Abstract
Background and purpose
Hereditary myopathies with limb‐girdle muscular weakness (LGW) are a genetically heterogeneous group of disorders, in which molecular diagnosis remains challenging. Our aim was to present a detailed clinical and genetic characterization of a large cohort of patients with LGW.
Methods
This nationwide cohort study included patients with LGW suspected to be associated with hereditary myopathies. Parameters associated with specific genetic aetiologies were evaluated, and we further assessed how they predicted the detection of causative variants by conducting genetic analyses.
Results
Molecular diagnoses were identified in 62.0% (75/121) of the cohort, with a higher proportion of patients diagnosed by next‐generation sequencing (NGS) than by single‐gene testing (77.3% vs. 22.7% of solved cases). The median (interquartile range) time from onset to genetic diagnosis was 8.9 (3.7–19.9) and 17.8 (7.9–27.8) years for single‐gene testing and NGS, respectively. The most common diagnoses were myopathies associated with variants in CAPN3 (n = 9), FKRP (n = 9), ANO5 (n = 8), DYSF (n = 8) and SGCA (n = 5), which together accounted for 32.2% of the cohort. Younger age at disease onset (p = 0.043), >10× elevated creatine kinase activity levels (p = 0.024) and myopathic electromyography findings (p = 0.007) were significantly associated with the detection of causative variants.
Conclusions
Our findings suggest that an earlier use of NGS in patients with LGW is needed to avoid long diagnostic delays. We further present parameters predictive of a molecular diagnosis that may help to select patients for genetic analyses, especially in centres with limited access to sequencing.
Keywords: Austria, limb‐girdle muscular dystrophy, limb‐girdle muscular weakness, myopathy, next‐generation sequencing
We provide a nationwide characterization of Austrian patients with limb‐girdle weakness. A molecular diagnosis was found in 62%, with CAPN3, FKRP, ANO5, DYSF and SGCA representing the five most common subtypes. Our data suggest an earlier use of NGS is needed to avoid diagnostic delays.
INTRODUCTION
Hereditary myopathies with a limb‐girdle pattern of weakness (LGW), also referred to as limb‐girdle muscular dystrophies (LGMD), are a group of rare and genetically heterogeneous neuromuscular disorders clinically characterized by weakness of shoulder and pelvic girdle muscles. Disease onset ranges from early childhood to adulthood with slow to rapid progression rates. The clinical spectrum often comprises other manifestations, with respiratory insufficiency and cardiomyopathy representing serious and potentially life‐threatening complications [1]. Substantial phenotypic overlaps with other neuromuscular diseases are common [2], and as hereditary myopathies with LGW are rare diseases with a total prevalence of 2–3/100,000 [3, 4], in‐depth genotype–phenotype correlations are difficult to establish. These factors, together with the vast genetic heterogeneity, pose diagnostic challenges to clinicians, and despite the application of next‐generation sequencing (NGS) techniques, a significant proportion of patients with LGW remain genetically undiagnosed [5, 6]. Systematic clinico‐genetic studies in large cohorts are therefore important, as they have the potential to guide diagnostic testing strategies, optimize the clinical management and improve genetic counselling of patients with LGW [7].
In this cohort study, we provide a detailed clinical and genetic characterization of a large Austrian cohort of patients with LGW. Different parameters associated with specific genetic aetiologies were evaluated, and we further assessed how these parameters predicted the detection of causative variants by conducting genetic analyses. The conclusions drawn from this study could be useful in directing genetic testing and clinical management of patients with neuromuscular conditions associated with LGW.
METHODS
Ethical approval and consent to participate
This study was approved by the Ethics Committee of the Medical University of Vienna (EK 1635/2017), and informed consent was obtained from all patients.
Study design and patient ascertainment
In this multicentre cohort study, all registered Austrian neurologists and all neuromuscular centres were contacted via email by the Austrian Society of Neurology (ÖGN) and provided with study information for patient enrolment during the study period between 1 January 2018 and 31 December 2020. Patients had to be ≥18 years of age with i) unexplained LGW together with elevated levels of creatine kinase (CK) activity and/or myopathic abnormalities in muscle histology, electromyography (EMG) or magnetic resonance imaging or ii) LGW and a previously established genetic diagnosis of LGMD. Clinical information and strength assessment of proximal and distal limb muscles according to the Medical Research Council (MRC) scale, ranging from 0 (i.e., complete paralysis) to 5 (i.e., normal), had to be provided. CK activity levels were grouped into: i) normal; ii) elevated to 5–10 times the upper limit of normal (ULN); and iii) elevated to >10 times the ULN. Collaborating centres were contacted and asked for data completion in cases of missing data. If available, data on previous genetic tests were collected, and patients with suspected but genetically undiagnosed disease were offered exome sequencing as described previously [6, 8]. All genetic variants that were reported prior to study enrolment were re‐analysed, and only pathogenic or likely pathogenic variants according to the standards of the American College of Medical Genetics and Genomics [9] were accepted for molecular diagnosis, with the mode of inheritance classified according to the identified variants. Inherent to the multicentric nature of this study and the performance of genetic tests in different laboratories, the sequencing of gene panels and targeted exomes may encompass different sets of genes. The different NGS approaches were therefore pooled and compared as a group to the results obtained by sequential gene‐by‐gene sequencing.
