Abstract
Background
Multi‐phenotype analysis of genetically correlated phenotypes can increase the statistical power to detect loci associated with multiple traits, leading to the discovery of novel loci. This is the first study to date to comprehensively analyze the shared genetic effects within different hemostatic traits, and between these and their associated disease outcomes.
Objectives
To discover novel genetic associations by combining summary data of correlated hemostatic traits and disease events.
Methods
Summary statistics from genome wide‐association studies (GWAS) from seven hemostatic traits (factor VII [FVII], factor VIII [FVIII], von Willebrand factor [VWF] factor XI [FXI], fibrinogen, tissue plasminogen activator [tPA], plasminogen activator inhibitor 1 [PAI‐1]) and three major cardiovascular (CV) events (venous thromboembolism [VTE], coronary artery disease [CAD], ischemic stroke [IS]), were combined in 27 multi‐trait combinations using metaUSAT. Genetic correlations between phenotypes were calculated using Linkage Disequilibrium Score Regression (LDSC). Newly associated loci were investigated for colocalization. We considered a significance threshold of 1.85 × 10−9 obtained after applying Bonferroni correction for the number of multi‐trait combinations performed (n = 27).
Results
Across the 27 multi‐trait analyses, we found 4 novel pleiotropic loci (XXYLT1, KNG1, SUGP1/MAU2, TBL2/MLXIPL) that were not significant in the original individual datasets, were not described in previous GWAS for the individual traits, and that presented a common associated variant between the studied phenotypes.
Conclusions
The discovery of four novel loci contributes to the understanding of the relationship between hemostasis and CV events and elucidate common genetic factors between these traits.
Keywords: blood coagulation, cardiovascular diseases, genetic pleiotropy, genome‐wide association study, hemostasis
Essentials.
Multi‐phenotype analysis of genetically correlated phenotypes may lead to novel discoveries.
Summary statistics of hemostatic traits and cardiovascular events were combined with metaUSAT.
We identified four novel associations with a shared variant between the studied phenotypes.
Our results shed light on the relationship between hemostatic traits and cardiovascular events.
1. INTRODUCTION
Genome‐wide association studies (GWAS) have identified dozens of loci underlying the variability of plasma levels for individual hemostatic traits. 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 Further, GWAS for venous thromboembolism (VTE), 9 , 10 coronary artery disease (CAD) 11 , 12 , 13 and ischemic stroke (IS), 11 , 14 have discovered 34, 169, and 20 genetic risk loci associated with these cardiovascular (CV) events, respectively.
Results from GWAS indicate that several of these hemostatic traits are genetically correlated with each other, sharing genetic loci that regulate their plasma levels. 1 , 4 , 5 , 6 , 7 , 8 There are also shared genetic loci between hemostatic traits and CV events, again suggesting common regulators and possibly a causal pathway between the hemostatic trait and the CV event. 4 , 7 , 8 , 9 , 12 , 14 The common regulatory loci between traits—even if the traits are not causally associated with each other—can be used to advance discovery of novel genetic loci common to the traits. This discovery can be accomplished with multi‐phenotype methods that incorporate summary statistics from several GWAS, increasing the statistical power to detect loci affecting two or more phenotypes by increasing the effective sample size. 15 , 16 , 17
In the present study, we used summary statistics of published GWAS from 7 hemostatic traits (FVII, FVIII, VWF, FXI, fibrinogen, PAI‐1, tPA), and 3 CV events (VTE, CAD, IS) to calculate their genetic correlations and to conduct multi‐phenotype meta‐analyses to detect new genetic loci not previously known to be associated with these phenotypes.
2. METHODS
2.1. Study design and resources
The setting of the project is the Cohorts of Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium Hemostasis Working Group. 18 We used GWAS summary statistics from seven hemostatic traits (FVII (N = 27 495), FVIII (N = 32 610), VWF (N = 46 354), FXI (N = 16 169), fibrinogen (N = 120 246), PAI‐1 (N = 19 599), tPA (N = 26 929)), and three CV events (VTE (N cases = 30 234, N controls = 172 122), CAD (N cases = 172 122, N controls = 566 864), IS (N cases = 60 341, N controls = 454 450)) to perform multi‐phenotype analyses. Summary statistics of FVII, 1 FVIII, 4 VWF, 4 VTE, 9 CAD 12 , 13 , 19 and IS, 14 come from trans‐ethnic analyses, while summary statistics of FXI, 6 fibrinogen, 5 tPA 8 and PAI‐1 7 are European ancestry only (additional information, including the sample sizes, detailed ancestry groups, confounders considered and data access URLs, of the phenotypes that have been used, is available in Supplementary Table S1). Summary statistics for FVII, FVIII, VWF, FXI, fibrinogen, tPA, PAI‐1, were obtained from the most recent CHARGE meta‐analysis data 18 and are available on dbGaP 20 (Appendix A). Data for VTE was obtained from INVENT 9 consortium and is available on request from corresponding authors (Appendix B). Data from IS were obtained from the MEGASTROKE Consortium, 14 and is available at https://www.megastroke.org/ (Appendix C). For CAD, we used METAL to perform an inverse variance weighted meta‐analysis between previously combined data from CARDIoGRAMplusC4D Consortium 19 and UK Biobank datasets 12 (available at https://data.mendeley.com/datasets/2zdd47c94h/1), and the Biobank Japan dataset available at https://humandbs.biosciencedbc.jp/en/hum0014‐v21 using METAL. All the included data have been published between December 2012 and October 2020. The overlap of individuals observed in our combinations of phenotypes ranges between 0 and 0.58. The overlap between the CARDIoGRAMplusC4D Consortium and UK Biobank datasets that were combined by other authors, and used in this project, was estimated to be <0.1 %. 12
2.2. Study of heritability and genetic correlation
We determined the heritability of each phenotype and genetic correlations (rg ) between all pairs of hemostatic traits, between each hemostatic factor and the CV events and between all pairs of CV events, using linkage disequilibrium (LD) score regression (LDSC). 21 LDSC uses a regression analysis between LD scores and the summary statistics of GWAS to provide an estimate of the shared heritability between phenotypes. 22 We used pre‐computed LD scores from the European population of 1000G project. 23 A subset of the European‐ancestry summary statistics was used in this step for each trait except for CAD where the European‐only meta‐analysis was not available. Alleles were merged with the HapMap3 single nucleotide polymorphisms (SNPs) list, 24 to avoid incompatibilities between phenotypes, as recommended by the authors, and missing variants were removed.
For the threshold of statistical significance for each genetic correlation, we applied a Bonferroni correction for multiple comparisons, considering all pairwise genetic correlations calculated (p <.05/45 = .001).
2.3. Multi‐phenotype analysis
We performed multi‐phenotype analyses using GWAS summary statistics from different combinations of traits using the metaUSAT R package. 17 metaUSAT is a statistical approach for testing genetic association with one or more phenotypes simultaneously, using only common variants between the phenotypes. metaUSAT allows summary data as input that includes overlapping samples, which can be a source of bias with other methods; further, it does not assume homogeneity of trait effects across studies. 17 Compared to similar methods, metaUSAT performs similarly while requiring less computational time. 25
In total, we performed 27 multi‐phenotype analyses, considering all pairs of hemostatic traits that showed significant genetic correlations (p <.001) (Supplementary Table S2), pairs of combinations between each hemostatic trait and each of the three CV events, and other combinations included based on biological aspects of the analyzed proteins: Fibrinogen‐FVII‐FXI‐tPA were analyzed because all of them are synthesized in the liver —although tPA is mostly produced by endothelial cells, recent studies that focused on the basal plasma tPA activity have also demonstrated the effects of hepatic produced tPA in fibrinolysis—. 26 Secondly, tPA was combined with FVIII and VWF, that are highly correlated, because it is known that these three phenotypes share loci like STXBP5 that are involved in endothelial exocytosis. 4 , 8 Finally, the combination of fibrinogen and FVII was included to potentially discover genetic insights to the antithrombin (AT) pathway. It is known that AT deficiency is a strong risk factor for VTE, and that AT inhibits the FVIIa‐tissue factor complex's activation of FX. 27 , 28 Moreover, AT modifies prothrombin's conversion of fibrinogen to fibrin. 29 Given this evidence, we hypothesize that potential common loci that regulate AT, FVII and fibrinogen might arise from this combination. Figure 1 shows all combinations that were analyzed.
FIGURE 1.
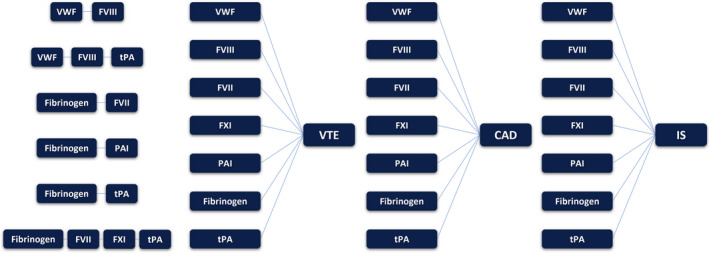
Schematic representation of the 27 multi‐phenotype combinations
For a metaUSAT p‐value (pvaluemultivariate) to be declared statistically significant, it needed to exceed a Bonferroni correction of the traditional GWAS statistical significance threshold to account for multiple testing for 27 multi‐trait combinations: 5 × 10−8/27 = 1.85 × 10−9. For those variants with statistically significant metaUSAT p‐values, we defined a locus as the genomic region +/− 500 kb around the variant with the lowest p‐value and any other variants that were in LD of r 2 > 0.2. We used HaploR R package to retrieve variants in LD with the lead variant in each locus (the variant with the lowest p‐value).
In order to identify novel associated loci, we considered the following steps (Figure 2): (1) we identified all loci that were statistically significant in the multi‐phenotype analysis (p‐valuemultivariate <1.85 x 10−9) (significant loci); (2) among these, we identified all loci with a lead variant that was at least nominally significant for two of the individual datasets used in each combination of phenotypes (lead variant p‐valueunivariate <.005) 30 , 31 , 32 (significant loci driven by more than one phenotype); (3) among the loci from step 2, we then narrowed it down to loci that were new for at least one of the phenotypes used in the combination, defined as loci where no other variant in the locus had a p‐value lower than 5 × 10−8 (p‐valueunivariate <5 × 10−8) in the GWAS we used, and the locus had not been previously detected in another GWAS for the same phenotypes (new loci for one of the phenotypes used in the combination); and (4) among the loci from step 3, we identified loci that were new for all the phenotypes used in the combination (new loci for all the phenotypes used in the combination). We used the GWAS catalog database, 33 (available at https://www.ebi.ac.uk/gwas/docs/file‐downloads) to detect loci that were published in previous GWAS. We used HaploReg v4 34 to retrieve previous results and biological annotations from the lead variants.
FIGURE 2.
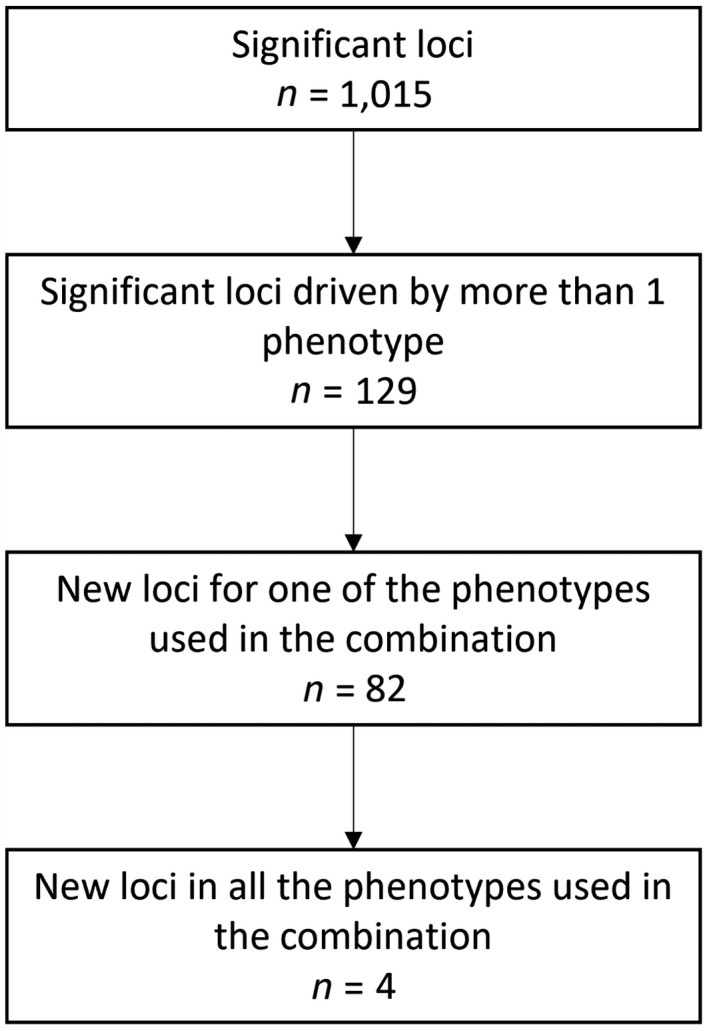
Schematic representation of the analysis plan for multi‐phenotype analyses
2.4. Trait‐trait colocalization
For novel loci that were new for all traits, we then performed additional colocalization analysis to look for the existence of common associated variants across multiple traits. We used COLOC 35 for loci associated with pairs of traits, and the R package HyPrColoc 36 for loci associated with more than two traits. We considered windows of +/− 500 kb around the lead variant to define loci. For each locus, COLOC returns posterior probabilities (PP) for 4 different hypotheses (Hn): PPH0 (the locus is not associated with any of the traits), PPH1/PPH2 (the locus is only associated with one of the traits), PPH3 (the locus is associated with both traits but there is no evidence of them sharing a causal variant), PPH4 (the locus is associated with both traits and LD patterns suggest the existence of a causal variant). We considered pleiotropic loci those that reached a conditional probability of colocalization (CPC) (PPH4 / (PPH4 + PPH3)) > 0.8, which is defined as the conditional probability of colocalization with one causal variant, assuming the existence of a signal in both traits. To consider pleiotropic loci in multiple traits, we performed colocalization using HyPrColoc, where posterior probabilities of colocalization > 0.7 were required. Regional plots for significant colocalizing loci were done using LocusCompare R package.
