Abstract
C4 photosynthesis is thought to have evolved via intermediate stages, with changes towards the C4 phenotype gradually enhancing photosynthetic performance. This hypothesis is widely supported by modelling studies, but experimental tests are missing. Mixing of C4 components to generate artificial intermediates can be achieved via crossing, and the grass Alloteropsis semialata represents an outstanding study system since it includes C4 and non‐C4 populations. Here, we analyse F1 hybrids between C3 and C4, and C3+C4 and C4 genotypes to determine whether the acquisition of C4 characteristics increases photosynthetic performance. The hybrids have leaf anatomical characters and C4 gene expression profiles that are largely intermediate between those of their parents. Carbon isotope ratios are similarly intermediate, which suggests that a partial C4 cycle coexists with C3 carbon fixation in the hybrids. This partial C4 phenotype is associated with C4‐like photosynthetic efficiency in C3+C4 × C4, but not in C3 × C4 hybrids, which are overall less efficient than both parents. Our results support the hypothesis that the photosynthetic gains from the upregulation of C4 characteristics depend on coordinated changes in anatomy and biochemistry. The order of acquisition of C4 components is thus constrained, with C3+C4 species providing an essential step for C4 evolution.
Keywords: C3–C4 intermediates, C4 photosynthesis, complex traits, hybrids, leaf anatomy, Poaceae, transcriptome
Summary statement
We analyse hybrids between C3 and C4, and C3+C4 and C4 accessions of the grass Alloteropsis semialata. While hybrids have upregulated C4 features, these do not consistently increase photosynthetic efficiency, highlighting the importance of the order of acquisition of components during C4 evolution.
1. INTRODUCTION
C4 photosynthesis is a complex adaptation that allows plants to sustain high photosynthetic rates in conditions that limit CO2 availability in the leaf, such as hot, dry and saline environments (Sage, 2004). The C4 metabolism relies on a biochemical cycle operating within a specialized leaf anatomy, which concentrates CO2 around Rubisco, suppressing the energetically costly photorespiratory pathway (Hatch, 1987). The C4 trait is highly polyphyletic, but its multiple origins are mostly clustered in a few groups (Sage et al., 2011). The increased evolutionary accessibility to a C4 physiology in these groups has been associated with a number of ecological, anatomical and genetic features that acted as evolutionary enablers (Christin et al., 2013; Edwards, 2019; Moreno‐Villena et al., 2018; Sage, 2001). The classic model of C4 evolution hypothesizes that, once these enablers are in place, an intermediate metabolism that relies on a photorespiratory CO2 pump is established (in ‘type I' C3–C4 intermediates or ‘C2' plants), and gradually complemented by a weak C4 cycle (in ‘type II' C3–C4 sensu Edwards & Ku, 1987, or ‘C3+C4' plants sensu Dunning et al., 2017), which is subsequently optimized (Hylton et al., 1988; Monson & Moore, 1989; Sage, 2004). The transition through the intermediate stages has been predicted by mechanistic modelling studies, which support the idea that the sequential acquisition of components of the C4 trait successively increases fitness through higher photosynthetic output (Heckmann et al., 2013, 2016; Mallmann et al., 2014). This model however currently lacks experimental support. Efforts to engineer the C4 trait into C3 crops have provided the first opportunities to study the effects of C4 components in isolation (Ermakova et al., 2021; Ishimaru et al., 1997; Taniguchi et al., 2008; Wang et al., 2017), but the recipient species do not necessarily possess the anatomical properties required for an efficient C4 metabolism. An alternative strategy is to segregate the components of the C4 trait via crosses between C4 and non‐C4 plants (Simpson et al., in press).
Successful interspecific crosses between C4 and non‐C4 plants have been reported particularly for the eudicot genera Atriplex (e.g., Björkman et al., 1969; Oakley et al., 2014) and Flaveria (e.g., Araus et al., 1990; Byrd et al., 1992; reviewed by Brown & Bouton, 1993), with a recent interest in exploring this strategy to dissect the C4 trait (Lin et al., 2021; Oakley et al., 2014; Simpson et al., in press; Sultmanis, 2018). Most of the diversity of C4 lineages however lies in monocots, and especially the grass family, with at least 22 independent origins of C4 photosynthesis (Grass Phylogeny Working Group II [GPWG II], 2012). The only existing crosses between photosynthetic types in monocots concerned C3 and C3–C4 parents (Bouton et al., 1986), and the grass Alloteropsis semialata offers a particularly suitable system to complement these studies, as it includes not only C3 and C4 (Ellis, 1974a), but also C3+C4 individuals (Lundgren et al., 2016). While different photosynthetic types of A. semialata can hybridize (Bianconi et al., 2020), the effects of different crosses remain to be described. Alloteropsis semialata has a paleotropical distribution, with C3 and C3+C4 populations in Southern and Central/Eastern regions of Africa, respectively, and C4 individuals occurring across Africa, South and Southeast Asia, and Oceania (Bianconi et al., 2020; Lundgren et al., 2015; Olofsson et al., 2016, 2021). The photosynthetic types in A. semialata are associated with distinct genetic lineages (Lundgren et al., 2015; Olofsson et al., 2016), which initially diverged around 3 Mya (Bianconi et al., 2020). Although the ranges of C4 and non‐C4 lineages geographically overlap in some regions of Africa, with C3+C4 and C4 in Central/Eastern Africa, and C3 and C4 in Southern Africa, genetic analyses have shown that natural hybrids are rare (Bianconi et al., 2020; Olofsson et al., 2016, 2021). The lack of a clear hybrid zone has been partially explained by the differences in ploidy levels among photosynthetic types where they are found in sympatry (Olofsson et al., 2021). However, the broad ranges of C4 and C3+C4 diploids overlap, and they have been coexisting for at least one million years (Bianconi et al., 2020; Lundgren et al., 2015). Besides ploidy differences, the low frequency of natural hybridization might be associated with pre‐ and postzygotic barriers, such as possible hybrid depression associated with pleiotropic costs of upregulating components of C4 photosynthesis without a fully functional C4 metabolism in place (Olofsson et al., 2021). Experimental crosses between C4 and non‐C4 individuals of A. semialata provide an opportunity to test this hypothesis.
Alloteropsis semialata has a paleotropical distribution, with C3 and C3 + C4 populations in Southern and Central/Eastern regions of Africa, respectively, and C4 individuals occurring across Africa, South and Southeast Asia, and Oceania (Bianconi et al., 2020; Lundgren et al., 2015; Olofsson et al., 2016, 2021). The photosynthetic types in A. semialata are associated with distinct genetic lineages (Lundgren et al., 2015; Olofsson et al., 2016), which initially diverged around 3 Mya (Bianconi et al., 2020). Although the ranges of C4 and non‐C4 lineages geographically overlap in some regions of Africa, with C3 + C4 and C4 in Central/Eastern Africa, and C3 and C4 in Southern Africa, genetic analyses have shown that natural hybrids are rare (Bianconi et al., 2020; Olofsson et al., 2016, 2021). The lack of a clear hybrid zone has been partially explained by the differences in ploidy levels among photosynthetic types where they are found in sympatry (Olofsson et al., 2021). However, the broad ranges of C4 and C3 + C4 diploids overlap, and they have been coexisting for at least one million years (Bianconi et al., 2020; Lundgren et al., 2015). Besides ploidy differences, the low frequency of natural hybridization might be associated with pre‐ and postzygotic barriers, such as possible hybrid depression associated with pleiotropic costs of upregulating components of C4 photosynthesis without a fully functional C4 metabolism in place (Olofsson et al., 2021). Experimental crosses between C4 and non‐C4 individuals of A. semialata provide an opportunity to test this hypothesis.
Here we generate hybrids between C3 and C4 (C3 × C4), and C3+C4 and C4 (C3+C4 × C4) accessions of A. semialata and analyse their phenotype to test the hypotheses that components of the C4 trait are additive, and that hybrids rank between their non‐C4 and C4 parents in terms of photosynthetic performance. Our study confirms that crosses between photosynthetic types in A. semialata are viable in experimental conditions, and shows that the C4 metabolism is disrupted in the hybrids despite the significant upregulation of anatomical and biochemical components of the C4 trait.
