Abstract
Listeria innocua 743 produces an inhibitory activity demonstrating broad-spectrum inhibition of Listeria monocytogenes isolates. Gel-electrophoretic analysis of culture supernatants indicated that two inhibitors with different molecular weights were produced by this strain. Insertion of Tn917 into a 2.9 Kb plasmid (pHC743) generated mutants with either an impaired ability or a loss in ability to produce one of the inhibitors. Sequence analysis of the transposon insertion regions revealed the presence of two continuous open reading frames, the first encoding a new pediocin-like bacteriocin (lisA) and the second encoding a protein homologous with genes involved in immunity toward other bacteriocins (lisB). Translation of the bacteriocin gene (lisA) initiates from a noncanonical start codon and encodes a 71-amino-acid prebacteriocin which lacked the double glycine leader peptidase processing site common in other type II bacteriocins. Alignment of the sequence with the processed N termini of related bacteriocins suggests that the mature bacteriocin consists of 43 amino acids, with a predicted molecular mass of 4,484 Da. Mutants containing insertions into lisA were sensitive to the inhibitor, indicating that lisAB forms a single operon and that lisB represents the immunity protein. Cloning of an amplicon containing the lisAB operon into Escherichia coli resulted in expression and export of the bacteriocin. This finding confirms that the phenotype is dependent on the structural and immunity gene only and that export of this bacteriocin is sec dependent. This is the first confirmation of bacteriocin production in a Listeria spp., and it is of interest that this bacteriocin is closely related to the pediocin family of bacteriocins produced by lactic acid bacteria.
Listeria monocytogenes represents a serious food-borne pathogen responsible for spontaneous abortions, as well as for mortality in infants and immunocompromised persons (11). Major food-borne outbreaks involving this species have been associated with the ingestion of contaminated fermented and nonfermented dairy products, processed meats, fish, and a variety of other food products (11, 31). Over the last decade, there has been considerable interest in the use of bacteriocins for the creation of additional barriers to control the growth of L. monocytogenes in a wide variety of processed foods (25). The presence of bacteriocin-producing bacteria (10, 42) or the addition of bacteriocins (8, 15) may create an additional hurdle to contribute to the inhibition of L. monocytogenes in processed foods. Bacteriocins may also find application for the inhibition of other nonpathogenic bacterial species associated with the spoilage of processed foods (22).
Ribosome-encoded peptide antibiotics fall within two broad classes, the lantibiotics and the bacteriocins (16, 26), with both classes consisting of small peptides ranging in size from 30 to 60 amino acid residues. The bacteriocins are a diverse collection of hydrophobic heat-stable peptides and fall within a number of distinct groups, i.e., IIa, IIb, and IIc (26). The class IIa bacteriocins, also referred to as the pediocin family, share significant homology and demonstrate broad-spectrum antilisterial activity (9, 26). In class IIb bacteriocins the inhibitory activity is dependent on the presence of two peptides (21, 26). The third class of bacteriocins (IIc) may also share significant homology with other class II bacteriocins; however, export of these bacteriocins occurs via the sec-dependent pathway (5, 29), in contrast to other bacteriocins, which require a dedicated transport system (26). The bacteriocins produced by a wide variety of lactic acid bacteria (LAB) have been intensively studied (9, 26), although related compounds are also produced by other gram-positive bacteria (18, 35).
Inhibitory activities have previously been reported among various species of Listeria (44), the majority of which appear to represent defective bacteriophage particles (17, 35, 44). Historically, interest in the inhibitors produced by Listeria spp. was primarily with regard to their usefulness for the development of phage typing schemes for L. monocytogenes (7, 19, 27, 41, 45). To date, there are only a few reports documenting the occurrence of isolates which produce bacteriocin-like activities (17, 24). Recently, a survey of 300 strains of Listeria spp. for bacteriocin-like activity suggested that the occurrence of the phenotype was relatively rare (17). Four isolates producing bacteriocin-like activity were identified and could be separated into two groups based on cross-immunity. Two isolates of L. innocua produced an inhibitory activity with very broad spectrum activity against the various serotypes of L. monocytogenes.
In this study, we report on the characterization of a bacteriocin-like activity produced by Listeria innocua 743. It was determined that this isolate produces two different inhibitors, one of which represents a new plasmid-encoded sec-dependent bacteriocin (Listeriocin 743A) with homology with the pediocin family of LAB bacteriocins. This is the first report describing bacteriocin production in a Listeria spp. It is also of interest that this bacteriocin falls within the pediocin family of type II bacteriocins widely produced by LAB; these bacteriocins are well known as effective inhibitors of L. monocytogenes.
MATERIALS AND METHODS
Bacterial cultures.
All cultures were maintained frozen as glycerol stocks at −80°C. Isolates of Listeria spp. were obtained from the culture collection of J. M. Farber in the Bureau of Microbial Hazards. The growth medium utilized for all culturing was brain heart infusion (BHI; Difco Laboratories, Detroit, Mich.) liquid and/or agar. Routine sensitivity testing was carried out using Listeria ivanovii 27 as the indicator. This culture was previously shown to be a very sensitive indicator of various inhibitory activities previously reported in other Listeria spp. (17). All incubations were carried out at a temperature of 37°C unless stated otherwise.
Inhibitory activity assays.
