Abstract
BACKGROUND
Microsporidia, a group of obligate intracellular fungal‐related parasites, have been used as efficient biocontrol agents for agriculture and forestry pests due to their host specificity and transovarial transmission. They mainly infect insect pests through the intestinal tract, but the interactions between microsporidia and the gut microbiota of the host have not been well demonstrated.
RESULTS
Based on the microsporidia–Bombyx mori model, we report that the susceptibility of silkworms to exposure to the microsporidium Nosema bombycis was both dose and time dependent. Comparative analyses of the silkworm gut microbiome revealed substantially increased abundance of Enterococcus belonging to Firmicutes after N. bombycis infection. Furthermore, a bacterial strain (LX10) was obtained from the gut of B. mori and identified as Enterococcus faecalis based on 16S rRNA sequence analysis. E. faecalis LX10 reduced the N. bombycis spore germination rate and the infection efficiency in vitro and in vivo, as confirmed by bioassay tests and histopathological analyses. In addition, after simultaneous oral feeding with E. faecalis LX10 and N. bombycis, gene (Akirin, Cecropin A, Mesh, Ssk, DUOX and NOS) expression, hydrogen peroxide and nitric oxide levels, and glutathione S‐transferase (GST) activity showed different degrees of recovery and correction compared with those under N. bombycis infection alone. Finally, the enterococcin LX protein was identified from sterile LX10 fermentation liquid based on liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis.
CONCLUSION
Altogether, the results revealed that E. faecalis LX10 with anti‐N. bombycis activity might play an important role in protecting silkworms from microsporidia. Removal of these specific commensal bacteria with antibiotics and utilization of transgenic symbiotic systems may effectively improve the biocontrol value of microsporidia. © 2022 The Authors. Pest Management Science published by John Wiley & Sons Ltd on behalf of Society of Chemical Industry.
Keywords: Bombyx mori, microsporidia, Enterococcus faecalis, anti‐Nosema bombycis activity, protein
According to the microsporidia–Bombyx mori model, we revealed that Enterococcus with anti‐Nosema bombycis activity might play an important role in protecting silkworms from microsporidia. The present study provides a clear profile of the complex interplay among microsporidia, silkworms, and gut microbiome.
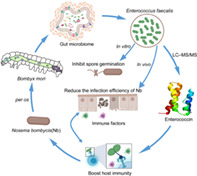
1. INTRODUCTION
The silkworm is an important model for studies on insect physiology and control of lepidopterous pests due to its relatively small body size and ease of rearing. 1 Microsporidians are a group of widely distributed pathogens that can invade a broad range of hosts, including immunocompromised humans. 2 Nosema bombycis is the first described microsporidium species recognized as the etiological agent of silkworm pebrine disease. 3 To date, pebrine disease is the only disease with a mandatory quarantine in the silk production industry due to its destructive consequences. 4 The microsporidian pathogens are also considered effective biological control agents for mosquitoes, gypsy moth, locust, and corn borer red imported fire ants. 5 , 6 , 7 In 1980, Paranosema locustae became the first microsporidium to be registered and successfully commercially marketed in the United States (USEPA 2000). It is still being sold for bait‐and‐spray applications for Mormon cricket and grasshopper control due to the high grasshopper mortality associated with products, such as Nolo Bait™ and Semaspore™. 8 , 9 Similar results have been reported in Argentina and China where government‐subsidized, large‐scale applications were deemed adequate by the growers because of wide host ranges and high transmission efficiency. 10 , 11
As an important intestinal pathogen, N. bombycis can be transmitted horizontally through per os infection as well as vertically from adults to offspring. 12 The pathogen N. bombycis is well adapted to the silkworm gut environment, where it germinates and penetrates the gut epithelium, absorbing nutrients and energy from the host and exporting cytotoxic compounds through the cell membrane. 13 This species has a sophisticated and unique mechanism for infecting host cells, involving a rigid spore wall and a coiled polar tube joined to an anchoring disc at the apical part of the spore. 14 With appropriate stimuli, mature microsporidian spores rapidly discharge the infective polar tube, come into contact with the surface of the cell membrane, and transfer the sporoplasm into the cell cytoplasm, where proliferation and the next round of spore production occurs. N. bombycis then proliferates inside the host, leading to chronic damage to larval organs and tissues, including the gut, muscles, silk glands, Malpighian tubules, and ultimately to death. 15 Nevertheless, it is not surprising that silkworms have evolved effective anti‐N. bombycis strategies, including its innate immune system as well as the chemical and physical barriers present in the gut, such as a strong alkaline digestion mechanism. 16 N. bombycis infection of Bombyx mori larvae activates the JAK/STAT and Toll pathways in the midgut, which induces the up‐regulation of the expression of innate immune‐related defense proteins involved in humoral and cellular immunity, including moricins, lebocins and lysozymes, to resist invasion by microsporidia. 17 , 18 , 19 , 20 In addition, the production of the prophenoloxidase (PPO) by the gut epidermal cells of the silkworm is also suppressed after microsporidian infection due to the induction of melanization, thus preventing pathogens from entering the surrounding habitat. 1 , 21
Similar to most lepidopteran larvae, silkworm larvae possess a relatively simple gut without specialized substructures; however, a sufficient gut bacterial population exceeding 107 CFU/mL is clearly present. 22 Recent surveys of the taxonomic composition of the gut bacteria reveal a relatively simple but specific community dominated by Enterococcus, Lactococcus, Bacillus, Stenotrophomonas and Pseudomonas. 23 The indigenous microbiota is being increasingly recognized as an important component associated with biological fitness, detoxication, vitamin synthesis, reproductive regulation and pathogen resistance. 22 , 24 Comparative metabolomics has revealed that Stenotrophomonas species provide essential amino acids to the host and hence increases host tolerance to organophosphate insecticides. 22 A study confirmed that Enterococcus spp. decreased the toxicity of Bacillus thuringiensis δ‐endotoxin in the silkworm larva gut. 25 , 26 Some Enterococcus species have been identified in the gut of insects, fish, birds, and mammals, and many of these Enterococcus species have been utilized as unique livestock probiotics for decades. 27 , 28 Additionally, these commensal intestinal bacteria can assist different insect hosts with pathogen resistance in different insects, such as by secretion of antimicrobial substances and activation of host defense pathways. 29 For example, for Aedes aegypti and Anopheles gambiae, the creation of microbe‐free aseptic strain with antibiotics renders the mosquitoes more susceptible to infection by the apicomplexan parasite Plasmodium falciparum. 30 In addition, the intestinal bacterium Pseudomonas aeruginosa BGf‐2 has been shown to decrease the mortality of cockroaches after treatment with Beauveria bassiana and to restrict conidial germination and hyphal growth by producing antifungal proteins. 31 Moreover, a stable class IIa bacteriocin (mundticin KS) from the gut bacterium Enterococcus mundtii of Spodoptera littoralis suppressed the proliferation of pathogenic bacteria in vitro. 32
Furthermore, 70% of pests are lepidopteran species, so exploration of the relationship between the commensal bacteria of silkworms and the fungal parasites N. bombycis in the lepidopteran model, B. mori is urgently needed. The aim of the current study was therefore to investigate the direct effects of N. bombycis infection on the community structure of the gut microbiota. We isolated the bacterial strain E. faecalis LX10 with anti‐N. bombycis activity from the B. mori gut. Moreover, we investigated the effect of supplementation with E. faecalis LX10 on immune gene expression, hydrogen peroxide (H2O2) and nitric oxide (NO) levels and glutathione S‐transferase (GST) activity of silkworm larvae challenged with N. bombycis.
2. MATERIALS AND METHODS
2.1. Silkworms and microsporidia
The hybrid B. mori strain Haoyue×Jingsong was provided by the Silkworm Germplasm Bank of the Cathaya Group, Chun an, Zhejiang, China (118°71′N, 29°36′E). All insects were reared under standard conditions (25 ± 1 °C, 70 ± 5% humidity) in plastic boxes (50 cm × 25 cm × 10 cm) using fresh mulberry leaves collected from a mulberry field at Zhejiang University, China (30°18′N, 120°04′E). Additionally, to prepare germ‐free (GF) larvae, silkworm eggs were surface sterilized using 1% sodium hypochlorite (NaClO) solution and then rinsed with sterile water. The eggs were subsequently immersed in 70% ethanol followed by sterile water. The GF larvae were reared with an autoclaved artificial diet. To validate the GF conditions, fecal samples were examined by Petri dish culture. 22
N. bombycis ZJU1 spores originally obtained from infected silkworms in Zhejiang, China, were produced and harvested at the Institute of Sericulture and Apiculture, Zhejiang University, and propagated and purified in laboratory‐reared silkworms adults at a temperature of 25 ± 1 °C by centrifugation over Percoll (pH=7). 33
2.2. Susceptibility of silkworms to N. bombycis applied through feeding
Haoyue×Jingsong silkworms were equally divided into four groups [control (CK), 103, 106, and 109] after the fourth exuviation (fifth‐instar larvae). Newly molted silkworm larvae were challenged by feeding on mulberry leaves (15 cm × 15 cm) that were previously artificially besmeared with N. bombycis spores at different concentrations (103 spores/mL, 106 spores/mL, and 109 spores/mL; 25 μL/larvae). The CK group larvae were reared with fresh mulberry leaves with the same dosage of sterile water. Briefly, 15 samples (n = 15) were collected from each group for DNA extraction and gut microbiota analysis (a single individual insect for each sample) at four time points (0, 2, 4, and 6 days) post‐infection (dpi). The gut samples were homogenized in a Precellys‐24 homogenizer (Bertin Technologies, Aixen, France) at 6000 × g for 120 s. Bacterial DNA was extracted using a MasterPure™ Purification Kit (Epicentre, Madison, WI, USA) following the manufacturer's protocol. Moreover, a negative control without gut tissue was processed simultaneously with the same DNA extraction method to control for contamination with the reagent and other substances. The concentration and quality of the DNA were measured by a 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA).
