Abstract
The tecB gene, located downstream of tecA and encoding tetrachlorobenzene dioxygenase, in Ralstonia sp. strain PS12 was cloned into Escherichia coli DH5α together with the tecA gene. The identity of the tecB gene product as a chlorobenzene dihydrodiol dehydrogenase was verified by transformation into the respective catechols of chlorobenzene, the three isomeric dichlorobenzenes, as well as 1,2,3- and 1,2,4-trichlorobenzenes, all of which are transformed by TecA into the respective dihydrodihydroxy derivatives. Di- and trichlorotoluenes were either subject to TecA-mediated dioxygenation (the major or sole reaction observed for the 1,2,4-substituted 2,4-, 2,5-, and 3,4-dichlorotoluenes), resulting in the formation of the dihydrodihydroxy derivatives, or to monooxygenation of the methyl substituent (the major or sole reaction observed for 2,3-, 2,6-, and 3,5-dichloro- and 2,4,5-trichlorotoluenes), resulting in formation of the respective benzyl alcohols. All of the chlorotoluenes subject to dioxygenation by TecA were transformed, without intermediate accumulation of dihydrodihydroxy derivatives, into the respective catechols by TecAB, indicating that dehydrogenation is no bottleneck for chlorobenzene or chlorotoluene degradation. However, only those chlorotoluenes subject to a predominant dioxygenation were growth substrates for PS12, confirming that monooxygenation is an unproductive pathway in PS12.
Chlorotoluenes are important intermediates in many chemical processes and are still produced in large amounts (9, 28). Dichlorotoluenes, known as moderately toxic chemicals (2, 3, 10), are used as precursors for the production of pesticides, dyes, and peroxides (9). Their high chemical stability causes their accumulation in the environment (10).
Only a few reports on the degradation of chlorotoluenes have appeared. Whereas organisms capable of mineralizing 3- or 4-chlorotoluene have been described in detail (8, 11), degradation routes for dichlorotoluenes have not been analyzed. Vandenbergh et al. described a pseudomonad able to utilize several chloraromatics, including 2,4-dichlorotoluene (24DCT) and 3,4-dichlorotoluene (34DCT), but no further indications of the metabolic pathway were given (33). Ralstonia sp. strain PS12 (formerly Pseudomonas, Burkholderia), capable of mineralizing various chlorobenzenes, e.g., 1,2,4,5-tetrachlorobenzene, was also reported to mineralize various dichlorotoluenes, again without indication of a metabolic sequence (5, 27).
Two distinct metabolic routes were reported for the mineralization of 4-chlorotoluene. Brinkmann et al. (8) constructed bacterial strains mineralizing 4-chlorotoluene (as well as 3-chlorotoluene and 3,5-dichlorotoluene [35DCT]) via the corresponding chlorinated benzoates and catechols by combining the TOL plasmid (12) with genes encoding enzymes of the chlorocatechol pathway (25). However, 2-chloro-substituted toluenes were no substrates for the TOL plasmid-encoded xylene monooxygenase; thus, they cannot be degraded by such a catabolic route. In Pseudomonas sp. strain JS6, 4-chlorotoluene is subject to dioxygenation and 3-chloro-6-methylcatechol is formed after dehydrogenation of the intermediate dihydrodiol (11). Enzymes of the chlorocatechol degradation pathway are responsible for further metabolism of this compound. Ralstonia sp. strain PS12 (27) was shown to be capable of growing with several chlorinated benzenes and, similar to JS6, to initialize the degradation by dioxygenolytic activation (5). The broad substrate spectrum tetrachlorobenzene dioxygenase TecA of PS12, catalyzing this initial step, has been extensively described (5, 6). The most prominent feature of this enzyme system is its ability to transform 1,2,4,5-tetrachlorobenzene and thereby to attack a chloro-substituted carbon atom. Such an attack results in the formation of an unstable diol intermediate which spontaneously rearomatizes with concomitant chloride elimination. Like the lower substituted chlorobenzenes, 4-chlorotoluene, which is used as a growth substrate by PS12, is subject to dioxygenation at two unsubstituted carbon atoms. In contrast, 2- and 3-chlorotoluenes were subject to monooxygenation of the methyl substituents with 2- and 3-chlorobenzyl alcohols as the main products (18).
Apparently, TecA can catalyze dioxygenating, dechlorinating, and monooxygenating reactions. In the present study, we analyzed which of these three types of reaction was performed with higher chloro-substituted toluenes and which of those substrates can be transformed by PS12-derived activities into chlorocatechols as central intermediates of chloroaromatic degradation.
MATERIALS AND METHODS
Organisms and culture conditions.
Escherichia coli DH5α(pSTE7), containing the tecA tetrachlorobenzene dioxygenase gene (5), and E. coli DH5α(pSTE44), containing the tetrachlorobenzene dioxygenase and dehydrogenase genes tecAB, were grown at 37°C in Luria broth medium containing ampicillin at 0.1 mg/ml and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
Cloning of the tecAB genes.
First, the 3′ end of the tecB gene was PCR amplified from template plasmid pCR12, containing the region located downstream of the tecA gene (7), with the primers prSTB86 (5′-CGTTCTCTACACCGCGGGC-3′) and prSTB68 (5′-GCCTTTGAGAGCTCATGTTGTC-3′), the latter containing an artificial SacI site (in boldface). For sequence confirmation, the resulting 0.4-kb fragment was cloned into plasmid pCR2.1 (Invitrogen), resulting in pCR17. A 0.3-kb BamHI-SacI fragment of pCR17 was then cloned into the BamHI-SacI site of pSTE7 encoding TecA, resulting in pSTE44 carrying the tecA and tecB genes.