Inhibitory activity was determined using both deferred antagonism and direct antagonism plate tests (35). Inhibitory activity in liquid cultures and crude extracts was determined by critical point dilution of samples using a 1-in-2 dilution series (35). The reciprocal of the highest dilution which gave an obvious clearing zone was defined as the value for the units of inhibitory activity per milliliter. Crude extracts for gel electrophoresis and plating consisted of 5× concentrates of ammonium sulfate precipitations of autoclaved early-stationary-phase culture supernatant.
Growth curves.
Production of the inhibitory activity over the course of batch culture growth was carried out as follows. Twelve tubes, each containing 5.0 ml of BHI, were inoculated with 10 μl of a fresh overnight culture of L. innocua 743 and then incubated at a temperature of 12°C. Two tubes per day were sampled, one in the morning and one at night. Then, 1.0 ml of culture was removed, and the absorbance at 600 nm determined. A total of 100 μl of acetic acid was added to the remaining 4.0 ml, and the culture was sterilized using a 0.22-μm (pore-size) filter. Activity present within the acidified sterile supernatant was determined by critical point dilution as stated above.
Gel electrophoresis.
The inhibitory activity present in culture were separated by electrophoresis using 16% Tricine–sodium dodecyl sulfate (SDS)-polyacrylamide gels (33). Samples were boiled for a 5-min period prior to loading; the solubilization buffer contained no β-mercaptoethanol. After electrophoresis, the gels were fixed in a solution containing acetic acid, isopropanol, and water (5:20:75, vol/vol/vol) for 30 min and then washed using two changes of distilled water for an additional 30 min (4). After completion of the final wash, the gels were placed onto a clean glass sheet, overlaid with 0.5% BHI agar containing the indicator strain and then incubated overnight at a temperature of 37°C. After incubation, the gels were examined for zones of clearing.
Insertion mutagenesis.
Insertion mutagenesis using Tn917 was carried out by transformation of L. innouca 743 with the broad-host-range thermolabile transposon delivery vector pTV1-0K (13). Electrotransformation was carried out as previously described (1). Transformants were grown for 48 h at 30°C on BHI plates containing kanamycin (50 μg/ml).
For generation of insertion mutants, five individual transformants were inoculated into separate tubes of BHI broth containing both kanamycin (50 μg/ml) and a subinhibitory amount of erythromycin (0.3 μg/ml) and then grown overnight at 24°C. Aliquots of 100 μl from each culture were plated onto BHI plates containing erythromycin (5 μg/ml) only and incubated for 48 to 72 h at 44°C. Colonies were picked, regrown at 37°C on BHI containing erythromycin (5 μg/ml) only, and screened to confirm loss of the kanamycin resistance (Kanr) marker by patching onto a separate plate containing kanamycin (50 μg/ml). A total of three hundred insertion mutants (Eryr Kans; Eryr = erythromycin resistance) were generated and screened using plate testing for the loss in production of the inhibitory activity.
DNA isolation.
For the isolation of genomic DNA, cells were grown overnight in 100 ml of BHI broth. The cells were pelleted by centrifugation (10,000 × g, 10 min), resuspended into 5.0 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 7.5), and placed on ice. Lysozyme (10 mg/ml) was added to the suspension and incubated on ice for 30 min, after which Triton X-100 was added to a level of 1% to lyse the cells. After lysis, 1.0 ml of 7.5 M ammonium acetate (pH 4.8) was added to the solution and mixed by gentle inversion. The crude mixture was extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), mixed by gentle inversion, and centrifuged to separate the phases (10,000 × g, 30 min). The aqueous phase was recovered by using a wide-bore 10-ml pipette and extracted a second time with an equal volume of chloroform. After centrifugation, the upper aqueous phase was recovered and transferred into a clean 30-ml centrifuge tube. The genomic DNA was precipitated from solution by the addition of 2 volumes of cold isopropanol, and the DNA recovered by spooling onto a glass rod. The spooled DNA was washed by immersion into 70% (vol/vol) ethanol solution and then allowed to air dry. The spooled DNA was resolubilized into 0.4 ml of TE buffer and then stored at 4°C.
Southern blotting.
Genomic DNA for Southern blotting was digested with EcoRI. Restricted DNA was electrophoresed through 0.7% agarose gels. Unblots of the DNA gels for Southern hybridizations were prepared as previously described (40) and hybridized in a solution consisting of 5× SSC (1× SSC is 0.15 M NaCl plus 0.15 M sodium citrate), 0.05 M potassium phosphate buffer (pH 6.8), and 0.01% SDS. Hybridization with radiolabeled nick translated probes was carried out at a temperature of 60°C, and washed at the same temperature in a solution consisting of 0.5× SSC and 0.1% (wt/vol) SDS. Unblots were exposed to X-ray film for up to a 24 h.
Nick translation of probes for Southern blotting was carried out using the method of Sambrook et al. (32). Probes were labeled using [α-32P]CTP.
Analysis of insertion mutants.
The site of Tn917 insertion in the L. innocua 743 mutants was mapped by probing EcoRI-digested genomic DNA prepared from each mutant. The probe consisting of nick translated 32P-labeled pTV1-OK. A 3.2-kb amplicon encompassing pHC743-100 from one mutant (L. innocua 743-100) was generated by PCR using the opposing proximal and distal oligonucleotides (P1 and P2) homologous with each end of Tn917 (13). The position of the transposon insertion into the plasmid within each mutant was determined by sequencing the 800-bp flanking amplicons using an oligonucleotide homologous to a region located 37 nucleotides downstream of open reading frame 2 (ORF2) (BacR, 5′-AAAATAACCAAGTAGCC-3′) and the Tn917 distal oligonucleotide P2.
