Abstract
Two genes, accB and accE, that form part of the same operon, were cloned from Streptomyces coelicolor A3(2). AccB is homologous to the carboxyl transferase domain of several propionyl coezyme A (CoA) carboxylases and acyl-CoA carboxylases (ACCases) of actinomycete origin, while AccE shows no significant homology to any known protein. Expression of accB and accE in Escherichia coli and subsequent in vitro reconstitution of enzyme activity in the presence of the biotinylated protein AccA1 or AccA2 confirmed that AccB was the carboxyl transferase subunit of an ACCase. The additional presence of AccE considerably enhanced the activity of the enzyme complex, suggesting that this small polypeptide is a functional component of the ACCase. The impossibility of obtaining an accB null mutant and the thiostrepton growth dependency of a tipAp accB conditional mutant confirmed that AccB is essential for S. coelicolor viability. Normal growth phenotype in the absence of the inducer was restored in the conditional mutant by the addition of exogenous long-chain fatty acids in the medium, indicating that the inducer-dependent phenotype was specifically related to a conditional block in fatty acid biosynthesis. Thus, AccB, together with AccA2, which is also an essential protein (E. Rodriguez and H. Gramajo, Microbiology 143:3109–3119, 1999), are the most likely components of an ACCase whose main physiological role is the synthesis of malonyl-CoA, the first committed step of fatty acid synthesis. Although normal growth of the conditional mutant was restored by fatty acids, the cultures did not produce actinorhodin or undecylprodigiosin, suggesting a direct participation of this enzyme complex in the supply of malonyl-CoA for the synthesis of these secondary metabolites.
Malonyl coenzyme A (CoA) is an essential metabolite in most living organisms. It is a substrate for fatty acid synthases (4, 16), for polyketide synthases in plants, fungi, and bacteria (19), and for fatty acid chain elongation systems (37). It also plays a role as a modulator of the activity of some proteins (8). Since malonyl-CoA is used in the production of many of the pharmaceutically important polyketides made by streptomycetes (19), there is considerable interest in understanding the pathway(s) that leads to its synthesis. Thus, knowledge of the enzyme(s) involved in the supply of this key metabolite will not only provide a better understanding of primary metabolism in streptomycetes but will potentially allow for the development of more rational approaches for improving the level of production of many useful secondary metabolites.
Biosynthesis of malonyl-CoA occurs in most species through ATP-dependent carboxylation of acetyl-CoA by an acetyl-CoA carboxylase (45). The reaction catalyzed by this enzyme is a two-step process that involves ATP-dependent formation of carboxybiotin, followed by transfer of the carboxyl moiety to acetyl-CoA. Acetyl-CoA carboxylase expression is essential for the normal growth of bacteria (27, 28, 32), yeasts (17), and isolated animal cells in culture (33), reflecting the importance of this biosynthetic pathway.
Several complexes with acyl-CoA carboxylase (ACCase) activity have been purified from a number of actinomycetes. These complexes also possess the ability to carboxylate other substrates, including propionyl- and butyryl-CoA (12, 18, 20). Consequently, these enzymes are referred to as ACCases, and all of them consist of two subunits, a larger one (the α chain) with the ability to carboxylate its covalently bound biotin group and a smaller one (the β chain) bearing the carboxyl transferase activity. Little is known about the physiological role of these enzymes.
The pathway for the biosynthesis of malonyl-CoA in Streptomyces coelicolor has not been established yet. However, acetyl-CoA carboxylase activity has been readily measured in crude extracts of S. coelicolor (7, 36), confirming the presence of this enzyme activity in this microorganism. Attempts to purify a complex with acetyl-CoA carboxylase activity from streptomycetes have been unsuccessful, probably reflecting its high instability in vitro (7). An alternative pathway for the biosynthesis of malonyl-CoA was described in Streptomyces aureofaciens (2, 25) and involved the anaplerotic enzymes phosphoenolpyruvate carboxylase and oxaloacetate dehydrogenase. However, oxaloacetate dehydrogenase could not be detected in S. coelicolor A3(2) (6), where malonyl-CoA synthesis appears to occur exclusively through the acetyl-CoA carboxylase complex.
Attempts to identify enzymes with carboxylase activity in S. coelicolor led to the characterization of two complexes exhibiting exclusively propionyl-CoA carboxylase (PCCase) activity. The PCCase purified by Bramwell et al. (7) consisted of a biotinylated protein, PccA, of 88 kDa and a nonbiotinylated component, the carboxyl transferase, of 66 kDa. More recently, we characterized, genetically and biochemically, the components of a second PCCase in this bacterium. In vitro reconstitution experiments showed that an active complex could be obtained by mixing a carboxyl transferase component of 65 kDa, PccB, with either of the two almost identical biotinylated components, AccA1 and AccA2 (36).
Here we present a detailed genetic and biochemical characterization of an essential ACCase from S. coelicolor. The enzyme complex possesses unique characteristics and appears to be the main pathway for malonyl-CoA synthesis in this microorganism.
MATERIALS AND METHODS
Bacterial strains, culture, and transformation conditions.
S. coelicolor strain M145 (SCP1− SCP2−) was manipulated as described by Hopwood et al. (19). The strain was grown on SFM, R2, and R5 agar media and in 50 ml of SMM or YEME liquid medium. Escherichia coli strain DH5α was used for routine subcloning and was transformed according to the method of Hanahan (15). Transformants were selected on media supplemented with the appropriate antibiotics at the following concentrations: ampicillin, 100 μg ml−1; apramycin (APR), 100 μg ml−1; chloramphenicol, 25 μg ml−1; and kanamycin, 30 μg ml−1. Strain BL21(DE3) is an E. coli B strain lysogenized with λDE3, a prophage that expresses the T7 RNA polymerase from the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lacUV5 promoter (43). ET12567/pUZ8002 (a gift from M. Paget, John Innes Centre, Norwich, United Kingdom) was used for E. coli-S. coelicolor conjugation experiments (3). For selection of Streptomyces transformants and exconjugants, media were overlaid with thiostrepton (TH) (300 μg per plate), hygromycin (HYG) (1 mg per plate), or APR (1 mg per plate), respectively. Strains and recombinant plasmids are listed in Table 1. Fatty acid supplementation studies were performed in SMM containing APR (10 μg ml−1) and 0.075% (vol/vol) Brij 58. The different fatty acids were added at a final concentration of 100 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| S. coelicolor | ||
| M145 | Parental strain SCP1− SCP2− | 22 |
| T124 | M145 (accB/pTR124); Thr Hygr | This work |
| T149 | T124 containing pTR149 integrated in the att site of φC31; Thr Hygr Amr | This work |
| T149A | T149 with the wild-type accB copy of the chromosome replaced by the accB::hyg mutant allele; Hygr Amr | This work |
| E. coli | ||
| DH5α | ΔlacU169 (φ80lacZΔM15) endA1 recA1 hsdR17 deoR supE44 thi-1 λ− gyrA96 relA1 | 15 |
| BL21λ(DE3) | ompT(DE3) | 43 |
| ET 12567 | supE44 hsdS20 ara-14 proA2 lacY galK2 rpsL20 xyl-5 mtl-1 Δdam Δdcm ΔhsdM Cmr | 29 |
| RG7 | DH5α carrying pCL1 and pBA11 plasmids | 43 |
| Plasmids | ||
| pBluescript SK(+) | Phagemid vector (AprlacZ′) | Stratagene |
| PCR-Blunt | Used for cloning PCR products | Invitrogen |
| pGEM-T Easy | Used for cloning PCR products | Promega |
| pIJ2925 | pUC18 derivative (AprlacZ′) | 22 |
| pSET151 | Used for the conjugal transfer of DNA from E. coli to Streptomyces spp. (Apr ThrlacZ′) | 3 |
| pET22b(+) | Phagemid vector (AprlacZ′) for expression of recombinant proteins under control of strong T7 transcription and translation signals | Novagen |
| pUZ8002 | RK2 derivative with defective oriT (Kmr) | 31 |
| pIJ8600 | Used for the conjugal transfer of DNA from E. coli to Streptomyces spp. and for expression of recombinant proteins under tipA promoter | 44 |
| pBA11 | Vector containing E. coli birA gene | 1 |
| pTR45 | pSK(+) with a chromosomal Pstl insert carrying accA2 | 36 |
| pRM08 | pSK(+) with an Sstl insert carrying accBE | This work |
| pTR82 | pSK(+) carrying accBE with an NdeI site in the translation start site of accB | This work |
| pTR88 | pET22b(+) with accBE under control of strong T7 transcription and translation signals | This work |
| pTR90 | pET22b(+) with accB under control of strong T7 transcription and translation signals | This work |
| pTR94 | pIJ8600 derivative with a deletion of the int and att sites and carrying the accBE genes under tipAp control | This work |
| pTR107 | pET22b(+) with accE under control of strong T7 transcription and translation signals | This work |
| pTR124 | pSET151 with a hyg (Hygr) gene inserted in the accB coding region | This work |
| pTR141 | pIJ8600 derivative carrying oriT RK2, ori pUC18, attP site, int φC31, and aac(3)IV (Amr) | This work |
| pTR149 | pTR141 with a KpnI insert carrying accBE | This work |
| pTR204 | pET21a(+) with accA2 under control of strong T7 transcription and translation signals | This work |
| pTR237 | pET28a(+) with an accE His tag fusion gene | This work |
Growth conditions, protein production, and preparation of cell extracts.
