Abstract
When cultivated in the presence of trypsin, the Ruminococcus gnavus E1 strain, isolated from a human fecal sample, was able to produce an antibacterial substance that accumulated in the supernatant. This substance, called ruminococcin A, was purified to homogeneity by reverse-phase chromatography. It was shown to be a 2,675-Da bacteriocin harboring a lanthionine structure. The utilization of Edman degradation and tandem mass spectrometry techniques, followed by DNA sequencing of part of the structural gene, allowed the identification of 21 amino acid residues. Similarity to other bacteriocins present in sequence libraries strongly suggested that ruminococcin A belonged to class IIA of the lantibiotics. The purified ruminococcin A was active against various pathogenic clostridia and bacteria phylogenetically related to R. gnavus. This is the first report on the characterization of a bacteriocin produced by a strictly anaerobic bacterium from human fecal microbiota.
It has been hypothesized previously that the capability to produce antibacterial compounds could play a key role in colonization and protection processes that take place in complex environments (25). Bacteriocin-producing bacteria have been isolated from mammalian digestive ecosystems (2, 13, 14, 17, 29), thus suggesting that they might be involved in colonization resistance, a fundamental function of gut microbiota.
Bacteriocins are proteinaceous compounds ribosomally synthesized. Their production by gram-positive bacteria has been documented extensively in the last few years, and they can be sorted into four distinct classes (15). Class I comprises lantibiotics which are characterized by (i) small size, (ii) activity toward the membrane, and (iii) the presence of dehydrated or unusual residues resulting from posttranslation modification events.
Lanthionine (Lan) and 3-methyllanthionine (Me-Lan) are the most common modified residues. They result from the formation of a thioether bond between a cysteine (Cys) residue and either a serine (Ser) or a threonine (Thr) residue, previously dehydrated as didehydroalanine (Dha) or didehydrobutyrine (Dhb), respectively. The dehydration step is catalyzed by a specific enzyme encoded by a gene included in the bacteriocin biosynthetic cluster. However, the number of didehydro residues generated is often larger than the number of potential Cys residues capable of further reacting with them. Therefore, many lantibiotics contain one or more didehydroamino acids (26). It was suggested that lantibiotics be grouped into type A and type B peptides according to their structure and activity. Type A lantibiotics are elongated and flexible peptides forming pores in the cytoplasmic membrane, resulting in, among other things, the loss of ion gradients, whereas type B lantibiotics are more globular and inhibit enzymatic functions (12). The increasing number of novel characterized lantibiotics with intermediate behavior makes this classification more and more difficult.
In the initial definition, bacteriocins were defined by inhibitory activity against closely related bacteria. Results obtained more recently have tended to show that this criterion should not be taken into account anymore (15).
In a previous work, we had demonstrated that strain E1, isolated from the fecal microbiota of a healthy man, was able to produce a trypsin-dependent antibacterial substance active against Clostridium perfringens (24). Initially, the E1 strain was identified as a Peptostreptococcus sp. (24) according to phenotypic characterization. Here we report on the isolation and characterization of ruminococcin A (RumA), a lantibiotic produced by the E1 strain in liquid culture medium and in the presence of trypsin. RumA is of particular interest since it is active against C. perfringens and various other enteric pathogens, suggesting that it could play an essential role in the protection of the host.
MATERIALS AND METHODS
Bacterial strains and media.
The E1 strain had been isolated from the predominant fecal microbiota of a healthy adult (24). Other bacterial strains used in this study are listed in Table 1. The C. perfringens CpA strain (24) was used as the reference target strain. Bacillus cereus strains were isolated from mashed vegetables (Collection INRA, Avignon, France). Clostridium difficile strains 79-685 (32), C253 (4), ATCC 43598 (5), and M-1 and B-1 (31) were kindly provided by T. Karjalainen (Université Paris Sud, Chatenay-Malabry, France), P. Mastrantonio (Istituto Superiore di Sanita, Rome, Italy), M. Delmée (Leuwen Universitate, Brussels, Belgium), and P. Borriello (Central Public Health Laboratory, London, United Kingdom), respectively. Other strains were from culture collections: ATCC (American Type Culture Collection, Manassas, Va.), CIP (Collection Institut Pasteur, Paris, France), NCTC (National Collection of Type Cultures, London, United Kingdom), and VPI (Virginia Polytechnic Institute, Blacksburg).
TABLE 1.