2.5. Trait‐tissues colocalization
In order to prioritize candidate causal genes, we used novel pleiotropic loci identified in previous steps and performed an additional trait‐expression colocalization analysis using RNAseq data from the Genotype‐Tissue Expression (GTEx) project. 37 First, we identified the lead variants that were significant expression quantitative trait loci (eQTL) and splicing quantitative trait loci (sQTL) for all tissues in GTEx V8 (available at https://www.gtexportal.org/home/datasets). Then, we performed colocalization with HyPrColoc, 36 between the two or more phenotypes and the GTEx eQTL and sQTL results, using the complete GTEx V8 files (available at https://console.cloud.google.com/storage/browser/gtex‐resources), in order to identify the functional tissue and elucidate on the biological mechanism causing the associations. We required a probability of colocalization > 0.7 to consider significant colocalization between traits and tissue expression.
We restricted eQTL and sQTL analyses to a subset of GTEx tissues that could be of interest in relation to CV events and hemostatic traits: vascular tissues (artery aorta, artery coronary, artery tibial) lipid metabolism related tissues (adipose subcutaneous, adipose visceral omentum), blood (whole blood) and liver. All loci that showed significant colocalization between at least two traits were analyzed for colocalization with tissue expression in those tissues.
3. RESULTS
3.1. Linkage disequilibrium score regression (LDSC)
Genetic correlations were calculated for every pair of phenotypes used, including hemostatic traits and CV events. In total, 45 genetic correlations were calculated, 24 of which presented nominal significant p‐values (p <.05) and seven were significant after applying multiple testing correction (p <.001). Among the seven genetic correlations that were significant, three were between hemostatic traits (VWF‐FVIII (rg = 0.86, p = 1.25 × 10−15), fibrinogen‐PAI‐1 (rg = 0.4, p = 9.29 × 10−5), fibrinogen‐tPA (rg = 0.28, p = 0.001)) and were used for multi‐phenotype analyses, 3 were between a CV event and a hemostatic trait (CAD‐fibrinogen (rg = 0.19, p = 6.6 × 10−6), CAD‐tPA (rg = 0.48, p = 4.9 × 10−7), CAD‐PAI‐1 (rg = 0.52, p = 4.55 × 10−6)) and one was between two CV events (CAD‐IS (rg = 0.5, p = 2.23 × 10−22)). All genetic correlations are shown in a heatmap in Figure 3 and are available at Supplementary Table S2.
FIGURE 3.
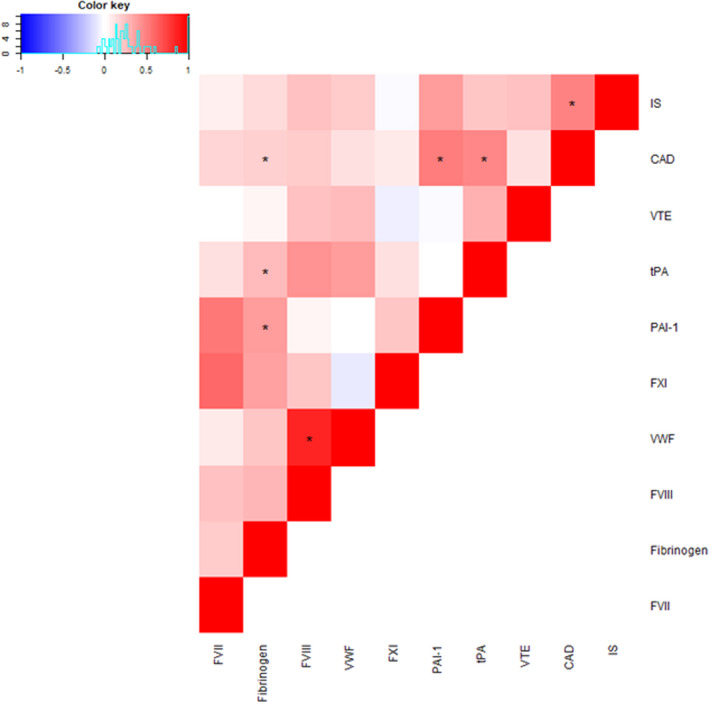
Heatmap of the genetic correlations between the two traits used in the multi‐phenotype analyses. * Indicates traits are significantly correlated with a p‐value <.001.
3.2. Multi‐phenotype analysis results
Overall, we performed 27 multi‐phenotype analyses: three (FVIII‐VWF, fibrinogen‐tPA and fibrinogen‐PAI‐1) based on significant genetic correlations between pairs of phenotypes, three (fibrinogen‐FVII‐tPA‐FXI, fibrinogen‐FVII and VWF‐FVIII‐tPA) due to previously known common regulatory biological pathways, and 21 between combinations of each of the seven hemostatic traits (FVII, FVIII, VWF, FXI, fibrinogen, tPA, PAI‐1) and each of the three CV events (VTE, CAD, IS).
The number of significant loci remaining in each step is represented in Figure 2. In total, we found 1 015 significant loci across the 27 multi‐phenotype combinations (Supplementary Table S3). Among them, 129 loci were driven by more than one of the phenotypes used in the combination (Supplementary Table S4), and 82 of them were new associations for one of phenotypes of the combination (Supplementary Table S5).
We found four novel associations that were not significant in the original individual datasets and had not been described in previous GWAS of the same traits (Table 1). Additional information on these loci, including the complete COLOC and HyPrColoc results are available at Supplementary Table S6. Figure 4 contains graphic representations of the p‐values and regional plots for each of these 4 loci.
TABLE 1.
Summary of the four novel associations identified in the 27 multi‐phenotype analyses that were not significant in the individual datasets and previous GWAS
| Marker Name | Traits | Variant | Effect Allele | MAF | Effect 1 | Effect 2 | Effect 3 | Effect 4 | Locus Name | p‐valuemultivariate | p‐valueunivariate 1 | p‐valueunivariate 2 | p‐valueunivariate 3 | p‐valueunivariate 4 | CPC b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3:194790434 | VTE‐FVII | rs3796159 | C | 0.2684 | 0.0605 | −0.0112 | ‐ | ‐ | XXYLT1 | 1.78 × 10−9 | 6.77 × 10−6 | 5.15 × 10−6 | ‐ | ‐ | 0.9727 |
| 3:186459927 | VTE‐FVIII | rs710446 | T | 0.4136 | −0.0601 | −0.012 | ‐ | ‐ | KNG1 | 4.12 × 10−11 | 3.71 × 10−7 | 1.1 × 10−6 | ‐ | ‐ | 0.9962 |
| 19:19407718 | CAD‐FIBR | rs10401969 | T | 0.0768 | 0.0386 | −0.0089 | ‐ | ‐ | SUGP1/MAU2 | 8 × 10−11 | 2.22 × 10−5 | 2.99 × 10−7 | ‐ | ‐ | 0.9827 |
| 7:72989390 | FIBR‐FVII‐FXI‐TPA | rs11974409 | A | 0.1816 | 0.0035 | 0.0154 | 0.0079 | 0.023 | TBL2/MLXIPL | 3.22 × 10−11 | .0019 | 1.87 x 10‐7 | 0.014737 | 1.71 x 10‐5 | 0.9896 a |
Posterior probability FVII‐tPA.
Conditional probability of colocalization.
FIGURE 4.
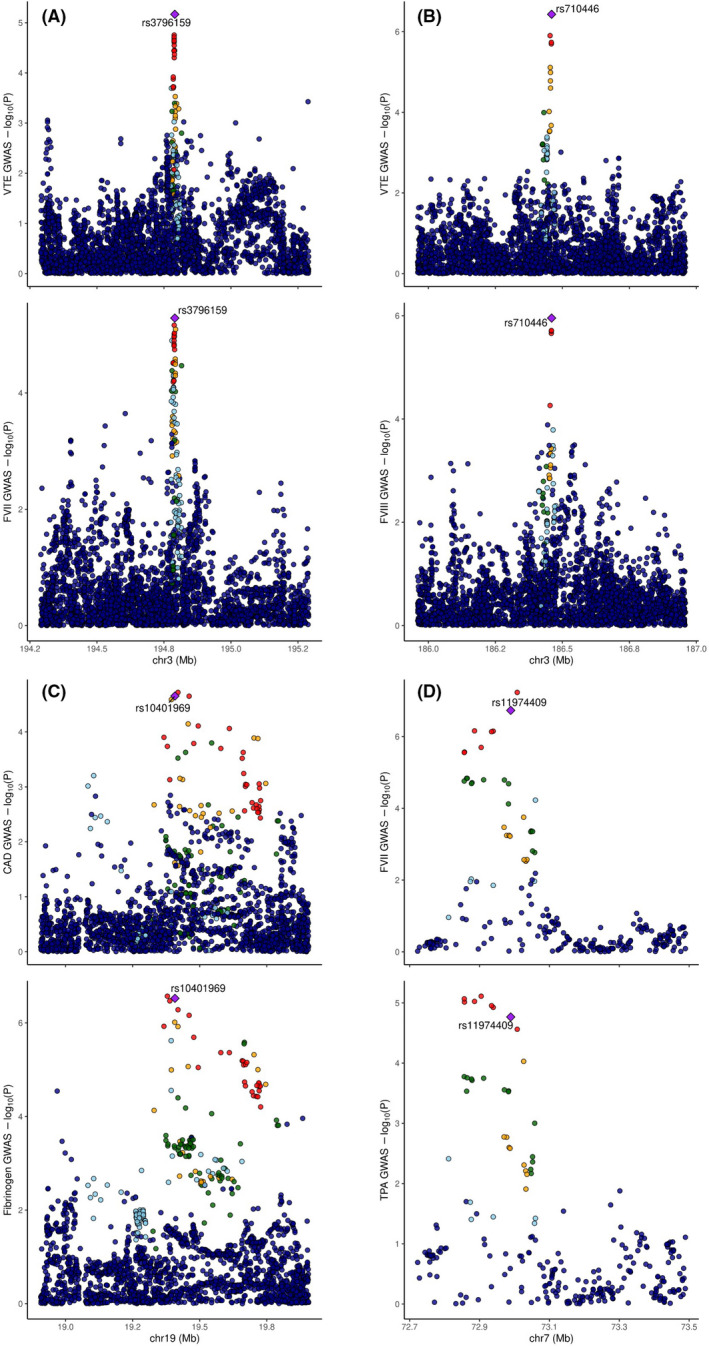
A: Regional plots for rs3796159 variant on XXYLT1 gene on VTE (top) and FVII (bottom). B: Regional plots for rs710446 variant on KNG1 gene on VTE (top) and FVIII (bottom). C: Regional plots for rs10401969 on SUGP1 gene on CAD (top) and fibrinogen (bottom). D: Regional plots for rs11974409 on TBL2 gene on FVII (top) and TPA (bottom).
3.3. XXYLT1
We detected a newly associated locus, with lead variant rs3796159, a 3’ UTR variant in XXYLT1 gene, in the multi‐phenotype analysis between VTE and FVII. Significant colocalization analysis (CPC > 0.8) (Figure 4A) in this locus suggested the existence of a common variant as a regulator of both phenotypes, VTE and FVII. Colocalization analysis in tissues indicated that rs3796159 is a significant eQTL for XXYLT1 in five different tissues (adipose subcutaneous, adipose visceral omentum, artery aorta, artery coronary, artery tibial) and HyPrColoc results (Supplementary Table S7) showed a triple significant colocalization (Posterior probability > 0.7) in adipose subcutaneous, adipose visceral omentum, artery aorta and artery tibial tissues in the XXLYT1 gene.
