Abstract
The diversity of methanotrophic bacteria associated with roots of submerged rice plants was assessed using cultivation-independent techniques. The research focused mainly on the retrieval of pmoA, which encodes the α subunit of the particulate methane monooxygenase. A novel methanotroph-specific community-profiling method was established using the terminal restriction fragment length polymorphism (T-RFLP) technique. The T-RFLP profiles clearly revealed a more complex root-associated methanotrophic community than did banding patterns obtained by pmoA-based denaturing gradient gel electrophoresis. The comparison of pmoA-based T-RFLP profiles obtained from rice roots and bulk soil of flooded rice microcosms suggested that there was a substantially higher abundance of type I methanotrophs on rice roots than in the bulk soil. These were affiliated to the genera Methylomonas, Methylobacter, Methylococcus, and to a novel type I methanotroph sublineage. By contrast, type II methanotrophs of the Methylocystis-Methylosinus group could be detected with high relative signal intensity in both soil and root compartments. Phylogenetic treeing analyses and a set of substrate-diagnostic amino acid residues provided evidence that a novel pmoA lineage was detected. This branched distinctly from all currently known methanotrophs. To examine whether the retrieval of pmoA provided a complete view of root-associated methanotroph diversity, we also assessed the diversity detectable by recovery of genes coding for subunits of soluble methane monooxygenase (mmoX) and methanol dehydrogenase (mxaF). In addition, both 16S rRNA and 16S ribosomal DNA (rDNA) were retrieved using a PCR primer set specific to type I methanotrophs. The overall methanotroph diversity detected by recovery of mmoX, mxaF, and 16S rRNA and 16S rDNA corresponded well to the diversity detectable by retrieval of pmoA.
The atmospheric trace gas methane (CH4) is a prominent “greenhouse” gas. Its atmospheric concentration has been increasing until recently at a rate of about 1% a year (8). Up to 70 to 80% of atmospheric CH4 is biogenic (55). Flooded rice fields are one of the major sources of biogenic CH4 (34, 50). Estimations of the annual emission rate from flooded rice fields range between 60 and 110 Tg (8, 21, 45). The upper limit of this emission rate accounts for approximately 25% of the total annual CH4 emission into the atmosphere (8, 21).
Approximately 90% of the CH4 that is emitted from rice paddies escapes through the aerenchyma of the rice plants, whereas only 10% escapes through the floodwater (19, 52). However, the aerenchyma does not merely function as a gas transport system but rather constitutes a dynamic, oxygenated biofilter. The diffusive input of oxygen into the below-ground plant surface area enables aerobic methanotrophs to oxidize CH4. Gilbert and Frenzel (22) showed that the activities of methanotrophs were directly dependent on the oxygen availability in the rice root environment. It was shown that up to 30% of the CH4 produced in rice paddy soil is oxidized by root-associated methanotrophs (5, 9, 15).
Based on phylogenetic, physiological, morphological, and biochemical characteristics, methanotrophs are divided into two major subgroups (27). The γ-proteobacterial type I methanotroph group comprises the genera Methylomonas, Methylocaldum, Methylomicrobium, Methylobacter, Methylosarcina, Methylosphaera, and Methylococcus (also classified as type X) (4, 6, 27, 58), while the α-proteobacterial type II methanotroph group consists of the genera Methylocystis and Methylosinus (27) and one more distant species, Methylocella palustris (14).
The methanotrophic diversity in rice field soil has been assessed in detail (28, 30), but knowledge about the diversity of methanotrophic populations associated with rice roots is still limited. Type II strains were isolated from the terminal positive-dilution steps of a most-probable-number dilution series (23). However, whether these results reflect the natural situation on rice roots, i.e., predominance of type II methanotrophs, or instead were the consequence of cultivation bias, is unclear.
We assessed the methanotrophic diversity associated with roots of submerged rice plants using various cultivation-independent techniques. This assessment was carried out in relation to the methanotrophic diversity detectable in rice paddy bulk soil. Despite the phylogenetic distance between type I and type II methanotrophs, almost all known methanotrophs possess a pmoA gene, which encodes the α subunit (PmoA) of the particulate methane monooxygenase (pMMO). The only exception is Methylocella palustris (13, 14). Consequently, this study focused mainly on the retrieval of pmoA using PCR primers described previously (32). These primers also target amoA, which encodes the α subunit (AmoA) of the ammonia monooxygenase in autotrophic ammonia oxidizers. Based on the pmoA sequence database created in this study, we established a novel methanotroph-specific community-profiling method using the terminal restriction fragment length polymorphism (T-RFLP) technique (37, 39). The methanotroph diversity detectable by pmoA-based T-RFLP profiling was compared with those detectable by comparative sequence analysis of cloned pmoA and by pmoA-based denaturing gradient gel electrophoresis (DGGE).
To examine the meaningfulness of the pmoA-based results, we also assessed the methanotrophic diversity detectable by retrieval of mmoX (25) and mxaF (42). The mmoX gene encodes the α subunit (MmoX) of the hydroxylase component of the soluble methane monooxygenase (sMMO). This monooxygenase is present in most type II methanotrophs, in members of the genus Methylococcus, and in some Methylomonas strains (53) but not in most of the other type I methanotrophs (27). The mxaF gene codes for the α subunit of the methanol dehydrogenase, which is present in all methylotrophs. In addition, both 16S ribosomal DNA (rDNA) and 16S rRNA were retrieved using PCR primers specific to type I methanotrophs (57).
MATERIALS AND METHODS
Rice microcosms.
Rice (Oryza sativa var. Roma, type japonica) was grown in three flooded, unfertilized microcosms for 70, 84, or 90 days using conditions described previously (20, 26).
Rice root samples obtained from the three microcosms were used as source material for the molecular analyses. This material, which we will refer to as root samples M70, M84, and M90, was washed by careful shaking in phosphate-buffered saline (7 mM Na2HPO4, 3 mM NaH2PO4, 130 mM NaCl [pH 7.2]) to remove adhering soil particles. Cores were obtained from the bulk soil between the plants by pressing a plastic corer into the soil to a depth of about 15 cm. Because of the dense surface root mat, only the lower 10-cm portions of the cores were used. The three microcosms were cultivated using soil sampled in 1995 (M90), 1997 (M84), and 1999 (M70) from drained rice fields of the Italian Rice Research Institute in Vercelli, Italy.
Extraction of total DNA.
Total DNA from the bulk soil of the flooded rice microcosms was extracted and purified using a protocol reported previously (38). In this protocol, microbial cells were lysed directly in the soil matrix by bead beating.
For extraction of total DNA from rice roots, the samples M84 and M90 were lyophilized, and the dried root material was subsequently pulverized with a mortar under liquid N2. The pulverized root material was resuspended in 1 ml of extraction buffer. Enzymatic lysis of microbial cells, as well as isolation and purification of total DNA, was performed using an extraction protocol described previously (26).
Simultaneous extraction of total DNA and RNA.