Evaluation of parameters predictive of the detection of causative variants and clinico‐genetic associations
To determine predictive parameters that were associated with the detection of (likely) pathogenic variants, we compared several parameters between patients with and without molecular diagnoses in an unbiased approach. The different methodologies used for genetic testing among the involved national and international laboratories were pooled for this purpose to assess the spectrum of genetic variants in a real‐world scenario. To evaluate clinico‐genetic associations, the parameters were also analysed across the most common molecular diagnoses that included ≥5 cases.
Statistical analysis
Descriptive statistics were performed using means and standard deviations or medians and interquartile ranges (IQRs). Evaluation of clinico‐genetic associations was performed for the most common genetic aetiologies by pairwise comparison with the remainder of the cohort using the chi‐squared test for each clinical feature. Bonferroni correction was applied to account for multiple testing. Comparison between more than two means was performed using one‐way analysis of variance, and significant main effects were followed by Dunnett's multiple comparison test. Parameters were compared between patients with and without a molecular diagnosis using the chi‐squared test, Mann–Whitney U‐test or independent t‐test (with Welch's correction in case of unequal standard deviations between the groups), as appropriate. Binary logistic regression models with molecular diagnosis as the dependent variable were calculated by stepwise inclusion of independent variables as defined by univariate associations with p values <0.1 after adjusting for sex and disease duration. The model's goodness of fit was tested by the omnibus test of fit, and Nagelkerke’s R‐squared was used to assess the contribution of each variable to the explanation of variance within the overall model. All variables were tested for normal distribution by the Lilliefors test and for collinearity by the variance inflation factor (VIF), with exclusion of variables from the regression analysis if the VIF was >2.0, corresponding to an R 2 of 0.60. Missing values were handled by multiple (20 times) imputation using the missing‐not‐at‐random approach, with pooling of estimates according to Rubin's rules [10]. Two‐sided p values <0.05 were taken to indicate statistical significance, unless otherwise stated. Data processing was performed using the statistical package SPSS v25 (released 2017, IBM Corp.) and Prism, version 9.1.0 (GraphPad Software Inc.).
RESULTS
Demographic and clinical characteristics
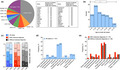
A total of 121 patients with LGW were enrolled during the study period (Table 1 and Figure 1a), with females accounting for 52.1% of the cohort. The mean (±SD) age at onset was 28.8 (±17.7) years (Figure 1b), with adult onset (i.e., onset at ≥18 years of age) in the majority of cases (65.8%). Lower limbs were affected more often (χ2 (1, n = 120) = 80.6; p ≤ 0.001) and more severely (χ2 (2, n = 121) = 9.4; p = 0.009 [Figure 1c]) than upper limbs, but the majority of patients (60.3%) could still walk independently at the time of their last visit (Table 1). The most common symptoms associated with LGW were Trendelenburg's sign (36.8%), winged scapula (34.2%), cardiomyopathy (11.7%), respiratory insufficiency (10.7%) and facial weakness (9.9%; Figure 1d). Three patients (LMNA, DES and RBCK1) had undergone heart transplantation and five patients depended on non‐invasive ventilation (two CAPN3, one FKRP, one SGCA, and one patient without a molecular diagnosis). Contractures were reported in patients with variants in CAPN3 (n = 3), FKRP (n = 2), SGCA (n = 1), FHL1 (n = 1) and COL6A1 (n = 1).
TABLE 1.