3.4. KNG1
Although previously identified in a candidate gene experiment as a risk factor for thrombosis 38 and suggestively associated in other GWAS for VTE, 39 this work represents the first time that the KNG1 locus, with lead variant rs710446, has been significantly associated to risk of VTE and FVIII in GWAS. rs710446 is a missense variant in the KNG1 gene that causes an amino acid change at the position 581 (Ile581Thr). 40 Colocalization results between VTE and FVIII (CPC > 0.8) suggest that rs710446 in KNG1 gene is associated both with FVIII and VTE (Figure 4B). Our results could not provide evidence for this variant being a significant eQTL or sQTL in any of the analyzed tissues which suggest an effect through protein function.
3.5. SUGP1/MAU2
CAD and fibrinogen multi‐trait analysis resulted in the identification of a novel association on SUGP1 gene (lead variant rs10401969, intronic). Colocalization analysis between CAD and fibrinogen (Figure 4C) implicated that there is a shared associated variant at this locus (CPC > 0.8), while the analysis in GTEx tissues (Supplementary Table S7) and colocalization using HyPrColoc indicated a significant triple colocalization between CAD, fibrinogen and the GTEx dataset in blood in MAU2 gene (Posterior probability > 0.7).
3.6. TBL2/MLXIPL
The multi‐phenotype combination of hemostatic proteins that are synthesized in the liver (fibrinogen‐FVII‐FXI‐tPA), revealed a new association on TBL2 gene (lead variant rs11974409, intronic). Although not reaching the significance threshold for the 4 phenotypes (Posterior probability > 0.8), significant colocalization results (Figure 4D) between FVII and tPA suggest that a shared causal variant regulates both phenotypes in this locus.
rs11974409 variant is an eQTL in adipose subcutaneous, adipose visceral omentum, artery aorta, and whole blood, and an sQTL in adipose subcutaneous and adipose visceral omentum tissues. HyPrColoc analysis results suggested the existence of a common causal variant that regulates FVII, tPA and the expression of three different genes (AC005089.1, MLXIPL, BCL7B) in adipose subcutaneous, adipose visceral omentum and blood tissues (Posterior probability > 0.7) (Supplementary Table S7), and a common causal variant that regulates FVII, tPA and the splicing of MLXIPL gene in adipose subcutaneous and adipose visceral omentum tissues (Posterior probability > 0.7) (Supplementary Table S8).
3.7. MYRF/TEMEM258/FADS1/FADS2
Additionally, a novel association on MYRF/TMEM258/FADS1/FADS2 locus was detected in the multi‐phenotype analysis between IS‐VWF. Although just below the limit of significance (p‐valuemultivariate = 1.64 × 10−8) (Supplementary Table S6), MYRF/TMEM8/FADS1/FADS2 locus has also been identified in the combinations between VWF and the other two CV events (VTE and CAD), (lead variant rs174528, intronic in MYRF gene). This association was reported for VTE in a previous European GWAS 10 and also for CAD, 13 but has not been associated with VWF or IS before. The three colocalization analyses between VWF and the CV events (Supplementary Table S6) in this locus suggest the existence of a variant regulating all traits (CPC > 0.8). HyPrColoc analysis revealed significant colocalization (Posterior probability > 0.7) between VWF, VTE and FADS1 gene expression in artery tibial and liver tissues (Supplementary Table S7), while also suggested —not significantly— (0.7 > Posterior probability > 0.5) an effect in splicing regulation in adipose visceral omentum in FADS2 and FEN1 genes with VTE and CAD (Supplementary Table S8). We were unable to identify triple colocalizations between VWF, IS and gene regulation.
4. DISCUSSION
We performed a multi‐phenotype approach using correlated hemostatic traits and three CV events and detected four novel pleiotropic loci that had not been previously described in association with these hemostatic traits or CV events.
Given a common locus between two or more phenotypes, three scenarios are possible: (1) there are different causal variants associated with the different traits, (2) the same variant associates with the different traits separately (horizontal pleiotropy), or (3) the variant associates with one trait, which in turn causes association with another trait (vertical pleiotropy). 41 While our analyses did not allow to differentiate between horizontal and vertical pleiotropy, we have found evidence of common variants in the four new pleiotropic loci, which agrees with the previous notion that pleiotropy is common between variants associated with correlated disease traits. 42 Common genetic regulators, however, do not mean that the associated phenotypes are causally associated. Causal associations between related phenotypes can be explored through Mendelian randomization (MR) methods.
We found a total of 1015 significant loci across all the multi‐phenotype combinations. Among these, 129 were driven by more than one phenotype, of which 46 were found in combinations with CAD, 38 with VTE, 28 in combinations between hemostatic traits, and 16 with IS. Finally, among the 82 loci that were new for at least one of the phenotypes used in the combination, 30 were identified in combinations with CAD, 23 with VTE, 15 between hemostatic traits, and 13 with IS.
4.1. XXYLT1 and regulation of FVII
XXYLT1 codes for xyloside xylotransferase 1, an enzyme that elongates O‐linked glycans in the epidermal growth factor (EGF) repeats of O‐linked glycosylated proteins like FVII. 43 However, the direction of the effect of this variant suggests a decrease in FVII levels for allele C and an increase in the risk of VTE. In addition, MR analyses previously performed between VTE and FVII did not conclusively identify FVII as a cause of VTE, 1 which suggests that the common variant at this locus on xyloside xylotransferase 1 enzyme could be affecting both phenotypes independently, through expression of XXYLT1 in adipose subcutaneous, adipose visceral omentum, artery aorta or artery tibial tissues. In this direction, it is known that other coagulation factors, like factor IX, are also glycosylated in the EGF repeats. 44 It would be plausible to speculate that XXYLT1 could affect FVII levels and also other EGF‐glycosylated proteins that would eventually modify VTE risk. Therefore, further research in this locus is recommended to fully understand the possible relationship between XXYLT1, FVII and VTE, and the possible effect that other hemostatic proteins could have in this association.
4.2. KNG1 and risk of VTE through FVIII levels
The protein encoded by this gene, Kininogen‐1 (KNG1), is the precursor of two other proteins, obtained through alternative splicing: high‐molecular‐weight kininogen (HMWK) and low‐molecular‐weight kininogen (LMWK). Through a process facilitated by Factor XII (FXII), the peptide bradykinin is cleaved from HMWK by the enzyme kallikrein. 45
There is strong biological evidence that associate KNG1 gene with the coagulation system. 40 , 46 , 47 HMWK, along with FXII and prekallikrein (PK) complex, conform the plasma kallikrein‐kinin system (KKS), that plays an important role in human physiology. The activation of KKS components results in the induction of genes and biomolecules that participate in blood coagulation, among other processes. 48 , 49 Bradykinin, on its turn, is an important molecule involved in vascular permeability and also in mechanism of pain. 45
We have previously shown 6 , 50 , 51 that the lead variant on KNG1 (rs710446) is also strongly associated to activated partial thromboplastin time (aPTT), prekallirein, FXI and coagulation activity of FVIII in a candidate gene experiment, indicating a pleiotropic effect of this gene on regulating the intrinsic pathway of coagulation, 52 resulting in modified risk of VTE.
Although the association with VTE has been demonstrated in candidate‐gene studies, 38 the combination between VTE‐FVIII, enhanced the association, suggesting a plausible functional relationship between KNG1 and FVIII that had never been reported in GWAS. This association could be explained by a putative regulation of KNG1 also on FVIII, which would imply an effect of KNG1 on the common pathway of coagulation. Associations of KNG1 with the entire coagulation cascade, and not just the intrinsic pathway, have been proposed by others.72 The significant colocalization analysis between VTE and FVIII in this locus aligns with previous evidence and suggests that rs710446 affects the regulation of both phenotypes along with other related hemostatic phenotypes.
4.3. SUGP1/MAU2 and CAD risk through fibrinogen levels
CAD and fibrinogen multi‐trait analysis resulted in the identification of a new pleiotropic locus on SUGP1/MAU2 genes, with lead variant rs10401969. SUGP1/MAU2 is a pleiotropic locus that has been associated to lipid's metabolism traits levels (total cholesterol, apoliprotein B, triglycerides), liver related proteins levels (alanine transaminase, asparatate aminotransferase, alanine aminotransferase), blood‐related phenotypes (red cell distribution width, mean reticulocyte volume), type 2 diabetes and cirrhosis. 33 This locus has also been associated to CAD in candidate gene studies in Chinese and Caucasian populations but never in GWAS studies. 53 , 54 SUGP1 codes for a protein called SURP and G patch domains‐containing protein 1 (SUGP1), that it is believed to function in pre‐mRNA splicing mechanisms. 55 This is the first time that SUGP1 is associated with coagulation factors and colocalization results suggest that the common variant at this locus is regulating both, CAD and fibrinogen. The identification of our lead variant, rs10401969, as a significant eQTL for MAU2 gene in blood, and the identification of this locus in a significant colocalization between CAD, fibrinogen and eQTL data in blood, also suggests the existence of a variant regulating both phenotypes that would take place through the MAU2 gene expression. MAU2 codes for MAU2 chromatid cohesion factor homolog and has an important role in loading the cohesion complex to DNA. 56 , 57 MAU2 has never been associated to coagulation factor levels.
Fibrinogen levels have been found significantly higher in cases of CAD in epidemiological studies, 58 , 59 although MR studies have only been able to demonstrate a small causal effect using multi‐variant MR approaches. 60 , 61 Further evidence is needed to clearly elucidate if the effect of this variant on SUGP1/MAU2 locus on CAD is through fibrinogen levels or if this locus influences both phenotypes in parallel.
4.4. Liver produced proteins and the TBL2/MLXIPL locus
The TBL2/MLXIPL locus has been associated with other phenotypes of interest related to lipids metabolism levels (triglycerides, high density lipoprotein, low density lipoprotein) and other liver related proteins levels (alkaline phosphatase, C‐reactive protein, gamma glutamyl transferase or alanine aminotransferase), 33 but this is the first time that a variant located on TBL2 reaches the significance threshold in a GWAS involving hemostatic factors. Also, in a previous candidate gene study for FVII levels, the variant rs7777102, located upstream the MLXIPL gene and ~70 kb away rs11974409 (D’ = 0.85, R 2 = 0.44, in 1000G project European population 51 ), was found associated to FVII. 51 TBL2 codes for an endoplasmic reticulum transmembrane protein called transducin (β)‐like 2 (TBL2) that, upon ER stress, interacts with PERK (PKR‐like ER‐resident kinase) and is able to regulate ATF4 (activation transcription factor 4) translation. It has also been demonstrated that TBL2 has a WD40 (beta‐transducin repeat) domain that is essential for the association with mRNA of ATF4. 62 , 63 MLXIPL codes for Carbohydrate response element binding protein (ChREBP), a transcription factor highly enriched in the liver with a key role in lipids metabolism. ChREBP has also shown a response for glucose metabolites that change its cellular location and stability and also imply post‐translational modifications. ChREBP binds to several proteins that are crucial to induce its nuclear translocation or biding to nuclear receptors. 64 Considering the previous results obtained in a candidate gene studies in FVII, 51 that associated this locus to FVII levels, together with the significant colocalization results between FVII and tPA, and the significant results also of HyPrColoc with several tissues, all suggest that the TBL2/MLXIPL locus has a pleiotropic effect on the expression of several hemostatic traits. The colocalization results in tissues suggest MLXIPL gene as a good candidate gene, with a common variant in this locus regulating MLXIPL expression and splicing in adipose visceral and adipose visceral omentum tissues, FVII levels, and TPA levels at the same time.
4.5. MYRF/TMEM258/FADS1/FADS2 and its effect on CV events
MYRF codes for a membrane‐associated transcription factor, that participates in the activation of myelin genes and that has been associated to brain development issues. 65 , 66 Previous GWAS have also associated the genomic region of MYRF and TMEM258 genes to hematologic and lipid metabolism traits. 33 TMEM258 codes for a protein with two predicted transmembrane domains, with no clear function in vivo, that has been associated with endoplasmic reticulum stress when knocked out and as an important regulator of intestinal hemostasis. TMEM258 has also been described as a potential causal gene of cardiovascular traits and as a regulatory site of abdominal visceral fat. 67 , 68
Although not new for VTE and CAD, the identification of the same locus in the multi‐phenotype analyses with all 3 CV events and a hemostatic trait supports the idea that the MYRF/TMEM258/FADS1/FADS2 locus is common regulator. Significant colocalization results between VWF and the cardiovascular outcomes, also support this idea. Our effort to prioritize a causal gene through the colocalization analysis in different tissues, revealed that a common variant at this locus regulates expression of FADS1 gene in artery tibial and liver and the splicing regulation of FADS2, although the colocalization was not significant. FADS1 and FADS2 code for members of the fatty acid desaturase gene family that catalyze several steps in the formation of omega‐3 and omega‐6 fatty acids. 69 The rs174547 variant located on FADS1, in high LD with our lead variant rs174528 (D’ = 1, R 2 = 0.84) in the European population of 1000 Genomes project, 70 has been implicated in the risk of suffering multiple CV events, including VTE, CAD and IS, in a previous MR study. 71 Our results clearly support a regulatory role of this locus on several CV events and suggest the involvement of VWF in the association between FADS1/FADS2 and CV events.