S. coelicolor M145 was grown at 30°C in shake flasks in YEME medium for 24 to 48 h. When necessary, 10 μg of APR ml−1 or 5 μg of TH ml−1 was added to the medium. Mycelia were harvested by centrifugation at 5,000 × g for 10 min at 4°C, washed in 100 mM potassium phosphate buffer, pH 8, containing 0.1 mM dithiothreitol (DTT), 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 10% glycerol (buffer A) and resuspended in 1 ml of the same buffer. The cells were disrupted by sonic treatment (4- or 5-s bursts) using a VibraCell Ultrasonic Processor (Sonics & Materials, Inc.). Cell debris was removed by centrifugation, and the supernatant was used as cell extract. For the expression of heterologous proteins, E. coli strains harboring the appropriate plasmids were grown at 37°C in shake flasks in Luria-Bertani medium in the presence of 25 μg of chloramphenicol ml−1 or 100 μg of ampicillin ml−1 for plasmid maintenance. In order to improve the biotinylation of AccA1 and AccA2 in E. coli, the strains containing pCL1 or pTR204 were also transformed with pBA11 (1), which overexpresses the E. coli biotin ligase; 10 μM d-biotin was also added to the medium. Overnight cultures were diluted 1:10 in fresh medium and grown to an A600 of 0.4 to 0.5 before the addition of IPTG to a final concentration of 0.1 mM. Induction was allowed to proceed for 4 h. The cells were harvested, washed, and resuspended in 1 ml of buffer A. Cell extracts were prepared as described above.
Protein methods.
Cell extracts were analyzed by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (26) using a Bio-Rad minigel apparatus. The final acrylamide monomer concentration was 12% (wt/vol) for the separating gel and 5% for the stacking gel. Coomassie brilliant blue was used to stain protein bands. Protein contents were determined by the method of Bradford (5) with bovine serum albumin as the standard. The relative concentration of soluble AccB and AccA2 overexpressed in E. coli was determined by densitometric scanning of the polyacrylamide-SDS gels.
Acetyl-CoA carboxylase and PCCase assay.
Acetyl-CoA carboxylase and PCCase activities in cell extracts were measured following the incorporation of HCO3− into acid nonvolatile material (7, 20). The reaction mixture contained 100 mM potassium phosphate (pH 8.0), 300 μg of bovine serum albumin, 3 mM ATP, 5 mM MgCl2, 50 mM NaH14CO3 (specific activity, 200 μCi mmol−1 [740 kBq mmol−1]), 1 mM substrate (acetyl-CoA or propionyl-CoA), and 100 μg of cell-free protein extract in a total reaction volume of 100 μl. The reaction was initiated by the addition of NaH14CO3, allowed to proceed at 30°C for 15 min, and stopped with 200 μl of 6 M HCl. The contents of the tubes were then evaporated to dryness at 95°C. The residue was resuspended in 100 μl of water, 1 ml of Optiphase scintillation liquid (Wallac Oy) was added, and the 14C radioactivity was determined in a Beckman liquid scintillation counter. Nonspecific CO2 fixation by crude extracts was assayed in the absence of substrate. One unit of enzyme activity catalyzed the incorporation of 1 μmol of 14C into acid-stable products per min. To confirm that the products of the reactions were malonyl- or methylmalonyl-CoA, samples were analyzed by high-performance liquid chromatography (24).
DNA manipulations.
Isolation of chromosomal and plasmid DNA, restriction enzyme digestion, and agarose gel electrophoresis were carried out by conventional methods (22, 38). Southern analyses were performed by using 32P-labeled probes made by random oligonucleotide priming (Prime-a-gene kit; Promega).
Gene cloning and plasmid construction.
The synthetic oligonucleotides TC1 (5′-CAGAATTCAAGCAGCACGCCAAGGGCAAG) and TC2 (5′-CAGAATTCGATGCCGTCGTGCTCCTGGTC) were used to amplify an internal fragment of the S. coelicolor pccB gene. The reaction mixture contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1 mM MgCl2, 6% glycerol, 25 μM each deoxynucleoside triphosphate, 2 ± 5 U of Taq or Pfu DNA polymerase, 20 pmol of each primer, and 50 ng of S. coelicolor chromosomal DNA in a final volume of 100 μl. Samples were subjected to 30 cycles of denaturation (95°C, 30 s), annealing (65°C, 30 s), and extension (72°C, 1 min). A 1-kb PCR fragment was used as a 32P-labeled probe to screen a size-enriched library. A 2.7-kb BamHI fragment containing an incomplete accB gene was cloned in BamHI-cleaved pBluescript SK(+), yielding pTR62. The synthetic oligonucleotides TC16 (5′-TATTCTAGACATATGACCGTTTTGGATGAGG), used to introduce an NdeI site at the translational start codon of the S. coelicolor accB gene, and TC17 (5′-ACCTCTAGACAACGCTCGTGGACC) were used to amplify an internal fragment of the S. coelicolor accB gene. The reaction mixture was the same as the one indicated above. Samples were subjected to 35 cycles of denaturation (95°C, 30 s), annealing (65°C, 30 s), and extension (72°C, 1 min). The PCR product was digested with XbaI and cloned in XbaI-cleaved pBluescript SK(−) in E. coli DH5α, yielding pTR82. This plasmid was digested with BstEII and SacI, ligated with a BstEII-SacI fragment cleaved from pMR08, and introduced by transformation into E. coli DH5α, yielding pTR87. An NdeI-SacI fragment from pTR87 was cloned in NdeI-SacI-cleaved pET22b(+) (Novagen) (pTR88), thus placing the accBE operon under the control of the powerful T7 promoter and ribosome-binding sequences. Expression of accB was achieved by eliminating part of the coding sequence of accE in pTR88. For this, pTR88 was digested with NotI and the large fragment was religated to obtain pTR90. The synthetic oligonucleotides NaccE (5′-TTATCTAGACATATGTCCCCTGCCGAC), used to introduce an NdeI site at the translational start codon of the S. coelicolor accE gene, and CaccE (5′-ATGAATTCTATGCATCGGGTCAGCGCCAGCTG) were used to amplify accE. The reaction mixture was the same as the one indicated above. Samples were subjected to 35 cycles of denaturation (95°C, 30 s), annealing (65°C, 30 s), and extension (72°C, 30 s). The PCR product was cloned using the pGEM-T easy vector (Promega) in E. coli DH5α, yielding pTR106. An NdeI-EcoRI fragment from pTR106 was cloned in NdeI-EcoRI-cleaved pET22b(+), yielding pTR107, thus placing the accE gene under the control of the T7 promoter and ribosome-binding sequences. To generate an accE His tag fusion gene (full-length accE fused to six His codons at its N terminus), the NdeI-EcoRI fragment from pTR107 was cloned in NdeI-EcoRI-cleaved pET28a(+), yielding pTR237. For the production of high levels of AccA2, we constructed pTR204. For that the synthetic oligonucleotides accANd (5′- CATATGCGAAAGGTGCTCATCGCCAATC) and accABa (5′-AAAGCGTTCTCCGAGAGGAATCCGTAGC) were used to amplify the N terminus of accA2 and to introduce an NdeI site at the translational start codon of the gene. The PCR fragment was cloned into PCR-Blunt (Invitrogen) to yield pTR200. A BamHI-KpnI fragment from pTR45 (36) was cloned into the BamHI-KpnI-digested pTR200, yielding pTR202 with a full-length accA2 gene. Finally, the NdeI-HindIII fragment from pTR202 was cloned into the NdeI-HindIII-digested pET21a(+), to yield pTR204.