Sequence analysis.
Plasmid DNA for sequencing was extracted with the Plasmid Maxi kit (Qiagen). Sequencing reactions on both strands were performed with the Applied Biosystems 373A DNA sequencer in accordance with the protocol of the manufacturer (Perkin-Elmer, Applied Biosystems) for Taq cycle sequencing with fluorescent-dye-labeled dideoxynuleotides, as described previously (14). The GeneWorks software package V2.45 (IntelliGenetics) was used for sequence evaluation. Similarity searches of nonredundant databases were done with the FASTA program. Sequence comparisons and calculations of evolutionary distances were carried out by using the PILEUP, DISTANCE, and UPGMA programs of the GCG software (Wisconsin Package, version 8; Genetics Computer Group, Madison, Wis.).
Resting cell assays.
Resting cell assays were performed as described by Beil et al. (5). For kinetic experiments, at each time point, 400-μl aliquots were removed and shock frozen in liquid nitrogen. The samples were stored at −20°C for subsequent analyses.
Extraction and derivatization of metabolites.
Metabolites were extracted as described previously (5). For subsequent gas chromatography-mass spectrometry (GC-MS) analysis, dihydrodiol and catechol intermediates were derivatized with butylburonic acid whereas benzyl alcohols were derivatized with Me3SOH (5).
Analytical methods.
For high-performance liquid chromatography (HPLC) analyses of metabolites, 10-μl samples were injected after removal of cells by centrifugation (20°C, 10 min, 15,000 × g). Product formation was analyzed with a Shimadzu HPLC system (LC-10AD liquid chromatograph, DGU-3A degasser, SPD-M10A diode array detector, and FCV-10AL solvent mixer) equipped with an SC125/Lichrospher 5-μm (Bischoff, Leonberg, Germany) column. The aqueous solvent system (flow rate, 1 ml/min) contained 0.1% (vol/vol) H3PO4 (87%) and 50 or 58% (vol/vol) methanol for the determination of metabolites or 80% (vol/vol) methanol for the determination of substrates. The alcohol intermediates were identified and quantified by comparison with authentic standards.
GC-MS analyses were performed as previously described (5).
For nuclear magnetic resonance (NMR) analyses, the extracted metabolites (about 2 mg) were dissolved in 1 ml of D6-acetone. 1H and 13C NMR spectra were recorded on a Bruker CXP 300 (Bruker, Rheinstetten, Germany) with Aspect 2000 software using tetramethylsilane as the internal standard.
Chemicals.
35DCT and 2,4,5-trichlorotoluene (245TCT) were synthesized and kindly provided by W. Reineke. Butylburonic acid was obtained from Acros organics (Geel, Belgium), trimethylsulfonium hydroxide was from Machery-Nagel (Dueren, Germany), 2,3-dichlorobenzyl alcohol was from TCI, and 3-chloro-, 4-chloro-, 3,4-dichloro-, and 3,4,5-trichlorocatechols were from Helix Biotech 3,5-, 3,6-, and 4,5-Dichlorocatechols were kindly provided by H.-A. Arfmann. All other chemicals were purchased from Aldrich Chemie (Steinheim, Germany), Fluka AG (Buchs, Switzerland), or Merck AG (Darmstadt, Germany).
Nucleotide sequence accession number.
The nucleotide sequence data of the TecB cis-chlorobenzene dihydrodiol dehydrogenase have been submitted to the GenBank sequence data bank and are available under accession number U78099.
RESULTS
The tecB gene encodes a chlorobenzene dihydrodiol dehydrogenase.
The deduced amino acid sequence of the 0.83-kb open reading frame downstream of the tecA gene, designated tecB, shows the greatest similarity (>99%) to the TcbB cis-chlorobenzene dihydrodiol dehydrogenase from Pseudomonas sp. strain P51 (34) and shows the typical features of the short-chain alcohol dehydrogenase family (23), including the short-chain alcohol dehydrogenase consensus pattern (23) and the binding site for the ADP moiety of the NAD+ coenzyme (4, 30). To verify that tecB encodes a chlorobenzene dihydrodiol dehydrogenase, the TecA dioxygenase and the tecB gene product were simultaneously produced in E. coli(pSTE44) cells, and the transformation of various chlorinated benzenes was analyzed by HPLC.
All of the chlorobenzenes analyzed and shown to be transformed into the respective dihydrodiols by TecA were converted by TecA plus TecB into the corresponding aromatic dihydroxy compounds (Table 1). The identities of the products were confirmed by comparison with authentic standards. A single catechol product was observed in each case, except for chlorobenzene transformation, where 3-chlorocatechol was the major product but 4-chlorocatechol was formed in minor amounts (Table 1). This indicates that TecB is the second enzyme in the degradative pathway of (chloro)benzenes and encodes a chlorobenzene dihydrodiol dehydrogenase. Accumulation of dihydrodiols was not detected, except for minute amounts in the case of 1,4-dichlorobenzene and 1,2,4-trichlorobenzene transformations. Thus, TecB dehydrogenase constitutes no pathway bottleneck for the transformation of products from TecA catalysis into the corresponding aromatic intermediates.
