Summary
Legumes usually have compound inflorescences, where flowers/pods develop from secondary inflorescences (I2), formed laterally at the primary inflorescence (I1). Number of flowers per I2, characteristic of each legume species, has important ecological and evolutionary relevance as it determines diversity in inflorescence architecture; moreover, it is also agronomically important for its potential impact on yield. Nevertheless, the genetic network controlling the number of flowers per I2 is virtually unknown.
Chickpea (Cicer arietinum) typically produces one flower per I2 but single flower (sfl) mutants produce two (double‐pod phenotype). We isolated the SFL gene by mapping the sfl‐d mutation and identifying and characterising a second mutant allele. We analysed the effect of sfl on chickpea inflorescence ontogeny with scanning electron microscopy and studied the expression of SFL and meristem identity genes by RNA in situ hybridisation.
We show that SFL corresponds to CaRAX1/2a, which codes a MYB transcription factor specifically expressed in the I2 meristem.
Our findings reveal SFL as a central factor controlling chickpea inflorescence architecture, acting in the I2 meristem to regulate the length of the period for which it remains active, and therefore determining the number of floral meristems that it can produce.
Keywords: chickpea breeding, chickpea double‐pod mutants, compound inflorescence, inflorescence architecture, meristem activity, R2R3‐MYB genes, RAX genes, seed yield
Introduction
Inflorescence architecture is a key trait, ecologically and evolutionary relevant, as it strongly influences pollination and fruit set and determines plant form (Wyatt, 1982; Weberling, 1992; Benlloch et al., 2007); it is an important relevant characteristic in agriculture, because it strongly influences fruit and seed production (Wang & Li, 2008). Inflorescence architecture depends on the identity of the meristems in the inflorescence apex, which determines the position in the inflorescence axes where flowers appear and on the activity of the inflorescence meristems, which controls how many flowers are produced (Prusinkiewicz et al., 2007; Teo et al., 2014; Benlloch et al., 2015).
For simple inflorescences, as in Arabidopsis, flowers are formed at the primary inflorescence (I1) stem; however, in compound inflorescences, as in legumes, flowers appear on secondary inflorescence (I2) stems, formed in lateral positions of the primary inflorescence (Fig. 1a,b; Supporting Information Fig. S1; Benlloch et al., 2015). Development of the legume compound inflorescence depends on the MADS domain transcription factor VEGETATIVE1/MtFULc (VEG1), which specifies I2 meristem identity (Berbel et al., 2012; Cheng et al., 2018), and PROLIFERATING INFLORESCENCE MERISTEM/MtPIM (PIM), a homologue to Arabidopsis APETALA1 (AP1), which specifies floral meristem identity (Berbel et al., 2001; Taylor et al., 2002; Benlloch et al., 2006). Legume I2 meristems are generally short‐lived meristems that produce some floral meristems before terminating as a stub (Fig. 1e).
Fig. 1.
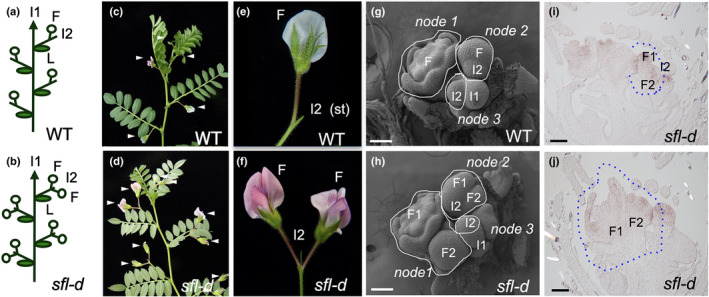
Double‐pod phenotype in chickpea (Cicer arietinum) caused by the mutation in the SINGLE FLOWER (SFL) gene and ontogeny of the inflorescence of the sfl‐d mutant. (a, b) Diagrams of wild‐type (WT) and sfl‐d chickpea plants. Flowers (F) develop at secondary inflorescences (I2) that are formed in the axil of the leaves (L) of the primary inflorescence (I1) stem. Wild‐type I2s (a) produce one flower, whereas sfl‐d I2s (b) produce two flowers. (c, d) Wild‐type and sfl‐d chickpea plants. Arrowheads mark individual flowers formed at the I2s of the wild‐type (c) and two flowers in the sfl‐d (d) I2s. (e) Close‐up of a wild‐type I2, in which the stub (st) is marked. (f) Close‐up of a sfl‐d I2. (g) Scanning electron micrograph (SEM) of the inflorescence apex of a wild‐type plant. In each I2 node one flower is found. (h) Scanning electron micrograph of the inflorescence apex of a sfl‐d plant. In the I2 nodes two flowers (at different developmental stages) are found. (i, j) In situ hybridisation of CaPIM mRNA in inflorescence apices of the sfl‐d mutant, in which each I2 node bears two flowers at different developmental stages. (g–j) Bar, 100 µm.
The number of flowers produced by the I2 meristem is an important developmental trait for at least two reasons. First, it is a key factor that creates diversity in inflorescence architecture, being the number of flowers per I2 characteristic of each legume species and variety. For instance, while Pisum sativum (pea) and Medicago truncatula I2s produce one or two flowers, Medicago sativa (alfalfa) I2s produce 8–12 flowers and Vicia cracca (cow vetch) I2s produce dozens of flowers (Fig. S1; Benlloch et al., 2015). Second, the number of flowers per I2 influences the number of pods produced by the plant, with the potential to positively impact on yield. In fact, in both pea and chickpea, some studies have found a correlation between pod number per I2 and seed yield, or with yield stability, supporting a positive effect of the multipod trait on crop performance (Milbourne & Hardwick, 1968; Sheldrake et al., 1978; French, 1990; Kumar et al., 2000; Rubio et al., 2004; Devi et al., 2018).
Mutations whose only effect is to increase the number of flowers/pods per I2 (multipod phenotype) have been described in several legumes, such as pea, Cicer arietinum (chickpea) and, more recently, in Lens culinaris (lentil) (White, 1917; Lamprecht, 1947; Singer et al., 1999; Srinivasan et al., 2006; Mishra et al., 2020), indicating the existence of genes that specifically regulate this trait.
Despite the importance of the number of flowers per I2 in legumes, virtually nothing is known about how it is genetically controlled, and no gene specifically related with this trait has been isolated. In chickpea, two loci, CYMOSE (CYM) and SINGLE FLOWER (SFL), have been described for which recessive mutations specifically increase the number of flowers in the I2 (Srinivasan et al., 2006). While most chickpea genotypes produce one flower per I2 (‘wild‐type’; Fig. 1a,c,e) (Prenner, 2012), mutations in the SFL gene lead to plants whose only evident phenotype is the production of two flowers/pods per I2 (double‐pod trait; Fig. 1b,d,f) (Srinivasan et al., 2006). SINGLE FLOWER was recently fine mapped to a 92.6 kb region of chromosome 6 (Ali et al., 2016).
