Abstract
Sex differences in lifespan remain an intriguing puzzle in evolutionary biology. While explanations range from sex differences in selection to sex differences in the expression of recessive lifespan‐altering mutations (via X‐linkage), little consensus has been reached. One unresolved issue is the extent to which genetic influences on lifespan dimorphism are modulated by the environment. For example, studies have shown that sex differences in lifespan can either increase or decrease depending upon the social environment. Here, we took an experimental approach, manipulating multiple axes of the social environment across inbred long‐ and short‐lived genotypes and their reciprocal F1s in the fly Drosophila serrata. Our results reveal strong genetic effects and subtle yet significant genotype‐by‐environment interactions for male and female lifespan, specifically due to both population density and mating status. Further, our data do not support the idea that unconditional expression of deleterious X‐linked recessive alleles in heterogametic males accounts for lower male lifespan.
Keywords: ageing, Drosophila, environment, genetic, lifespan, sex, unguarded X hypothesis
We compared homozygous founder line (DsGRP20 and DsGRP57) and F1 genotypes to assess genetic differences in lifespan, inbreeding and X chromosome genome influences under different social environments.

1. INTRODUCTION
The question of why males and females differ in lifespan has long fascinated evolutionary biologists. While exceptions exist, across many taxa females have higher mean lifespan than males (Austad, 2019). Despite a long history of ageing research, no proven or unifying theories have emerged, and studies still yield contradictory results. Sexual dimorphism in lifespan can arise in response to sex differences in selection on life histories. Males and females maximise reproductive fitness in different ways (Friberg, 2005; Maklakov et al., 2009) with males typically investing more in early reproduction than females, even at the cost of their own somatic maintenance and lifespan (Maklakov & Lummaa, 2013). In other words, if males invest more in early reproductive efforts than females, selection on survival to advanced ages may be weaker in males than females, leading to sex differences in age‐dependent mortality across the sexes. Selection, therefore, alters the overall costs of reproduction for each sex and affects the evolution of ageing by shaping sex‐specific mortality rates (Promislow 2003; Bonduriansky et al., 2008). Sexual dimorphism in lifespan may also be caused by asymmetric inheritance of uneven numbers of sex chromosomes between males and females. This hypothesis posits that for species where males are the hemizygous sex, harmful recessive mutations on the X chromosome will always be expressed in males whereas they will commonly be masked by dominance in females (Trivers, 1985). A general prediction of this hypothesis, coined the ‘unguarded X hypothesis’, is that males should on average have shorter lifespans than females. It should be noted that sex‐specific selection on life‐history strategies and the unguarded X are not necessarily mutually exclusive explanations and that each could contribute to lifespan differences between males and females.
Several studies have shown that variation in environmental or genetic background can influence sexual dimorphism in lifespan (Brengdahl et al., 2018b; Kimber & Chippindale, 2013; Sultanova et al., 2018). Species of the genus Drosophila have featured prominently in ageing research. In addition to D. melanogaster [see reviews by (Rogina, 2011) and (Piper & Partridge, 2018)], other species such as D. simulans (Ballard, 2005) have also been used as models for ageing research. With the development of the Drosophila serrata Genome Reference Panel, a panel of re‐sequenced lines (DsGRP) (Reddiex et al., 2018), D. serrata has now also emerged as a potential model for ageing research. Here, we describe the results of a systematic analysis of lifespan comparisons amongst two isogenic laboratory lines: DsGRP20 and DsGRP57. Using isogenic lines can provide insight into how the underlying genetic architecture of lifespan varies in response to genetic and social conditions. For instance, Swindell and Bouzat (2006) showed that stressful environments such as increased competition and temperature had pronounced effects on mitigating lifespan reducing effects of inbreeding depression in D. melanogaster. While the existence of inbreeding depression on lifespan is well documented, how heterozygous and homozygous genotypes respond to social (Brengdahl et al., 2018a; Carazo et al., 2016; Sultanova et al., 2018) and environmental conditions (Brengdahl et al., 2018b; Sultanova & Carazo, 2019; Tan et al., 2013) such as mating and density is less well understood.