Bacterial strains: sensitivity to RumA and metronidazolea
| Species | Strain | Culture medium | MIC (μg/ml) of agent:
|
|
|---|---|---|---|---|
| Prepurified RumA | Metronidazole | |||
| Bacteroides putredenis | ATCC 29800 | BHI-YH | NI∗ | 3.0 |
| Bacteroides thetaiotaomicron | ATCC 29148 | BHI-YH | NI∗ | 12.5 |
| Bacteroides vulgatus | ATCC 8482 | BHI-YH | 600 | 12.5 |
| Bacteroides fragilis | ATCC 25285 | BHI-YH | NI∗ | NI∗∗ |
| Bifidobacterium adolescentis | ATCC 15703 | BHI-YH | 150 | NI∗∗ |
| Bifidobacterium breve | ATCC 15700 | BHI-YH | 300 | NI∗∗ |
| Bifidobacterium catenulatum | ATCC 27539 | BHI-YH | 150 | NI∗∗ |
| B. longum | ATCC 15707 | BHI-YH | 75 | NI∗∗ |
| Enterococcus faecalis | CIP 76117 | BHI-YH | NI∗ | NT |
| Eubacterium contortum | ATCC 25540 | BHI-YH | 32.5 | 1.5 |
| Ruminococcus obeum | ATCC 29174 | BHI-YH | 32.5 | 1.5 |
| R. torques | ATCC 27756 | BHI-YH | 300 | NT |
| R. gnavus | ATCC 29149 | BHI-YH | 75 | 12.5 |
| C. nexile | ATCC 27757 | BHI-YH | 150 | 3.0 |
| C. oroticum | ATCC 13619 | BHI-YH | 600 | 6.25 |
| C. perfringens | CpA | BHI-YH | 75 | 25 |
| C. difficile serogroup A | B-1 | BHI-YH | 75 | 6.25 |
| C. difficile serogroup C | C253 | BHI-YH | 75 | 6.25 |
| C. difficile serogroup D | ATCC 43598 | BHI-YH | 150 | 6.25 |
| C. difficile serogroup F | M-1 | BHI-YH | 150 | 12.5 |
| C. difficile serogroup S3 | 79685 | BHI-YH | 75 | 6.25 |
| C. difficile | ATCC 43255 | BHI-YH | 150 | 6.25 |
| C. botulinum type A | CIP 38 | TGY | 75 | NT |
| C. botulinum type B | NCTC 7273 | TGY | 75 | NT |
| C. botulinum type E | NCTC 8266 | TGY | 75 | NT |
| C. sordellii | VPI 9048 | BHI-YH | 150 | NT |
| C. bifermentans | NCTC 506 | BHI-YH | 75 | NT |
| C. septicum | ATCC 12464 | BHI-YH | 150 | NT |
| C. sporogenes | ATCC 19404 | BHI-YH | 300 | NT |
| B. cereus | Z4222, TZ415 | J agar | 75 | NT |
| B. cereus | Z4234, K1231, P2101 | J agar | 150 | NT |
| B. cereus | K1166, L2104, P2174, Z421, P21S | J agar | NI∗ | NT |
In this test, a Sep-pack-purified RumA containing 600 μg of total protein per ml was used. The antibacterial activity of RumA resulting from two RP-HPLC purifications and tested against C. perfringens CpA was found to be 0.4 μg/ml. This result must be compared to the metronidazole MICs for CpA (25 μg/ml); C. difficile serogroups A, B, D, and S3 (6.25 μg/ml); and C. difficile serogroup F (12.5 μg/ml). NI∗, no inhibition halo could be detected with the Sep-pack-purified RumA; NI∗∗, no inhibition halo could be detected with metronidazole (100 μg/ml); NT, not tested.
Strain E1 was grown in an anaerobic cabinet in brain heart infusion broth supplemented with yeast extract and hemin (BHI-YH; BHI [Difco Laboratories, Detroit, Mich.] supplemented with 5 g of yeast extract [Difco Laboratories] per liter and 5 mg of hemin [Sigma-Aldrich Chimie, St. Quentin Fallavier, France] per liter). When required, trypsin from bovine pancreas (type XIII; l-1-tosyl-amide phenylalanyl chloride treated; Sigma-Aldrich Chimie) was added at a final concentration of 50 μg/ml.
B. cereus was plated on J agar (5 g of tryptone [Biokar Diagnostics, Beauvais, France] per liter, 15 g of yeast extract [Biokar Diagnostics] per liter, 3 g of K2HPO4 per liter, 2 g of glucose per liter, and 15 g of agar [Difco Laboratories] per liter). Clostridium botulinum was propagated on TGY medium (30 g of tryptone per liter, 20 g of yeast extract per liter, 5 g of glucose per liter, 0.5 g of cysteine-HCl per liter, 0.004 g of resazurine per liter, 15 g of agar per liter, pH 7.3). Other strains were plated on solid BHI-YH medium obtained by adding 15 g of agar per liter.
Bacteriocin purification.
R. gnavus E1 was inoculated at 1% in 200 ml of trypsin-supplemented BHI-YH broth and incubated at 37°C for 48 h under anaerobic conditions. After growth, bacterial cells were removed by centrifugation; the supernatant was lyophilized, resuspended in 35 ml of 0.05% trifluoroacetic acid, and loaded onto C18 Sep-Pack 35-cm3 reverse-phase cartridges (Waters; Millipore). The column was washed twice with 140 ml of 0.05% trifluoroacetic acid and then with 84 ml of 20 and 35% acetonitrile. The bacteriocin was eluted with 84 ml of 40% acetonitrile. Bacteriocin-containing fractions were pooled, evaporated in a Rotavapor R-134 (Büchi, Switzerland), and then resuspended in 9 ml of 36% acetonitrile before further purification by C18 reverse-phase high-pressure liquid chromatography (RP-HPLC). Five-hundred-microliter aliquots were loaded onto a C18 Novapak preparative column (300 by 7.8 mm). Elution was performed at a flow rate of 2 ml/min under 36% acetonitrile isocratic conditions and then at a flow rate of 0.5 ml/min using a linear gradient of acetonitrile from 36 to 60%. Peptide fractions were detected spectrophotometrically by measuring A214 and collected manually. The active fractions were pooled and concentrated under vacuum to a final volume of 9 ml. Protein content, estimated with the Folin reagent, and antagonistic activity were determined at each step. The bacteriocin was purified by a second RP-HPLC step performed under the same conditions. Active fractions were lyophilized and then resuspended in distilled water before further analysis.
Bacteriocin activity assays.