Here, we characterised the sfl‐d mutant, analysing the ontogeny of its inflorescence and the expression of a floral meristem gene. Then, we identified SFL as a homologue of the Arabidopsis RAX1/2 genes, encoding MYB transcription factors, and investigated how it functions by studying its expression in the chickpea inflorescence apex compared with other inflorescence meristem genes. Our findings revealed that SFL plays a central role in the control of inflorescence architecture of chickpea, specifically acting in the I2 meristem to control the time period for which it stays active, and therefore determining the number of floral meristems that it can produce.
Materials and Methods
Plant material
JG62 (syn. ICC 4951), an Indian double‐pod chickpea (C. arietinum) landrace, maintained by ICRISAT (Hyderabad, India, icrisat.org), was parental in a cross with CA2156 (single pod) to develop pairs of nearly isogenic lines (NILs) used to map SFL (Ali et al., 2016). Six double‐pod genotypes from USDA (Beltsville, MD, USA, https://npgsweb.ars‐grin.gov), AOS1, CA2969, ICC1083, LINE6560, LINE6581 and RPIP12‐069‐06223 were used to identify a second sfl mutant allele.
Single‐pod genotypes were kabuli type. CA2156 is a Spanish cultivar, ILC3279 a Russian landrace maintained by ICARDA and BT6‐17 is an advanced line from our breeding programme at IFAPA‐UCO.
Controlled crosses
Genetic crosses were performed as described previously (Caballo et al., 2018).
Genotyping and sequencing
DNA was isolated using the DNeasy Plant Mini Kit (Qiagen). PCRs were performed using Phusion High‐Fidelity DNA polymerase (Thermo Scientific™, Madrid, Spain). PCR products were analysed using nondenaturing polyacrylamide electrophoresis gels. Primers used in this work are listed in Table S1. PCR products were sequenced either directly after purification with SureClean (Bioline, London, UK) or after cloning in pGEMTeasy (Promega).
Bioinformatics analysis
To identify sequence differences between double‐pod and single‐pod genotypes, the genome of the double‐podded accession no. JG62 was re‐sequenced using Ion‐Torrent, and mapped against the chickpea reference genome (from the single‐podded cultivar CDC‐Frontier; Varshney et al., 2013). Binary Alignment Map (BAM) files were inspected using Geneious ® 8 software to detect polymorphism in relevant regions of the genome between both lines. To infer the extension of the deletion affecting CaRAX2‐like, all reads assembled to a region of chromosome 6 spanning 100 kb and containing genes LOC101505360, LOC101505694, LOC101506220 (CaRAX2‐like), LOC101506550, LOC101490413, LOC101490737 and LOC101507108 were extracted and re‐assembled using geneious mapper and the following options: minimum mapping quality = 30; maximum gaps per read = 10%; maximum gap size = 50 000; word length = 17 and Index word length = 14; maximum mismatches per read = 15%; maximum ambiguity = 2.
For phylogenetic trees, sequences of 117 Arabidopsis thaliana R2R3‐MYB proteins were used to retrieve 1110 R2R3‐MYB proteins from 13 plant species using Blastp with an E‐value threshold of 1e‐40 (Table S2). The Capsicum annuum BLIND protein (NP.001311565.1) was also included for subsequent analyses. The sequences of the R2 and R3 domains of these 1228 proteins were aligned in geneious prime 2020.2.2 (http://www.geneious.com) software using mafft (Katoh et al., 2002) with the following options: algorithm = FFT‐NSI‐i × 1000; scoring matrix = BLOSUM30; gap open penalty = 3; offset value = 0.128. An approximately‐maximum‐likelihood phylogenetic tree was then derived from this alignment using FastTree v.2.1.12 plugin (Price et al., 2010) on geneious prime, with the default settings and a bootstrap support of 1000 rep (Fig. S3). Based on this tree, the complete sequence of a subset of 29 proteins belonging to the S14 subfamily were aligned with mafft, and the evolutionary history was inferred in Mega X (Kumar et al., 2018) using the maximum‐likelihood method, Le and Gascuel model (Le & Gascuel, 2008), and a bootstrap of 1000 replications. Sequences of Arabidopsis MYB35 and MYB80 were used as outgroups. Accessions numbers and information on the sequences used in this analysis are available in Table S2.
Scanning electron microscopy
Inflorescence apices were fixed in FAE (50% ethanol, 3.7% formaldehyde, 5% glacial acetic acid) at 4°C overnight in the dark. Samples were dehydrated with ethanol and critical point dried in liquid CO2 (CPD300; Leica, Wetzlar, Germany). Dried samples were sputtercoated with argon–platinum plasma at a distance of 6–7 cm and 45 mA intensity for 15 s in a sputtering chamber (Leica Microsystems EM MED020). Scanning electron microscopy (SEM) micrographs were acquired using an AURIGA compact FIB‐SEM (Zeiss, http://www.zeiss.com/) at EHT = 1–2 kV.
Quantitative real‐time PCR
For expression analysis with quantitative real‐time PCR (RT‐qPCR) of the wild‐type and the sfl‐d mutant, samples collected from different tissues (roots, stem, leaves, vegetative apices) from plants of the NIL5‐1V line (nearly isogenic line 5, single pod), after producing three or four leaves, and from inflorescence apices of NIL5‐1V and NIL5‐2V (double pod), were used. Total RNA was extracted using the E.Z.N.A® Plant RNA Kit (OMEGA Bio‐Tek, Norcross, GA, USA). Next, 1.5 μg of total RNA, previously treated with DNase I (Turbo DNA‐free Kit; Thermo Fisher Scientific Baltics UAB, Vilnius, Lithuania) was retrotranscribed using the First Strand cDNA Synthesis Kit (Invitrogen), and RT‐qPCR was carried out using the Evagreen Master Mix (Cultex, Madrid, Spain). PCRs were run and analysed using the Quant Studio3 Real‐time PCR system (Applied Biosystems, Foster City, CA, USA). The C. arietinum UBIQUITIN (UBQ, AJ001901) gene was used as an internal reference (Castro et al., 2012). Calculations of each sample were done according the comparative ΔΔCT method (Livak & Schmittgen, 2001). Expression analyses were performed in three biological replicates of pooled samples, each with three technical replicates. Primers (Table S1) were designed with the Primer Express™ v.3.0 software (Applied Biosystems).