Studies of D. melanogaster have shown that lifespan is sensitive to both genetic and environmental manipulations of organismal condition and that both male‐ and female‐biased effects can occur (Burger & Promislow, 2004). This substantial variation in male and female responses emphasizes the importance of including not only both sexes, but also social environment when analysing lifespan. Amongst the different social effects that have an impact on adult lifespan, mating activity and adult population density have been shown to influence longevity (Iliadi et al., 2009; Malick & Kidwell, 1966). In species of Drosophila, such as D. virilis, mating status significantly affected lifespan, with male and female virgins being affected very differently (Aigaki & Ohba, 1984). In D. virilis, male sexual activity played the most important role amongst the complex interactions between both sexes. Mating status also affected lifespan of both female and male D. melanogaster flies, though males were less affected (Koliada et al., 2020). The few systematic studies conducted on the effects of high adult density have found increased male sensitivity to variation in density, erratic mortality rates and decreased mortality amongst higher density cohorts of middle‐aged D. melanogaster females (Khazaeli et al., 1996).
This study aims to clarify how genetic background, sex, inbreeding, mating and density act and interact with each other to shape lifespan in Drosophila serrata. In doing this, we also test the general predictions of the unguarded X hypothesis and evaluate their sensitivity to social backgrounds (Figure S1). To better characterise the effects of recessive mutations on lifespan, we have subjected each of our inbred long‐lived, short‐lived and outcrossed genotypes to a factorial design of varying social and/or demographic conditions resulting in 48 different treatments (Figure 1, Table S2). To quantify the effects of genotype on lifespan, we crossed fully inbred flies to generate outbred and reciprocal F1 flies (Vaiserman et al., 2013). To explore interactions with social contexts of mating (Aigaki & Ohba, 1984; Service, 1989; Zajitschek et al., 2013), we measured the lifespan of these flies as both virgins and non‐virgins. Furthermore, we varied the population density of flies held together in a vial, as this is also known to affect lifespan and mortality rates (Graves & Mueller, 1993; Joshi & Mueller, 1997; Khazaeli et al., 1995, 1996). This will ultimately bring us closer in our attempts to characterize sexual dimorphism in lifespan resulting from sex differences in selection as opposed to variation resulting from X‐linked deleterious recessive mutations. We present evidence of genetic interactions with sex and with mating and density on lifespan.
FIGURE 1.
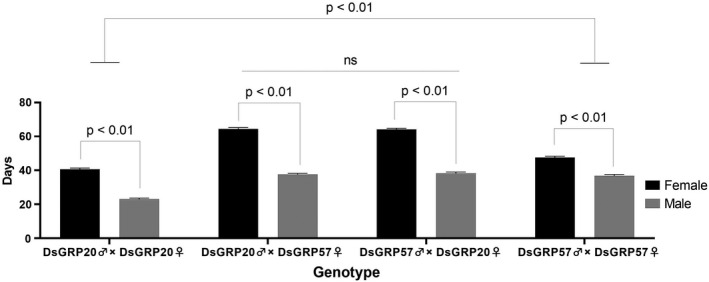
Genotype‐dependent effects on sex differences in lifespan in D. serrata. Sex differences in mean adult lifespan in the four genotypes resulting for our reciprocal cross between DsGRP20 and DsGRP57 (two parental lines plus alternate F1s). Bars represent the mean lifespan of each genotype pooled across the six density (low, medium and high) × mating status (mated and non‐mated) treatment combinations. Error bars represent 1 SE
2. MATERIALS AND METHODS
2.1. Fly stocks and culturing conditions
All analyses were carried out using fruit fly genotypes, DsGRP20 and DsGRP57, randomly chosen from the DsGRP (Reddiex et al., 2018). Flies were maintained in vials containing agar–sugar–yeast medium, in a temperature‐controlled room at 25°C and a 12/12‐h light/dark cycle. We then performed density‐controlled crosses between the two lines to produce inbred (DsGRP20♂ × DsGRP20♀ and DsGRP57♂ × DsGRP57♀) and outbred reciprocal crosses (DsGRP20♂ × DsGRP57♀, and DsGRP57♂ × DsGRP20♀) from here on referred to as genotypes. All experimental flies were collected as virgins within 24h after eclosion, and male and female offspring from each cross were randomly allocated into the experimental treatments in a factorial design including the effects of mating and density. For the mated treatment, flies were housed as two pairs with equal sex ratios and allowed to mate. After 48 hours, mated flies were collected using CO2, sorted by sex and transferred to single‐sex experimental vials for the lifespan trial. For each cross, virgin and mated treatments were maintained at three different vial densities. Vial densities were 5, 10 and 15 flies per vial (10 replicate vials per variant, per sex).