Rice roots of sample M70 were placed in a 2-ml reaction tube together with 1 g of glass beads (diameter, 0.17 to 0.18 mm) and 700 μl of precooled TPM buffer (50 mM Tris-HCl [pH 7.5], 1.7% [wt/vol] polyvinylpyrrolidone, 10 mM MgCl2) (18). This suspension was shaken for 60 s at maximum speed in a bead beater (Dismembrator-S; B. Braun Biotech GmbH, Melsungen, Germany). Glass beads, root particles, and cell debris were pelleted by centrifugation for 5 min at 4°C, and the supernatant was transferred to a new reaction tube. Seven hundred microliters of a phenol-based lysis buffer was added to the pellet, and the bead-beating procedure was repeated. After centrifugation, both supernatants were pooled and extracted three times with cold phenol-chloroform (1:1, vol/vol) followed by precipitation of total nucleic acids with 0.1 volume of sodium acetate (3 M; pH 5.2) and 2.5 volumes of ethanol. The pellet was dried and suspended in 100 μl of Tris-EDTA buffer. For subsequent DNA-based analyses a 50-μl aliquot was stored at −20°C. For preparation of total RNA, the other 50-μl aliquot was mixed with 1 volume of TMC buffer (10 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 0.1 mM CsCl2) (18) and 5 U of RNase-free DNase (Promega, Madison, Wis.) and incubated for 1 h at 37°C to remove the DNA. The reaction was stopped by extraction with 1 volume of chloroform. Precipitation and resuspension of total RNA were performed as described above.
PCR amplification.
The primer sets used in this study are listed in Table 1. For pmoA-based T-RFLP analysis, the 5′ primer A189 was labeled with the dye carboxyfluorescein. The reaction mixture contained 1 to 5 ng of DNA, 50 μl of MasterAmp PCR premix F (Epicentre Technologies, Madison, Wis.), 0.3 μM concentrations of each primer (MWG-Biotech, Ebersberg, Germany), and 2.5 U of Taq DNA polymerase (AmpliTaq; PE Applied Biosystems, Foster City, Calif.). Amplification was performed in a total volume of 100 μl in 0.2-ml reaction tubes, using a DNA thermal cycler (model 2400; PE Applied Biosystems). The thermal PCR profile was as follows: initial denaturation for 2 min at 94°C and 30 cycles consisting of denaturation at 94°C for 45 s, primer annealing for 60 s (annealing temperature specific to each target gene [Table 1]), and elongation at 72°C for 120 s. The final elongation step was 6 min. Aliquots of the amplicons (10 μl) were checked by electrophoresis on a 1% agarose gel.
TABLE 1.
PCR primers used in this study
| Target gene (fragment length in bp) | Primer set | Sequence (5′ to 3′)a | Annealing temperature (°C) | Reference |
|---|---|---|---|---|
| pmoA (531)b | A189c | GGNGACTGGGACTTCTGG | 62 to 52 touchdownd | 32 |
| A682 | GAASGCNGAGAAGAASGC | |||
| mmoX (863) | 534fe | CCGCTGTGGAAGGGCATGAA | 62 to 52 touchdownd | 25 |
| 1393re | CACTCGTAGCGCTCCGGCTC | |||
| mxaF (557) | f1003 | GCGGCACCAACTGGGGCTGGT | 55 | 42 |
| r1561 | GGGCAGCATGAAGGGCTCCC | |||
| 16S rDNA (922) and 16S rcDNA (556) | MethT1dF | CCTTCGGGMGCYGACGAGT | 56 | 57 |
| MethT1bR | GATTCYMTGSATGTCAAGG | |||
| MethT1cRf | ATCCAATCGAGTTCCCAGGTTAAGCCC |
N, bases A, C, T, or G; M, bases A or C; S, bases G or C; Y, bases C or T.
The pmoA-targeted primers also detect amoA, which encodes the α subunit of ammonia monooxygenase in autotrophic ammonia oxidizers.
GC-clamp for DGGE analysis: 5′-cccccccccccccgccccccgccccccgcccccgccgccc (28).
PCR profile according to Henckel et al. (28).
The primers target the nucleotide sequence positions 534 to 553 (534f) and 1374 to 1393 (1393r), according to the open reading frame of the mmoX sequence published by Cardy et al. (7). The primer 1393r targets almost the same stretch as primer r1403 of McDonald et al. (40).
MethT1cR was used in RT-PCR.
RT-PCR of 16S rRNA.
Ribosomal copy DNA (rcDNA) of type I methanotrophs was synthesized from total RNA using primer MethT1cR (Table 1), Moloney murine leukemia virus reverse transcriptase (RT) (RNase H minus; Promega, Mannheim, Germany), and a previously described protocol (38). PCR of 16S rcDNA was carried out as described above.
pmoA-based T-RFLP analysis.
T-RFLP analysis was performed for each total DNA extract in triplicate using a protocol reported previously (38, 39). The pmoA was amplified by PCR as described above. After purification with Qiaquick spin columns (Qiagen, Hilden, Germany), approximately 100 ng of the amplicons were digested with 10 U of the restriction endonuclease MspI (Promega). The digestions were carried out in a total volume of 10 μl for 3 h at 37°C. Aliquots (2.5 μl) of the digested amplicons were mixed with 2.0 μl of formamide and 0.5 μl of an internal lane standard (GeneScan-1000 ROX; PE Applied Biosystems). The mixtures were denatured at 100°C for 3 min and then chilled on ice. Electrophoresis on a polyacrylamide gel (6%) was performed using an automated DNA sequencer (model 373; PE Applied Biosystems) for 6 h at the following settings: 2,500 V, 40 mA, and 27 W (24-cm gel length). After electrophoresis, the sizes of the 5′-terminal restriction fragments (T-RFs) and the intensities of their fluorescence emission signals (i.e., signal intensities) were automatically calculated by the GeneScan Analysis software, version 2.1 (PE Applied Biosystems). The accuracy of size estimation between replicates was ±1 bp. The relative signal intensity of each T-RF was calculated based on the signal intensity of the individual T-RF in relation to the total signal intensity of all T-RFs (including the 531-bp fragment) detected in the respective T-RFLP community profile. The 531-bp fragment corresponds to pmoA amplicons without any restriction site for MspI.
pmoA-based DGGE.
PCR amplification of pmoA and DGGE in the Dcode System (Bio-Rad, Munich, Germany) were carried out as described by Henckel et al. (28). In brief, PCR products were separated in 1-mm-thick polyacrylamide gels (6.5% [wt/vol] acrylamide-bisacrylamide [37.5:1]) using a linear denaturing gradient that ranged from 35 to 80%. A denaturing gradient of 80% corresponded to 6.5% acrylamide, 5.6 M urea, and 32% deionized formamide. The electrophoresis was performed in 0.5× TAE buffer (0.04 M Tris-base, 0.02 M sodium acetate, 1 mM EDTA [pH 7.4]) for 15 h at a constant voltage of 74 V at 60°C. Gels were stained with 1:10,000 (vol/vol) SYBR-Green I (Biozym, Hessisch-Oldendorf, Germany) for 45 min and scanned with a Storm 860 Phosphorimager (Molecular Dynamics, Sunnyvale, Calif.).