In situ hybridisation
RNA in situ hybridisation with digoxigenin‐labelled probes was performed on 8‐µm longitudinal paraffin sections of chickpea shoot apices as described previously (Ferrándiz et al., 2000). RNA antisense and sense probes were generated using, as substrate, specific fragments of CaRAX1/2a, CaRAX1/2b, CaVEG1, CaPIM or CaUNI, amplified by PCR from chickpea inflorescence cDNA and cloned into the pGEM‐T Easy vector (Promega). Information on the primers used to generate the cDNA fragments for probes is provided in Table S1. The sequences of CaVEG1 (XM_004491849), CaPIM (XM_004509697) and CaUNI (XM_004501703), which share 86.4%, 96.2% and 87.9% amino acid identity with VEG1, PIM and UNI (Hofer et al., 1997; Taylor et al., 2002; Berbel et al., 2012) from pea, respectively, were retrieved from the chickpea genome database. For CaRAX1/2b, CaPIM, CaVEG1 and CaUNI, transcription of antisense and sense probes was carried with SP6 or T7 polymerases, after linearising with NcoI or SalI, respectively. Transcription of CaRAX1/2a AS probe was carried out using T7 polymerase, after linearising with SalI. Signal was viewed as a purple precipitate under a light microscope.
Results
Effect of the sfl‐d mutation on chickpea development
As previously mentioned, the I2 of wild‐type genotypes produce one flower before terminating into a stub (Fig. 1a,c,e). However, in genotypes with homozygous mutations in the SINGLE FLOWER (SFL) gene, the I2s produce two flowers/pods and a stub (Fig. 1b,d,f) (Srinivasan et al., 2006).
The production of two flowers in double‐pod genotypes suggests that the I2 meristem is either larger and divides to produce more flowers, or is active for longer and forms more flowers in a sequential manner. With SEM we analysed inflorescence ontogeny of the pair of nearly isogenic lines NIL5‐1V (single pod) and NIL5‐2V (double pod), derived from a cross with the sfl‐d parental JG62. In inflorescence apices of the single‐pod plants, each node showed an I2 meristem from which only a floral primordium initiated (Fig. 1g). In inflorescence apices of double‐pod plants, I2 meristems seemed to have a similar size to the wild‐type but, in each I2 node, two flowers at a different stages of development were observed (Fig. 1h).
In situ hybridisation of double‐pod inflorescence apices probed with the floral marker CaPIM (a homologue of the floral meristem identity genes PIM and AP1) showed the presence of two flowers at different developmental stages of the I2 nodes (Fig. 1i,j), confirming the result of the SEM analysis.
Therefore, the two floral meristems produced by the double‐pod I2s are initiated sequentially, indicating that these I2 meristems produce more flowers compared with the wild‐type I2 meristems because they are active for longer.
Identification of candidates for the SFL gene
The sfl‐d mutation had been previously mapped to a 92.6 kb region of chromosome 6, with seven annotated genes that code for two uncharacterised proteins, four enzymes and a MYB transcription factor, CaRAX2‐like (Fig. 2a,b,c) (Ali et al., 2016). When primers for these genes were tested using PCR in single‐pod lines they amplified fragments of the expected size (Fig. 2a). By contrast, in the double‐pod line JG62, no amplification of LOC101506550 (N‐lysine methyltransferase‐like) or LOC101506220 (CaRAX2‐like) genes was observed (Fig. 2a). As the genes encoding CaRAX2‐like and N‐lysine methyltransferase are adjacent, this suggested the existence of a deletion in the 92.6 kb SFL mapping interval that affects these genes in the sfl‐d mutants.
Fig. 2.
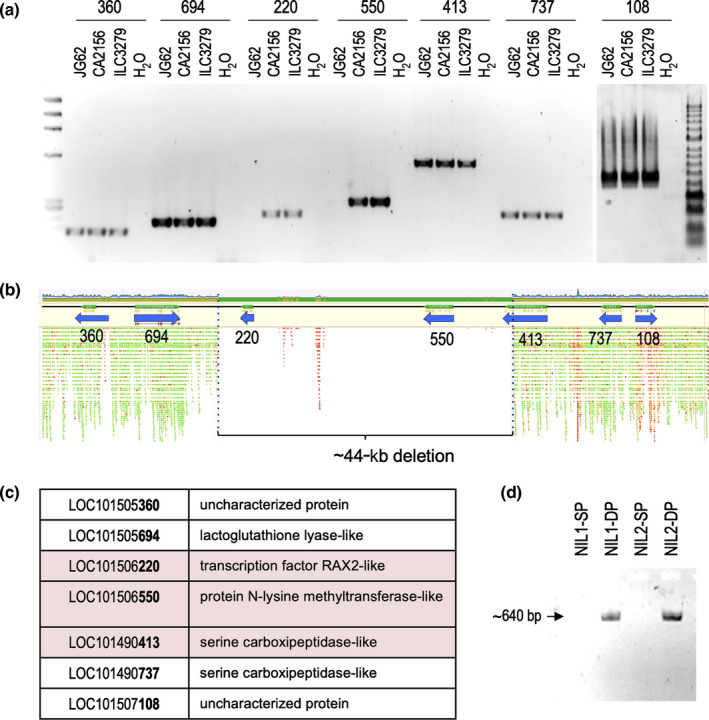
Deletion in the 92.6‐kb SFL mapping interval. (a) PCR amplification with primers for the seven genes in the 92.6‐kb SFL mapping in DNA from the chickpea (Cicer arietinum) double‐pod line JG62 and the single‐pod lines CA2156 and ILC3279. (b) Mapping of sequencing reads of the JG62 re‐sequencing against the genome of the reference single‐pod line CDC‐Frontier in the 92.6‐kb SFL mapping interval, showing a deletion affecting three genes. (c) List of the genes contained in the 92.6‐kb SFL mapping interval. Genes affected by the deletion are highlighted in pink. (d) PCR amplification with primers at the limits of the deletion in two pairs of single‐pod (SP) or double‐pod (DP) nearly isogenic lines (NIL1, NIL2).
Therefore, we examined the mapping quality of the JG62 (sfl‐d) re‐sequencing against the reference genome in this region of chickpea chromosome 6. We found a region of c. 44 kb with an unusually low density of reads (Fig. 2b). We then extracted all the reads mapped to a 100 kb region containing the SFL mapping interval (Ali et al., 2016), and performed a new mapping against the same reference but allowing a within‐read gap of 50 kb, which allowed us to infer the deletion breakpoints (Figs 2, S2). As expected, the predicted 44‐kb deletion included the genes CaRAX2‐like and N‐lysine methyltransferase‐like genes; it also includes part of the gene LOC101490413 (serine carboxypeptidase‐like) (Fig. 2b,c). To validate the existence of the deletion in the double‐pod genotype JG62, we tested PCR primers from the limits of the deletion in single/double‐pod genotypes. In double‐pod genotypes, a fragment of the expected size (642 bp) was amplified (Fig. 2d), which was latter sequenced to confirm the deletion borders. Conversely, as expected, no amplification product was obtained in single‐pod genotypes.