TABLE 1.
Retention volumes and absorption maxima in HPLC analyses of products formed by E. coli(pSTE7) and E. coli(pSTE44) cells from chlorobenzenes and di- and trichlorotoluenes
| Substrate | Proposed product(s) | [MeOH] (%)a | RV (ml)b | λmax (nm)c | Growth substrated |
|---|---|---|---|---|---|
| Chlorobenzene | 3-Chloro-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-dienea | 36 | 3.4 | 272 | + |
| 4-Chloro-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-dienee | 36 | 4.4 | 265 | ||
| 3-Chloro-1,2-dihydroxybenzenef | 58 | 2.6 | 210 | ||
| 4-Chloro-1,2-dihydroxybenzenef | 58 | 2.9 | 210 | ||
| 1,2-Dichlorobenzene | 3,4-Dichloro-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-dienee | 36 | 6.8 | 272 | + |
| 3,4-Dichloro-1,2-dihydroxybenzenef | 58 | 4.2 | 210 | ||
| 1,3-Dichlorobenzene | NDg | ||||
| 3,5-Dichloro-1,2-dihydroxybenzenef | 58 | 5.3 | 210 | + | |
| 1,4-Dichlorobenzene | 3,6-Dichloro-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-dieneef | 36 | 4.1 | 283 | + |
| 3,6-Dichloro-1,2-dihydroxybenzenee | 58 | 4.0 | 210 | ||
| 1,2,3-Trichlorobenzene | 3,4,5-Trichloro-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-dienee | 72 | 2.3 | 282 | − |
| 3,4,5-Trichloro-1,2-dihydroxybenzenef | 58 | 8.6 | 210 | ||
| 1,2,4-Trichlorobenzene | 3,4,6-Trichloro-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-dieneef | 58 | 3.4 | 288 | + |
| 3,4,6-Trichloro-1,2-dihydroxybenzenef | 58 | 8.3 | 210 | ||
| 1,2,4,5-Tetrachlorobenzene | 3,4,6-Trichloro-1,2-dihydroxybenzeneef | 58 | 8.3 | 210 | + |
| 23DCT | 2,3-Dichlorobenzylalcoholef | 60 | 5.4 | 201 | − |
| 24DCT | 2,4-Dichlorobenzylalcoholef | 60 | 6.2 | 201 | + |
| 4,6-Dichloro-3-methyl-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-dienee | 60 | 3.7 | 278 | ||
| 4,6-Dichloro-3-methyl-1,2-dihydroxybenzenef | 60 | 7.5 | 205 | ||
| 25DCT | 2,5-Dichlorobenzylalcoholef | 60 | 6.1 | 201 | + |
| 3,6-Dichloro-4-methyl-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-dienea | 60 | 2.9 | 280 | ||
| 3,6-Dichloro-4-methyl-1,2-dihydroxybenzenef | 60 | 6.0 | 204 | ||
| 26DCT | 2,6-Dichlorobenzylalcoholef | 60 | 3.9 | 203 | − |
| 3,5-Dichloro-4-methyl-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5.dienee | 60 | 3.7 | 275 | ||
| 3,5-Dichloro-4-methyl-1,2-dihydroxybenzenef | 60 | 6.3 | 203 | ||
| 34DCT | 3,5-Dichloro-6-methyl-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-dienee | 60 | 3.5 | 276 | + |
| 3,4-Dichloro-6-methyl-1,2-dihydroxybenzenef | 60 | 6.6 | 204 | ||
| 35DCT | 3,5-Dichlorobenzylalcoholef | 60 | 7.4 | 203 | − |
| 245DCT | 2,4,5-Trichlorobenzylalcoholef | 60 | 12.1 | 205 | − |
[MeOH], percentage of methanol used in the solvent system.
RV, retention volume.
λmax, wavelength of maximal absorption.
+, used as a growth substrate; −, not used as a growth substrate (27).
Proposed product derived by transformation of the indicated substrate with E. coli-(pSTE7) producing TecA dioxygenase.
Proposed product derived by transformation of the indicated substrate by E. coli(pSTE44) simultaneously producing TecA tetrachlorobenzene dioxygenase and TecB dehydrogenase.
ND, not done for E. coli(pSTE44).
Transformation of di- and trichlorotoluenes by TecA and TecB.
The potential of TecA tetrachlorobenzene dioxygenase to transform 2,3-dichlorotoluene (23DCT), 24DCT, 2,5-dichlorotoluene (25DCT), 2,6-dichlorotoluene (26DCT), 34DCT, 35DCT, and 245TCT was analyzed initially by HPLC using supernatant fluid of resting E. coli cells carrying plasmid pSTE7 incubated with the respective substrates. HPLC analysis revealed the formation of products from all of the substrates tested (Table 1). Formation of the respective benzyl alcohols from dichlorotoluenes could be directly verified by comparison of retention volumes and UV absorption spectra with those of authentic standards. The corresponding benzyl alcohol(s) was the only product formed from 23DCT and 35DCT, the apparent major product formed from 26DCT, and the apparent minor product from 24DCT and 25DCT and was obviously not produced from 34DCT.
The UV spectra of the other products formed during dichlorotoluene turnover were indicative for the formation of dihydrodiols (λmax = 275 to 280 nm). The only product formed during 245TCT transformation did not exhibit such UV spectrum.