Isolation of plasmid DNA.
Plasmid DNA from L. innocua 743 was isolated from cells previously grown up in 10 ml of BHI overnight. Cells were recovered by centrifugation (10,000 × g, 10 min). Cell pellets were resuspended into 1.0 ml of TE buffer, and lysozyme (10 mg/ml) was added, followed by incubation on ice for 30 min. Plasmid DNA was isolated using Wizard Mini-Prep kit (Promega).
DNA sequencing.
DNA for sequence analysis was generated as described above. PCR-generated amplicons were cloned into the sequencing vector pCR2.1-TOPO (Invitrogen) and transformed into Escherichia coli TOP10 (Invitrogen). Double-stranded sequencing was carried out directly on the plasmids containing the cloned amplicons by cycle sequencing using Taq polymerase and the M13 universal primers (both forward and reverse). Sequences were run using a Licor (LI-COR, Inc.) automated sequencing system. Sequences of homologous peptides and proteins were identified and aligned using Psi-BLAST (2).
Nucleotide sequence accession number.
The GenBank nucleotide accession number for the bacteriocin Listeriocin743A and the immunity protein is AF330821.
RESULTS
Initial characterization of culture inhibitory activity.
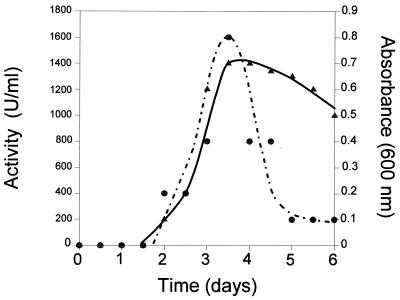
Low levels of inhibitory activity could be detected in overnight cultures by drop testing filtered culture supernatants onto overlays containing a sensitive indicator strain. Comparison of the spectrum of inhibitory activities from liquid cultures with that determined by deferred antagonism plate testing indicated that the identical inhibitory activity was produced under both growth conditions (results not shown). However, it was also noted that extended incubation of liquid cultures (>12 h at 37°C) resulted in significant losses in the total inhibitory activity present. The production of inhibitory activity during batch culturing was investigated, and results from experiments carried out at 12°C are presented in Fig. 1.
FIG. 1.
Production of inhibitory activity by L. innocua 743 during growth in batch culture (BHI) at 12°C. Symbols: ▴, absorbance; ●, inhibitory activity.
The production levels of inhibitory activity at both 12 and 37°C was identical and occurred during exponential growth, with maximum activity present during the early stationary phase. During the exponential growth phase in liquid cultures, the majority of inhibitory activity remained cell associated, a finding similar to that reported for other bacteriocins (15), requiring acidification of the medium to release the adsorbed inhibitors from the cell surface and to allow determination of the total inhibitory activity. After the cessation of growth, a rapid decline in inhibitory activity occurred (Fig. 1). The loss of activity could be prevented by autoclaving the culture, suggesting that it resulted from a cell-associated proteolytic activity.
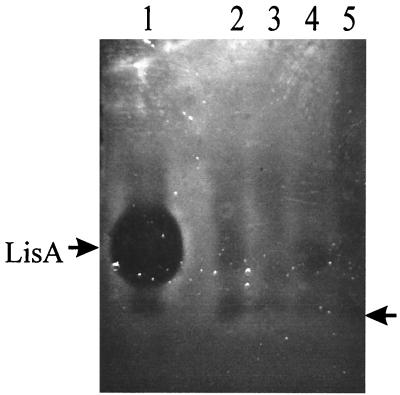
Gel-electrophoretic analysis (4) of ammonium sulfate-precipitated acidified culture supernatants from autoclaved early-stationary-phase cultures indicated that two inhibitors were present. The major inhibitor had a molecular mass of ca. 4,000 Da, whereas the minor inhibitor electrophoresed to a position on the gel slightly ahead of the methylene blue tracking dye (∼1 to 2 kDa; Fig. 2, lane 1).
FIG. 2.
Analysis on 16% Tricine–SDS-PAGE gels of autoclaved ammonium sulfate-precipitated early-stationary-phase culture supernatants. Lane 1, L. innocua 743; lane 2, L. innocua 743-48; lane 3, L. innocua 743-83; lane 4, L. innocua 743-100; lane 5, L. innocua 743-228. Gels were treated according to the method of Buhnia et al. (4) and overlaid with L. ivanovii 27. The position of Listeriocin 743A (LisA) is indicated with an arrow (left); the position of the low-molecular-mass inhibitor is also indicated with an arrow (right).