4.6. Strengths and limitations
This is the first systematic multi‐phenotype analysis using summary data for hemostatic traits related to CV events to increase power to detect loci associated with more than one related phenotype. We have leveraged data from the leading consortia worldwide analyzing genetics of hemostatic traits, VTE, CAD, and IS, often providing the largest datasets currently available. We consider this, one of the most major strengths of this work. Moreover, most phenotypes included trans‐ethnic population, which give a broader transferability of the results.
We are aware of the existence of several limitations in this work. First, there are notable differences between the sample sizes of the hemostatic traits used, the largest one being fibrinogen (N = 120 246 cases) and the smallest one PAI‐1 (N = 19 599), which leads to differences in statistical power between multi‐phenotype analyses. Second, we were limited to use summary statistics of mostly European origin to calculate genetic correlations, given the lack of good references in other populations to generate the LD scores, which implies that these results cannot be applied to global populations. LDSC filters out variants with low sample size. For some phenotypes this information was not available, and we used the maximum sample size of the phenotype. This could have created some error. Third, we are also aware that there are limitations associated with the use of GTEx data. This data has limited sample sizes that vary greatly from tissue to tissue. For example, the number of liver samples (N = 208) is considerably lower than the samples of tissues such as artery tibial (N = 584) or adipose subcutaneous (N = 581), which may end up in differences in power to detect associations. The lower numbers of liver samples may have affected our power to detect some of the identified variants as significant eQTL in the liver, and therefore the implication of causal tissues should be interpreted with caution. Finally, we are not providing functional validation of these results. Therefore, further experiments are needed to confirm the implication of the novel suggested loci in disease.
5. CONCLUSIONS
We have shown that the multi‐phenotype analysis of biologically related phenotypes expands discovery of newly associated loci. Using summary GWAS data from hemostasis and CV events, we identified four colocalizing novel loci that were not identified as statistically significant in the source datasets and have not been described in other GWAS of the phenotypes involved. Although our strategy does not allow to unequivocally identify the causal variant or variants at each locus, the colocalization results suggest the existence of common regulatory variants at the newly identified loci.
Some of these loci appear to represent genes that may simultaneously regulate more than one hemostatic trait (horizontal pleiotropy), and some seem to reflect the risk mechanism from a gene to one or more CV events through regulation of a hemostatic factor (vertical pleiotropy), 41 therefore revealing novel biological mechanisms. Both cases of pleiotropy are novel interesting insights that will help understand the pathophysiology of clinical CV events.
CONFLICT OF INTEREST
S.M. Damrauer is named as a co‐inventor on a government‐owned US Patent application related to the use of genetic risk prediction for venous thromboembolic disease filed by the US Department of Veterans Affairs in accordance with Federal regulatory requirements. All other authors have no conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
M. Sabater‐Lleal, N.L Smith and P.S. de Vries conceived and designed the study and provided access to the data. G. Temprano‐Sagrera performed, analyzed, and interpreted the data and drafted the manuscript. C. M. Sitlani, W. P. Bone, and M. Martin‐Bornez, analyzed data and contributed to method selection and interpretation. B. F. Voight, A. C. Morrison, S. M. Damrauer, P. S. de Vries N. L. Smith and M. Sabater‐Lleal contributed to writing and editing the manuscript. All the authors revised and approved the final version of the manuscript.
Supporting information
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
ACKNOWLEDGEMENTS
This study is supported in part by the National Heart, Lung, and Blood Institute grants HL134894, HL139553, and HL141291. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; the Department of Veterans Affairs; the United States Government or the US Department of Health and Human Services. G. Temprano‐Sagrera is supported by the Pla Estratègic de Recerca i Innovació en Salut (PERIS) grant from the Catalan Department of Health for junior research personnel (SLT017/20/000100). S.M. Damrauer is supported by IK2‐CX001780. M. Sabater‐Lleal is supported by a Miguel Servet contract from the ISCIII Spanish Health Institute (CP17/00142) and co‐financed by the European Social Fund.
Data on coronary artery disease has been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from www.CARDIOGRAMPLUSC4D.ORG. We thank the authors P. van der Harst and N. Verwejj who have made available the meta‐analysis between CARDIoGRAMplusC4D and UK Biobank data on Mendeley (https://data.mendeley.com/datasets/gbbsrpx6bs/1).
Part of the data used for this research was provided by M. Kubo and is available at the website of the National Bioscience Database Center (NBDC; https://humandbs.biosciencedbc.jp/en/hum0014‐v21#cad) of the Japan Science and Technology Agency (JST). We also thank the MEGASTROKE consortium for making the IS data available at https://www.megastroke.org/. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html. Appendix A contains a list of investigators belonging to the CHARGE consortium Hemostasis Working Group that contributed to the hemostatic summary data. Appendix B contains a list of investigators from the INVENT consortium that contributed to the VTE summary data. Appendix C contains a list of investigators from the MEGASTROKE consortium that contributed to the IS summary data.
APPENDIX A.
Abbas Dehghan1, Adam SHeath2, Alanna CMorrison2, Alex PReiner3, Andrew Johnson4, Anne Richmond5, Annette Peters6, Astrid van Hylckama Vlieg7 Barbara McKnight8, Bruce MPsaty9, Caroline Hayward5, Cavin Ward‐Caviness10, Christopher O’Donnell11, Daniel Chasman12, David PStrachan13, David ATregouet14, Dennis Mook‐Kanamori7, Dipender Gill1, Florian Thibord4, Folkert WAsselbergs15, Frank W.G. Leebeek16, Frits RRosendaal7, Gail Davies17, Georg Homuth18, Gerard Temprano19, Harry Campbell20, Herman ATaylor21, Jan Bressler2, Jennifer EHuffman22, Jerome IRotter21, Jie Yao21, James FWilson5, Joshua CBis23, Julie MHahn2, Karl CDesch24, Kerri LWiggins23, Laura MRaffield25, Lawrence FBielak26, Lisa RYanek27, Marcus EKleber28, Maria Sabater‐Lleal19, Martina Mueller6, Maryam Kavousi29, Massimo Mangino30, Melissa Liu4, Michael RBrown2, Matthew PConomos8, Min‐A Jhun26, Ming‐Huei Chen4, Moniek P.M. de Maat16, Nathan Pankratz31, Nicholas LSmith3, Patricia APeyser26, Paul Elliot1, Paul Sde Vries2, Peng Wei32, Philipp SWild33, Pierre EMorange34, Pim van der Harst35, Qiong Yang36, Ngoc‐Quynh Le19, Riccardo Marioni37, Ruifang Li7, Scott MDamrauer38, Simon RCox17, Stella Trompet39, Stephan BFelix40, Uwe Völker18, Weihong Tang41, Wolfgang Koenig42, J. Wouter Jukema43, Xiuqing Guo21.
1Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, London, UK. 2Human Genetics Center, Department of Epidemiology, Human Genetics, and Environmental Sciences; School of Public Health, The University of Texas Health Science Center at Houston, Houston, Texas, USA. 3Department of Epidemiology, University of Washington, Seattle, WA, USA. 4National Heart Lung and Blood Institute, Division of Intramural Research, Population Sciences Branch, The Framingham Heart Study, Framingham, MA, USA. 5MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh, Western General Hospital, Edinburgh, Scotland. 6Research Unit Molecular Epidemiology, Helmholtz Zentrum München, München, Germany. 7Department of Clinical Epidemiology, Leiden University Medical Center, the Netherlands. 8Department of Biostatistics, University of Washington, Seattle, WA. 9Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA, United States. 10Office of Research and Development, U.S. Environmental Protection Agency, Chapel Hill, NC, USA. 11Cardiology, VA Boston Healthcare System, Boston, MA, USA. 12Division of Preventive Medicine, Brigham and Women's Hospital, Boston, MA, USA. 13Population Health Research Institute, St George's University of London, London, UK. 14Bordeaux Population Health Research Center, University of Bordeaux, Bordeaux, France. 15Department of Cardiology, Division Heart & Lungs, University Medical Center Utrecht, Utrecht University, Utrecht, the Netherlands. 16Department of Hematology, Erasmus Medical Center, Rotterdam, the Netherlands. 17Lothian Birth Cohorts, Department of Psychology, University of Edinburgh, Edinburgh, UK. 18Department of Functional Genomics, Interfaculty Institute for Genetics and Functional Genomics, University Medicine Greifswald, Greifswald, Germany. 19Unit of Genomics of Complex Diseases. Sant Pau Biomedical Research Institute (IIB‐Sant Pau), Barcelona, Spain. 20Global Health Research, Usher Institute for Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, Scotland, UK. 21The Institute for Translational Genomics and Population Sciences, Department of Pediatrcs, The Lundquist Institute for Biomedical Innovation at Harbor‐UCLA Medical Center, Torrance, CA USA. 22Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC), VA Boston Healthcare System, Boston, MA, USA. 23Cardiovascular Health Research Unit Department of Medicine University of Washington Seattle Washington USA. 24Department of Pediatrics, University of Michigan, C.S. Mott Children's Hospital, Ann Arbor, MI, USA. 25Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA. 26Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI. 27Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, MD, USA. 28SYNLAB MVZ für Humangenetik Mannheim, Mannheim, Germany. 29Department of Epidemiology, Erasmus Medical Center, University Medical Center Rotterdam, Rotterdam, Netherlands. 30Department of Twin Research and Genetic Epidemiology, Kings College London, London, UK. 31Department of Laboratory Medicine and Pathology, University of Minnesota Medical School, Minneapolis, MN, USA. 32Department of Biostatistics, The University of Texas MD Anderson Cancer Center, Houston. 33Department of Cardiology, Cardiology I, University Medical Center, Johannes Gutenberg University Mainz, Mainz, Germany. 34Hematology Laboratory, La Timone University Hospital of Marseille, Marseille, France. 35Department of Cardiology, University Medical Center Utrecht, Utrecht, Netherlands. 36Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA. 37Centre for Genomic and Experimental Medicine, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK. 38Department of Surgery, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA. 39Section of Gerontology and Geriatrics, Department of Internal Medicine, Leiden University Medical Center, Leiden, The Netherlands. 40Department of Internal Medicine B, University Medicine Greifswald, Greifswald, Germany. 41School of Public Health, University of Minnesota, Minneapolis, MN, USA. 42DZHK (German Centre for Cardiovascular Research), partner site Munich Heart Alliance, Munich, Germany. 43Department of Cardiology, Leiden University Medical Center, The Netherlands.
APPENDIX B.
Sara Lindstrom, PhD1, Lu Wang, PhD2, Erin NSmith, PhD3, William Gordon, MS4, Astrid van Hylckama Vlieg, PhD5, Mariza de Andrade, PhD6, Jennifer ABrody, BA7, Jack WPattee, BA8, Jeffrey Haessler, MS9, Ben MBrumpton, PhD, MPH10, Daniel IChasman, PhD11, Pierre Suchon, MD‐PhD12, Ming‐Huei Chen, PhD13, Constance Turman, MS14, Marine Germain15, Kerri LWiggins, MS, RD16, James MacDonald, MS17, Sigrid KBraekkan, PhD18, Sebastian MArmasu, MS19, Nathan Pankratz, PhD20, Rabecca DJackson, MD21, Jonas BNielsen, MD, PhD22, Franco Giulianini, PhD23, Marja KPuurunen, MD, PhD24, Manal Ibrahim, MD25, Susan RHeckbert, MD, PhD26, Theo KBammler, PhD27, Kelly AFrazer, PhD28, Bryan MMcCauley, MS29, Kent Taylor, PhD30, James SPankow, PhD, MPH31, Alexander PReiner, MD, MPH32, Maiken EGabrielsen, PhD33, Jean‐François Deleuze, PhD34, Chris JO'Donnell, MD35, Jihye Kim, PhD, MPH36, Barbara McKnight, PhD37, Peter Kraft, PhD38, John‐Bjarne Hansen, MD, PhD39, Frits RRosendaal, MD, PhD40, John AHeit, MD41, Bruce MPsaty, MD, PhD42, Weihong Tang, MD, PhD43, Charles Kooperberg, PhD44, Kristian Hveem, MD, PhD45, Paul MRidker, MD, MPH46, Pierre‐Emmanuel Morange, MD‐PhD47, Andrew DJohnson, PhD48, Christopher Kabrhel, MD MPH49, David‐Alexandre Trégouët, PhD50, Nicholas LSmith, PhD51.
