Abstract
Rugose phenotypes, such as those observed in Vibrio cholerae, have increased resistance to chlorine, oxidative stress, and complement-mediated killing. In this study we identified and defined a rugose phenotype in Salmonella enterica serovar Typhimurium DT104 and showed induction only on certain media at 25°C after 3 days of incubation. Incubation at 37°C resulted in the appearance of the smooth phenotype. Observation of the ultrastructure of the rugose form and a stable smooth variant (Stv), which was isolated following a series of passages of the rugose cells, revealed extracellular substances only in cells from the rugose colony. Observation of the extracellular substance by scanning electron microscopy (SEM) was correlated with the appearance of corrugation during development of rugose colony morphology over a 4-day incubation period at 25°C. In addition, the cells also formed a pellicle in liquid broth, which was associated with the appearance of interlacing slime and fibrillar structures, as observed by SEM. The pellicle-forming cells were completely surrounded by capsular material, which bound cationic ferritin, thus indicating the presence of an extracellular anionic component. The rugose cells, in contrast to Stv, showed resistance to low pH and hydrogen peroxide and an ability to form biofilms. Based on these results and analogy to the rugose phenotype in V. cholerae, we propose a possible role for the rugose phenotype in the survival of S. enterica serovar Typhimurium DT104.
Salmonella enterica serovar Typhimurium is a causative agent of gastrointestinal salmonellosis in humans. Phage type 104 (definitive type 104 [DT104]) strain of S. enterica serovar Typhimurium has recently emerged with resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline in the United States (3), the United Kingdom (25), France, Denmark (2), and Japan (23). This multidrug resistance has posed a major problem in the treatment of complications resulting from these Salmonella infections. Moreover, new strains with resistance to additional antibiotics, such as trimethoprim and fluoroquinolones, are beginning to emerge (18, 26). The Centers for Disease Control and Prevention has reported that DT104 was identified in 32% of the S. enterica serovar Typhimurium strains isolated from humans in 1996, an increase from 28% in 1995 and 7% in 1990. DT104 is now only the second most prevalent type of Salmonella after serovar Enteritidis phage type 4 in the United Kingdom (13).
During our study on S. enterica serovar Typhimurium DT104, we have observed a rugose phenotype, which has also been described for Vibrio cholerae after serial passages in alkaline peptone media (14, 19, 28) or in response to starvation (17, 27, 29). This phenotype of V. cholerae was defined by corrugated colony morphology, which was associated with the formation of exopolysaccharide (EPS) and cell aggregation. Because there was a question of whether the rugose variant of V. cholerae was virulent, a study was performed and showed that the rugose cells were fully virulent when inoculated into human volunteers and resulted in full-scale diarrheal disease (19). In fact, compared with the smooth variant, rugose cells displayed increased resistance to chlorine, salt, and oxidative stress (22, 27, 29) and had the ability to form biofilms (17, 27, 29), which suggested that this phenotype might be an aid in the survival of V. cholerae between outbreaks.
Similarly, the increasing incidence of S. enterica serovar Typhimurium in food-borne disease suggested that it may possess an enhanced capacity to survive under adverse conditions. We have observed that this organism assumed the rugose phenotype only on certain media and under particular growth conditions. Here we identify and define the rugose phenotype in S. enterica serovar Typhimurium DT104 and show that it appears to enhance survival under unfavorable conditions.
MATERIALS AND METHODS
Strains, media, and growth conditions. (i) Strains and media.
S. enterica serovar Typhimurium DT104 strain 11601 was routinely cultured on Trypticase soy agar (TSA) (Difco) incubated at 25°C for the formation of rugose colonies (Rv/25) or at 37°C for the formation of smooth colonies (Rv/37). Other agar media, including nutrient agar (NA) (Difco), Luria-Bertani agar (LB), brilliant green agar (BG) (Difco), brain heart infusion agar (BHI) (Difco), xylose lysine tergitol-4 agar (XLT-4) (Difco), and MacConkey's agar (MAC) (Difco) were used to assess the ability of S. enterica serovar Typhimurium DT104 to induce the rugose morphotype on agar media other than TSA. Stock cultures were prepared by suspending individual colonies in 4.0-ml vials containing Trypticase soy broth (TSB) with 15% glycerol added, followed by storage at −80°C. To compare the ability of other Salmonella serovars and other S. enterica serovar Typhimurium DT104 strains to induce rugose phenotype on TSA, the following strains were used: S. enterica serovar Typhimurium DT104 strains 10931, G-10601, G-11704, G-10631, and G-11074 (a kind gift from B. Swaminathan) and S. enterica serovar Typhimurium (non-DT104) UMD597, serovar Thompson UMD1317, serovar Senftenberg UMD1408, serovar Enteritidis UMD352, serovar Cowdy UMD1321, serovar Hadar UMD598, and serovar Johannesburg UMD600 (S.W.J. collection).
(ii) Growth conditions.
Cells were grown in static 10-ml volumes of TSB (Difco) in 17- by 150-mm glass test tubes or on TSA for at least 3 days at 25°C to induce pellicle formation or rugose-colony formation, respectively. To induce the development of stable, smooth variants (Stv) at low frequency, serial passage (>30) of the rugose colonies was performed by sequentially streaking a single rugose colony every 3 days onto a fresh TSA plate at 25°C.
(iii) Biochemical and serological testing.