Selected demographic, clinical and genetic characteristics of the patients with limb‐girdle muscular weakness
|
Total cohort (n = 121) |
With molecular diagnosis (n = 75) |
Without molecular diagnosis (n = 46) |
p value | |
|---|---|---|---|---|
| Sex: female, n (%) | 63 (52.1) | 42 (56.0) | 21 (45.7) | 0.349 |
| Positive family history, n (%) | 24 (19.8) | 15 (20.0) | 9 (19.6) | 0.574 |
| Inheritance pattern, n (%) | ||||
| Recessive | 48 (39.7) | 48 (64.0) | – | |
| Dominant | 20 (16.5) | 20 (26.7) | – | |
| X‐linked | 4 (3.3) | 4 (5.3) | – | |
| Dual diagnosis | 2 (1.7) | 2 (2.7) | – | |
| Digenic | 1 (0.8) | 1 (1.3) | ||
| Age at onset in years, n = 118, mean (SD) | 28.8 (17.8) | 25.4 (16.4) | 34.1 (18.7) | 0.010* |
| Age group at onset, n (%) | ||||
| Infantile, ≤6 years | 7 (5.9) | 6 (8.2) | 1 (2.2) | 0.033* |
| Juvenile, 7–17 years | 34 (28.8) | 23 (31.5) | 11 (24.4) | |
| Adult, 18–60 years | 73 (61.9) | 44 (60.3) | 29 (64.4) | |
| Late, >60 years | 4 (3.4) | 0 (0) | 4 (8.9) | |
| Disease duration in years, median (IQR) | 18.1 (13.3) | 19.2 (13.3) | 16.1 (13.3) | 0.227 |
| Time to genetic diagnosis in years (n = 66), median (IQR) | – | 14.8 (6.5–26.1) | – | |
| Single gene testing | – | 8.9 (3.7–19.9) | – | |
| Next‐generation sequencing | – | 17.8 (7.9–27.8) | – | |
| Genetic analysis leading to molecular diagnosis, n (%) | ||||
| Single gene testing | ‐ | 17 (22.7) | – | |
| Next‐generation sequencing | ‐ | 58 (77.3) | – | |
| Creatine kinase (n = 116), n (%) | ||||
| Normal | 14 (12.1) | 6 (8.3) | 8 (18.2) | 0.093 |
| Elevated <10× ULN | 62 (53.4) | 37 (51.4) | 25 (56.8) | |
| Elevated >10× ULN | 40 (34.5) | 29 (40.3) | 11 (25.0) | |
| Muscle biopsy with signs of myopathy (n = 82), n (%) | 64 (78.0) | 37 (72.5) | 27 (87.1) | 0.171 |
| Myopathic EMG changes (n = 94), n (%) | 64 (68.1) | 46 (78.0) | 18 (51.5) | 0.011* |
| Mobility, n (%) | ||||
| Independent | 73 (60.3) | 40 (53.3) | 33 (71.7) | 0.098 |
| Walking with aid | 21 (17.4) | 14 (18.7) | 7 (15.2) | |
| Unable to walk | 27 (22.3) | 21 (28.0) | 6 (13.0) | |
| Region of onset (n = 120), n (%) | ||||
| Upper limbs | 18 (14.9) | 6 (8.0) | 12 (26.7) | 0.028* |
| Lower limbs | 87 (72.5) | 57 (76.0) | 30 (66.7) | |
| Other | 15 (12.5) | 12 (16.0) | 3 (6.6) | |
| Symmetric onset (n = 116), n (%) | 104 (88.1) | 68 (91.9) | 36 (81.8) | 0.141 |
Percentages in brackets refer to absolute subgroup numbers in the corresponding columns.
Abbreviations: EMG, electromyography; IQR, interquartile range; SD, standard deviation; ULN, upper limit of normal.
Statistically significant.
FIGURE 1.

Genetic spectrum and clinical characteristics of patients with limb‐girdle muscular weakness. (a) Causative variants located in 27 different genes could be identified in 75/121 patients. The five most frequent genotypes were CAPN3, FKRP, ANO5, DYSF and SGCA and accounted for over 50% of patients with a molecular diagnosis. (b) Age at onset was most frequent in the second decade with a wide range into late adulthood. (c) In patients with molecular diagnoses, legs were more often and more severely affected by muscle weakness than arms. In patients without molecular diagnoses, by contrast, muscle weakness was similarly distributed to both legs and arms. Frequency of clinical symptoms in the total cohort (d) and as compared between patients with and without molecular diagnoses (e), with none of the symptoms differing significantly between the groups. *CACNA1S was identified together with RYR1 in the same person, and SCN4A together with DMD in another person [Colour figure can be viewed at wileyonlinelibrary.com]
Distribution of molecular diagnoses
Pathogenic or likely pathogenic (i.e., causative) variants in 27 different genes were identified in 75 patients (62.0%; Table 1) with a diagnostic delay of 14.8 years from disease onset (IQR 6.5–26.1 years; genetic variant details provided in Table S1). The majority of molecular diagnoses were established by NGS approaches (77.3%), while only 22.7% of cases were solved by targeted single‐gene testing. Prior single‐gene analysis was performed in 31% of patients eventually diagnosed by NGS, and 6.9% of patients had had at least three genes sequentially screened before their referral to NGS. Reasons for the application of single‐gene analyses included specific muscle biopsy with findings suggestive of immunoblot and immunohistochemistry analyses in eight cases (three CAPN, three DYSF, one FKRP, one DMD), characteristic phenotypes in two cases (one DUX4, one DMD) and an already known causative variant within the family in one case (DNAJB6).
Variant types in solved cases comprised missense and frameshift variants in 51.6% and 21%, respectively, followed by structural (13.7%), nonsense (7.3%), splice‐site (3.2%) and near‐splice variants (3.2%). Uncertain findings were reported in 11 individuals (9.1% of the cohort). Ten of these patients harboured one or two variants of uncertain significance (VUSs) in candidate genes, while one patient carried one VUS and one likely pathogenic variant in ANO5 (details provided in Table S2).