2.2. Lifespan assay
We attempted to measure the lifespan of a total of 4800 flies. Vials were randomized and flies tipped into fresh food vials without anaesthesia every 3–4 days. On these occasions, dead flies were counted and removed without replacement. Care was taken to prevent dead flies from being tipped on again into the fresh food vials. Flies that escaped while tipping vials were excluded from further analysis (n = 194 flies). Survivorship was scored at the time of tipping until all flies had died. This factorial design enables us to quantify the effects of genotype, sex, social environment and their interactions on phenotypic variation for lifespan (Table S2).
2.3. Statistical analyses
To compare the effects of sex, genotype, mating and density on adult lifespan, we used a mixed model analysis of variance using restricted maximum likelihood (REML) estimates of the variance components (PROC GLIMMIX in SAS 9.2). Sex, DsGRP genotypes (DsGRP20♂ × DsGRP20♀, DsGRP57♂ × DsGRP57♀, DsGRP20♂ × DsGRP57♀, and DsGRP57♂ × DsGRP20♀), mated status (non‐mated/mated), density (5, 10 and 15 flies per vial) and their interactions were modelled as fixed factors and tested with F‐statistics. For tests of fixed effects, we applied a Satterthwaite approximation to calculate the denominator degrees of freedom via the ‘ddfm=SAT’ option in SAS. Vial was modelled as a random effect. Density was treated as a categorical factor. Models were simplified by backward single‐term deletions (p ≤ 0.05).
In our initial modelling, we used a four‐level ‘genotype’ effect that includes the homozygous founder lines (DsGRP20 and DsGRP57) and both reciprocal F1 crosses between these lines. Subsequent contrasts between these four levels allowed us to test multiple genetic effects. First, we compared homozygous line differences to assess genetic differences in lifespan. Second, contrasts between the F1 and homozygous genotypes permitted a test for the effect of inbreeding. Third, contrasts between the two F1 crosses allowed us to test for a reciprocal cross effect that includes X‐chromosome genome influences. We present effect sizes as least square means and used Tukey's HSD to correct for multiple testing.
3. RESULTS
After censoring 194 flies that escaped while being transferred to fresh holding vials (<5% of total flies), 4606 flies were available for analysis. Across the entire experiment, female‐biased longevity was apparent (Figure 1 and Figure S2). While female D. serrata lived on average 54 days (range 4–104 days), males lived an average of only 34 days (range 4–69 days) (Figure 2 and Figure S3). The initial and final linear models describing genetic and environmental influences on lifespan variation appear in Tables S1 and 1, respectively. While the final simplified model provided statistical support for sex differences in lifespan in D. serrata (Sex: F 1, 454.4 = 1798.3, p = 4.54e−160), males and females were influenced differently by genotype (Sex × Genotype: F 3,454.3 = 64.6, p = 8.36e−35), which was also a significant main effect in the model (Genotype: F 3,601.3 = 340.4, p = 3.83e−129). Here, three key results are of interest. First, reciprocal crossing did not affect the degree of sexual dimorphism with no lifespan differences found between the males of F1 genotypes (DsGRP20♂ × DsGRP57♀) and (DsGRP57♂ × DsGRP20♀) or between the females of these two F1 genotypes (Figure 1). Second, males and females were affected by outcrossing in different ways. F1 females lived at least 17 days longer than homozygous parental line females, and a similar degree of increase (~ 40%) was observed in F1 males compared with parental line DsGRP20 males (Figure 1). However, there was no difference in male lifespan between the F1s and parental line DsGRP57 (Figure 1) consistent with a lack of any outcrossing effect. Third, genetic differences were also apparent between the two parental lines with both males and females from line DsGRP57 living between 14 and 7 days longer than males and females from line DsGRP20, respectively.
FIGURE 2.