Biochemical tests using triple sugar iron agar slants, lysine iron agar, and flagella broth and subsequent serological tests using polyvalent A, group B, and factor 4, 5, and 12 somatic (O) and flagellar (H) Salmonella antisera were performed on Rv/25 and Stv. Other biochemical tests using API 20E strips (BioMerieux, Inc.) were also used to confirm the identity of Rv/25 and Stv.
SEM.
For scanning electron microscopy (SEM) observation, well-isolated colonies (Rv/25 or Stv) and a broth pellicle (Rv/25), grown on TSA and in TSB, respectively, were processed after growth for at least 4 days at 25°C. Sections of colonies were cut to about 1.0 cm2, with a 1.0-mm thickness of agar remaining (5). The colonies or pellicles were fixed in 2% glutaraldehyde in phosphate-buffered saline (PBS), pH 7.4, then postfixed with 1% osmium tetroxide, dehydrated with ethanol, critical-point dried, and coated with gold-palladium alloy. Finally, samples were examined with a Hitachi S-4700 SEM (Hitachi Scientific Inst., Gaithersburg, Md.) at 2.7- to 5-kV acceleration.
Transmission electron microscopy. (i) Cationic ferritin staining.
Rv/37 and Rv/25 grown in TSB for 7 days at 37 and 25°C, respectively, and the pellicle from Rv/25 grown in TSB for 7 days at 25°C, as well as a TSB cell suspension from colonies of Rv/25 or Rv/37 grown on TSA at 25°C for 4 days, were washed three times with PBS (pH 7.4). After fixation with 2% glutaraldehyde in the same buffer for 1 h at 25°C and overnight at 4°C, the samples were reacted with 1.0 mg of cationic ferritin (Sigma) per ml for 30 min (14). Cationic ferritin was included at a concentration of 0.25 mg/ml in subsequent washes and during postfixation in 1% osmium tetroxide and in 2% aqueous uranyl acetate. Samples were dehydrated in ethanol, embedded in Spurr resin, sectioned, and observed with a Zeiss EM10 CA (Leo Electron Microscope, Thornwood, N.Y.).
(ii) Ruthenium red staining.
Rv/25 and Stv grown on TSA for 4 days at 25°C were removed as 1.0-cm2 blocks, fixed with 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), for 15 to 30 min, transferred to a glass slide, and encased with molten 4% agar. Blocks of 1.0 mm2 were prepared from the encased colonies. This procedure was used to ensure the integrity of the extracellular substances (ES). The agar blocks were fixed with 0.075% ruthenium red, 75 mM lysine monohydrochloride (Sigma), 2% paraformaldehyde (from a 10% stock solution of methanol-free pure formaldehyde), and 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 1 h at room temperature and 18 h at 4°C (9). After a series of washes in the same buffer and postfixation with 1% osmium tetroxide followed by 2% aqueous uranyl acetate, samples were dehydrated with ethanol and embedded in Spurr resin. Thin sections were examined with a Zeiss EM10 CA.
LPS profiles.
Whole-cell lysates were prepared by adding 50.0 μl of lysing buffer (2% sodium dodecyl sulfate [SDS], 4% 2-mercaptoethanol, 10% glycerol, 1 M Tris [pH 6.8], 0.1% bromphenol blue) to a 1.0-ml suspension of a well-isolated colony in TSB. The lysate was then boiled for 10 min and treated with 5 μl of proteinase K (5 mg/ml) at 60°C for 1 h. The lipopolysaccharide (LPS) components were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by LPS silver staining (12). No LPS differences between Rv/25 and Stv were detected (data not shown).
Biofilms.
S. enterica serovar Typhimurium DT104 strains (Rv/25 and Stv) were grown separately in 500-ml Erlenmeyer flasks containing 25 ml of TSB at 25°C with shaking (50 rpm) in a Psycrotherm controlled environment incubator shaker (New Brunswick Scientific, Edison, N.J.). Spent medium was replaced with an equal volume of fresh, sterile TSB every 24 h for a total of 7 days. At day 7, the spent medium was replaced with 20 ml of a 1% crystal violet solution in PBS, and the flasks were incubated for another 5 min at 25°C with shaking (50 rpm) (20). The crystal violet solution was discarded, and the flasks were rinsed with PBS.
Susceptibility to osmotic, acidic, and oxidative stress.
Individual colonies of Rv/25 and Stv approximately 0.5 cm in diameter were taken from TSA incubated at 25°C for 4 days and suspended separately in 10 ml of TSB in 17- by 150-mm glass test tubes. Because the cells in rugose colonies tend to aggregate, glass beads were used to disperse the cells. Therefore, the cell counts included some aggregates as well as cells in the suspension. To assay resistance to oxidative stress, 1.0 ml of the culture, approximately 1 × 107 to 5 × 107 cells in TSB, was mixed with 9.0 ml of prewarmed TSB, and finally hydrogen peroxide was added to a final concentration of 10 mM. To assay resistance to acidic stress, the original 10.0-ml cell suspension was concentrated 20 times. One hundred microliters of this suspension was added to 9.9 ml of TSB to which concentrated HCl was added to a final pH of 3.0 ± 0.05. The reaction mixture was incubated either at 37°C (for the hydrogen peroxide experiment) or at 25°C (for the acid experiment). Aliquots were taken every 10 min for a total of 90 min, were serially diluted in either saline (for the H2O2 experiment) or PBS (for the acid experiment) to stop the reaction, and were plated on TSA to determine the viable cell counts. The plates were incubated at 25°C and were observed for up to 5 days to determine whether changes in colony morphology had occurred. The pH remained constant at 3.0 ± 0.05 throughout the acid experiment. Each experiment was repeated at least three times on different days, and corresponding results were obtained.