Cloning and sequencing.
PCR products of pmoA, mmoX, mxaF, 16S rDNA, and 16S rcDNA were cloned using the TOPO TA cloning kit (Invitrogen Corp., San Diego, Calif.) as recommended by the manufacturer. The preparation of plasmid DNA of randomly selected clones, PCR amplification of cloned inserts, and nonradioactive sequencing were carried out as described previously (48). In addition, oligonucleotide primers targeting internal regions of the cloned inserts were used for sequencing of mmoX and 16S rDNA and rcDNA.
Phylogenetic analysis.
Based on sequence information deposited either in public-domain databases or generated in the course of this study, we established sequence databases for pmoA, mxaF, and mmoX. Each of these sequence databases was integrated into the ARB program package (developed by O. Strunk and W. Ludwig; Technische Universität München [http://www.arb-home.de]) and was manually put into an aligned format. The 16S rDNA and rcDNA clone sequences were added to a database of about 14,000 complete or partial bacterial 16S rRNA sequences. Evolutionary distances (ARB and PHYLIP [17]) between pairs of inferred amino acid sequences (pmoA, mmoX, and mxaF) were calculated using various models (11, 36). Evolutionary-distance values between pairs of 16S rDNA and rcDNA clone sequences were calculated by applying the Jukes-Cantor correction (35). The trees were constructed using the neighbor-joining method (49). The statistical significance levels of interior nodes were determined by performing bootstrap analyses by the neighbor-joining method (500 data resamplings). To exclude obvious chimeric primary structures from the pmoA, mmoX, mxaF, and 16S rDNA and rRNA sequence databases, separate treeing analyses of the 5′ and 3′ halves of the respective sequence data sets were carried out.
Nucleotide sequence accession numbers.
The environmental pmoA (plus amoA and sequence types of uncertain affiliation), mmoX, mxaF, 16S rDNA, and 16S rcDNA clone sequences recovered in this study from rice roots of flooded rice microcosms have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. AJ299946 to AJ299968, AJ299515 to AJ29953, AJ299504 to AJ299514, AJ299969 to AJ299983, and AJ299984 to AJ299989, respectively.
RESULTS
The use of PCR primer sets with intended-target specificity for partial stretches of pmoA, mmoX, mxaF, and 16S rDNA resulted in amplicons of the predicted sizes (Table 1). Clone libraries were generated from the PCR products, and subsequently individual clones were randomly selected for comparative sequence analysis. In addition, methanotroph diversity was assessed by pmoA-based T-RFLP analysis.
pmoA-based cloning approach.
One clone library (each) was generated from samples M84 and M90. In total, 47 pmoA clones were randomly selected for comparative sequence analysis (28 and 19 clones from samples M84 and M90, respectively).
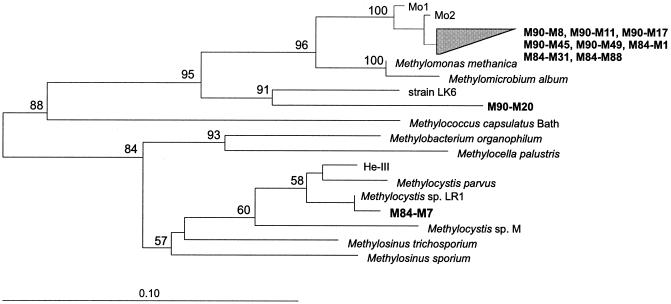
Thirty-six clones formed three distinct clusters within the phylogenetic radiation of type I methanotrophs (Fig. 1A). Cluster I (16 clones) was affiliated with Methylomonas methanica. Cluster II (10 clones) formed a separate lineage without any clear affiliation to any of the known genera. Cluster III (10 clones) exhibited a moderate relationship to Methylococcus capsulatus.
FIG. 1.
(A) Distance dendrogram constructed for partial pmoA and amoA gene sequences based on 165 derived amino acid sites in relation to pmoA-based T-RFLP (B) and DGGE (C) community patterns. The two patterns and most of the pmoA clone sequences were obtained from sample M84. (A) The dendrogram shows environmental pmoA and amoA (plus other putative monooxygenase) sequences retrieved from roots of submerged rice plants (M84, M90) in relation to pmoA of cultured type I and type II methanotrophs, environmental pmoA clone sequences, and amoA sequences of the β-proteobacterial Nirosomonas and Nitrosospira group. The environmental pmoA sequences used for reference were retrieved from various habitats as follows: beech forest in Denmark (RA14 [AF148521], RA21 [AF148522], Rold1 [AF148523], Rold4 [AF148526], Rold5 [AF148527]), rain forest in Brazil (Pantanal13 [AF148525]), mixed hardwood forest in the United States (Maine6 [AF148528], Maine9 [AF148531]) (33), deciduous forest soil near Marburg, Germany (MR2 [AF200726], MR16 [AF 200729] (29), MR1 [AF200729]) (29), rice soil incubations (He-I [AF126908], He-II [AF126909], He-III [AF126910], He-IV [AF126913], He-VI [AF126911]) (28), and blanket peat bog (PE9 [AF006050], PD2 [AF006047]) (41). The numbers I, II, and III refer to three distinct pmoA sequence clusters of type I methanotrophs, which have been retrieved from rice roots. The numbers at the nodes indicate the percentage of recovery in 500 bootstrap resamplings. Only bootstrap values ≥50 are shown. Scale bar, 0.1 substitution per amino acid site. Database accession numbers of reference organisms are as follows: Methylocystis sp. strain M, U81596; Methylocystis parvus, U31651; Methylosinus trichosporium, U31550; Methylobacter sp. strain BB5.1, AF016982; Methylomicrobium album, U31654; Methylomicrobium pelagicum, U31652; Methylomonas methanica, U31653; Methylocaldum gracile, U89301; Methylocaldum szegediense, U89303; Methylocaldum tepidum, U89304; Methylococcus capsulatus, L40804; strain HB, U89302; Nitrosospira multiformis, U89833; and Nitrosomonas europaea, AF037107. (B) pmoA-based T-RFLP profile. The x axis shows the lengths (in base pairs) of the T-RFs, and the y axis shows the intensities of the fragments in arbitrary units. The numbers in boxes indicate the sizes of T-RFs which could be assigned to phylogenetically defined methanotroph populations or to autotrophic ammonia oxidizers (see arrows). (C) pmoA-based DGGE pattern. For comparative sequence analysis, predominant DGGE bands were excised, reamplified, and reanalyzed by DGGE to verify band purity. Affiliation of these bands to distinct pmoA clusters is indicated (compare with Fig. 1A).
