia to the extracellular environment could provide temporary neutralisation of organic acids produced as a result of S. pneumoniae fermentation.
Our results reveal the importance of arginine in the polymicrobial environment and strongly suggest the presence of a modified arginine deiminase pathway in H. influenzae, necessitating future investigations for a candidate gene with orthologous function to the arginine deiminase.
Arginine and ornithine, a product of arginine catabolism through the arginine deiminase pathway, have been increasingly shown to play an important role in other bacterial inter-species interactions. In the context of oral bacteria, co-aggregation of Actinomyces naeslundii has been shown to enable Streptococcus gordonii to survive in a arginine restricted environment [29]. Ornithine has been shown to function as a signalling molecule, and to drive the formation of a polymicrobial biofilm of S. gordonii and Fusobacterium nucleatum [30]. Therefore, it is possible that arginine plays a broader role in bacterial communication and in driving transition from planktonic to biofilm state for the otitis media polymicrobial biofilm. Our results, particularly with strain 86-028NP, showed a less pronounced growth inhibition in polymicrobial environments in a biofilm state compared to the planktonic state (Fig 3), suggesting that arginine may have a distinct role in the polymicrobial biofilm.
Overall, our analysis of the triple-species polymicrobial environment of H. influenzae, S. pneumoniae and M. catarrhalis has shown time-dependent competitive interactions between these bacterial species, and a strong requirement for arginine in permitting H. influenzae survival in a dual-species environment with S. pneumoniae, and in a triple-species environment. Arginine was shown to drive ATP production in H. influenzae, and increased ATP production was seen in dual-species and triple-species environments. These results highlight the critical role of arginine in the polymicrobial community and strongly suggest the presence of an arginine deiminase pathway in H. influenzae, which should be investigated in future studies.
Supporting information
Differential gene expression of artM in H. influenzae 86-028NP (86), H. influenzae in co-culture with S. pneumoniae (86+11), H. influenzae in co-culture with M. catarrhalis (86+QC), and H. influenzae in triple-species culture (Triple). Gene expression in presented as % gene expression relative to artM expression in 86-028NP in mono-culture. The data represents the average of 2 independent experiments, each performed in triplicate (*, p<0.1).
(DOCX)
Planktonic growth of mono-, dual- and triple-species cultures of H. influenzae 86-028NP, S. pneumoniae 11 and M. catarrhalis QC at pH 8 over 18h, with (grey) and without (black) supplementation of exogenous arginine at a concentration of 4g/L. Growth is shown for H. influenzae 86-028NP in mono-culture (A), S. pneumoniae 11 in mono-culture (B), M. catarrhalis QC in mono-culture (C), dual-species culture of H. influenzae 86-028NP and S. pneumoniae 11 (D), dual-species culture of H. influenzae 86-028NP and M. catarrhalis QC (E), dual-species culture of S. pneumoniae 11 and M. catarrhalis QC (F), and triple-species culture (G).
(DOCX)
Intracellular pH of H. influenzae following 30min incubation in the presence of 0, 2 or 4g/L of supplemented arginine.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We acknowledge the contribution of Olivia Pei Ying Oh to assistance with initial growth experiments. We acknowledge Peter Sideris from SA Pathology for providing the reference strain of Moraxella sp.
Data Availability
All relevant data are within the paper and Supporting information files.