These results showed the existence of a deletion affecting three genes in the mapping region of the SFL gene, only in double‐pod genotypes. This points to these genes as the most likely candidates for the double‐pod phenotype in sfl‐d mutants.
Identification and analysis of a new double‐pod mutant allele confirms CaRAX2‐like as the SFL gene
To assess whether any of these three genes was in fact responsible for the double‐pod phenotype of the sfl‐d mutant lines, we looked for new sfl mutant alleles. We analysed six chickpea double‐pod accessions from the USDA collection (Fig. 3a). Two primers pairs designed for the 44‐kb deletion and for the CaRAX2‐like gene were used to test the double‐pod USDA genotypes. Three of the six USDA genotypes (ICC1083, RPIP12‐069‐06223 and CA2969) contained the 44‐kb deletion (Fig. 3b), indicating that, in these genotypes, the double‐pod mutation was the same as in JG62. However, the other three double‐pod genotypes, LINE6560, AOS1 and LINE6581, did not contain the deletion, but showed amplification with the CaRAX2‐like primers (Fig. 3b). This suggested that the double‐pod phenotype of these three last genotypes could be due to mutations in the SFL gene different to that in JG62.
Fig. 3.
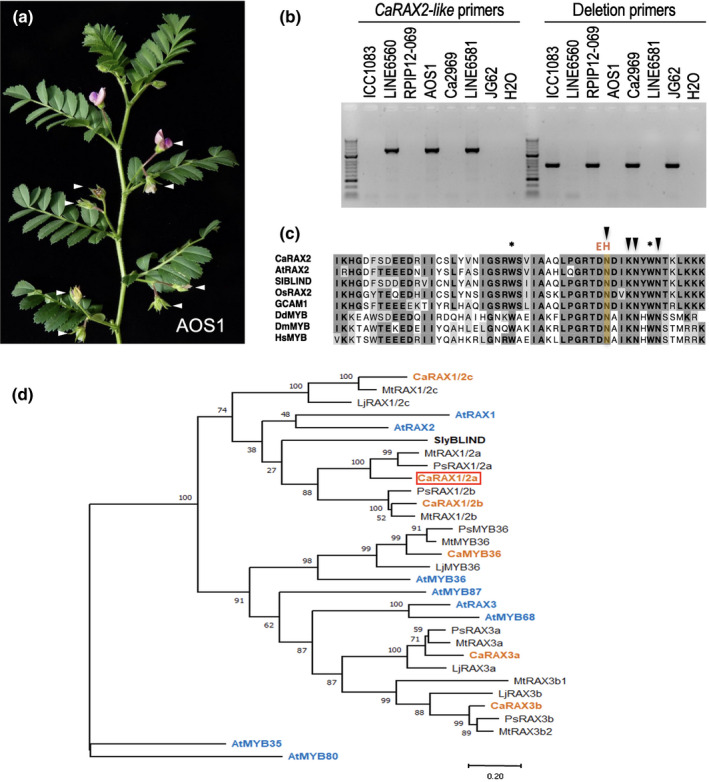
CaRAX2‐like is mutated in a new mutant allele of SFL and a phylogenetic tree of legume RAX proteins. (a) Double‐pod phenotype of a chickpea (Cicer arietinum) AOS1 plant. Arrowheads mark the flowers. (b) PCR amplification in the USDA double‐pod lines with primer pairs at CaRAX2‐like or at the limits of the deletion. (c) clustalw alignment of the R3 repeat of representative R2R3‐MYB proteins from plants, microorganisms and animals. At, Arabidopsis thaliana; Ca, Cicer arietinum; Dd, Dictyostelium discoideum; Dm, Drosophila melanogaster; GCMA1, Marchantia polymorpha; Hs, Homo sapiens; Os, Oriza sativa; Sl, Solanum lycopersicum. Asterisks mark conserved tryptophan residues. Arrowheads mark base‐contacting residues of the mouse homologue of HsMYB. (d) Phylogenetic tree of subgroup 14 MYB proteins from Arabidopsis and legumes. Legume proteins have been named after their Arabidopsis homologues. CaRAX2‐like/CaRAX1/2a is framed in red. At, Arabidopsis thaliana; Ca, Cicer arietinum (chickpea); Lj, Lotus japonicus; Mt, Medicago truncatula; Ps, Pisum sativum (pea); Sl, Solanum lycopersicum. Accession numbers of the genes in the phylogenetic tree can be found in Supporting Information Table S2.
To determine the allelic relationship of the double‐pod mutation in the USDA lines with the sfl‐d double‐pod mutation of JG62, we crossed JG62 with the USDA double‐pod lines AOS1 and LINE6560. F1 plants from these crosses exhibited a double‐pod phenotype (Fig. S3a,b), indicating no complementation. Moreover, F1 plants derived from the cross between the double‐pod line AOS1 × single‐pod line BT6‐17 exhibited a single‐pod phenotype (Fig. S3c), confirming that the double‐pod mutation in the AOS1 line is recessive. These results indicated that the USDA lines AOS1 and LINE6560 bore mutation(s) in the SFL gene allelic to the sfl‐d mutation in JG62; this new allele was named sfl‐3.
The three genes affected by the 44‐kb deletion present in JG62 and other related double‐pod genotypes were sequenced in the USDA lines and aligned against the chickpea reference sequence. In the three double‐pod USDA genotypes, both LOC101506550 (N‐lysine methyltransferase‐like) and LOC101490413 (serine carboxypeptidase‐like) genes had a sequence identical to the reference, while the LOC101506220 (CaRAX2‐like) gene had a sequence variant identical in the three lines. In the USDA lines, bases 306 and 307 of the CaRAX2‐like coding sequence replaced CA in the reference with AC in the double‐pod lines. This changes the amino acids 102 and 103 from aspartate–asparagine in the wild‐type reference to glutamate–histidine in the double‐pod lines (Fig. 3c).
