Abstract
A yellow-pigmented marine bacterium, designated strain SD-21, was isolated from surface sediments of San Diego Bay, San Diego, Calif., based on its ability to oxidize soluble Mn(II) to insoluble Mn(III, IV) oxides. 16S rRNA analysis revealed that this organism was most closely related to members of the genus Erythrobacter, aerobic anoxygenic phototrophic bacteria within the α-4 subgroup of the Proteobacteria (α-4 Proteobacteria). SD-21, however, has a number of distinguishing phenotypic features relative to Erythrobacter species, including the ability to oxidize Mn(II). During the logarithmic phase of growth, this organism produces Mn(II)-oxidizing factors of ≈250 and 150 kDa that are heat labile and inhibited by both azide and o-phenanthroline, suggesting the involvement of a metalloenzyme. Although the expression of the Mn(II) oxidase was not dependent on the presence of Mn(II), higher overall growth yields were reached in cultures incubated with Mn(II) in the culture medium. In addition, the rate of Mn(II) oxidation appeared to be slower in cultures grown in the light. This is the first report of Mn(II) oxidation within the α-4 Proteobacteria as well as the first Mn(II)-oxidizing proteins identified in a marine gram-negative bacterium.
The oxidation of soluble Mn(II) to insoluble Mn(III, IV) oxides and oxyhydroxides is an environmentally important process because the solid-phase products oxidize a variety of organic and inorganic compounds [e.g., humic substances, Cr(III), and Fe(II)], scavenge many metals (e.g., Cu, Co, Cd, Zn, Ni, and Pb), and serve as electron acceptors for anaerobic respiration. In most environments, Mn(II) oxidation is believed to be bacterially mediated (29). Over the years, Mn(II)-oxidizing bacteria have been isolated from a wide variety of environments, including marine and freshwaters, soils, sediments, water pipes, Mn nodules, and hydrothermal vents (10–12, 14, 18–20, 28, 32). Phylogenetically, Mn(II)-oxidizing bacteria appear to be quite diverse, with all isolates analyzed to date falling within either the low G+C gram-positive bacteria, the Actinobacteria, or the α, β, and γ subgroups of the Proteobacteria branch of the domain Bacteria (38).
The most well-characterized Mn(II)-oxidizing bacteria are Bacillus sp. strain SG-1, Leptothrix discophora strain SS-1, and Pseudomonas putida strains MnB1 and GB-1. Although distantly related phylogenetically, enzymes related to multicopper oxidases appear to be involved in Mn(II) oxidation in all of these organisms (3, 6, 41). Multicopper oxidases are a diverse family of proteins that utilize multiple copper ions as cofactors in the oxidation of a wide variety of substrates (35). In each of the model systems, Mn(II)-oxidizing activity is inhibited by azide (1, 30, 31), a potent inhibitor of multicopper oxidases, and stimulated by the presence of copper (3, 4, 41). These findings suggest the possibility of a universal mechanism for bacterial Mn(II) oxidation which is dependent on copper as an essential enzymatic cofactor.
Relative to the model Mn(II)-oxidizing organisms described above, very little is known regarding the mechanisms of Mn(II) oxidation within the α subgroup of the Proteobacteria (α-Proteobacteria). Despite numerous reports of Mn(II) oxidation by various prosthecate bacteria (e.g., Pedomicrobium, Hyphomicrobium, and Caulobacter) within the α-Proteobacteria (13, 14, 16, 18, 34, 39), few studies have directly addressed the biochemical mechanisms of Mn(II) oxidation in these organisms (15, 16). One recent study, however, demonstrated that Mn(II) oxidation by Pedomicrobium sp. strain ACM 3067 appears to be catalyzed by a copper-dependent enzyme (25), consistent with the possible involvement of a multicopper oxidase in this organism. However, no further purification of the Mn(II)-oxidizing enzyme or identification of the gene(s) involved has been reported. The Mn(II)-oxidizing strain SI85-9A1 is a novel marine α-proteobacterium that possesses genes for both the large and small subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) (5). Although this was the first report of RubisCO genes in a Mn(II)-oxidizing bacterium, the molecular and biochemical mechanisms of Mn(II) oxidation in this organism have yet to be explored. Considering the abundance and diversity of α-Proteobacteria in the marine environment (17), it is important to determine both the diversity of organisms capable of Mn(II) oxidation within this group as well as the mechanisms underlying this environmentally important process.
In the present study, we describe the isolation and characterization of an organism, strain SD-21, which has a number of features that make it an attractive candidate as a new model Mn(II)-oxidizing α-proteobacterium.
MATERIALS AND METHODS
Sample collection and strain isolation.