The five most common molecular diagnoses (CAPN3, FKRP, ANO5, DYSF and SGCA) accounted for 32.2% of the cohort (Table 2), with a contrasting distribution in children as compared to adults. In patients with disease onset at <18 years of age, CAPN3 was the most commonly mutated gene, accounting for 17.5% of the group, followed by FKRP and SGCA, accounting for 12.5% and 10%, respectively. In adult patients with disease onset at ≥18 years of age, by contrast, ANO5 was the most common diagnosis, at 9.1%, followed by DYSF with 6.5%.
TABLE 2.
Main demographic and clinical characteristics of the five most commonly diagnosed genotypes
| CAPN3 | FKRP | ANO5 | DYSF | SGCA | |
|---|---|---|---|---|---|
| Number | 9 | 9 | 8 | 8 | 5 |
| Sex, female, n (%) | 8 (88.9) | 6 (66.7) | 4 (50.0) | 3 (37.5) | 4 (80.0) |
| Family history, n (%) | 2 (22.2) | 1 (11.1) | 3 (37.5) | 1 (12.5) | 1 (20.0) |
| Age at onset in years, mean (SD) | 14.3 (7.7) | 16.6 (12.9) | 39.1 (13.2) | 24.2 (10.1) | 12.5 (10.1) |
| Age group at onset, n (%) | |||||
| Infantile, ≤6 years | 1 (11.1) | 3 (33.3) | 0 | 0 | 1 (20.0) |
| Juvenile, 7–17 years | 6 (66.7) | 2 (22.2) | 1 (12.5) | 2 (28.6) | 3 (60.0) |
| Adult, 18–60 years | 2 (22.2) | 4 (44.4) | 7 (87.5) | 5 (71.4) | 1 (20.0) |
| Late, >60 years | 0 | 0 | 0 | 0 | 0 |
| Time to molecular diagnosis in years, median (IQR) | 23.5 (11.5–29.0) | 17.3 (5.8–29.7) | 8.0 (6.1–14.0) | 7.6 (1.8–19.1) | 28.6 (5.9–42.4) |
| Genetic analysis leading to molecular diagnosis, n (%) | |||||
| Single gene sequencing | 5 (55.6) | 3 (33.3) | 0 | 3 (37.5) | 0 |
| Next‐generation sequencing | 4 (44.4) | 6 (66.7) | 8 (100) | 5 (62.5) | 5 (100) |
| Creatine kinase, n (%) | |||||
| Normal | 1 (12.5) | 0 | 0 | 0 | 1 (20.0) |
| Elevated <10x ULN | 3 (37.5) | 4 (44.4) | 0 | 3 (37.5) | 2 (40.0) |
| Elevated >10x ULN | 4 (50.0) | 5 (55.6) | 8 (100) | 5 (62.5) | 2 (40.0) |
| Myopathic EMG changes, n (%) | 4/5 (80.0) | 6/6 (100) | 4/6 (66.7) | 6/7 (85.7) | 2/3 (66.7) |
| Muscle biopsy with signs of myopathy, n (%) | 6/8 (75.0) | 4/5 (80.0) | 3/6 (50.0) | 6/6 (100) | 1/2 (50.0) |
| Mobility, n (%) | |||||
| Independent | 2 (22.2) | 3 (33.3) | 7 (87.5) | 3 (37.5) | 1 (20.0) |
| Walking with aid | 0 | 2 (22.2) | 1 (12.5) | 2 (25.0) | 1 (20.0) |
| Unable to walk | 7 (77.8) | 4 (44.4) | 0 | 3 (37.5) | 3 (60.0) |
| Region of onset, n (%) | |||||
| Upper limbs | 1 (11.1) | 0 | 1 (12.5) | 0 | 0 |
| Lower limbs | 7 (77.8) | 8 (88.9) | 5 (62.5) | 7 (87.5) | 5 (100) |
| Other | 1 (11.1) | 1 (11.1) | 2 (25.0) | 1 (12.5) | 0 |
| Symmetric onset, n (%) | 8 (88.9) | 9 (100) | 6 (75.0) | 8 (100) | 5 (100) |
Abbreviations: EMG, electromyography; IQR, interquartile range; SD, standard deviation; ULN, upper limit of normal.
Further clinico‐genetic analyses revealed that CAPN3 was associated with significantly earlier disease onset (p = 0.038; Figure 2a) and that these patients were more likely to lose independent ambulation when compared to other aetiologies in the cohort (p < 0.001; Figure 2b). CK activity levels, by contrast, were significantly higher in ANO5 and DYSF (p < 0.001 and p = 0.003, respectively; Figure 2d). Time to molecular diagnosis, disease duration and walking ability were not significantly associated with any of the molecular diagnoses (Figure 2d–f). Moreover, none of the other clinical parameters (listed in Figure 1d) was significantly associated with any of the five common molecular diagnoses.
FIGURE 2.