CaRAX2‐like encodes a R2R3‐MYB transcription factor with sequence similarity to REGULATOR OF AXILLARY MERISTEMS (RAX) genes from Arabidopsis (Keller et al., 2006; Müller et al., 2006). Alignment of the sequence of CaRAX2‐like from the sfl‐3 mutant lines with other related MYB proteins showed that the mutation in sfl‐3 mutants was located in the R3 repeat of the conserved MYB DNA‐binding domain (Fig. 3c; Dubos et al., 2010). The amino acids affected by the sfl‐3 mutation in CaRAX2‐like, aspartate–asparagine, are conserved in plant RAX proteins, but are also conserved in MYB proteins from other organisms, from yeast to humans (Fig. 3c). Moreover, in the mouse homologue of the human c‐MYB protein, this asparagine residue was shown to directly contact DNA (Ogata et al., 1994; Martin & Paz‐Ares, 1997) and therefore is presumably critical for function, strongly suggesting that the sfl‐3 mutation significantly affected the activity of the CaRAX2‐like protein.
Phylogenetic analysis confirmed that CaRAX2‐like groups with the subgroup 14 of the R2R3‐MYB family of transcription factors (Fig. S4a), in Arabidopsis include three RAX proteins, RAX1, 2 and 3, and AtMYB36, AtMYB87 and AtMYB68. (Figs 3d, S4b; Dubos et al., 2010). R2R3‐MYB transcription factors, and RAX proteins in particular, are a family and a subgroup, respectively, with a high number of members (Feng et al., 2017; Romani & Moreno, 2021). The subgroup 14 includes six proteins in chickpea, analogous to other legume species analysed in our tree and similar to the number of proteins present in Arabidopsis, indicating conservation in copy number between these species (Fig. 3d). CaRAX2‐like belongs to the same clade as Arabidopsis AtRAX1 and AtRAX2 and the tomato Blind protein (Figs 3d, S4b; Schmitz et al., 2002; Dubos et al., 2010), although it groups with a separate legume specific clade. As CaRAX2‐like seems similarly close to both Arabidopsis proteins, CaRAX2‐like was renamed as CaRAX1/2a (Fig. 3d). In addition, the chickpea genome codes for two homologues to CaRAX1/2a /SFL: CaRAX1/2b and CARX1/2c. These three CaRAX proteins show high amino acid identity in the MYB domain (84–89%) although lower in the rest of the protein (13–30%) (Table S3).
In summary, the fact that two independent allelic mutations in the CaRAX1/2a gene, a complete deletion and a probable loss‐of‐function, associate with the double‐pod mutant phenotype strongly indicates that CaRAX1/2a corresponds to the SFL gene, responsible for the double‐pod trait.
CaRAX1/2a/SFL acts in the I2 meristem controlling its activity
SINGLE FLOWER regulates the number of flowers produced by the I2. To learn about where the CaRAX1/2a/SFL gene acts, we analysed its expression in different chickpea tissues using RT‐qPCR. This analysis showed that, among aerial tissues, the highest expression was in floral apices. High expression was also found in roots (Fig. S5a).
To know in detail where the CaRAX1/2a/SFL gene is expressed in inflorescence apices, we performed in situ hybridisation with CaRAX1/2a on wild‐type and sfl‐d apices. Inflorescence apices of the sfl‐d mutant showed no signal when hybridised with a probe for CaRAX1/2a (Fig. 4a,b), as expected because of the deletion in sfl‐d. In wild‐type inflorescence apices hybridised with CaRAX1/2a, the signal was observed in I2 meristems, as confirmed with markers for I2 and floral meristems. Contiguous sections hybridised with probes for CaRAX1/2a or CaVEG1 (orthologue of the I2 meristem gene VEG1; Berbel et al., 2012), essentially exhibited the same pattern, showing that CaRAX1/2a is expressed throughout the I2 meristem (Fig. 4a). Moreover, VEG1 expression was not affected in the sfl mutant, suggesting that SFL does not affect specification of I2 meristem identity.
Fig. 4.
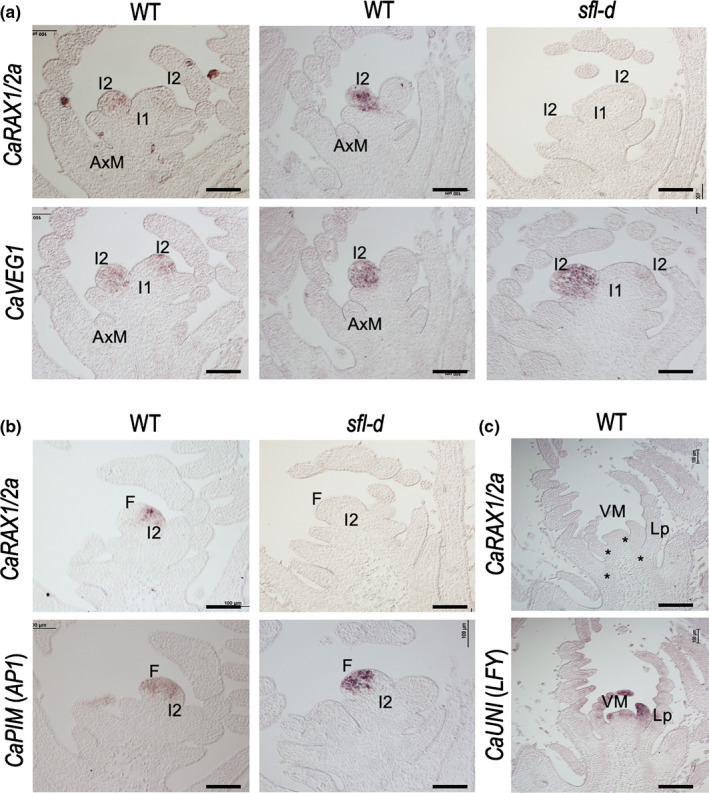
Expression pattern of the CaRAX1/2a/SFL gene in shoot apices of chickpea (Cicer arietinum). (a) In situ hybridisation of CaRAX1/2a mRNA (upper row) and of the secondary inflorescence meristem (I2) marker CaVEG1 mRNA (lower row) in contiguous sections of inflorescence shoot apices of wild‐type (WT) or sfl‐d plants. AxM, vegetative axillary meristem; I1, primary inflorescence meristem; I2, secondary inflorescence. (b) In situ hybridisation of CaRAX1/2a mRNA (upper row) and of the floral meristem (F) marker CaPIM mRNA (lower row) in contiguous sections of inflorescence shoot apices of wild‐type or sfl‐d plants. (c) In situ hybridisation of CaRAX1/2a mRNA (upper) and of the leaf (and floral) marker CaUNI mRNA (lower) in contiguous sections of vegetative shoot apices of a wild‐type plant. Asterisks in the top image mark leaf axils. Lp, leaf primordium; VM, vegetative shoot apical meristem. Bar, 100 μm.