Surface sediments were collected from San Diego Bay and Mission Bay (San Diego, Calif.), diluted in sterile seawater, and spread onto K plates (40) containing 20 mM HEPES buffer (pH 7.8) and 100 μM MnCl2. Mn(II)-oxidizing strains were isolated based on their ability to produce brown Mn oxide-encrusted colonies on plates. The presence of Mn oxides was confirmed using the colorimetric dye Leucoberbelin blue (LBB) (22). Additional strains used in this study were Erythrobacter litoralis ATCC 700002T, Erythrobacter longus strain Och101 ATCC 33941T, and a yellow-pigmented Mn(II)-oxidizing Sphingomonas strain isolated by our laboratory from pulp mill effluent.
Physiological characterization.
The growth temperature range of strain SD-21 was determined by incubating 5-ml aliquots of K cultures over a range of temperatures (4, 12, 18, 25, 30, 37, and 42°C) for 2 weeks in the dark and measuring the optical densities at 600 nm (OD600) in a Perkin-Elmer spectrophotometer. The pH range for SD-21 growth was determined in K media of pH ranging from 5 to 9. The salt tolerance or requirement was determined by incubating SD-21 in K media made with artificial seawater containing a range of NaCl concentrations (0 to 15%). For pigment analysis, bacterial pellets (0.2 g wet weight) of dark-grown cultures of SD-21, E. longus, E. litoralis, and the Sphingomonas isolate were extracted with 2 ml of acetone-methanol (7:2, vol/vol). Absorption spectra (200 to 900 nm) were then obtained using a Perkin-Elmer UV-Vis spectrophotometer (Lambda Bio 20).
Growth experiments.
Strain SD-21 was grown in 1-liter flasks containing 500 ml of K media, on an orbital shaker (150 rpm), in the presence and absence of both Mn(II) and fluorescent light (5 to 10 μE/m2). The OD600 of duplicate cultures was measured at 12-h intervals for 7 days. The production of Mn oxides in cultures was quantified spectrophotometrically with LBB (620 nm) relative to a standard curve of KMnO4 as described previously (26). The effect of Mn oxides on the OD of Mn(II)-grown cultures was determined by remeasuring the OD600 of cultures after removal of the oxides with 200 μM ascorbate.
SDS-PAGE and analysis of Mn(II)-oxidizing activity.
Cell suspensions (in 10 mM HEPES [pH 7.6]) were passed through a French press cell three times at 20,000 lb/in2 followed by centrifugation for 10 min at 14,000 × g to remove cell debris. Cell lysis was confirmed microscopically. Cell extracts were assayed for Mn(II)-oxidizing activity in 10 mM HEPES containing 200 μM Mn(II), followed by LBB detection. The effect of the potential enzyme inhibitors, sodium dodecyl sulfate (SDS; 0.1 to 1%) and sodium azide (10 μM to 1 mM) on cell-free activity was also determined. Supernatants were mixed with 2× Laemmli buffer and run in 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels in a mini-Protean II (Bio-Rad) electrophoresis unit under standard conditions (23). For staining of total protein, gels were incubated in Coomassie blue. To assay for in-gel Mn(II) oxidation activity, gels were first incubated for 30 min in 10% glycerol–0.1% Triton X-100 to remove SDS, followed by incubation in 10 mM HEPES (pH 7.6) buffer containing 200 μM MnCl2. Mn(II)-oxidizing activity was visualized by the formation of brown Mn oxide bands in gels after several hours of incubation. To assay for in-gel organic oxidation, the colorimetric substrate p-phenylenediamine (Sigma) was substituted for MnCl2 at a concentration of 1 mM. The temperature stability of the Mn(II)-oxidizing protein was determined by incubating cell extracts at room temperature, 37, 42, 45, 55, 65, and 95°C for 15 min prior to running the gels. The sensitivity of the Mn(II)-oxidizing activity to copper chelators was assayed by incubating gels in HEPES buffer (pH 7.6) containing o-phenanthroline for 15 min prior to the addition of 200 μM Mn(II).
DNA extraction, PCR, cloning, and sequencing.
DNA was extracted from cultures using the DNeasy DNA extraction kit (Qiagen). The 16 rRNA genes were amplified using the primers 27F and 1492R (24) in a standard 30-cycle PCR using Taq polymerase (Roche) with an annealing temperature of 50°C. PCR products were cloned into the vector pCR2.1 by using a TOPO-TA cloning kit (Invitrogen). Plasmid DNA was purified using a Qiagen mini-prep kit, and both strands of the cloned PCR products were sequenced using an ABI 373A automated sequencer.
Phylogenetic analysis.
16S rRNA gene sequences were aligned manually using Sequencher 3.1, compared to alignments generated using CLUSTAL W and the Ribosomal Database Project (RDP) Sequence Aligner, and edited to remove ambiguously aligned regions and gaps. Phylogenetic trees were generated by neighbor-joining, using Jukes-Cantor corrected distances, or by maximum parsimony within the PAUP (version 4.0b3) software package. Bootstrap analysis was used to estimate the the reliability of phylogenetic reconstructions (1,000 replicates).