Only two pmoA clones could be assigned to the Methylocystis-Methylosinus group (type II methanotrophs). Four sequence types grouped with ammonia oxidizers of the β-proteobacterial Nitrosomonas-Nitrosospira group. Five sequences formed the lineages A, B, and C (Fig. 1A), which branched distinctly from all currently known methanotrophs or autotrophic ammonia oxidizers.
Separate treeing analysis of the 5′ and 3′ halves of the respective sequence types suggested that the clone sequences assigned to the lineages A, B, and C were of natural origin, i.e., separate treeing analysis did not provide evidence that any of these clones were chimeric. We therefore determined amino acid signature residues for the inferred peptide sequences of the lineages A, B, and C (Table 2) (33). These are either universal to PmoA (methanotrophs) and AmoA (autotrophic ammonia oxidizers) or specific for either PmoA or AmoA (substrate-diagnostic residues). Based on this approach, Holmes et al. (33) assigned a newly detected cluster of sequence types (forest clones) (Table 2) to an uncharacterized group of methanotrophs. The percentage distribution of substrate-diagnostic residues suggests that lineage A corresponds to a novel pmoA cluster rather than to an amoA cluster. By contrast, due to the relatively low number of conserved substrate-diagnostic residues, sequence types of the lineages B and C could not be assigned to either PmoA or AmoA.
TABLE 2.
Signature residues of predicted amino acid sequencesa
| Sequence typesb | No. of signature residues (%)
|
||
|---|---|---|---|
| MMO (n = 21) | AMO (n = 21) | Universalc (n = 52) | |
| Lineage A | 17 (81) | 1 (5) | 49 (94) |
| Lineage B | 7 (33) | 4 (19) | 43 (83) |
| Lineage C | 9 (43) | 6 (29) | 40 (77) |
| Forest clones | 16 (76) | 2 (10) | 49 (94) |
Amino acids were considered to be putative substrate-diagnostic signature residues only if they fulfilled the following two criteria: (i) residues were conserved in all currently known pmoA sequences of type I and type II methanotrophs (putative MMO signature residues), and (ii) all amoA sequence types also exhibited at the same alignment positions a conserved amino acid residue but different from that of the pmoA sequence types (putative AMO signature residues). Twenty-one of 165 amino acid positions met these two criteria. MMO, methane monooxygenase; AMO, ammonia monooxygenase.
Lineage A, clone M84-P3; lineage B, clones M84-P105 and M90-P69; lineage C, clones M84-P22 and M84-P36. Forest clones represent a distinct cluster of environmental pmoA sequences characterized by the clones RA14, Pantanal13, Rold1, Rold5, and Maine6 (Fig. 1A). The values for the respective signature residues were taken from Holmes et al. (33).
Residues which are universally conserved in both pmoA and amoA sequence types.
pmoA-based T-RFLP analysis.
Based on our pmoA sequence database derived from cultured methanotrophs and environmental samples, we predicted that the tetrameric restriction enzyme MspI would be the restriction enzyme most appropriate to analyze the genetic diversity of methanotrophic communities in a single electrophoretic profile. Defined mixtures of genomic DNA from cultured methanotrophs subjected to MspI-based T-RFLP analysis exactly produced those T-RFs predicted based on our pmoA sequence database (data not shown). Consequently, MspI was used for pmoA-based T-RFLP analysis of methanotrophic communities. Extraction of total DNA from the same root sample in triplicate followed by PCR amplification of pmoA and T-RFLP analysis produced highly similar T-RFLP community profiles. The coefficients of variation of the relative signal intensities between these profiles were between 5.4 and 12.3% for the major peaks, i.e., those with sizes of 80, 245, 350, 440, 505, and 531 bp (Fig. 1B; see also Fig. 2). The coefficients of variation for the major peaks between T-RFLP community profiles generated in triplicate from individual DNA extracts ranged from 1.7 to 7.6%. This analysis showed that the T-RFLP technique was reliable for a rapid PCR-based fingerprinting of methanotrophic communities. High reproducibility of the T-RFLP technique has been reported previously (e.g., for 16S rDNA-based T-RFLP analysis [39, 44]).
FIG. 2.
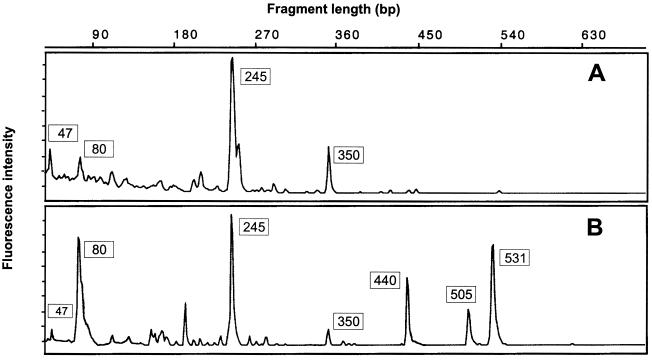
Comparison of pmoA-based T-RFLP profiles obtained from bulk soil (A) and rice roots (B) of the flooded rice microcosm M84. See Fig. 1 for assignment of the major T-RFs to defined methanotrophic populations and to autotrophic ammonia oxidizers.
The comparison of T-RFLP community profiles obtained from rice root sample M84 with T-RFs of individual pmoA and amoA sequence types allowed the differentiation among type I methanotrophs, type II methanotrophs, and autotrophic ammonia oxidizers (Fig. 1). Type I methanotrophs were further differentiated into five distinct T-RFs, which correspond to phylogenetically distinct sublineages affiliated with the following: (i) pmoA cluster III plus members of the genera Methylococcus and Methylocaldum (80-bp T-RF); (ii) Methylomicrobium album (350-bp T-RF); (iii) pmoA cluster I plus Methylomonas methanica (440-bp T-RF); (iv) Methylobacter sp. strain BB5.1 (505-bp T-RF); and (v) pmoA cluster II (531-bp peak; no recognition site for MspI). The type II methanotrophs of the Methylocystis-Methylosinus group were characterized by the 245-bp T-RF, while the 47-bp T-RF was indicative of amoA (Nitrosomonas-Nitrosospira group). Three further minor peaks could be assigned to the lineages A (280-bp T-RF), B (228-bp T-RF), and C (113-bp T-RF). Several peaks of mainly lower relative signal intensity could be assigned to none of the pmoA or amoA sequence types retrieved from rice roots or deposited in public-domain databases. These T-RFs may correspond to novel pmoA or amoA sequence types.
Root-associated methanotroph diversity detectable by T-RFLP analysis was compared to that detectable by DGGE using the same extract of total DNA as the starting material. DGGE revealed the presence of only pmoA sequence types affiliated with type II methanotrophs and pmoA cluster III (Fig. 1C).