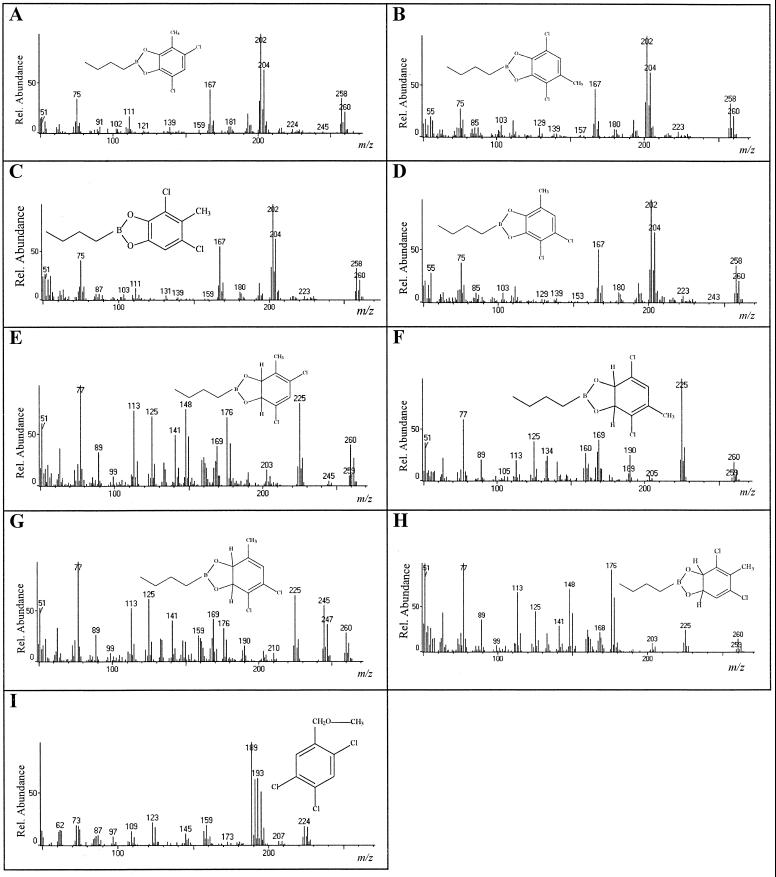
Confirmation of the identities of the intermediates formed from dichlorotoluenes as the corresponding dihydrodiols was obtained by GC-MS analysis of the boronated derivatives. Whereas no such products were detected from 23DCT and 35DCT, prominent signals showing the expected molecular ion of m/z 260, 262, and 264 (relative intensities, 100:62:9) were observed from 34DCT, 24DCT, 25DCT, and 26DCT. The products showed the fragmentation pattern typical for n-butylboronated chlorobenzene or methylbenzene dihydrodiols (5, 16), i.e., loss of C4H9 [M-57]+, O-B-C4H9 [M-84]+, as well as loss of one or two chlorine atoms [M-35]+ (Fig. 1).
FIG. 1.
Mass spectra of the boronated (A to H) or methylated (I) products formed by E. coli(pSTE44) (A to D) and E. coli(pSTE7) (E to I) from di- and trichlorotoluenes. A, 4,6-dichloro-3-methylcatechol; B, 3,6-dichloro-4-methylcatechol; C, 3,5-dichloro-4-methylcatechol; D, 3,4-dichloro-6-methylcatechol; E, 4,6-dichloro-3-methyl-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-diene; F, 3,6-dichloro-4-methyl-1,2,-dihydroxy-1,2-dihydrocyclohexa-3,5-diene; G, 3,5-dichloro-4-methyl-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-diene; H, 3,5-dichloro-6-methyl-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-diene; I, 2,4,5-trichlorobenzyl alcohol. Rel. relative.
1H and 13C NMR data of the product formed after transformation of 245TCT indicated the formation of 2,4,5-trichlorobenzyl alcohol [1H NMR, δ = 7.76 ppm (bs, H-6), 7.62 ppm (bs, H-3), 4.68 ppm (bs, 2 H, CH2); 13C NMR, δ = 141.6 ppm (s, C-1), 131.6 ppm (s, C-4), 131.4 ppm (2∗s, C-2/C-5), 131.1 ppm, 129.9 ppm (2∗d, C-3/C-6), 61.0 ppm (t, C, CH2)] due to a monooxygenolytic attack by TecA. GC-MS analysis of the methylated product confirmed the identity of the product as 2,4,5-trichlorobenzyl alcohol (Fig. 1). Molecular ion signals at m/z 224, 226, 228, and 230 (relative intensities, 100:98:31:4) showed the expected mass peak of the benzyl alcohol with its three chlorines. Intense signals at m/z 193 or 189 resulted from the loss of O-CH3 [M-31]+ or a chlorine atom [M-35]+. Analysis of the boronated extract gave no indication for the formation of dihydroxylated products. Thus, attack on 245TCT is clearly different from that on 1,2,4,5-tetrachlorobenzene, where dioxygenolytic attack on an unsubstituted and a chlorosubstituted carbon atom leads to an unstable intermediate, which spontaneously rearranges to form 3,5,6-trichlorocatechol.