2. MATERIALS AND METHODS
2.1. Plant material and growth conditions
Crosses between accessions of Alloteropsis semialata (R. Br.) Hitchc. (Poaceae) from distinct geographical origins were generated from parental plants collected in the wild as seeds or cuttings (Table S1) and grown in greenhouse conditions at the Arthur Willis Environment Centre, University of Sheffield (UK). Initial attempts of controlled cross‐pollination using pollination bags to isolate hand‐pollinated inflorescences were unsuccessful. For this reason, we adopted a noncontrolled pollination strategy, in which plants were allowed to receive pollen from any other plant in the greenhouse, with subsequent determination of the pollen parent via genotyping (see below). The resulting seeds were collected while still attached to the mother plant, and subsequently germinated on Petri dishes before being potted. Here, a total of 31 seedlings from seven mother plants (two C3, two C3+C4 and three C4) were obtained. Subsequent genotyping of the seedlings identified multiple F1 hybrids between parents with distinct photosynthetic types, including 14 C3 × C4 and 6 C3+C4 × C4 individuals (Table S1). The remaining seedlings consisted of 10 C4 × C4 crosses, and an additional C3+C4 plant resulting from self‐pollination (Table S2). Five C3 × C4 individuals grew to maturity and were sampled for DNA analyses but died before being phenotyped, and another two died before being sampled for RNA sequencing and leaf gas‐exchange analyses (Table S1). For the phenotypic analyses, we included the mother plants, the known and possible pollen parents (i.e., similar C4 accessions that could not be distinguished by the genotyping analyses; see Table S1), and seven additional accessions of A. semialata (three C3, two C3+C4 and two C4) that were growing alongside them were added to increase the phenotypic diversity within photosynthetic types (Table S1). All plants were grown in 11‐L, free‐draining pots containing a 2:1 mix of M3 compost (Levington) and perlite (Sinclair) under well‐watered conditions, and were fertilized once every 3 months with an NK fertilizer 16‐0‐5 containing Iron (Evergreen Extreme Green Lawn Food). Plants were grown on a 12‐h day/night cycle, with metal halide lamps providing supplementary light (additional 200 μmol m−2 s−1 at bench level), 25/20°C day/night temperature, ambient CO2 concentration and relative humidity between 30% and 60%.
2.2. Genotyping
We used three genetic markers to determine the genetic lineage of the pollen parent of each plant. These markers correspond to regions of the nuclear genome (~500 bp long) with sufficient variation to distinguish between different photosynthetic types and geographical groups, and were selected from a set of markers previously assembled to determine the origin of allopolyploids in A. semialata (Bianconi et al., 2020). Custom primers were designed to PCR amplify the target genes (Table S3). Genomic DNA (gDNA) of the putative crosses was isolated from fresh leaves using the DNeasy Plant Mini Kit (Qiagen). PCR reactions contained ca. 10–40 ng of gDNA template, 5 µl of 5× GoTaq Flexi reaction buffer (Promega), 2 mM of MgCl2, 0.08 mM of dNTPs, 0.2 µM of each primer and 0.5 unit of GoTaq polymerase (Promega) in a total volume of 25 µl. The PCR mixtures were initially incubated in a thermocycler for 2 min at 94°C followed by 35 cycles consisting of 30 s at 94°C (denaturation), 1 min at 48°C (annealing) and 1 min at 72°C (elongation). Amplicons were cleaned using Exo‐SAP‐IT (Affymetrix), and Sanger‐sequenced at the Core Genomic Facility at the University of Sheffield. Sequencing chromatograms were individually inspected for heterozygous sites that are polymorphic among genetic groups corresponding to distinct photosynthetic types, since the presence of such heterozygous sites indicates an F1 hybrid between photosynthetic types. Because the maternal origin was known in advance (the maternal plant was the one on which the seed was collected), the information from the alternative bases was used to narrow down the pollen parent to a genetic lineage (Table S2). The hybrid origin of the plants was subsequently confirmed using RNAseq data (see below).
2.3. Stable carbon isotopes
Carbon isotope composition was determined from the central portion of fully expanded leaf blades. Samples were dried in silica gel, and subsequently ground to a fine powder using a TissueLyzer II (Qiagen). Carbon isotope analysis was conducted on 1–2 mg of leaf sample using an ANCA GSL preparation module coupled to a Sercon 20‐20 stable isotope ratio mass spectrometer (PDZ Europa). Carbon isotopic ratios (δ13C, in ‰) were reported relative to the standard Pee Dee Belemnite (PDB). Values of δ13C higher than −16‰ indicate that the plants grew using C4 photosynthesis (O'Leary, 1988; Stata et al., 2019). δ13C values of additional accessions of A. semialata that were previously phenotyped and identified as C3, C3+C4 or C4 (Dunning et al., 2017, 2019a; Lundgren et al., 2016, 2019) were retrieved from Lundgren et al. (2015, 2016).
2.4. Leaf anatomy
Leaf cross sections were obtained from the central portion of fully expanded leaf blades. Fresh samples were initially dehydrated in an ethanol series from 70% to 100% EtOH, and resin‐infiltrated with Technovit 7100 (Heraeus Kulzer GmbH) following the manufacturer's instructions. Cross sections 7–10‐μm‐thick were obtained using a microtome (Leica RM 2245; Leica Biosystems Nussloch GmbH), and stained with Toluidine Blue O (Sigma‐Aldrich). An Olympus BX51 microscope coupled to an Olympus DP71 camera (Olympus Corporation) was used to photograph the cross sections. Leaf anatomical traits were measured using ImageJ v.1.51q (Schneider et al., 2012) in one cross section per individual. Measurements were taken following Lundgren et al. (2019) within segments, where the segment was defined as the area between two consecutive secondary veins (i.e., veins with large metaxylem vessels; see Figure S1). Segments that were adjacent to the midvein or to the lateral margins of the cross section were avoided to maintain consistency between samples. The variables measured here included leaf thickness (at the leftmost secondary vein in the segment), the number of minor veins (4th and 5th order veins), vein density, distance between edges of consecutive bundle sheaths (BSD), width of inner (IS) and outer sheath (OS), and the areas of epidermis (including bulliform cells, Epd), mesophyll (M), extraxylary fibres (Fb), bundle sheath (separately for IS and OS) and vascular tissue (V; see Figure S1 for an illustrated key for each variable). Areas were expressed relative to the total area of the segment before analysis, except IS and OS, which were expressed relative to the mesophyll area (IS/M and OS/M). A principal component analysis (PCA) was performed on these 12 leaf anatomical variables using the function prcomp in R v. 3.6.3 (R Core Team, 2020).
2.5. Leaf transcriptome
Leaf mRNA was isolated and sequenced as previously described (Dunning et al., 2019a). In short, the distal halves of fully expanded leaves were sampled in the middle of the light period, flash‐frozen in liquid N2 and kept at −80°C. All samples were collected on the same day. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) with an on‐column DNA digestion step (RNase‐Free DNase Set; Qiagen). A total of 20 RNA‐Seq libraries (one per individual) were prepared with the TruSeq RNA Library Preparation Kit v2 (Illumina) using 0.5 μg of starting RNA and aiming at a median insert size of ~155 bp (standard fragmentation protocol). Libraries were paired‐end sequenced (read length = 100 bp) on 1/24 of a single lane of an Illumina HiSeq 2500 flow cell in rapid mode (with four additional samples from an unrelated project) at the Sheffield Diagnostic Genetics Service. Raw sequence reads were filtered to remove adaptor contamination and low‐quality reads (i.e., <80% of bases with Phred score >20) using NGSQCToolkit v. 2.3.3 (Patel & Jain, 2012). Reads were further trimmed from the 3′ end to remove bases with Phred score <20. The quality of the filtered data was assessed using FastQC v. 0.11.9 (Andrews, 2010).
Transcript abundance was quantified by mapping the filtered reads to a reference data set consisting of coding sequences from multiple transcriptomes of A. semialata retrieved from Dunning et al. (2017) and modified by Bianconi et al. (2018). In short, this data set consists of 5540 groups of homologous genes that are common to the Panicoideae grasses (the subfamily that includes Alloteropsis), each group containing all paralogs detected in the transcriptomes of A. semialata (total of 12 234 groups of co‐orthologs). We used this data set as it includes manually curated sets of co‐orthologs of 23 gene families known to have a function in the C4 biochemistry (Bianconi et al., 2018). This curated gene set increases the read mapping accuracy when paralogs exist with a high sequence similarity. Furthermore, this data set includes the sequences of paralogs of core C4 genes that are absent from the reference genome of A. semialata, such as four different genes for the enzyme phosphoenolpyruvate carboxylase (PEPC) known to be highly expressed in other A. semialata accessions (Dunning et al., 2017, 2019a). Filtered reads were mapped to the reference data set using Bowtie2 v. 2.3.5 (Langmead & Salzberg, 2012) with default parameters. Read counts are reported here in reads per million mapped reads (RPM). To visualize the diversity among samples, a PCA on the global patterns of gene expression was conducted using the function prcomp in R after log2‐transforming the RPM values. Only genes with transcript abundance >10 RPM in at least five samples were used for the PCA. To test whether the global patterns of gene expression were consistent when the whole gene set of A. semialata was considered, we repeated the analysis using the chromosome‐level genome assembly of this species (Dunning et al., 2019b) as reference for read mapping. Finally, to verify whether the expression level differences between photosynthetic types were consistent with previous reports for A. semialata, we quantified transcript abundance (using the co‐orthologous gene set as reference) for 20 additional accessions (four C3, six C3+C4 and 10 C4) retrieved from two previous RNAseq studies (Dunning et al., 2017, 2019a).