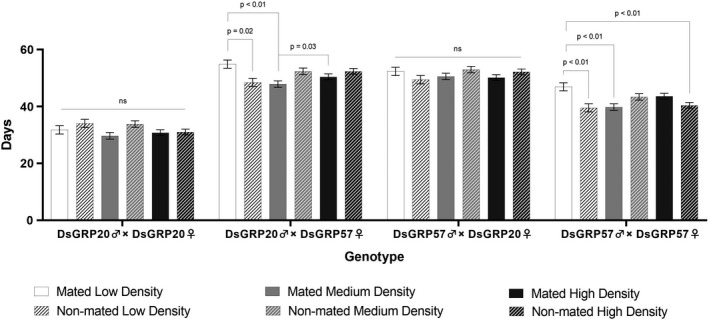
Genotype‐dependent effects of the social environment on lifespan in D. serrata. Shown are pooled adult, male and female lifespan for the homozygous founder lines DsGRP20♂ × DsGRP20♀ and DsGRP57♂ × DsGRP57♀, as well as both reciprocal F1 crosses DsGRP20♂ × DsGRP57♀ and DsGRP57♂ × DsGRP20♀ between these lines. Each bar represents the mean of each genotype measured in one of six different densities (low, medium and high) × mating status (mated and non‐mated) treatment combinations. Error bars represent the 1 S.E. of the mean
Our analysis also indicated a genotype‐by‐environment interaction for lifespan. Genotype‐dependent effects were observed for both density and mating via a significant three‐way interaction (Table 1: Genotype × Density × Mating: F 6,552.8 = 2.45, p = 0.024). The social environmental effects underlying this significant interaction were, however, typically more subtle than the effects seen in the interaction between sex and genotype (Figure 2) Considering this interaction further, post hoc comparisons revealed significant differences between density and mating within only two of the four genotypes the parental (DsGRP 57♂ × DsGRP 57♀) and the reciprocal F1 (DsGRP 20♂ × DsGRP 57♀). For these genotypes, an effect of mating was detected but only in the low‐density treatments, with the lifespan of mated flies on average, 6 days higher than unmated flies (Figure 2).
TABLE 1.
F‐tests of fixed effects for the reduced model examining the significance of contributions of sex, genotype, mating and density to D. serrata lifespan
| Effect | df | F | p |
|---|---|---|---|
| Sex | 1, 454.4 | 1798.3 | 4.54e−160 |
| Genotype | 3, 601.3 | 340.4 | 3.83e−129 |
| Sex × Genotype | 3, 454.3 | 64.6 | 8.36e−35 |
| Density | 2, 552.9 | 1.18 | 0.308 |
| Genotype × Density | 6, 552.7 | 0.84 | 0.539 |
| Mating | 1, 601.5 | 0.09 | 0.764 |
| Genotype × Mating | 3, 601.3 | 3.28 | 0.021 |
| Density × Mating | 2, 552.9 | 15.0 | 4.53e−07 |
| Genotype × Density × Mating | 6, 552.8 | 2.45 | 0.024 |
4. DISCUSSION
4.1. Unguarded X and female‐biased lifespan in D. serrata
In all treatment combinations, female D. serrata lived longer than males, a result consistent with a wide range of wild and captive species, where on average, the homogametic sex lives longer than its heterogametic counterpart (Xirocostas et al., 2020). Our result is also consistent with two previous studies of Drosophila serrata both of which indicate female‐biased longevity (Robson et al., 2006; Wit et al., 2015). One prominent hypothesis for reduced male lifespan is the unguarded X hypothesis (Trivers, 1985). This hypothesis predicts that reduced male lifespan is a result of the unconditional expression of recessive deleterious alleles on the single X chromosome. To date, the few studies that have explicitly tested predictions arising from the unguarded X hypothesis, conducted in Drosophila melanogaster (Brengdahl et al., 2018a; Carazo et al., 2016; Sultanova et al., 2018), have produced inconsistent results.
Here, we used two randomly selected inbred lines with differing lifespans to create outbred and reciprocal F1’s to test for reduced lifespan in males in response to the crossing as predicted by the unguarded X hypothesis. Despite differences in parental lifespan, we found no differences in lifespan between the outbred and reciprocal male F1’s that could be attributed to the accumulation of recessive deleterious mutations on the X chromosome as predicted by the unguarded X hypothesis (Figure 1). Under the unguarded X hypothesis, outbred male F1 offspring of the shorter‐lived maternal line inherit deleterious mutations on their X chromosome, resulting in lower lifespan than offspring from the longer‐lived maternal line without recessive deleterious mutations on the X chromosome. Although the effects of recessive deleterious mutations may be underestimated in crosses between highly inbred lines due to higher expected levels of purging during the inbreeding process (Hedrick, 1994), similar to studies in D. melanogaster (Brengdahl et al., 2018a), the unguarded X hypothesis appears to be insufficient to explain sexual dimorphism in D. serrata lifespan, at least in the small number of genotypes we consider here. Our results, thus, point to sex‐specific selection (Bonduriansky et al., 2008; Maklakov et al., 2009; Maklakov & Lummaa, 2013) as a factor driving the higher mortality observed in D. serrata males.