Author Affiliations: 1Department of Epidemiology, University of Washington, Seattle, Washington, USA; Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA. 2Department of Environmental and Occupational Health Sciences, University of Washington, Seattle, Washington, USA. 3Department of Pediatrics and Rady Children's Hospital, University of California San Diego, La Jolla, USA; K.G. Jebsen Thrombosis Research and Expertise Center, Department of Clinical Medicine, UiT – The Arctic University of Norway, Tromsø, Norway. 4Department of Epidemiology, University of Washington, Seattle, WA, USA. 5Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands. 6Department of Health Sciences Research, Mayo Clinic, Rochester, MN USA. 7Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle WA USA. 8Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN USA. 9Division of Public Health, Fred Hutchinson Cancer Research Center, Seattle WA, USA. 10K.G. Jebsen Center for Genetic Epidemiology, Department of Public Health and Nursing, NTNU, Norwegian University of Science and Technology, Trondheim, Norway; MRC Integrative Epidemiology Unit, University of Bristol, Bristol, UK; Clinic of Thoracic and Occupational Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway. 11Division of Preventive Medicine, Brigham and Women's Hospital, Boston, USA; Harvard Medical School, Boston, USA. 12Laboratory of Haematology, La Timone Hospital, Marseille, France; C2VN, Aix Marseille University, INSERM, INRA, C2VN, Marseille, France. 13Population Sciences Branch, Division of Intramural Research, National Heart, Lung and Blood Institute, Bethesda, MD, USA; NHLBI and Boston University's The Framingham Heart Study, Framingham, MA, USA. 14Program in Genetic Epidemiology and Statistical Genetics, Harvard T.H. Chan School of Public Health, Boston, MA, USA. 15INSERM UMR_S 1219, Bordeaux Population Health Research Center, University of Bordeaux, France. 16Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle WA USA. 17Department of Environmental and Occupational Health Sciences, University of Washington, Seattle, Washington, USA. 18K.G. Jebsen Thrombosis Research and Expertise Center, Department of Clinical Medicine, UiT – The Arctic University of Norway, Tromsø, Norway; Division of Internal Medicine, University Hospital of North Norway, Tromsø, Norway. 19Health Sciences Research, Mayo Clinic, Rochester, MN USA. 20Department of Laboratory Medicine and Pathology, School of Medicine, University of Minnesota, Minneapolis, MN, USA. 21Division of Endocrinology, Diabetes and Metabolism, The Ohio State University, Columbus OH, USA. 22Department of Internal Medicine, Division of Cardiology, University of Michigan Medical School, Ann Arbor, Michigan, USA. 23Division of Preventive Medicine, Brigham and Women's Hospital, Boston, USA. 24NHLBI and Boston University's The Framingham Heart Study, Framingham, MA, USA. 25Laboratory of Haematology, La Timone Hospital, Marseille, France.; C2VN, Aix Marseille University, INSERM, INRA, C2VN, Marseille, France. 26Department of Epidemiology, University of Washington, Seattle, Washington, USA; Kaiser Permanente Washington Health Research Institute, Kaiser Permanente Washington, Seattle WA USA. 27Department of Environmental and Occupational Health Sciences, University of Washington, Seattle, Washington, USA. 28Department of Pediatrics and Rady Children's Hospital, University of California San Diego, La Jolla, USA; K.G. Jebsen Thrombosis Research and Expertise Center, Department of Clinical Medicine, UiT – The Arctic University of Norway, Tromsø, Norway; Institute of Genomic Medicine, University of California San Diego, La Jolla, California, USA. 29Health Sciences Research, Mayo Clinic, Rochester, MN USA. 30Los Angeles Biomedical Research Institute and Department of Pediatrics, Harbor‐UCLA Medical Center, Torrence CA 90502, USA. 31Division of Epidemiology and Community Health, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 32Department of Epidemiology, University of Washington, Seattle WA, United States; Division of Public Health, Fred Hutchinson Cancer Research Center, Seattle WA, United States. 33K.G. Jebsen Center for Genetic Epidemiology, Department of Public Health and Nursing, NTNU, Norwegian University of Science and Technology, Trondheim, Norway. 34Centre National de Recherche en Génomique Humaine, Direction de la Recherche Fondamentale, CEA, 91057 Evry, France; CEPH, Fondation Jean Dausset, Paris, France. 35Million Veteran's Program, Veteran's Administration, Boston, MA; Population Sciences Branch, Division of Intramural Research, National Heart, Lung and Blood Institute, Bethesda, MD, USA; NHLBI and Boston University's The Framingham Heart Study, Framingham, MA, USA. 36Program in Genetic Epidemiology and Statistical Genetics, Harvard T.H. Chan School of Public Health, Boston, MA, USA. 37Department of Biostatistics, University of Washington, Seattle WA USA; Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA. 38Program in Genetic Epidemiology and Statistical Genetics, Harvard T.H. Chan School of Public Health, Boston, MA, USA. 39K.G. Jebsen Thrombosis Research and Expertise Center, Department of Clinical Medicine, UiT – The Arctic University of Norway, Tromsø, Norway; Division of Internal Medicine, University Hospital of North Norway, Tromsø, Norway. 40Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands. 41Health Sciences Research, Mayo Clinic, Rochester, MN USA. 42Cardiovascular Health Research Unit, Departments of Medicine, Epidemiology, and Health Services, University of Washington, Seattle WA USA; Kaiser Permanente Washington Health Research Institute, Kaiser Permanente Washington, Seattle WA USA. 43Division of Epidemiology and Community Health, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 44Division of Public Health, Fred Hutchinson Cancer Research Center, Seattle WA, United States. 45K.G. Jebsen Center for Genetic Epidemiology, Department of Public Health and Nursing, NTNU, Norwegian University of Science and Technology, Trondheim, Norway. 46Division of Preventive Medicine, Brigham and Women's Hospital, Boston, USA; Harvard Medical School, Boston, USA. 47C2VN, Aix Marseille Univ, INSERM, INRA, C2VN, Marseille, France; Laboratory of Haematology, La Timone Hospital, Marseille, France; CRB Assistance Publique ‐ Hôpitaux de Marseille, HemoVasc (CRB AP‐HM HemoVasc), Marseille, France. 48Population Sciences Branch, Division of Intramural Research, National Heart, Lung and Blood Institute, Bethesda, MD, USA; NHLBI and Boston University's The Framingham Heart Study, Framingham, MA, USA. 49Center for Vascular Emergencies, Department of Emergency Medicine, Massachusetts General Hospital; Channing Division of Network Medicine, Brigham and Women's Hospital; Harvard Medical School. 50INSERM UMR_S 1219, Bordeaux Population Health Research Center, University of Bordeaux, France. 51Department of Epidemiology, University of Washington, Seattle, Washington, USA; Kaiser Permanente Washington Health Research Institute, Kaiser Permanente Washington, Seattle WA USA; Seattle Epidemiologic Research and Information Center, Department of Veterans Affairs Office of Research and Development, Seattle WA USA.
APPENDIX C.
Rainer Malik1, Ganesh Chauhan2, Matthew Traylor3, Muralidharan Sargurupremraj4,5, Yukinori Okada6,7,8, Aniket Mishra4,5, Loes Rutten‐Jacobs3, Anne‐Katrin Giese9, Sander W van der Laan10, Solveig Gretarsdottir11, Christopher D Anderson12,13,14,14, Michael Chong15, Hieab HH Adams16,17, Tetsuro Ago18, Peter Almgren19, Philippe Amouyel20,21, Hakan Ay22,13, Traci M Bartz23, Oscar R Benavente24, Steve Bevan25, Giorgio B Boncoraglio26, Robert D Brown, Jr.27, Adam S Butterworth28,29, Caty Carrera30,31, Cara L Carty32,33, Daniel I Chasman34,35, Wei‐Min Chen36, John W Cole37, Adolfo Correa38, Ioana Cotlarciuc39, Carlos Cruchaga40,41, John Danesh28,42,43,44, Paul IW de Bakker45,46, Anita L DeStefano47,48, Marcel den Hoed49, Qing Duan50, Stefan T Engelter51,52, Guido J Falcone53,54, Rebecca F Gottesman55, Raji P Grewal56, Vilmundur Gudnason57,58, Stefan Gustafsson59, Jeffrey Haessler60, Tamara B Harris61, Ahamad Hassan62, Aki S Havulinna63,64, Susan R Heckbert65, Elizabeth G Holliday66,67, George Howard68, Fang‐Chi Hsu69, Hyacinth I Hyacinth70, M Arfan Ikram16, Erik Ingelsson71,72, Marguerite R Irvin73, Xueqiu Jian74, Jordi Jiménez‐Conde75, Julie A Johnson76,77, J Wouter Jukema78, Masahiro Kanai6,7,79, Keith L Keene80,81, Brett M Kissela82, Dawn O Kleindorfer82, Charles Kooperberg60, Michiaki Kubo83, Leslie A Lange84, Carl D Langefeld85, Claudia Langenberg86, Lenore J Launer87, Jin‐Moo Lee88, Robin Lemmens89,90, Didier Leys91, Cathryn M Lewis92,93, Wei‐Yu Lin28,94, Arne G Lindgren95,96, Erik Lorentzen97, Patrik K Magnusson98, Jane Maguire99, Ani Manichaikul36, Patrick F McArdle100, James F Meschia101, Braxton D Mitchell100,102, Thomas H Mosley103,104, Michael A Nalls105,106, Toshiharu Ninomiya107, Martin J O'Donnell15,108, Bruce M Psaty109,110,111,112, Sara L Pulit113,45, Kristiina Rannikmäe114,115, Alexander P Reiner65,116, Kathryn M Rexrode117, Kenneth Rice118, Stephen S Rich36, Paul M Ridker34,35, Natalia S Rost9,13, Peter M Rothwell119, Jerome I Rotter120,121, Tatjana Rundek122, Ralph L Sacco122, Saori Sakaue7,123, Michele M Sale124, Veikko Salomaa63, Bishwa R Sapkota125, Reinhold Schmidt126, Carsten O Schmidt127, Ulf Schminke128, Pankaj Sharma39, Agnieszka Slowik129, Cathie LM Sudlow114,115, Christian Tanislav130, Turgut Tatlisumak131,132, Kent D Taylor120,121, Vincent NS Thijs133,134, Gudmar Thorleifsson11, Unnur Thorsteinsdottir11, Steffen Tiedt1, Stella Trompet135, Christophe Tzourio5,136,137, Cornelia M van Duijn138,139, Matthew Walters140, Nicholas J Wareham86, Sylvia Wassertheil‐Smoller141, James G Wilson142, Kerri L Wiggins109, Qiong Yang47, Salim Yusuf15, Najaf Amin16, Hugo S Aparicio185,48, Donna K Arnett186, John Attia187, Alexa S Beiser47,48, Claudine Berr188, Julie E Buring34,35, Mariana Bustamante189, Valeria Caso190, Yu‐Ching Cheng191, Seung Hoan Choi192,48, Ayesha Chowhan185,48, Natalia Cullell31, Jean‐François Dartigues193,194, Hossein Delavaran95,96, Pilar Delgado195, Marcus Dörr196,197, Gunnar Engström19, Ian Ford198, Wander S Gurpreet199, Anders Hamsten200,201, Laura Heitsch202, Atsushi Hozawa203, Laura Ibanez204, Andreea Ilinca95,96, Martin Ingelsson205, Motoki Iwasaki206, Rebecca D Jackson207, Katarina Jood208, Pekka Jousilahti63, Sara Kaffashian4,5, Lalit Kalra209, Masahiro Kamouchi210, Takanari Kitazono211, Olafur Kjartansson212, Manja Kloss213, Peter J Koudstaal214, Jerzy Krupinski215, Daniel L Labovitz216, Cathy C Laurie118, Christopher R Levi217, Linxin Li218, Lars Lind219, Cecilia M Lindgren220,221, Vasileios Lioutas222,48, Yong Mei Liu223, Oscar L Lopez224, Hirata Makoto225, Nicolas Martinez‐Majander172, Koichi Matsuda225, Naoko Minegishi203, Joan Montaner226, Andrew P Morris227,228, Elena Muiño31, Martina Müller‐Nurasyid229,230,231, Bo Norrving95,96, Soichi Ogishima203, Eugenio A Parati232, Leema Reddy Peddareddygari56, Nancy L Pedersen98,233, Joanna Pera129, Markus Perola63,234, Alessandro Pezzini235, Silvana Pileggi236, Raquel Rabionet237, Iolanda Riba‐Llena30, Marta Ribasés238, Jose R Romero185,48, Jaume Roquer239,240, Anthony G Rudd241,242, Antti‐Pekka Sarin243,244, Ralhan Sarju199, Chloe Sarnowski47,48, Makoto Sasaki245, Claudia L Satizabal185,48, Mamoru Satoh245, Naveed Sattar246, Norie Sawada206, Gerli Sibolt172, Ásgeir Sigurdsson247, Albert Smith248, Kenji Sobue245, Carolina Soriano‐Tárraga240, Tara Stanne249, O Colin Stine250, David J Stott251, Konstantin Strauch229,252, Takako Takai203, Hideo Tanaka253,254, Kozo Tanno245, Alexander Teumer255, Liisa Tomppo172, Nuria P Torres‐Aguila31, Emmanuel Touze256,257, Shoichiro Tsugane206, Andre G Uitterlinden258, Einar M Valdimarsson259, Sven J van der Lee16, Henry Völzke255, Kenji Wakai253, David Weir260, Stephen R Williams261, Charles DA Wolfe241,242, Quenna Wong118, Huichun Xu191, Taiki Yamaji206, Dharambir K Sanghera125,169,170, Olle Melander19, Christina Jern171, Daniel Strbian172,173, Israel Fernandez‐Cadenas31,30, W T Longstreth, Jr174,65, Arndt Rolfs175, Jun Hata107, Daniel Woo82, Jonathan Rosand12,13,14, Guillaume Pare15, Jemma C Hopewell176, Danish Saleheen177, Kari Stefansson11,178, Bradford B Worrall179, Steven J Kittner37, Sudha Seshadri180,48, Myriam Fornage74,181, Hugh S Markus3, Joanna MM Howson28, Yoichiro Kamatani6,182, Stephanie Debette4,5, Martin Dichgans1,183,184.