The GenBank, EMBL, and DDBJ accession numbers for the 16S rRNA sequences that were used for comparison are as follows: Acidiphilium cryptum, D30773; Agrobacterium tumefaciens, D14500; Azospirillum lipoferum, X79730; Beijerinckia indica, M59060; Caulobacter crescentus, AJ227757; Citromicrobium bathyomarinum, Y16267; E. litoralis, AB013354; E. longus, L01786; Erythromicrobium ramosum, X72909; Escherichia coli, M24828; Magnetospirillum gryphiswaldense, Y10109; Mesorhizobium loti, D12791; Methylobacterium extorquens, D32224; Paracoccus denitrificans, X69159; Pedomicrobium manganicum, X97691; Porphyrobacter neustonensis, L01785; Rhodobacter capsulatus, D16427; Rhodophila globiformis, D86513; Rhodopseudomonas palustris, D84187; Rhodospirillum salinarum, D14432; Roseobacter litoralis, X78312; Roseobacter denitrificans, M96746; Roseococcus thiosulfatophilus, X72908; Sphingomonas adhaesiva, D13722; Sphingomonas yanoikuyae, D13728; strain RE35F/1, AF118020; and strain SI85-9A1, U53824.
Nucleotide sequence accession numbers.
The 16S rRNA sequences of strains SD-21 and MB-16 determined in this study have been deposited in GenBank under accession numbers AF325445 and AF325446, respectively.
RESULTS AND DISCUSSION
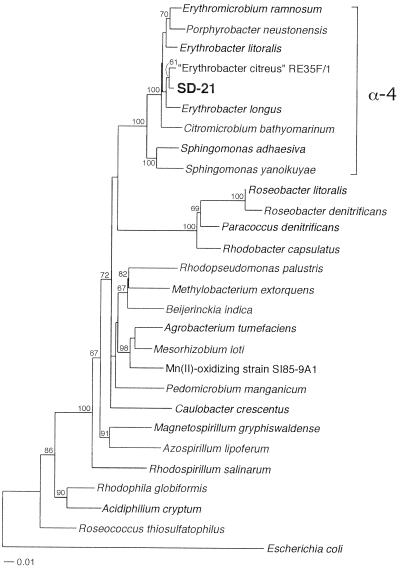
A yellow-pigmented Mn(II)-oxidizing bacterium, designated strain SD-21, was isolated from surface sediments of San Diego Bay (San Diego, Calif.). When grown on Mn(II)-containing plates for over 1 week, colonies become dark brown due to the formation of Mn oxides on the cell surface. A phenotypically similar Mn(II)-oxidizing bacterium, strain MB-16, was also isolated from sediments of Mission Bay (San Diego, Calif.). Comparison of the 16S rRNA sequences of strains SD-21 and MB-16 revealed that these organisms were very closely related, sharing 99.7% identity over 1,440 bases (differing by only 4 bases). Due to the striking similarity between these organisms, only one of the strains, SD-21, was fully characterized in this study. Database searches (using BLAST and RDP) demonstrated that the 16S rRNA sequence of SD-21 was most closely related to that of “Erythrobacter citreus” strain RE35F/1 (98.9% identity, 1,403 bases considered), a yellow-pigmented organism recently isolated from the 0.2-μm-pore-size filterable fraction of Mediterranean seawater samples (35-m depth) (42). The 16S rRNA sequence of SD-21 was 97.4 and 97.3% identical (over 1,403 bases) to the sequences of the type strains E. litoralis and E. longus, respectively. The next most closely related sequences (1,402 bases considered) were from P. neustonensis (96.2%), E. ramosum (96.0%), and C. bathyomarinum (95.5%), indicating that small differences (<1%) at the 16S rRNA level may correspond to genus-level physiological differences in this group of aerobic anoxygenic phototrophic bacteria (43). A phylogenetic tree (Fig. 1) based on closely related sequences in the databases and diverse representatives of the α-Proteobacteria indicated that strain SD-21 clusters within the α-4 subgroup of Proteobacteria, forming an additional clade with RE35F/1. From this tree, it is also clear that SD-21 is not closely related to other known Mn(II)-oxidizing α-proteobacteria (e.g., SI85-9A1, P. manganicum), providing further evidence that Mn(II) oxidation is not confined to a single group or lineage within the α-Proteobacteria.
FIG. 1.
Neighbor-joining phylogenetic tree showing the relationship of strain SD-21 to closely related members of the α-4 subgroup of the Proteobacteria as well as diverse representatives of α-Proteobacteria. Percentages of bootstrap support (>60%) from 1,000 replicates are indicated at the branch points. The scale bar represents the number of substitutions per nucleotide position.