The absolute quantification of N. bombycis (n = 15) was conducted by real‐time quantitative polymerase chain reaction (qPCR) analysis for each silkworm gut. 34 The primers ssu1092F and ssu1227R were designed to amplify the N. bombycis small‐subunit rRNA gene fragment from position 1092 to position 1227 (GenBank accession no. EU864525.1). Then, a standard curve was constructed with pure N. bombycis DNA (1 × 100 to 1 × 1010 copies/μL) as the standard substance. All runs were performed using SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China) and a Roche LightCycler 480 system (Roche, Basel, Switzerland). In addition, sterile water served as a negative control. The qPCR was conducted using the following program: 95 °C for 5 min and 40 cycles of 95 °C for 10 s, 60 °C for 10 s and 72 °C for 10 s. A melting curve (from 65 to 92 °C, 0.5 °C/s) was analyzed to ensure that the reaction produced a single product. The gene copies of N. bombycis were calculated by comparing Cq values to the standard curve.
An analytical balance (Mettler Toledo, Greifensee, Switzerland) was used to measure the larval mass at 2, 4, and 6 dpi for the experimental (N. bombycis treatment, Nb) and CK groups (n = 15). The pH measurements were performed using a pH microelectrode (Unisense, Aarhus, Denmark) with a tip diameter of 20 to 30 μm (Unisense). As soon as the larvae pupated, they were transferred into labeled 2 cm‐wide and 5 cm‐long sterilized cages at a temperature of 25 ± 1 °C and humidity of 70% ± 1%. The whole cocoon weight, cocoon length, cocoon width and cocoon shell weight were recorded after cocooning (n = 60).
2.3. Gut microbiota analysis by 16S rRNA gene sequencing
Gut DNA was extracted by following the method described in Section 2.2. To prepare an Illumina sequencing library, a region encompassing the V3–V4 hypervariable regions of the 16S rRNA gene was amplified from 1 μL of purified DNA using the universal primers 338F and 806R, which are shown in Supporting Information Table S2. Agarose gel electrophoresis was used to estimate the product size (~450 bp). All 16S rRNA products were purified and pooled together after gel purification by a MiniBEST Agarose Gel DNA Extraction Kit (Takara, Dalian, China) for the Illumina MiSeq platform (San Diego, CA, USA), and paired‐end sequencing was performed by Shanghai Biotechnology (Majorbio, Shanghai, China). 35 In total, 8 908 622 raw reads were obtained and preprocessed as previously described. 36 In brief, the demultiplexed paired‐end sequence reads were assembled by DADA2. 37 Thus, 16S rRNA gene sequencing generated 120 000–450 000 sequences per sample after preprocessing. Sequences assigned to chloroplasts and mitochondria were removed from the amplicon sequence variant (ASV) tables. We obtained high‐quality processed sequences with a mean length of 410 bp and an average of 49 492 sequences per sample. To minimize bias due to sequencing depth, we normalized the sequence number to the lowest sequence number by rarefaction. High coverage values (average = 99%) were obtained for sequences in all samples, indicating that the sequencing depth was sufficient. To investigate the gut bacterial community structure, Shannon and Faith’s Phylogenetic Diversity (Faith's PD) indices were estimated. 38 Bray–Curtis dissimilarities were determined and visualized via principal coordinate analysis (PCoA) for community comparison. 39 The relationships between gut microbiota structure and environmental variables (N. bombycis concentration, inoculation time) were determined with the use of multivariate redundancy analysis (RDA) and canonical correspondence analysis (CCA). Network analyses were used to explore the cooccurrence patterns of gut bacterial microorganisms in each niche. The Spearman's correlation between two ASVs was considered statistically robust if the P value was < 0.01 and Spearman's correlation coefficient (r) was > 0.6. The resulting adjacency matrices were converted to network objects by the R package igraph. Network summary statistics and node centrality measures were calculated to compare network properties across microbial species (degree, average degree, average path length, modularity, diameter, and transitivity). Networks were visualized using the interactive Gephi platform. 40
Receiver operator characteristic (ROC) curve analysis for sample classification was performed using the random forest algorithm. The area under the curve (AUC) was calculated to measure the predictive accuracy. Moreover, linear discriminant analysis (LDA) effect size (LEfSe) analysis was performed using the normalized ASV table. Variation analyses were performed using Fisher's least significant difference (LSD) followed by one‐way analysis of variance (ANOVA) or the non‐parametric Kruskal–Wallis test. Finally, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) was employed for stringent predictions of microbial functional metabolic pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. 41 Sequencing data were deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) under accession number PRJNA763083.
Absolute quantification of the number of bacteria was conducted by qPCR of 16S rRNA gene copies of total bacteria (n = 15) and Enterococcus (n = 15) using the DB200Q (all bacteria) and EN144Q (Enterococcus) primers. The number of copies was routinely deduced by comparing the Cq values to a standard curve generated from a ten‐fold serial dilution series of the full‐length 16S rRNA genes.
2.4. Enterococcus isolation and anti‐N . bombycis effects in vitro and in vivo
Enterococcus bacteria were isolated from the normal fifth‐instar B. mori larval gut using Enterococcus selective agar with 1% 2,3,5‐triphenyl tetrazolium chloride (TTC) in the medium. Molecular analysis was performed by isolating genomic DNA and amplifying the bacterial 16S rRNA genes using universal primers 27F/1492R (Galkiewicz & Kellogg, 2008). The synthesis and sequencing of PCR products were performed by Sangon Biotech (Shanghai, China). The phylogenetic trees were constructed by MEGA 11 using a statistical neighbor‐joining method by applying the following parameters: bootstrap method (1000 replicates), Kimura 2‐parameter model, substitutions to include (d: Transitions + Transversions), Uniform Rates, and pairwise deletion.
The Enterococcus strain was cultured in de Man–Rogosa–Sharpe (MRS) broth (Hopebio, Qingdao, China) at 37 °C for 36 h. The supernatant (500 mL) was collected after centrifugation at 10 000 × g for 10 min and filtered through a 0.22 μm polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA) to obtain cell‐free fermentation liquid. Crude proteins were precipitated with 20–80% saturated ammonium sulfate ((NH4)2SO4). The proteins were dialyzed with regenerated cellulose membrane tubing (3.5 kDa; Spectrum) in distilled water. Then, the crude proteins from E. faecalis LX10 supernatants (SMP) were concentrated via ultrafiltration with Amicon Ultra 3 kDa (< 3 K and > 3 K SMP) and 10 kDa (< 10 K and > 10 K SMP) cellulose ultrafiltration device (Millipore) to remove proteins < 3 kDa in size, according to the manufacturer's instructions. Crude protein from MRS medium (MRS) was obtained in the same way as a negative control. The protein concentration was quantified using a BCA protein detection kit (Thermo Fisher Scientific, Waltham, MA, USA) and adjusted for equal loading. The effect of pH on the inhibitory substance was tested by adjusting each of the crude protein samples to pH 5.0 to 10.0 with 1 mol/L soidum hydroxide (NaOH) or 1 mol/L hydrochloric acid (HCl). The samples were readjusted to pH 6.0 after 30 min of incubation and tested for antimicrobial activity. Moreover, the effect of temperature was tested by heating the crude protein at 40, 60, 80 and 100 °C for 30 min, and the residual activity was measured.
Anti‐N. bombycis activity assays were performed both in vitro and in vivo. In the in vitro study, 20 μL of N. bombycis ZJU1 spore suspension (107 spores/mL) was aliquoted into 1.5 mL centrifuge tubes, and 30 μL of crude protein (2 mg/mL) with different molecular weight ranges (< 3 K, > 3 K, < 10 K, and > 10 K SMP) was added to the tubes in the treatment group. Additionally, 30 μL of phosphate buffer (pH 6) was used as a control. The control and treatment group samples were treated with 150 μL of GKK germination buffer [0.05 mol/L glycine, 0.05 mol/L potassium hydroxide (KOH), and 0.375 mol/L potassium chloride (KCl); pH 10.5], and the samples were incubated at 27 °C for 60 min. The germination rate was monitored by measuring the absorbance at 625 nm (OD625). 42 The calculation formula was as follows: germination percentage = [(germination initiation liquid OD625 − germination termination liquid OD625)/germination initiation liquid OD625] × 100%.
For in vivo assessment of anti‐N. bombycis activity, fifth‐instar GF silkworm larvae were divided into four groups (CK, Ef, Nb, Nb + Ef) (n = 15). Each silkworm was fed 25 μL of E. faecalis LX10 cell suspension (1 × 107 CFU/mL) and 25 μL of N. bombycis suspension (1 × 109 spores/mL) (Ef + Nb); 25 μL of E. faecalis LX10 cell suspension (1 × 107 CFU/mL) and 25 μL of sterile water (Ef); or 25 μL of N. bombycis suspension (1 × 109 spores/mL) and 25 μL of sterile water (Nb), with GF larvae serving as a negative control (CK). After 6 days, randomly selected gut samples were processed for scanning electron microscopy (SEM) and histopathological analysis, whereas the remaining sample were homogenized for assessment of the N. bombycis burden via qPCR after 2, 4, and 6 days. For SEM analysis, the gut samples were fixed in 2.5% glutaraldehyde at 4 °C overnight. Then several gut samples were washed in phosphate‐buffered saline (PBS) three times and postfixed in 1% osmium tetroxide for 2 h. Then, the tissue with serial dilutions of ethanol (30% to 100%) were dehydrated. The samples were dried using a critical point dryer, mounted, and sputter coated with gold/palladium. SEM was conducted on gold‐sputtered samples with a Hitachi SU8010 scanning electron microscope (Tokyo, Japan). Moreover, the gut epithelial samples were fixed in polyformaldehyde fixing solution for histopathological analysis. Periodic acid–Schiff (PAS) staining was performed to evaluate pathological injury.