To provide an additional copy of accB, pIJ8600 was digested with BglII and EcoRI and the fragment containing oriT from RK2, ori from pUC18, the attP site and int of φC31, and the aac(3)IV (Amr) gene was ligated with a linker containing sites for the following restriction enzymes to yield pTR141 (Mike Butler, personal communication): BglII, AseI, EcoRI, BglII, NdeI, KpnI, XbaI, PstI, HindIII, BamHI, SstI, and NotI. A 4.0-kb KpnI fragment containing the complete accBE operon from pRM08 was cloned into KpnI-cleaved pTR141, yielding pTR149. To place the chromosomal copy of accBE under the control of the TH-inducible tipA promoter, the synthetic oligonucleotides TC16 (5′-ATTCTAGACATATGACCGTTTTGGATGAGG), used to introduce an NdeI site at the translational start codon of the S. coelicolor accB gene, and TC17 (5′-ACCTCTAGACAACGCTCGTGGACC) were used to amplify an internal fragment of S. coelicolor accB. The reaction mixture contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1 mM MgCl2, 6% glycerol, 25 μM (each) deoxynucleoside triphosphate, 2 to 5 U of Taq DNA polymerase, 20 pmol of each primer, and 50 ng of S. coelicolor chromosomal DNA in a final volume of 100 μl. Samples were subjected to 30 cycles of denaturation (95°C, 30 s), annealing (65°C, 30 s), and extension (72°C, 1 min). The 1-kb PCR product was digested with XbaI (these sites were introduced by the 5′ end of the oligonucleotides TC16 and TC17) and cloned in XbaI-cleaved pBluescript SK(+), yielding pTR82. An NdeI-XbaI fragment from the plasmid pTR82 was cloned in NdeI-XbaI-cleaved pIJ8600, yielding pTR93. In order to place the chromosomal copy of the accBE operon under the tipA promoter we removed from pTR93 a HindIII fragment containing the int gene and att of φC31, yielding pTR94.
Protein purification protocols.
The His6-tagged fusion protein H6AccE was purified from cultures of RG12 [strain BL21(DE3) harboring pTR237] after the addition of 0.1 mM IPTG to induce the DE3-encoded T7 RNA polymerase. Cells were pelleted, resuspended in 50 mM phosphate buffer (pH 7.2)–300 mM NaCl–0.75 mM dithiothreitol–10% glycerol, and disrupted by sonication. Cell debris was removed by centrifugation, and the supernatant was passed through a Ni2+-nitrilotriacetic acid-agarose affinity column equilibrated with the same buffer. The H6AccE protein was recovered by elution with 100 mM imidazole and dialyzed against a solution containing 100 mM sodium phosphate (pH 7.2), 1 mM dithiothreitol, 1 mM EDTA, and 20% glycerol.
Nucleotide sequencing.
The sequence of the SstI fragment containing accB was determined by subcloning ApaI fragments from pRM08 in pSKBluescript SK(+). Synthetic oligonucleotides were used where needed to complete the sequence. Dideoxy sequencing (39) was carried out using the Promega TaqTrack sequencing kit and double-stranded DNA templates.
S1 nuclease mapping.
For each S1 nuclease reaction, 30 μg of RNA was hybridized in trichloroacetic acid-sodium salt (NaTCA) buffer (solid NaTCA [Aldrich] was dissolved to 3 M in 50 mM PIPES, 5 mM EDTA, pH 7.0) to about 0.002 pmol (approximately 104 cpm) of the following probes. For accA2, the oligonucleotide 5′-GCTTTGAGGACCTTGGCGATG (accA2down) corresponding to a sequence within the coding region of accA2 was uniquely labeled at the 5′ end with [32P]ATP using T4 polynucleotide kinase and then used in PCR with the unlabeled oligonucleotide 5′-GAAGTACAGGCCGAAGACCAC (accA2up), which corresponds to a sequence upstream of the accA2 promoter region, to generate a 766-bp probe. For accA1, the oligonucleotide 5′-GCGATTTCGCCACGATTGGCG (accA1down) corresponding to a sequence within the coding region of accA1 was uniquely labeled at the 5′ end and used in a PCR with the unlabeled oligonucleotide 5′-CCGATATCAGCCCCTGATGAC (accA1down), which corresponds to a sequence upstream of the accA1 promoter, to generate a 563-bp probe. For accB, the oligonucleotide 5′-CGTCAGCTTGCCCTTGGCGTG (accBdown) corresponding to a sequence within the coding region of accB was labeled at the 5′ end and then used in a PCR with the unlabeled oligonucleotide 5′-CTACGCTCCGGGTGAGCGAAC (accBup), which corresponds to a sequence upstream of the accB promoter, to generate a 483-bp probe. For accBE, the oligonucleotide 5′-GGAGGGCCGTGATGGCGGCGACTTCCTCGGG (accBEdown) corresponding to a sequence within the coding region of accE was labeled at the 5′ end and used in a PCR with the unlabeled oligonucleotide 5′-GAGGAACTGGTACGCGCGGGCG[GTACAAGCAAGCT] (accBEup) corresponding to a sequence in the coding region of accB (bracketed oligonucleotides constitute a tail added to the probe to differentiate probe reannealing from full-length protection) to generate a 563-bp probe. Subsequent steps were performed as described by Strauch et al. (41).
Nucleotide sequence accession number.
The accB and accE genes were identified in cosmid SC1C2 (S. coelicolor genome project [http://www.sanger.ac.uk/Projects/S_coelicolor/]; nucleotide accession number AL031124).
RESULTS
Cloning the accBE genes.
Since pccB mutants of S. coelicolor produce wild-type levels of acetyl-CoA carboxylase (36), we foresaw that a second gene encoding a different carboxyl transferase β subunit capable of recognizing acetyl-CoA as a substrate should exist in this organism. Based on the high level of sequence homology shown by genes encoding putative carboxyl transferases in the same species (e.g., in Mycobacterium tuberculosis [10]), we attempted to clone this alternative β subunit gene using pccB as a hybridization probe. When a BamHI digest of S. coelicolor DNA was probed with pccB under conditions of low stringency, a second poorly hybridizing band was readily detected (data not shown). This hybridizing sequence was cloned from a size-enriched library as a 2.5-kb BamHI fragment. Sequencing revealed the presence of an incomplete open reading frame (ORF) with high homology to pccB; the complete gene was subsequently cloned on a 6-kb SstI fragment, yielding pRM08 (Fig. 1). Sequencing of this fragment revealed a putative protein with end-to-end similarity to a likely decarboxylase of Streptomyces cyanogenus (76% identity [46]), to PccB from S. coelicolor (57% identity [36]), and to the β subunit (PccB) of the Saccharopolyspora erythraea PCCase (56% identity [11]). The gene encoding this new putative carboxyl transferase was called accB.
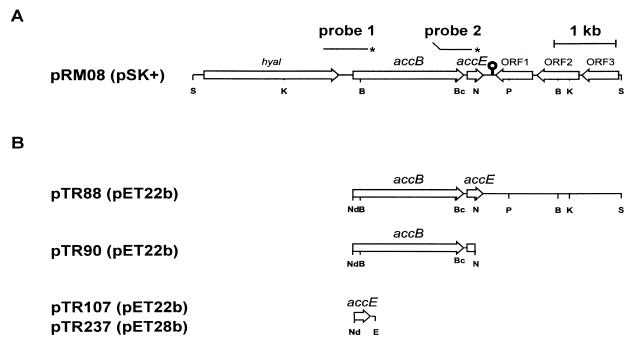
FIG. 1.