To explore the global differences in gene expression between hybrids and the parental types, we performed a differential expression analysis on the data set that was generated using the complete genome of A. semialata as reference for read mapping. Here, due to an insufficient number of replicates for C3 and C3+C4 plants, we restricted our analyses to the comparisons between the two hybrid types, and between each of these and the C4 group. We then investigated the main metabolic functions associated with the set of differentially expressed genes from each comparison (see full description of the global RNAseq analyses on the Supporting Information Methods).
Finally, to confirm the paternity of the putative hybrids, we genotyped the plants using the RNAseq data set mapped to the reference genome of A. semialata. Here we also included individuals of A. semialata from the RNAseq studies of Dunning et al. (2017, 2019a) to increase the genetic diversity of the data set. Read alignment files were filtered to remove duplicates using Picard Tools v. 1.102 (https://broadinstitute.github.io/picard/), and variants were called on all individuals combined using BCFtools v. 1.9 (Li et al., 2009). Variant sites were filtered using VCFtools v. 0.1.17 (Li et al., 2009) to remove low‐quality genotype calls (i.e., quality score <30 and read depth <3), indels, and sites with more than 10% missing data, which resulted in an initial set of 521 929 single‐nucleotide polymorphisms (SNPs). BCFtools was then used to retain only multiallelic sites in which all hybrids were heterozygous and all potential parents were homozygous, which resulted in 15 406 SNPs that were then used for the genotyping tests. First, to determine the type of cross (i.e., whether C3 × C4 or C3+C4 × C4), we identified SNPs that were common to all individuals of the same photosynthetic type and exclusive to them. Sites with more than 25% missing data within each photosynthetic group were removed, and a total of 114 SNPs were retained (Table S4). We then tested, for each SNP, whether hybrid individuals had one C4 allele and one allele that was either C3 or C3+C4, according to the type of cross (Table S4). With the confirmation of the type of cross, we then narrowed down the genetic lineage of the pollen parent (as the mother is already known). Only one individual representing each genetic lineage was retained for this analysis (Table S5). First, we used the 15 406 SNP set to identify all singletons of each potential pollen parent, which in this case were defined as homozygous genotypes that were unique to a single individual among all individuals with the same photosynthetic type. A total of 7779 singletons were identified (minimum of 287 per potential pollen parent; Table S5). We then verified, for each singleton, which hybrid individuals carried that same allele, and counted the number of positive matches (Table S5). All genotyping analyses were conducted using R (scripts are provided as Supplementary Files).
2.6. Leaf gas‐exchanges
The photosynthetic response to intercellular CO2 concentration (A/Ci curve) was measured using two portable gas‐exchange systems Li‐6400XT (Li‐Cor). A/Ci curves were measured within the first 6 h of the photoperiod on the widest fully expanded leaf at a block temperature (T block) of 25°C, flow rate = 300 μmol s−1, photosynthetic photon flux density (PPFD) = 1500 μmol m−2 s−1, and were started after both net CO2 uptake (A) and stomatal conductance (g S) reached steady state at reference CO2 = 400 μmol mol−1. Reference CO2 was then changed in a stepwise manner to 250, 150, 120, 100, 85, 70, 50, 35, 400, 600, 800, 1000 and 1200 μmol mol−1. Readings were automatically logged after 2–3 min of leaf acclimation to each CO2 level. The CO2 compensation point (CCP) and maximum carboxylation efficiency (CE) were calculated following Bellasio et al. (2016). In this approach, the CO2‐dependence of A is described by an empirical nonrectangular hyperbola, and allows for the estimation of parameters irrespective of the photosynthetic physiology, which is therefore a suitable approach for hybrids (Bellasio et al., 2016). Water‐use efficiency (WUE) was calculated as the ratio between A and g S at steady state at reference CO2 = 400 μmol mol−1.
Stomatal density on the abaxial side of the leaves was quantified using leaf impressions. Dental resin (ImpressPlus Wash Light Body, Perfection Plus Ltd.) was applied to the central portion of fully expanded leaves, and clear nail varnish was applied to the set resin impression. Images were captured from the nail varnish impressions at 20× using an Olympus BX51 microscope coupled to an Olympus DP71 camera, and stomatal density was quantified on 0.38 mm² fields using ImageJ.
3. RESULTS
3.1. Carbon isotopic ratios are intermediate between the parents
Carbon isotopic ratios (δ13C) of all hybrids between C4 and non‐C4 accessions were below the range of C4 values in A. semialata (Figure 1 and Table S1). However, δ13C was consistently higher in the hybrids than in their non‐C4 parents, with average increases of 2.8‰ in C3 × C4 and 5.4‰ in C3+C4 × C4 hybrids relative to their C3 and C3+C4 parents, respectively. The δ13C ranged between −27.7 and −25.3‰ in C3 × C4 and −21.4 and −18.5‰ in C3+C4 × C4 hybrids.
Figure 1.
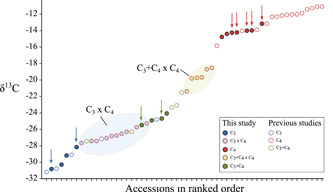
Distribution of carbon isotopic ratios (δ13C) in Alloteropsis semialata. δ13C values of accessions of A. semialata that are not part of this study were retrieved from Bianconi et al. (2020). Blue, green and red arrows indicate δ13C values of C3, C3+C4 and C4 known or potential parents of the hybrids analysed in this study [Color figure can be viewed at wileyonlinelibrary.com]
3.2. Leaf anatomy is additive in the hybrids
C4 and non‐C4 leaves in A. semialata are mainly distinguished by the presence of minor veins in C4 accessions (Lundgren et al., 2019). Here, minor veins were observed in all hybrids, except two C3 × C4 individuals (Table S6). The presence of chloroplasts in the inner sheath of all hybrids is suggested by the strong and consistent staining patterns in these cells, which are similar to the patterns observed in C4 and C3+C4 A. semialata, but not in C3 accessions, where the staining is very weak (Figure 2a). The first component of a PCA on 12 quantitative anatomical traits explains 57.1% of the total variation and separates the accessions by photosynthetic type (Figure 2b). C4 individuals are associated with negative values of the first component, which correspond to increased vein density (including minor veins) and increased abundance of bundle sheath tissue (IS and OS) relative to the mesophyll (Figure 2b). C3+C4 and C3 accessions partially overlap in the PCA space, and are associated with positive values of the first component. Hybrid individuals are intermediate between their respective parents along this first component. The second principal component explains 13.9% of the variance, and is correlated with leaf thickness (r = 0.88, p < 0.001), width of inner bundle sheath cells (ISW; r = 0.64, p < 0.001) and epidermis area (Epd; r = −0.54, p < 0.001), but these variables do not differentiate photosynthetic or cross types. The proportion of inner bundle sheath relative to mesophyll (IS/M) was on average doubled in C3 × C4 hybrids compared to C3 accessions, and was within the range of C3+C4, but the values were still 62% lower than the C4 average (Figure 2c). The distance between consecutive bundle sheaths (BSD) in C3 × C4 and C3+C4 × C4 hybrids was on average 49% and 55% smaller than in C3 and C3+C4 plants, respectively (Figure 2d). The mean width of IS cells was similar between C3+C4 and C4 accessions, and values were on average 48% higher than in C3, but their differences among photosynthetic types or between hybrids and the respective parental types were not significant (Figure 2e). OS cells were however significantly smaller in C4 than in C3 accessions, with hybrids having intermediate values that were not significantly different from those of C3+C4 accessions (Figure 2f). In the hybrids, the variation in BSD was largely explained by vein density (R 2 = 0.92, p < 0.001), with no significant effects of OS or IS widths. The IS/M ratio, on the other hand, was not significantly associated with vein density, but with both OS and IS width (R 2 = 0.75, p < 0.001). Overall, C3 × C4 hybrids were not significantly different from C3+C4 in relation to the quantitative anatomical traits analysed here, except for BSD, which was significantly reduced in C3 × C4 as a result of the presence of some minor veins in most accessions. In C3+C4 × C4 hybrids, a quantitatively distinct phenotype was generated, with trait values ranking between C3+C4 and C4 plants. Overall, our results show that leaf anatomical traits in A. semialata are mostly inherited in an additive manner.