RESULTS
Rugose and smooth colonies.
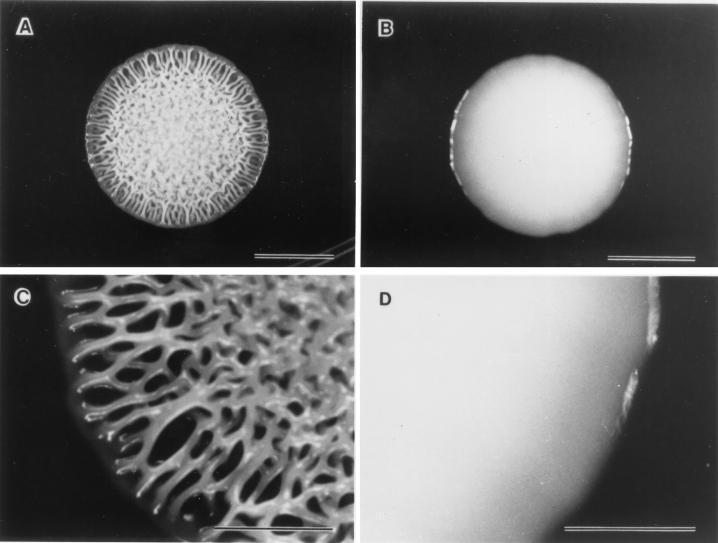
The temperature-dependent rugose phenotype (Rv/25) was observed when S. enterica serovar Typhimurium DT104 was incubated for at least 3 days on TSA at 19 to 28°C (Fig. 1A). Even prolonged incubation on TSA at a higher temperature (37°C) showed smooth colonies only (Rv/37) (Fig. 1B). Rv/25 colonies were approximately 5 to 10 mm in diameter, raised, and corrugated, had entire edges, which became even more irregular after 7 days of incubation, and had a grayish white pigmentation. Rv/37 colonies were raised with an even appearance and a buttery texture, had entire edges, and were grayish white. The Rv/25 colonies were tenacious, and cells in the rugose colonies aggregated such that the whole colony could be lifted easily with an inoculating loop. The colony remained as an aggregate when suspended, unlike cells from the smooth colonies, which dispersed easily and formed a homogenous suspension in broth. In contrast to the rugose colonies exhibited by V. cholerae, the corrugations of S. enterica serovar Typhimurium DT104 rugose colonies were very regular and symmetrical, with a circular pattern in the center of the colonies. Figure 1C shows how the corrugations formed an interlacing pattern, in contrast to the smooth surfaces of Rv/37 colonies (Fig. 1D). Spontaneous smooth variants (Stv), which appear to be similar in morphology to Rv/37 and did not turn rugose even after a 3-day incubation period at 25°C, were obtained after repeated passages on TSA. Rv/25 and Stv demonstrated the same reactions typical of S. enterica serovar Typhimurium after biochemical and serological tests. Rugose colonies were also formed on NA, LB, and BG but not on BHI, XLT-4, or MAC. Formation of the rugose characteristic on NA, BG, or LB sometimes required more than 3 days of incubation (data not shown).
FIG. 1.
Morphology of rugose (Rv/25) (A and C) and smooth (Rv/37) (B and D) colonies of S. enterica serovar Typhimurium DT104 grown for 4 days on TSA at 25 and 37°C, respectively. Scale bars, 5 mm (A and B) and 2 mm (C and D).
A few other Salmonella serovars and other strains of S. enterica serovar Typhimurium DT104 were also tested for their ability to induce rugose formation on TSA. All strains of DT104 tested demonstrated the rugose phenotype, except G-10601, which showed only slight corrugation in the middle of the colony. Salmonella serovars Typhimurium non-DT104, Thompson, Senftenberg, and Enteritidis all displayed rugose growth, but serovars Cowdy, Hadar, and Johannesburg remained smooth under the conditions tested.