Organization of the region of the S. coelicolor M145 chromosome containing the accB and accE genes. (A) Genetic and physical map of the 6.2-kb insert in pRM08. The secondary structure downstream of accE may represent a factor-independent transcriptional terminator. Probes 1 and 2 were generated by PCR using the oligonucleotides accBup-accBdown and accBEup-accBEdown, respectively, uniquely labeled at the 5′ end (∗) and were used in transcriptional analysis of the accBE operon. (B) Map of the DNA fragments cloned in pET22b that were used for expression of accB and/or accE in E. coli. Only relevant restriction sites are shown: B, BamHI; Bc, BclI; E, EcoRI; K, KpnI; Nd, NdeI; N, NotI; S, SpHI.
The sequence also revealed the presence of a small ORF, accE, whose start codon was only 17 bp downstream of the termination codon of accB. A 17-nucleotide (nt) inverted repeat which could function as a factor-independent bidirectional transcriptional terminator separates accE from three convergent ORFs with homology to putative proteins of M. tuberculosis of unknown function. The putative AccE protein has a deduced molecular mass of 7.5 kDa and does not resemble any other known protein. The region upstream of accB encodes a putative protein which is highly homologous to several known hyaluronidases.
Heterologous expression of accB, accE, and in vitro reconstitution of an ACCase complex.
Recently, we achieved reconstitution of a PCCase complex activity by mixing E. coli cell extracts containing PccB (the carboxyl transferase) with cell extracts containing the biotinylated subunits AccA1 and AccA2 (36). To assess whether AccB and AccE were components of a previously uncharacterized carboxylase complex, we attempted similar in vitro reconstitution experiments with crude extracts containing these proteins. Since E. coli does not contain PCCase and acetyl-CoA carboxylase activity cannot be assayed directly by carboxylation of acetyl-CoA (34), the acetyl-CoA carboxylase activity measured in these crude extracts represents the activity of heterologous complexes reconstituted in vitro.
Overexpression of accB and accE in E. coli was attempted with strain RG8, a BL21(DE3) strain containing pTR88 (Fig. 1). SDS-PAGE of crude extracts of RG8, prepared from IPTG-induced cultures, revealed overexpression of a 57-kDa protein, corresponding to the predicted size of AccB. In the same electrophoretic analysis no clearly identifiable AccE band was observed. In vitro reconstitution of ACCase activity was then obtained by mixing a crude extract prepared from an IPTG-induced culture of RG8 with a cell extract of E. coli strain RG11, which overproduces the biotinylated protein AccA2 and the E. coli biotin ligase BirA, harbored in plasmids pTR204 and pBA11, respectively. After incubation for 1 h at 4°C, the mixture was assayed for acetyl-CoA carboxylase and PCCase activities. As shown in Table 2, an enzyme complex with both acetyl-CoA carboxylase and PCCase activities was readily detected, confirming that AccB was the carboxyl transferase component of an ACCase complex. Similar results were obtained when the reconstitution experiments were performed using cell extracts of strain RG7, a BL21(DE3) strain containing pCL1 that provides AccA1 instead of AccA2 as the biotinylated component of the ACCase. The lower levels of both acetyl-CoA carboxylase and PCCase activity are due to the lower level of expression of accA1 by pCL1 (36). These results confirmed that either AccA1 or AccA2 could be used efficiently, at least in vitro, as the α subunit of the enzyme complex (Table 2).
TABLE 2.
Heterologous expression of ACCase components in cell extracts of E. coli and in vitro reconstitution of enzyme activity
| E. coli straina | Protein(s) induced by IPTG | Protein expression (mU mg of protein−1) in cell extractsb
|
|
|---|---|---|---|
| Acetyl-CoA carboxylase | PCCase | ||
| RG7c | AccA1 | <0.02e | <0.02e |
| RG11c | AccA2 | <0.02e | <0.02e |
| RG8 | AccB, AccE | <0.02e | <0.02e |
| RG11-RG8d | AccA2-AccB, AccE | 7.53 ± 0.18 | 9.10 ± 0.20 |
| RG7-RG8d | AccA1-AccB, AccE | 1.85 ± 0.12 | 2.25 ± 0.13 |
All the RG strains are derived from E. coli BL21(DE3), except RG7, which derives from DH5α.
Results are the means of three determinations ± standard errors.
Contains plasmid pBA11 that expresses BirA constitutively.
Mix of equal amounts of proteins from cell extracts of each of the strains indicated.
The amount of 14C fixed into acid-stable products was not significantly higher than background levels (10 cpm, equivalent to 0.02 mU).
Is AccE a functional component of the ACCase complex?
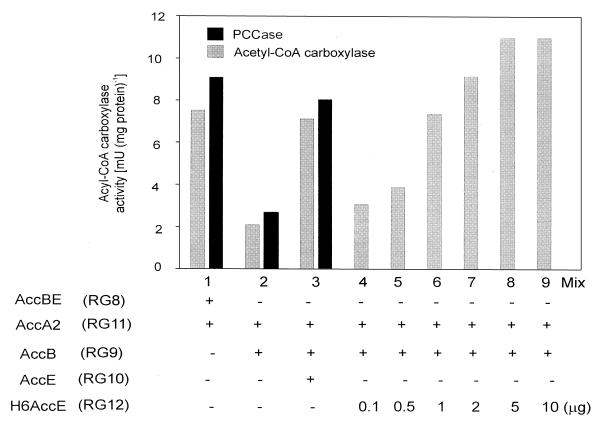
The genetic organization of accB and accE as members of the same transcription unit suggested that AccE could also be a functional component of the ACCase complex. To investigate this hypothesis we assayed acetyl-CoA carboxylase and PCCase activities in a mixture of cell extracts that contained AccB [strain RG9, a BL21(DE3) strain containing pTR90] and AccA2 [strain RG11, a BL21(DE3) strain containing pTR204] but not AccE. Although ACCase activity was readily detected in this mixture, indicating that AccE is not catalytically necessary for the successful reconstitution of an active complex in vitro, the levels of acetyl-CoA carboxylase and PCCase activities were considerably lower (approximately 30%) than those obtained with cell extracts that contained AccB and AccE (Fig. 2, compare mixes 1 and 2). Since the levels of AccB in the cell extracts of RG8 and RG9 were essentially the same, we inferred from these experiments that AccE was necessary to obtain a fully active ACCase complex. To confirm that the absence of AccE was responsible for the lower ACCase activity observed, we studied the effect that the addition of cell extracts containing high levels of soluble AccE [strain RG10, a BL21(DE3) strain containing pTR107] had on the ACCase activity present in a mix of crude extracts containing AccB and AccA2. As shown in Fig. 2 (mixes 2 and 3) the specific activities of both acetyl-CoA carboxylase and PCCase were almost 3.5 times higher in the presence of AccE than in the control experiment that lacked this protein and resembled those values obtained by mixing RG8 (AccBE) and RG11 (AccA2) cell extracts. Similar results were obtained when purified H6AccE was added to the AccB-AccA2 mix (Fig. 2, mixes 4 to 9). The addition of different amounts of H6AccE (from values ranging from 0.1 to 10 μg of pure protein) increased the levels of acetyl-CoA carboxylase activity, reaching saturation when more than 2 μg of AccE was present in the reaction mix. The fact that the maximum level of enzyme activity was obtained at high concentrations of AccE proposed a direct participation of this protein in the activation of the complex formed by AccB and AccA2. Although the results presented in this section suggest that AccE increases the rate of the ACCase reaction, kinetic analysis using purified components will be necessary to understand the precise role played in enzyme activity by this small polypeptide.
FIG. 2.