Figure 2.
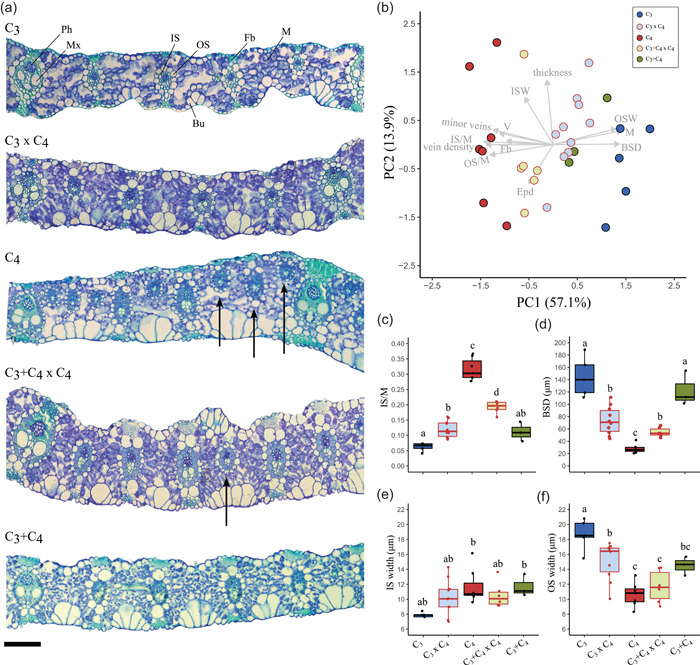
Leaf anatomy of F1 hybrids and the parental photosynthetic types in Alloteropsis semialata. (a) Representative cross sections of C3 (RSA9), C3 × C4 (H08), C4 (TPE1‐10), C3+C4 × C4 (H11) and C3+C4 (TAN1602‐03c) accessions. Arrows indicate minor veins (i.e., fourth‐ and fifth‐order veins). Scale bar = 100 µm. (b) Principal component analysis of selected leaf anatomical variables. (c) Proportion of inner sheath to mesophyll area (IS/M). (d) Distance between consecutive bundle sheaths (BSD). (e) Mean inner sheath cell width. (f) Mean outer sheath cell width. For (c–f), different lower‐case letters indicate statistical differences between groups (ANOVA, p < 0.05 post hoc Tukey HSD; n ≥ 4). Bu, bulliform cells; BSD, distance between consecutive bundle sheaths; Epd, proportion of epidermis area; Fb, extraxylary fibres (=sclerenchyma girder); IS, inner bundle sheath (=mestome sheath); ISW, IS width; M, mesophyll; Mx, metaxylem; OS, outer bundle sheath; OSW, OS width; Ph, phloem; V, proportion of vascular tissue area
3.3. Gene expression
Global gene expression patterns were assessed via a PCA on 7482 genes (Figure 3a). The first principal component mostly separated three sister C3+C4 × C4 plants (H06, H11, and H23) from the other individuals, and accounted for 24.7% of the variation. Photosynthetic types were clearly separated by the second principal component, which accounted for 16.8% of the variation and placed the hybrids as intermediate to the photosynthetic types of their parents. Similar clustering patterns were observed when transcript abundance was quantified for the complete gene set extracted from the genome of A. semialata (Figure S2).
Figure 3.
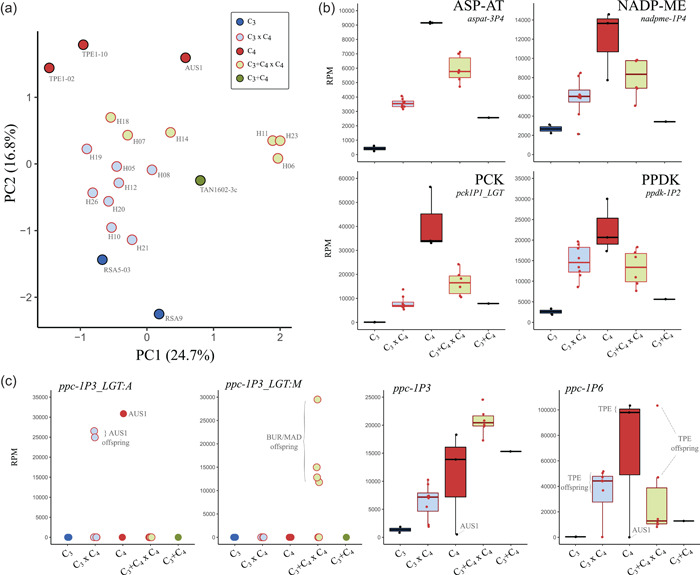
Leaf gene expression of F1 hybrids and the parental photosynthetic types in Alloteropsis semialata. (a) Principal component analysis on 7482 genes. (b) Transcript abundance in reads per million mapped reads (RPM) of selected genes encoding core C4 enzymes: aspartate aminotransferase (ASP‐AT, gene aspat‐3P4), NADP‐dependent malic enzyme (NADP‐ME, gene nadpme‐1P4), phosphoenolpyruvate carboxykinase (PCK, gene pck1P1_LGT), pyruvate phosphate dikinase (PPDK, gene ppdk‐1P2). (c) Transcript abundance of selected phosphoenolpyruvate carboxylase (PEPC) genes [Color figure can be viewed at wileyonlinelibrary.com]
Transcript abundance of five genes encoding key enzymes for the C4 biochemistry, and which are known to be upregulated in C4 plants of A. semialata (Dunning et al., 2019a), were analysed in detail (Figure 3b,c and Table S7). The patterns of transcript abundance of C3, C3+C4 and C4 plants are highly similar to those reported by Dunning et al. (2017, 2019a), where a larger sample size was used (Figure S3). Transcript abundance of the genes encoding the C4‐specific forms of the enzymes aspartate aminotransferase (ASP‐AT, gene aspat‐3P4), NADP‐malic enzyme (NADP‐ME, gene nadpme‐1P4) and pyruvate phosphate dikinase (PPDK, gene ppdk‐1P2) were 4‐ to 17‐fold higher in C4 than in C3 plants, with the transcript abundances of the C3+C4 plant slightly above those of C3 plants (Figure 3b). In each case, the transcript abundance of the hybrids was intermediate to the abundance of the photosynthetic types of the parents, showing that these expression patterns are mostly additive. C3 accessions of A. semialata lack the gene copy encoding phosphoenolpyruvate carboxykinase (PCK) that is highly expressed in C4 and C3+C4 accessions (pck‐1P1_LGT; Olofsson et al., 2016); the transcript abundance of this gene reaches 20% of the mean C4 values in C3 × C4 hybrids, and 40% in C3+C4 × C4.
Out of the nine genes encoding the key C4 enzyme phosphoenolpyruvate carboxylase (PEPC) that have been identified in A. semialata accessions (Dunning et al., 2017), only five were reported to be highly expressed in leaves of some C4 accessions (Christin et al., 2012; Dunning et al., 2017, 2019a). One of these genes, ppc‐1P3_LGT:A is only present in the C4 lineage from Oceania, which is represented here by an Australian accession (AUS1) that is the pollen parent of two C3 × C4 hybrids (H19 and H21; Table S5); both plants have expression levels of this gene that are similar to the Australian parent (Figure 3c). Only one of the other two laterally acquired PEPC copies is present in the plants analysed here (ppc‐1P3_LGT:M), and it is highly expressed (>10 000 RPM) in four C3+C4 × C4 individuals that are offspring from the C4 accessions from Burkina Faso or Madagascar (H06, H11, H14 and H23; Table S5 and Figure 3c), which have been previously shown to be part of the only C4 lineage to carry this gene (Olofsson et al., 2016). The expression levels of ppc‐1P3 are similar across C3+C4 and C4 accessions, and are on average 10‐fold higher than in C3 accessions (except the C4 AUS1, which has a nonfunctional copy of this gene; Olofsson et al., 2016); in the C3+C4 × C4 hybrids, however, the transcript abundance of this gene is on average 42% higher than the median of C3+C4 and C4 accessions (Figure 3c). Finally, the gene ppc‐1P6 reaches high expression levels in C4 accessions (except in AUS1, where it is absent; Dunning et al., 2019b), but is disproportionally upregulated in the C4 Taiwanese lineage (TPE; Dunning et al., 2017), where it reaches levels that are up to 100‐fold higher than in C3 plants, and at least twice higher than the values in other C4 and C3+C4 accessions. Here, the ppc‐1P6 copy reaches high levels in the C3 × C4 and C3+C4 × C4 offspring of TPE, mostly ranging from 10 000 to 60 000 RPM, except for one C3+C4 × C4 which has a transcript abundance similar to TPE (~100 000 RPM).