Production and characterization of BLIS− mutants
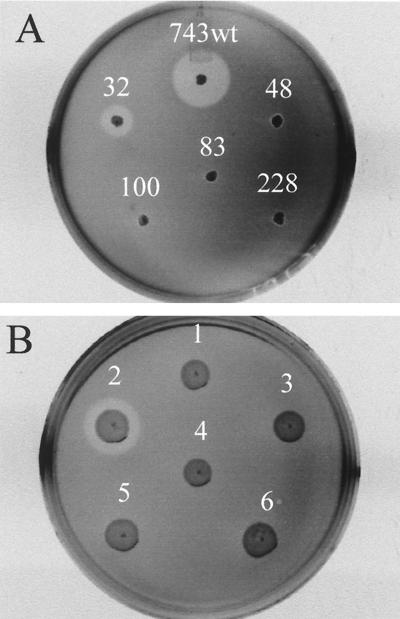
Tn917 transposon mutants of L. innocua 743 were generated by transformation of the strain using the broad-host-range heat-labile transposon delivery vector pTV1-OK (13). Three hundred mutants (Eryr Kans) were initially screened by direct antagonism plate testing (35) with L. ivanovii 27 as the indicator species. Five mutants that demonstrated either a loss in activity (L. innocua 743-48, 743-83, 743-100, and 743-228) or a reduced level of production (L. innocua 743-32) were identified. Results showing a direct antagonism plate test for the wild type and each of the mutants are shown in Fig. 3A. The sensitivity of each mutant to inhibition by the wild-type strain was also determined using deferred antagonism plate testing. Four of the mutants (L. innocua 743-48, 743-83, 743-100, and 743-228) were sensitive to inhibition, whereas L. innocua 743-32, which produced reduced levels of inhibitory activity, was not (results not shown). Tricine–SDS-polyacrylamide gel electrophoresis (PAGE) of spent culture fluids from each of the mutants demonstrated no impairment in the ability to produce the low-molecular-weight inhibitory activity (Fig. 2, lanes 2 to 5).
FIG. 3.
Inhibitory activity in L. innocua 743 and insertion mutants. (A) Direct antagonism plate test demonstrating production of inhibitory activity in L. innocua 743 and each transposon insertion mutant. (B) Inhibitory activity of E. coliTOP10 recombinants as determined by direct antagonism testing. Spots: 1, pCR2.1-TOPO; 2, pCR2.1-32; 3, pCR2.1-48; 4, pCR2.1-83; 5, pCR2.1-100; 6, pCR2.1-228. The cloned amplicons were each generated using the P2 and BacR primers. In each case the indicator organism using in the agar overlay was L. ivanovii 27.
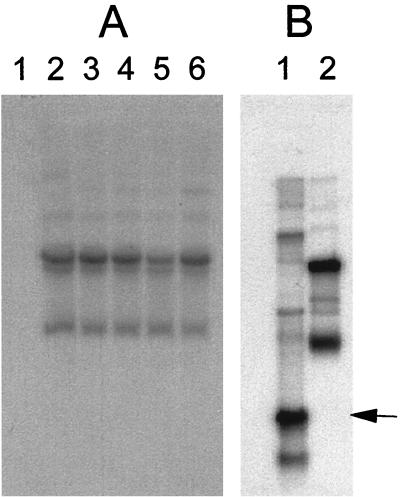
The location of transposon insertion in each of the five mutants was determined by Southern blotting. Initially, EcoRI-digested genomic DNA from each mutant was probed using nick-translated radiolabeled pTV1-OK. Results are presented in Fig. 4A. As expected, the probe failed to hybridize any sequences within the wild-type strain (Fig. 4A, lane 1) but did hybridize to a series of three identical bands in each of the five mutants (Fig. 4A, lanes 2 to 6). It was obvious that the positive hybridization signals did not represent restriction fragments but rather corresponded to unrestricted plasmid DNA. In all five mutants, transposon insertion had occurred into a small previously undetected cryptic plasmid.
FIG. 4.
Southern blot analysis of transposon insertion mutants. (A) EcoRI-digested genomic DNA from L. innocua 743 wild-type (lane 1), 743-32 (lane 2), 743-48 (lane 3), 743-83 (lane 4) 743-100 (lane 5), and 743-228 (lane 6) probed with nick-translated radiolabeled pTV1-OK. (B) Plasmid preparation from L. innocua 743 wild type (lane 1) and mutant 743-83 (lane 2) probed with the 3.2-kb nick-translated radiolabeled P1/P2 amplicon. The position of the wild-type 2.9-kb plasmid is indicated with an arrow.
An amplicon encompassing the entire plasmid was generated by PCR using a set of opposing oligonucleotides (P1 and P2) homologous with each end of the transposon (13). The PCR yielded a linear DNA fragment of ca. 3.2 kb. To confirm that the 3.2-kb amplified DNA represented a plasmid, the nick-translated radiolabeled amplicon was reprobed back onto the wild-type strain and one of the Tn917 mutants (L. innocua 742-83). The results are presented in Fig. 4B. The nick-translated amplicon hybridized a 2.9-kb plasmid in the wild type and an 8.0-kb plasmid in L. innocua 743-83; the difference in molecular weight results from the insertion of Tn917. The 3.2-kb amplicon was cloned into pCR2.1-TOPO, and both ends were sequenced. The 2.9-kb wild-type plasmid was named pHC743.
DNA sequence analysis.
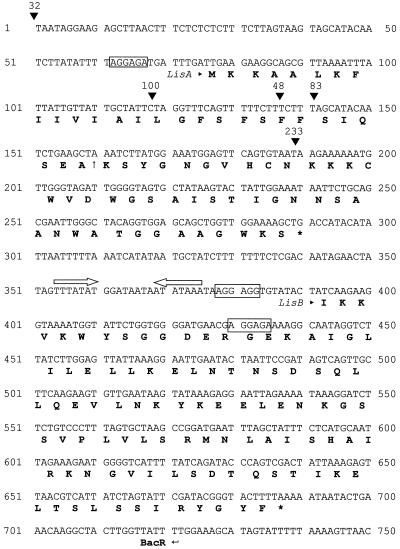
Sequencing of one end of the cloned amplified plasmid (pHC743-83) revealed the presence of two continuous ORFs, ORF1 and -2. The predicted product encoded downstream of the insertion site in L. innocua 742-83 (Fig. 5) demonstrated significant homology with other type IIa bacteriocins. In order to map the location of each transposon insertion, a second primer (BacR) was designed and used in combination with P2 to amplify the region spanning the inserted transposon and 37 bp downstream of the termination codon for ORF2. The position of each transposon insertion site was determined by sequencing the respective DNA generated using these primer sets. In four of the mutants (743–48, 743–83, 743–100, and 743–228) transposon insertion occurred at four different positions within ORF1 (Fig. 5). In the case of the leaky mutant 743-32, insertion occurred 78 bp upstream of the encoded bacteriocin (Fig. 5).