Funding Statement
This work was supported by the Garnett Passe and Rodney Williams Memorial Foundation Research Training Fellowship to A.T., and a National Health and Medical Research Council (NHMRC) Investigator Grant 1174876 to J.C.P. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vergison A, Dagan R, Arguedas A, Bonhoeffer J, Cohen R, DHooge I, et al. Otitis media and its consequences: beyond the earache. Lancet Infect Dis. 2010; 10(3):195–203. doi: 10.1016/S1473-3099(10)70012-8 [DOI] [PubMed] [Google Scholar]
- 2.Williamson I. Otitis media with effusion in children. BMJ Clin Evidence. 2015;2015:0502. [PMC free article] [PubMed] [Google Scholar]
- 3.Williams RL, Chalmers TC, Stange KC, Chalmers FT, Bowlin SJ. Use of antibiotics in preventing recurrent acute otitis media and in treating otitis media with effusion: A meta-analytic attempt to resolve the brouhaha. JAMA. 1993; 270(11): 1344–51. doi: 10.1001/jama.1993.03510110084037 [DOI] [PubMed] [Google Scholar]
- 4.Steele DW, Adam GP, Di M, Halladay CH, Balk EM, Trikalinos TA. Effectiveness of Tympanostomy Tubes for Otitis Media: A Meta-analysis. Pediatrics. 2017: e20170125. doi: 10.1542/peds.2017-0125 [DOI] [PubMed] [Google Scholar]
- 5.Smith-Vaughan HC, Binks MJ, Marsh RL, Kaestli M, Ward L, Hare KM, et al. Dominance of Haemophilus influenzae in ear discharge from Indigenous Australian children with acute otitis media with tympanic membrane perforation. 2013; 13(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006; 296(2): 202–11. doi: 10.1001/jama.296.2.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMuri GP, Gern JE, Eickhoff JC, Lynch SV, Wald ER. Dynamics of bacterial colonization with Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis during symptomatic and asymptomatic viral upper respiratory tract infection. Clin Infect Dis. 2018; 66(7): 1045–53. doi: 10.1093/cid/cix941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chochua S, D’Acremont V, Hanke C, Alfa D, Shak J, Kilowoko M, et al. Increased nasopharyngeal density and concurrent carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are associated with pneumonia in febrile children. PLOS One. 2016; 11(12): e0167725. doi: 10.1371/journal.pone.0167725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tikhomirova A, Trappetti C, Paton JC, Kidd SP. The outcome of H. influenzae and S. pneumoniae inter-species interactions depends on pH, nutrient availability and growth phase. Int J Med Microbiol. 2015; 305(8): 881–92. doi: 10.1016/j.ijmm.2015.09.003 [DOI] [PubMed] [Google Scholar]
- 10.Tikhomirova A, Trappetti C, Standish AJ, Zhou Y, Breen J, Pederson S, et al. Specific growth conditions induce a Streptococcus pneumoniae non-mucoidal, small colony variant and determine the outcome of its co-culture with Haemophilus influenzae. Pathog Dis. 2018; 76(7): fty074. doi: 10.1093/femspd/fty074 [DOI] [PubMed] [Google Scholar]
- 11.Tikhomirova A, Kidd SP. Haemophilus influenzae and Streptococcus pneumoniae: living together in a biofilm. Pathog Dis. 2013; 69(2): 114–26. doi: 10.1111/2049-632X.12073 [DOI] [PubMed] [Google Scholar]
- 12.Andrade DC, Borges IC, Bouzas ML, Oliveira JR, Käyhty H, Ruuskanen O, et al. Antibody responses against Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis in children with acute respiratory infection with or without nasopharyngeal bacterial carriage. Infect Dis. 2018; 50(9): 705–13. doi: 10.1080/23744235.2018.1463451 [DOI] [PubMed] [Google Scholar]
- 13.Jacoby P, Watson K, Bowman J, Taylor A, Riley TV, Smith DW, et al. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine. 2007; 25(13): 2458–64. doi: 10.1016/j.vaccine.2006.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettigrew MM, Gent JF, Revai K, Patel JA, Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerging Infect Dis. 2008;14(10): 1584–91. Epub 2008/10/02. doi: 10.3201/eid1410.080119 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Littorin N, Rünow E, Ahl J, Resman F, Riesbeck K. Decreased prevalence of Moraxella catarrhalis in addition to Streptococcus pneumoniae in children with upper respiratory tract infection after introduction of conjugated pneumococcal vaccine: a retrospective cohort study. Clin Microbiol Infect. 2021; 27(4): 630.e1–.e6. doi: 10.1016/j.cmi.2020.04.033 [DOI] [PubMed] [Google Scholar]