Demographic and clinical characterization of common genetic aetiologies in patients with limb‐girdle muscular weakness. (a) Disease onset was significantly earlier for CAPN3 (F(5,115) = 4.1, p = 0.002, one‐way analysis of variance with Dunnett's multiple comparison test), and (b) patients were more often unable to walk (χ2 (10, n = 121) = 33.5, p < 0.001 with Bonferroni correction for multiple testing) as compared to other genes. (c) Creatine kinase was higher in the FKRP, ANO5 and DYSF subgroups (χ2 (10, n = 116) = 29.0, p = 0.001 with Bonferroni correction for multiple testing). Time to molecular diagnosis (d), disease duration (e) and region of onset (f), by contrast, were similar between all groups. CAPN3: n = 9; FKRP: n = 9; ANO5: n = 8; DYSF: n = 8; SGCA: n = 5; Other: n = 82. ULN, upper limit of normal [Colour figure can be viewed at wileyonlinelibrary.com]
Parameters associated with the detection of causative variants
Univariate comparison revealed that age at disease onset (p = 0.010), the degree of lower limb weakness (MRC ≤3; p = 0.038) and myopathic EMG changes (p = 0.011) were associated with a molecular diagnosis. After adjusting for sex and disease duration in a multivariate model, however, younger age at onset (odds ratio [OR] 0.96, 95% confidence interval [CI] 0.93–0.99), >10 times elevated CK activity levels (OR 5.3, 95% CI 1.2–10.5) and myopathic EMG changes (OR 3.3, 95% CI 1.4–17.4) were significantly associated with the detection of causative variants in pooled genetic analyses. By contrast, an isolated upper limb manifestation at disease onset predicted the lack of a molecular diagnosis (OR 0.25, 95% CI 0.08–0.94; Table 3).
TABLE 3.
Likelihood of a molecular diagnosis depending on different parameters
| OR (95% CI) | p value | Change in R 2 (overall R 2) | |
|---|---|---|---|
| Age at onset (per year) | 0.96 (0.93–0.99) | 0.043* | 0.095 (0.115) |
| Region of onset | |||
| Lower limbs only | Reference category | 0.174 (0.289) | |
| Upper limbs only | 0.25 (0.08–0.94) | 0.039* | |
| Other | 1.8 (0.34–9.9) | 0.489 | |
| Symmetric onset | 1.0 (0.26–4.0) | 0.979 | 0.001 (0.290) |
| Degree of lower limb weakness (KG ≤3) | 1.3 (0.37–4.5) | 0.694 | 0.003 (0.293) |
| Cardiomyopathy | 3.4 (0.57–20.1) | 0.184 | 0.021 (0.314) |
| Mobility | |||
| Independent | Reference category | 0.001 (0.315) | |
| Walking with aid | 1.1 (0.25–4.5) | 0.949 | |
| Immobile | 1.1 (0.28–4.2) | 0.912 | |
| Creatine kinase | |||
| Normal | Reference category | 0.092 (0.407) | |
| Elevated <10x ULN | 2.5 (1.1–8.3) | 0.050 | |
| Elevated >10x ULN | 5.3 (1.2–10.5) | 0.024* | |
| Myopathic EMG changes | 3.3 (1.4–17.4) | 0.007* | 0.112 (0.519) |
Abbreviations: CI, confidence interval; MRC, Medical Research Council; OR, odds ratio; R 2, regression coefficient; ULN, upper limit of normal.
Statistically significant.
DISCUSSION
Neuromuscular disorders with LGW represent a challenging entity for molecular diagnosis due to a significant phenotypic overlap and genetic heterogeneity. The implementation of exome sequencing and other NGS techniques in clinical routine has helped to improve the diagnostic outcome, but these applications are still associated with highly variable hit rates, ranging from 27% to 76% [6, 11, 12, 13, 14, 15, 16]. Differences in the extent of genetic prescreening and the application of gene panels and exome sequencing covering different numbers of genes have been proposed to contribute to divergent detection rates of causative variants [6, 8, 14]. The association of founder mutations with specific ethnicities [17] and higher rates of consanguinity in some study populations are additional factors potentially resulting in higher rates of molecular diagnoses [15, 18]. In accordance with these data, the vast majority of solved cases (77.3%) in our study could also be diagnosed by NGS‐based approaches. However, the time from symptom onset to molecular diagnosis was found to be markedly longer in cases diagnosed by NGS when compared to targeted genetic testing. As NGS has become cost‐effective and widely available for an increasing number of Austrian centres in the past few years, most patients in our cohort were initially tested by single‐gene analyses before referral to exome sequencing, which might explain the difference in diagnostic latencies.
We further sought to determine parameters that were predictive for the identification of molecular diagnoses, and found younger age at onset, more than 10‐fold elevated CK activity levels and myopathic EMG changes to be associated with a higher detection rate of causative variants, whereas isolated upper limb weakness at onset was inversely associated with a molecular diagnosis. The definition of such predictive parameters could be helpful to select patients for genetic analyses in centres with limited access to sequencing.