FIG. 5.
Nucleotide sequence of the Listeriocin 743A operon. The position of each transposon insertion is indicated with a solid arrow above the nucleotide sequence. Sequences bearing homology to ribosome-binding sequences are boxed, and the predicted protein sequence is indicated below the nucleotide sequence. The presumptive N-terminal cleavage site within listeriocin 743A is indicated with an arrow. An inverted repeat falling within the intergenic region of lisAB is indicated above the nucleotide sequence with opposing arrows (ΔG° = −4.7 kcal/mol). The position of the BacR primer used to generate PCR products for cloning is indicated.
ORF1 potentially encoded a peptide of 71 amino acids. A sequence bearing homology with other eubacterial ribosome-binding sites (AGGAGA) was present upstream of ORF1 at a position 62 bp downstream of the transposon insertion site in mutant 743-32 (Fig. 5). No methionine residue was present downstream of this putative ribosome-binding sequence, suggesting that translation initiation takes place at the lysine residue located at a position −10 from the potential ribosome-binding site. The predicted product encoded by ORF1 demonstrated significant homology with a variety of previously reported bacteriocin sequences produced by various LAB (Fig. 6).
FIG. 6.
Alignment of the predicted amino acid sequence of Listeriocin 743A with homologous bacteriocin sequences. Alignment was carried out using Psi-BLAST (2). The sources for the homologous sequences are Sakacin P (36), Mundticin (3), Piscicolin 126 (15), and Divercin V41 (23). Identical residues are indicated in black, and a conserved substitution is indicated in gray.
The predicted product of ORF1 (Listeriocin743A) shared significant homology with other type IIa bacteriocins, all of which contain the YGNG and CXXXXCXV consensus sequence motifs in the N terminus of the mature peptide, a finding common among the pediocin family of bacteriocins (26). The bacteriocins demonstrating the highest degree of homology to Listeriocin 743A included Sakacin P (36), Mundticin (3), Pisciolin 126 (15), and Divercin V41 (23). A Psi-BLAST alignment (2) of these bacteriocin sequences is shown in Fig. 6.
Alignment of LisA with the processed N terminus of homologous mature bacteriocins suggested that the leader sequence is cleaved at amino acid position 28 of ORF1 (NH2- … SIQSEA↓KSY…), which would result in a predicted molecular mass of 4,484 Da. The putative leader peptide, although not closely homologous with the sec-dependent leader sequences of other bacteriocins, has all of the characteristics found in other sec-dependent leader sequences (39). First, it is of an appropriate size (20 to 30 amino acids) and contains the requisite hydrophobic core sequence. Second, the predicted cleavage site is consistent with other leader peptides with the preferred residues (A, G, S, C, T, or Q) at positions −1 and −3, followed by a charged residue at the +1 position and allowed residues at both the −2 and −4 positions (39).
ORF2 was separated from ORF1 by an intervening region of 16 bp and could potentially encode a protein of 127 amino acids. An inverted repeat of 7 bp was located 63 bp downstream of the termination codon for ORF1 (ΔG° = −4.7 kcal/mol). The first methionine within ORF2 occurred 29 bp downstream of the termination codon for ORF1. However, no obvious ribosome-binding sequences were found within this region. Two sequences bearing close homology with the putative ribosome-binding site of ORF1 were located 88 and 138 bp downstream of the termination codon of ORF1 (AGGAGG and AGGAGA, respectively). Translation initiation might occur at either isoleucine residue located at positions −9 and −7 from these putative ribosome-binding sites. Initiation from either residue would result in proteins of 98 or 81 amino acids, with approximate molecular masses of 9 to 11 kDa. Comparison of homologous GenBank sequences with the predicted product of ORF2 indicated significant homology over the entire protein with the immunity proteins from the Sakacin P operon (47% identical and 74% similar [14]) and the carnobacteriocin B2 operon (51% identical and 70% similar [29]).
Heterologous expression of Listeriocin 743A.
One transposon mutant (L. innocua 743-32) was found to produce reduced quantities of Listerocin 743A compared to the wild-type strain (Fig. 3A). Transposon insertion within this mutant occurred at a position 78 bp upstream of the predicted translation initiation site for the lisA (Fig. 5). No obvious promoter sequences were found within the 78-bp 5′ region of ORF1. The reduced level of production likely results from a polar effect from the upstream insertion of Tn917, as has been previously described for other Tn917 mutants (6, 30). Cloning the PCR-generated amplicon encompassing the entire lisAB operon, generated using the P2 and BacR primers, into the sequencing vector pCR2.1-TOPO, and subsequent transformation into E. coli TOP10, was found to result in the production of the inhibitor (Fig. 3B). Neither E. coli TOP10/pCR2.1-TOPO nor the cloned amplicons derived from the additional four mutants produced an inhibitory activity (Fig. 3B). In this case, sequencing confirmed that the lisAB amplicon was cloned in the same orientation as the lacZ promoter of pCR2.1-TOPO, which is constitutively expressed E. coli TOP10.