Effect of AccE on the catalytic activity of the ACCase complex. In mixes 1, 2, and 3, acetyl-CoA carboxylase and PCCase activities were measured after mixing equal amounts of proteins from cell extracts from each of the strains indicated. In mixes 4 to 9, ACCase activity was determined using a mix of RG9 and RG11 cell extracts containing different amounts of purified H6AccE. Results are the means of three determinations. When ACCase activity was measured in individual cell extracts, the amount of 14C fixed into acid-stable products was not significantly higher than background levels (10 cpm, equivalent to 0.02 mU).
accB is an essential gene in S. coelicolor
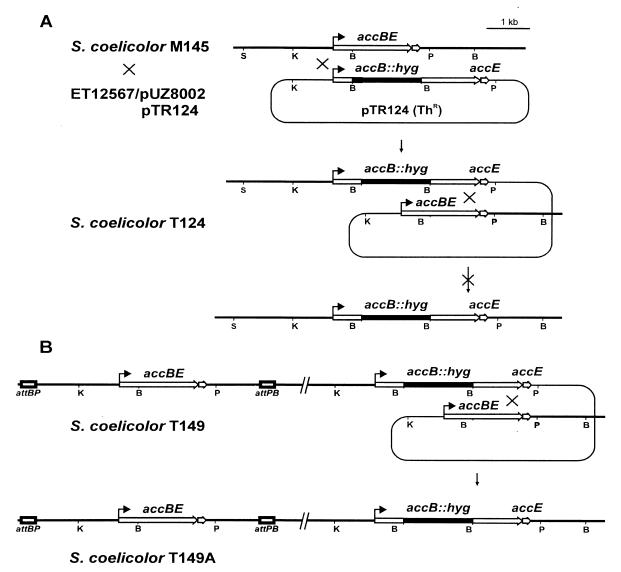
To study the role of AccB in vivo, we attempted to construct an accB mutant by gene replacement (Fig. 3A). A HYG resistance cassette was cloned in the unique BamHI site present in the coding sequence of accB contained in pTR80. After an intermediate cloning step in pIJ2925, a BglII fragment containing the mutated allele was inserted in the conjugative E. coli vector pSET151. The resulting plasmid, pTR124, was introduced into the E. coli donor strain ET12567/pUZ8002 and transferred by conjugation into M145. Thr Hygr exconjugants were selected in which the plasmid had integrated into the chromosome at the accB locus by a single crossover. One of the exconjugants, T124, was taken through four rounds of sporulation on SFM medium with HYG to allow for a second crossover and replacement of the wild-type accB with the mutant allele. Although several thousand colonies were screened for TH sensitivity (which would have reflected successful gene replacement), none were obtained, suggesting that accB could be an essential gene in S. coelicolor. If this were true, the presence of a second copy of accB in the chromosome of T124 ought to permit a second crossover event, leading to the replacement of the wild-type accB gene by the Hygr mutant allele. To confirm this hypothesis, we first integrated pTR149 (see Materials and Methods; Fig. 3B) containing accBE and the native promoter into the φC31 attB site of T124 (the presence of accE in this construct would also cater for any polar effect on the expression of accE caused by disruption of the native copy of accB). The resulting strain, T149 (Hygr Thr Amr), was subjected to three rounds of sporulation on SFM agar containing HYG and APR, and after screening approximately 500 colonies, 20 were found to be Amr Hygr Ths; one of these was designated T149A. Disruption of accB, but only in the presence of an additional copy of the gene (i.e., in strain T149A), was confirmed by Southern analysis using an internal fragment of accB as a hybridization probe. These results confirmed the essentialness of AccB for S. coelicolor viability.
FIG. 3.
Attempted disruption of accB. (A) Diagram showing integration of pTR124 through one of the accBE flanking regions and resolution of the cointegrate by a second crossover event. The × on top of the arrow indicates the inability to obtain the replacement of the wild-type accB by the Hygr mutant allele. (B) Integration of a second copy of accBE at the φC31 att site of T124 (to yield strain T149) allowed replacement of the wild-type accB by the mutant allele.
Construction and characterization of an accBE conditional mutant.
In order to regulate the expression of the putative accBE operon and study its effect on the physiology of S. coelicolor, we constructed a conditional mutant strain in which the expression of these genes was under the control of the TH-inducible tipA promoter (30). For this, pTR94 was transformed into the E. coli strain ET12567/pUZ8002 and conjugated into the S. coelicolor strain M145. Integration of pTR94 by Campbell recombination through the accBE homologous sequences left the accBE operon under tipAp. The strain obtained was named M94 and the genetic modification introduced was confirmed by Southern blot experiments (data not shown).
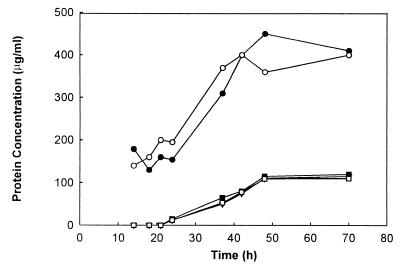
Normal growth of strain M94 on SMM depended on the presence of 5 μg of TH/ml, which derepresses the expression of the accBE operon. In the absence of TH growth was strongly affected, and the low growth levels observed were probably due to a leakiness of the control system (Fig. 4) (E. Takano and M. Bibb, unpublished data); no antibiotic production was observed in these cultures. To determine the effect of TH on the acetyl-CoA carboxylase and PCCase enzyme levels, both activities were measured in 38-h cultures grown in SMM with or without the addition of 5 μg of TH/ml. We used this time point because both cultures were still in their exponential phase and we expected, at least for the acetyl-CoA carboxylase activity, its maximal levels. As observed in Table 3, the acetyl-CoA carboxylase activity present in crude extracts prepared from the uninduced cultures was almost 10 times lower than that found in the TH-induced cultures. This difference was not observed in the levels of PCCase, a result that was expected considering that the ACCase containing AccB as a β subunit is only one of the three known complexes with PCCase activity in S. coelicolor (7, 36). These results correlate the growth deficiency of the M94 conditional mutant with the low levels of acetyl-CoA carboxylase activity in the absence of the inducer and strongly support the hypothesis that AccB is an essential protein for S. coelicolor viability.
FIG. 4.
Effects of TH and various fatty acids on growth of S. coelicolor M94 bearing the tipAp-accBE fusion. Cultures of strain M94 were grown in SMM medium containing 10 mg of APR ml−1 (□) or the same medium supplemented with TH (5 μg/ml) (○) or with the following fatty acids at 0.01%: octanoic acid (▪), palmitic acid (▿), and oleic acid (●).
TABLE 3.
Acetyl-CoA carboxylase and PCCase activities in cell extracts of S. coelicolor M94
| Growth medium | Activity (mU mg of protein−1)a of:
|
|
|---|---|---|
| Acetyl-CoA carboxylase | PCCase | |
| SMM | 0.12 ± 0.03 | 2.20 ± 0.06 |
| SMM + THb | 1.24 ± 0.06 | 3.90 ± 0.07 |
| SMM + Oleateb | 0.15 ± 0.03 | 1.40 ± 0.05 |
Results are means of three determinations ± standard errors.
TH, 5 μg/ml; oleate, 0.01% (wt/vol).
Since acetyl-CoA carboxylase catalyzes the synthesis of malonyl-CoA, the primer for the elongation step of fatty acids, we investigated whether the growth defect showed by M94 could be corrected by growing it in the presence of different fatty acids. M94 grew very poorly in SMM (Fig. 4); however, when the SMM medium was supplemented with oleic acid, a straight-chain unsaturated fatty acid, growth was restored to normal levels (Fig. 4). The growth of the mutant was not stimulated by the straight-chain octanoic acid or palmitic acid, indicating that these saturated fatty acids are not incorporated efficiently into S. coelicolor membrane phospholipids or that the resulting membranes are not functional. Similar results were also described for an accBC conditional mutant of Bacillus subtilis (32). Interestingly, although growth was restored in oleate-supplemented medium, the cultures were still impaired in antibiotic production and the levels of acetyl-CoA carboxylase activity were the same as those found in the absence of the inducer (Table 3). All these results strongly suggest that AccB is the carboxyl transferase component of an essential ACCase complex whose main physiological role appears to be the supply of malonyl-CoA for both fatty acid and polyketide biosynthesis.