2.5. Anti‐N . bombycis protein identification for LC–MS/MS
The active crude proteins (3–10 K) were analyzed by 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) to determine the components of the proteins samples and to estimate their molecular masses. Protein binding was visualized by staining with Coomassie Blue 250. The gel bands were subjected to in‐gel digestion followed by alkylation and trypsin digestion at 37 °C for 20 h. An aliquot of the tryptic digest (in 0.1% formic acid–0.1% acetonitrile in acetonitrile–water) was analyzed by liquid chromatography tandem mass spectrometry (LC–MS/MS) on a Q Exactive Orbitrap Fusion Tribrid mass spectrometer that was coupled to an Easy LLC system (Thermo Fisher Scientific, Waltham, MA, USA). All raw MS and MS/MS files were processed and analyzed using MASCOT 2.2 software (Matrix Science, London, UK) against the Protein Homology/Analogy Recognition Engine version 2.0 (Phyre2) database. The search parameters were set as follows: variable modification, oxidation; fixed modification, carbamidomethylation; peptide tolerance, ± 20 ppm; number of missed cleavages, 2; fragment mass tolerance, ± 0.1 Da; and filtered by score ≥ 20. The MS/MS data of the anti‐N. bombycis proteins were retrieved and aligned against both the NCBI BLAST database and the protein sequence database of E. faecalis (NCBI, UNIPROT). The three‐dimensional structure prediction models were illustrated using Phyre2 database (http://www.sbg.bio.ic.ac.uk/phyre2/htmL/page.cgi?id=index).
2.6. Gene expression analysis and enzyme activity assays
To evaluate the effect of N. bombycis infection (CK, 103, 106, and 109 spores/mL) on host immunity and whether E. faecalis LX10 has a protective effect on silkworm infection, B. mori larvae were orally inoculated with an E. faecalis LX10 suspension (1 × 107 CFU/mL, Ef), N. bombycis (1 × 109 spores/mL, Nb), or E. faecalis LX10 and N. bombycis (Ef + Nb), with healthy B. mori GF larvae serving as a negative control (CK). Then, the expression levels of immune‐related genes (cecropin, Akirin, Mesh, Ssk, DUOX, and NOS) were determined in the gut at 2, 4, and 6 days post‐exposure (n = 5). Total RNA was extracted with a Promega Eastep® Total RNA Super Extraction Kit (Beijing Biotec. Co., Ltd, Beijing, China) according to the manufacturer's protocol. Total RNA (1 μg) was reverse transcribed to complementary DNA (cDNA) using HiScript® II Q RT SuperMix R223‐01 (Vazyme Biotech) according to the manufacturer's guidelines. The reverse transcription qPCR (RT‐qPCR) was performed as described earlier. The expression level of the housekeeping gene RP49 was used as an endogenous control. The sequences of gene‐specific primers used for transcript quantification are listed in Table S2. The 2−∆∆ct method was used to analyze the relative transcript levels of the genes of interest.
The H2O2 and NO levels and GST enzyme‐mediated protective response in the silkworm gut were determined (n = 5). The silkworm gut was isolated and immediately homogenized in ice‐cold PBS (10 mmol/L). The gut homogenates were centrifuged at 10 000 × g for 10 min at 4 °C, and then, the supernatants were collected for different measurements. The H2O2 and NO levels were assayed using a Hydrogen Peroxide Assay Kit (serial no. S0038; Beyotime Biotechnology, Shanghai, China) and Total Nitric Oxide Assay Kit (serial no. S0023) (Beyotime Biotechnology). The protein content was determined using a BCA kit according to the manufacturer's protocol (Thermo Fisher Scientific, Waltham, MA, USA). GST activity was analyzed with a GST Activity Detection Kit (Sangon Biotech, Shanghai, China).
2.7. Statistical analysis
Statistical analyses were carried out using GraphPad Prism (version 9.0) and Statistical Package for the Social Sciences, version 20.0 (SPSS, Chicago, IL, USA). All the results are presented as the mean ± standard error (SE). Parametric/non‐parametric statistical analyses were performed after the data were checked for normality and homogeneity of variance. One‐way ANOVA, Tukey's post hoc test or Student's t test was performed to compare the absolute abundances of bacteria and N. bombycis, larval mass, pH, whole cocoon weight, cocoon length, cocoon width, cocoon shell weight, relative germination rate, gene expression changes, and enzyme activities between the experimental and control groups. In each case, the type of test is stated before the P value, and P ≤ 0.05 was considered statistically significant. All experiments were repeated at least three times. The software used included GraphPad Prism 9.0 and Python and R packages, run as standalone software on Windows 10.
3. RESULTS
3.1. Susceptibility of silkworms to N. bombycis
The susceptibility of silkworms to N. bombycis treatment was both dose and time dependent, and this phenomenon was more pronounced when the silkworms were treated with high concentrations of N. bombycis (Fig. 1(A)). N. bombycis was detected in only the gut tissues of 106 and 109 spore/mL‐treated silkworms but not in those in the CK group and 103 spore/mL‐treated group. For example, at 6 dpi, the number of spores in the gut reached 0.89 × 105 and 5.25 × 108 spores/gut compared to that at 2 days (0 versus 5.37 × 104 spores/gut) at 106 and 109 spores/mL (t = −40.18, df = 14, P = 0.006 for 106, t = 18.68, df = 14, P < 0.001 for 109) (Fig. 1(A)).
Figure 1.
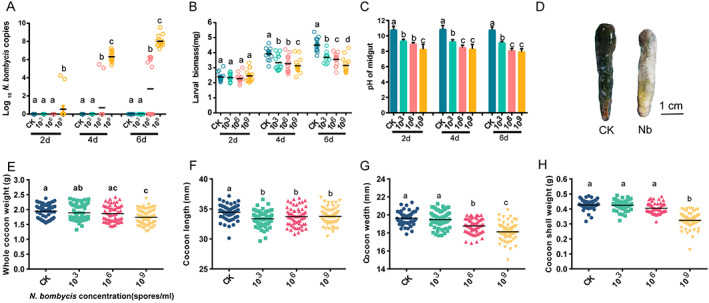
Effect of N. bombycis exposure on silkworms. (A) DNA was extracted from the intestinal epithelium, and the N. bombycis burden was estimated by absolute quantification in gut samples at different times post‐infection (2, 4, and 6 days) (mean with 95% confidence interval, n = 15). (B) Larval masses of the different groups (mean with 95% confidence interval, n = 15 for each group). (C) pH of the silkworm gut. The pH was measured, and the values are shown as the mean ± SE (n = 10). (D) Gut morphology of silkworms infected with N. bombycis after 6 days. (E,H) Whole‐cocoon weight (E), cocoon length (F), cocoon width (G) and cocoon shell weight (H) of normal and N. bombycis‐treated silkworms. CK, healthy silkworm; 103, 106, and 109 groups: N. bombycis treatment (Nb) with 103, 106, and 109 spores/mL concentrations, respectively. Different letters represent significant differences between groups according to Tukey's honestly significant difference (HSD, P < 0.05).
The larval mass and pH are shown in Fig. 1(B,C). After treatment with N. bombycis, the larval mass decreased significantly (P < 0.05), especially at concentrations of 1 × 106 and 1 × 109 spores/mL (Fig. 1(B)). There was no significant difference in larval mass between the N. bombycis‐fed and CK groups on Day 2 (P = 0.4916, F (3,56) = 0.8138, Fig. 1(B)). With increasing N. bombycis concentrations and inoculation time, the difference gradually increased (P < 0.0001, F (3,56) = 8.534 for 4 days, P < 0.0001, F (3,56) = 27.74 for 6 days) (Fig. 1(B)). The gut pH showed a similar trend (F (11,48) = 40.37, P < 0.0001) (Fig. 1(C)). The gut exhibited macroscopic white swelling and wrinkles at 6 days after the treatment Fig. 1(D). The fitness‐related parameters reported in Fig. 1(E–H) show that the whole‐cocoon weight, cocoon length, cocoon width, and cocoon shell weight decreased significantly after N. bombycis treatment (P < 0.05).
3.2. N. bombycis infection causes dysbiosis of the gut microbiota
A total of 180 intestinal samples were prospectively collected, and 18 samples failed to generate data and were removed from the following analyses. In total, 1081 species, 754 genera, 453 family, 261 order, 118 class, and 39 phyla were detected. To characterize the effect of N. bombycis on gut microbiome diversity, different measures of alpha diversity were applied based on 16S rRNA sequences. Briefly, the Shannon and Faith's PD indices were higher in the non‐infected silkworms (CK group) than in the N. bombycis‐infected silkworms at 2, 4, and 6 dpi (P < 0.05). Overall, N. bombycis infection resulted in decreased gut microbial richness and diversity (Fig. 2(A,B)). With prolonged infection, PCoA showed that the gut bacterial communities of gut regions with different inoculation concentrations clustered separately (Fig. 2(C–E)). A random distribution of the samples was found despite the gut microbiota being somewhat similar among individuals at 2 days after inoculation Fig. 2(C).
Figure 2.
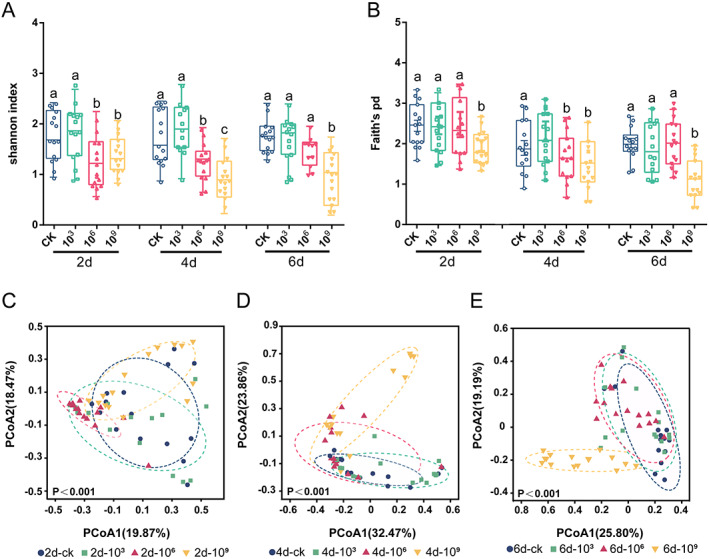
Spatial structure of the B. mori gut microbiota. (A,B) Alpha diversity and (D,E) beta diversity were investigated. Alpha diversity (Shannon and Faith's PD) of bacterial communities (based on ASVs) at different spore concentrations (CK, 103, 106, and 109 spores/mL) and infection times (2, 4, and 6 days). Different letters describe significant differences at P = 0.05. One‐way ANOVA with Tukey's multiple comparisons test and unpaired Student's t test. PCoA plot showing variation in community structure among different infection concentrations [permutational multivariate analysis of variance (PERMANOVA) test with 999 permutations, P > 0.05] based on Bray–Curtis distance. Each point represents an individual sample.