DISCUSSION
Over the last decade there has been considerable interest in the application of bacteriocins for the control of L. monocytogenes in foods (16, 25). Bacteriocins produced by a wide variety of LAB and other gram-positive bacteria have been purified and genetically characterized (16, 18, 26). Since peptide antibiotics represent competitive factors (12, 35), we investigated the possibility that new broad-spectrum antilisterial inhibitors might be produced by isolates within the genus itself. To date, there are only a few examples of what may constitute bacteriocin production among isolates of Listeria spp. (17, 24), the majority of previously described inhibitors representing either bacteriophage (44) or replication-defective phage particles (17). Recently, we identified four Listeria isolates which produced bacteriocin-like inhibitory activities, two of which demonstrated very broad spectrum activity against isolates of L. monocytogenes (17). On this basis, one of these producing strains, L. innocua 743, was selected for further study.
Gel-electrophoretic analysis of concentrated spent culture fluids indicated that L. innocua 743 produced two inhibitors of different molecular mases. The major inhibitor had an approximate molecular mass of 4,000 Da. The molecular mass of the second inhibitor was estimated to be on the order of 1,000 to 2,000 Da based on estimates derived from gel electrophoresis and on the finding that the material could be washed through a 3-kDa cutoff membrane (results not shown). The production of multiple bacteriocins is not unusual and has been reported in a variety of LAB bacteria, including L. lactis (38), Leuconostoc sp. (28), and Carnobacterium sp. (29). The unusual feature of the minor inhibitory activity was that its molecular mass falls within a size range normally associated with antibiotics produced via the nonribosomal pathway (34).
Mutants unable to produce the major inhibitory activity were generated using the broad-host-range heat-labile transposon delivery vector pTV1-OK (13). Analysis of these mutants yielded a number of findings. First, transposon insertion into a 2.9-kb cryptic plasmid (pHC743) resulted in the loss of the of the major inhibitor, indicating that production of the inhibitor was plasmid mediated. This finding was reflected in terms of the high frequency of mutants isolated, given that only 300 insertions were screened. Plasmid-mediated bacteriocin production is common among a great variety of LAB (16, 26), although chromosome-encoded bacteriocins have also been reported in LAB and other non-LAB species (18, 36). Second, gel-electrophoretic analysis of these mutants revealed that the minor inhibitory activity was still produced, indicating that the inhibitors were distinct from one another. Finally, in the four insertion mutants no longer producing the major activity, the insertions occurred in a single gene with significant homology to other bacteriocins. One can therefore conclude that this gene (lisA) was responsible for the production of the major inhibitory activity.
Analysis of the predicted amino acid sequence from lisA indicated significant homology with other type IIa or pediocin-like bacteriocins, bacteriocins which are potent inhibitors of L. monocytogenes (9, 26). However, the prebacteriocin did have a number of unusual features. First, the leader peptide lacked the double glycine signal processing site common to the majority of class IIa bacteriocins, suggesting that export occurs via the sec-dependent export pathway (26). Both the length and sequence features of the putative leader peptide also support this hypothesis and suggest that cleavage occurs between the alanine and lysine residues located at positions 28 and 29 in the predicted protein sequence. In fact, the presence of a sec-dependent leader sequence was confirmed by production and export of the functional bacteriocin in a laboratory E. coli strain. Other examples of class II bacteriocins exported via the sec-dependent pathway include Divergicin A (43), Acidocin B (20), Enterocin P (5), and Bacteriocin 31 (37). On this basis, Listeriocin743A represents a new class IIc bacteriocin.
Immediately downstream of lisA was a second ORF with significant homology to other reported immunity proteins (14, 29). With many type II bacteriocins, the immunity protein is directly linked with the bacteriocin gene forming a single transcriptional unit (26). Both the direct linkage and the role as an immunity protein was confirmed through analysis of our transposon mutants. L. innocua 743-32, which produced a reduced level of the bacteriocin, retained immunity. In contrast, mutants containing insertions directly into lisA were no longer immune. These findings indicate that both genes are linked into a single transcriptional unit and that lisB functions as the immunity protein. An unusual feature of the lisAB operon was the presence of an inverted repeat within the intergenic region spanning both genes. A similar structural feature was recently reported to follow the gene encoding Lactococcin 972 (21).
The Listeriocin 743A operon is relatively simple compared to the bacteriocin production operons in other species of bacteria (16, 26). Generally, the production of a type II bacteriocin requires a number of accessory genes encoding both regulatory and transport functions (26). However, in this case there were no additional bacteriocin-related sequences present on pHC743, and heterologous expression of the bacteriocins required only the structural and immunity genes. This represents the first example of bacteriocin production in a Listeria spp. Perhaps the most interesting result from this study was that this new bacteriocin (Listeriocin 743A) is closely related to other pediocin-like bacteriocins, which are well known to be effective inhibitors of L. monocytogenes.
ACKNOWLEDGMENTS
We acknowledge the gift of pTV1-OK from A. S. Bleiweis, University of Florida, Gainesville. In addition, we thank Dominique Elien, who carried out the batch kinetics as part of a fourth-year undergraduate thesis while at the University of Ottawa, and the excellent technical assistance of S. D'Aoust, J.-C. Ethier, and N. Corneau.