We examined transcript abundance of selected protein‐coding genes with a role in photorespiratory metabolism. We observed reduced transcript levels of the genes encoding the peroxisomal enzymes flavin mononucleotide‐dependent glycolate oxidase (GOX, gene glo‐1P2) and glutamate:glyoxylate aminotransferase (GGT, gene ggat‐1P6) in C3+C4 × C4 hybrids relative to C3 and C3+C4 accessions, with average values that are similar to or below those of C4 accessions (Figure S4). The genes encoding the proteins P‐, T‐, L‐ and H‐ that compose the mitochondrial multienzyme system glycine decarboxylase (GDC, genes gldp‐1P1, gcvt‐1P1, lpd‐1P2 and gdh‐2P2, respectively) had on average 63% lower transcript levels in C4 than in C3 accessions; in C3 × C4 and C3+C4 × C4 hybrids, transcript levels of these four genes were on average 12% and 33% lower than in C3, respectively, suggesting additivity (Figure S4 and Table S8). Multiple genes encoding a serine hydroxymethyltransferase (SHMT) are found in the genome of A. semialata (genes shm‐1P1, shm‐2P2, shm‐3P3 and shm‐3P4; Dunning et al., 2019a), but none had patterns of transcript abundance similar to those observed for GDC; in fact, one of the genes (shm‐3P3) had higher average transcript abundance in C4 than in C3 accessions. The chloroplast genes glycerate 3‐kinase (GLYK, gene glyk‐1P1) and 2‐phosphoglycolate (2‐PG) phosphatase (PGLP, gene pglp‐2P2) had overlapping transcript abundance ranges across hybrid and photosynthetic types. Finally, a gene encoding a GOLDEN2‐LIKE transcription factor that has been previously demonstrated to be involved in the redistribution of organelles among cell types (Wang et al., 2017) is expressed at higher levels in C4 than in C3 and C3+C4 accessions (Figure S4L and Table S8). This gene is at intermediate levels in the C3 × C4 hybrids, but largely variable among C3+C4 × C4 individuals.
We identified genes differentially expressed (DE) between hybrid types, and between these and C4 accessions (Table S9). We found a total of 4104 genes differentially expressed between C3 × C4 and C3+C4 × C4 hybrids (adjusted p value <0.05), out of which 21 were related to C4 photosynthesis and 4 to photorespiration (Table S9). This complete DE gene set is reduced to 206 when only higher count genes (base mean >500) and with larger differences (log2fold change >1) are considered (Table S9). Among this reduced gene set, we found seven C4‐related genes, but none related to photorespiration (Figure S5 and Table S9). The individual comparisons between C3 × C4 or C3+C4 × C4 and C4 accessions identified 1925 and 2727 significant DE genes, respectively. These numbers are reduced to 110 and 181 when the same filters described above are applied (Table S9). In this reduced set, we found 13 C4‐related genes that were differentially expressed between hybrids and C4 accessions, 3 of these common to both C3 × C4 vs. C4 and C3+C4 × C4 vs. C4 comparisons, namely PCK (pck‐1P1_LGT), NADP‐malate dehydrogenase (NADP‐MDH, gene nadpmdh‐1P2) and soluble inorganic pyrophosphatase (PPA, gene ppa‐2P1). While nadpmdh‐1P2 was downregulated in the C4 group, the two others had increased transcript abundance in C4 relative to both hybrid types (Figure S5). Finally, we found four genes related to photorespiration in the DE gene set between C3 × C4 and C4 accessions, three of which were upregulated in C3 × C4, namely the genes encoding the GDC proteins T‐ (gcvt‐1P1), P‐ (gldp1P) and L‐ (lpd‐1P2; Figure S5). The remaining gene, which encode the enzyme SHMT (shm‐3P3) was downregulated in both C3 × C4 and C3+C4 × C4 hybrids relative to C4 accessions. Gene ontology (GO) analyses did not detect any significantly enriched cellular processes for any of the full DE gene sets. However, the reduced DE gene set from the comparison between hybrid types was significantly enriched with genes related to the photosystems I and II, oxidoreductase activity, and 1,3‐beta‐d‐glucan synthesis (Table S10). In the reduced gene set from comparisons between each of the hybrid types and the C4 type, GO analyses indicated a significant enrichment for genes associated with C4 photosynthesis (particularly malate metabolism), transmembrane transport, and photosystems I and II (Table S10).
Overall, the analyses of expression profiles show that the inheritance of expression levels of C4 genes and those involved in the photorespiratory pathway is largely additive. In almost all cases, the transcript abundance of the hybrids ranks between those of the parental lineages.
3.4. Evidence of reduced photosynthetic performance in the hybrids
Photosynthetic performance was assessed from CO2 response curves (A/Ci) and steady‐state measurements at ambient CO2 (Figure S6; Tables S11 and S12). Photosynthetic rates in the hybrids ranked between the parents at low Ci levels, but the differences between hybrids and parents were progressively reduced as CO2 levels increased, until they were equal to their non‐C4 parents at Ci of 100 in C3 × C4, and 250 µmol mol−1 in C3+C4 × C4 hybrids (Figure 4). The C3 and C3+C4 accessions had on average similar values of maximum CE, and these were 57% lower than in C4 accessions (Figure 5a and Table S11). Despite the larger variation among individuals, C3+C4 × C4 hybrids had CE values that were on average 83% and 96% higher than in C3 and C3+C4 accessions, respectively, and these were not significantly different from the average values of C4 accessions. In C3 × C4 hybrids, however, CE was consistently lower than in C3 accessions, and reached the lowest mean values of all photosynthetic types/crosses. The CCP ranged between 43 and 48 µmol mol−1 in C3 accessions, and between 5 and 15 µmol mol−1 in C3+C4 individuals (Figure 5b). All C4 had CCP below 10 µmol mol−1. In the hybrids, CCP was below 13 µmol mol−1 in C3+C4 × C4 individuals, and ranged between 6 and 40 µmol mol−1 in C3 × C4 hybrids. CCP estimates varied substantially among A/Ci curves in C3 × C4 individuals (Figure 5b), but mean values were 52% lower than in C3 plants.
Figure 4.
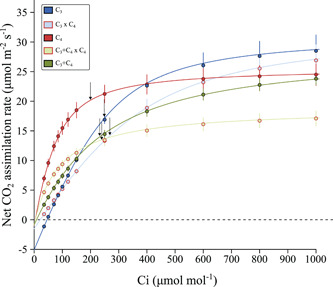
Response of net CO2 assimilation rate (A) to intercellular CO2 (Ci) of F1 hybrids and the parental photosynthetic types in Alloteropsis semialata. Data points are predicted values from an empirical nonrectangular hyperbola model (Bellasio et al., 2016) based on the mean parameter estimates per individual within each cross/photosynthetic type. Error bars show standard errors for the predicted values (n ≥ 3). Arrows indicate Ci values at reference CO2 = 400 µmol mol−1 for each cross/photosynthetic type [Color figure can be viewed at wileyonlinelibrary.com]
Figure 5.