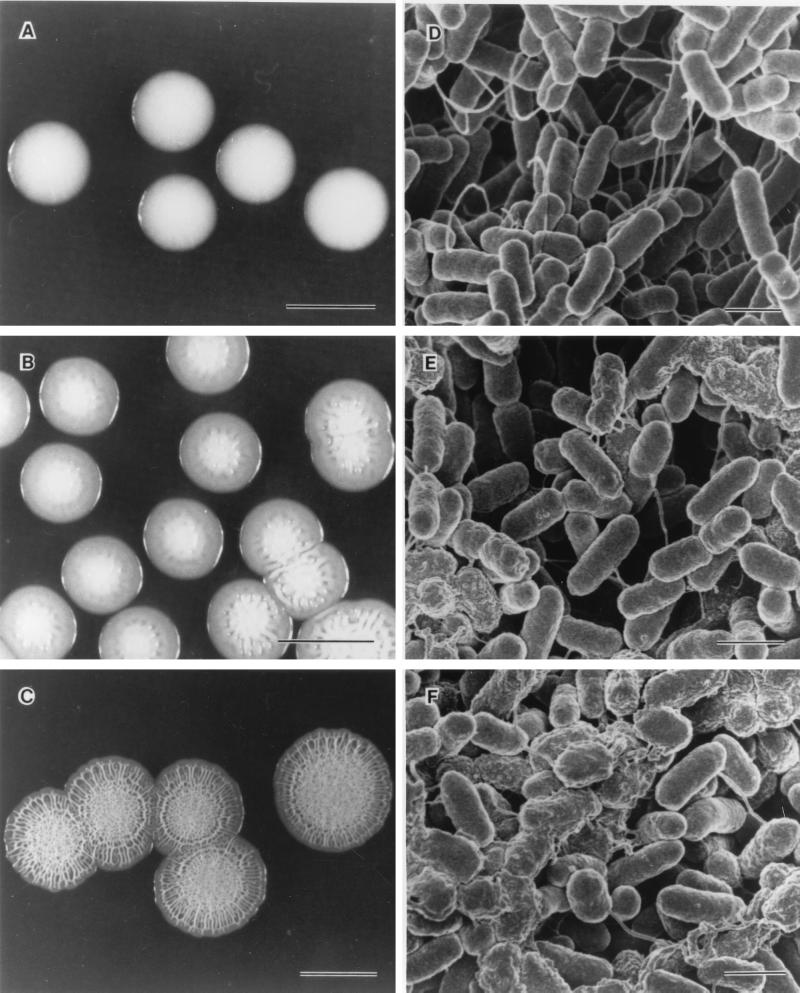
After initial inoculation of Rv/25 on TSA, the colonies appeared smooth on the first 2 days of incubation at 25°C (Fig. 2A), thus resembling Rv/37 and Stv. At day 3, some wrinkles started to appear on the colonies (Fig. 2B), and the rugose appearance (Rv/25) was fully evident at day 4 (Fig. 2C), thus demonstrating that observable rugose formation in S. enterica serovar Typhimurium DT104 occurs progressively over a 3- to 4-day period.
FIG. 2.
Progression of rugose formation at day 2 through day 4 at 25°C on TSA. Panels A to C show Rv/25 colonies at day 2, 3, and 4, while panels D to F show scanning electron micrographs of cells from the respective colonies. Expression of ES is apparent in cells starting at day 3 when the colonies just become wrinkled. Scale bars, 5 mm (A to C) and 1 μm (D to F).
SEM.
As shown in Fig. 2D, individual Rv/25 cells grown for only 2 days at 25°C appeared to have smooth surfaces, and the cells seemed to be connected to each other by long fibrils that averaged 40 nm. The 3-day-old colonies contained cells that were surrounded by an amorphous extracellular substance (ES), as shown by SEM (Fig. 2E). In the absence of further analysis, we refer to this material as ES. At 4 days, a large proportion of the cells was encapsulated by ES (Fig. 2F). Therefore, the presence of ES correlated with the change to wrinkled colony appearance. The cells that were surrounded by the ES seemed to form aggregates, thus providing an indication that the substance may be responsible for the aggregation. The Stv cells showed characteristics similar those of to 2-day-old Rv/25, and no ES was observed (data not shown).
The method of using intact colonies on agar for electron microscopic observation allowed preservation of the cellular surface of the individual cells in the colony. This was an essential method for this purpose, because the ES around the cells could not be detected by SEM when the colonies were suspended in buffer and then washed and centrifuged (data not shown).
Pellicle examination.
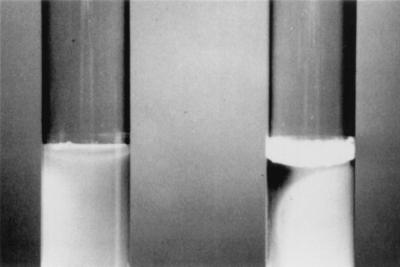
The rugose colony morphology seemed to correlate with formation of a surface pellicle by Rv/25 in TSB at 25°C (Fig. 3). Conversely, Stv cells showed growth in turbid suspension with only a slight appearance of biofilm on the glass at the surface of the growth. Incubation of Rv/37 cells at 37°C in TSB did not induce pellicle formation. The cells encased in the pellicle were also shown to possess ES (Fig. 4) and a more extensive formation of the matrix of the cross-linked fibrils. The ES and fibrillar network resulted in the formation of a very tenacious structure, which seemed to bind the cells together.
FIG. 3.
Pellicle formation was produced by Rv/25 (right) but not by Stv (left) in TSB after 7 days of incubation at 25°C.
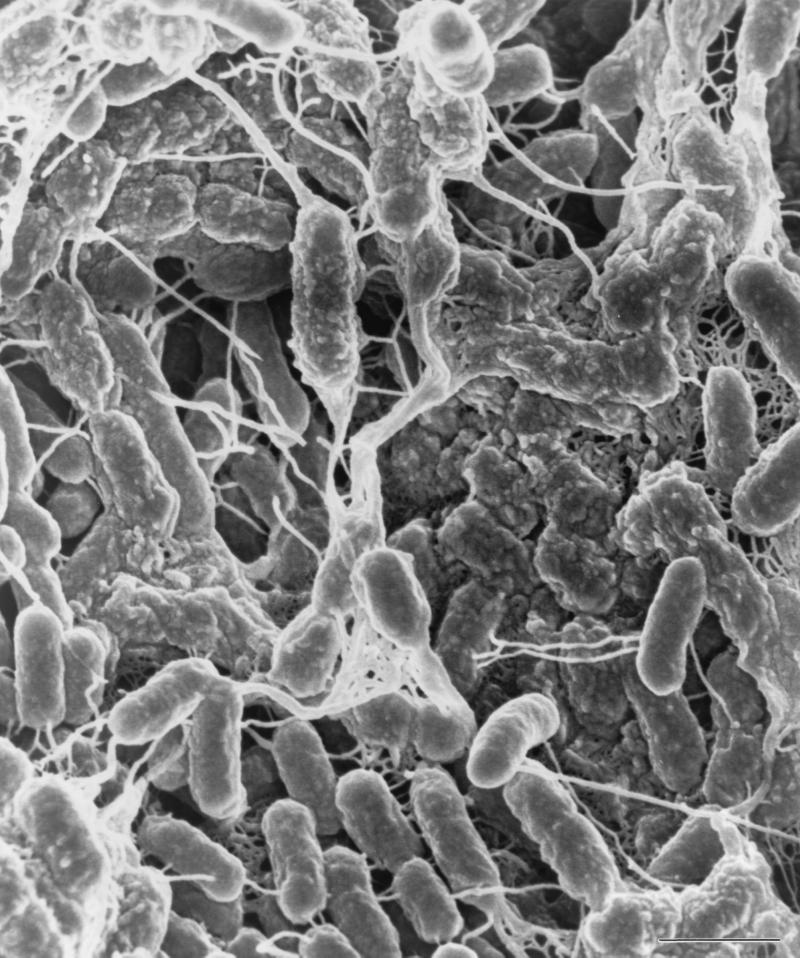
FIG. 4.