Antibacterial activity present in the fractions was determined at each step of purification by a modification of the critical dilution assay (11). One hundred fifty microliters of a BHI-YH broth culture (37°C, 16 to 24 h) of the sensitive indicator strain C. perfringens CpA was added to 15 ml of BHI-YH agar medium and poured in a sterile plate. Six-millimeter-diameter wells were then prepared with a vacuum pump. Five hundred microliters of each fraction was concentrated under vacuum to a 50-μl final volume. Then, 30 μl of a serial twofold dilution of the samples was deposited into the wells, and the plates were incubated at 37°C for 16 to 24 h in an anaerobic cabinet. Bacteriocin titers were expressed as the reciprocal highest dilution exhibiting an inhibition halo and were reported in arbitrary units (AU) per milliliter, thus allowing the determination of the total AU present in a given fraction.
For determining the activity spectrum, a solution of Sep-pack-prepurified ruminococcin A with a total protein concentration of 600 μg/ml was used. Ten microliters of a fresh broth culture (containing approximately 106 CFU) of the target strain was spread on appropriate agar plates. After 90 min of incubation at room temperature, 10 μl of a serial twofold dilution of ruminococcin A was spotted onto the surface of the plates. The plates were then incubated until confluent growth of the target strain. The MIC of ruminococcin A was compared to that of metronidazole (Sigma-Aldrich Chimie), an antibiotic frequently used in the case of anaerobic infections of the digestive tract.
Amino acid composition analysis.
Purified bacteriocin was hydrolyzed in 6 N HCl under vacuum at 110°C for 24 h. The amino acid composition was determined on an LC3000 analyzer (Eppendorf Biotronik, Munich, Germany) using a ninhydrin postcolumn derivative system (20).
Mass spectrometry.
Mass measurement of the purified bacteriocin was performed by electron spray mass spectrometry, using a VG Bio-Q quadrupole (Bio-Tech, Manchester, United Kingdom) in the positive mode. The protein was dissolved in H2O-CH3CN (50/50 [vol/vol] ratio) with 1% acetic acid at a concentration of about 5 pmol/μl (by volume); 10-μl aliquots were introduced into the ion source at a flow rate of 4 μl/min. Scanning was usually performed from m/z = 500 to m/z = 1,500 in 10 s with the resolution adjusted so that the peak at m/z = 998 from horse heart myoglobin was 1.5 to 2 wide on the base. Calibration was performed by using the multiply charged ions produced by a separate introduction of horse heart myoglobin (16,950.4 Da) (10).
Amino acid sequence analysis.
The amino acid sequence of the purified bacteriocin was determined by Edman degradation using an Applied Biosystems model 477A (Foster City, Calif.) pulsed liquid sequencer, connected to a 120A analyzer (Applied Biosystems) (6). Sequencing reagents were purchased from Applied Biosystems.
The amino acid sequence was also determined by tandem mass spectrometry (MS/MS) with a Sciex API III Plus triple-quadrupole apparatus (Sciex Instruments, Thornhill, Canada). The spectrometer was fitted with an articulated pneumatically assisted nebulization probe and an atmospheric pressure ionization source. Briefly, the sample dissolved in H2O-CH3CN (50/50 ratio)–0.2% formic acid was infused at 5 μl/min in the positive mode. During the first step, the molecular mass was determined from the measured m/z values for the protonated molecules. Then, the collision-assisted dissociation of selected ions was performed using argon as a collision gas (Q2 quadrupole) with a collision energy of 60 eV. The first quadrupole (Q1) was set to transmit the selected precursor ion, while the third quadrupole (Q3) was set to scan the masses of produced ions.
Cyanogen bromide (CNBr) cleavage.
CNBr (0.5 mg) was added to 0.2 mg of the purified bacteriocin resuspended in 70% formic acid diluted in acetonitrile and then incubated for 72 h to selectively cleave at Met residues. The reaction mixture was then subjected to electron spray mass spectrometry analysis.
Resistance to trypsin.
An 0.01-mg amount of the purified bacteriocin resuspended in 5 μl of acetonitrile was diluted with 35 μl of PBS (10 mM KH2PO4-K2HPO4, 0.9% NaCl, pH 7) containing variable amounts of trypsin (0, 50, or 500 μg/ml). The reaction mixture was then incubated for 2 h at 37°C and checked for antibacterial activity by the critical dilution assay as described in the previous paragraph.
DNA isolation.
R. gnavus E1 chromosomal DNA was isolated using the Nucleospin kit (Machery-Nagel GmbH & Co., Düren, Germany).
Amplification and cloning of E1 16S rDNA.
Amplification of 16S ribosomal DNA (rDNA) was performed using oligonucleotide primers F515 [5′-GTGCCAGC(AC)GCCGCGG-3′], R930 [5′-G(CT)CCCCGTCAATTC(AC)T-3′], F915 [5′-A(GT)GAATTGACGGGG(GA)C-3′], and R1406 [5′-ACGGGCGGTGTGT(GA)C-3′], which corresponded to bacterial 16S rRNA gene conserved sequences (from positions 515 to 530, 930 to 915, 915 to 930, and 1406 to 1392 on the Escherichia coli 16S rRNA, respectively) (19). PCR conditions used included annealing at 42°C (30 s) for fragment A (position 515 to position 930) or 55°C (30 s) for fragment B (position 915 to position 1406), polymerization at 72°C (45 s), and denaturation at 94°C (2 min). Amplification reactions (30 cycles) were carried out in a Gene Amp PCR system 2400 cycler (Perkin-Elmer Cetus, Norwalk, Conn.) using 75 ng of the E1 strain chromosomal DNA as a template. Fragments A and B were then cloned using the LigATor kit (R&D Systems, Abingdon, United Kingdom) according to the manufacturer's recommendations.
Amplification and cloning of part of the ruminococcin structural gene.