We next investigated the variability in bacterial communities in relation to N. bombycis infection. The bacterial phyla Proteobacteria, Actinobacteria, Firmicutes, Bacteroidota and Desulfobacterota, together accounting for up to 90% of sequences on average, were the five most dominant taxa (Fig. 3(A)). Compared with that in healthy individuals, the abundance of Firmicutes generally decreased in a time‐ and dose‐dependent manner in the treatment groups. At 6 days after inoculation, the abundances of Firmicutes in the CK, 103, 106, and 109 groups showed a decreasing trend (58.33%, 58.51%, 56.29% and 17.41%) (P < 0.05, Fig. 3(A)). The qPCR confirmed that the total bacterial load in infected silkworms after infection (1.21 × 1011 copies/gut) was significantly higher than that in the CK group (0.58 × 109 copies/gut) and that in silkworms in the 109 group at 6 dpi (t = 6.199, df = 28, P < 0.0001) (Supporting Information Fig. S1). Correlation analysis of the microbial abundance at the phylum level was used to construct networks, wherein the network parameters represented the microbial communities and the correlations of their abundances. Compared to the CK network, those of the treatment groups, especially the 109 group, showed a higher average path length (6.10), modularity (0.80), and diameter (10.25) (Fig. S2(A)), while the CK network showed a higher degree (496), higher average degree (3.16), shorter average path length (3.37), and lower network diameter (6.99). These finding, together with the network global properties, indicated that the CK group exhibited much closer interconnections than the treatment group at the phylum level (Fig. S2(A)).
Figure 3.
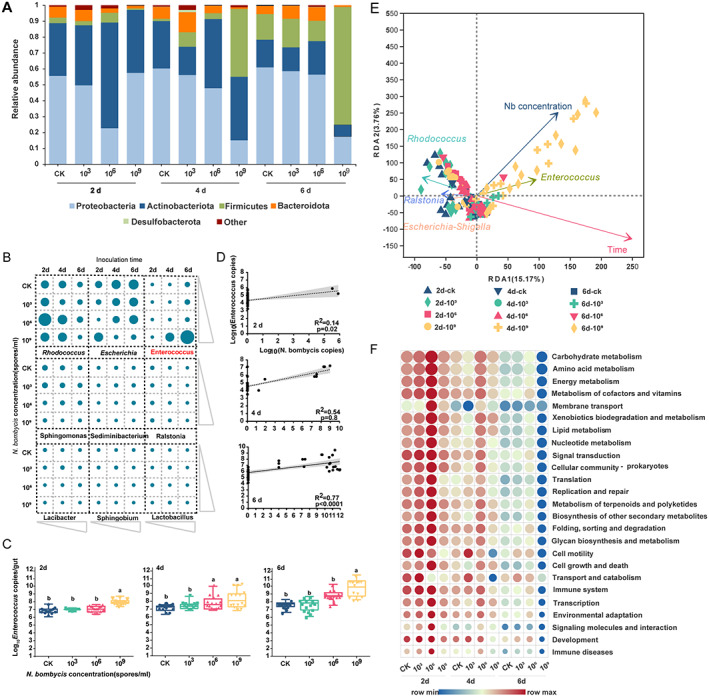
N. bombycis infection alters the composition of the gut microbiota in silkworms. (A) Relative abundance of bacterial phyla in different samples. Taxa with abundances < 1% are included in ‘others’. (B) The nine most abundant genera in bacterial communities detected in the 168 samples. Each point represents the relative abundance of bacterial genera in different samples. (C) Absolute quantification of a specific gene in Enterococcus in silkworms infected with N. bombycis. Enterococcus gene transcript numbers expressed as copies per gram sample of B. mori at 2, 4, and 6 days. (D) Correlation between Enterococcus and N. bombycis at 2, 4, and 6 dpi with N. bombycis. Dotted lines denote the linear regressions. Each point represents an individual silkworm. (E) Distance‐based RDA using three explanatory variables (group, time, and spore concentration). The time and spore concentration of the subjects are represented by color gradients and the shape of each dot, respectively. Arrows in the distance‐based RDA biplot denote the magnitudes and directions of the effects of explanatory variables. (F) The functional content of the gut microbiota was inferred by using PICRUSt based on the baseline and endpoint 16S amplicon sequencing data. Changes in the functional KEGG pathways during each N. bombycis intervention were calculated by comparing the relative abundance of the KEGG orthologs and pathways (Level 2) at baseline versus the endpoint data and were interpreted in the form of the mean log2‐fold change.
At the genus level, the most dominant genera detected within the gut microbiome were Rhodococcus, Escherichia‐Shigella, Enterococcus, Sphingomonas, Sediminibacterium, Ralstonia, Lacibacter, Sphingobium and Lactobacillus, which accounted for 80% of the overall abundance of bacteria in the silkworm gut (Figs 3(B) and S2(B)). Coexistence and competition relationships between gut bacteria were found in the silkworm. The abundance of Enterococcus was negatively correlated with that of the most predominant genera (Fig. S2(C)). LEfSe analysis confirmed that the gut microbiota composition at different inoculation times was consistent, but the abundance differed. For example, the relative abundances of Enterococcus, Escherichia‐Shigella, Staphylococcus and Glutamicibacter were higher at 6 dpi (Fig. S2(D)). With prolonged infection, the abundance of Enterococcus increased gradually (F (3,56) = 40.04, P < 0.0001 for 2 days; F (3,56) = 5.408, P < 0.0024 for 4 days; F (3,56) = 25.35, P < 0.0001 for 6 days). With treatment at a concentration of 109 spores/mL (6 days), 1010 16S rRNA gene copies of Enterococcus were observed in silkworm intestinal contents (Fig. 3(C)).
There was a statistically significant positive correlation between the total N. bombycis number and the Enterococcus number in the gut (R 2 = 0.14, P = 0.002 for 2 days; R 2 = 0.54, P = 0.8 for 4 days; R 2 = 0.77, P < 0.001 for 6 days; Fig. 3(D)). The results of RDA and CCA of the environmental factors and the bacterial microbial community in all the treatments are shown in Fig 3(E) and S3. The Enterococcus abundance showed a positive correlation with infection time (R 2 = 0.4398, P = 0.001) and N. bombycis concentration (R 2 = 0.3121, P = 0.001) (P < 0.05) (Fig. S3, Table S1). In addition, we explored the predicted functions of the gut microbiota across all groups in our study by PICRUSt analyses Fig. 3(F). Across all samples, the enriched KEGG pathways included carbohydrate metabolism, amino acid metabolism and energy metabolism. Interestingly, almost all the KEGG pathways of the N. bombycis treatment group were significantly down‐regulated compared with those of the CK group (P < 0.01) (Fig. 3(F)). Finally, the Sloan neutral model showed a more niche‐based process of the gut microbiome in N. bombycis‐treated silkworm larvae (Fig. S4(A)). The AUC at 4 dpi (0.84) and 6 dpi (0.82) was significantly greater than that at 2 dpi (0.61) (P < 0.001) (Fig. S4(B–D)).
3.3. Anti‐N . bombycis assay of Enterococcus
Amplification of the LX10 16S rRNA sequence yielded a 1200‐bp PCR product, and the sequence (GenBank) showed 98% similarity with that of E. faecalis Feb‐67 (MH385355) (Fig. S5). The proteins (3–10 K SMP) precipitated by (NH4)2SO4 from the supernatant showed strong inhibitory activity (Fig. 4(A)). The protein concentration was quantified using the Bradford reagent and adjusted to 2 mg/mL. The inhibitory activity of Enterococcus showed that the spore germination rate was in the order CK (GKK) > proteinase K > (< 3 K SMP) > MRS > (> 10 K SMP) > (> 3 K SMP) > (< 10 K SMP) (Fig. 4(A)). The germination rate with the precipitated proteins (> 3 K SMP, < 10 K SMP) was significantly decreased to 10.44 ± 0.63% (t = 8.804, df = 10, P = 0.0009) and 9.45 ± 1.22% (t = 8.887, df = 10, P = 0.0009) compared to that of the CK group (42.06 ± 2.72%) (Fig. 4(A)). The inhibitory activity was absent in supernatants treated with proteinase K.
Figure 4.
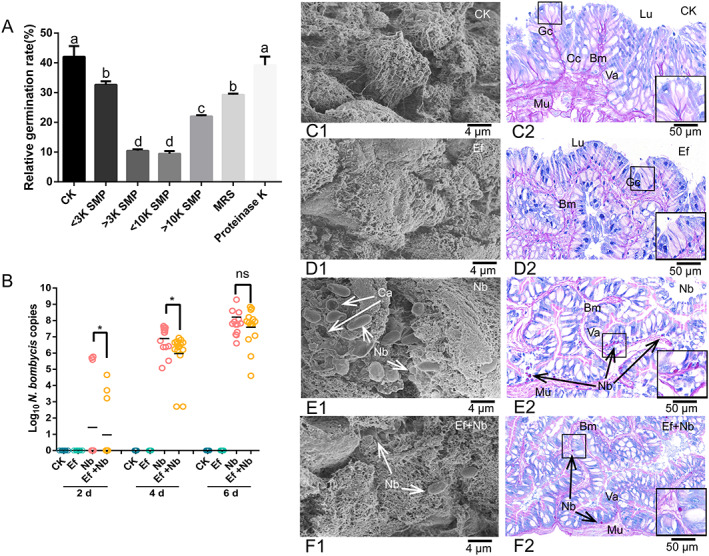
Anti‐N. bombycis assay of En. faecalis LX10 in vitro and in vivo. (A) In vitro inhibitory activity of the crude proteins in E. faecalis LX10 supernatants determined by measuring the relative germination rate of N. bombycis spores. After size exclusion centrifugation, inhibition from the crude proteins (SMP) was preserved between 3 and 10 kDa. Crude protein from MRS medium (MRS) was obtained as a negative control in the same way. (B) Inhibitory activity of E. faecalis LX10 in vivo. DNA was extracted from the intestinal epithelium, and the N. bombycis burden was estimated by qPCR amplification after treatment with E. faecalis LX10 (107 spores/mL) (Ef), N. bombycis (109 spores/mL) (Nb), or N. bombycis and E. faecalis LX10 (Ef + Nb). Healthy silkworms with the same volume of sterilized water were used as the control group (CK). (C–F) The gut of silkworms was excised for SEM and histological examination (PAS staining) of the CK (C1, C2), Ef (D1, D2), Nb (E1, E2), and Ef + Nb (F1, F2) group silkworms. The arrows indicate that the N. bombycis spores proliferated in epithelial cells. Cc, columnar cell; Gc, goblet cell; Lu, lumen; Mu, muscle layer; Mi, microvilli; Va, vacuoles. Statistically significant differences were calculated by using one‐way ANOVA (*P < 0.05).