Microscopic examination of SD-21 cultures revealed motile, gram-negative rods that were quite small, approximately 0.2 to 0.5 μm by 0.9 to 3.0 μm. Growth occurred over a wide range of conditions, including temperature (12 to 37°C), pH (6 to 9), and NaCl concentration (1 to 8%). Optimal growth occurred at 25 to 30°C, pH 6.5 to 7.5, and 1.5 to 3.5% NaCl, respectively. SD-21 did not grow in freshwater K media, with or without added NaCl. This organism required NaCl as well as other artificial seawater constituents for growth, indicating that, like Erythrobacter species, it appears to be a true marine (i.e., seawater-requiring) bacterium.
One of the defining and rather striking characteristics of Erythrobacter species is their dark orange-red (“erythrus” means red) pigmentation, which is due to the presence of extremely high amounts of carotenoids (33, 44). The yellow pigmentation of strain SD-21, however, differed greatly from the more reddish pigmentation of both E. longus and E. litoralis when grown under the same conditions. Another defining characteristic of Erythrobacter species, and of all aerobic anoxygenic phototrophic bacteria, is the presence of bacteriochlorophyll a (Bchl-a) (43), an essential component of the light-harvesting complexes of these organisms. Unexpectedly, such Bchl-a-containing organisms have recently been reported to be significant contributors to photosynthetic electron transport in the upper ocean (21). To determine whether Bchl-a and carotenoids were present in SD-21, methanol-acetone extracts were obtained from dark-grown cells of SD-21, as well as of E. longus, E. litoralis, and a yellow-pigmented Sphingomonas isolate as reference strains. The extracts of both Erythrobacter species were dark orange, while those of SD-21 and the Sphingomonas isolate were pale yellow. Absorption spectra revealed not only that SD-21 had considerably lower amounts of carotenoids than the Erythrobacter species, but also that Bchl-a was undetectable in this organism while clearly present in the Erythrobacter strains (data not shown). These findings are particularly interesting in light of the fact that Bchl-a was recently reported to be absent in the closest known relative of SD-21, the yellow-pigmented strain, RE35F/1 (42). In fact, that organism was described as being chemotaxonomically more similar to the genus Sphingomonas than to Erythrobacter (42), despite its close phylogenetic affiliation with the genus Erythrobacter. Finally, unlike SD-21, neither E. longus nor E. litoralis oxidized Mn(II) when grown on liquid or solid K media containing 100 μM MnCl2. Overall, the significant phylogenetic and phenotypic differences between SD-21 and the two established Erythrobacter type strains suggest that SD-21 (and RE35F/1) may represent a new species or possibly genus of bacteria.
Regardless of the precise taxonomic placement of SD-21, this organism is without question the first Mn(II) oxidizer described within the α-4-Proteobacteria. The formation of visible brown Mn oxides in SD-21 cultures generally occurred around the onset of stationary phase, after 3 to 4 days of growth in K media (Fig. 2). Although growth was essentially identical in Mn(II)-containing cultures incubated in light or darkness (Fig. 2A), the rate of Mn(II) oxidation appeared to be slower in the presence of light (Fig. 2B). This is interesting in that both photoinhibition of bacterial Mn(II) oxidation (27, 36) and photoreductive dissolution of Mn oxides (37) have been reported to occur within near-surface waters of the ocean. Cultures grown in the presence of Mn(II) also reached higher overall cell densities than those grown without Mn(II), suggesting that either Mn(II) itself or Mn(II) oxidation somehow enhances the overall growth yield of SD-21. Although Mn(II) serves as a cofactor for many cellular enzymes, for most organisms Mn(II) is generally only required in trace amounts and, thus, would not be expected to limit bacterial growth. A more intriguing possibility is that SD-21 is capable of coupling growth to Mn(II) oxidation, although there have been no conclusive reports of this phenomenon in the literature. Another possibility is that the Mn oxides encrusting the cells may enhance the overall growth yield by providing protection against various harmful agents (e.g., toxic oxygen species, proteases, metabolic byproducts) which may accumulate in the culture media over time. In the natural environment, a number of different biological functions have been proposed for the Mn oxides which encrust Mn(II)-oxidizing organisms, including the following: protection against UV light, toxic metals, viral attack, and predation; oxidation of refractory organic matter into utilizable substrates; scavenging of trace metals required for growth; and storage of an electron acceptor for anaerobic respiration (38). However, most of these functions are unlikely to explain the different growth yields of SD-21 cultures grown in the presence and absence of Mn(II).
FIG. 2.
Growth (A) and Mn(II) oxidation (B) by strain SD-21 under different light and metal regimens. Duplicate cultures were incubated in the presence or absence of 100 μM MnCl2 and/or fluorescent light. The production of Mn oxides in cultures was measured using LBB.