Amplification of the DNA fragment encoding the active bacteriocin was performed with degenerate oligonucleotide primers Ol1 and Ol2. The Ol1 sequence (5′-CAGGAAACAGCTATGACCGGNAAYGGNGTNYTNAA-3′) was based on the pBSK+ reverse primer followed by the degenerate codons (underlined) encoding the six amino acid residues identified at the N-terminal end of the bacteriocin. The Ol2 sequence (5′-TGTAAAACGACGGCCAGTRAANARRAAYTGCCA-3′) was based on the pBSK+ −21 M13 primer sequence and the degenerate codons (underlined) encoding the five amino acid residues identified at the C-terminal end. PCR conditions used included annealing at 50°C (30 s), polymerization at 72°C (1 min), and denaturation at 94°C (1 min).
Amplification reactions (30 cycles) were carried out in a Gene Amp PCR system 2400 (Perkin-Elmer Cetus) using 500 ng of the E1 strain chromosomal DNA as a template.
The 100-bp amplified fragment was extracted from an agarose gel, polished with the PCR polishing kit (Stratagene, La Jolla, Calif.), and subsequently cloned at the blunt-end EcoRV restriction site of the pBR322 vector, using standard molecular genetics techniques (27).
DNA sequencing and analysis.
Nucleotide sequences were determined by the dideoxy chain terminator method (28) using the Prism Ready Reaction d-Rhodamine Terminator sequencing kit (Applied Biosystems Division) in an ABI Prism 310 Genetic Analyzer (Perkin-Elmer Cetus). DNA or protein homology searches (GenBank, EMBL, and SWISS-PROT) were carried out with the programs included in the GCG sequence analysis software package (University of Wisconsin). Multiple alignments were edited by the Genedoc program.
RESULTS
Identification of the E1 strain.
Two fragments, A and B, of the E1 16S rDNA were amplified, cloned, and sequenced as described in Materials and Methods. The total DNA sequence was 861 bp long and exhibited 99.40% sequence identity with the corresponding fragment of the ATCC 29149 R. gnavus reference strain. DNA-DNA hybridization between the E1 and ATCC 29149 strains showed that they were 80% homologous (F. Gavini, personal communication). These results demonstrated that the E1 strain belonged to the species R. gnavus, a member of the Clostridium coccoides phylogenetically defined group (Ribosomal Database Project registration no. 2.16.4.1) (18), which includes some of the predominant bacterial genera found in the human large bowel (30).
Purification of the bacteriocin produced by the E1 strain.
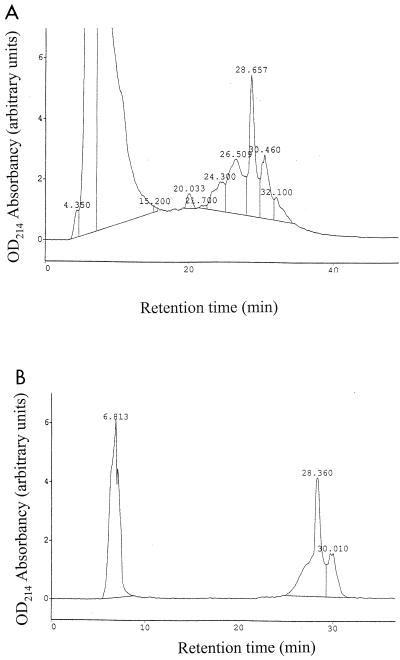
Previous studies had shown that antibacterial activity was present in the supernatant of strain E1 cultivated in the presence of trypsin (24). The purification of the antibacterial substance was achieved by reverse-phase chromatography (Sep-Pack followed by HPLC) (3). The first step of purification removed a significant amount of contaminating proteins (Table 2). The biologically active fraction eluted with 40% acetonitrile and was loaded onto a C18 Novapak preparative column. The antibacterial substance eluted with fractions collected after a 27- to 32-min retention time (Fig. 1A). A second RP-HPLC purification performed under the same conditions yielded two peaks corresponding to fractions F1 and F2 that eluted from 27.4 to 29.3 min and from 29.3 to 31.15 min, respectively (Fig. 1B). Both fractions were active against C. perfringens.
TABLE 2.
Purification yield
| Purification stage | Total biological activity (AU) | Total proteins (mg) | Sp act (AU/mg) | Increase in sp act (fold) | Yield (%) |
|---|---|---|---|---|---|
| Supernatant | 21,333 | 4,150 | 5.1 | 1 | 100 |
| Sep-pack | 8,960 | 3.4 | 2,635 | 517 | 40 |
| RP-HPLC 1 | 2,238 | 0.3 | 6,662 | 1,306 | 10 |
| RP-HPLC 2 | 900 | 0.0133 | 67,669 | 13,268 | 4.21 |
FIG. 1.
Purification of RumA by RP-HPLC. Active fractions eluted after a 27- to 32-min retention time (A). They were repurified under the same conditions. Two active fractions, F1 and F2, eluted after 27.4 to 29.3 and 29.3 to 31.15 min, respectively (B). OD214, optical density at 214 nm.
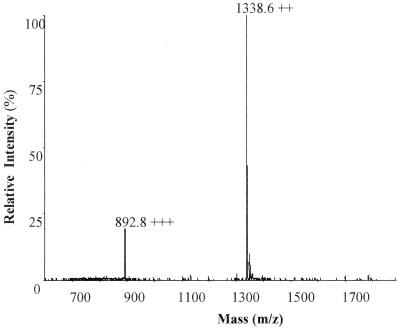
Samples corresponding to fractions F1 and F2 were subjected to mass spectrometric analysis. The molecular mass deduced from fraction F1 was 2,675 Da (Fig. 2). Fraction F2 appeared to contain a major product with a molecular mass of 2,675 Da, contaminated with other molecules (data not shown). Further analysis was performed only on fraction F1, which contained the antibacterial substance now called ruminococcin A (RumA).