1Institute for Stroke and Dementia Research (ISD), University Hospital, LMU Munich, Munich, Germany. 2Centre for Brain Research, Indian Institute of Science, Bangalore, India. 3Stroke Research Group, Division of Clinical Neurosciences, University of Cambridge, UK. 4INSERM U1219 Bordeaux Population Health Research Center, Bordeaux, France. 5University of Bordeaux, Bordeaux, France. 6Laboratory for Statistical Analysis, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan. 7Department of Statistical Genetics, Osaka University Graduate School of Medicine, Osaka, Japan. 8Laboratory of Statistical Immunology, Immunology Frontier Research Center (WPI‐IFReC), Osaka University, Suita, Japan. 9Department of Neurology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA. 10Laboratory of Experimental Cardiology, Division of Heart and Lungs, University Medical Center Utrecht, University of Utrecht, Utrecht, Netherlands. 11deCODE genetics/AMGEN inc, Reykjavik, Iceland. 12Center for Genomic Medicine, Massachusetts General Hospital (MGH), Boston, MA, USA. 13J. Philip Kistler Stroke Research Center, Department of Neurology, MGH, Boston, MA, USA. 14Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA. 15Population Health Research Institute, McMaster University, Hamilton, Canada. 16Department of Epidemiology, Erasmus University Medical Center, Rotterdam, Netherlands. 17Department of Radiology and Nuclear Medicine, Erasmus University Medical Center, Rotterdam, Netherlands. 18Department of Medicine and Clinical Science, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan. 19Department of Clinical Sciences, Lund University, Malmö, Sweden. 20Univ. Lille, Inserm, Institut Pasteur de Lille, LabEx DISTALZ‐UMR1167, Risk factors and molecular determinants of aging‐related diseases, F‐59000 Lille, France. 21Centre Hosp. Univ Lille, Epidemiology and Public Health Department, F‐59000 Lille, France. 22AA Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA. 23Cardiovascular Health Research Unit, Departments of Biostatistics and Medicine, University of Washington, Seattle, WA, USA. 24Division of Neurology, Faculty of Medicine, Brain Research Center, University of British Columbia, Vancouver, Canada. 25School of Life Science, University of Lincoln, Lincoln, UK. 26Department of Cerebrovascular Diseases, Fondazione IRCCS Istituto Neurologico "Carlo Besta", Milano, Italy. 27Department of Neurology, Mayo Clinic Rochester, Rochester, MN, USA. 28MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK. 29The National Institute for Health Research Blood and Transplant Research Unit in Donor Health and Genomics, University of Cambridge, UK. 30Neurovascular Research Laboratory, Vall d'Hebron Institut of Research, Neurology and Medicine Departments‐Universitat Autònoma de Barcelona, Vall d’Hebrón Hospital, Barcelona, Spain. 31Stroke Pharmacogenomics and Genetics, Fundacio Docència i Recerca MutuaTerrassa, Terrassa, Spain. 32Children's Research Institute, Children's National Medical Center, Washington, DC, USA. 33Center for Translational Science, George Washington University, Washington, DC, USA. 34Division of Preventive Medicine, Brigham and Women's Hospital, Boston, MA, USA. 35Harvard Medical School, Boston, MA, USA. 36Center for Public Health Genomics, Department of Public Health Sciences, University of Virginia, Charlottesville, VA, USA. 37Department of Neurology, University of Maryland School of Medicine and Baltimore VAMC, Baltimore, MD, USA. 38Departments of Medicine, Pediatrics and Population Health Science, University of Mississippi Medical Center, Jackson, MS, USA. 39Institute of Cardiovascular Research, Royal Holloway University of London, UK & Ashford and St Peters Hospital, Surrey UK. 40Department of Psychiatry, The Hope Center Program on Protein Aggregation and Neurodegeneration (HPAN), Washington University, School of Medicine, St. Louis, MO, USA. 41Department of Developmental Biology, Washington University School of Medicine, St. Louis, MO, USA. 42NIHR Blood and Transplant Research Unit in Donor Health and Genomics, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK. 43Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK. 44British Heart Foundation, Cambridge Centre of Excellence, Department of Medicine, University of Cambridge, Cambridge, UK. 45Department of Medical Genetics, University Medical Center Utrecht, Utrecht, Netherlands. 46Department of Epidemiology, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, Netherlands. 47Boston University School of Public Health, Boston, MA, USA. 48Framingham Heart Study, Framingham, MA, USA. 49Department of Immunology, Genetics and Pathology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden. 50Department of Genetics, University of North Carolina, Chapel Hill, NC, USA. 51Department of Neurology and Stroke Center, Basel University Hospital, Switzerland. 52Neurorehabilitation Unit, University and University Center for Medicine of Aging and Rehabilitation Basel, Felix Platter Hospital, Basel, Switzerland. 53Department of Neurology, Yale University School of Medicine, New Haven, CT, USA. 54Program in Medical and Population Genetics, The Broad Institute of Harvard and MIT, Cambridge, MA, USA. 55Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD, USA. 56Neuroscience Institute, SF Medical Center, Trenton, NJ, USA. 57Icelandic Heart Association Research Institute, Kopavogur, Iceland. 58University of Iceland, Faculty of Medicine, Reykjavik, Iceland. 59Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden. 60Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA. 61Laboratory of Epidemiology and Population Science, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA. 62Department of Neurology, Leeds General Infirmary, Leeds Teaching Hospitals NHS Trust, Leeds, UK. 63National Institute for Health and Welfare, Helsinki, Finland. 64FIMM ‐ Institute for Molecular Medicine Finland, Helsinki, Finland. 65Department of Epidemiology, University of Washington, Seattle, WA, USA. 66Public Health Stream, Hunter Medical Research Institute, New Lambton, Australia. 67Faculty of Health and Medicine, University of Newcastle, Newcastle, Australia. 68School of Public Health, University of Alabama at Birmingham, Birmingham, AL, USA. 69Department of Biostatistical Sciences, Wake Forest School of Medicine, Winston‐Salem, NC, USA. 70Aflac Cancer and Blood Disorder Center, Department of Pediatrics, Emory University School of Medicine, Atlanta, GA, USA. 71Department of Medicine, Division of Cardiovascular Medicine, Stanford University School of Medicine, CA, USA. 72Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala, Sweden. 73Epidemiology, School of Public Health, University of Alabama at Birmingham, USA. 74Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center at Houston, Houston, TX, USA. 75Neurovascular Research Group (NEUVAS), Neurology Department, Institut Hospital del Mar d'Investigació Mèdica, Universitat Autònoma de Barcelona, Barcelona, Spain. 76Department of Pharmacotherapy and Translational Research and Center for Pharmacogenomics, University of Florida, College of Pharmacy, Gainesville, FL, USA. 77Division of Cardiovascular Medicine, College of Medicine, University of Florida, Gainesville, FL, USA. 78Department of Cardiology, Leiden University Medical Center, Leiden, the Netherlands. 79Program in Bioinformatics and Integrative Genomics, Harvard Medical School, Boston, MA, USA. 80Department of Biology, East Carolina University, Greenville, NC, USA. 81Center for Health Disparities, East Carolina University, Greenville, NC, USA. 82University of Cincinnati College of Medicine, Cincinnati, OH, USA. 83RIKEN Center for Integrative Medical Sciences, Yokohama, Japan. 84Department of Medicine, University of Colorado Denver, Anschutz Medical Campus, Aurora, CO, USA. 85Center for Public Health Genomics and Department of Biostatistical Sciences, Wake Forest School of Medicine, Winston‐Salem, NC, USA. 86MRC Epidemiology Unit, University of Cambridge School of Clinical Medicine, Institute of Metabolic Science, Cambridge Biomedical Campus, Cambridge, UK. 87Intramural Research Program, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA. 88Department of Neurology, Radiology, and Biomedical Engineering, Washington University School of Medicine, St. Louis, MO, USA. 89KU Leuven – University of Leuven, Department of Neurosciences, Experimental Neurology, Leuven, Belgium. 90VIB Center for Brain & Disease Research, University Hospitals Leuven, Department of Neurology, Leuven, Belgium. 91Univ.‐Lille, INSERM U 1171. CHU Lille, Lille, France. 92Department of Medical and Molecular Genetics, King's College London, London, UK. 93SGDP Centre, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London, UK. 94Northern Institute for Cancer Research, Paul O'Gorman Building, Newcastle University, Newcastle, UK. 95Department of Clinical Sciences Lund, Neurology, Lund University, Lund, Sweden. 96Department of Neurology and Rehabilitation Medicine, Skåne University Hospital, Lund, Sweden. 97Bioinformatics Core Facility, University of Gothenburg, Gothenburg, Sweden. 98Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden. 99University of Technology Sydney, Faculty of Health, Ultimo, Australia. 100Department of Medicine, University of Maryland School of Medicine, MD, USA. 101Department of Neurology, Mayo Clinic, Jacksonville, FL, USA. 102Geriatrics Research and Education Clinical Center, Baltimore Veterans Administration Medical Center, Baltimore, MD, USA. 103Division of Geriatrics, School of Medicine, University of Mississippi Medical Center, Jackson, MS, USA. 104Memory Impairment and Neurodegenerative Dementia Center, University of Mississippi Medical Center, Jackson, MS, USA. 105Laboratory of Neurogenetics, National Institute on Aging, National institutes of Health, Bethesda, MD, USA. 106Data Tecnica International, Glen Echo MD, USA. 107Department of Epidemiology and Public Health, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan. 108Clinical Research Facility, Department of Medicine, NUI Galway, Galway, Ireland. 109Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA, USA. 110Department of Epidemiology, University of Washington, Seattle, WA. 111Department of Health Services, University of Washington, Seattle, WA, USA. 112Kaiser Permanente Washington Health Research Institute, Seattle, WA, USA. 113Brain Center Rudolf Magnus, Department of Neurology, University Medical Center Utrecht, Utrecht, The Netherlands. 114Usher Institute of Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, UK. 115Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, UK. 116Fred Hutchinson Cancer Research Center, University of Washington, Seattle, WA, USA. 117Department of Medicine, Brigham and Women's Hospital, Boston, MA, USA. 118Department of Biostatistics, University of Washington, Seattle, WA, USA. 119Nuffield Department of Clinical Neurosciences, University of Oxford, UK. 120Institute for Translational Genomics and Population Sciences, Los Angeles Biomedical Research Institute at Harbor‐UCLA Medical Center, Torrance, CA, USA. 121Division of Genomic Outcomes, Department of Pediatrics, Harbor‐UCLA Medical Center, Torrance, CA, USA. 122Department of Neurology, Miller School of Medicine, University of Miami, Miami, FL, USA. 123Department of Allergy and Rheumatology, Graduate School of Medicine, the University of Tokyo, Tokyo, Japan. 124Center for Public Health Genomics, University of Virginia, Charlottesville, VA, USA. 125Department of Pediatrics, College of Medicine, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA. 126Department of Neurology, Medical University of Graz, Graz, Austria. 127University Medicine Greifswald, Institute for Community Medicine, SHIP‐KEF, Greifswald, Germany. 128University Medicine Greifswald, Department of Neurology, Greifswald, Germany. 129Department of Neurology, Jagiellonian University, Krakow, Poland. 130Department of Neurology, Justus Liebig University, Giessen, Germany. 