In the in vivo study, a significant decrease in the silkworm gut N. bombycis burden was evident in the E. faecalis LX10‐treated (Nb) group relative to the group inoculated only with N. bombycis (Fig. 4(B)). In addition, the CK group (Fig. 4(C1,C2)) and E. faecalis LX10‐treated group ((Fig. 4(D1,D2)) showed clear microvilli of superficial epithelial cells and gut morphology. N. bombycis‐treated silkworms showed classical signs of N. bombycis infection, including extensive invasion of the epithelium by spores, disruption of the basement membrane, distended distal ends of goblet cells and columnar cells, and vacuolization of intracellular organelles (Fig. 4(E1,E2)). In contrast, silkworms that were treated with E. faecalis LX10 showed significantly reduced N. bombycis invasion in vivo, and epithelial cells still rested on muscle fibers and the basement membrane. Although the vacuoles increased in size, there were no signs of damage at this stage (Fig. 4(F1,F2)).
3.4. Antimicrobial protein identification
The molecular weight of the crude protein (enterococcin LX) (3–10 K) was determined by SDS‐PAGE (Fig. S6(A)). Through matrix‐assisted laser desorption/ionization‐time‐of‐flight‐mass spectrometry (MALDI‐TOF‐MS) analysis, the protein was confirmed as enterococcin EntV (E. faecalis) (NCBI reference sequence: QHN67019.1), an antimicrobial peptide of the streptococcin A‐M57 family with a molecular mass of 7.2 KDa and translated from the EF1097 locus gene in E. faecalis (strain V583) (Fig. 5(A,D)). The bactericidal substance (3–10 K) retained its activity after being incubated over a wide pH range (5–10) (F (6,14) = 35.53, P < 0.0001) and tolerated heat treatment (40–100 °C) (F (4,10) = 29.83, P < 0.0001), even at 100 °C for 30 min (Fig. 5(B,C)). These characteristics reveal the high stability of this antimicrobial compound, suggesting that it can function under harsh gut conditions. The templates with the highest quality were then selected for model building, and a Phyre2 model for enterococcin LX showed sequence identity (44.4%) (Fig. S6(B)).
Figure 5.
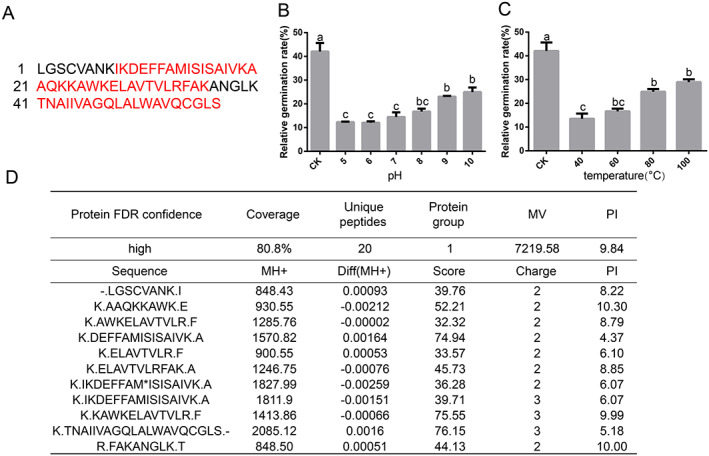
Identification of the E. faecalis LX10 protein (enterococcin LX) by LC–MS/MS. (A) Analysis of the amino acid sequence of the enterococcin LX protein by MS/MS. Amino acid sequence of the enterococcin LX protein deduced from the draft Enterococcus genome sequence. Peptide sequences matching the LC–MS/MS results are highlighted in red. (B) Effect of pH (5‐10) on the activity of crude proteins (3–10 K). (C) The effect of temperature on the inhibitory substance was tested by heating the supernatant at 40, 60, 80, and 100 °C. (D) Peptides identified with LC–MS/MS.
3.5. Gene expression changes and enzyme activities
The results showed that the Akirin, Cecropin A, Mesh, and ssk genes were gradually significantly up‐regulated after treatment with N. bombycis (P < 0.05) (Fig. 6(A–D)). Nevertheless, the expression levels of DUOX and NOS showed gradual up‐regulation during the early stages of infection (2 days) and then gradual down‐regulation in later stages of infection (4 and 6 days) (P < 0. 0001) (Fig. 6(E,F)). After feeding with E. faecalis LX10, the gene expression level of the N. bombycis‐treated silkworms showed different degrees of recovery and correction compared with the simple N. bombycis‐infected silkworms (Fig. 6(G–L)).
Figure 6.
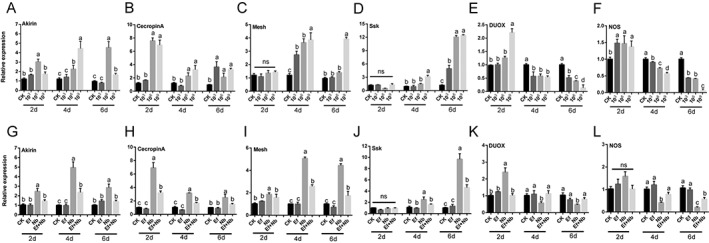
Immune‐related gene expression level. (A–F) The expression of each gene in the gut of infected larvae was compared to that in the gut of uninfected larvae (CK) after inoculation with different concentrations of N. bombycis (103, 106, and 109 spores/mL). (G–L) Expression levels of each gene after treatment with E. faecalis LX10 (107 spores/mL) (Ef), N. bombycis (109 spores/mL) (Nb), or N. bombycis and E. faecalis LX10 (Ef + Nb). The healthy silkworms were fed the same volume of sterile water as a control (CK). The messenger RNA (mRNA) levels of Akirin (A, G), Cecropin (B, H), Mesh (C, I), Ssk (D, J), DUOX (E, K) and NOS (F, L) were quantified by RT‐qPCR. Each bar represents the mean fold‐change ± SE of five independent experiments. Different letters represent significant differences at P < 0.05 (one‐way ANOVA with Tukey's post hoc test).
We further investigated the host gut H2O2 and NO levels and GST enzyme activities. The H2O2 and NO levels displayed complex temporal profiles, initially increasing after 2 days (F (3,8) = 42.08, P < 0.001 for H2O2; F (3,8) = 9.548, P = 0.005 for NO) and then decreasing after 4 days (F (3,8) = 1.888, P = 0.2101 for H2O2; F (3,8) = 10.06, P = 0.0043 for NO) and 6 days (F (3,8) = 7.633, P = 0.009 for H2O2; F (3,8) = 36.47, P < 0.001 for NO) (Fig. 7(A,B)). Similarly, GST activity was clearly inhibited after 4 and 6 days by N. bombycis (F (3,8) = 43.10, P < 0.001 for 4 days, F (3,8) = 4.311, P = 0.0437 for 6 days) (Fig. 7(C)). Notably, after treatment with E. faecalis LX10, the enzyme activity level showed a certain degree of correction, but there was no significant difference compared with that in the group with N. bombycis infection alone (Fig. 7(D–F)).
Figure 7.
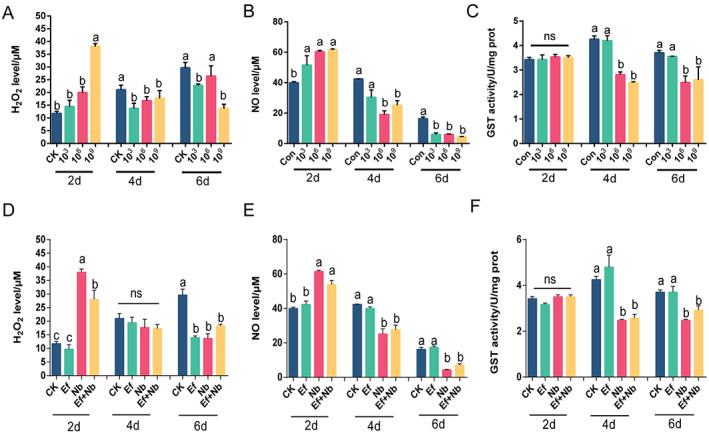
Hydrogen peroxide (H2O2) and NO levels and GST activity. (A–C) Effect of acute N. bombycis exposure on H2O2 level (A) and NO level (B) and GST activity (C). The vertical axis represents the relative gene expression ratios, and the horizontal axis represents the times (2, 4, and 6 days) and N. bombycis concentrations (CK, 103, 106, and 109 spores/mL). (D–F) H2O2 level (D) and NO level (E) and GST activity (F) after treatment with E. faecalis LX10 (107 spores/mL) (Ef), N. bombycis (109 spores/mL) (Nb), or N. bombycis and E. faecalis LX10 (Ef + Nb). Healthy silkworms were fed the same volume of sterile water as a control (CK). Each bar represents the mean fold‐change ± SE of five independent experiments. Different letters represent significant differences at P < 0.05 (one‐way ANOVA with Tukey's post hoc test).