Transcriptional analysis of accBE, accA1, and accA2
Biosynthesis of malonyl-CoA in S. coelicolor should occur not only during exponential phase, when the synthesis of fatty acids is essential, but also during transition and stationary phase to provide the elongation units for the synthesis of actinorhodin and undecylprodigiosin. Genetics and biochemical data propose that AccB forms part of the main ACCase of S. coelicolor involved in the biosynthesis of malonyl-CoA and that either AccA2 or AccA1 could function as the biotinylated components of this enzyme complex. In order to study the levels of transcription of the enzyme components and hopefully gain more information into the subunit composition of the complex throughout growth we performed transcriptional studies of the accBE, accA1, and accA2 genes.
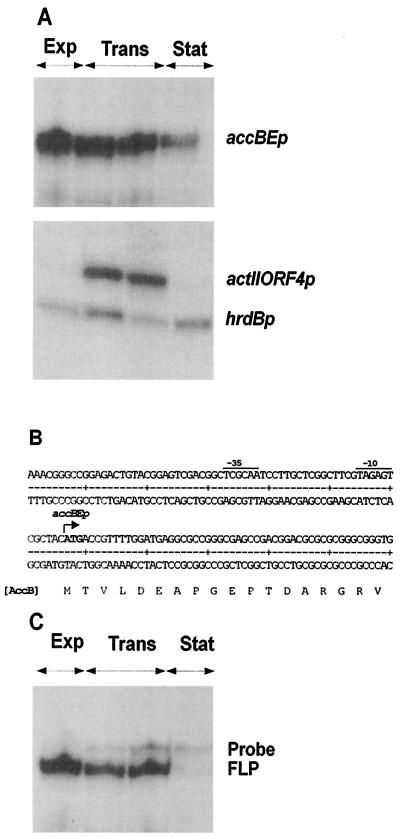
S. coelicolor A3(2) strain M145 was grown in SMM medium and RNA was extracted during the exponential, transition, and stationary phases of growth. S1 nuclease protection analysis of accB mRNA was performed using a 483-bp PCR product, uniquely labeled at the 5′ end of the downstream oligonucleotide. Transcription of accB occurred primarily during active growth (exponential and transition phases) and then declined significantly upon entry into stationary phase (Fig. 5A). The transcripts of the major and essential sigma factor gene of S. coelicolor, hrdB, and of the pathway-specific activator gene for actinorhodin biosynthesis, actII-ORF4, were monitored as controls. As expected from previous work, hrdB was expressed throughout growth (9), while the actII-ORF4 transcript peaked during transition phase and disappeared in stationary phase (13).
FIG. 5.
Growth phase-dependent expression and transcription start site of the accBE operon. (A) S1 nuclease mapping of accB, actII-ORF4, and hrdB, using RNA isolated from a liquid-grown culture of S. coelicolor M145. Exp, Trans, and Stat indicate the exponential, transition, and stationary phases of growth, respectively. (B) The nucleotide sequence of both strands of the accB promoter region is shown. The arrow indicates the most likely transcription start point for the accBE promoter, as determined by S1 nuclease mapping. Potential −10 and −35 regions for accBEp are underlined. (C) S1 nuclease mapping of the accB-accE intergenic region using a 563-nt probe. FLP, full-length protection of the probe reflecting transcription across the intergenic region.
The RNA-protected fragment identified for accB corresponds to a transcript that would start 1 bp upstream of or at the adenine of the most likely translation start codon of accB. Putative −10 and −35 promoter regions similar to those likely to be recognized by ςhrdB (42) are located upstream of the transcription initiation site (Fig. 5B).
To determine if accB and accE were cotranscribed, a 563-bp probe was generated by PCR that spanned the intergenic region. For this we used a 5′ oligonucleotide corresponding to a sequence within the coding region of accB and a 3′ oligonucleotide corresponding to a sequence within accE. The addition of a 13-nt tail to the 5′ oligonucleotide allowed for facile discrimination of full-length protection (reflecting cotranscription) of the probe from probe reannealing. The results clearly showed that accB and accE were part of the same transcript (Fig. 5C). The pattern of transcription of accBE during the different growth phases corresponded well to the profile observed with the accB probe.
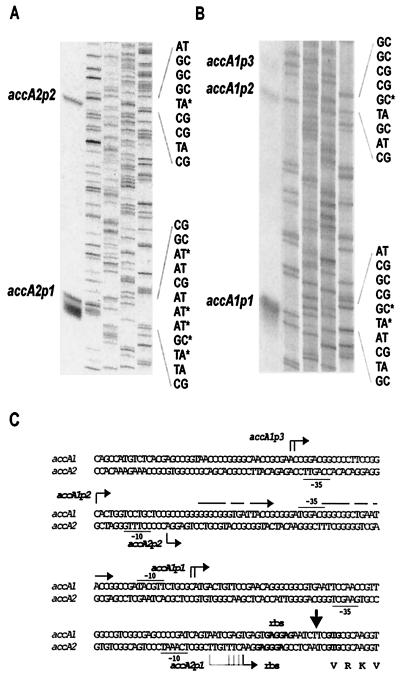
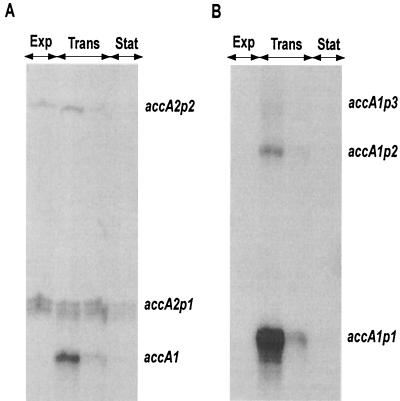
The transcripts of accA2 present during exponential phase were studied by high-resolution S1 mapping. The probe used was a 766-bp PCR-generated DNA fragment uniquely labeled at the 5′ end of the oligonucleotide corresponding to a sequence within accA2. The experimental data revealed the presence of two RNA protected fragments, consistent with transcripts initiated 25 bp (from accA2p1) and 153 bp (from accA2p2) upstream of the putative translation start site of accA2 (Fig. 6A and C). The putative −10 and −35 regions of these promoters also show some similarity to the consensus sequence of promoters that are likely to be recognized by ςhrdB. The growth phase-dependent expression of accA2 from these two putative promoters closely resembled that observed for the accBE operon; i.e., there was a constant and high level of expression during the exponential and transition phases of growth that declined markedly upon entry into stationary phase (Fig. 7A). However, a new RNA protected fragment of 185 bp was also detected during transition phase. Since the nucleotide sequences of accA1 and accA2 are identical from nt −2 to nt +200 with respect to the coding sequence (36), 185 bp of this probe should also be protected by the accA1 mRNA. Thus, although the existence of a third promoter for accA2 that is regulated in a different manner cannot be ruled out, this transcript could also correspond to accA1.
FIG. 6.
Mapping of the accA2 and accA1 transcription start points. (A and B) High-resolution S1 nuclease mapping of the 5′ end of accA2 transcripts. Lanes 1, RNA protected products of the S1 nuclease protection assay; lanes 2 to 5, A, C, G, and T lanes of a dideoxy sequencing ladder using the same oligonucleotide that was used to make the S1 probe (accA2down for accA2 and accA1down for accA1). ∗, uniquely labeled with 32P at the 5′ end. (C) Sequence of the accA2 and accA1 upstream regions, indicating the most likely transcription start point(s) for the accA1 and accA2 promoters (bent arrows). Potential −10 and −35 regions are underlined. Potential ribosomal binding sites (rbs) are in bold. The 17-nt direct repeats found upstream of the transcription start point of accA1p1 are indicated with straight arrows.
FIG. 7.
Growth phase-dependent expression of accA2 and accA1. S1 nuclease mapping of accA2 (A) and accA1 (B), using RNA isolated from a liquid-grown culture of S. coelicolor M145 harvested at different stages of growth, is shown. Exp, Trans, and Stat indicate the exponential, transition, and stationary phases of growth, respectively.