Scanning electron micrograph of pellicle cells. Note the presence of ES in both slime and the fibrillar structure. Scale bar, 1 μm.
Transmission electron microscopy.
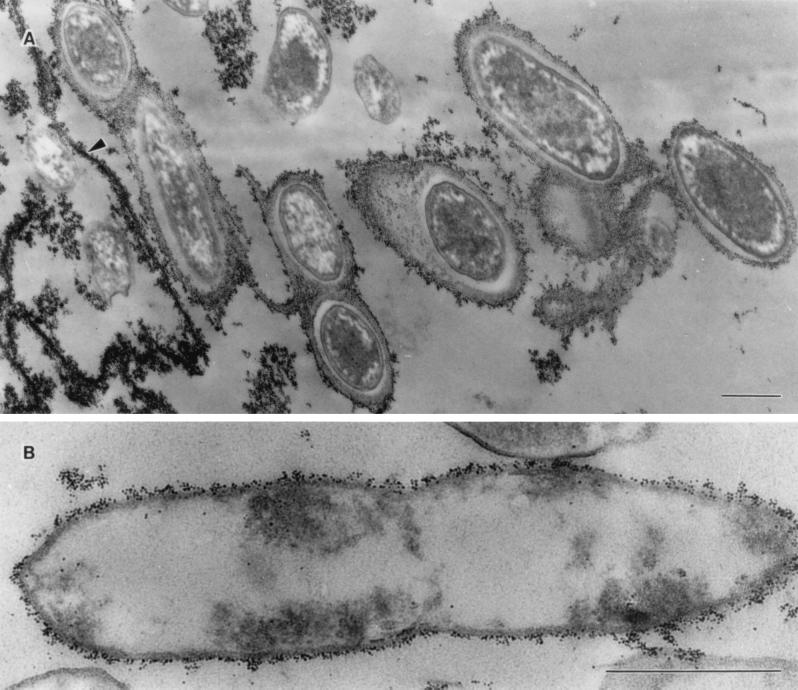
Binding of the ES in the pellicle cells from TSB incubated at 25°C to cationic ferritin (Fig. 5A) demonstrated that it contained an acidic component(s) (7). Cells sampled from the broth under the pellicle in TSB also showed ferritin binding (Fig. 5B), although the ES was not as apparent as that seen in the pellicle cells. In contrast, Rv/37 grown in TSB at 37°C had obvious diminished binding (data not shown). We also observed the slime-like ES present between the cells from the Rv/25 colonies using ruthenium red staining (data not shown), as was shown with V. cholerae O1 E1 Tor (29). This ES was preserved only in closely packed rugose colonies during preparation for electron microscopy. Because of the shrinkage and detachment of the hydrated ES during normal electron microscopy preparation, the ES was lost, as reported previously (9). However, the ES in cells from the pellicle were preserved.
FIG. 5.
Transmission electron micrographs of cells from the pellicle (A) and from broth under the pellicle (B) grown in TSB for 7 days, showing the presence of capsular material and a thin layer of ES, respectively, that binds to cationic ferritin. The fibrillar material was also bound to cationic ferritin (arrow in panel A). Scale bars, 0.5 μm.
Biofilms.
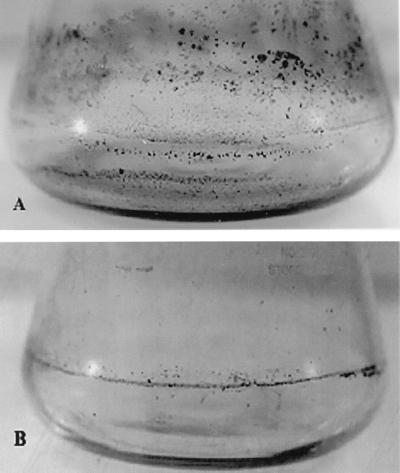
Rv/25 was more capable than Stv of forming biofilms on the inner surfaces of shaken glass flasks when cultured in TSB at 25°C (Fig. 6). The biofilms, which retained crystal violet after treatment, were then immediately observable.
FIG. 6.
Formation of biofilm matrix on the inner surface of glass flasks. The cultures of Rv/25 (A) and Stv (B) in TSB were incubated with shaking (50 rpm) at 25°C for 7 days with the TSB changed every 24 h. The resulting biofilms were stained with crystal violet for immediate observation.