4. DISCUSSION
This study showed that the sensitivity of the silkworm to N. bombycis treatment was both dose‐ and time‐dependent. Similar results have been observed for Plutella xylostella 43 and Apis mellifera. 44 Moreover, the growth of silkworms and food consumption were significantly inhibited, leading to substantial loss in cocoon yield after infection with N. bombycis. Many studies have found negative impacts of microsporidian infections on foraging efficiency, wing deformity, lifespan, colony fitness, fecundity and survival in honeybees and bumblebees. 45 , 46 Other studies have also explored whether microsporidian infection reduced the weight of the silk gland and growth rate due to the reduced intake of food and its utilization compared to those of healthy silkworms. 47 Many factors, such as initial spore dose, silkworm species, physiological resistance, nutrition status and immune response, may influence the virulence of N. bombycis to silkworms, which may contribute to the detoxification of B. mori. 48 , 49
Comparative analyses of the silkworm gut microbiome revealed that the gut microbiota compositions and abundance were changed dramatically after N. bombycis infection, and these differences might be associated with the gut health of the host. 50 , 51 The occurrence and codiversity pattern of the silkworm gut microbiota was significantly altered by N. bombycis. High microbial diversity can improve community stability, which may explain why a more intertwined and complex network was present in samples with healthy silkworms. 52 Importantly, we also found a strong positive correlation of Enterococcus with inoculation time and infection concentration. It has been reported that the genus Enterococcus is the most dominant bacterial genus in the silkworm gut, implying that the resistance of the silkworm to infection with N. bombycis can be increased by the use of probiotics and optimization of the configuration of the gut bacterial microbiota. 53 , 54 Enterococci isolated from the gastrointestinal tract of humans and other diseased animals have been considered important opportunistic pathogens with strong antibiotic resistance. 55 However, Enterococcus sp. have also been found to be probiotic and beneficial in several biological models, including in protecting honey bees from American foulbrood disease and improving fish health, so we hypothesize that the gut microbiota might be an important factor driving differences in resistance to N. bombycis infection. 56 , 57 Consistent with the KEGG prediction, the changes in gut bacteria caused by microspores may contribute to intestinal dysfunction.
E. faecalis LX10 not only reduced the spore germination rate of N. bombycis in vitro but also ameliorated gut injury in vivo, as confirmed by histopathological analyses. It is worth noting that E. faecalis LX10 reduced but did not eliminate N. bombycis colonization. Similar to our research, Suraporn and Terenius demonstrated that feeding with Lactobacillus bacteria could increase the survival rate of silkworm larvae challenged by N. bombycis. 58 In addition, the presence of E. faecalis could inhibit fungal morphogenesis and thereby prevent tissue damage caused by hypha expansion in nematode and mouse gut infection models. 59 Using a liquid probiotic form of E. faecalis L3 in infants has a positive impact on overall health and can increase resistance to acute respiratory infections. 60 Some species of Enterococcus produce lactic acid, and accumulation of lactic acid might reduce the pH of digestive juice in grasshoppers and silkworms. 16 , 61 The highest germination rate of Nosema occurs at pH 9–10. 26 , 62 Thus, the decrease in intestinal injury in silkworms treated with Enterococcus could be related to a reduction in pH in the gut, which possibly led to lowered infectivity of the spores. An alternative inhibitory mechanism of action that could affect pathogen infection might be competitive adhesion of E. faecalis to the intestinal tissue, which would inhibit N. bombycis adhesion. We speculated that in the presence of E. faecalis, N. bombycis reverts to a benign commensal interaction with the host. In addition, the antimicrobial protein enterococcin LX with anti‐N. bombycis activities was confirmed as enterococcin EntV. Graham et al. identified that the E. faecalis bacteriocin EntV, produced from the entV (ef1097) locus, is both necessary and sufficient for the reducing in Candida albicans virulence and biofilm formation through the inhibition of hyphal formation, a critical virulence trait. 63 Recent studies have also suggested that members of the gut bacterial genus Enterococcus that produce the peptidoglycan hydrolase SagA improve checkpoint inhibitor cancer immunotherapy in mouse melanoma models. 64 Nevertheless, whether metabolic products of intestinal bacteria can also directly inhibit silkworm infection by pathogens deserves further exploration. We speculate that after the mixing of Enterococcus with N. bombycis, an enzymatic reaction occurs to produce enterococcin, which is an antimicrobial substance, thus limiting the germination of N. bombycis.
We also demonstrated that E. faecalis LX10 can alter host resistance by influencing host physiology, such as via the host immune system and enzyme activity. In our study, after simultaneous feeding with E. faecalis LX10 and N. bombycis, the gene expression level and enzyme activity showed different degrees of recovery and correction compared with those with simple N. bombycis infection. Recent research has revealed that the gut microbiota plays an important role in combating systemic Salmonella infection by inducing local immune responses in mice. 65 In addition, the intestinal symbiotic genus Bacteroides has been found to directly reset the h'st's innate immune system through surface carbohydrate structures, increasing tolerance toward pathogens. 66 , 67 Modulation of the immune system by E. faecalis could potentially lead to the production of antimicrobial peptides and modulate key signaling pathways, such as the NF‐ĸB and MAPK pathways, to confer an advantage to the host. 68 Nishida et al. demonstrated that lactic acid‐producing bacteria stimulated innate immunity in silkworms. 69 In addition, the B. mori immune system quickly activates immune defense mechanisms after infection with N. bombycis, e.g. increased expression of Akirin, cecropinA, Mesh and Ssk. N. bombycis exposure has been shown to induce changes at the molecular level of the innate immune system. 70 For example, the relative expression of the antibacterial peptides abaecin, hymenoptaecin, and defensin significantly increases after Nosema apis infection of honey bees. 71 Moreover, conserved components of smooth septate junction transmembrane proteins (Mesh, Snakeskin: Ssk) in Drosophila and B. mori have been implicated in immune signaling functions and maintenance of the midgut barrier and homeostasis. 72 In contrast, N. bombycis infection seems to suppress the immune response by reducing protection against oxidative stress genes (DUOX, NOS) with prolonged infection. During most gut–pathogen interactions, intestinal redox homeostasis mediated via infection‐induced ROS generation by the dual oxidase enzyme and subsequent ROS elimination by immune‐regulated catalase is critical for host survival. 73
In conclusion, the present study provides a clear profile of the complex interplay among N. bombycis, silkworms, and the gut microbiome. From the perspective of biological control, there have been many successful precedents based on the removal of certain commensal bacteria and transgenic symbiosis, especially for vector pests. 74 , 75 , 76 For example, the symbiont, Rhodococcus rhodnii, has been genetically transformed to express cecropin A and a functional single‐chain antibody in the intestinal track of the host vector, Rhodnius prolixus, thereby producing stable paratransgenic insects to liminate or reduce the number of Trypanosoma cruzi. 74 The present study paves the way for the expression of molecules with anti‐microsporidia activity via genetically transformed Enterococcus symbionts of disease‐transmitting insects, or direct construction of transgenic insects via mobile DNA elements, which will greatly improve the practical application of microsporidia in lepidopteran pest groups.
CONFLICTS OF INTEREST
All authors compiled, wrote and approved this version of the article, and no part of this article has been published or submitted elsewhere. No conflict of interest exists in the submission of this article.
Supporting information
Figure S1.. Total bacterial 16S rRNA gene‐based abundances in the normal and N. bombycis treatment groups at different spore concentrations (CK, 103, 106, and 109 spores/mL) and infection times (2, 4, and 6 days).
Figure S2. Gut microbiota composition and correlations of dominant taxa. (A) The correlation‐based network of gut bacteria‐associated microbial phyla was detected at different infection times (CK, 2, 4, and 6 days). Co‐occurrence networks in topological properties include degree, average degree, average path length, modularity, diameter, and transitivity. (B) Heatmap showing the relative abundances of dominant taxa. Each row in the heatmap has been standardized to have a mean of zero (blue) and a standard deviation of one (red). Bacteria sorted from top to bottom according to the microbial abundance profiles. (C) Pairwise Pearson's correlation matrix distances of gut microbiota composition. Significant correlations (P < 0.05) are shown by asterisks; blue indicates a positive correlation, and red indicates a negative correlation. Color intensity and shape represent the correlation strength. (D) Linear discriminant analysis (LDA) effect size (LEfSe) analysis identifying taxonomic differences in the gut microbiota at different infection times. Histograms of LDA scores of 16S gene sequences at CK, 2, 4, and 6 days are shown, with a cut‐off LDA score (log10) above 3.0. *P < 0.05, **P < 0.01, ****P < 0.0001
Figure S3. Distance‐based redundancy analysis (RDA) and canonical correspondence analysis (CCA) using time and spore concentration as explanatory variables. The time (A) and spore concentration (B) of the subjects are represented by color gradients andthe shape of each dot, respectively. Arrows in the distance‐based‐RDA/CCA biplot denote the magnitudes and directions of the effects of explanatory variables.
Table S1. Statistically significant differences in different environmental variables are shown with distance‐based redundancy analysis (RDA) and canonical correspondence analysis (CCA)
Figure S4. Interactions between the gut microbiota of silkworms and N. bombycis. (A) Sloan neutral model predictions of the gut microbiota. CK, healthy silkworms; Nb, N. bombycis treatment group. (B–D) ROC curves of the proposed model and the baselines, (B) 103 group versus CK, AUC = 0.61, (C) 106 group versus CK, AUC = 0.84, and (D) 109 group versus CK, AUC = 0.82.
Figure S5. Phylogenetic tree indicating the relationship of the E. faecalis strain LX10 16S rRNA gene sequences compared with the sequences of related bacterial species from GenBank. The phylogenetic trees were constructed by MEGA 11 using a statistical neighbor‐joining method by applying the following parameters: bootstrap method (1000 replicates), Kimura 2‐parameter model, substitution to include (d: transitions + transversions), uniform rates, and pairwise deletion
Figure S6. SDS‐PAGE of the crude proteins (3–10 K) and identification of the enterococcin LX protein. (A) The crude protein component between 3 and 10 kDa was retained after size exclusion centrifugation. Samples were subjected to 15% SDS‐PAGE, stained with Coomassie Brilliant Blue protein gel stain, and visualized using a 300 nm UV transilluminator. Experiments were repeated at least three times. (B) Tertiary structure prediction for enterococcin LX by Phyre2. (C) Identification of the enterococcin LX protein (Excel table).
Table S2. Primers used in this study.
ACKNOWLEDGEMENTS
This work was supported by China Agriculture Research System of MOF and MARA, and the National Natural Science Foundation of China (81572027).
Contributor Information
Fan Zhang, Email: zhangfan0531@163.com.
Xingmeng Lu, Email: xmlu@zju.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number PRJNA763083.