The NGS applications were performed in different national and international laboratories and thus covered heterogeneous numbers of genes. We were therefore not able to clearly differentiate and perform a direct comparison between gene panel and exome sequencing. However, previous studies compared the diagnostic yield between different NGS applications and found higher yields associated with sequencing of comprehensive panels or targeted exomes covering large numbers of genes [8, 19], while whole‐exome sequencing did not further improve the diagnostic yield [15]. Exome sequencing has nevertheless been deemed superior, as it enables both the identification of novel disease genes and the reiteration of analyses when new disease genes are discovered [6]. As exome sequencing has also become cost‐effective and thus widely available for an increasing number of centres, we propose that patients with LGW should generally be referred to exome sequencing early in the disease course.
In spite of comprehensive diagnostic efforts, a significant number of patients remain undiagnosed. These unsolved cases could be caused by somatic mosaicism, repeat expansions or by mutations in intronic or intergenic regions that are commonly not detected by gene panel or exome sequencing, while other patients may have acquired disorders such as inflammatory or drug‐induced myopathies [6]. A subset of patients may also have monogenic conditions with causative variants in genes not yet associated with human disease. Moreover, some myopathies are increasingly recognized as digenic or oligogenic disorders, in which the combination of multiple variants in different genes contributes to disease pathogenesis [11]. Hence, the clinical presentation could result from the combined effect of variants providing a polygenic background for disease susceptibility with marked inter‐ and intrafamilial phenotypic variability [11, 20, 21]. Variants of uncertain significance have also been detected in a significant proportion of unsolved cases in our study and could potentially have contributed to the phenotype in affected individuals.
Causative variants in the five most commonly found genes were identified in 32.2% of cases in our cohort, representing a higher proportion than previously reported. The largest study to date on 1001 undiagnosed patients with LGW reported pathogenic or likely pathogenic variants in the most common eight genes (i.e., CAPN3, DYSF, ANO5, DMD, RYR1, TTN, COL6A2 and SGCA) accounting for 27.1% of their cohort [6], and another study on 504 patients with hereditary myopathies found mutations in six common genes (i.e., RYR1, CAPN3, ANO5, DYSF, SGCA and GAA) accounting for 19.2% of cases [5]. As those studies usually included preselected patients, some of which had undergone extensive testing before being referred to exome sequencing, such studies do not represent the genetic landscape in corresponding populations, as findings from targeted analyses prior to the application of gene panel or exome sequencing would have been missed. Our approach, by contrast, was driven by the clinical phenotype and included all patients irrespective of the applied diagnostic approach and molecular diagnosis, and is therefore considered to provide an overview of the genetic spectrum that is representative of patients with LGW in Austria. As a result, we also identified neuromuscular diseases including facioscapulohumeral muscular dystrophy type 1 (FSHD1) or spinal muscular atrophy that may display LGW but were not reported in previous studies with preselected cohorts. These entities should therefore be considered in the diagnostic evaluation of patients with LGW, especially as chromosomal deletions causing inadequate repression of DUX4 that underlie FSHD1 are commonly not detected by exome sequencing. However, it should be noted that our approach could have been limited by reporting bias, as cases with molecular diagnoses could have been more likely to be reported. This would also have contributed to the high rate of molecular diagnoses in our study but is unlikely to have obscured the reported genetic landscape.
Some of the patients with LGW in other cohorts were shown to carry variants in genes that resulted in a specific treatment. Pompe disease with variants in GAA was reported in 1.0%–7.5% of large cohorts of patients with LGW [5, 6, 22, 23, 24, 25], the early treatment of whom has been associated with better clinical outcomes [26]. No causative GAA variants were identified in the present study, which was unexpected as Pompe disease in Austria was proposed to be underdiagnosed considering the low overall prevalence as compared to other European countries [27]. As patients with Pompe disease generally present with limb‐girdle or axial weakness and/or hyperCKaemia, we would have expected to identify GAA variants in our cohort as reported in previous studies [22, 23]. Highly effective treatments are also available for congenital myasthenic syndromes (CMS). There are now over 30 causative genes described in the context of CMS, 10 of which have been reported to generally present with a limb‐girdle pattern of weakness [28]. CMS patients can therefore often be found in large cohorts of patients with LGW [6]. One CMS patient with pathogenic variants in GMPPB was also identified in our study, the subsequent treatment of whom with salbutamol led to a significant clinical improvement. Moreover, although specific therapies were unavailable for the remainder of the cohort, the identification of molecular diagnoses enabled genetic counselling of patients and their families, as well as the inclusion of patients in registries to facilitate trial readiness, or helped to improve the clinical management, for example, by reducing the risk of malignant hyperthermia in RYR1‐associated disease.