4.2. Genotype‐by‐social environment interactions for lifespan
In addition to sex‐ and genotype‐biased longevity, we also found interactions of genotype with mating and density, our two experimentally manipulated axes of social background. Across a range of taxa, sexual dimorphism is a result of complex relationships between environmental conditions and sex‐specific reproductive costs (Lemaître et al., 2020). Mean lifespan did not differ significantly between density treatments within genotypes (Figure 2), even though large sex and genotype effects were detected. Although we detected no Genotype × Density or Sex × Density interaction, there was a highly significant interaction between density and mating that appeared to be driven by a change in rank‐order lifespan between low and medium density, which was highest for low density in the mated treatment but lowest for the unmated treatment (Figure 2). The absence of lifespan reduction at high density was somewhat unexpected given its potential influence on individual condition. We note that survivorship experiments with high densities at the beginning can sometimes produce high mortality rates at young ages and that this in turn creates a low‐density environment for the remainder of the assay (Graves & Mueller, 1993); however, we observed no such effect in our high‐density assays. Another possibility is that perhaps 15 flies per vial were not sufficiently high to detect any high‐density effects that exist under regular husbandry conditions.
In our study, mating had no effect on mean lifespan. While we did detect a significant Genotype × Mating interaction, this can be explained by idiosyncratic effects of genotype on mating and density (Figure 1). Adverse effects of multiple mating on lifespan in D. melanogaster males have also been reported in several laboratory‐based studies, as have toxic effects of male accessory gland proteins on female fitness and lifespan (Chapman et al., 1995; Fowler & Partridge, 1989). In female D. serrata, continued male courtship and harassment also lead to decreased female fitness (Chenoweth et al., 2015). However, in the wild, mated D. melanogaster females tend to live longer than their virgin counterparts, without any adverse mating effects on lifespan (Markow, 2011). In other species of Drosophila such as D. pseudoobscura, females showed no difference in longevity when mated one or multiple times in the laboratory (Gowaty et al., 2010). Intermittent and short‐term mating in the laboratory could explain similar patterns of longevity with no observed differences to the lifespan component of fitness, except at low density in two genotypes where unmated flies lived on average 6 days longer. These results add to a growing number of studies showing that although widespread, trade‐offs between longevity and reproduction are hardly ubiquitous, can be highly plastic, to the extent that the traits are uncorrelated under certain environmental or genetic conditions (Flatt, 2011).
5. CONCLUSION
Here, we show that the pattern of sexual dimorphism in D. serrata is consistent with females living longer than males across all genotypes and treatments. As expected, outbred genotypes lived longer, and female lifespan was more adversely affected by inbreeding. However, outbred male lifespan for the outbred F1 genotypes did not differ as expected from a cross between parental genotypes with significantly different lifespans. Overall, our findings converge with existing evidence to suggest that sex‐specific selection may be an important factor driving sexual dimorphism in lifespan (Bonduriansky et al., 2008; Maklakov et al., 2009; Maklakov & Lummaa, 2013) and that physiological differences resulting from strategies developed amongst sexes to maximize fitness can be independent of the effects of mating and/or density (Harvanek et al., 2017; Kimber & Chippindale, 2013; Maklakov et al., 2017; Sultanova et al., 2020; Vermeulen & Bijlsma, 2004a, 2004b; Ziehm et al., 2013). As the first study dissecting contributions of both genetic background and social environment on lifespan in D. serrata, the robustness of these findings will no doubt be revealed by further testing effects on lifespan across a much larger number of genotypes, which was not logistically feasible in the current study owing to the large number of environmental conditions considered (48 different treatments). Going forward, it will be possible to increase genotypic replication and reduce the number of different environments considered—the genetic effects observed here were much larger than the social effects. It is, however, reasonable to conclude that lifespan in D. serrata is best viewed as a condition‐dependent environmental modulation of a genetically determined trait.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
PEER REVIEW
We thank three anonymous reviewers for their very helpful comments on the manuscript https://publons.com/publon/10.1111/jeb.13992.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
We thank NC Appleton for assistance with fly work. Open access publishing facilitated by The University of Queensland, as part of the Wiley ‐ The University of Queensland agreement via the Council of Australian University Librarians. [Correction added on 18 May 2022, after first online publication: CAUL funding statement has been added.]