REFERENCES
- 1. Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B et al., A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306:1937–1940 (2004). [DOI] [PubMed] [Google Scholar]
- 2. Didier ESDP, Snowden KF and Shadduck JA, Microsporidiosis in mammals. Microbes Infect 2:709–720 (2000). [DOI] [PubMed] [Google Scholar]
- 3. Bhat SA, Bashir I and Kamili AS, Microsporidiosis of silkworm, Bombyx mori L. (Lepidoptera‐bombycidae): a review. Afr J Agric Res 4:1519–1523 (2009). [Google Scholar]
- 4. Li Z, Wang Y, Wang L and Zhou Z, Molecular and biochemical responses in the midgut of the silkworm, Bombyx mori, infected with Nosema bombycis . Parasit Vectors 11:147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bjornson SOD, Microsporidia biological control agents and pathogens of beneficial insects, in Microsporidia Biological Control Agents and Pathogens of Beneficial Insects[M]. John Wiley & Sons, Ltd: (2014). [Google Scholar]
- 6. Nattoh G, Maina T, Makhulu EE, Mbaisi L, Mararo E, Otieno FG et al., Horizontal transmission of the symbiont microsporidia MB in anopheles arabiensis. Front Microbiol 12:647183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Down RE, Bell HA, Kirkbride‐Smith AE and Edwards JP, The pathogenicity of Vairimorpha necatrix (Microspora: Microsporidia) against the tomato moth, Lacanobia oleracea (Lepidoptera: Noctuidae) and its potential use for the control of lepidopteran glasshouse pests. Pest Manag Sci 60:755–764 (2004). [DOI] [PubMed] [Google Scholar]
- 8. Bardi C, Mariottini Y, Plischuk S and Lange CE, Status of the alien pathogen Paranosema locustae(microsporidia) in grasshoppers (Orthoptera: Acridoidea) of the Argentine Pampas . Biocontrol Sci Technol 22:497–512 (2012). [Google Scholar]
- 9. Wang‐Peng S, Zheng X, Jia WT, Li AM, Camara I, Chen HX et al., Horizontal transmission of Paranosema locustae (microsporidia) in grasshopper populations via predatory natural enemies. Pest Manag Sci 74:2589–2593 (2018). [DOI] [PubMed] [Google Scholar]
- 10. Lange CE and Azzaro FG, New case of long‐term persistence of Paranosema locustae (microsporidia) in melanopline grasshoppers (Orthoptera: Acrididae: Melanoplinae) of Argentina. J Invertebr Pathol 99:357–359 (2008). [DOI] [PubMed] [Google Scholar]
- 11. Miao J, Guo Y and Shi W, The persistence of Paranosema locustae after application in Qinghai plateau, China. Biocontrol Sci Technol 22:733–735 (2012). [Google Scholar]
- 12. Zheng S, Huang Y, Huang H, Yu B, Zhou N, Wei J et al., The role of NbTMP1, a surface protein of sporoplasm, in Nosema bombycis infection. Parasit Vectors 14:81 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heinz E, Hacker C, Dean P, Mifsud J, Goldberg AV, Williams TA et al., Plasma membrane‐located purine nucleotide transport proteins are key components for host exploitation by microsporidian intracellular parasites. PLoS Pathog 10:e1004547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han B and Weiss LM, Microsporidia: obligate intracellular pathogens within the fungal kingdom. Microbiol Spectr 5:2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han B, Takvorian PM and Weiss LM, Invasion of host cells by microsporidia. Front Microbiol 11:172 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang X, Sun C, Chen B, Du K, Yu T, Luang‐In V et al., Insect symbionts as valuable grist for the biotechnological mill: an alkaliphilic silkworm gut bacterium for efficient lactic acid production. Appl Microbiol Biotechnol 102:4951–4962 (2018). [DOI] [PubMed] [Google Scholar]
- 17. Wang Q, Ren M, Liu X, Xia H and Chen K, Identification and characterization of novel short‐type BmPGRP‐S4 from the silkworm, Bombyx mori, involved in innate immunity. Z Naturforsch C J Biosci 75:13–21 (2020). [DOI] [PubMed] [Google Scholar]
- 18. Sun YX, Chen C, Xu WJ, Abbas MN, Mu FF, Ding WJ et al., Functions of Bombyx mori cathepsin L‐like in innate immune response and anti‐microbial autophagy. Deve Comp Immunol 116:103927 (2021). [DOI] [PubMed] [Google Scholar]
- 19. Wang Q, Wang J, Ren M, Ma S, Liu X, Chen K et al., Peptidoglycan recognition protein‐S1 acts as a receptor to activate AMP expression through the IMD pathway in the silkworm Bombyx mori . Deve Comp Immunol 115:103903 (2021). [DOI] [PubMed] [Google Scholar]
- 20. Xin ZZ, Liu QN, Liu Y, Zhang DZ, Wang ZF, Zhang HB et al., Transcriptome‐wide identification of differentially expressed genes in Chinese oak silkworm Antheraea pernyi in response to Lead challenge. J Agric Food Chem 65:9305–9314 (2017). [DOI] [PubMed] [Google Scholar]
- 21. Ma Z, Li C, Pan G, Li Z, Han B, Xu J et al., Genome‐wide transcriptional response of silkworm (Bombyx mori) to infection by the microsporidian Nosema bombycis . PLoS One 8:e84137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen B, Zhang N, Xie S, Zhang X, He J, Muhammad A et al., Gut bacteria of the silkworm Bombyx mori facilitate host resistance against the toxic effects of organophosphate insecticides. Environ Int 143:105886 (2020). [DOI] [PubMed] [Google Scholar]
- 23. Kumar D, Sun Z, Cao G, Xue R, Hu X and CJJoA‐PE G, Study of gut bacterial diversity of Bombyx mandarina and Bombyx mori through 16S rRNA gene sequencing. J Asia Pac Entomol 22:522–530 (2019). [Google Scholar]
- 24. Wang D, Dong Z, Zhang Y, Guo K, Guo P, Zhao P et al., Proteomics provides insight into the interaction between mulberry and silkworm. J Proteome Res 16:2472–2480 (2017). [DOI] [PubMed] [Google Scholar]
- 25. Brummel T, Ching A, Seroude L, Simon AF and Benzer S, Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A 101:12974–12979 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu H, Chen B, Hu S, Liang X, Lu X and Shao Y, Quantitative proteomic analysis of germination of Nosema bombycis spores under extremely alkaline conditions. Front Microbiol 7:1459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peralta‐Sanchez JM, Martin‐Platero AM, Ariza‐Romero JJ, Rabelo‐Ruiz M, Zurita‐Gonzalez MJ, Banos A et al., Egg production in poultry farming is improved by probiotic bacteria. Front Microbiol 10:1042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eveno M, Savard P, Belguesmia Y, Bazinet L, Gancel F, Drider D et al., Compatibility, cytotoxicity, and gastrointestinal tenacity of bacteriocin‐producing bacteria selected for a consortium probiotic formulation to be used in livestock feed. Probiotics Antimicrob Proteins 13:208–217 (2021). [DOI] [PubMed] [Google Scholar]
- 29. Nairz M, Dichtl S, Schroll A, Haschka D, Tymoszuk P, Theurl I et al., Iron and innate antimicrobial immunity‐depriving the pathogen, defending the host. J Trace Elem Med Biol 48:118–133 (2018). [DOI] [PubMed] [Google Scholar]
- 30. Xi Z, Ramirez JL and Dimopoulos G, The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog 4:e1000098 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang F, Yang C, Zhang X, Zhu H, Zhao D and Huang Y, Isolation of an anti‐entomopathogenic fungal protein secreted from Pseudomonas aeruginosa BGf‐2: an intestinal bacteriam of Blattella germanica (L.). J Invertebr Pathol 173:107371 (2020). [DOI] [PubMed] [Google Scholar]
- 32. Shao Y, Chen B, Sun C, Ishida K, Hertweck C and Boland W, Symbiont‐derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Cell Chem Biol 24:66–75 (2017). [DOI] [PubMed] [Google Scholar]
- 33. Zhang F, Lu X, Kumar VS, Zhu H, Chen H, Chen Z et al., Effects of a novel anti‐exospore monoclonal antibody on microsporidial Nosema bombycis germination and reproduction in vitro . Parasitology 134:1551–1558 (2007). [DOI] [PubMed] [Google Scholar]
- 34. Fu Z, He X, Cai S, Liu H, He X, Li M et al., Quantitative PCR for detection of Nosema bombycis in single silkworm eggs and newly hatched larvae. J Microbiol Methods 120:72–78 (2016). [DOI] [PubMed] [Google Scholar]
- 35. Chen B, Du K, Sun C, Vimalanathan A, Liang X, Li Y et al., Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry‐feeding relatives. ISME J 12:2252–2262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He J, Zhang N, Muhammad A, Shen X, Sun C, Li Q et al., From surviving to thriving, the assembly processes of microbial communities in stone biodeterioration: a case study of the West Lake UNESCO World Heritage area in China. Sci Total Environ 805:150395 (2022). [DOI] [PubMed] [Google Scholar]
- 37. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ and Holmes SP, DADA2: high‐resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al., Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McMurdie PJ and Holmes S, Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bastian M, Heyman S and Jacomy M, An Open Source Software for Exploring and Manipulating Networks, Vol. 17. AAI Publications, San Jose, CA, p. 20 (2009). [Google Scholar]
- 41. Douglas GMBR and Langille MGI, Predicting the functional potential of the microbiome from marker genes using PICRUS. Methods Mol Biol 1849:169–177 (2018). [DOI] [PubMed] [Google Scholar]
- 42. Sinai L, Rosenberg A, Smith Y, Segev E and Ben‐Yehuda S, The molecular timeline of a reviving bacterial spore. Mol Cell 57:695–707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kermani N, Abu‐Hassan ZA, Dieng H, Ismail NF, Attia M and Abd Ghani I, Pathogenicity of Nosema sp. (microsporidia) in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). PLoS One 8:e62884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rubanov A, Russell KA, Rothman JA, Nieh JC and McFrederick QS, Intensity of Nosema ceranae infection is associated with specific honey bee gut bacteria and weakly associated with gut microbiome structure. Sci Rep 9:3820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Osterman J, Wintermantel D, Locke B, Jonsson O, Semberg E, Onorati P et al., Clothianidin seed‐treatment has no detectable negative impact on honeybee colonies and their pathogens. Nat Commun 10:692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li T, Fang Z, He Q, Wang C, Meng X, Yu B et al., Characterizing the Xenoma of Vairimorpha necatrix provides insights into the Most efficient mode of microsporidian proliferation. Front Cell Infect Microbiol 11:699239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gupta SK, Hossain Z, Nanu MM and Mondal K, Impact of microsporidian infection on growth and development of silkworm Bombyx mori L (Lepidoptera: Bombycidae). Agri Nat Resour 50:388–395 (2016). [Google Scholar]