The HPLC analyses of TecA-plus-TecB-mediated transformation of dichlorotoluenes showed, as expected, no difference in the amounts of benzyl alcohols formed, compared to TecA-mediated transformation. However, in no case was accumulation of dihydrodiols detected, but new products were observed to be formed from 34DCT (only product), 24DCT and 25DCT (major product), and 26DCT (minor product). GC-MS analysis of the boronated derivatives confirmed the hypothesis that those products are identical to dichloro-substituted methylcatechols (Fig. 1). The recorded intensities of molecular ion signals at m/z 258, 260, and 262 were always in close agreement with the theoretical pattern for dichloro-substituted catechols. Intense signals at m/z 202, 204, and 206 result from the loss of butene [M-C4H8]+, as described for the butylboronates of 3-chlorocatechol, 3,6-dichlorocatechol (16), and 3,4,6-trichlorocatechol (5). To our knowledge, this is the first report about MS spectra of boronated dichloromethyl dihydrodiols, dichloro-substituted methylcatechols, and methylated 2,4,5-trichlorobenzyl alcohol.
Quantification of product ratios and product formation rates for TecA- and TecB-catalyzed turnover of di- and trichlorotoluenes.
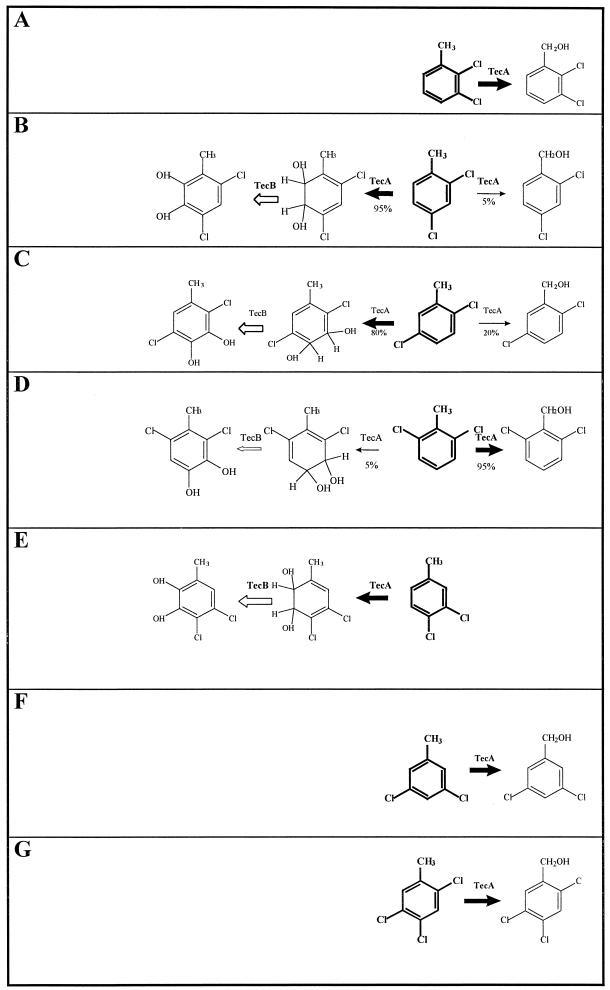
Dichlorobenzyl alcohols are available commercially, and their production rate could thus be quantified. No standards are available for 2,4,5-trichlorobenzyl alcohol, dihydrodiols, or dichloromethylcatechols; therefore, neither product formation rates nor the ratio in which dioxygenation versus monooxygenation occurred could be quantified. However, as the absorption at λ = 210 nm of all dichloro-substituted catechols varied by a factor of only 0.05, the absorption of dichloromethylcatechols can be assumed to be similar. Under this assumption, the dichloromethylcatechols can be proposed to constitute 5% ± 0.25% of the products formed during the transformation of 26DCT, 80% ± 4% of those formed during the transformation of 25DCT, and 90% ± 4.5% of those formed during the transformation of 24DCT (Fig. 2).
FIG. 2.
Proposed transformation of different di- and trichlorotoluenes by tetrachlorobenzene dioxygenase TecA and dehydrogenase TecB. Substrates: A, 23DCT; B, 24DCT; C, 25DCT; D, 26DCT; E, 34DCT; F, 35DCT; G, 245TCT. ➞, major reaction catalyzed by TecA; →, minor reaction catalyzed by TecA; ➩, reaction catalyzed by TecB.
To verify the above assumption, the ratio of monooxygenation versus dioxygenation was quantified by 1H NMR analysis in the case of TecA- and TecB-catalyzed 25DCT transformation. Analysis of the product mixture showed the expected signals for 2,5-dichlorobenzyl alcohol [1H NMR, δ = 7.63 ppm (dt, H-6, JH-6,H-4 2.6 Hz, JH-6,H-7 1.3 Hz), 7.39 ppm (d, H-3, JH-3,H-4 8.6 Hz), 7.31 ppm (ddt, H-4, JH-4,H-7 0.6 Hz), 4.70 ppm (bs, H-7)] and 3,6-dichloro-4-methylcatechol [1H NMR, δ = 8.31 ppm, 8.40 ppm (3-OH, 4-OH), 6.84 ppm (bs, H-6), 2.25 ppm (d, H-7, JH-6,H-7 0.7 Hz)]. Comparison of the integrals of the signals of the respective H-6 protons showed that these products were present in the mixture at a 1:4 ratio. This confirms the previous result of 80% formation of 3,6-dichloro-4-methylcatechol during transformation of 25DCT. For quantification of 2,4,5-trichlorobenzyl alcohol, 245TCT was transformed by TecAB. The product prepared for 1H NMR analysis was spiked with a defined concentration of 2,3-dichlorobenzaldehyde (used as an internal standard); thus, the concentration of 2,4,5-trichlorobenzyl alcohol in the mixture could be determined by 1H NMR analysis. This sample served as an HPLC standard.