FIG. 2.
Electron spray ionization mass spectrometry analysis of RumA. The recorded multiply charged ions are consistent with a molecular mass estimate of 2,675.17 ± 0.14 Da.
RumA antagonistic activity was not affected by high temperatures (10 min of incubation at 100°C).
Peptide analysis and sequencing.
Analysis of the amino acid composition of RumA highlighted the presence of both Dhb and Lan residues (data not shown). This latter observation suggested that RumA was a lantibiotic.
The peptide sequence obtained by the Edman degradation technique identified the respective positions of 17 residues, 4 remaining unknown (Table 3). The reaction was blocked after the 21st cycle.
TABLE 3.
RumA peptide sequencea
| Origin | Residue at position:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
| Ed. | G | N | G | V | L | K | NI | I | NI | H | E | NI | N | M | N | NI | W | Q | F | L | F |
| MS/MS | G | N | G | V | I/L | K | Dhb | I | M | N | Dhb | W | Q | F | I/L | F | |||||
| DNA | G | N | G | V | L | K | T | I | S | H | E | C | N | M | N | T | W | Q | F | L | F |
Comparison of the peptide sequence information obtained by the Edman degradation technique (Ed.) or MS/MS or deduced from the gene sequence (DNA). Residues present at positions 7, 9, 12, and 16 could not be identified by Edman degradation (NI). Residues at positions 5 and 20 appeared as Leu or Ile (I/L) by MS/MS.
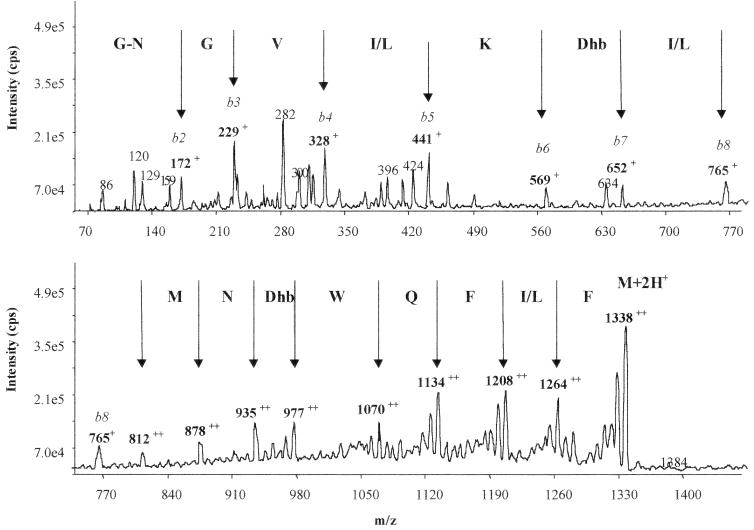
RumA was thus subjected to MS/MS. Fragmentation of the doubly charged ion (m/z 1338) of RumA yielded two b series ions. The first series corresponded to the successive cleavage of the sequence from the N-terminal part (Fig. 3). The observed singly charged product ions, i.e., m/z 172, 229, 328, 441, 569, 652, and 765, corresponded to the sequence GNGV(I/L)KDhb(I/L), consistent with the RumA N-terminal sequence obtained by the Edman degradation method (Table 3). It made it possible to identify the seventh residue as a Dhb (Dhb7). The second series (Fig. 3), constituted by doubly charged product ions (m/z 812, 878, 935, 977, 1070, 1134, 1208, and 1264), corresponded to a successive loss of MNDhbWQF(I/L)F residues from the C-terminal region of the precursor ion (m/z 1338). Again, these data were consistent with the RumA C-terminal sequence obtained by the Edman degradation method (Table 3). They made it possible to identify the 16th residue as a Dhb (Dhb16). It was noticeable that the signals corresponding to Dhb7 and DHb16 were comparable, suggesting that the two residues were at the same stage. From these results, it could also be deduced that this last fragment was an internal sequence of RumA, since the molecular mass difference between m/z 1338 and 1264, i.e., 148 Da, corresponded to the Mr of Phe minus 17 Da (OH group), underlining the fact that this amino acid, i.e., Phe21, was naturally involved in two peptide bonds and hence demonstrating that it was not the last C-terminal residue of RumA.
FIG. 3.
MS/MS analysis of RumA. The peaks representing cleavage at peptide bonds are identified by the vertical arrows. It is noticeable that the masses of the two Phe residues present at the C-terminal region of the molecule are identical.
DNA amplification and sequence.
As residues 9 and 12 still remained unidentified, two degenerate oligonucleotide primers were designed on the basis of residues 1 through 6 and 17 through 21 of the active peptide, respectively; all the possible encoding sequences were represented. These two primers, Ol1 and Ol2, were then used to amplify total DNA extracted from the E1 strain. One main amplified fragment could be visualized, sizing the expected value at around 100 bp (data not shown). As direct sequencing with the −21 and reverse primers was not possible, this fragment was cloned onto the pBR322 vector and the nucleotide sequence was obtained.
The peptide sequence deduced from the DNA sequence exhibited high levels of homology with the sequences obtained previously by biochemical techniques (Table 3), demonstrating that the amplified fragment was part of the structural gene coding for the active RumA. Two amino acids not previously identified by biochemical techniques could be deduced from the DNA sequence. They corresponded to one Ser and one Cys residue, Ser9 and Cys12, respectively.
Compared with databases, the RumA peptide sequence exhibited high similarity to various other lantibiotics belonging to group AII of the lantibiotics (21) (Fig. 4). This result was in agreement with the previous detection of a Lan residue.