131Department of Clinical Neurosciences/Neurology, Institute of Neuroscience and Physiology, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden. 132Sahlgrenska University Hospital, Gothenburg, Sweden. 133Stroke Division, Florey Institute of Neuroscience and Mental Health, University of Melbourne, Heidelberg, Australia. 134Austin Health, Department of Neurology, Heidelberg, Australia. 135Department of Internal Medicine, Section Gerontology and Geriatrics, Leiden University Medical Center, Leiden, the Netherlands. 136INSERM U1219, Bordeaux, France. 137Department of Public Health, Bordeaux University Hospital, Bordeaux, France. 138Genetic Epidemiology Unit, Department of Epidemiology, Erasmus University Medical Center Rotterdam, Netherlands. 139Center for Medical Systems Biology, Leiden, Netherlands. 140School of Medicine, Dentistry and Nursing at the University of Glasgow, Glasgow, UK. 141Department of Epidemiology and Population Health, Albert Einstein College of Medicine, NY, USA. 142Department of Physiology and Biophysics, University of Mississippi Medical Center, Jackson, MS, USA. 143A full list of members and affiliations appears in the Supplementary Note. 144Department of Human Genetics, McGill University, Montreal, Canada. 145Department of Pathophysiology, Institute of Biomedicine and Translation Medicine, University of Tartu, Tartu, Estonia. 146Department of Cardiac Surgery, Tartu University Hospital, Tartu, Estonia. 147Clinical Gene Networks AB, Stockholm, Sweden. 148Department of Genetics and Genomic Sciences, The Icahn Institute for Genomics and Multiscale Biology Icahn School of Medicine at Mount Sinai, New York, NY, USA. 149Department of Pathophysiology, Institute of Biomedicine and Translation Medicine, University of Tartu, Biomeedikum, Tartu, Estonia. 150Integrated Cardio Metabolic Centre, Department of Medicine, Karolinska Institutet, Karolinska Universitetssjukhuset, Huddinge, Sweden. 151Clinical Gene Networks AB, Stockholm, Sweden. 152Sorbonne Universités, UPMC Univ. Paris 06, INSERM, UMR_S 1166, Team Genomics & Pathophysiology of Cardiovascular Diseases, Paris, France. 153ICAN Institute for Cardiometabolism and Nutrition, Paris, France. 154Department of Biomedical Engineering, University of Virginia, Charlottesville, VA, USA. 155Group Health Research Institute, Group Health Cooperative, Seattle, WA. USA. 156Seattle Epidemiologic Research and Information Center, VA Office of Research and Development, Seattle, WA, USA. 157Cardiovascular Research Center, Massachusetts General Hospital, Boston, MA, USA. 158Department of Medical Research, Bærum Hospital, Vestre Viken Hospital Trust, Gjettum, Norway. 159Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore. 160National Heart and Lung Institute, Imperial College London, London, UK. 161Department of Gene Diagnostics and Therapeutics, Research Institute, National Center for Global Health and Medicine, Tokyo, Japan. 162Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA, USA. 163Department of Cardiology, University Medical Center Groningen, University of Groningen, Netherlands. 164MRC‐PHE Centre for Environment and Health, School of Public Health, Department of Epidemiology and Biostatistics, Imperial College London, London, UK. 165Department of Epidemiology and Biostatistics, Imperial College London, London, UK. 166Department of Cardiology, Ealing Hospital NHS Trust, Southall, UK. 167National Heart, Lung and Blood Research Institute, Division of Intramural Research, Population Sciences Branch, Framingham, MA, USA. 169Department of Phamaceutical Sciences, Collge of Pharmacy, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA. 170Oklahoma Center for Neuroscience, Oklahoma City, OK, USA. 171Department of Pathology and Genetics, Institute of Biomedicine, The Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden. 172Department of Neurology, Helsinki University Hospital, Helsinki, Finland. 173Clinical Neurosciences, Neurology, University of Helsinki, Helsinki, Finland. 174Department of Neurology, University of Washington, Seattle, WA, USA. 175Albrecht Kossel Institute, University Clinic of Rostock, Rostock, Germany. 176Clinical Trial Service Unit and Epidemiological Studies Unit, Nuffield Department of Population Health, University of Oxford, Oxford, UK. 177Department of Genetics, Perelman School of Medicine, University of Pennsylvania, PA, USA. 178Faculty of Medicine, University of Iceland, Reykjavik, Iceland. 179Departments of Neurology and Public Health Sciences, University of Virginia School of Medicine, Charlottesville, VA, USA. 180Department of Neurology, Boston University School of Medicine, Boston, MA, USA. 181Human Genetics Center, University of Texas Health Science Center at Houston, Houston, TX, USA. 182Center for Genomic Medicine, Kyoto University Graduate School of Medicine, Kyoto, Japan. 183Munich Cluster for Systems Neurology (SyNergy), Munich, Germany. 184German Center for Neurodegenerative Diseases (DZNE), Munich, Germany. 185Boston University School of Medicine, Boston, MA, USA. 186University of Kentucky College of Public Health, Lexington, KY, USA. 187University of Newcastle and Hunter Medical Research Institute, New Lambton, Australia. 188Univ. Montpellier, Inserm, U1061, Montpellier, France. 189Centre for Research in Environmental Epidemiology, Barcelona, Spain. 190Department of Neurology, Università degli Studi di Perugia, Umbria, Italy. 191Department of Medicine, University of Maryland School of Medicine, Baltimore, MD, USA. 192Broad Institute, Cambridge, MA, USA. 193Univ. Bordeaux, Inserm, Bordeaux Population Health Research Center, UMR 1219, Bordeaux, France. 194Bordeaux University Hospital, Department of Neurology, Memory Clinic, Bordeaux, France. 195Neurovascular Research Laboratory. Vall d'Hebron Institut of Research, Neurology and Medicine Departments‐Universitat Autònoma de Barcelona. Vall d’Hebrón Hospital, Barcelona, Spain. 196University Medicine Greifswald, Department of Internal Medicine B, Greifswald, Germany. 197DZHK, Greifswald, Germany. 198Robertson Center for Biostatistics, University of Glasgow, Glasgow, UK. 199Hero DMC Heart Institute, Dayanand Medical College & Hospital, Ludhiana, India. 200Atherosclerosis Research Unit, Department of Medicine Solna, Karolinska Institutet, Stockholm, Sweden. 201Karolinska Institutet, Stockholm, Sweden. 202Division of Emergency Medicine, and Department of Neurology, Washington University School of Medicine, St. Louis, MO, USA. 203Tohoku Medical Megabank Organization, Sendai, Japan. 204Department of Psychiatry, Washington University School of Medicine, St. Louis, MO, USA. 205Department of Public Health and Caring Sciences / Geriatrics, Uppsala University, Uppsala, Sweden. 206Epidemiology and Prevention Group, Center for Public Health Sciences, National Cancer Center, Tokyo, Japan. 207Department of Internal Medicine and the Center for Clinical and Translational Science, The Ohio State University, Columbus, OH, USA. 208Institute of Neuroscience and Physiology, the Sahlgrenska Academy at University of Gothenburg, Goteborg, Sweden. 209Department of Basic and Clinical Neurosciences, King's College London, London, UK. 210Department of Health Care Administration and Management, Graduate School of Medical Sciences, Kyushu University, Japan. 211Department of Medicine and Clinical Science, Graduate School of Medical Sciences, Kyushu University, Japan. 212Landspitali National University Hospital, Departments of Neurology & Radiology, Reykjavik, Iceland. 213Department of Neurology, Heidelberg University Hospital, Germany. 214Department of Neurology, Erasmus University Medical Center. 215Hospital Universitari Mutua Terrassa, Terrassa (Barcelona), Spain. 216Albert Einstein College of Medicine, Montefiore Medical Center, New York, NY, USA. 217John Hunter Hospital, Hunter Medical Research Institute and University of Newcastle, Newcastle, NSW, Australia. 218Centre for Prevention of Stroke and Dementia, Nuffield Department of Clinical Neurosciences, University of Oxford, UK. 219Department of Medical Sciences, Uppsala University, Uppsala, Sweden. 220Genetic and Genomic Epidemiology Unit, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK. 221The Wellcome Trust Centre for Human Genetics, Oxford, UK. 222Beth Israel Deaconess Medical Center, Boston, MA, USA. 223Wake Forest School of Medicine, Wake Forest, NC, USA. 224Department of Neurology, University of Pittsburgh, Pittsburgh, PA, USA. 225BioBank Japan, Laboratory of Clinical Sequencing, Department of Computational biology and medical Sciences, Graduate school of Frontier Sciences, The University of Tokyo, Tokyo, Japan 226Neurovascular Research Laboratory, Vall d'Hebron Institut of Research, Neurology and Medicine Departments‐Universitat Autònoma de Barcelona. Vall d’Hebrón Hospital, Barcelona, Spain. 227Department of Biostatistics, University of Liverpool, Liverpool, UK. 228Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK. 229Institute of Genetic Epidemiology, Helmholtz Zentrum München ‐ German Research Center for Environmental Health, Neuherberg, Germany. 230Department of Medicine I, Ludwig‐Maximilians‐Universität, Munich, Germany. 231DZHK (German Centre for Cardiovascular Research), partner site Munich Heart Alliance, Munich, Germany. 232Department of Cerebrovascular Diseases, Fondazione IRCCS Istituto Neurologico “Carlo Besta”, Milano, Italy. 233Karolinska Institutet, MEB, Stockholm, Sweden. 234University of Tartu, Estonian Genome Center, Tartu, Estonia, Tartu, Estonia. 235Department of Clinical and Experimental Sciences, Neurology Clinic, University of Brescia, Italy. 236Translational Genomics Unit, Department of Oncology, IRCCS Istituto di Ricerche Farmacologiche Mario Negri, Milano, Italy. 237Department of Genetics, Microbiology and Statistics, University of Barcelona, Barcelona, Spain. 238Psychiatric Genetics Unit, Group of Psychiatry, Mental Health and Addictions, Vall d’Hebron Research Institute (VHIR), Universitat Autònoma de Barcelona, Biomedical Network Research Centre on Mental Health (CIBERSAM), Barcelona, Spain. 239Department of Neurology, IMIM‐Hospital del Mar, and Universitat Autònoma de Barcelona, Spain. 240IMIM (Hospital del Mar Medical Research Institute), Barcelona, Spain. 241National Institute for Health Research Comprehensive Biomedical Research Centre, Guy's & St. Thomas’ NHS Foundation Trust and King's College London, London, UK. 242Division of Health and Social Care Research, King's College London, London, UK. 243FIMM‐Institute for Molecular Medicine Finland, Helsinki, Finland. 244THL‐National Institute for Health and Welfare, Helsinki, Finland. 245Iwate Tohoku Medical Megabank Organization, Iwate Medical University, Iwate, Japan. 246BHF Glasgow Cardiovascular Research Centre, Faculty of Medicine, Glasgow, UK. 247deCODE Genetics/Amgen, Inc., Reykjavik, Iceland. 248Icelandic Heart Association, Reykjavik, Iceland. 249Institute of Biomedicine, the Sahlgrenska Academy at University of Gothenburg, Goteborg, Sweden. 250Department of Epidemiology, University of Maryland School of Medicine, Baltimore, MD, USA. 251Institute of Cardiovascular and Medical Sciences, Faculty of Medicine, University of Glasgow, Glasgow, UK. 252Chair of Genetic Epidemiology, IBE, Faculty of Medicine, LMU Munich, Germany. 253Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Japan. 254Department of Epidemiology, Nagoya University Graduate School of Medicine, Nagoya, Japan. 255University Medicine Greifswald, Institute for Community Medicine, SHIP‐KEF, Greifswald, Germany. 256Department of Neurology, Caen University Hospital, Caen, France. 257University of Caen Normandy, Caen, France. 258Department of Internal Medicine, Erasmus University Medical Center, Rotterdam, Netherlands. 259Landspitali University Hospital, Reykjavik, Iceland. 260Survey Research Center, University of Michigan, Ann Arbor, MI, USA. 261University of Virginia Department of Neurology, Charlottesville, VA, USA.