Resistance to acid and oxidative stress.
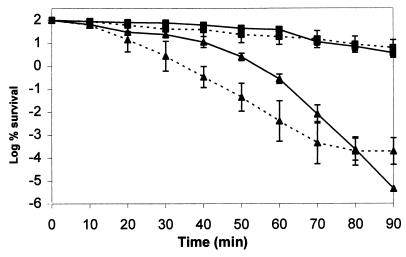
The cells from Rv/25 were shown to resist exposure to hydrogen peroxide (10 mM) and acid (pH 3) to a much greater extent than Stv (Fig. 7). A 90-min exposure to either condition resulted in a 2-log reduction of rugose cells, whereas all of the Stv cells (approximately 107) were killed in the same time frame. After a 50-min exposure to pH 3, a small proportion of the Stv cells turned rugose and thus became more resistant to the acid. This rugose colony morphology developed after 3 days of incubation at 25°C on TSA. At the end of the assay (90-min exposure to acid), all cells that survived eventually formed rugose colonies on TSA. Interestingly, none of the Stv cells converted to the rugose form after 10 to 80 min of exposure to H2O2.
FIG. 7.
Survival of S. enterica serovar Typhimurium DT104 cells from 4-day-old Rv/25 (■) and Stv (▴) upon a 90-min exposure to 10 mM hydrogen peroxide at 37°C (—) or pH 3 at 25°C (- - - ). Each data point represents the mean with standard deviation for three separate experiments.
DISCUSSION
While rugose colonies are observed in both S. enterica serovar Typhimurium DT104 and V. cholerae serogroup O139, O1 El Tor, and non-O1 strains, there are critical differences in the conditions under which rugosity occurs. First, formation of the rugose colony morphology in S. enterica serovar Typhimurium DT104 progresses over 3 to 4 days at room temperature (19 to 30°C) in TSA, while V. cholerae formation of rugose colony morphology occurs either after repeated passage of a smooth variant in alkaline peptone at 37°C (19) or under starved conditions at 16°C for 2 to 3 weeks (17, 27, 29). Secondly, with S. enterica serovar Typhimurium DT104, under the same growth conditions the phenotype remained constant, while the rugose and smooth variants in V. cholerae could revert back to the opposite phenotype at low frequency after 3 to 5 days of incubation in broth (1, 29). Third, unlike V. cholerae, the expression of the rugose variant of S. enterica serovar Typhimurium DT104 was temperature dependent.
Observation of adequately prepared S. enterica serovar Typhimurium DT104 with SEM revealed the presence of extracellular substance closely associated with the cells in the pellicle and in Rv/25 colonies. Formation of pellicles composed of closely packed cells has also been reported for rugose V. cholerae O1 E1 Tor (29). The observation that Rv/25 formed rugose colonies on TSA and pellicles in TSB and that Rv/37 formed neither rugose colonies on TSA nor pellicles in TSB further reinforced the temperature-dependent nature of the morphogenesis of S. enterica serovar Typhimurium DT104. For Stv, neither rugose colonies nor pellicles were observed at either 25 or 37°C. The ES in the pellicle seems to be responsible for the formation of cell aggregates (microcolonies). In contrast, the thinner layer of ES that surrounds the cells under the pellicle does not appear to play a role in aggregation. The ES appears to be composed of an anionic component(s). Whether the ES in the pellicle and that in the rugose colony have the same composition remains to be determined.
Although there are fibrils that seem to connect the cells in smooth colonies, these structures do not seem to provide a very strong support for cell aggregation, since cells in these colonies disperse very easily. These fibrils have been shown in wild-type S. enterica serovar Typhimurium during growth in the laboratory, and they are lost during subsequent exposure to epithelial cells and replaced by shorter appendages (10). Morphologically similar types of fibrils in Myxococcus xanthus were shown to be composed of equal amounts of protein and carbohydrate and seemed to play a role in communication among bacteria and between bacteria and their host (8). As shown in Fig. 5, the fibrils in S. enterica serovar Typhimurium DT104 are stained with ferritin, indicating the presence of an anionic substance. In Aeromonas veronii biovar sobria BC88, similar filamentous structures were suggested to be bundle-forming pili, which mediate interactions between bacteria and initial attachment to the intestinal cells (15). However, the diameter of the individual pilus would argue against the fibrils being pili. Because these fibrils appear to connect the bacteria, they might function in signaling for communications among the bacteria rather than for structural purposes, as suggested by Dworkin for Myxococcus (8).
Stable, crenated colonies were also observed in the marine bacterium Hyphomonas strain MHS-3 when grown on marine agar at 25°C for 2 weeks. Similar to rugose colonies of V. cholerae, the cells in these colonies also produced capsular EPS for attachment to surfaces and participated in matrix formation in biofilms. Smooth regions called papillae were shown on the outer edges of the colonies in older plates (21). In addition, a wrinkled colony variation has also been reported for S. enterica serovar Enteritidis as a lacy phenotype and was shown to be associated with a cell surface matrix composed of flagellin and 35-kDa protein components that bound to LPS high-molecular-weight O antigen. The serovar Enteritidis lacy colonies were induced after 16 h of growth at 42°C and an additional 24-h incubation at 25°C on BG. Prolonged incubation for 72 h resulted in a loss of matrix from the colonies, which correlates with the loss of O antigen (11). In contrast, we observed that in S. enterica serovar Typhimurium DT104, the rugose phenotype was stable in the colonies at 25°C, except that starting around 10 days, additional growth on the outer edge of the colony was smooth and spreading, similar to the papillae of Hyphomonas (21). Furthermore, there were no apparent differences in LPS profiles between the rugose and smooth phenotype in S. enterica serovar Typhimurium DT104 (data not shown), similar to the observations made with the rugose form of V. cholerae (19, 27, 28). The lack of noticeable differences in the LPS profiles between Rv/25 and Stv as well as the apparent presence of ES further reinforces the notion of rugosity, as opposed to the formation of rough colonies, which results from the expression of truncated LPS (28).