Narayan, V. P. , Wilson, A. J. , & Chenoweth, S. F. (2022). Genetic and social contributions to sex differences in lifespan in Drosophila serrata . Journal of Evolutionary Biology, 35, 657–663. 10.1111/jeb.13992
DATA AVAILABILITY STATEMENT
All data are publicly available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.jdfn2z3cq.
REFERENCES
- Aigaki, T. , & Ohba, S. (1984). Effect of mating status on Drosophila virilis lifespan. Experimental Gerontology, 19, 267–278. [DOI] [PubMed] [Google Scholar]
- Austad, S. N. (2019). Sex differences in health and aging: a dialog between the brain and gonad? Geroscience, 41, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard, J. W. O. (2005). Drosophila simulans as a novel model for studying mitochondrial metabolism and aging. Experimental Gerontology, 40, 763–773. [DOI] [PubMed] [Google Scholar]
- Bonduriansky, R. , Maklakov, A. , Zajitschek, F. , & Brooks, R. (2008). Sexual selection, sexual conflict and the evolution of ageing and life span. Functional Ecology, 22, 443–453. [Google Scholar]
- Brengdahl, M. , Kimber, C. M. , Maguire‐Baxter, J. , & Friberg, U. (2018a). Sex differences in life span: Females homozygous for the X chromosome do not suffer the shorter life span predicted by the unguarded X hypothesis. Evolution, 72, 568–577. [DOI] [PubMed] [Google Scholar]
- Brengdahl, M. , Kimber, C. M. , Maguire‐Baxter, J. , Malacrino, A. , & Friberg, U. (2018b). Genetic quality affects the rate of male and female reproductive aging differently in Drosophila melanogaster . American Naturalist, 192, 761–772. [DOI] [PubMed] [Google Scholar]
- Burger, J. M. , & Promislow, D. E. (2004). Sex‐specific effects of interventions that extend fly life span. Science of Aging Knowledge Environment, 2004, pe30. [DOI] [PubMed] [Google Scholar]
- Carazo, P. , Green, J. , Sepil, I. , Pizzari, T. , & Wigby, S. (2016). Inbreeding removes sex differences in lifespan in a population of Drosophila melanogaster . Biology Letters, 12(6), 20160337. 10.1098/rsbl.2016.0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T. , Liddle, L. F. , Kalb, J. M. , Wolfner, M. F. , & Partridge, L. (1995). Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature, 373, 241–244. [DOI] [PubMed] [Google Scholar]
- Chenoweth, S. F. , Appleton, N. C. , Allen, S. L. , & Rundle, H. D. (2015). Genomic evidence that sexual selection impedes adaptation to a novel environment. Current Biology, 25, 1860–1866. [DOI] [PubMed] [Google Scholar]
- Flatt, T. (2011). Survival costs of reproduction in Drosophila. Experimental Gerontology, 46, 369–375. [DOI] [PubMed] [Google Scholar]
- Fowler, K. , & Partridge, L. (1989). A cost of mating in female fruitflies. Nature, 338, 760–761. [Google Scholar]
- Friberg, U. (2005). Genetic variation in male and female reproductive characters associated with sexual conflict in Drosophila melanogaster. Behavior Genetics, 35, 455–462. [DOI] [PubMed] [Google Scholar]
- Gowaty, P. A. , Kim, Y. K. , Rawlings, J. , & Anderson, W. W. (2010). Polyandry increases offspring viability and mother productivity but does not decrease mother survival in Drosophila pseudoobscura. Proceedings of the National Academy of Sciences of the United States of America, 107, 13771–13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves, J. L. Jr , & Mueller, L. D. (1993). Population density effects on longevity. Genetica, 91, 99–109. [DOI] [PubMed] [Google Scholar]
- Harvanek, Z. M. , Lyu, Y. , Gendron, C. M. , Johnson, J. C. , Kondo, S. , Promislow, D. E. L. , & Pletcher, S. D. (2017). Perceptive costs of reproduction drive ageing and physiology in male Drosophila. Nature Ecology Evolution, 1, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, P. W. (1994). Purging inbreeding depression and the probability of extinction: full‐sib mating. Heredity (Edinb), 73(Pt 4), 363–372. [DOI] [PubMed] [Google Scholar]