S1 nuclease protection analysis of accA1 was performed using a 563-bp PCR product uniquely labeled at the 5′ end of the downstream oligonucleotide corresponding to a sequence within accA1. Two major RNA protected fragments were identified (as well as a faint band designated accA1p3 in Fig. 6B), with the most abundant representing a putative transcriptional start site located 88 bp upstream of the GTG initiation codon of AccA1. Putative −10 and −35 regions resembling those likely to be recognized by ςhrdB are again located upstream of each of the two more prominent start sites (Fig. 6B and C). Two direct repeat sequences of 16 bp containing only two mismatches flank the putative −35 region of accA1p1 and the transcription start point of accA1p2 and might represent the binding sites of a putative transcriptional regulator (Fig. 6C). S1 nuclease protection experiments with RNA from different growth phases revealed accA1 transcripts exclusively during transition phase (Fig. 7B), showing a completely different regulation than accA2 and suggesting that the smallest RNA protected fragment detected for accA2 during transition phase most probably reflects transcription of accA1.
DISCUSSION
In Streptomyces malonyl-CoA is not only an essential metabolite used as the main elongation unit for fatty acid biosynthesis (4, 8) but also one of the most common building blocks utilized in the synthesis of several pharmaceutically important polyketide compounds (19). Therefore, the interest in establishing the pathway(s) leading to the biosynthesis of this metabolic intermediate in this microorganism has relevance not only from a fundamental view but also from a more applied point of view.
In most species, malonyl-CoA is synthesized through carboxylation of acetyl-CoA by an acetyl-CoA carboxylase (45), and this enzyme complex has been shown to be essential for many microorganisms, such as E. coli, B. subtilis, and Saccharomyces cerevisiae (17, 28, 32). Based on this knowledge and in an attempt to characterize the malonyl-CoA biosynthetic pathway in S. coelicolor we searched for a carboxyl tranferase component that could function as the β subunit of an acetyl-CoA carboxylase complex. Thus, by using pccB (36) as a hybridization probe we isolated the accBE operon of S. coelicolor. Expression of accB and accE in E. coli and subsequent in vitro reconstitution of enzyme activity in the presence of the biotinylated proteins AccA1 and AccA2 confirmed that AccB was the carboxyl transferase subunit of an ACCase. The additional presence of AccE considerably enhanced the activity of the enzyme complex (Table 2), suggesting that this small polypeptide is a functional component of the ACCase. Whether this protein plays a role as an allosteric regulator of the enzyme or as a structural component of the complex remains to be elucidated. All the actinomycete ACCases studied so far contain three functional domains located in two polypeptides (18, 20). Thus, AccE, for which there are no known homologues, might be a distinctive feature of ACCases from Streptomyces spp.
Based on these biochemical studies we decided to prove in vivo whether AccB was the carboxyl transferase component of an essential ACCase. The impossibility of obtaining an accB null mutant and the TH growth dependency of a tipAp-accB conditional mutant (Fig. 3A and 4) confirmed that AccB is essential for S. coelicolor viability. A normal growth phenotype in the absence of the inducer was restored in the conditional mutant by the addition of exogenous long-chain fatty acids in the medium (Fig. 4), indicating that the inducer-dependent phenotype was specifically related to a conditional block in fatty acid biosynthesis and that the acetyl-CoA carboxylase activity of the ACCase complex, containing AccB as the carboxyl transferase subunit, is the main pathway of malonyl-CoA biosynthesis in S. coelicolor. Although normal growth was restored by unsaturated fatty acids in liquid SMM medium, we were unable to obtain an accB mutant of T124 in the presence of oleate after several rounds of sporulation in SFM medium (41) supplemented with oleate and APR. We suggest that de novo fatty acid synthesis may be essential for an efficient sporulation of this microorganism, as was shown in B. subtilis in which fatty acid synthesis is essential to couple the activation of the mother cell transcription factors with the formation of differentiating cells (40). If this hypothesis was correct accB mutants would not be able to sporulate, even in the presence of oleate, and would be lost in the isolation procedure utilized.
Considering the essential role played by AccB and taking into account the apparent inviability of accA2 mutants in S. coelicolor (36), we postulate that AccA2 and AccB are the α and β components of an ACCase, whose main physiological role is the synthesis of malonyl-CoA. Transcriptional studies of accBE and accA2 showed that the expression of these genes occurred principally during the exponential and transition phases of growth (Fig. 5A and 6A), in agreement with their essential role in this organism. Consistent with these results the levels of acetyl-CoA carboxylase and PCCase activity throughout growth were also found to be maximal during exponential phase (data not shown).
In S. coelicolor, in addition to the need for malonyl-CoA synthesis during vegetative growth, there is also a requirement for this metabolite during transition and stationary phase. At least two of the secondary metabolites produced by S. coelicolor, undecylprodigiosin and actinorhodin, are synthesized during these growth phases and require malonyl-CoA for their synthesis. If the essential ACCase characterized in this work is the only enzyme capable of synthesizing malonyl-CoA, then it will also be required during the production of these two antibiotics. In agreement with this hypothesis fatty acid-supplemented cultures of the M94 conditional mutant, for which ACCase activity was barely detectable, were unable to produce actinorhodin or undecylprodigiosin. Based on the proposed composition of the enzyme complex and on the transcriptional studies reported here, we suggest that the low level of expression of accA2 and accBE that occurs during stationary phase provides enough of the α and β components to produce sufficient ACCase for secondary metabolism. If this assumption is correct, the biosynthesis of polyketide antibiotics in S. coelicolor should be improved by overproduction of the ACCase components during stationary phase.
While the burst of accA1 transcription during transition phase could provide a new biotinylated component for the ACCase complex during stationary phase, mutation of accA1 did not change the level of acetyl-CoA carboxylase or PCCase throughout growth. Moreover, this mutation has no deleterious effect on antibiotic production in S. coelicolor (36); consequently, the physiological role of AccA1 remains uncertain (although its location in cosmid AH10 [35] adjacent to a new putative PKS cluster might suggest a role in the synthesis of a hitherto unknown polyketide). For instance, a gene, jadJ, whose deduced amino acid sequence showed a high degree of similarity with that of AccA1 (70% identity) has been recently located in the gene cluster associated with jadomycin B biosynthesis in Streptomyces venezuelae (14). Disruption of jadJ had no effect on growth or morphology of the organism, implying that the product of this gene was not essential for fatty acid biosynthesis, but the mutant did show a reduced production of jadomycin.
Recently, a two-component acetyl-CoA carboxylase was partially characterized in Myxococcus xanthus (23). The biotinylated component of this enzyme complex, AccA, also contains the BC domain, resembling the organization of the biotinylated component of the ACCases. Interestingly, AccA of M. xanthus was not essential for the viability of this microorganism and mutants in this subunit only affected the intracellular levels of acetyl-CoA carboxylase but not of the PCCase activity, showing a sharp difference with our findings.
ACKNOWLEDGMENTS
We are grateful to R. Cabrera for cloning the 5′ end of accB and E. Takano for assistance with the S1 mapping experiments. We are also grateful to Diego de Mendoza and E. Ceccarelli for helpful discussions and useful comments on the manuscript.
This work was supported by the National Research Council of Argentina (CONICET), ANPCyT grants N:01-00078-01686 and 01-06622, the Universidad Nacional de Rosario, and the John Innes Foundation.