By contrast, contiguous sections of wild‐type inflorescence apices hybridised with probes for CaRAX1/2a or CaPIM (orthologue of PIM, floral meristem gene; Taylor et al., 2002) exhibited a complementary pattern, in which CaPIM was detected in the floral meristem and CaRAX1/2a was detected in the I2 meristem (Fig. 4b). Expression of CaRAX1/2a and CaPIM was not completely exclusive, as some overlap of the CaRAX1/2a and CaPIM signal was found at the boundary between the I2 and the floral meristems (Fig. 4b).
In contrast with RAX genes from Arabidopsis and tomato (Müller et al., 2006; Busch et al., 2011), we did not detect the expression of CaRAX1/2a in vegetative axillary meristems or leaf axils at the vegetative apex (Fig. 4a,c).
These results indicated that CaRAX1/2a was specifically expressed in the I2 meristems, further supporting that CaRAX1/2a corresponded to the SFL gene, which specifically regulated I2 meristem activity.
As the homology of CaRAX1/2b and CaRAX1/2c with CaRAX1/2a/SFL could suggest functional redundancy among these genes, we also analysed the expression of these homologues. For both genes we observed a tissue expression pattern similar to that of CaRAX1/2a/SFL (Fig. S5a). In situ hybridisation showed that the CaRAX1/2b homologue was also expressed in the I2 meristems, in domains that partly overlapped with CaRAX1/2a/SFL (Fig. S5b). We also observed that the expression of CaRAX1/2b or CaRAX1/2c did not increase in inflorescence apices of sfl‐d mutant plants, in which the CaRAX1/2a/SFL gene is deleted (Fig. S5c). These results are compatible with the possibility that these genes act redundantly in the control of chickpea inflorescence development.
Discussion
Our results show that the SFL gene, responsible for the chickpea double‐pod phenotype, corresponds to CaRAX1/2a, which encodes a R2R3‐MYB transcription factor. Among aerial tissues, the highest expression of CaRAX1/2a was observed in inflorescence apices, in agreement with its role in inflorescence development. High expression of CaRAX1/2a was observed in roots. Interestingly, expression, and in some cases also function, associated with roots and nodules has also been described for some legume genes that regulate floral development (Zucchero et al., 2001; Couzigou et al., 2012). It will be worth studying the possible function of CaRAX1/2a in roots. In accordance with its role in regulating I2 meristem activity, in the inflorescence apex, CaRAX1/2a is specifically expressed in the I2 meristem, overlapping with CaVEG1, a homologue of legume I2 meristem identity genes (Berbel et al., 2012; Cheng et al., 2018). CaVEG1 can be placed upstream of CaRAX1/2a, as supported by its unchanged expression in the sfl‐d mutant. Interestingly, that CaRAX1/2a is not expressed in the I1 meristem, together with the fact that sfl mutations seem to specifically affect I2 activity, suggests that genetic networks that control meristem activity differ for I1 and I2.
CaRAX1/2a expression patterns have similarities, but also marked differences, with that of RAX1/3 (REGULATORS OF AXILLARY MERISTEMS1 and 3) and Blind genes, Arabidopsis and tomato homologues, respectively (Keller et al., 2006; Müller et al., 2006; Busch et al., 2011). The Arabidopsis and tomato genes have been shown to be transiently expressed in the axils of vegetative leaves, at axillary meristem initiation and RAX1 also in stage 1 floral meristems. CaRAX1/2a is expressed in the I2 meristems, formed at the axils of inflorescence leaves, but we did not detect its expression in vegetative axillary meristems or at the axils of vegetative leaves. Moreover, expression of CaRAX1/2a was not transient, but was observed in the I2 meristem throughout its development, supporting its role in controlling its activity.
The Arabidopsis RAX1/2/3 and tomato Blind genes regulate axillary meristems, promoting their initiation (Keller et al., 2006; Müller et al., 2006; Busch et al., 2011). The Marchantia polymorpha GCAM1 (GEMMA CUP‐ASSOCIATED MYB1) gene, encodes a R2R3‐MYB from subgroup 14, as RAX and Blind proteins (Dubos et al., 2010). Overexpression of GCAM1 in M. polymorpha promotes the formation of cell clumps with low differentiation levels and with competence to proliferate, somehow resembling the Arabidopsis and tomato RAX proteins, which promote initiation of meristems (Yasui et al., 2019). The chickpea CaRAX1/2a protein also regulates meristems but, in contrast with Arabidopsis and tomato RAX proteins, CaRAX1/2a acts to limit the proliferative phase of the I2 meristems, suggesting that the chickpea RAX protein interacts in a different way with some central regulatory components of the genetic machinery for meristem functioning. Interestingly, the expression of GCAM1 in the Arabidopsis rax1 rax2 rax3 mutant did not promote meristem formation, but inhibited it. However, the expression of a truncated version of the GCAM1 protein in which a N‐terminal domain, upstream of the R2R3‐MYB domain, not present in Arabidopsis, tomato or chickpea proteins, was deleted notably recovered axillary meristem formation in the triple mutant (Yasui et al., 2019). Therefore, a change in the M. polymorpha RAX protein sequence turned it from a negative into a positive meristem regulator, supporting the idea that the different inhibitory activity of CaRAX1/2a on meristem regulation might be due to differences in its protein sequence.
What could be the CaRAX1/2a contribution to controlling the number of flowers per I2 in nature? The double‐pod phenotype, increasing from one to two flowers, is moderate. In addition, although sfl‐d is a null mutation, not every I2 in a sfl‐d plant produced two flowers, and the expression of the double‐pod phenotype depends on environmental conditions (Kumar et al., 2000). As chickpea RAX genes are duplicated, it is possible that redundancy may exist among CaRAX genes for regulation of I2 meristem activity, as occurs between Arabidopsis RAX genes for regulation of axillary meristems (Müller et al., 2006). Indeed, the tissue expression pattern of CaRAX1/2a/SFL and of its homologues, CaRAX1/2b and CaRAX1/2c, looks similar; CaRAX1/2b is expressed in I2 meristems, partly overlapping with CaRAX1/2a/SFL, which support this hypothesis. Nevertheless, expression in the inflorescence apex of the CaRAX1/2a/SFL homologues apparently did not change to compensate its absence in the sfl‐d mutant. Thus, the CaRAX1/2c expression level is not affected in the sfl‐d mutant and CaRAX1/2b expression seems to show a moderate decrease. This might reflect mutual positive regulation between CaRAX1/2a/SFL and CaRAX1/2b, but further studies would be required to test this possibility. In addition, the three CaRAX1/2 proteins showed low amino acid identity outside the conserved MYB domain. Therefore, additional studies are required to assess any possible redundancy between the chickpea RAX genes. Another gene, CYM, has been shown to also repress the production of flowers by the chickpea I2 (Srinivasan et al., 2006). It is likely that the number of flowers at the I2 is determined by the combined action of CaRAX1/2a with CYM and maybe also other CaRAX genes.