The transformation rates, expressed as the amount of product formed per time unit, were of the same order of magnitude as that of 1,2,4,5-tetrachlorobenzene (5) (Table 2). The product formation rates of dichlorotoluenes that were mainly subject to dioxygenation were significantly higher than those of dichlorotoluenes that were subject to monooxygenation (Table 2). The transformation rate of 245TCT was about threefold the rate observed with 1,2,4,5-tetrachlorobenzene and two- to threefold the rate observed with dichlorotoluenes. Comparisons of the product formation rates as catalyzed by E. coli(pSTE7) and E. coli(pSTE44) showed that the rates of E. coli(pSTE7) expressing TecA were twofold higher than the rates of E. coli(pSTE44) expressing TecAB (Table 2).
TABLE 2.
Absolute and relative rates of transformation of chlorinated toluenes and tetrachlorobenzene catalyzed by E. coli DH5α(pSTE7) and E. coli DH5α (pSTE44)
| Substrate | Product formation (μM/min) (relative transformation rate [%])
|
|
|---|---|---|
| pSTE7 | pSTE44 | |
| 245 TCT | 3.7 (100) | 1.9 (100) |
| 1,2,4,5-Tetrachlorobenzenea | 1.0 (27) | NDb |
| 23DCT | 1.3 (35) | 0.6 (32) |
| 24DCT | ND | 1.0 (53) |
| 25DCT | ND | 1.0 (53) |
| 26DCT | 1.3 (35) | 0.7 (37) |
| 34DCT | ND | 1.3 (68) |
| 35DCT | 1.4 (38) | 0.6 (32) |
The data are from Beil et al. (5). All rates were determined at a cell density producing an A600 of 1.8.
ND, not done.
DISCUSSION
Dioxygenation, as carried out by multicomponent dioxygenases, is the initial step in the degradation of various aromatic compounds and commonly results in the formation of the respective cis-dihydrodiols, which are further transformed into dihydroxy aromatics by the action of cis-dihydrodiol dehydrogenases (20, 26, 29). Among these dioxygenases, naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4 and toluene dioxygenase (TOD) from Pseudomonas putida F1 have been extensively studied (17, 31, 35, 36). These enzymes were reported to have relaxed substrate specificities and, in addition to a stereospecific cis-dihydroxylation, they catalyze monooxygenation, desaturation, dealkylation, and sulfoxidation reactions (21). The TecA tetrachlorobenzene dioxygenase of Ralstonia sp. strain PS12 carries out cis-dihydroxylations as well and has been shown to transform toluene and various chlorinated benzenes in a dioxygenolytic manner (5). The enzyme differs from the TOD of P. putida F1 in its failure to attack benzene but is superior in its ability to transform 1,2,4,5-tetrachlorobenzene. In addition to chlorobenzenes, TecA dioxygenase, like TOD, also transforms 4-chlorotoluene by a dioxygenolytic attack, whereas 2- and 3-chlorotoluenes were subject mainly to monooxygenation of the methyl side chain (18). All of the dichlorotoluenes recently indicated to be growth substrates for Ralstonia sp. strain PS12, i.e., 24DCT, 25DCT, and 34DCT, were mainly or exclusively subject to dioxygenation (Fig. 2). It is interesting that obviously all 1,2,4-substituted dichlorotoluenes are subject to mainly dioxygenation (like 1,4-disubstituted 4-chlorotoluene), whereas chlorinated toluenes with substitutions at the 1,2-, 1,3- 1,2,3-, or 1,3,5-positions were subject to mainly monooxygenation (Fig. 2). As already outlined by Sander et al. (27), 1,3,5-trichlorobenzene is not subject to dioxygenolytic attack by PS12; thus, a dioxygenolytic attack on 35DCT could be restricted for similar yet unknown reasons. Surprisingly, 245TCT was also exclusively transformed into the corresponding benzyl alcohol, even though a dioxygenation followed by spontaneous chloride elimination, as observed for the structural analogue 1,2,4,5-tetrachlorobenzene (5, 27), was expected. As differences in the electrical properties of the different chlorinated toluenes seem to be negligible for the specificity of the attack, it is more likely that the reaction depends on the position of the substrate with respect to the active site and on the active site's structure. It has been reported that even small amino acid sequence differences lead to major differences in enzymatic properties, such as substrate range and regioselectivity (19). The 2-nitrotoluene dioxygenase from Pseudomonas sp. strain JS42 (1) catalyzes predominantly a dioxygenation of 2-nitrotoluene, whereas the closely related 2,4-dinitrotoluene dioxygenase from Burkholderia sp. strain DNT (32) catalyzes monooxygenation. TOD of P. putida F1, like dinitrotoluene dioxygenase, catalyzes a monooxygenation of 2-nitrotoluene, whereas 4-nitrotoluene is subject to dioxygenation. Evidently, regiospecificity depends in a complex fashion on both the substitution pattern and the active-site structure. Based on its crystal structure, several amino acids were identified near the active site of naphthalene dioxygenase (15) and this information has been used to identify amino acids that control the regioselectivity and enantioselectivity of naphthalene dioxygenase. However, only poor information is available on the amino acids governing the selectivity of benzyl alcohol versus cis-dihydrodiol formation (22).