Analogies of rugose phenotypes are found in other bacteria that produce EPSs, such as alginate-expressing Pseudomonas spp., which also produce biofilms (4). Similar to these bacteria, S. enterica serovar Typhimurium DT104 also formed biofilms in liquid media. Biofilms are known to be composed of mostly EPS with channels for water circulation. This structure enables attachment of the cells for the formation of bacterial communities to obtain nutrients and to provide protection (6). EPS was also shown to promote biofilm formation in rugose V. cholerae (27), suggesting that EPS plays an important role in the survival ability of these bacteria in aquatic environments, thus promoting the spread of this pathogenic species.
The occurrence of the phenotypic variation in S. enterica serovar Typhimurium DT104 in the natural environment remains hypothetical. However, by analogy with the conclusions drawn for V. cholerae, the aggregation of cells by the ES might provide protection for S. enterica serovar Typhimurium DT104 cells in the environment. In fact, the results from our susceptibility studies with Rv/25 and Stv showed that the ES in rugose cells appeared to have a unique function, allowing the cells to survive in the presence of acid and hydrogen peroxide and possibly other adverse conditions compared to Stv. In addition, the increasing proportion of surviving Stv cells demonstrating a phenotypic switch from smooth to rugose in the presence of acid in the later stages of incubation lends further credence to the importance of this phenotype during acid stress (Fig. 7). Typically, these cells did not exhibit the rugose phenotype until 3 days of incubation at 25°C. Although this finding might suggest the possibility of contamination, we discarded this possibility because (i) there was no evidence of rugose colonies in the 0- to 50-min plates after 5 days of incubation at 25°C and (ii) corresponding results were obtained from the three different experiments.
The unique expression of the rugose phenotype that is temperature dependent is intriguing. It is possible that ES may be more beneficial for the cells for survival in the environment outside the host. We speculate that temperature may provide cues for the cells to prevent the formation of ES in a warmer environment such as the human host, since this structure might prevent Salmonella cells from being phagocytized and multiplying inside macrophages (16). Such modification of surface polysaccharide expression has been shown in Haemophilus influenzae type b capsule (24). Studies are under way to determine if ES is associated with the virulence of the rugose cells in S. enterica serovar Typhimurium DT104. It may be possible that the cells with the rugose phenotype are better able to establish initial infection than their smooth counterparts. The rugose variant of V. cholerae can resist complement-mediated killing (14), although the virulence of rugose V. cholerae and that of smooth V. cholerae are comparable in terms of clinical response when these bacteria are introduced into human volunteers (19). Regardless of these possible functions of rugosity, there are other means for S. enterica serovar Typhimurium DT104 to survive, since this organism is also capable of transitioning to the viable-but-nonculturable state (unpublished data).
Rugosity is not a unique characteristic of S. enterica serovar Typhimurium DT104 11601, since several other DT104 strains and several other serovars of S. enterica tested also exhibited this phenotype. Therefore, the rugosity might be a more universal characteristic in Salmonella. The rugose state has been observed in O1 and non-O1 groups of V. cholerae (1, 14, 17, 19, 27, 29). Regardless of the differences observed between the formation of the rugose phenotype in S. enterica serovar Typhimurium DT104 and in V. cholerae, we now know that this condition is not exclusive to V. cholerae. This phenomenon may be more widespread in other bacteria than heretofore recognized. Hence, knowledge of rugosity may be more important to our understanding of survival of bacteria in their environments than previously appreciated.
ACKNOWLEDGMENTS
This work was supported in part by the Joint Institute for Food Safety and Applied Nutrition, University of Maryland, College Park.
We are grateful to Tim K. Maugel for expert advice on electron microscopy. We also thank Phares Okelo for assistance in serotyping the strains.
Footnotes
The electron microscopy work described herein is contribution number 96 from the Laboratory of Biological Ultrastructure at the University of Maryland, College Park.