Temprano‐Sagrera G, Sitlani CM, Bone WP, et al. Multi‐phenotype analyses of hemostatic traits with cardiovascular events reveal novel genetic associations. J Thromb Haemost. 2022;20:1331–1349. doi: 10.1111/jth.15698
Manuscript handled by: Sigrid Braekkan
Final decision: Sigrid Braekkan, 8 March 2021
REFERENCES
- 1. de Vries PS, Sabater‐Lleal M, Huffman JE, et al. A genome‐wide association study identifies new loci for factor VII and implicates factor VII in ischemic stroke etiology. Blood. 2019;133:967‐977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Junker R, Heinrich J, Schulte H, van de Loo J, Assmann G. Coagulation factor VII and the risk of coronary heart disease in healthy men. Arterioscler Thromb Vasc Biol. 1997;17:1539‐1544. [DOI] [PubMed] [Google Scholar]
- 3. Olson NC, Raffield LM, Lange LA, et al. Associations of activated coagulation factor VII and factor VIIa‐antithrombin levels with genome‐wide polymorphisms and cardiovascular disease risk. J Thromb Haemost. 2018;16:19‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sabater‐Lleal M, Huffman JE, de Vries PS, et al. Genome‐wide association transethnic meta‐analyses identifies novel associations regulating coagulation factor VIII and von Willebrand factor plasma levels. Circulation. 2019;139:620‐635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Vries PS, Chasman DI, Sabater‐Lleal M, et al. A meta‐analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration. Hum Mol Genet. 2016;25:358‐370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sennblad B, Basu S, Mazur J, et al. Genome‐wide association study with additional genetic and post‐transcriptional analyses reveals novel regulators of plasma factor XI levels. Hum Mol Genet. 2017;26:637‐649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang J, Sabater‐Lleal M, Asselbergs FW, et al. Genome‐wide association study for circulating levels of PAI‐1 provides novel insights into its regulation. Blood. 2012;120:4873‐4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang J, Huffman JE, Yamkauchi M, et al. Genome‐wide association study for circulating tissue plasminogen activator (tPA) levels and functional follow‐up implicates endothelial STXBP5 and STX2. Arterioscler Thromb Vasc Biol. 2014;34:1093‐1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindström S, Wang L, Smith EN, et al. Genomic and transcriptomic association studies identify 16 novel susceptibility loci for venous thromboembolism. Blood. 2019;134:1645‐1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klarin D, Busenkell E, Judy R, et al. Genome‐wide association analysis of venous thromboembolism identifies new risk loci and genetic overlap with arterial vascular disease. Nat Genet. 2019;51:1574‐1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishigaki K, Akiyama M, Kanai M, et al. Large scale genome‐wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet. 2020;52:669‐679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122:433‐443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koyama S, Ito K, Terao C, et al. Population‐specific and trans‐ancestry genome‐wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat Genet. 2020;52:1169‐1177. [DOI] [PubMed] [Google Scholar]
- 14. Malik R, Chauhan G, Traylor M, et al. Multiancestry genome‐wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524‐537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diggle P, Heagerty P, Liang K‐Y, Zeger S. Analysis of Longitudinal Data. OUP; 2013. [Google Scholar]
- 16. Turley P, Walters RK, Maghzian O, et al. Multi‐trait analysis of genome‐wide association summary statistics using MTAG. Nat Genet. 2018;50:229‐237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ray D, Boehnke M. Methods for meta‐analysis of multiple traits using GWAS summary statistics. Genet Epidemiol. 2018;42:134‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Psaty BM, O'Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium: design of prospective meta‐analyses of genome‐wide association studies from five cohorts. Circ Cardiovasc Genet. 2009;2:73‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nikpay M, Goel A, Won H‐H, et al. A comprehensive 1000 Genomes–based genome‐wide association meta‐analysis of coronary artery disease. Nat Genet. 2015;47:1121‐1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rich SS, Wang ZY, Sturke A, et al. Rapid evaluation of phenotype, SNP and summary results through the dbGaP Charge Summary site. Nat Genet. 2016;48:702‐703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bulik‐Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236‐1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bulik‐Sullivan BK, Loh P‐R, Finucane H, et al. LD score regression distinguishes confounding from polygenicity in genome‐wide association studies. Nat Genet. 2015;47:291‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Auton A, Abecasis GR, Altshuler DM, et al. A global reference for human genetic variation. Nature. 2015;526:68‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sitlani CM, Baldassari AR, Highland HM, Hodonsky CJ, McKnight B, Avery CL. Comparison of adaptive multiple phenotype association tests using summary statistics in genome‐wide association studies. Hum Mol Genet. 2021;30:1371‐1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng Z, Nayak L, Wang W, et al. An ATF6‐tPA pathway in hepatocytes contributes to systemic fibrinolysis and is repressed by DACH1. Blood. 2019;133:743‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spiezia L, Rossetto V, Campello E, et al. Factor VIIa‐antithrombin complexes in patients with arterial and venous thrombosis. Thromb Haemost. 2010;103:1188‐1192. [DOI] [PubMed] [Google Scholar]
- 28. Spiezia L, Campello E, Valle FD, Woodhams B, Simioni P. Factor VIIa‐antithrombin complex: a possible new biomarker for activated coagulation. Clin Chem Lab Med. 2017;55:484‐488. [DOI] [PubMed] [Google Scholar]
- 29. Patnaik MM, Moll S. Inherited antithrombin deficiency: a review. Haemophilia. 2008;14:1229‐1239. [DOI] [PubMed] [Google Scholar]
- 30. Greenwald AG, Gonzalez R, Harris RJ, Guthrie D. Effect sizes and p values: what should be reported and what should be replicated? Psychophysiology. 1996;33:175‐183. [DOI] [PubMed] [Google Scholar]
- 31. Johnson VE. Revised standards for statistical evidence. Proc Natl Acad Sci USA. 2013;110:19313‐19317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benjamin DJ, Berger JO, Johannesson M, et al. Redefine statistical significance. Nat Hum Behav. 2018;2:6‐10. [DOI] [PubMed] [Google Scholar]
- 33. Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI‐EBI GWAS Catalog of published genome‐wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005‐D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877‐D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Foley CN, Staley JR, Breen PG, et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat Commun. 2021;12:764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lonsdale J, Thomas J, Salvatore M, et al. The Genotype‐Tissue Expression (GTEx) project. Nat Genet. 2013;45:580‐585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morange P‐E, Oudot‐Mellakh T, Cohen W, et al. KNG1 Ile581Thr and susceptibility to venous thrombosis. Blood. 2011;117:3692‐3694. [DOI] [PubMed] [Google Scholar]
- 39. Hinds DA, Buil A, Ziemek D, et al. Genome‐wide association analysis of self‐reported events in 6135 individuals and 252 827 controls identifies 8 loci associated with thrombosis. Hum Mol Genet. 2016;25:1867‐1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Q, Cheng G, Wang X, et al. Genetic effects of BDKRB2 and KNG1 on deep venous thrombosis after orthopedic surgery and the potential mediator. Sci Rep. 2018;8:17332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27:R195‐R208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sivakumaran S, Agakov F, Theodoratou E, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89:607‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sethi MK, Buettner FFR, Krylov VB, et al. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O‐glucosylated notch epidermal growth factor repeats. J Biol Chem. 2010;285:1582‐1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R‐56R. [DOI] [PubMed] [Google Scholar]
- 45. Golias C, Charalabopoulos A, Stagikas D, Charalabopoulos K, Batistatou A. The kinin system ‐ bradykinin: biological effects and clinical implications. Multiple role of the kinin system ‐ bradykinin. Hippokratia. 2007;11:124‐128. [PMC free article] [PubMed] [Google Scholar]
- 46. Houlihan LM, Davies G, Tenesa A, et al. Common variants of large effect in F12, KNG1, and HRG are associated with activated partial thromboplastin time. Am J Hum Genet. 2010;86:626‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu Z, Liu J, Song Z, Hou Q, Fan X, Hou D. Variants in the Atherogenic ALOX5AP, THBD, and KNG1 genes potentiate the risk of ischemic stroke via a genetic main effect and epistatic interactions in a Chinese population. J Stroke Cerebrovasc Dis. 2015;24:2060‐2068. [DOI] [PubMed] [Google Scholar]
- 48. Bryant JW, Shariat‐Madar Z Human plasma kallikrein‐kinin system: physiological and biochemical parameters. Cardiovasc Hematol Agents Med Chem. 2009;7:234‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Madeddu P, Emanueli C, El‐Dahr S. Mechanisms of Disease: the tissue kallikrein–kinin system in hypertension and vascular remodeling. Nat Rev Nephrol. 2007;3:208‐221. [DOI] [PubMed] [Google Scholar]
- 50. Sabater‐Lleal M, Martinez‐Perez A, Buil A, et al. A genome‐wide association study identifies KNG1 as a genetic determinant of plasma factor XI level and activated partial thromboplastin time. Arterioscler Thromb Vasc Biol. 2012;32:2008‐2016. [DOI] [PubMed] [Google Scholar]
- 51. Taylor KC, Lange LA, Zabaneh D, et al. A gene‐centric association scan for coagulation factor VII levels in European and African Americans: the Candidate Gene Association Resource (CARe) Consortium. Hum Mol Genet. 2011;20:3525‐3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chaudhry R, Usama SM, Physiology BHM. Coagulation Pathways [Internet]. En: StatPearls. StatPearls Publishing; 2021. [citado 2021 oct 18]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK482253/ [Google Scholar]
- 53. Zhou L, Ding H, Zhang X, et al. Genetic variants at newly identified lipid loci are associated with coronary heart disease in a Chinese Han population. PLoS One. 2011;6:e27481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhuang K, Zhang W, Zhang X, Wu F, Cheng L. Effects of SNPs at newly identified lipids loci on blood lipid levels and risk of coronary heart disease in Chinese Han population: a case control study. J Huazhong Univ Sci Technolog Med Sci. 2011;31:452. [DOI] [PubMed] [Google Scholar]
- 55. Sampson ND, Hewitt JE. SF4 and SFRS14, two related putative splicing factors on human chromosome 19p13.11. Gene. 2003;305:91‐100. [DOI] [PubMed] [Google Scholar]
- 56. Bot C, Pfeiffer A, Giordano F, Edara DM, Dantuma NP, Ström L. Independent mechanisms recruit the cohesin loader protein NIPBL to sites of DNA damage. J Cell Sci. 2017;130:1134‐1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bermudez VP, Farina A, Higashi TL, et al. In vitro loading of human cohesin on DNA by the human Scc2‐Scc4 loader complex. Proc Natl Acad Sci USA. 2012;109:9366‐9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kannel WB, Wolf PA, Castelli WP, D’Agostino RB. Fibrinogen and risk of cardiovascular disease: the Framingham study. JAMA. 1987;258:1183‐1186. [PubMed] [Google Scholar]
- 59. Stec JJ, Silbershatz H, Tofler GH, et al. Association of fibrinogen with cardiovascular risk factors and cardiovascular disease in the Framingham offspring population. Circulation. 2000;102:1634‐1638. [DOI] [PubMed] [Google Scholar]
- 60. Ward‐Caviness CK, de Vries PS, Wiggins KL, et al. Mendelian randomization evaluation of causal effects of fibrinogen on incident coronary heart disease. PLoS One. 2019;14:e0216222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Keavney B, Danesh J, Parish S, et al. Fibrinogen and coronary heart disease: test of causality by ‘Mendelian randomization’. Int J Epidemiol. 2006;35:935‐943. [DOI] [PubMed] [Google Scholar]
- 62. Tsukumo Y, Tsukahara S, Furuno A, Iemura S, Natsume T, Tomida A. TBL2 associates With ATF4 mRNA via its WD40 domain and regulates its translation during ER stress. J Cell Biochem. 2016;117:500‐509. [DOI] [PubMed] [Google Scholar]
- 63. Tsukumo Y, Tsukahara S, Furuno A, Iemura S, Natsume T, Tomida A. The endoplasmic reticulum‐localized protein TBL2 interacts with the 60S ribosomal subunit. Biochem Biophys Res Comm. 2015;462:383‐388. [DOI] [PubMed] [Google Scholar]
- 64. Ortega‐Prieto P, Postic C. Carbohydrate sensing through the transcription factor ChREBP. Front Genet. 2019;10:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kurahashi H, Azuma Y, Masuda A, et al. MYRF is associated with encephalopathy with reversible myelin vacuolization. Ann Neurol. 2018;83:98‐106. [DOI] [PubMed] [Google Scholar]
- 66. An H, Fan C, Sharif M, Kim D, Poitelon Y, Park Y. Functional mechanism and pathogenic potential of MYRF ICA domain mutations implicated in birth defects. Sci Rep. 2020;10:814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Graham DB, Lefkovith A, Deelen P, et al. TMEM258 is a component of the oligosaccharyltransferase complex controlling ER stress and intestinal inflammation. Cell Rep. 2016;17:2955‐2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Taylor K, Davey Smith G, Relton CL, Gaunt TR, Richardson TG. Prioritizing putative influential genes in cardiovascular disease susceptibility by applying tissue‐specific Mendelian randomization. Genome Med. 2019;11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Park WJ, Kothapalli KSD, Lawrence P, Tyburczy C, Brenna JT. An alternate pathway to long‐chain polyunsaturates: the FADS2 gene product Δ8‐desaturates 20:2n–6 and 20:3n–3. J Lipid Res. 2009;50:1195‐1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Machiela MJ, Chanock SJ. LDlink: a web‐based application for exploring population‐specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555‐3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yuan S, Bäck M, Bruzelius M, Mason AM, Burgess S, Larsson S. Plasma phospholipid fatty acids, FADS1 and risk of 15 cardiovascular diseases: a Mendelian randomisation study. Nutrients. 2019;11:3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brunel H, Massanet R, Martinez‐Perez A, Ziyatdinov A, Martin‐Fernandez L, Souto JC, Perera A, Soria JM. The Central Role of KNG1 Gene as a Genetic Determinant of Coagulation Pathway‐Related Traits: Exploring Metaphenotypes. PLOS ONE. 2016;11(12):e0167187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8