The community profiles obtained by pmoA-based T-RFLP analysis from root samples M70, M84, and M90 showed similar relative abundances of the major T-RFs, i.e., of T-RFs with sizes of 80, 245, 350, 440, 505, and 531 bp. The T-RFLP profiles generated from the anoxic bulk soil were characterized mainly by the 245-bp T-RF of type II methanotrophs, but minor peaks indicative of type I methanotrophs (80- and 350-bp T-RFs) and ammonia oxidizers (47-bp T-RF) were also present. The comparison of T-RFLP profiles obtained from bulk soil versus rice roots is shown for rice microcosm M84 (Fig. 2).
mmoX.
Five of 15 clones analyzed were closely related to the mmoX of Methylocystis sp. strain LR1 (Fig. 3). This type II methanotroph was isolated in Canada (16). Three mmoX clones were assigned to Methylocystis sp. strain M. The treeing analysis suggested that clone M84-S38 was affiliated to the mmoX of the acidophile Methylocella palustris (13, 14). However, the dissimilarity values of the predicted peptide sequence of clone M84-S38 with MmoX of Methylocella palustris (15.7%), the Methylocystis-Methylosinus group (17.1 to 18.5%), and Methylococcus capsulatus Bath (17.5%) were all similar. The MmoX sequence types of the phylogenetically distinct type I methanotrophs Methylomonas sp. strain KSWIII and Methylococcus capsulatus Bath differ by 11.3% (53). Taking into account these dissimilarity values, it can only be speculated that clone M84-S38 corresponds to a novel methanotrophic bacterium which harbors sMMO. The remaining six clones were false positives containing non-mmoX sequence types.
FIG. 3.
Distance dendrogram constructed for partial mmoX sequences based on 286 derived amino acid sites. The dendrogram shows environmental mmoX sequences retrieved from roots of submerged rice plants (M84, M90) in relation to mmoX sequences of representative type I and type II methanotrophs. The environmental mmoX sequence designated peat bog clone 24 (AF004555) was retrieved from an acidic Sphagnum peat bog (12). The numbers at the nodes indicate the percentage of recovery in 500 bootstrap resamplings. Only bootstrap values ≥50 are shown. Scale bar, 0.1 substitution per amino acid site. Database accession numbers of reference organisms are as follows: Methylocystis sp. strain LR1, Y18440; Methylocystis sp. strain M, U81594; Methylosinus trichosporium, X55394; Methylocella palustris, AF004554; Methylococcus capsulatus, M90050; and Methylomonas sp. strain KSWIII, AB025022.
mxaF.
Twenty-four of 50 clones analyzed formed a coherent mxaF sequence cluster related to Methylomonas methanica and Methylomicrobium album. One sequence type each could be assigned to Methylocystis sp. strain LR1 (16), strain LK6 (Fig. 4), and Hyphomicrobium sp. strain CM2 (data not shown). The remaining 23 clones were false positives containing non-mxaF sequence types.
FIG. 4.
Distance dendrogram constructed for partial mxaF sequences based on 172 derived amino acid sites. The dendrogram shows environmental mxaF sequences retrieved from roots of submerged rice plants (M84, M90) in relation to mxaF sequences of representative type I and type II methanotrophs, and Methylobacterium organophilum. The mxaF sequence He-III [AF126296] was retrieved from rice soil incubations (28), while Mo1 [AF283243] and Mo2 [AF283244] were detected in a methanotrophic consortium enriched with a high CH4/low O2 mixing ratio (31). The numbers at the nodes indicate the percentage of recovery in 500 bootstrap resamplings. Only bootstrap values ≥50 are shown. Scale bar, 0.1 substitution per amino acid site. Database accession numbers of reference organisms are as follows: Methylocystis sp. strain LR1, Y18441; Methylocystis sp. strain M, U70517; Methylocystis parvus, U70515; Methylosinus sporium, U70514; Methylosinus trichosporium, U70516; Methylocella palustris, AJ27831; Methylomicrobium album, U70513; Methylomonas methanica, U70512; Methylococcus capsulatus, U70511; strain LK6, U86503; and Methylobacterium organophilum, M22629.
16S rDNA and 16S rRNA analysis of type I methanotrophs.
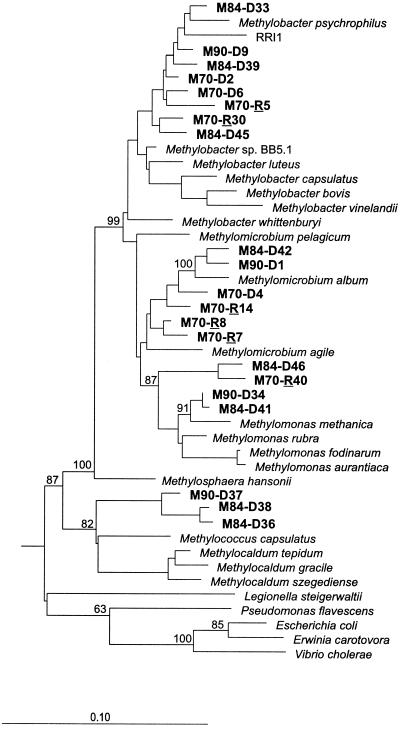
16S rDNA clone libraries were generated from samples M70, M84, and M90. An rcDNA clone library was created only from freshly prepared roots of sample M70. Because RT-PCR was unsuccessful with the primer set MethT1dF and MethT1bR, we replaced MethT1bR with primer MethT1cR (Table 1). The use of MethT1cR resulted in an amplicon of the predicted size (556 bp). The retrieval of both 16S rDNA and 16S rRNA led to the identification of a diverse community of type I methanotrophs (Fig. 5). No false-positive clone sequences were detected in a set of 23 16S rDNA and 6 16S rcDNA clones randomly selected for analysis. This underlines the target specificity of these PCR primers for type I methanotrophs (57).
FIG. 5.
Distance dendrogram showing the 16S rDNA clone sequences retrieved from roots of submerged rice plants (samples M70, M84, and M90) in relation to type I methanotrophs and nonmethanotrophic members of γ-Proteobacteria. The environmental 16S rDNA sequences encompass the clones M70-D2 to M90-D34. Due to limited sequence length (556 bp), the 16S rcDNA clones M70-R5 to M70-R40 (R = 16S ribosomal copy DNA recovered from total RNA of sample M70) have been inserted into the distance dendrogram using parsimony methods. RRI1 (AF179603) was retrieved from rhizosphere soil of a flooded rice microcosm (3). The 16S rDNA sequences of α-proteobacterial type II methanotrophs were used to root the tree. The numbers at the nodes indicate the percentage of recovery in 500 bootstrap resamplings. Only bootstrap values ≥50 are shown. Scale bar, 0.1 substitution per nucleotide sequence position. Database accession numbers of reference organisms are as follows: Methylocystis sp. strain M, U81595; Methylocystis parvus, Y18945; Methylobacter sp. strain BB5.1, AF016981; Methylobacter bovis, L20839; Methylobacter capsulatus, L20843; Methylobacter luteus, M95657; Methylobacter psychrophilus, AF152597; Methylobacter vinelandii, L20841; Methylobacter whittenburyi, X72773; Methylomicrobium agile, X72767; Methylomicrobium album, M95659; Methylomicrobium pelagicum, L35540; Methylomonas aurantiaca, X72776; Methylomonas methanica, AF50806; Methylomonas fodinarum, X72778; Methylomonas rubra, M95662; Methylosphaera hansonii, U77533; Methylocaldum gracile, U89298; Methylocaldum szegediense, U89300; Methylocaldum tepidum, U89297; Methylococcus capsulatus, L20842; Escherichia coli, V00348; Erwinia carotovora, M59149; Vibrio cholerae, O11197; Pseudomonas flavescens, U01916; and Legionella steigerwaltii, X73400.