FIG. 4.
Multiple sequence alignment of RumA with homologous type IIA lantibiotics. RumA was compared to the peptide sequences of butyrivibriocin OR79A (Bvi79A) (13), lacticin 481 (LctA) (22), variacin (VarA) (23), streptococcin A-M49 and A-FF22 (ScnA) (8, 9), and mutacin II (MutA)
Cyanogen bromide cleavage.
RumA was incubated for 72 h in the presence of CNBr and then analyzed by mass spectrometry. The action of CNBr resulted in a single peptide with a 31-Da loss compared to the 2,675-Da original molecule. This result was typical of the presence of covalent linkages spanning the Met residue and holding peptide fragments together.
Resistance of RumA to trypsin.
It had been previously demonstrated that the presence of trypsin was necessary just before the inoculation of strain E1, to promote the generation of antibacterial activity (24). However, a Lys residue (known to be a specific cleavage site for trypsin) was evidenced in the N-terminal region of RumA (Lys6). The solution containing the purified RumA was adjusted to neutral pH, and various amounts of trypsin were added before incubation. The activity of RumA against C. perfringens was not affected despite 2 h of incubation in the presence of concentrations of trypsin as high as 500 μg/ml. Control experiments demonstrated that trypsin was active despite the presence of acetonitrile in the reaction mixture. Mass spectrometry analysis of RumA incubated with the highest amount of trypsin revealed the presence of only one peak, with a 2,675-Da mass, confirming that RumA had not been cleaved, probably because Lys6 was not accessible to trypsin.
Activity spectrum.
The activity of RumA was tested against different target strains chosen as follows: (i) anaerobic gram-negative bacteria representative of the human predominant intestinal microbiota (Bacteroides spp.), (ii) anaerobic gram-positive bacteria representative of the human predominant intestinal microbiota (Bifidobacterium spp.) or phylogenetically closely related to the E1 strain (Ruminococcus spp., Eubacterium sp.; Clostridum oroticum and Clostridium nexile), and (iii) pathogenic clostridia (C. difficile, C. perfringens, C. sordellii, C. bifermentans, C. sporogenes, C. septicum, C. histolyticum, and C. botulinum) and the food-borne pathogen B. cereus. Complete results are summarized in Table 1. All the Bacteroides sp. strains tested were resistant to RumA. The Bifidobacterium sp. strains displayed intermediate sensitivity, except for Bifidobacterium longum, which was as sensitive as C. perfringens to RumA. Strains phylogenetically related to R. gnavus were mainly sensitive to RumA, except for Ruminococcus torques and C. oroticum. Pathogenic Clostridium species were sensitive to RumA with the exception of C. sporogenes. Most of the B. cereus strains tested were rather insensitive or resistant to RumA activity. Metronidazole, an antibiotic commonly used in intestinal infections due to anaerobic bacteria (7), was also tested in order to compare the RumA antibacterial activities (Table 1).
DISCUSSION
Ruminococcin A (RumA) is the first bacteriocin produced by strictly anaerobic bacteria (R. gnavus) of the human gut so far characterized at the molecular level. Purification was achieved by reverse-phase chromatography, leading to a 10,000-fold increase in specific activity. The presence of Dhb and Lan modified residues, evidenced by amino acid composition, allowed us to conclude that RumA was a lantibiotic.
The first attempt at peptide sequencing was realized by the Edman degradation technique. Seventeen out of the 21 N-terinal amino acids were identified, and then the reaction was blocked. Thus, RumA was subjected to MS/MS analysis. Two series of b ions were obtained. The first one corresponded to the N-terminal cleavage of the molecule. It confirmed seven out of the first eight residues previously identified by Edman degradation and evidenced that the residue at position 7 was a Dhb (Dhb7). The second series, corresponding to the cleavage of residues present in the C-terminal part of the molecule, also confirmed previous results obtained by Edman degradation and highlighted the presence of another Dhb residue (Dhb16). It had been shown previously that MS/MS detection signals corresponding to Dhb may vary, depending on whether the residues are involved in Me-Lan structures or remain unreacted (16). In our experiments, MS/MS signals corresponding to both Dhb7 and Dhb16 were comparable. As the presence of Me-Lan structures generally leads to blockages during recurrent degradation of lantibiotics (26), these observations strongly suggested that both didehydro residues remained unreacted in RumA.
At this step, Edman degradation and MS/MS techniques made it possible to identify 19 amino acids but neither Cys nor Ser residues capable of generating the lanthionine structure evidenced previously. Thus, degenerate oligonucleotides deduced on the basis of the first six N-terminal residues and the last five C-terminal residues identified, respectively, were used to amplify, clone, and sequence part of the structural gene encoding RumA. The deduced peptide sequence was in agreement with the already known RumA sequence and made it possible to identify two new residues present in the central part of the molecule, Ser9 and Cys12, that had not been detected by biochemical techniques.
However, two observations suggested that the sequence that we were able to obtain was still only partial: (i) the RumA molecular mass measured (2,675 Da) was 288 Da higher than the one calculated from experimental data (2,387 Da), and (ii) Phe21, the last residue identified by MS/MS, appeared to be involved in two peptide bonds, highlighting the presence of other residues at the C-terminal end of the molecule.
The comparison of the RumA sequence with bacteriocins described already highlighted high levels of homology to group AII of the lantibiotics (21). It was noticeable that the TCCS or TCC motif present at their C-terminal end had not been detected in RumA. DNA sequence data (A. Gomez et al., submitted for publication) confirmed that the TCC motif is also present at the C-terminal part of RumA, and so a most probable structure of the molecule can be proposed (Fig. 5). In this structure, containing two unreacted Dhb residues and both a Lan and a Me-Lan residue (resulting from the Ser9-Cys23 and Thr22-Cys24 associations, respectively), the theoretical molecular mass would fit in perfectly with the measured one. One covalent bond due to the Lan structure would span the Met residue, keeping the CNBr-derived peptides together as observed in other lantibiotics (16).