- 48. He Q, Luo J, Xu JZ, Wang CX, Meng XZ, Pan GQ et al., Morphology and transcriptome analysis of Nosema bombycis Sporoplasm and insights into the initial infection of microsporidia. mSphere 5:1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dong Z, Zheng N, Hu C, Deng B, Fang W, Wu Q et al., Nosema bombycis microRNA‐like RNA 8 (Nb‐milR8) increases fungal pathogenicity by modulating BmPEX16 gene expression in its hos, Bombyx mori . Microbiol Spectr 9:e0104821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin L, Du Y, Song J, Wang W and Yang C, Imaging commensal microbiota and pathogenic bacteria in the gut. Acc Chem Res 54:2076–2087 (2021). [DOI] [PubMed] [Google Scholar]
- 51. Chang PV, Chemical mechanisms of colonization resistance by the gut microbial metabolome. ACS Chem Biol 15:1119–1126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schirmer M, Denson L, Vlamakis H, Franzosa EA, Thomas S, Gotman NM et al., Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe 24:600–610 e604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiang HLM, Zhao Y, Zhao LP, Zhang YH and Huang YP, Bacterial community in midguts of the silkworm larvae estimated by PCR/DGGE and 16S rDNA gene library analysis. Acta Entomologica Sinica 50:222–233 (2007). [Google Scholar]
- 54. Sun Z, Lu Y, Zhang H, Kumar D, Liu B, Gong Y et al., Effects of BmCPV infection on silkworm Bombyx mori intestinal bacteria. PLoS One 11:e0146313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Torres C, Alonso CA, Ruiz‐Ripa L, Leon‐Sampedro R, Del Campo R and Coque TM, Antimicrobial resistance in enterococcus spp. of animal origin. Microbiol Spectr 6:4 (2018). [DOI] [PubMed] [Google Scholar]
- 56. Du Y, Luo S and Zhou X, Enterococcus faecium regulates honey bee developmental genes. Int J Mol Sci 22:12105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Costa Sousa N, Couto MVS, Abe HA, Paixão PEG, Cordeiro CAM, Monteiro Lopes E et al., Effects of an Enterococcus faecium‐based probiotic on growth performance and health of Pirarucu, Arapaima gigas . Aquac Res 50:3720–3728 (2019). [Google Scholar]
- 58. Suraporn S and Terenius O, Supplementation of Lactobacillus casei reduces the mortality of Bombyx mori larvae challenged by Nosema bombycis . BMC Res Notes 14:398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Garsin DA and Lorenz MC, Candida albicans and Enterococcus faecalis in the gut: synergy in commensalism? Gut Microbes 4:409–415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gonchar NVSA, Maryshev VP, Sorokina TM, Churkova TV and Kharit SM, Probiotics, nutritional status and resistance to respiratory infections in infants. Eksp Klin Gastroenterol 1:48–54 (2015). [PubMed] [Google Scholar]
- 61. Mead LJ, Khachatourians GG and Jones GA, Microbial ecology of the gut in laboratory stocks of the migratory grasshopper, Melanoplus sanguinipes (Fab.) (Orthoptera: Acrididae). Appl Environ Microbiol 54:1174–1181 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Singh TBM and Khan MA, Microsporidiosis in the silkworm, Bombyx mori L (Lepidoptera: Bombycidae). Pertanika J Trop Agric Sci 35:387–406 (2012). [Google Scholar]
- 63. Graham CE, Cruz MR, Garsin DA and Lorenz MC, Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans . Proc Natl Acad Sci U S A 114:4507–4512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Griffin ME, Espinosa J, Becker JL, Luo JD, Carroll TS, Jha JK et al., Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 373:1040–1046 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M et al., Gut microbiota‐induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 44:647–658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mazmanian SK, Liu CH, Tzianabos AO and Kasper DL, An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107–118 (2005). [DOI] [PubMed] [Google Scholar]
- 67. Edelman SM and Kasper DL, Symbiotic commensal bacteria direct maturation of the host immune system. Curr Opin Gastroenterol 24:720–724 (2008). [DOI] [PubMed] [Google Scholar]
- 68. Bermudez‐Brito M, Plaza‐Diaz J, Munoz‐Quezada S, Gomez‐Llorente C and Gil A, Probiotic mechanisms of action. Ann Nutr Metab 61:160–174 (2012). [DOI] [PubMed] [Google Scholar]
- 69. Nishida S, Ono Y and Sekimizu K, Lactic acid bacteria activating innate immunity improve survival in bacterial infection model of silkworm. Drug Discov Ther 10:49–56 (2016). [DOI] [PubMed] [Google Scholar]
- 70. He X, Fu Z, Li M, Liu H, Cai S, Man N et al., Nosema bombycis (Microsporidia) suppresses apoptosis in BmN cells (Bombyx mori). Acta Biochim Biophys Sin (Shanghai) 47:696–702 (2015). [DOI] [PubMed] [Google Scholar]
- 71. Antúnez K, Martín‐Hernández R, Prieto L, Meana A, Zunino P and MJEM H, immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ Microbiol 11:2284–2290 (2009). [DOI] [PubMed] [Google Scholar]
- 72. Chen HJ, Li Q, Nirala NK and Ip YT, The snakeskin‐mesh complex of smooth septate junction restricts Yorkie to regulate intestinal homeostasis in drosophila. Stem Cell Reports 14:828–844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang Y, Liu C, Jin R, Wang Y, Cai T, Ren Z et al., Dual oxidase‐dependent reactive oxygen species are involved in the regulation of UGT overexpression‐mediated clothianidin resistance in the brown planthopper, Nilaparvata lugens. Pest Manag Sci 77:4159–4167 (2021). [DOI] [PubMed] [Google Scholar]
- 74. Durvasula RV, Gumbs A, Panackal A, Kruglov O, Aksoy S, Merrifield RB et al., Prevention of insect‐borne disease: an approach using transgenic symbiotic bacteria. Proc Natl Acad Sci U S A 94:3274–3278 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang S, Dos‐Santos AL, Huang W, Liu KC, Oshaghi MA, Wei G et al., Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 357:1399–1402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang F, Sun XX, Zhang XC, Zhang S, Lu J, Xia YM et al., The interactions between gut microbiota and entomopathogenic fungi: a potential approach for biological control of Blattella germanica (L.). Pest Manag Sci 74:438–447 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.. Total bacterial 16S rRNA gene‐based abundances in the normal and N. bombycis treatment groups at different spore concentrations (CK, 103, 106, and 109 spores/mL) and infection times (2, 4, and 6 days).
Figure S2. Gut microbiota composition and correlations of dominant taxa. (A) The correlation‐based network of gut bacteria‐associated microbial phyla was detected at different infection times (CK, 2, 4, and 6 days). Co‐occurrence networks in topological properties include degree, average degree, average path length, modularity, diameter, and transitivity. (B) Heatmap showing the relative abundances of dominant taxa. Each row in the heatmap has been standardized to have a mean of zero (blue) and a standard deviation of one (red). Bacteria sorted from top to bottom according to the microbial abundance profiles. (C) Pairwise Pearson's correlation matrix distances of gut microbiota composition. Significant correlations (P < 0.05) are shown by asterisks; blue indicates a positive correlation, and red indicates a negative correlation. Color intensity and shape represent the correlation strength. (D) Linear discriminant analysis (LDA) effect size (LEfSe) analysis identifying taxonomic differences in the gut microbiota at different infection times. Histograms of LDA scores of 16S gene sequences at CK, 2, 4, and 6 days are shown, with a cut‐off LDA score (log10) above 3.0. *P < 0.05, **P < 0.01, ****P < 0.0001
Figure S3. Distance‐based redundancy analysis (RDA) and canonical correspondence analysis (CCA) using time and spore concentration as explanatory variables. The time (A) and spore concentration (B) of the subjects are represented by color gradients andthe shape of each dot, respectively. Arrows in the distance‐based‐RDA/CCA biplot denote the magnitudes and directions of the effects of explanatory variables.
Table S1. Statistically significant differences in different environmental variables are shown with distance‐based redundancy analysis (RDA) and canonical correspondence analysis (CCA)
Figure S4. Interactions between the gut microbiota of silkworms and N. bombycis. (A) Sloan neutral model predictions of the gut microbiota. CK, healthy silkworms; Nb, N. bombycis treatment group. (B–D) ROC curves of the proposed model and the baselines, (B) 103 group versus CK, AUC = 0.61, (C) 106 group versus CK, AUC = 0.84, and (D) 109 group versus CK, AUC = 0.82.
Figure S5. Phylogenetic tree indicating the relationship of the E. faecalis strain LX10 16S rRNA gene sequences compared with the sequences of related bacterial species from GenBank. The phylogenetic trees were constructed by MEGA 11 using a statistical neighbor‐joining method by applying the following parameters: bootstrap method (1000 replicates), Kimura 2‐parameter model, substitution to include (d: transitions + transversions), uniform rates, and pairwise deletion
Figure S6. SDS‐PAGE of the crude proteins (3–10 K) and identification of the enterococcin LX protein. (A) The crude protein component between 3 and 10 kDa was retained after size exclusion centrifugation. Samples were subjected to 15% SDS‐PAGE, stained with Coomassie Brilliant Blue protein gel stain, and visualized using a 300 nm UV transilluminator. Experiments were repeated at least three times. (B) Tertiary structure prediction for enterococcin LX by Phyre2. (C) Identification of the enterococcin LX protein (Excel table).
Table S2. Primers used in this study.
Data Availability Statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number PRJNA763083.