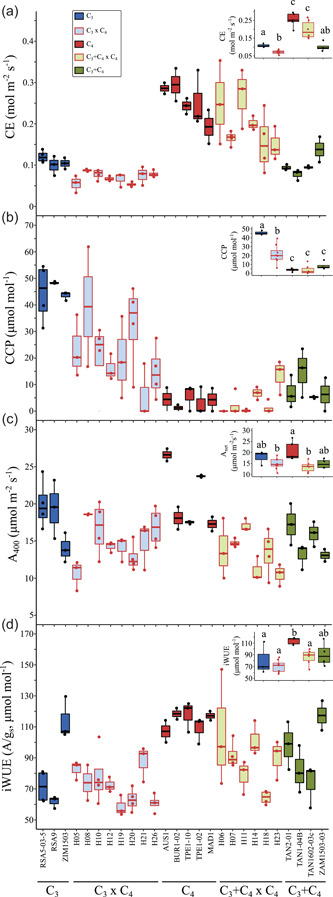
Photosynthetic performance of F1 hybrids and the parental photosynthetic types in Alloteropsis semialata. (a) Maximum carboxylation efficiency (CE), (b) CO2 compensation point (CCP), and steady‐state (c) net photosynthetic rate (A 400), and (d) intrinsic water use efficiency (iWUE, A/g s) at reference CO2 = 400 µmol mol−1. Different lower‐case letters indicate statistical differences between groups (ANOVA, p < 0.05 post hoc Tukey HSD; n ≥ 4) [Color figure can be viewed at wileyonlinelibrary.com]
The steady‐state net CO2 assimilation rate at ambient CO2 (400 µmol mol−1; A 400) was highest in two C4 accessions, but the differences between C3, C3+C4 and C4 plants were not significant (Figure 5c). C3+C4 × C4 hybrids had the lowest A 400, but mean values were not significantly different from C3 × C4, C3 and C3+C4 accessions. Stomatal conductance (g s) had the highest median values in C3 accessions, but the differences between groups were not significant (Figure S7A). C3 accessions had on average 50% higher stomatal density than C3+C4 and C4 accessions and their hybrids (Figure S7B). We found a positive correlation between g s and stomatal density (R² = 0.31, p < 0.01), but only after excluding the outlier C3 accession ZIM1503, which had the highest stomatal density, but one of the lowest g s. The correlation between stomatal density and vein density was only marginally significant (R² = 0.01, p = 0.06). The intrinsic water use efficiency (iWUE) at ambient CO2 was on average 38% higher in C4 than in other accessions, although a few C3, C3+C4 and C3+C4 × C4 individuals also showed C4‐like values (Figure 5d). The lowest iWUE values were observed in C3 and C3 × C4 individuals, but the means were not significantly different from C3+C4 and C3+C4 × C4 hybrids. To investigate whether changes in leaf temperature (T leaf) could explain the observed patterns, we inspected the variation in T leaf during the A/Ci curves. Although T leaf values up to 3°C above the median (26.3°C) were observed in a few accessions, 81% of all data points were collected within a 2°C interval (Figure S8). There were no significant differences between groups when all points were considered, nor when only above‐ or below‐ambient CO2 levels were analysed (Figure S8). Similar patterns among cross/photosynthetic types were observed in all photosynthetic parameters after removing individuals for which the median T leaf was 1°C above the median of all curves (Figure S9).
Overall, these results show that the physiological characters are not consistently additive in the hybrids, with some individuals performing below their two parents for several traits.
4. DISCUSSION
4.1. Partial contribution of the C4 cycle to carbon fixation in hybrids
In this study, we analyse the phenotypes of hybrids between individuals of the grass A. semialata with different photosynthetic types. The C3 × C4 quantitatively resemble the naturally occurring C3+C4 in most anatomical and gene expression traits (Figures 2 and 3), as previously noticed in other taxa (Kadereit et al., 2017). They however differ qualitatively, and minor veins and high expression of some C4‐related genes are especially restricted to C3 × C4 individuals (Figure 3 and Table S6). These findings therefore provide additional support for the hypothesis that naturally occurring C3+C4 in A. semialata do not result from recent crosses between photosynthetic types (Lundgren et al., 2015, 2016), although genome analyses support a role for introgression in their distant history (Bianconi et al., 2020).
Across all crosses, we show that hybrids between C4 and non‐C4 individuals of the grass A. semialata do not exhibit a full C4 physiology, as indicated by their δ13C, which are clearly outside the range of C4 values in the species (Figure 1). This indicates that a significant fraction of CO2 assimilation in these hybrids occurs directly via Rubisco in the mesophyll, which implies that the genetic contribution of the C4 parent was insufficient to (1) completely suppress Rubisco expression and/or activity in the mesophyll cells and/or (2) create an effective C4 cycle. However, the CCP of the C3 × C4 hybrids is substantially reduced compared to the C3 parents (Figure 5b). While this change could theoretically result from an increase of the photorespiratory shuttle observed in many C3–C4 plants (called ‘C2' plants; Keerberg et al., 2014; Khoshravesh et al., 2016), the expression of photorespiration‐related genes is rather unchanged or decreased in our hybrids compared to the C4 parents. Changes in CCP are therefore more likely linked to an increase in the C4 activity, which is further supported by significantly higher δ13C values compared to their non‐C4 parents (Figure 1). Our results therefore suggest that, in most hybrids, a fraction of the CO2 that enters the leaf is fixed by PEPC in the mesophyll and follows the C4 route to be refixed by Rubisco in the inner bundle sheath. Furthermore, given the C4‐like CCP and CE values, and δ13C above −20‰ (in most cases), the degree of C4 activity is higher in C3+C4 × C4 than in C3 × C4 hybrids. Our findings therefore recapitulate what has been shown for interspecific hybrids between photosynthetic types in several eudicot and monocot genera (Araus et al., 1990; Björkman et al., 1969; Brown & Bouton, 1993; Brown et al., 1985; Byrd et al., 1992; Oakley et al., 2014; Sultmanis, 2018). The causes for this only partial contribution of the C4 pathway to the overall carbon assimilation in the hybrids are potentially the same in all these cases, and are probably related to the insufficient expression of components of C4 metabolism (Brown & Bouton, 1993).
Two interrelated leaf anatomical traits were significantly different from the C4 individuals in both C3 × C4 and C3+C4 × C4 hybrids, namely the proportion of inner sheath to mesophyll tissue (IS/M) and the distance between consecutive bundle sheaths (BSD). These traits are mostly determined by the presence of minor veins in C4 leaves, which has been previously shown to be the major anatomical difference between C4 and non‐C4 A. semialata (Lundgren et al., 2019). Here, significant changes towards the C4 phenotype in IS/M and BSD were observed in the hybrids, but realized values were still far from those observed in C4 accessions (Figure 2c,d). This is particularly clear for IS/M, where values were below the mean of both parents in the two hybrid types. While minor veins are observed in the hybrids, their number per segment remains below that observed in C4 accessions (Table S6). This indicates that the proliferation of minor veins is a continuous trait, which can sustain a C4 cycle only over a threshold in this species. Since CO2 refixation by Rubisco takes place in the IS cells in C4 A. semialata (Ellis, 1974b; Ueno & Sentoku, 2006), the leaf anatomy of the hybrids might prevent an optimal coupling between carbon assimilation and reduction reactions, therefore decreasing the efficiency of the C4 pathway. The distribution of organelles among cell types was not quantified here, but a gene previously linked to the relocation of organelles to BS cells (Wang et al., 2017) is more highly expressed in C4 accessions and upregulated in some hybrids compared to their non‐C4 parents, which might suggest that organelles are partially relocated to their BS. The size of the OS cells might also play a role in creating an effective C4 cycle, as suggested by the large differences between C3 and C4 accessions (Figure 2e). Few other C4 species have an extra layer of BS cells outside of the BS cells containing chloroplasts, and the presence of an OS may represent an increased resistance to the C4 acid shuttle (Lundgren et al., 2014). In the grass genus Neurachne that presents a similar C4 anatomy, the smallest OS is also observed in C4 individuals, with intermediate species presenting intermediate OS cell sizes (Khoshravesh et al., 2020), supporting the idea that OS cells must be reduced during C4 evolution. Here, the OS cells were significantly reduced in the hybrids relative to their non‐C4 parents, with most C3+C4 × C4 individuals and a few C3 × C4 having OS cell sizes within the C4 range (Figure 2f), which suggests that this character might have played a minor role, if any, in reducing the efficiency of the C4 acid shuttle in the hybrids.
C4 activity could also have been limited by gene expression and protein activity. Despite significant increases in the transcript abundance of core C4 genes, particularly the genes encoding PEPC (Figure 4), it is possible that the insufficient expression levels of some genes might have reduced the efficiency of the C4 cycle, for example, by decreasing the rate of carboxylation and/or decarboxylation reactions. Furthermore, the lack of proper compartmentation of the expression/activity of these genes due to a non‐tissue‐specific expression of C3 or C3+C4 alleles could have impaired the formation of intermediate metabolite pools, therefore preventing the operation of an efficient C4 cycle (Brown & Bouton, 1993; Ermakova et al., 2021). Finally, the lack or insufficiency of posttranscriptional and posttranslational modifications might also have affected the realized enzyme activity. This might be particularly relevant in the case of hybrids, where the cell environment may be substantially modified by pleiotropic effects emerging from the expression of two divergent genomes. Detailed analyses of cellular localization and activity of C4 enzymes coupled to CO2‐labelling studies are necessary to identify the major determinants of the phenotype observed in the hybrids reported here.