FIG. 5.
Hypothetical structure of RumA. Considering that the presence of the TCC motif at the C terminus of RumA was confirmed, the measured molecular mass of the molecule (2,675 Da) makes it necessary to consider the presence of two unreacted Dhb, one Lan, and one Me-Lan residue. Since Me-Lan residues are well known for being responsible for blockages during Edman degradation, Dhb7, Dhb167, and Cys12 cannot be involved in such a structure. The association between Ser9 and Cys12 in a Lan structure is not possible, since the CNBr reaction highlighted the presence of a covalent bond spanning the Met14 residue. Thus, it is highly probable that (i) Ser9 and the putative Cys23 and (ii) the putative His22 and Thr24 are associated in a Lan and a Me-Lan residue, respectively. Unreacted Dhb7 and Dhb16 residues are noted as T*; circled numerals 1 and 2 indicate the putative Lan and Me-Lan structures, respectively.
RumA antibacterial activity is targeted against two groups of microorganisms: phylogenetically R. gnavus-related bacteria, including the reference strain ATCC 29149, and pathogenic Clostridium spp. All lantibiotic-producing strains harbor self-protection mechanisms preventing cell death by the action of their own bacteriocin (1). The genes coding for immunity factors are included in the clusters encoding the biosynthetic machinery (26). As R. gnavus ATCC 29149 is sensitive to RumA and was not shown to produce an antibacterial substance, one can conclude that this strain probably does not carry the genetic system responsible for the biosynthesis of RumA. Comparison with metronidazole shows that the activity spectrum of RumA is narrower, since Bacteroides spp. are mainly resistant. However, metronidazole and RumA activities against C. perfringens and C. difficile are comparable, suggesting that RumA could be used as a therapeutic agent targeting these pathogens.
This observation gives reason to believe that RumA could play a double role in gut colonization processes and its protection against invasion by pathogens. It is generally hypothesized that, if produced in the digestive tract, bacteriocins might play a local role. Since their proteinaceous nature makes them sensitive to proteases, a close relationship between the producer and the target strain is necessary, or otherwise they would be destroyed. RumA resistance to trypsin, one of the major host proteases, indicates that it could constitute a powerful weapon in bacterium-bacterium antagonisms potentially applicable to human health.
Previous results suggested that trypsin was involved in the induction of bacteriocin synthesis rather than in the hypothetical activation of the secreted peptide by proteolysis (24). Additional studies aiming to characterize the genetic determinants of RumA biosynthesis and determine the exact role of trypsin are in progress.
ACKNOWLEDGMENTS
We are indebted to M. Laurière and C. Beauvallet for constructive discussion and to A. M. Wall for correcting the manuscript.
This work was supported by the grants Actions Concertées Coordonnées Sciences du Vivant from the French Ministry for Research and Technology and FAIR CT 95-0433 from the European Community.
REFERENCES
- 1.Abee T. Pore-forming bacteriocins of gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol Lett. 1995;129:1–10. doi: 10.1016/0378-1097(95)00137-T. [DOI] [PubMed] [Google Scholar]
- 2.Balla E, Dicks L M, Du Toit M, Van Der Merwe M J, Holzapfel W H. Characterization and cloning of the genes encoding enterocin 1071A and enterocin 1071B, two antimicrobial peptides produced by Enterococcus faecalis BFE 1071. Appl Environ Microbiol. 2000;66:1298–1304. doi: 10.1128/aem.66.4.1298-1304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhugaloo-Vial P, Dousset X, Metivier A, Sorokine O, Anglade P, Boyaval P, Marion D. Purification and amino acid sequences of piscicocins V1a and V1b, two class IIa bacteriocins secreted by Carnobacterium piscicola V1 that display significantly different levels of specific inhibitory activity. Appl Environ Microbiol. 1996;62:4410–4416. doi: 10.1128/aem.62.12.4410-4416.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerquetti M, Pantosti A, Stefanelli P, Mastrantonio P. Purification and characterization of an immunodominant 36 kDa antigen present on the cell surface of Clostridium difficile. Microb Pathog. 1992;13:271–279. doi: 10.1016/0882-4010(92)90037-o. [DOI] [PubMed] [Google Scholar]
- 5.Collignon A, Ticchi L, Depitre C, Gaudelus J, Delmee M, Corthier G. Heterogeneity of Clostridium difficile isolates from infants. Eur J Pediatr. 1993;152:319–322. doi: 10.1007/BF01956743. [DOI] [PubMed] [Google Scholar]
- 6.Cornwell G G, III, Sletten K, Johansson B, Westermark P. Evidence that the amyloid fibril protein in senile systemic amyloidosis is derived from normal prealbumin. Biochem Biophys Res Commun. 1988;154:648–653. doi: 10.1016/0006-291x(88)90188-x. [DOI] [PubMed] [Google Scholar]
- 7.Freeman C D, Klutman N E, Lamp K C. Metronidazole. A therapeutic review and update. Drugs. 1997;54:679–708. doi: 10.2165/00003495-199754050-00003. [DOI] [PubMed] [Google Scholar]