REFERENCES
- 1.Alexander J E, Andrew P W, Jones D, Roberts I S. Development of an optimized system for electroporation of Listeria species. Lett Appl Microbiol. 1990;10:179–181. doi: 10.1111/j.1472-765x.1990.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J I, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennik M H J, Vanloo B, Brasseur R, Goris L G M, Smid E J. A novel bacteriocin with a YGNGV motif from vegetable-associated Enterococcus mundtii: full characterization and interaction with target organisms. Biochim Biophys Acta. 1998;1373:47–58. doi: 10.1016/s0005-2736(98)00086-8. [DOI] [PubMed] [Google Scholar]
- 4.Buhnia A K, Johnson M C, Ray B. Direct detection of an antimicrobial peptide of Pediococcus acidilactici in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Ind Microbiol. 1987;2:319–322. [Google Scholar]
- 5.Cintas L M, Casaus P, Havarstein L S, Hernandez P E, Nes I F. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol. 1997;63:4321–4330. doi: 10.1128/aem.63.11.4321-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coburn P S, Hancock L E, Booth M C, Gilmore M S. A novel means of self-protection, unrelated to toxin activation, confers immunity to the bactericidal effects of the Enterococcus faecalis cytolysin. Infect Immun. 1999;67:3339–3347. doi: 10.1128/iai.67.7.3339-3347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis G W D, Mitchell R G. Bacteriocin (monocin) interactions among Listeria monocytogenes strains. Int J Food Microbiol. 1992;16:283–292. doi: 10.1016/0168-1605(92)90030-7. [DOI] [PubMed] [Google Scholar]
- 8.Davies E A, Bevis H E, Delves-Broughton J. The use of the bacteriocin, nisin, as a preservative in ricotta-type cheeses to control the food-borne pathogen Listeria monocytogenes. Lett Appl Microbiol. 1997;24:343–346. doi: 10.1046/j.1472-765x.1997.00145.x. [DOI] [PubMed] [Google Scholar]
- 9.Eijsink V G, Skeie N, Middelhoven P H, Brurberg N B, Nes I F. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl Environ Microbiol. 1998;64:3275–3281. doi: 10.1128/aem.64.9.3275-3281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eppert I, Valdés-Stauber N, Götz H, Busse M, Scherer S. Growth reduction of Listeria spp. caused by undefined industrial red smear cheese cultures and bacteriocin-producing Brevibacterium linens as evaluated in situ on soft cheese. Appl Environ Microbiol. 1997;63:4812–4817. doi: 10.1128/aem.63.12.4812-4817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gronroos L, Saarela M, Matto J, Tanner-Salo U, Vuorela A, Alaluusua S. Mutacin production by Streptococcus mutans may promote transmission of bacteria from mother to child. Infect Immun. 1998;66:2595–2600. doi: 10.1128/iai.66.6.2595-2600.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez J A, Crowley P J, Brown D P, Hillman J D, Youngman P, Bleiweis A S. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J Bacteriol. 1996;178:4166–4175. doi: 10.1128/jb.178.14.4166-4175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huhne K, Axelsson L, Holck A, Krockel L. Analysis of the sakacin P gene cluster from Lactobacillus sake Lb674 and its expression in sakacin-negative Lb. sake strains. Microbiology. 1996;142:1437–1448. doi: 10.1099/13500872-142-6-1437. [DOI] [PubMed] [Google Scholar]
- 15.Jack R W, Wan J, Gordon J, Harmark K, Davidson B E, Hillier A J, Wettenhall R E, Hickey M W, Coventry M J. Characterization of the chemical and antimicrobial properties of piscicolin 126, a bacteriocin produced by Carnobacterium piscicola JG126. Appl Environ Microbiol. 1996;62:2897–28903. doi: 10.1128/aem.62.8.2897-2903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jack R W, Tagg J R, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalmokoff M L, Daley E, Austin J W, Farber J M. Bacteriocin-like inhibitory activities among various species of Listeria. Int J Food Microbiol. 1999;50:191–201. [Google Scholar]
- 18.Kalmokoff M L, Lu D, Whitford M F, Teather R M. Evidence for production of a new lantibiotic (butyrivibriocin OR79A) by the ruminal anaerobe Butyrivibrio fibrisolvens OR79: characterization of the structural gene encoding butyrivibriocin OR79A. Appl Environ Microbiol. 1999;65:2128–2135. doi: 10.1128/aem.65.5.2128-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebek G, Teysseire P, Baumgartner A. A method for typing Listeria monocytogenes strains by classification of listeriocins and phage receptors. Zentbl Bakteriol. 1993;278:58–68. doi: 10.1016/s0934-8840(11)80279-3. [DOI] [PubMed] [Google Scholar]
- 20.Leer R J, van der Vossen J M B M, van Giezen M, van Noort J M, Pouwels P H. Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology. 1995;141:1629–1635. doi: 10.1099/13500872-141-7-1629. [DOI] [PubMed] [Google Scholar]
- 21.Martínez B, Fernández N, Suárez J E, Rodríguez A. Synthesis of lactococcin 972, a bacteriocin produced by Lactococcus lactis IPLA 972, depends on the expression of a plasmid-encoded bicistronic operon. Microbiology. 1999;145:3155–3161. doi: 10.1099/00221287-145-11-3155. [DOI] [PubMed] [Google Scholar]