REFERENCES
- 1.Barker D, Campbell A. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J Mol Biol. 1981;146:89–102. doi: 10.1016/0022-2836(81)90043-7. [DOI] [PubMed] [Google Scholar]
- 2.Behal V, Jechova V, Vanek Z, Hostalek Z. Alternative pathways of malonyl-CoA formation in Streptomyces aureofaciens. Phytochemistry. 1977;16:347–350. [Google Scholar]
- 3.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli and Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 4.Bloch K, Vance D. Control mechanisms in the synthesis of saturated fatty acids. Annu Rev Biochem. 1977;46:263–298. doi: 10.1146/annurev.bi.46.070177.001403. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Bramwell H, Nimmo H G, Hunter I S, Coggins J R. Phosphoenolpyruvate carboxylase from Streptomyces coelicolor A3(2); purification of enzyme, cloning of the ppc gene and overexpression of the protein in a streptomycete. Biochem J. 1993;293:131–136. doi: 10.1042/bj2930131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bramwell H, Hunter I S, Coggins J R, Nimmo H G. Propionyl-CoA carboxylase from Streptomyces coelicolor A3(2): cloning of the gene encoding the biotin-containing sub-unit. Microbiology. 1996;142:649–655. doi: 10.1099/13500872-142-3-649. [DOI] [PubMed] [Google Scholar]
- 8.Brownsey R W, Zhande R, Boone A N. Isoforms of acetyl-CoA carboxylase: structures, regulatory properties and metabolic functions. Biochem Soc Trans. 1997;25:1232–1238. doi: 10.1042/bst0251232. [DOI] [PubMed] [Google Scholar]
- 9.Buttner M J, Chater K F, Bibb M J. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 11.Donadio S, Staver M, Katz L. Erythromycin production in Saccharopolyspora erythraea does not require a functional propionyl-CoA carboxylase. Mol Microbiol. 1996;19:977–984. doi: 10.1046/j.1365-2958.1996.439969.x. [DOI] [PubMed] [Google Scholar]
- 12.Erfle J D. Acetyl-CoA and propionyl-CoA carboxylation by Mycobacterium phlei. Biochim Biophys Acta. 1973;316:143–155. doi: 10.1016/0005-2760(73)90004-0. [DOI] [PubMed] [Google Scholar]
- 13.Gramajo H C, Takano E, Bibb M J. Studies of the growth-phase dependent expression of genes for actinorhodin production in Streptomyces coelicolor. Mol Microbiol. 1993;7:837–845. doi: 10.1111/j.1365-2958.1993.tb01174.x. [DOI] [PubMed] [Google Scholar]
- 14.Han L, Yang K, Kulowski K, Wendt-Pienkowski E, Hutchinson C R, Vining L C. An acyl-coenzyme A carboxylase encoding gene associated with jadomycin biosynthesis in Streptomyces venezuelae ISP5230. Microbiology. 1999;146:903–910. doi: 10.1099/00221287-146-4-903. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D. Studies of transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 16.Harwood J L. Fatty acid metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:101–138. [Google Scholar]
- 17.Hasselacher M, Ivessa A S, Paltauf F, Kohlwein S D. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 18.Henrikson K P, Allen S H G. Purification and sub-unit structure of propionyl coenzyme A carboxylase of Mycobacterium smegmatis. J Biol Chem. 1979;254:5888–5891. [PubMed] [Google Scholar]
- 19.Hopwood D A, Sherman D H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- 20.Huanaiti A R, Kolattukudy P E. Isolation and characterization of an acyl-coenzyme A carboxylase from an erythromycin-producing Streptomyces erythraeus. Arch Biochem Biophys. 1982;216:362–371. doi: 10.1016/0003-9861(82)90222-3. [DOI] [PubMed] [Google Scholar]
- 21.Janssen G R, Bibb M J. Derivatives of pUC18 that have bglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 22.Kieser T, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical Streptomyces genetics. Norwich, United Kingdom: The John Innes Foundation; 2000. [Google Scholar]
- 23.Kimura Y, Miyake R, Tokumasu Y, Sato M. Molecular cloning and characterization of two genes for the biotin carboxylase and carboxyltransferase sub-units of acetyl-coenzyme A carboxylase in Myxococcus xanthus. J Bacteriol. 2000;182:5462–5469. doi: 10.1128/jb.182.19.5462-5469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King M T, Reiss P D. Separation and measurement of short-chain coenzyme-A compounds in rat liver by reverse-phase high-performance liquid chromatography. Anal Biochem. 1985;146:173–179. doi: 10.1016/0003-2697(85)90412-9. [DOI] [PubMed] [Google Scholar]
- 25.Laakel M, Lebrihi A, Khaoua S, Schneider F, Lefebvre G, Germain P. A link between primary and secondary metabolism: malonyl-CoA formation in Streptomyces ambofaciens growing on ammonium ions or valine. Microbiology. 1994;140:1451–1456. [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Li S J, Cronan J E., Jr The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992;267:855–863. [PubMed] [Google Scholar]
- 28.Li S J, Cronan J E., Jr Growth-rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step in lipid biosynthesis. J Bacteriol. 1993;175:332–340. doi: 10.1128/jb.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacNiel D J, Occi J L, Gewain K M, MacNeil T, Gibbons P H, Rudy C L, Danis S J. Complex organisation of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene. 1992;115:119–125. doi: 10.1016/0378-1119(92)90549-5. [DOI] [PubMed] [Google Scholar]
- 30.Murakami T, Holt T, Thompson C J. Thiostrepton-induced gene expression in Streptomyces lividans. J Bacteriol. 1989;171:1459–1466. doi: 10.1128/jb.171.3.1459-1466.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paget M S, Leibovitz E, Buttner M J. A putative two-component signal transduction system regulates sigmaE, a sigma factor required for normal cell wall integrity in Streptomyces coelicolor A3(2) Mol Microbiol. 1999;3:97–107. doi: 10.1046/j.1365-2958.1999.01452.x. [DOI] [PubMed] [Google Scholar]
- 32.Perez C A, Marini P, de Mendoza D. Effects on Bacillus subtilis of conditional expression of the accBC operon encoding sub-units of acetyl coenzyme A carboxylase, the first enzyme of fatty acid synthesis. Microbiology. 1998;144:895–903. doi: 10.1099/00221287-144-4-895. [DOI] [PubMed] [Google Scholar]
- 33.Pizer E S, Jackisoh C, Wood F D, Paternack G R, Davidson N E, Kuhajda F P. Inhibition of fatty acid synthesis induces programmed cell death in human breast cancer cells. Cancer Res. 1996;56:2745–2747. [PubMed] [Google Scholar]
- 34.Polakis S, Guchhait R, Lane M. On the possible involvement of a carbonyl phosphate group intermediate in the adenosine triphosphate-dependent carboxylation of biotin. J Biol Chem. 1972;247:1335–1337. [PubMed] [Google Scholar]
- 35.Redenbach M, Kieser M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:77–95. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez E, Gramajo H. Genetic and biochemical characterization of the α and β components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2) Microbiology. 1999;145:3109–3119. doi: 10.1099/00221287-145-11-3109. [DOI] [PubMed] [Google Scholar]
- 37.Saggerson E D, Ghadiminejad I, Awan M. Regulation of mitochondrial carnitine palmitoyl transferases from liver and extrahepatic tissues. Adv Enzyme Regul. 1992;32:285–306. doi: 10.1016/0065-2571(92)90023-s. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shujman G, Grau R, Gramajo H, Ornella L, de Mendoza D. De novo fatty acid synthesis is required for establishment of cell type specific gene transcription during sporulation in B. subtilis. Mol Microbiol. 1998;29:1215–1224. doi: 10.1046/j.1365-2958.1998.01004.x. [DOI] [PubMed] [Google Scholar]
- 41.Strauch E, Takano E, Baylis H A, Bibb M J. The stringent response in Streptomyces coelicolor A(3)2. Mol Microbiol. 1991;5:289–298. doi: 10.1111/j.1365-2958.1991.tb02109.x. [DOI] [PubMed] [Google Scholar]
- 42.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Studier F W, Moffatt B A. Use of the bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 44.Sun J, Kelemen G H, Fernandez-Abalos M, Bibb M J. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2221–2227. doi: 10.1099/00221287-145-9-2221. [DOI] [PubMed] [Google Scholar]
- 45.Toh H, Kondo H, Tanabe T. Molecular evolution of biotin-dependent carboxylases. Eur J Biochem. 1993;215:687–696. doi: 10.1111/j.1432-1033.1993.tb18080.x. [DOI] [PubMed] [Google Scholar]
- 46.Westrich L, Domann S, Faust B, Bedford D, Hopwood D A, Bechthold A. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol Lett. 1999;170:381–387. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]