- Iliadi, K. G. , Iliadi, N. N. , & Boulianne, G. L. (2009). Regulation of Drosophila life‐span: effect of genetic background, sex, mating and social status. Experimental Gerontology, 44, 546–553. [DOI] [PubMed] [Google Scholar]
- Joshi, A. , & Mueller, L. D. (1997). Adult crowding effects on longevity in Drosophila melanogaster: Increase in age‐independent mortality. Current Science, 72, 255–260. [Google Scholar]
- Khazaeli, A. A. , Xiu, L. , & Curtsinger, J. W. (1995). Effect of adult cohort density on age‐specific mortality in Drosophila melanogaster. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 50, B262–B269. [DOI] [PubMed] [Google Scholar]
- Khazaeli, A. A. , Xiu, L. , & Curtsinger, J. W. (1996). Effect of density on age‐specific mortality in Drosophila: a density supplementation experiment. Genetica, 98, 21–31. 10.1007/BF00120215 [DOI] [PubMed] [Google Scholar]
- Kimber, C. M. , & Chippindale, A. K. (2013). Mutation, condition, and the maintenance of extended lifespan in Drosophila. Current Biology, 23, 2283–2287. [DOI] [PubMed] [Google Scholar]
- Koliada, A. , Gavrilyuk, K. , Burdylyuk, N. , Strilbytska, O. , Storey, K. B. , Kuharskii, V. , Lushchak, O. , & Vaiserman, A. (2020). Mating status affects Drosophila lifespan, metabolism and antioxidant system. Comparative Biochemistry and Physiology Part A Molecular Integrative Physiology, 246, 110716. [DOI] [PubMed] [Google Scholar]
- Lemaître, J. F. , Ronget, V. , Tidière, M. , Allainé, D. , Berger, V. , Cohas, A. , Colchero, F. , Conde, D. A. , Garratt, M. , Liker, A. , Marais, G. , Scheuerlein, A. , Székely, T. , & Gaillard, J. M. (2020). Sex differences in adult lifespan and aging rates of mortality across wild mammals. Proceedings of the National Academy of Sciences of the United States of America, 117(15), 8546–8553. 10.1073/pnas.1911999117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklakov, A. A. , Bonduriansky, R. , & Brooks, R. C. (2009). Sex differences, sexual selection, and ageing: an experimental evolution approach. Evolution, 63, 2491–2503. 10.1111/j.1558-5646.2009.00750.x [DOI] [PubMed] [Google Scholar]
- Maklakov, A. A. , Carlsson, H. , Denbaum, P. , Lind, M. I. , Mautz, B. , Hinas, A. , & Immler, S. (2017). Antagonistically pleiotropic allele increases lifespan and late‐life reproduction at the cost of early‐life reproduction and individual fitness. Proceedings of the Royal Society B: Biological Sciences, 284(1856), 20170376. 10.1098/rspb.2017.0376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklakov, A. A. , & Lummaa, V. (2013). Evolution of sex differences in lifespan and aging: causes and constraints. BioEssays, 35, 717–724. [DOI] [PubMed] [Google Scholar]
- Malick, L. E. , & Kidwell, J. F. (1966). The effect of mating status, sex and genotype on longevity in Drosophila melanogaster. Genetics, 54, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow, T. A. (2011). "Cost" of virginity in wild Drosophila melanogaster females. Ecology and Evolution, 1, 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, M. D. W. , & Partridge, L. (2018). Drosophila as a model for ageing. Biochim Et Biophysica Acta Molecular Basis of Disease, 1864, 2707–2717. [DOI] [PubMed] [Google Scholar]
- Promislow, D. (2003). Mate choice, sexual conflict, and evolution of senescence. Behavior genetics, 33(2), 191–201. 10.1023/a:1022562103669 [DOI] [PubMed] [Google Scholar]
- Reddiex, A. J. , Allen, S. L. , & Chenoweth, S. F. (2018). A genomic reference panel for drosophila serrata. G3 (Bethesda), 8, 1335‐1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson, S. K. , Vickers, M. , Blows, M. W. , & Crozier, R. H. (2006). Age determination in individual wild‐caught Drosophila serrata using pteridine concentration. Journal of Experimental Biology, 209, 3155–3163. [DOI] [PubMed] [Google Scholar]