Genotypes with a specific increase in the number of flowers in the I2 are also found in other legume species (Murfet, 1985; Mishra et al., 2020), which suggests that the function of CaRAX1/2a/SFL could be conserved in other legumes. This would agree with the fact that the function of genes regulating other aspects of inflorescence development is generally conserved in legumes (Benlloch et al., 2015; Cheng et al., 2018; Roque et al., 2018).
SUPERMAN (SUP), encoding a C2H2 zinc‐finger transcriptional repressor, restricts the proliferation of floral organs in Arabidopsis flowers (Hiratsu et al., 2002). MtSUPERMAN (Mtsup), its M. truncatula orthologue, recently described, restricts the proliferation of floral organs as well, but also regulates I2 meristem activity. mtsup mutant plants produced an increased number of abnormal flowers in their I2s, whose stubs were converted into terminal flowers (Rodas et al., 2020). Therefore, although it is not its only role, MtSUP, as CaRAX1/2a, restricts I2 meristem activity, suggesting that both genes might cooperate in this function.
Further analysis of legume RAX genes promises to lead to valuable knowledge for designing useful tools to improve seed yield, but should also help in understanding the basis of form variety among different legume inflorescences to generate morphological diversity.
Author contributions
CC, AB, TM, JR, JG, RO and FM developed and designed the experiments and interpreted the data; CC, AB, TM and JR performed the experiments; RO carried out the bioinformatics and the phylogenetic analyses; CC and FM wrote the manuscript with the input of all the authors. CC and AB contributed equally to this work.
Supporting information
Fig. S1 Legume species with different numbers of flowers in the I2.
Fig. S2 Sequence of chromosome 6 containing the deletion in the sfl‐d mutant.
Fig. S3 Allelic relationship between double‐pod USDA mutants and the JG62 double‐pod mutant.
Fig. S4 Phylogenetic tree of the R2R3‐MYB proteins from representative eudicot species.
Fig. S5 Expression pattern of the homologue genes CaRAX1/2b and CaRAX1/2c.
Table S1 List of primers used in this study.
Table S2 Accession numbers of the genes in the phylogenetic trees in Figs 3(d), S3.
Table S3 Percentage of amino acid identity between chickpea RAX1/2 proteins.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Ludovico Dreni for advice on phylogenetic analysis and M. J. Domenech for efficient technical assistance. We also thank C. Ferrándiz, M. A. Blázquez and J. L. Weller for helpful discussions and critical reading. Work at the FM laboratory was financed through grants from the Spanish Ministerio de Ciencia Innovación y Universidades, FEDER and Generalitat Valenciana (BIO2015‐64307‐R, PGC2018‐099232‐B‐I00 and Prometeo/2019/004). Work at IFAPA and UCO has been supported by INIA project RTA2017‐00041‐00‐00, co‐financed by the European Union through the ERDF2014–2020 ‘Programa Operativo de Crecimiento Inteligente’. CC acknowledges her PhD fellowship INIA‐CCAA.
Contributor Information
Cristina Caballo, Email: cristinacaballolinares@gmail.com.
Francisco Madueño, Email: madueno@ibmcp.upv.es.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Ali L, Deokar A, Caballo C, Tar’an B, Gil J, Chen W, Millan T, Rubio J. 2016. Fine mapping for double podding gene in chickpea. Theoretical and Applied Genetics 129: 77–86. [DOI] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Serrano‐Mislata A, Madueño F. 2007. Floral initiation and inflorescence architecture: a comparative view. Annals of Botany 100: 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Ali L, Gohari G, Millán T, Madueño F. 2015. Genetic control of inflorescence architecture in legumes. Frontiers in Plant Science 6: 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R, d'Erfurth I, Ferrandiz C, Cosson V, Beltrán JP, Cañas LA, Kondorosi A, Madueño F, Ratet P. 2006. Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1‐like functions in legumes. Plant Physiology 142: 972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel A, Ferrándiz C, Hecht V, Dalmais M, Lund OS, Sussmilch FC, Taylor SA, Bendahmane A, Ellis THN, Beltrán JP et al. 2012. VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nature Communications 3: 659–714. [DOI] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrandiz C, Cañas LA, Madueno F, Beltrán JP. 2001. Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1‐like genes controlling both floral meristem and floral organ identity in different plant species. The Plant Journal 25: 441–451. [DOI] [PubMed] [Google Scholar]
- Busch BL, Schmitz G, Rossmann S, Piron F, Ding J, Bendahmane A, Theres K. 2011. Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell 23: 3595–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballo C, Castro P, Gil J, Izquierdo I, Millán T, Rubio J. 2018. STMS (sequence tagged microsatellite site) molecular markers as a valuable tool to confirm controlled crosses in chickpea (Cicer arietinum L.) breeding programs. Euphytica 214: 231. [Google Scholar]
- Castro P, Román B, Rubio J, Die JV. 2012. Selection of reference genes for expression studies in Cicer arietinum L.: analysis of cyp81E3 gene expression against Ascochyta rabiei . Molecular Breeding 29: 261–274. [Google Scholar]
- Cheng X, Li G, Tang Y, Wen J. 2018. Dissection of genetic regulation of compound inflorescence development in Medicago truncatula. Development 145: dev158766. [DOI] [PubMed] [Google Scholar]
- Couzigou JM, Zhukov V, Mondy S, Abu el Heba G, Cosson V, Ellis THN, Ambrose M, Wen J, Tadege M, Tikhonovich I et al. 2012. NODULE ROOT and COCHLEATA maintain nodule development and are legume orthologs of Arabidopsis BLADE‐ON‐PETIOLE genes. Plant Cell 24: 4498–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi J, Mishra GP, Sanwal SK, Dubey RK, Singh PM, Singh B. 2018. Development and characterization of penta‐flowering and triple‐flowering genotypes in garden pea (Pisum sativum L. var. hortense). PLoS ONE 13: e0201235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science 15: 573–581. [DOI] [PubMed] [Google Scholar]
- Feng G, Burleigh JG,Braun EL, Mei W, Barbazuk WB. 2017. Evolution of the 3R‐MYB gene family in plants. Genome Biology and Evolution 9: 1013–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. 2000. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER . Development 127: 725–734. [DOI] [PubMed] [Google Scholar]