All of the cis-dihydrodiols shown to be formed by TecA dioxygenase in the present study are obviously transformed at a high rate by the TecB cis-dihydrodiol dehydrogenase. TecB evidently does not constitute a bottleneck for chlorobenzene or chlorotoluene transformation. The tecB gene product, belonging to the family of short-chain alcohol dehydrogenases (23), is closely related to the TcbB cis-chlorobenzene dihydrodiol dehydrogenase from Pseudomonas sp. strain 51 (34), an enzyme recently described to be of broad substrate specificity (24). However, it still remains to be elucidated whether those gene products, like the respective tcbA and tecA gene products, are specifically adapted for the metabolism of chloro-substituted derivatives.
Dichlorotoluenes subject to monooxygenation of the methyl function cannot be used by PS12 as growth substrates (27). This is similar to the observation that 2- and 3-chlorotoluenes cannot be mineralized by this strain (18). The growth failure was attributed to further oxidation of the benzyl alcohols produced into the corresponding benzoates at rates too low to support growth and, in the case of 2-chlorobenzoate, to the restricted substrate range of the probably chromosomally encoded benzoate dioxygenase. Like 2-chlorobenzyl alcohol, 2,3-, 2,4-, 2,5-, and 2,6-dichlorobenzyl alcohols are transformed by PS12 into the corresponding benzoates, which cannot be mineralized by PS12 due to, at least, the absence of a broad-spectrum 2-halobenzoate dioxygenase (data not shown). Thus, monooxygenation in this strain results in the channeling of all (in case of 23DCT), the major part (26DCT), or a minor part of the substrate into an unproductive pathway, explaining the failure of the strain to grow with 23DCT and 26DCT. Similarly, no effective pathway is present for 35DCT degradation. Complementation of a degradative pathway for 2-chlorotoluene by recruitment of different pathway modules from various bacteria has been proposed recently (18), and respective experiments have recently been performed (13). This strategy can be adapted, at least theoretically, for the mineralization of dichlorotoluenes. Alternatively, the search for new chlorobenzene dioxygenases with new substrate specificities and regioselectivities of attack, as well as protein engineering strategies, will lead to the isolation of enzymes with capabilities more suited to dichlorotoluene mineralization.
ACKNOWLEDGMENTS
This work was supported by contract BIO4-CT972040 of the BIOTECH program of the EC.
We thank Victor Wray for helpful discussion of the NMR data.
REFERENCES
- 1.An D, Gibson D T, Spain J C. Oxidative release of nitrite from 2-nitrotoluene by a three-component enzyme system from Pseudomonas sp. strain. JS42. J Bacteriol. 1994;176:7462–7467. doi: 10.1128/jb.176.24.7462-7467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2,4-Dichlortoluol. Toxikologische Bewertung. Heidelberg, Germany: Berufsgenossenschaft der chemischen Industrie; 1995. [Google Scholar]
- 3.Anonymous. 3,4-Dichlortoluol. Toxikologische Bewertung. Heidelberg, Germany: Berufsgenossenschaft der chemischen Industrie; 1995. [Google Scholar]
- 4.Baker M E. Sequence similarity between Pseudomonas dihydrodiol dehydrogenase, part of the gene cluster that metabolizes polychlorinated biphenyls, and dehydrogenases involved in the metabolism of ribitol and glucidol and synthesis of antibiotics and 17b-oestradiol, testosterone and corticosterone. Biochem J. 1990;267:839–841. doi: 10.1042/bj2670839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beil S, Happe B, Timmis K N, Pieper D H. Genetic and biochemical characterization of the broad spectrum chlorobenzene dioxygenase from Burkholderia sp. strain PS12—dechlorination of 1,2,4,5-tetrachlorobenzene. Eur J Biochem. 1997;247:190–199. doi: 10.1111/j.1432-1033.1997.00190.x. [DOI] [PubMed] [Google Scholar]
- 6.Beil S, Mason J R, Timmis K N, Pieper D H. Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J Bacteriol. 1998;180:5520–5528. doi: 10.1128/jb.180.21.5520-5528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beil S, Timmis K N, Pieper D H. Genetic and biochemical analyses of the tec operon suggest a route for evolution of chlorobenzene degradation genes. J Bacteriol. 1999;181:341–346. doi: 10.1128/jb.181.1.341-346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinkmann U, Reineke W. Degradation of chlorotoluenes by in vivo constructed hybrid strains: problems of enzyme specificity, induction and prevention of meta-pathway. FEMS Microbiol Lett. 1992;75:81–87. doi: 10.1016/0378-1097(92)90460-6. [DOI] [PubMed] [Google Scholar]
- 9.Drotleff J, Fluthwedel A, Pohle H, Spilok K. Handbuch Chlorchemie. II. Umweltbundesamt, Berlin, Germany: Ausgewählte Produktlinien.; 1992. [Google Scholar]
- 10.Gerhartz W. Ceramic to chlorohydrins. In: Gerhartz W, editor. Ullmann's encyclopedia of industrial chemistry. A6. Weinheim, Germany: VCH; 1986. pp. 344–347. [Google Scholar]
- 11.Haigler B E, Spain J C. Degradation of p-chlorotoluene by a mutant of Pseudomonas sp. strain JS6. Appl Environ Microbiol. 1989;55:372–379. doi: 10.1128/aem.55.2.372-379.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harayama S, Rekik M, Wubbolts M, Rose K, Leppik R A, Timmis K N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989;171:5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haro M, de Lorenzo V. Metabolic engineering of bacteria for environmental applications: construction of Pseudomonas strains for biodegradation of 2-chlorotoluene. J Biotechnol. 2001;85:103–113. doi: 10.1016/s0168-1656(00)00367-9. [DOI] [PubMed] [Google Scholar]