- 16.Dagan R, Leibovitz E, Greenberg D, Bakaletz L, Givon-Lavi N. Mixed pneumococcal–nontypeable Haemophilus influenzae otitis media is a distinct clinical entity with unique epidemiologic characteristics and pneumococcal serotype distribution. J Infect Dis. 2013;208(7):1152–60. doi: 10.1093/infdis/jit289 [DOI] [PubMed] [Google Scholar]
- 17.Krishnamurthy A, McGrath J, Cripps AW, Kyd JM. The incidence of Streptococcus pneumoniae otitis media is affected by the polymicrobial environment particularly Moraxella catarrhalis in a mouse nasal colonisation model. Microbes Infec. 2009; 11(5): 545–53. doi: 10.1016/j.micinf.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 18.Schaar V, Nordström T, Mörgelin M, Riesbeck K. Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrobial Agents Chemother. 2011; 55(8): 3845–53. doi: 10.1128/AAC.01772-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikryannikova L, Malakhova M, Lominadze G, Karpova IY, Kostryukova E, Mayansky N, et al. Inhibitory effect of Streptococci on the growth of M. catarrhalis strains and the diversity of putative bacteriocin-like gene loci in the genomes of S. pneumoniae and its relatives. AMB Express. 2017; 7(1): 218. doi: 10.1186/s13568-017-0521-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tikhomirova A, Trappetti C, Paton JC, Watson-Haigh N, Wabnitz D, Jervis-Bardy J, et al. A single nucleotide polymorphism in an IgA1 protease gene determines Streptococcus pneumoniae adaptation to the middle ear during otitis media. Pathog Dis. 2021; 79(1): ftaa077. doi: 10.1093/femspd/ftaa077 [DOI] [PubMed] [Google Scholar]
- 21.Cai L-l, Hu H-j, Lu Q, Wang H-h, Xu X-l, Zhou G-h, et al. Morphophysiological responses of detached and adhered biofilms of Pseudomonas fluorescens to acidic electrolyzed water. Food Microbiol. 2019; 82: 89–98. doi: 10.1016/j.fm.2019.01.007 [DOI] [PubMed] [Google Scholar]
- 22.McLean KT, Tikhomirova A, Brazel EB, Legendre S, Haasbroek G, Minhas V, et al. Site-Specific mutations of GalR affect galactose metabolism in Streptococcus pneumoniae. J Bacteriol. 2020; 203(1):e00180–20. doi: 10.1128/JB.00180-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, Mushegian AR, Bork P, Brown NP, Hayes WS, Borodovsky M, et al. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr Biol. 1996; 6(3): 279–91. doi: 10.1016/s0960-9822(02)00478-5 [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Reitzer L, Rasko DA, Pearson MM, Blick RJ, Laurence C, et al. Metabolic analysis of Moraxella catarrhalis and the effect of selected in vitro growth conditions on global gene expression. Infect Immun. 2007; 75(10): 4959–71. doi: 10.1128/IAI.00073-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloosterman TG, Kuipers OP. Regulation of arginine acquisition and virulence gene expression in the human pathogen Streptococcus pneumoniae by transcription regulators ArgR1 and AhrC. J Biol Chem. 2011; 286(52): 44594–605. doi: 10.1074/jbc.M111.295832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong L, Teng JLL, Watt RM, Kan B, Lau SKP, Woo PCY. Arginine deiminase pathway is far more important than urease for acid resistance and intracellular survival in Laribacter hongkongensis: a possible result of arc gene cassette duplication. BMC Microbiol. 2014; 14:42-. doi: 10.1186/1471-2180-14-42 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zúñiga M, Pérez G, González-Candelas F. Evolution of arginine deiminase (ADI) pathway genes. Mol Phylogenet Evol. 2002; 25(3): 429–44. doi: 10.1016/s1055-7903(02)00277-4 [DOI] [PubMed] [Google Scholar]
- 28.Fibriansah G, Veetil VP, Poelarends GJ, Thunnissen A-MWJB. Structural basis for the catalytic mechanism of aspartate ammonia lyase. Biochemistry. 2011; 50(27): 6053–62. doi: 10.1021/bi200497y [DOI] [PubMed] [Google Scholar]
- 29.Jakubovics NS, Gill SR, Iobst SE, Vickerman M, Kolenbrander PE. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J Bacteriol. 2008; 190(10): 3646–57. doi: 10.1128/JB.00088-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakanaka A, Kuboniwa M, Takeuchi H, Hashino E, Amano A. Arginine-ornithine antiporter ArcD controls arginine metabolism and interspecies biofilm development of Streptococcus gordonii. J Biol Chem. 2015; 290(35): 21185–98. doi: 10.1074/jbc.M115.644401 [DOI] [PMC free article] [PubMed] [Google Scholar]