- French RF. 1990. The contribution of pod numbers to field pea (Pisum sativum L.) yields in a short growing season environment. Australian Journal of Agricultural Research 41: 853–862. [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme‐Takagi M. 2002. The SUPERMAN protein is an active repressor whose carboxy‐terminal repression domain is required for the development of normal flowers. FEBS Letters 514: 351–354. [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N. 1997. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Current Biology 7: 581–587. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Abbot J, Moritz T, Doener P. 2006. Arabidopsis REGULATOR OF AXILLARY MERISTEMS 1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Srivastava RK, Ganesh M. 2000. Penetrance and expressivity of the gene for double podding in chickpea. Journal of Heredity 91: 234–236. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. Mega X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht H. 1947. The inheritance of the number of flowers per inflorescence and the origin of Pisum, illustrated by polymeric genes. Agri Hortique Genetica 5: 16–25. [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Molecular Biology and Evolution 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Martin C, Paz‐Ares J. 1997. MYB transcription factors in plants. Trends in Genetics 13: 67–73. [DOI] [PubMed] [Google Scholar]
- Milbourne G, Hardwick RC. 1968. The growth of vining peas, 1. The effect of time of sowing. Journal of Agricultural Science 70: 393–402. [Google Scholar]
- Mishra GP, Dikshit HK, Kumari J, Priti, Tripathi K, Devi J, Aski M, Mehra R, Sarker A, Kumar S. 2020. Identification and characterization of novel penta‐podded genotypes in the cultivated lentil. Crop Science 60: 1974–1985. [Google Scholar]
- Müller D, Schmitz G, Theres K. 2006. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfet IC. 1985. Pisum sativum. In: Halevy AH, ed. Handbook of flowering, vol. IV. Boca Raton, FL, USA: CRC Press, 97–126. [Google Scholar]
- Ogata K, Morikawa S, Nakamur H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. 1994. Solution structure of a specific DNA complex of the Myb DNA‐binding domain with cooperative recognition helices. Cell 79: 639–648. [DOI] [PubMed] [Google Scholar]
- Prenner G. 2012. Papilionoid inflorescences revisited (Leguminosae‐Papilionoideae). Annals of Botany 112: 1567–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum‐likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. 2007. Evolution and development of inflorescence architectures. Science 316: 1452–1456. [DOI] [PubMed] [Google Scholar]
- Rodas AL, Roque R, Hamza R, Gómez‐Mena C, Minguet EG, Wen J, Mysore KS, Beltrán JP, Cañas LA. 2020. MtSUPERMAN plays a key role in compound inflorescence and flower development in Medicago truncatula . The Plant Journal 105: 816–830. [DOI] [PubMed] [Google Scholar]
- Romani F, Moreno JE. 2021. Molecular mechanisms involved in functional macroevolution of plant transcription factors. New Phytologist 230: 1345–1353. [DOI] [PubMed] [Google Scholar]
- Roque E, Gómez‐Mena C, Ferrándiz C, Beltrán JP, Cañas LA. 2018. Functional genomics and genetic control of flower and fruit development in Medicago truncatula: an overview. Methods in Molecular Biology 1822: 273–290. [DOI] [PubMed] [Google Scholar]
- Rubio J, Flores F, Moreno MT, Cubero JI, Gil J. 2004. Effects of the erect/bushy habit, single/double pod and late/early flowering genes on yield and seed size and their stability in chickpea. Field Crops Research 90: 255–262. [Google Scholar]
- Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K. 2002. The tomato blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proceedings of the National Academy of Sciences, USA 99: 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrake AR, Saxena NP, Krishnamurthy L. 1978. The expression and influence on yield of the “double‐podded” character in chickpeas (Cicer arietinum L.). Field Crops Research 1: 243–253. [Google Scholar]
- Singer S, Sollinger J, Maki S, Fishbach J, Short B, Reinke C, Fick J, Cox L, McCall A, Mullen H. 1999. Inflorescence architecture: a developmental genetics approach. The Botanical Review 65: 385–410. [Google Scholar]
- Srinivasan S, Gaur PM, Chaturvedi SK, Rao BV. 2006. Allelic relationships of genes controlling number of flowers per axis in chickpea. Euphytica 152: 331–337. [Google Scholar]
- Taylor SA, Hofer JMI, Murfet IC, Sollinger JD, Singer SR, Knox MR, Ellis THN. 2002. PROLIFERATING INFLORESCENCE MERISTEM, a MADS‐box gene that regulates floral meristem identity in pea. Plant Physiology 129: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo ZWN, Song S, Wang YQ, Liu J, Yu H. 2014. New insights into the regulation of inflorescence architecture. Trends in Plant Science 19: 158–165. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar'an B et al. 2013. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nature Biotechnology 31: 240–248. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J. 2008. Molecular basis of plant architecture. Annual Review of Plant Biology 59: 253–279. [DOI] [PubMed] [Google Scholar]
- Weberling F. 1992. Morphology of flowers and inflorescences. Cambridge, UK: Cambridge University Press. [Google Scholar]
- White OE. 1917. Studies of inheritance in Pisum. II. The present state of knowledge of heredity and variation in pea. Proceedings of the American Philosophical Society 56: 487–589. [Google Scholar]
- Wyatt R. 1982. Inflorescence architecture: how flower number, arrangement, and phenology affect pollination and fruit‐set. American Journal of Botany 69: 585–594. [Google Scholar]
- Yasui Y, Tsukamoto S, Sugaya T, Nishihama R, Wang Q, Kato H, Yamato KT, Fukaki H, Mimura T, Kubo H et al. 2019. GEMMA CUP‐ASSOCIATED MYB1, an ortholog of axillary meristem regulators, is essential in vegetative reproduction in Marchantia polymorpha . Current Biology 29: 3987–3995. [DOI] [PubMed] [Google Scholar]
- Zucchero JC, Caspi M, Dunn K. 2001. ngl9: a third MADS box gene expressed in alfalfa root nodules. Molecular Plant–Microbe Interactions 14: 1463–1467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Legume species with different numbers of flowers in the I2.
Fig. S2 Sequence of chromosome 6 containing the deletion in the sfl‐d mutant.
Fig. S3 Allelic relationship between double‐pod USDA mutants and the JG62 double‐pod mutant.
Fig. S4 Phylogenetic tree of the R2R3‐MYB proteins from representative eudicot species.
Fig. S5 Expression pattern of the homologue genes CaRAX1/2b and CaRAX1/2c.
Table S1 List of primers used in this study.
Table S2 Accession numbers of the genes in the phylogenetic trees in Figs 3(d), S3.
Table S3 Percentage of amino acid identity between chickpea RAX1/2 proteins.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