4.2. Photosynthetic performance of plants expressing partial C4 traits and implications for the model of C4 evolution
The current model of C4 evolution assumes that the sequential acquisition of certain features, such as Rubisco activity in bundle sheath cells and upregulation of core C4 enzymes, increases photosynthetic output and consequently fitness in an environment that favours the C4 physiology (Heckmann et al., 2013; Malmmann et al., 2014; Monson & Moore, 1989; Sage, 2004). However, this Mt. Fuji‐like fitness landscape (Heckmann et al., 2013) is conditional on the presence of anatomical enablers; when these are lacking, the C4 phenotype is not accessible because intermediate states have lower fitness than the C3 state (Heckmann, 2016). The two groups of intraspecific hybrids generated in this study provide an opportunity to examine this prediction while controlling for phylogenetic effects, with C3 × C4 and C3+C4 × C4 plants being proxies for the upregulation of C4 traits at an early and intermediate stage, respectively, along C4 evolution. Here, we show that the CE of C3 × C4 hybrids is lower than that of both parents (Figures 4 and 5a), despite significant changes towards the C4 phenotype in leaf anatomy and gene expression. In C3+C4 × C4 hybrids however, CE was increased in comparison to the C3+C4 parent, and was not significantly different from C4 accessions. Such disparate effects of upregulating C4 components in a non‐C4 background are compatible with the initial prediction that fitness gains are conditional on the presence of enabling factors, which here might have been present in C3+C4 parents, but lacking in the C3 plants. Such changes would have acted as a switch that permitted a full C4 pathway to operate, and therefore increase photosynthetic performance. Note however that such gains in photosynthetic performance are restricted to conditions of low CO2 concentration in the leaf (Figure 4). In fact, at ambient CO2 levels, several individuals of both hybrid groups had lower photosynthetic rates than both parents, suggesting that the partial upregulation of C4 components, all at once, might have negative consequences, possibly due to pleiotropic effects (e.g., C4 enzymes competing for reducing power and ATP with other cell reactions). This in turn supports the idea that the changes towards a full C4 physiology must be built sequentially upon pre‐existing characteristics that enable the next change to provide a fitness advantage (Heckmann, 2016). The fertility of the F1 hybrids reported here is not known yet, although evidence of rare gene flow between photosynthetic types in the wild suggest that backcrossing is possible (Bianconi et al., 2020; Olofsson et al., 2016, 2021). If a F2 population can be produced, characterization of the photosynthetic performance of such individuals expressing C4 components independently, particularly under conditions expected to favour C4 plants, including long‐term exposure to low CO2 levels, should provide further insights on whether C4 evolution must follow a particular sequence of events for it to be viable.
5. CONCLUSIONS
In this study, we characterize the phenotype of hybrids between different photosynthetic types in the grass A. semialata to investigate how the upregulation of components of the C4 trait affects photosynthetic performance. We show that the hybrids have in most cases anatomical traits and gene expression patterns that are intermediate between those of the parents, and this leads to C4 activity that is equally intermediate between the two parents. The physiological benefits of a partial C4 metabolism in the hybrids appear only at low CO2 levels; at ambient CO2, there is no evidence of enhanced photosynthetic performance in the hybrids relative to their non‐C4 parents. Some hybrid individuals in fact perform worse than both parents at ambient CO2, and this possible hybrid depression could explain the lack of a clear hybrid zone in regions where the distributions of C4 and non‐C4 A. semialata lineages overlap. Overall, our results support the hypothesis that photosynthetic gains arising from the upregulation of C4 features are conditional on coordinated changes in leaf anatomy and biochemistry. Future studies with the F2 offspring, where the different C4 components can be segregated, will be able to pinpoint the key genetic and phenotypic changes that lead to fitness gains, providing a unique opportunity to experimentally test long‐standing hypotheses about the evolution of C4 photosynthesis.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGEMENTS
This study was funded by the European Research Council (grant ERC‐2014‐STG‐638333) and the Royal Society (grant RGF\EA\181050). P. A. C. is supported by a Royal Society University Research Fellowship (grant URF\R\180022).
Bianconi, M. E. , Sotelo, G. , Curran, E. V. , Milenkovic, V. , Samaritani, E. , Dunning, L. T. et al. (2022) Upregulation of C4 characteristics does not consistently improve photosynthetic performance in intraspecific hybrids of a grass. Plant, Cell & Environment, 45, 1398–1411. 10.1111/pce.14301
DATA AVAILABILITY STATEMENT
Newly generated RNA sequencing datasets were deposited in the NCBI SRA database under Bioproject PRJNA752516. Scripts used for the genotyping analysis are available at https://github.com/matheusbianconi/RNAgenotyping_hybrids
REFERENCES
- Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Araus, J.L. , Brown, R.H. , Bouton, J.H. & Serret, M.D. (1990) Leaf anatomical characteristics in Flaveria trinervia (C4), Flaveria brownii (C4‐like) and their F1 hybrid. Photosynthesis Research, 26, 49–57. [DOI] [PubMed] [Google Scholar]
- Bellasio, C. , Beerling, D.J. & Griffiths, H. (2016) An Excel tool for deriving key photosynthetic parameters from combined gas exchange and chlorophyll fluorescence: theory and practice. Plant, Cell & Environment, 39, 1180–1197. [DOI] [PubMed] [Google Scholar]
- Bianconi, M.E. , Dunning, L.T. , Curran, E.V. , Hidalgo, O. , Powell, R.F. , Mian, S. et al. (2020) Contrasted histories of organelle and nuclear genomes underlying physiological diversification in a grass species. Proceedings of the Royal Society B: Biological Sciences, 287, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianconi, M.E. , Dunning, L.T. , Moreno‐Villena, J.J. , Osborne, C.P. & Christin, P.‐A. (2018) Gene duplication and dosage effects during the early emergence of C4 photosynthesis in the grass genus Alloteropsis . Journal of Experimental Botany, 69, 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman, O. , Gauhl, E. & Nobs, M.A. (1969) Comparative studies of Atriplex species with and without ß‐carboxylation photosynthesis. Carnegie Institution of Washington, Yearbook, 68, 620–633. [Google Scholar]
- Bouton, J.H. , Brown, R.H. , Evans, P.T. & Jernstedt, J.A. (1986) Photosynthesis, leaf anatomy, and morphology of progeny from hybrids between C3 and C3/C4 Panicum species. Plant Physiology, 80, 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.H. , Bouton, J.H. , Evans, P.T. , Malter, H.E. & Rigsby, L.L. (1985) Photosynthesis, morphology, leaf anatomy, and cytogenetics of hybrids between C3 and C3/C4 Panicum species. Plant Physiology, 77, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, H.R. & Bouton, J.H. (1993) Physiology and genetics of interspecific hybrids between photosynthetic types. Annual Review of Plant Biology, 44, 435–456. [Google Scholar]
- Byrd, G.T. , Brown, R.H. , Bouton, J.H. , Bassett, C.L. & Black, C.C. (1992) Degree of C4 photosynthesis in C4 and C3‐C4 Flaveria Species and their hybrids: I. CO2 assimilation and metabolism and activities of phosphoeno/pyruvate carboxylase and NADP‐malic enzyme. Plant Physiology, 100, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin, P.‐A. , Edwards, E.J. , Besnard, G. , Boxall, S.F. , Gregory, R. , Kellogg, E.A. et al. (2012) Adaptive evolution of C4 photosynthesis through recurrent lateral gene transfer. Current Biology, 22, 445–449. [DOI] [PubMed] [Google Scholar]
- Christin, P.‐A. , Osborne, C.P. , Chatelet, D.S. , Columbus, J.T. , Besnard, G. , Hodkinson, T.R. , Garrison, L.M. , Vorontsova, M.S. & Edwards, E.J. (2013) Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proceedings of the National Academy of Sciences, 110, 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning, L.T. , Lundgren, M.R. , Moreno‐Villena, J.J. , Namaganda, M. , Edwards, E.J. , Nosil, P. et al. (2017) Introgression and repeated co‐option facilitated the recurrent emergence of C4 photosynthesis among close relatives. Evolution, 71, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning, L.T. , Moreno‐Villena, J.J. , Lundgren, M.R. , Dionora, J. , Salazar, P. , Adams, C. et al. (2019a) Key changes in gene expression identified for different stages of C4 evolution in Alloteropsis semialata . Journal of Experimental Botany, 70, 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning, L.T. , Olofsson, J.K. , Parisod, C. , Choudhury, R.R. , Moreno‐Villena, J.J. , Yang, Y. et al. (2019b) Lateral transfers of large DNA fragments spread functional genes among grasses. Proceedings of the National Academy of Sciences of the United States of America, 116, 4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, E.J. (2019) Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. New Phytologist, 223, 1742–1755. [DOI] [PubMed] [Google Scholar]