- Rogina, B. (2011). For the special issue: aging studies in Drosophila melanogaster . Experimental Gerontology, 46, 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service, P. M. (1989). The effect of mating status on lifespan, egg laying, and starvation resistance in Drosophila melanogaster in relation to selection on longevity. Journal of Insect Physiology, 35, 447–452. [Google Scholar]
- Sultanova, Z. , Andic, M. , & Carazo, P. (2018). The "unguarded‐X" and the genetic architecture of lifespan: Inbreeding results in a potentially maladaptive sex‐specific reduction of female lifespan in Drosophila melanogaster. Evolution, 72, 540–552. [DOI] [PubMed] [Google Scholar]
- Sultanova, Z. , & Carazo, P. (2019). Sex ratio at mating does not modulate age fitness effects in Drosophila melanogaster. Ecology and Evolution, 9, 6501–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultanova, Z. , Garcia‐Roa, R. , & Carazo, P. (2020). Condition dependent mortality exacerbates male (but not female) reproductive senescence and the potential for sexual conflict. Journal of Evolutionary Biology, 33(8), 1086–1096. 10.1111/jeb.13636 [DOI] [PubMed] [Google Scholar]
- Swindell, W. R. , & Bouzat, J. L. (2006). Selection and inbreeding depression: effects of inbreeding rate and inbreeding environment. Evolution, 60, 1014–1022. [PubMed] [Google Scholar]
- Tan, C. K. , Pizzari, T. , & Wigby, S. (2013). Parental age, gametic age, and inbreeding interact to modulate offspring viability in Drosophila melanogaster. Evolution, 67, 3043–3051. [DOI] [PubMed] [Google Scholar]
- Trivers, R. (1985). Social evolution. Menlo Park, California: The Benjamin/Cummings Publishing Company, Inc. [Google Scholar]
- Vaiserman, A. M. , Zabuga, O. G. , Kolyada, A. K. , Pisaruk, A. V. , & Kozeretska, I. A. (2013). Reciprocal cross differences in Drosophila melanogaster longevity: an evidence for non‐genomic effects in heterosis phenomenon? Biogerontology, 14, 153–163. 10.1007/s10522-013-9419-6 [DOI] [PubMed] [Google Scholar]
- Vermeulen, C. J. , & Bijlsma, R. (2004a). Changes in mortality patterns and temperature dependence of lifespan in Drosophila melanogaster caused by inbreeding. Heredity (Edinb), 92, 275–281. [DOI] [PubMed] [Google Scholar]
- Vermeulen, C. J. , & Bijlsma, R. (2004b). Characterization of conditionally expressed mutants affecting age‐specific survival in inbred lines of Drosophila melanogaster: lethal conditions and temperature‐sensitive periods. Genetics, 167, 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wit, J. , Loeschcke, V. , & Kellermann, V. (2015). Life span variation in 13 Drosophila species: a comparative study on life span, environmental variables and stress resistance. Journal of Evolutionary Biology, 28, 1892–1900. [DOI] [PubMed] [Google Scholar]
- Xirocostas, Z. A. , Everingham, S. E. , & Moles, A. T. (2020). The sex with the reduced sex chromosome dies earlier: a comparison across the tree of life. Biology Letters, 16, 20190867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajitschek, F. , Zajitschek, S. R. , Friberg, U. , & Maklakov, A. A. (2013). Interactive effects of sex, social environment, dietary restriction, and methionine on survival and reproduction in fruit flies. Age, 35, 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehm, M. , Piper, M. D. , & Thornton, J. M. (2013). Analysing variation in Drosophila aging across independent experimental studies: a meta‐analysis of survival data. Aging Cell, 12, 917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All data are publicly available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.jdfn2z3cq.