- 22.McMullen L M, Stiles M E. Potential for use of bacteriocin-producing lactic acid bacteria in the preservation of meats. J Food Prot Suppl. 1996;64:71. doi: 10.4315/0362-028X-59.13.64. [DOI] [PubMed] [Google Scholar]
- 23.Metivier A, Pilet M F, Dousset X, Sorokine O, Anglade P, Zagorec M, Piard J C, Marion D, Cenatiempo Y, Fremaux C. Divercin V41, a new bacteriocin with two disulphide bonds produced by Carnobacterium divergens V41: primary structure and genomic organization. Microbiology. 1998;144:2837–2844. doi: 10.1099/00221287-144-10-2837. [DOI] [PubMed] [Google Scholar]
- 24.Mollerach M E, Ogueta S B, De Torres R A. Production of linnocuicina 819, a bacteriocin produced by Listeria innocua. Microbiologica. 1988;11:219–224. [PubMed] [Google Scholar]
- 25.Muriana P. Bacteriocins for control of Listeria spp. in foods. J Food Prot Suppl. 1996;54:63. doi: 10.4315/0362-028X-59.13.54. [DOI] [PubMed] [Google Scholar]
- 26.Nes I F, Diep D B, Håvarstein L S, Brurberg M B, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 27.Ortel S. Listeriocins (monocins) Int J Food Microbiol. 1989;8:249–250. doi: 10.1016/0168-1605(89)90021-4. [DOI] [PubMed] [Google Scholar]
- 28.Papathanasopoulos M A, Krier F, Revol-Junelles A M, Lefebvre G, Le-Caer J P, von Holy A, Hastings J W. Multiple bacteriocin production by Leuconostoc mesenteroides TA33a and other Leuconostoc/Weissella strains. Curr Microbiol. 1997;35:331–335. doi: 10.1007/s002849900264. [DOI] [PubMed] [Google Scholar]
- 29.Quadri L E N, Sailor M, Roy K L, Vederas J C, Stiles M E. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J Biol Chem. 1994;269:12204–12211. [PubMed] [Google Scholar]
- 30.Rouquette C, Bolla J M, Berche P. An iron-dependent mutant of Listeria monocytogenes of attenuated virulence. FEMS Microbiol Lett. 1995;133:77–83. doi: 10.1111/j.1574-6968.1995.tb07864.x. [DOI] [PubMed] [Google Scholar]
- 31.Ryser E L. Food borne listeriosis. In: Ryser E T, Marth E H, editors. Listeria, listeriosis, and food safety. 2nd ed. New York, N.Y: Marcel Dekker, Inc; 1999. pp. 299–358. [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schägger H, von Jagow G. Tricine-sodium dodecyl sulphate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 34.Stachelhaus T, Marahiel M A. Modular structure of genes encoding multifunctional peptide sythetases required for non-ribosomal peptide synthesis. FEMS Microbiol Lett. 1995;125:3–14. doi: 10.1111/j.1574-6968.1995.tb07328.x. [DOI] [PubMed] [Google Scholar]
- 35.Tagg J R, Dajani A S, Wannamaker L W. Bacteriocins of gram-positive bacteria. Bacteriol Rev. 1976;40:722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tichaczek P S, Vogel R F, Hammes W P. Cloning and sequencing of sakP encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH 673. Microbiology. 1994;140:361–367. doi: 10.1099/13500872-140-2-361. [DOI] [PubMed] [Google Scholar]
- 37.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J Bacteriol. 1996;178:3585–3593. doi: 10.1128/jb.178.12.3585-3593.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Belkum M J, Kok J, Venema G. Cloning, sequencing, and expression in Escherichia coli of lcnB, a third bacteriocin determinant from the lactococcal bacteriocin plasmid p9B4-6. Appl Environ Microbiol. 1992;58:572–577. doi: 10.1128/aem.58.2.572-577.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Heijne G. Patterns of amino acids near signal sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 40.Wallace R B, Mayada C G. Oligonucleotide probes for screening of recombinant libraries. Methods Enzymol. 1987;157:432–442. doi: 10.1016/0076-6879(87)52050-x. [DOI] [PubMed] [Google Scholar]
- 41.Wilhelms D, Sandow D. Preliminary studies on monocine typing of Listeria monocytogenes strains. Acta Microbiol Hung. 1989;36:235–238. [PubMed] [Google Scholar]
- 42.Winkowski K, Crandall A D, Montville T J. Inhibition of Listeria monocytogenes by Lactobacillus bavaricus MN in beef systems at refrigeration temperatures. Appl Environ Microbiol. 1993;59:2552–2557. doi: 10.1128/aem.59.8.2552-2557.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Worobo R W, van Belkum M J, Sailer M, Roy K L, Vederas J C, Stiles M E. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol. 1995;177:3143–3149. doi: 10.1128/jb.177.11.3143-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zink R, Loessner M J, Scherer S. Characterization of cryptic prophages (monocins) in Listeria and sequence analysis of a holin/endolysin gene. Microbiology. 1995;141:2577–2584. doi: 10.1099/13500872-141-10-2577. [DOI] [PubMed] [Google Scholar]
- 45.Zink R, Loessner M J, Glas I, Scherer S. Supplementary Listeria-typing with defective Listeria phage particles (monocins) Lett Appl Microbiol. 1994;19:99–101. doi: 10.1111/j.1472-765x.1994.tb00915.x. [DOI] [PubMed] [Google Scholar]