REFERENCES
- 1.Ali A, Johnson J A, Franco A A, Metzger D J, Connell T D, Morris J G, Jr, Sozhamannan S. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect Immun. 2000;68:1967–1974. doi: 10.1128/iai.68.4.1967-1974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baggesen D L, Sandvang D, Aarestrup F M. Characterization of Salmonella enterica serovar Typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J Clin Microbiol. 2000;38:1581–1586. doi: 10.1128/jcm.38.4.1581-1586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besser T E, Gay C C, Gay J M, Hancock D D, Rice D, Pritchett L C, Erickson E D. Salmonellosis associated with S. typhimurium DT104 in the USA. Vet Rec. 1997;140:75. [PubMed] [Google Scholar]
- 4.Boyd A, Chakrabarty A M. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J Ind Microbiol. 1995;15:162–1688. doi: 10.1007/BF01569821. [DOI] [PubMed] [Google Scholar]
- 5.Cole G T. Preparation of microfungi for scanning electron microscopy. In: Aldrich H C, Todd W J, editors. Ultrastructure techniques for microorganisms. New York, N.Y: Plenum Press; 1986. pp. 1–38. [Google Scholar]
- 6.Costerton J W, Cheng K J, Geesey G G, Ladd T I, Nickel J C, Dasgupta M, Marrie T J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 7.Dannon D, Goldstein L, Marikovsky Y, Skutelsky E. Use of cationized ferritin as a label of negative charges on cell surfaces. J Ultrastruct Res. 1972;38:500–501. doi: 10.1016/0022-5320(72)90087-1. [DOI] [PubMed] [Google Scholar]
- 8.Dworkin M. Fibrils as extracellular appendages of bacteria: their role in contact-mediated cell-cell interactions in Myxococcus xanthus. Bioessays. 1999;21:590–595. doi: 10.1002/(SICI)1521-1878(199907)21:7<590::AID-BIES7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 9.Fassel T A, Edminston C E., Jr Ruthenium red and the bacterial glycocalyx. Biotech Histochem. 2000;74:194–212. doi: 10.3109/10520299909047974. [DOI] [PubMed] [Google Scholar]
- 10.Ginocchio C C, Olmsted S B, Wells C L, Galan J E. Contact with epithelial cells induces the formation of surface appendages on Salmonella Typhimurium. Cell. 1994;76:717–724. doi: 10.1016/0092-8674(94)90510-x. [DOI] [PubMed] [Google Scholar]
- 11.Guard-Petter J, Keller L H, Rahman M M, Carlson R W, Silvers S. A novel relationship between O-antigen variation, matrix formation, and invasiveness of Salmonella enteritidis. Epidemiol Infect. 1996;117:219–231. doi: 10.1017/s0950268800001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosek G, Leschinsky D, Irons S, Safranek T J. Multidrug resistant Salmonella serotype Typhimurium—United States, 1996. Morb Mortal Wkly Rep. 1997;46:308–310. [PubMed] [Google Scholar]
- 14.Johnson J A, Panigrahi P, Morris J G., Jr Non-O1 Vibrio cholerae NRT36S produces a polysaccharide capsule that determines colony morphology, serum resistance, and virulence in mice. Infect Immun. 1992;60:864–869. doi: 10.1128/iai.60.3.864-869.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirov S, Donovan L A, Sanderson K. Functional characterization of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect Immun. 1999;67:5447–5454. doi: 10.1128/iai.67.10.5447-5454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C A, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizunoe Y, Wai S N, Takade A, Yoshida S. Isolation and characterization of rugose form of Vibrio cholerae O139 strain MO10. Infect Immun. 1999;67:958–963. doi: 10.1128/iai.67.2.958-963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molbak K, Baggesen D L, Aarestrup F M, Ebbesen J M, Engberg J, Frydendahl K, Gerner-Smidt P, Peterson A M, Wegener H C. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurium DT104. N Engl J Med. 1999;341:1420–1425. doi: 10.1056/NEJM199911043411902. [DOI] [PubMed] [Google Scholar]
- 19.Morris J G, Jr, Sztein M B, Rice E W, Nataro J P, Losonsky G A, Panigrahi P, Tacket C O, Johnson J A. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J Infect Dis. 1996;174:1364–1368. doi: 10.1093/infdis/174.6.1364. [DOI] [PubMed] [Google Scholar]
- 20.O'Toole G A, Pratt L A, Watnick P I, Newman D K, Weaver V B, Kolter R. Genetic approaches to study biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 21.Quintero E J, Weiner R M. Evidence for the adhesive function of the exopolysaccharide of Hyphomonas strain MHS-3 in its attachment to surfaces. Appl Environ Microbiol. 1995;61:1897–1903. doi: 10.1128/aem.61.5.1897-1903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice E W, Johnson C J, Clark R M, Fox K R, Reasoner D J, Dunnigan M E, Panigrahi P, Johnson J A, Morris J G., Jr Chlorine and survival of “rugose” Vibrio cholerae. Lancet. 1992;340:740. doi: 10.1016/0140-6736(92)92289-r. [DOI] [PubMed] [Google Scholar]
- 23.Sameshima T, Akiba M, Izumiya H, Terajima J, Tamura K, Watanabe H, Nakazawa M. Salmonella typhimurium DT104 from livestock in Japan. Jpn J Infect Dis. 2000;53:15–16. [PubMed] [Google Scholar]
- 24.St Geme J W, 3rd, Cutter D. Influence of pili, fibrils, and capsule on in vitro adherence by Haemophilus influenzae type b. Mol Microbiol. 1996;21:21–31. doi: 10.1046/j.1365-2958.1996.6241331.x. [DOI] [PubMed] [Google Scholar]
- 25.Threlfall E J, Frost J A, Ward L R, Rowe B. Epidemic in cattle and humans of Salmonella typhimurium DT104 with chromosomally integrated multiple drug resistance. Vet Rec. 1994;134:577. doi: 10.1136/vr.134.22.577. [DOI] [PubMed] [Google Scholar]
- 26.Threlfall E J, Frost J A, Ward L R, Rowe B. Increasing spectrum of resistance in multiresistant Salmonella typhimurium. Lancet. 1996;347:1053–1054. doi: 10.1016/s0140-6736(96)90199-3. [DOI] [PubMed] [Google Scholar]
- 27.Wai S N, Mizunoe Y, Takade A, Kawabata S I, Yoshida S I. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White P B. The rugose variant of vibrios. J Pathol Bacteriol. 1938;46:1–6. [Google Scholar]
- 29.Yildiz F H, Schoolnik G K. Vibrio cholerae El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]