- 8.Hynes W L, Ferretti J J, Tagg J R. Cloning of the gene encoding streptococcin A-FF22, a novel lantibiotic produced by Streptococcus pyogenes, and determination of its nucleotide sequence. Appl Environ Microbiol. 1993;59:1969–1971. doi: 10.1128/aem.59.6.1969-1971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hynes W L, Friend V L, Ferretti J J. Duplication of the lantibiotic structural gene in M-type 49 group A streptococcus strains producing streptococcin A-M49. Appl Environ Microbiol. 1994;60:4207–4209. doi: 10.1128/aem.60.11.4207-4209.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaquinod M, Potier N, Klarskov K, Reymann J M, Sorokine O, Kieffer S, Barth P, Andriantomanga V, Biellmann J F, Van Dorsselaer A. Sequence of pig lens aldose reductase and electrospray mass spectrometry of non-covalent and covalent complexes. Eur J Biochem. 1993;218:893–903. doi: 10.1111/j.1432-1033.1993.tb18445.x. [DOI] [PubMed] [Google Scholar]
- 11.Joerger M C, Klaenhammer T R. Characterization and purification of helveticin J and evidence for a chromosomally determined bacteriocin produced by Lactobacillus helveticus 481. J Bacteriol. 1986;167:439–446. doi: 10.1128/jb.167.2.439-446.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung G. Lantibiotics: a survey. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: Kluwer Academic Publishers; 1991. pp. 1–34. [Google Scholar]
- 13.Kalmokoff M L, Lu D, Whitford M F, Teather R M. Evidence for production of a new lantibiotic (butyrivibriocin OR79A) by the ruminal anaerobe Butyrivibrio fibrisolvens OR79: characterization of the structural gene encoding butyrivibriocin OR79A. Appl Environ Microbiol. 1999;65:2128–2135. doi: 10.1128/aem.65.5.2128-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalmokoff M L, Teather R M. Isolation and characterization of a bacteriocin (butyrivibriocin AR10) from the ruminal anaerobe Butyrivibrio fibrisolvens AR10: evidence in support of the widespread occurrence of bacteriocin-like activity among ruminal isolates of B. fibrisolvens. Appl Environ Microbiol. 1997;63:394–402. doi: 10.1128/aem.63.2.394-402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 16.Krull R E, Chen P, Novak J, Kirk M, Barnes S, Baker J, Krishna N R, Caufield P W. Biochemical structural analysis of the lantibiotic mutacin II. J Biol Chem. 2000;275:15845–15850. doi: 10.1074/jbc.275.21.15845. [DOI] [PubMed] [Google Scholar]
- 17.Laukova A, Marekova M. Antimicrobial spectrum of bacteriocin-like substances produced by rumen staphylococci. Folia Microbiol. 1993;38:74–76. doi: 10.1007/BF02814554. [DOI] [PubMed] [Google Scholar]
- 18.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Farris R J, Garrity G M, Olsen G J, Schmidt T M, Tiedje J M. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metivier A, Pilet M F, Dousset X, Sorokine O, Anglade P, Zagorec M, Piard J C, Marion D, Cenatiempo Y, Fremaux C. Divercin V41, a new bacteriocin with two disulphide bonds produced by Carnobacterium divergens V41: primary structure and genomic organization. Microbiology. 1998;144(Pt. 10):2837–2844. doi: 10.1099/00221287-144-10-2837. [DOI] [PubMed] [Google Scholar]
- 21.Nes I F, Tagg J R. Novel lantibiotics and their pre-peptides. Antonie Leeuwenhoek. 1996;69:89–97. doi: 10.1007/BF00399414. [DOI] [PubMed] [Google Scholar]
- 22.Piard J C, Kuipers O P, Rollema H S, Desmazeaud M J, de Vos W M. Structure, organization, and expression of the lct gene for lacticin 481, a novel lantibiotic produced by Lactococcus lactis. J Biol Chem. 1993;268:16361–16368. [PubMed] [Google Scholar]
- 23.Pridmore D, Rekhif N, Pittet A C, Suri B, Mollet B. Variacin, a new lanthionine-containing bacteriocin produced by Micrococcus varians: comparison to lacticin 481 of Lactococcus lactis. Appl Environ Microbiol. 1996;62:1799–1802. doi: 10.1128/aem.62.5.1799-1802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramare F, Nicoli J, Dabard J, Corring T, Ladire M, Gueugneau A M, Raibaud P. Trypsin-dependent production of an antibacterial substance by a human Peptostreptococcus strain in gnotobiotic rats and in vitro. Appl Environ Microbiol. 1993;59:2876–2883. doi: 10.1128/aem.59.9.2876-2883.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riley M A. Molecular mechanisms of bacteriocin evolution. Annu Rev Genet. 1998;32:255–278. doi: 10.1146/annurev.genet.32.1.255. [DOI] [PubMed] [Google Scholar]
- 26.Sahl H G, Bierbaum G. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu Rev Microbiol. 1998;52:41–79. doi: 10.1146/annurev.micro.52.1.41. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siragusa G R. Production of bacteriocin inhibitory to Listeria species by Enterococcus hirae. Appl Environ Microbiol. 1992;58:3508–3513. doi: 10.1128/aem.58.11.3508-3513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suau A, Bonnet R, Sutren M, Godon J J, Gibson G R, Collins M D, Dore J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tasteyre A, Barc M-C, Karjalainen T, Dodson P, Hyde S, Bourlioux P, Borriello P. Flagellin gene of Clostridium difficile. Microbiology. 2000;146:957–966. doi: 10.1099/00221287-146-4-957. [DOI] [PubMed] [Google Scholar]
- 32.Waligora A J, Barc M C, Bourlioux P, Collignon A, Karjalainen T. Clostridium difficile cell attachment is modified by environmental factors. Appl Environ Microbiol. 1999;65:4234–4238. doi: 10.1128/aem.65.9.4234-4238.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]