DISCUSSION
Root-associated methanotroph diversity assessed by comparative analysis of pmoA, mmoX, mxaF, and 16S rRNA and rDNA sequences.
The branching pattern of the pmoA tree showed a remarkable congruence with that of the 16S rDNA and rcDNA tree (Fig. 1A and 5). Therefore, it is highly likely for example that pmoA cluster III corresponds to the 16S rDNA branch characterized by the clones M90-D37, M84-D38, and M84-D36. Although it is difficult to deduce a close phylogenetic correspondence between distinct clusters of environmental sequence types from two different gene markers, the similarity in the tree topologies suggests that members of the same type I sublineages were detected by both the pmoA and 16S rDNA approaches. This finding agrees well with the results of a cultivation-independent characterization of methanotrophic populations in the sediment of Lake Washington (Seattle, Wash.) (10). The comparison of environmental libraries constructed for methanotroph 16S rRNA and pmoA genes also showed that the two different genes cover very similar ranges of diversity.
The mxaF gene represents a universal marker for methylotrophs. Its retrieval should allow detection of a range of methanotroph diversity similar to that detected by recovery of pmoA and 16S rRNA and rDNA. However, almost all mxaF clones formed only one distinct cluster which might phylogenetically correspond to members of pmoA cluster I (Fig. 1A and 4). This finding suggests that the currently available mxaF PCR assay is only of limited value for the assessment of methanotroph and methylotroph diversity and should be applied only in conjunction with other gene markers.
Both the pmoA-based T-RFLP profiles (Fig. 1 and 2) and the retrieval of mmoX confirmed the presence of type II methanotrophs on rice roots. The mmoX-targeted primers used in this study can be considered to be complementary to primer sets previously published for the specific detection of mmoX (2, 40, 43). Their applicability in environmental studies is demonstrated especially by the retrieval of clone M84-S38 (Fig. 3). This sequence type expands our current knowledge about mmoX-based methanotroph diversity. Overall, comparative sequence analysis of cloned pmoA, mmoX, mxaF, and 16S rDNA and rcDNA revealed a genetically highly diverse methanotrophic community associated with rice roots.
Comparison of various pmoA-based approaches to assessing methanotroph diversity (cloning, DGGE, and T-RFLP analysis).
The frequency distribution of methanotroph subgroups in the clone library generated from sample M84 was not consistent with the relative signal intensities of the corresponding T-RFs observed in the T-RFLP community profiles (Fig. 1). For instance, pmoA sequence types affiliated with Methylomicrobium album (350-bp T-RF) and Methylobacter sp. strain BB.1 (505-bp T-RF) were not detected by the cloning approach. This finding might be indicative of cloning bias and/or sampling error (i.e., analysis of too few pmoA clones). Similarly, the proportion of type II methanotroph pmoA sequence types in the clone library was lower (2 of 28 clones analyzed) than one would expect based on the high relative signal intensity of the 245-bp T-RF (35.4%). This observation indicates either some bias against cloning of pmoA genes from type II methanotrophs or errors in the strict correlation of the 245-bp T-RF with type II methanotrophs of the Methylocystis-Methylosinus group. However, both T-RFLP analysis of individual pmoA clones retrieved from rice roots and a thorough in silico analysis of our current database of approximately 130 pmoA sequences suggest such a strict correlation of the 245-bp T-RF with type II methanotrophs.
The disparity between the frequency distribution of methanotroph subgroups in the clone library and the relative signal intensities of the corresponding T-RFs stresses a major methodological advantage of MspI-based T-RFLP analysis—that it provides an exact quantitative measure of the T-RF composition of pmoA PCR products. Also, pmoA-based methanotroph diversity detectable by T-RFLP analysis was clearly higher than that detectable by DGGE (Fig. 1B versus C). Explanations for this finding might be that sequence types are separated in T-RFLP analysis and DGGE by different methodological principles and staining of DGGE gels is less sensitive than fluorescence detection in T-RFLP analysis. Taking these observations together, it can be concluded that for cultivation-independent assessment of methanotroph diversity pmoA-based T-RFLP analysis represents an important tool to complement the pmoA-based cloning approach and DGGE.
Ecological significance of root-associated methanotroph diversity.
The analysis of phospholipid ester-linked fatty acids (PLFA) recovered from rhizosphere soil of flooded rice microcosms indicated an increased abundance of type I methanotrophs after NH4+ fertilization (ninefold increase in the type-I-specific PLFA biomarker), while the type-II-specific PLFA biomarker increased only two- to threefold after NH4+ fertilization (3). These results led to the conclusion that a high ammonium concentration is essential for growth of type I methanotrophs. In a control experiment based on molecular retrieval of 16S rDNA and DGGE, type I methanotrophs were also detected in rhizosphere soil of unfertilized rice microcosms. However, type I methanotrophs were not detected in the bulk soil of these microcosms. Based on these preliminary data, it was concluded that the rice plant itself also favors growth of type I methanotrophs (3).
The latter conclusion is clearly supported by this study. The T-RFLP profiles suggest a substantially higher relative abundance of type I methanotrophs on rice roots than in the bulk soil (Fig. 2). The detection of rRNA from type I methanotrophs (Fig. 5) provides evidence that these species are metabolically active and thus further supports the idea that rice roots are an important habitat for type I methanotrophs. The promotion of methanotrophic activity by rice plants might be of twofold nature. The diffusional input of oxygen into the root environment directly affects the activities of the obligately aerobic methanotrophs. An indirect effect on NH4+ availability might be mediated by the escape of oxygen from the root tissue into the rhizosphere soil, which can increase the redox potential in the root vicinity and, as a consequence, lead to the desorption of fixed NH4+ ions from clay minerals (51). Increased availability of ammonium should especially favor proliferation of type I methanotrophs (3, 27).
The T-RFLP profiles obtained from rice roots and bulk soil are characterized by high relative signal intensities of the type II methanotroph-specific 245-bp T-RF (35.4 and 67%, respectively). In the anoxic bulk soil, 16S rRNA genes extracted from desiccation-resistant exospores and lipid cysts formed by Methylosinus spp. and Methylocystis spp. (56), respectively, as well as from vegetative cells present in a stage of anaerobic dormancy (46, 47), might have contributed to the high relative signal intensity.