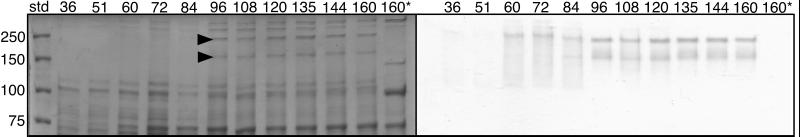
SDS-PAGE analysis of protein extracts from cells collected throughout a 7-day time course revealed that active Mn(II)-oxidizing factors of ≈250 and 150 kDa were detectable after 60 h of growth (Fig. 3). This timing of expression corresponds to the mid-logarithmic growth phase [prior to the onset of detectable Mn(II) oxidation in cultures] as well as the point after which growth begins to deviate between cultures grown in the presence or absence of Mn(II). Thus, there may be a potential link between Mn(II) oxidation and the observed differences in growth.
FIG. 3.
Expression of the Mn(II)-oxidizing protein(s) during the time course shown in Fig. 2. Duplicate gels of cell extracts collected from time points throughout the growth curve were stained for total protein (left) or assayed for in-gel Mn(II)-oxidizing activity (right). Arrows correspond to the Coomassie-stained proteins responsible for Mn(II) oxidation. The last sample in both gels (lane 160*) was heat-treated at 65°C for 5 min prior to electrophoresis.
Although expression of the Mn(II)-oxidizing proteins in SD-21 occurs during mid-logarithmic growth, Mn(II)-oxidizing activity is not observed in cultures until about 24 h later, just before the onset of stationary phase (Fig. 2). This suggests that at 60 to 72 h the Mn(II) oxidase may be present in active form within the cell, but it is not yet localized to the outer membrane. In P. putida strains MnB1 and GB-1, Mn(II) oxidation is believed to be induced by starvation (7) and/or the onset of stationary phase (19, 30). In P. putida GB-1, Brouwers et al. (2) identified genes involved in a two-step protein secretion pathway essential for transporting the Mn(II)-oxidizing factor across the outer membrane. Transposon mutants with insertions in these genes were incapable of Mn(II) oxidation in cultures, yet the Mn(II)-oxidizing protein could be released from cells by French pressure and recovered in active form in gels (9). In SD-21, the first detectable Mn(II)-oxidizing proteins (at 60 to 72 h) consistently appeared to be slightly larger than those found in protein extracts from subsequent time points, possibly due to the presence of a signal peptide which is cleaved during localization or secretion to the cell surface. In fact, signal peptides have been identified in the sequences of the Mn(II) oxidation-associated multicopper oxidases CumA from P. putida GB-1 (3) and MofA from L. discophora SS-1 (6).
A final point regarding the timing of Mn(II) oxidation in SD-21 is that the onset of Mn(II) oxidation in cultures varied depending on the type of peptone (e.g., trypticase, Casamino Acids, or proteose) used in the medium (data not shown), possibly indicating that Mn(II) oxidation [expression of the Mn(II) oxidase] is influenced by the relative concentrations of specific amino acids or other trace contaminants present in the different peptones. The specific factors involved in the regulation of Mn(II) oxidation in SD-21 require further investigation.
The activities of the Mn(II)-oxidizing factors produced by strain SD-21 were found to be extremely stable, capable of withstanding exposure to a variety of harsh conditions, including multiple freeze-thaw cycles, high concentrations of SDS (1%) and NaCl (1 M), and SDS-PAGE under denaturing conditions (without boiling). In addition, several lines of evidence suggest that these high-molecular-weight Mn(II)-oxidizing factors are actually multiprotein complexes. First, exposure of cell extracts to temperatures above 45°C results in the disappearance of both the Mn(II)-oxidizing bands and the corresponding Coomassie bands in gels (Fig. 3, lane 160*), with the concomitant appearance of several distinct Coomassie-bands of ≈100 and ≈140 kDa. This suggests heat inactivation of the enzyme as well as the dissociation of the Mn(II)-oxidizing complexes into smaller protein components which lack detectable activity. Secondly, lower-molecular-weight Mn(II)-oxidizing bands (from 140 kDa to as small as 50 to 60 kDa) have occasionally been observed to have activity in gels (data not shown). Thus, a single smaller protein is likely to be directly responsible for Mn(II) oxidation but may need to be present in multimeric form or in association with additional proteins (and possibly cofactors) for optimal activity.
The involvement of multiprotein complexes in Mn(II) oxidation has also been reported in the model systems of P. putida GB-1 and the Bacillus sp. strain SG-1. In P. putida GB-1, Mn(II)-oxidizing complexes with estimated molecular masses of 180 and 250 kDa (30), which were identified in native polyacrylamide gradient gels, are quite similar in size to the Mn(II)-oxidizing factors of strain SD-21. Although the product of the multicopper oxidase gene, cumA, is believed to be a key component of the P. putida Mn(II)-oxidizing complexes (3, 9), no activity has been observed outside of the complexes in a single protein of the appropriate size (≈50 kDa). In spores of the marine Bacillus sp. strain SG-1, Mn(II)-oxidizing activity has only been consistently recovered in the form of a high-molecular-weight complex which barely enters SDS-PAGE gels that have a low concentration of polyacrylamide (C. A. Francis, K. L. Casciotti, and B. M. Tebo, unpublished data). As in P. putida GB-1, a multicopper oxidase, MnxG, appears to be the key protein involved in Mn(II) oxidation by SG-1 spores (41), but it apparently requires the direct association with other proteins for activity.