In conclusion, our study provides the first nationwide clinico‐genetic characterization of a large cohort of patients with LGW in Austria. We present parameters predictive of the identification of molecular diagnoses that could be helpful for the selection of patients for genetic analyses in centres with limited access to sequencing facilities. Our data support the early application of NGS‐based methods because of the extensive phenotypic overlap and the wide spectrum of different genetic aetiologies associated with LGW.
CONFLICT OF INTEREST
The authors report no conflict of interest related to this article.
AUTHOR CONTRIBUTION
Martin Krenn: Data curation (supporting); Formal analysis (lead); Methodology (equal); Visualization (supporting); Writing – original draft (equal). Matthias Tomschik: Data curation (supporting); Writing – review and editing (supporting). Matias Wagner: Formal analysis (supporting); Methodology (supporting); Writing – review and editing (supporting). Gudrun Zulehner: Data curation (supporting); Writing – review and editing (supporting). Rosa Weng: Data curation (supporting); Writing – review and editing (supporting). Jakob Rath: Data curation (supporting); Writing – review and editing (supporting). Sigrid Klotz: Data curation (supporting); Methodology (supporting); Writing – review and editing (supporting). Ellen Gelpi: Data curation (supporting); Formal analysis (supporting); Methodology (supporting); Writing – review and editing (supporting). Gabriel Bsteh: Formal analysis (supporting); Methodology (supporting); Writing – review and editing (supporting). Omar Keritam: Data curation (supporting); Writing – review and editing (supporting). Isabella Colonna: Data curation (supporting); Writing – review and editing (supporting). Chiara Paternostro: Data curation (supporting); Writing – review and editing (supporting). Fiona Jaeger: Data curation (supporting); Writing – review and editing (supporting). Elisabeth Lindeck‐Pozza: Data curation (supporting); Writing – review and editing (supporting). Stephan Iglseder: Data curation (supporting); Writing – review and editing (supporting). Susanne Grinzinger: Data curation (supporting); Writing – review and editing (supporting). Martina Schönfelder: Data curation (supporting); Writing – review and editing (supporting). Christina Hohenwarter: Data curation (supporting); Writing – review and editing (supporting). Manfred Freimüller: Data curation (supporting); Writing – review and editing (supporting). Norbert Embacher: Data curation (supporting); Writing – review and editing (supporting). Julia Wanschitz: Data curation (supporting); Writing – review and editing (supporting). Raffi Topakian: Data curation (equal); Writing – review and editing (supporting). Ana Töpf: Data curation (supporting); Formal analysis (supporting); Writing – review and editing (supporting). Volker Straub: Formal analysis (equal); Methodology (equal); Writing – review and editing (equal). Stefan Quasthoff: Data curation (equal); Writing – review and editing (supporting). Fritz Zimprich: Data curation (equal); Writing – review and editing (equal). Wolfgang Loescher: Conceptualization (supporting); Data curation (equal); Formal analysis (equal); Project administration (supporting); Writing – review and editing (equal). Hakan Cetin: Conceptualization (lead); Data curation (equal); Formal analysis (equal); Funding acquisition (lead); Methodology (equal); Project administration (lead); Visualization (lead); Writing – original draft (supporting); Writing – review and editing (lead).
Supporting information
ACKNOWLEDGEMENTS
The authors would like to thank the Austrian Society of Neurology (ÖGN) for the nationwide coordination and distribution of study‐related information between collaborating neuromuscular centres.
Krenn M, Tomschik M, Wagner M, et al. Clinico‐genetic spectrum of limb‐girdle muscular weakness in Austria: A multicentre cohort study. Eur J Neurol. 2022;29:1815–1824. doi: 10.1111/ene.15306
Funding information
The study was financially supported by Sanofi Genzyme
DATA AVAILABILITY STATEMENT
Anonymized data not published in this article will be made available by request from the corresponding author.