The cultivation-independent characterization of type I methanotrophs in unfertilized rhizosphere soil by Bodelier et al. (3) resulted in the detection of only one distinct cluster of highly similar 16S rDNA sequence types related to Methylobacter spp. (Fig. 5, sequence type RRI1). By contrast, our results of comparative sequence analysis of cloned pmoA and T-RFLP community profiling revealed a more complex population structure encompassing type I methanotrophs affiliated with the genera Methylomonas, Methylobacter, Methylomicrobium, and Methylococcus and a novel type I methanotroph sublineage. The considerable number of distinct methanotrophic populations that colonized the root compartment is also illustrated by the recovery of various 16S rDNA sublineages of type I methanotrophs. The presence of such a highly diverse methanotrophic community might indicate that there are a large number of ecological microniches characterized by spatiotemporal variations in the mixing ratios of CH4 and O2 (1, 30, 31) and in the availability of nitrogen (3, 24).
The hypothesis that the CH4/O2 mixing ratio is an important regulator of root-associated methanotroph diversity is supported by the close correspondence between mxaF sequence types obtained in a previous study (clones Mo1/Mo2 [31]) and the mxaF cluster detected on rice roots (Fig. 4). Mo1 and Mo2 were detected by pmoA-based DGGE in a methanotrophic consortium enriched from rice field soil under a high CH4/low O2 mixing ratio, while enrichment conditions using low CH4/high O2 and low CH4/low O2 mixing ratios did not favor growth of these type I methanotrophs.
Final conclusions.
The comparison of data obtained by retrieval of pmoA with those obtained by recovery of mmoX, mxaF, and 16S rDNA and rRNA clearly indicates that pmoA represents an excellent functional gene marker for cultivation-independent analysis of methanotrophic diversity. However, the data also indicate that a comprehensive view of methanotrophic diversity can be obtained only by a combined use of various molecular techniques, i.e., by cloning and sequencing and by cloning-independent fingerprinting. Within this framework, the pmoA-based T-RFLP analysis proved to be a suitable tool to rapidly assess methanotrophic diversity. Taking all molecular data together, a highly diverse community of type I and type II methanotrophs was detected. Except for Methylomicrobium album, type I methanotroph populations were detected in the root environment with clearly higher relative signal intensities than in the bulk soil. Thus, the data as a whole agree well with the hypothesis that type I methanotrophs are predominant in environments that allow rapid growth of methanotrophic bacteria, while type II methanotrophs are more abundant in environments where growth rates are periodically restricted (27, 54).
ACKNOWLEDGMENTS
We are grateful to Sonja Fleissner for excellent technical assistance.
This work was supported by a grant from the European Community RTD Programme Biotechnology (contract BIO-CT96-0419). M.T.Y. thanks the Deutschen Akademischen Austauschdienst for financial support in the form of a PhD scholarship.
REFERENCES
- 1.Amaral J A, Knowles R. Growth of methanotrophs in methane and oxygen counter gradients. FEMS Microbiol Lett. 1995;126:215–220. [Google Scholar]
- 2.Auman A J, Stolyar S, Costello A M, Lidstrom M E. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol. 2000;66:5259–5266. doi: 10.1128/aem.66.12.5259-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodelier P L E, Roslev P, Henckel T, Frenzel P. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature. 2000;403:421–424. doi: 10.1038/35000193. [DOI] [PubMed] [Google Scholar]
- 4.Bodrossy L, Holmes E M, Holmes A J, Kovács K L, Murrell J C. Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch Microbiol. 1997;168:493–503. doi: 10.1007/s002030050527. [DOI] [PubMed] [Google Scholar]
- 5.Bosse U, Frenzel P. Activity and distribution of methane-oxidizing bacteria in flooded rice soil microcosms and in rice plants (Oryza sativa) Appl Environ Microbiol. 1997;63:1199–1207. doi: 10.1128/aem.63.4.1199-1207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman J P, McCammon S A, Skerratt J H. Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology. 1997;143:1451–1459. doi: 10.1099/00221287-143-4-1451. [DOI] [PubMed] [Google Scholar]
- 7.Cardy D L N, Laidler V, Salmond G P C, Murrell J C. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991;5:335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 8.Cicerone R J, Oremland R S. Biogeochemical aspects of atmospheric methane. Global Biogeochem Cycles. 1988;2:299–327. [Google Scholar]
- 9.Conrad R, Rothfuss F. Methane oxidation in the soil surface layer of a flooded rice field and the effect of ammonium. Biol Fertil Soils. 1991;12:28–32. [Google Scholar]
- 10.Costello A M, Lidstrom M E. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl Environ Microbiol. 1999;65:5066–5074. doi: 10.1128/aem.65.11.5066-5074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dayhoff M O, Schwartz R M, Orcutt B C. A model of evolutionary change in proteins. In: Dayhoff M O, editor. Atlas of protein sequence and structure. 5, suppl. 3. Silver Spring, Md: National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- 12.Dedysh S N, Panikov N S, Tiedje J M. Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl Environ Microbiol. 1998;64:922–929. doi: 10.1128/aem.64.3.922-929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dedysh S N, Panikov N S, Liesack W, Groβkopf R, Zhou J, Tiedje J M. Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science. 1998;282:281–284. doi: 10.1126/science.282.5387.281. [DOI] [PubMed] [Google Scholar]
- 14.Dedysh S N, Liesack W, Khmelenina V N, Suzina N E, Trotsenko Y A, Semrau J D, Bares A M, Panikov N S, Tiedje J M. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol. 2000;50:955–969. doi: 10.1099/00207713-50-3-955. [DOI] [PubMed] [Google Scholar]
- 15.Denier van der Gon H A C, Neue H U. Oxidation of methane in the rhizosphere of rice plants. Biol Fertil Soils. 1996;22:359–366. [Google Scholar]
- 16.Dunfield P F, Liesack W, Henckel T, Knowles R, Conrad R. High-affinity methane oxidation by a soil enrichment culture containing a type II methanotroph. Appl Environ Microbiol. 1999;65:1009–1014. doi: 10.1128/aem.65.3.1009-1014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 18.Felske A, Engelen B, Nübel U, Backhaus H. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl Environ Microbiol. 1996;62:4162–4167. doi: 10.1128/aem.62.11.4162-4167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frenzel P. Plant-associated methane oxidation in rice fields and wetlands. In: Schink B, editor. Advances in microbial ecology. New York, N.Y: Kluwer Academic/Plenum Publishers; 2000. pp. 85–114. [Google Scholar]