The results of inhibitor assays revealed additional parallels with the model Mn(II)-oxidizing organisms as well as multicopper oxidases. The Mn(II)-oxidizing activity of SD-21 cell extracts was inhibited by azide concentrations greater than 1 mM (data not shown). This is significant because azide strongly inhibits multicopper oxidases by bridging the type 2 and type 3 copper sites (35), and it also inhibits Mn(II) oxidation by P. putida GB-1 (30), L. discophora SS-1 (1), and Bacillus sp. strain SG-1 (31). In addition, the in-gel Mn(II)-oxidizing activity of SD-21 was completely inhibited by the copper chelator o-phenanthroline (data not shown) at a concentration (50 μM) well below the Mn(II) concentration (200 μM). These results are similar to previous findings in P. putida GB-1 (30) and Pedomicrobium sp. strain ACM 3067 (25), in which Mn(II) oxidation was also inhibited by the copper chelators o-phenanthroline and diethyldithiocarbamate, respectively, consistent with the involvement of Cu-dependent oxidases.
Finally, like all known multicopper oxidases (e.g., laccase, ceruloplasmin, and ascorbate oxidase) (35), the Mn(II)-oxidizing proteins of SD-21 are also capable of directly oxidizing various organic compounds, including p-phenylenediamine, in gels (Fig. 4). This is particularly analogous to the Fe(II)-oxidizing multicopper oxidase FET3 from yeast, which is capable of oxidizing both Fe(II) and p-phenylenediamine but has a much higher affinity (450-fold) for Fe(II) than the organic compound (8). Although the Mn(II)-oxidizing protein of SD-21 clearly has a number of properties in common with multicopper oxidases, definitive proof of this awaits the purification and characterization of the active enzyme, as well as cloning and sequencing of the underlying gene(s) involved. This protein is particularly well-suited for both biochemical and spectroscopic studies, due to its stability over a wide range of conditions. Overall, strain SD-21 may serve as a useful model for Mn(II)-oxidizing bacteria not only for studying the mechanism of Mn(II) oxidation within the α-Proteobacteria but also for studying the biological function of bacterial Mn(II) oxidation, which remains enigmatic.
FIG. 4.
In-gel oxidation of both Mn(II) and the organic substrate, p-phenylenediamine, by the 150- and 250-kDa proteins of strain SD-21. Gels were incubated in either Coomassie blue (lane 1), Mn(II) buffer (lane 2), or HEPES-buffered p-phenylenediamine (lane 3). The arrows highlight the two bands present in all three lanes.
ACKNOWLEDGMENTS
We thank Deeanne Edwards for her assistance in characterizing the Mn(II)-oxidizing protein of strain SD-21.
This research was funded in part by the National Science Foundation (MCB98-08915), the Collaborative UC/Los Alamos Research Program, and the Superfund Basic Research Program (NIEHS grant ES10337) of the National Institutes of Health. C.A.F. was supported in part by a STAR Graduate Fellowship from the U.S. Environmental Protection Agency and the University of California Toxic Substances Research and Training Program. E.-M.C. acknowledges support from the Howard Hughes Undergraduate Research Program at UCSD.