REFERENCES
- 1. Mercuri E, Muntoni F. Muscular dystrophies. Lancet. 2013;381(9869):845‐860. [DOI] [PubMed] [Google Scholar]
- 2. Thompson R, Straub V. Limb‐girdle muscular dystrophies ‐ international collaborations for translational research. Nat Rev Neurol. 2016;12(5):294‐309. [DOI] [PubMed] [Google Scholar]
- 3. Liu W, Pajusalu S, Lake NJ, et al. Estimating prevalence for limb‐girdle muscular dystrophy based on public sequencing databases. Genet Med. 2019;21(11):2512‐2520. [DOI] [PubMed] [Google Scholar]
- 4. Norwood FL, Harling C, Chinnery PF, Eagle M, Bushby K, Straub V. Prevalence of genetic muscle disease in Northern England: in‐depth analysis of a muscle clinic population. Brain. 2009;132(Pt 11):3175‐3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Savarese M, Di Fruscio G, Torella A, et al. The genetic basis of undiagnosed muscular dystrophies and myopathies: results from 504 patients. Neurology. 2016;87(1):71‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Topf A, Johnson K, Bates A, et al. Sequential targeted exome sequencing of 1001 patients affected by unexplained limb‐girdle weakness. Genet Med. 2020;22(9):1478‐1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chu ML, Moran E. The limb‐girdle muscular dystrophies: is treatment on the horizon? Neurotherapeutics. 2018;15(4):849‐862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krenn M, Tomschik M, Rath J, et al. Genotype‐guided diagnostic reassessment after exome sequencing in neuromuscular disorders: experiences with a two‐step approach. Eur J Neurol. 2020;27(1):51‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17(5):405‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Council NR. The prevention and treatment of missing data in clinical trials. National Academies Press; 2010. [PubMed] [Google Scholar]
- 11. Fichna JP, Macias A, Piechota M, et al. Whole‐exome sequencing identifies novel pathogenic mutations and putative phenotype‐influencing variants in Polish limb‐girdle muscular dystrophy patients. Hum Genomics. 2018;12(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghaoui R, Cooper ST, Lek M, et al. Use of whole‐exome sequencing for diagnosis of limb‐girdle muscular dystrophy: outcomes and lessons learned. JAMA Neurol. 2015;72(12):1424‐1432. [DOI] [PubMed] [Google Scholar]
- 13. Harris E, Topf A, Barresi R, et al. Exome sequences versus sequential gene testing in the UK highly specialised service for limb girdle muscular dystrophy. Orphanet J Rare Dis. 2017;12(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuhn M, Glaser D, Joshi PR, et al. Utility of a next‐generation sequencing‐based gene panel investigation in German patients with genetically unclassified limb‐girdle muscular dystrophy. J Neurol. 2016;263(4):743‐750. [DOI] [PubMed] [Google Scholar]
- 15. Monies D, Alhindi HN, Almuhaizea MA, et al. A first‐line diagnostic assay for limb‐girdle muscular dystrophy and other myopathies. Hum Genomics. 2016;10(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nallamilli BRR, Chakravorty S, Kesari A, et al. Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients. Ann Clin Transl Neurol. 2018;5(12):1574‐1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hicks D, Sarkozy A, Muelas N, et al. A founder mutation in Anoctamin 5 is a major cause of limb‐girdle muscular dystrophy. Brain. 2011;134(Pt 1):171‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dardas Z, Swedan S, Al‐Sheikh Qassem A, Azab B. The impact of exome sequencing on the diagnostic yield of muscular dystrophies in consanguineous families. Eur J Med Genet. 2020;63(4):103845. [DOI] [PubMed] [Google Scholar]
- 19. Ankala A, da Silva C, Gualandi F, et al. A comprehensive genomic approach for neuromuscular diseases gives a high diagnostic yield. Ann Neurol. 2015;77(2):206‐214. [DOI] [PubMed] [Google Scholar]
- 20. Bharucha‐Goebel DX, Neil E, Donkervoort S, et al. Intrafamilial variability in GMPPB‐associated dystroglycanopathy: broadening of the phenotype. Neurology. 2015;84(14):1495‐1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zima J, Eaton A, Pal E, et al. Intrafamilial variability of limb‐girdle muscular dystrophy, LGMD1D type. Eur J Med Genet. 2020;63(2):103655. [DOI] [PubMed] [Google Scholar]
- 22. Lukacs Z, Nieves Cobos P, Wenninger S, et al. Prevalence of Pompe disease in 3,076 patients with hyperCKemia and limb‐girdle muscular weakness. Neurology. 2016;87(3):295‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gutierrez‐Rivas E, Bautista J, Vilchez JJ, et al. Targeted screening for the detection of Pompe disease in patients with unclassified limb‐girdle muscular dystrophy or asymptomatic hyperCKemia using dried blood: a Spanish cohort. Neuromuscul Disord. 2015;25(7):548‐553. [DOI] [PubMed] [Google Scholar]
- 24. Musumeci O, la Marca G, Spada M, et al. LOPED study: looking for an early diagnosis in a late‐onset Pompe disease high‐risk population. J Neurol Neurosurg Psychiatry. 2016;87(1):5‐11. [DOI] [PubMed] [Google Scholar]
- 25. Johnson K, Topf A, Bertoli M, et al. Identification of GAA variants through whole exome sequencing targeted to a cohort of 606 patients with unexplained limb‐girdle muscle weakness. Orphanet J Rare Dis. 2017;12(1):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Toscano A, Schoser B. Enzyme replacement therapy in late‐onset Pompe disease: a systematic literature review. J Neurol. 2013;260(4):951‐959. [DOI] [PubMed] [Google Scholar]
- 27. Loscher WN, Huemer M, Stulnig TM, et al. Pompe disease in Austria: clinical, genetic and epidemiological aspects. J Neurol. 2018;265(1):159‐164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Connor E, Topf A, Zahedi RP, et al. Clinical and research strategies for limb‐girdle congenital myasthenic syndromes. Ann N Y Acad Sci. 2018;1412(1):102‐112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized data not published in this article will be made available by request from the corresponding author.