- Edwards, G. & Ku, M.B. (1987) Biochemistry of C3–C4 intermediates. In: Hatch, M.D. & Boardman, N.K. (Eds.) The biochemistry of plants: a comprehensive treatise, Vol. 14: Photosynthesis. Academic Press, pp. 275–325. [Google Scholar]
- Ellis, R.P. (1974a) Significance of the occurrence of both Kranz and non‐Kranz leaf anatomy in the grass species Alloteropsis semialata . South African Journal of Science, 70, 169–173. [Google Scholar]
- Ellis, R.P. (1974b) Anomalous vascular bundle sheath structure in Alloteropsis semialata leaf blades. Bothalia, 11, 273–275. [Google Scholar]
- Ermakova, M. , Arrivault, S. , Giuliani, R. , Danila, F. , Alonso‐Cantabrana, H. , Vlad, D. et al. (2021) Installation of C4 photosynthetic pathway enzymes in rice using a single construct. Plant Biotechnology Journal, 19, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group II (GPWG II) . (2012) New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytologist, 193, 304–312. [DOI] [PubMed] [Google Scholar]
- Hatch, M.D. (1987) C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta, 895, 81–106. [Google Scholar]
- Heckmann, D. (2016) C4 photosynthesis evolution: the conditional Mt. Fuji. Current Opinion in Plant Biology, 31, 149–154. [DOI] [PubMed] [Google Scholar]
- Heckmann, D. , Schulze, S. , Denton, A. , Gowik, U. , Westhoff, P. , Weber, A.P.M. et al. (2013) Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell, 153, 1579–1588. [DOI] [PubMed] [Google Scholar]
- Hylton, C.M. , Rawsthorne, S. , Smith, A.M. , Jones, D.A. & Woolhouse, H.W. (1988) Glycine decarboxylase is confined to the bundle‐sheath cells of leaves of C3‐C4 intermediate species. Planta, 175, 452–9. [DOI] [PubMed] [Google Scholar]
- Ishimaru, K. , Ichikawa, H. , Matsuoka, M. & Ohsugi, R. (1997) Analysis of a C4 maize pyruvate, orthophosphate dikinase expressed in C3 transgenic Arabidopsis plants. Plant Science, 129, 57–64. [Google Scholar]
- Kadereit, G. , Bohley, K. , Lauterbach, M. , Tefarikis, D.T. & Kadereit, J.W. (2017) C3–C4 intermediates may be of hybrid origin—a reminder. New Phytologist, 215, 70–76. [DOI] [PubMed] [Google Scholar]
- Keerberg, O. , Pärnik, T. , Ivanova, H. , Bassüner, B. & Bauwe, H. (2014) C2 photosynthesis generates about 3‐fold elevated leaf CO2 levels in the C3‐C4 intermediate species Flaveria pubescens . Journal of Experimental Botany, 65, 3649–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshravesh, R. , Stinson, C.R. , Stata, M. , Busch, F.A. , Sage, R.F. , Ludwig, M. et al. (2016) C3‐C4 intermediacy in grasses: organelle enrichment and distribution, glycine decarboxylase expression, and the rise of C2 photosynthesis. Journal of Experimental Botany, 67, 3065–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshravesh, R. , Stata, M. , Busch, F.A. , Saladie, M. , Castelli, J.M. , Dakin, N. et al. (2020) The evolutionary origin of C4 photosynthesis in the grass subtribe Neurachninae. Plant Physiology, 182, 566–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. & Salzberg, S.L. (2012) Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M. , Schlüter, U. , Stich, B. & Weber, A.P.M. (2021) Cis‐regulatory divergence underpins the evolution of C3‐C4 intermediate photosynthesis in Moricandia . bioRxiv [Preprint]Available at 10.1101/2021.05.10.443365 [DOI] [Google Scholar]
- Lundgren, M.R. , Besnard, G. , Ripley, B.S. , Lehmann, C.E. , Chatelet, D.S. , Kynast, R.G. et al. (2015) Photosynthetic innovation broadens the niche within a single species. Ecology Letters, 18, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Lundgren, M.R. , Osborne, C.P. & Christin, P.A. (2014) Deconstructing Kranz anatomy to understand C4 evolution. Journal of Experimental Botany, 65, 3357–3369. [DOI] [PubMed] [Google Scholar]
- Lundgren, M.R. , Christin, P.‐A. , Escobar, E.G. , Ripley, B.S. , Besnard, G. , Long, C.M. et al. (2016) Evolutionary implications of C3–C4 intermediates in the grass Alloteropsis semialata . Plant, Cell and Environment, 39, 1874–1885. [DOI] [PubMed] [Google Scholar]
- Lundgren, M.R. , Dunning, L.T. , Olofsson, J.K. , Moreno‐Villena, J.J. , Bouvier, J.W. , Sage, T.L. et al. (2019) C4 anatomy can evolve via a single developmental change. Ecology Letters, 22, 302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallmann, J. , Heckmann, D. , Bräutigam, A. , Lercher, M.J. , Weber, A.P.M. , Westhoff, P. et al. (2014) The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. eLife, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson, R.K. & Moore, B.D. (1989) On the significance of C3‐C4 intermediate photosynthesis to the evolution of C4 photosynthesis. Plant, Cell & Environment, 12, 689–699. [Google Scholar]
- Moreno‐Villena, J.J. , Dunning, L.T. , Osborne, C.P. & Christin, P.A. (2018) Highly expressed genes are preferentially co‐opted for C4 Photosynthesis. Molecular Biology and Evolution, 35, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary, M.H. (1988) Carbon isotopes in photosynthesis. BioScience, 38, 328–336. [Google Scholar]
- Oakley, J.C. , Sultmanis, S. , Stinson, C.R. , Sage, T.L. & Sage, R.F. (2014) Comparative studies of C3 and C4 Atriplex hybrids in the genomics era: physiological assessments. Journal of Experimental Botany, 65, 3637–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson, J.K. , Bianconi, M. , Besnard, G. , Dunning, L.T. , Lundgren, M.R. , Holota, H. et al. (2016) Genome biogeography reveals the intraspecific spread of adaptive mutations for a complex trait. Molecular Ecology, 25, 6107–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson, J.K. , Curran, E.V. , Nyirenda, F. , Bianconi, M.E. , Dunning, L.T. , Milenkovic, V. et al. (2021) Low dispersal and ploidy differences in a grass maintain photosynthetic diversity despite gene flow and habitat overlap. Molecular Ecology, 30, 1–15. [DOI] [PubMed] [Google Scholar]
- Patel, R.K. & Jain, M. (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One, 7, e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team .(2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing.
- Sage, R.F. (2001) Environmental and evolutionary preconditions for the origin and diversification of the C4 photosynthetic syndrome. Plant Biology, 3, 202–213. [Google Scholar]
- Sage, R.F. (2004) The evolution of C4 photosynthesis. New Phytologist, 161, 341–370. [DOI] [PubMed] [Google Scholar]
- Sage, R.F. , Christin, P.‐A. & Edwards, E.J. (2011) The C4 plant lineages of planet Earth. Journal of Experimental Botany, 62, 3155–3169. [DOI] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. & Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, C.J.C. , Reeves, G. , Tripathi, A. , Singh, P. & Hibberd, J.M. Using breeding and quantitative genetics to understand the C4 pathway. Journal of Experimental Botany (in press). 10.1093/jxb/erab486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stata, M. , Sage, T.L. & Sage, R.F. (2019) Mind the gap: the evolutionary engagement of the C4 metabolic cycle in support of net carbon assimilation. Current Opinion in Plant Biology, 49, 27–34. [DOI] [PubMed] [Google Scholar]
- Sultmanis, S.D. (2018). Developmental and expression evolution in C3 and C4 Atriplex and their hybrids. PhD thesis, Canada, 178p, University of Toronto.
- Taniguchi, Y. , Ohkawa, H. , Masumoto, C. , Fukuda, T. , Tamai, T. , Lee, K. et al. (2008) Overproduction of C4 photosynthetic enzymes in transgenic rice plants: an approach to introduce the C4‐like photosynthetic pathway into rice. Journal of Experimental Botany, 59, 1799–1809. [DOI] [PubMed] [Google Scholar]
- Ueno, O. & Sentoku, N. (2006) Comparison of leaf structure and photosynthetic characteristics of C3 and C4 Alloteropsis semialata subspecies. Plant, Cell & Environment, 29, 257–268. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Khoshravesh, R. , Karki, S. , Tapia, R. , Balahadia, C.P. & Bandyopadhyay, A. et al. (2017) Re‐creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Current Biology, 27, 3278–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
Newly generated RNA sequencing datasets were deposited in the NCBI SRA database under Bioproject PRJNA752516. Scripts used for the genotyping analysis are available at https://github.com/matheusbianconi/RNAgenotyping_hybrids