REFERENCES
- 1.Boogerd R C, de Vrind J P M. Manganese oxidation by Leptothrix discophora SS-1. J Bacteriol. 1987;169:489–494. doi: 10.1128/jb.169.2.489-494.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brouwers G J, de Vrind J P M, Corstjens P L A M, de Vrind-de Jong E W. Genes of the two-step proteins secretion pathway are involved in the transport of the manganese-oxidizing factor across the outer membrane of Pseudomonas putida GB-1. Am Mineral. 1998;83:1573–1582. [Google Scholar]
- 3.Brouwers G J, de Vrind J P M, Corstjens P L A M, Cornelis P, Baysse C, de Vrind-de Jong E W. cumA, a gene encoding a multicopper oxidase, is involved in Mn2+-oxidation in Pseudomonas putida GB-1. Appl Environ Microbiol. 1999;65:1762–1768. doi: 10.1128/aem.65.4.1762-1768.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouwers G J, Corstjens P L A M, de Vrind J P M, Verkamman A, de Kuyper M, de Vrind-de Jong E W. Stimulation of Mn2+-oxidation in Leptothrix discophora SS-1 by Cu(II) and sequence analysis of the region flanking the gene encoding putative multicopper oxidase MofA. Geomicrobiol J. 2000;17:25–33. [Google Scholar]
- 5.Caspi R, Haygood M G, Tebo B M. Unusual ribulose-1,5-bisphosphate carboxylase/oxygenase genes from a marine manganese-oxidizing bacterium. Microbiology. 1996;142:2549–2559. doi: 10.1099/00221287-142-9-2549. [DOI] [PubMed] [Google Scholar]
- 6.Corstjens P L A M, de Vrind J P M, Goosen T, de Vrind-de Jong E W. Identification and molecular analysis of the Leptothrix discophora SS-1 mofA gene, a gene putatively encoding a manganese-oxidizing protein with copper domains. Geomicrobiol J. 1997;14:91–108. [Google Scholar]
- 7.DePalma S R. Manganese oxidation by Pseudomonas putida. Ph.D. thesis. Cambridge, Mass: Harvard University; 1993. [Google Scholar]
- 8.de Silva D, Davis-Kaplan S, Fergestad J, Kaplan J. Purification and characterization of Fet3 protein, a yeast homologue of ceruloplasmin. J Biol Chem. 1997;272:14208–14213. doi: 10.1074/jbc.272.22.14208. [DOI] [PubMed] [Google Scholar]
- 9.de Vrind J P M, Brouwers G J, Corstjens P L A M, den Dulk J, de Vrind-de Jong E W. The cytochrome c maturation operon is involved in manganese oxidation in Pseudomonas putida GB-1. Appl Environ Microbiol. 1998;64:3556–3562. doi: 10.1128/aem.64.10.3556-3562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douka C. Study of the bacteria from manganese concretions. Soil Biol Biochem. 1977;9:89–97. [Google Scholar]
- 11.Douka C. Kinetics of manganese oxidation by cell-free extracts of bacteria isolated from manganese concretions from soil. Appl Environ Microbiol. 1980;39:74–80. doi: 10.1128/aem.39.1.74-80.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrlich H L. Bacteriology of manganese nodules. II. Manganese oxidation by cell-free extract from a manganese nodule bacterium. Appl Microbiol. 1968;16:197–202. doi: 10.1128/am.16.2.197-202.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebers R. Enrichment, isolation, and emended description of Pedomicrobium ferrugineum Aristovskaya and Pedomicrobium manganicum Aristovskaya. Int J Syst Bacteriol. 1981;31:302–316. [Google Scholar]
- 14.Ghiorse W C. Biology of iron- and manganese-depositing bacteria. Annu Rev Microbiol. 1984;38:515–550. doi: 10.1146/annurev.mi.38.100184.002503. [DOI] [PubMed] [Google Scholar]
- 15.Ghiorse W C, Hirsch P. An ultrastructural study of iron and manganese deposition associated with extracellular polymers of Pedomicrobium-like budding bacteria. Arch Microbiol. 1979;123:213–226. [Google Scholar]
- 16.Ghiorse W C, Hirsch P. Isolation and properties of ferromanganese-depositing budding bacteria from Baltic Sea ferromanganese concretions. Appl Environ Microbiol. 1982;43:1464–1472. doi: 10.1128/aem.43.6.1464-1472.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giovannoni S, Rappé M. Evolution, diversity and molecular ecology of marine prokaryotes. In: Kirchman D L, editor. Microbial ecology of the oceans. New York, N.Y: Wiley-Liss; 2000. pp. 47–84. [Google Scholar]
- 18.Gregory E, Staley J T. Widespread distribution of ability to oxidize manganese among freshwater bacteria. Appl Environ Microbiol. 1982;44:509–511. doi: 10.1128/aem.44.2.509-511.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung W E, Schweissfurth R. Manganese oxidation by an intracellular protein of a Pseudomonas species. Zentralbl Allg Mikrobiol. 1979;19:107–115. [PubMed] [Google Scholar]
- 20.Juniper S K, Tebo B M. Microbe-metal interactions and mineral deposition at hydrothermal vents. In: Karl D M, editor. The microbiology of deep-sea hydrothermal vents. Boca Raton, Fla: CRC Press; 1995. pp. 219–254. [Google Scholar]
- 21.Kolber Z S, Van Dover C L, Niederman R A, Falkowski P G. Bacterial photosynthesis in surface waters of the open ocean. Nature. 2000;407:177–179. doi: 10.1038/35025044. [DOI] [PubMed] [Google Scholar]