- 14.Karlson U, Rojo F, van Elsas J D, Moore E. Genetic and serological evidence for the recognition of four pentachlorophenol-degrading bacterial strains as a species of the genus Sphingomonas. Syst Appl Microbiol. 1995;18:539–548. [Google Scholar]
- 15.Kauppi B, Lee K, Carredano E, Parales R E, Gibson D T, Eklund H, Ramaswamy S. Structure of an aromatic-ring-hydroxylating dioxygenase—naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 16.Kirsch N H, Stan H-J. Gas chromatographic-mass spectrometric determination of chlorinated cis-1,2-dihydroxycyclohexadienes and chlorocatechols as their boronates. J Chromatogr. 1994;A684:277–287. [Google Scholar]
- 17.Kurkela S, Lehvaslaiho H, Palva E T, Teeri T H. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene. 1988;73:355–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- 18.Lehning A, Fock U, Wittich R-M, Timmis K N, Pieper D H. Metabolism of chlorotoluenes by Burkholderia sp. strain PS12 and toluene dioxygenase of Pseudomonas putida F1: evidence for monooxygenation by toluene and chlorobenzene dioxygenases. Appl Environ Microbiol. 1996;63:1974–1979. doi: 10.1128/aem.63.5.1974-1979.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mondello F J, Turcich M P, Lobos J H, Erickson B D. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl Environ Microbiol. 1997;63:3096–3103. doi: 10.1128/aem.63.8.3096-3103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino S F, Spain J C, Belcher L A, Litchfield C D. Chlorobenzene degradation by bacteria isolated from contaminated groundwater. Appl Environ Microbiol. 1992;58:1719–1726. doi: 10.1128/aem.58.5.1719-1726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parales R E, Emig M D, Lynch N A, Gibson D T. Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J Bacteriol. 1998;180:2337–2344. doi: 10.1128/jb.180.9.2337-2344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parales R E, Lee K, Resnick S M, Jiang H, Lessner D J, Gibson D T. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J Bacteriol. 2000;182:1641–1649. doi: 10.1128/jb.182.6.1641-1649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Persson B, Krook M, Jornvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991;200:537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- 24.Raschke H, Fleischmann T, Van Der Meer J R, Kohler H-P E. cis-Chlorobenzene dihydrodiol dehydrogenase (TcbB) from Pseudomonas sp. strain P51, expressed in Escherichia coli DH5α(pTCB149), catalyzes enantioselective dehydrogenase reactions. Appl Environ Microbiol. 1999;65:5242–5246. doi: 10.1128/aem.65.12.5242-5246.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reineke W. Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu Rev Microbiol. 1998;52:287–331. doi: 10.1146/annurev.micro.52.1.287. [DOI] [PubMed] [Google Scholar]
- 26.Reineke W, Knackmuss H J. Microbial metabolism of haloaromatics: isolation and properties of a chlorobenzene-degrading bacterium. Appl Environ Microbiol. 1984;47:395–402. doi: 10.1128/aem.47.2.395-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sander P, Wittich R-M, Fortnagel P, Wilkes H, Francke W. Degradation of 1,2,4-trichloro- and 1,2,4,5-tetrachlorobenzene by Pseudomonas strains. Appl Environ Microbiol. 1991;57:1430–1440. doi: 10.1128/aem.57.5.1430-1440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt T. Chlortoluole—Schluesselintermediate der Feinchemie. GIT Labor-Fachzeitschrift. 2000;5:638–639. [Google Scholar]
- 29.Schraa G, Boone M L, Jetten M S, van Neerven A R, Colberg P J, Zehnder A J. Degradation of 1,4-dichlorobenzene by Alcaligenes sp. strain A175. Appl Environ Microbiol. 1986;52:1374–1381. doi: 10.1128/aem.52.6.1374-1381.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scrutton N S, Berry A, Perham R N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990;343:38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- 31.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W C, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816–4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 32.Suen W C, Haigler B E, Spain J C. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J Bacteriol. 1996;178:4926–4934. doi: 10.1128/jb.178.16.4926-4934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandenbergh P A, Olsen R H, Colaruotolo J F. Isolation and genetic characterization of bacteria that degrade chloroaromatic compounds. Appl Environ Microbiol. 1981;42:737–739. doi: 10.1128/aem.42.4.737-739.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werlen C, Kohler H P, van der Meer J R. The broad substrate chlorobenzene dioxygenase and cis-chlorobenzene dihydrodiol dehydrogenase of Pseudomonas sp. strain P51 are linked evolutionarily to the enzymes for benzene and toluene degradation. J Biol Chem. 1996;271:4009–4016. doi: 10.1074/jbc.271.8.4009. [DOI] [PubMed] [Google Scholar]
- 35.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
- 36.Zylstra G J, McCombie W R, Gibson D T, Finette B A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988;54:1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]