- 20.Frenzel P, Bosse U. Methyl fluoride, an inhibitor of methane oxidation and methane production. FEMS Microbiol Ecol. 1996;21:25–36. [Google Scholar]
- 21.Galchenko V F, Lein A, Ivanov M. Biological sinks of methane. In: Andreae M O, Schimel D S, editors. Exchange of trace gases between terrestrial ecosystems and the atmosphere. Chichester, United Kingdom: John Wiley and Sons; 1989. pp. 59–71. [Google Scholar]
- 22.Gilbert B, Frenzel P. Methanotrophic bacteria in the rhizosphere of rice microcosms and their effect on porewater methane concentration and methane emission. Biol Fertil Soils. 1995;20:93–100. [Google Scholar]
- 23.Gilbert B, Frenzel P. Rice roots and CH4 oxidation: the activity of bacteria, their distribution and the microenvironment. Soil Biol Biochem. 1998;30:1903–1916. [Google Scholar]
- 24.Graham D W, Chaudhary J A, Hanson R S, Arnold R G. Factors affecting competition between type-I and type-II methanotrophs in 2-organism, continuous-flow reactors. Microb Ecol. 1993;25:1–17. doi: 10.1007/BF00182126. [DOI] [PubMed] [Google Scholar]
- 25.Groβkopf R. Entwicklung eines molekularen Nachweissystems (PCR) für methanoxidierende Bakterien basierend auf Gensequenzen der löslichen Methanmonooxygenase. Diploma thesis. Marburg, Germany: Philipps-Universität Marburg; 1994. [Google Scholar]
- 26.Groβkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henckel T, Jäckel U, Schnell S, Conrad R. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl Environ Microbiol. 2000;66:1801–1808. doi: 10.1128/aem.66.5.1801-1808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henckel T, Jäckel U, Conrad R. Vertical distribution of the methanotrophic community after drainage of rice field soil. FEMS Microbiol Ecol. 2000;34:279–291. doi: 10.1111/j.1574-6941.2001.tb00778.x. [DOI] [PubMed] [Google Scholar]
- 31.Henckel T, Roslev P, Conrad R. Effects of O2 and CH4 on presence and activity of the indigenous methanotrophic community in rice field soil. Environ Microbiol. 2000;2:666–679. doi: 10.1046/j.1462-2920.2000.00149.x. [DOI] [PubMed] [Google Scholar]
- 32.Holmes A J, Costello A, Lidstrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 33.Holmes A J, Roslev P, McDonald I R, Iversen N, Henriksen K, Murrell J C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol. 1999;65:3312–3318. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Intergovernmental Panel on Climate Change (IPCC) Climate change, the supplementary report to the IPCC scientific assessment. New York, N.Y: Cambridge University Press; 1992. [Google Scholar]
- 35.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 36.Kimura M. The neutral theory of molecular evolution. In: Nei M, Koehn R K, editors. Evolution of genes and proteins. Sunderland, Mass: Sinauer; 1983. pp. 208–233. [Google Scholar]
- 37.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lüdemann H, Arth I, Liesack W. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl Environ Microbiol. 2000;66:754–762. doi: 10.1128/aem.66.2.754-762.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukow T, Dunfield P F, Liesack W. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol Ecol. 2000;32:241–247. doi: 10.1111/j.1574-6941.2000.tb00717.x. [DOI] [PubMed] [Google Scholar]
- 40.McDonald I R, Kenna E M, Murrell J C. Detection of methanotrophic bacteria in environmental samples with PCR. Appl Environ Microbiol. 1995;61:116–121. doi: 10.1128/aem.61.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald I R, Murrell J C. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol Lett. 1997;156:205–210. doi: 10.1111/j.1574-6968.1997.tb12728.x. [DOI] [PubMed] [Google Scholar]
- 42.McDonald I R, Murrell J C. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol. 1997;63:3218–3224. doi: 10.1128/aem.63.8.3218-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miguez C B, Bourque D, Sealy J A, Greer C W, Groleau D. Detection and isolation of methanotrophic bacteria possessing soluble methane monooxygenase (sMMO) genes using the polymerase chain reaction (PCR) Microb Ecol. 1997;33:21–31. doi: 10.1007/s002489900004. [DOI] [PubMed] [Google Scholar]
- 44.Osborne A M, Moore E R B, Timmis K N. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 45.Prinn R G, editor. Global atmospheric-biospheric chemistry. New York, N.Y: Plenum Press; 1994. pp. 1–18. [Google Scholar]
- 46.Roslev P, King G M. Survival and recovery of methanotrophic bacteria starved under oxic and anoxic conditions. Appl Environ Microbiol. 1994;60:2602–2608. doi: 10.1128/aem.60.7.2602-2608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roslev P, King G M. Aerobic and anaerobic starvation metabolism in methanotrophic bacteria. Appl Environ Microbiol. 1995;61:1563–1570. doi: 10.1128/aem.61.4.1563-1570.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Sass R L. Short summary chapter for methane. In: Minami T, Mosier K A, editors. CH4 and N2O: global emission and controls from rice fields and other agricultural and industrial sources. Tokyo, Japan: Yokendo; 1994. pp. 1–8. [Google Scholar]
- 51.Schneiders M, Scherer H W. Fixation and release of ammonium in flooded rice soils as affected by redox potential. Eur J Agron. 1998;8:179–187. [Google Scholar]
- 52.Schütz H, Seiler W, Conrad R. Processes involved in formation and emission of methane in rice paddies. Biogeochemistry. 1989;7:33–53. [Google Scholar]
- 53.Shigematsu T, Hanada S, Eguchi M, Kamagata Y, Kanagawa T, Kurane R. Soluble methane monooxygenase gene clusters from trichloroethylene-degrading Methylomonas sp. strains and detection of methanotrophs during in situ bioremediation. Appl Environ Microbiol. 1999;65:5198–5206. doi: 10.1128/aem.65.12.5198-5206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vecherskaya M S, Galchenko V F, Sokolova E N, Samarkin V A. Activity and species composition of aerobic methanotrophic communities in tundra soils. Curr Microbiol. 1993;27:181–184. doi: 10.1007/BF01576018. [DOI] [PubMed] [Google Scholar]
- 55.Wahlen M, Tanaka N, Henry R, Deck B, Zeglen J, Vogel J S, Southon J, Shemesh A, Fairbanks R, Broecker W. Carbon-14 in methane sources and in atmospheric methane: the contribution from fossil carbon. Science. 1989;245:286–290. doi: 10.1126/science.245.4915.286. [DOI] [PubMed] [Google Scholar]
- 56.Whittenbury R, Davies S L, Davey J F. Exospores and cysts formed by methane-utilizing bacteria. J Gen Microbiol. 1970;61:219–226. doi: 10.1099/00221287-61-2-219. [DOI] [PubMed] [Google Scholar]
- 57.Wise M G, McArthur J V, Shimkets L J. Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis. Appl Environ Microbiol. 1999;65:4887–4897. doi: 10.1128/aem.65.11.4887-4897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wise M G, McArthur J V, Shimkets L J. Methylosarcina fibrata gen. nov., sp. nov. and Methylosarcina quisquiliarum sp. nov., novel type I methanotrophs. Int J Syst Evol Microbiol. 2001;51:611–621. doi: 10.1099/00207713-51-2-611. [DOI] [PubMed] [Google Scholar]