- 22.Krumbein W E, Altman H J. A new method for detection and enumeration of manganese-oxidizing and -reducing micoorganisms. Helgol Wiss Meeresunters. 1973;25:347–356. [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 25.Larsen E I, Sly L I, McEwan A G. Manganese(II) adsorption and oxidation by whole cells and a membrane fraction of Pedomicrobium sp. ACM 3067. Arch Microbiol. 1999;171:257–264. [Google Scholar]
- 26.Lee Y, Tebo B M. Cobalt oxidation by the marine manganese(II)-oxidizing Bacillus sp. strain SG-1. Appl Environ Microbiol. 1994;60:2949–2957. doi: 10.1128/aem.60.8.2949-2957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffett J W, Ho J. Oxidation of cobalt and manganese in seawater via a common microbially catalyzed pathway. Geochim Cosmochim Acta. 1996;60:3415–3424. [Google Scholar]
- 28.Nealson K H. The isolation and characterization of marine bacteria which catalyze manganese oxidation. In: Krumbein W E, editor. Environmental biogeochemistry and geomicrobiology: methods, metals and assessment. Ann Arbor, Mich: Ann Arbor Science; 1978. pp. 847–858. [Google Scholar]
- 29.Nealson K H, Tebo B M, Rosson R A. Occurrence and mechanisms of microbial oxidation of manganese. Adv Appl Microbiol. 1988;33:279–318. [Google Scholar]
- 30.Okazaki M, Sugita T, Shimizu M, Ohode Y, Iwamoto K, de Vrind-de Jong E W, de Vrind J P M, Corstjens P L A M. Partial purification and characterization of manganese-oxidizing factors of Pseudomonas fluorescens GB-1. Appl Environ Microbiol. 1997;63:4793–4799. doi: 10.1128/aem.63.12.4793-4799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosson R A, Nealson K H. Manganese binding and oxidation by spores of a marine bacillus. J Bacteriol. 1982;151:1027–1034. doi: 10.1128/jb.151.2.1027-1034.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schutt C, Ottow J C. Distribution and identification of manganese-precipitating bacteria from noncontaminated ferroomanganese nodules. In: Krumbein W E, editor. Environmental biogeochemistry and geomicrobiology: methods, metals and assessment. Ann Arbor, Mich: Ann Arbor Science; 1978. pp. 869–878. [Google Scholar]
- 33.Shiba T, Simidu U. Erythrobacter longus gen. nov., sp. nov., an aerobic bacterium which contains bacteriochlorophyll a. Int J Syst Bacteriol. 1982;32:211–217. [Google Scholar]
- 34.Sly L I, Arunpairojana V, Hodgkinson M C. Pedomicrobium manganicum from drinking-water distribution systems with manganese-related “dirty water” problems. Syst Appl Microbiol. 1988;11:75–84. [Google Scholar]
- 35.Solomon E I, Sundaram U M, Machonkin T E. Multicopper oxidases and oxygenases. Chem Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 36.Sunda W G, Huntsman S A. Effect of sunlight on redox cycles of manganese in the southwestern Sargasso Sea. Deep-Sea Res. 1988;35:1297–1317. [Google Scholar]
- 37.Sunda W G, Huntsman S A. Photoreduction of manganese oxides in seawater. Mar Chem. 1994;46:133–152. [Google Scholar]
- 38.Tebo B M, Ghiorse W C, van Waasbergen L G, Siering P L, Caspi R. Bacterially mediated mineral formation: insights into manganese(II) oxidation from molecular genetic and biochemical studies. Rev Mineral. 1997;35:225–266. [Google Scholar]
- 39.Tyler P A. Hyphomicrobia and the oxidation of manganese in aquatic systems. Antonie Leeuwenhoek J Microbiol Serol. 1970;36:567–578. doi: 10.1007/BF02069059. [DOI] [PubMed] [Google Scholar]
- 40.van Waasbergen L G, Hoch J A, Tebo B M. Genetic analysis of the marine manganese-oxidizing Bacillus sp. strain SG-1: protoplast transformation Tn917: mutagenesis and identification of chromosomal loci involved in manganese oxidation. J Bacteriol. 1993;175:7594–7603. doi: 10.1128/jb.175.23.7594-7603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Waasbergen L G, Hildebrand M, Tebo B M. Identification and characterization of a gene cluster involved in manganese oxidation by spores of the marine Bacillus sp. strain SG-1. J Bacteriol. 1996;12:3517–3530. doi: 10.1128/jb.178.12.3517-3530.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vybiral D, Denner E B M, Haller C M, Busse H, White A, Hofle M G, Velimirov B. Polyphasic classification of 0.2 μM filterable bacteria from the Western Mediterranean Sea. Syst Appl Microbiol. 1999;22:635–646. doi: 10.1016/s0723-2020(99)80017-4. [DOI] [PubMed] [Google Scholar]
- 43.Yurkov V W, Beatty J T. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev. 1998;62:695–724. doi: 10.1128/mmbr.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yurkov V, Stackebrandt E, Holmes A, Fuerst J A, Hugenholtz P, Golecki J, Gad'on N, Gorlenko V M, Kompantseva E I, Drews G. Phylogenetic positions of novel aerobic, bacteriochlorophyll a-containing bacteria and description of Roseococcus thiosulfatophilus gen. nov., sp. nov., Erythromicrobium ramosum gen. nov., sp. nov., and Erythrobacter litoralis sp. nov. Int J Syst Bacteriol. 1994;44:427–434. doi: 10.1099/00207713-44-3-427. [DOI] [PubMed] [Google Scholar]