Abstract
Most Escherichia coli overexpression vectors used for recombinant protein production (RPP) depend on organic inducers, for example, sugars or simple conjugates. However, these can be expensive and, sometimes, chemically unstable. To simplify this and to cut the cost of RPP, we have developed vectors controlled by the Escherichia coli nitrate‐responsive NarL transcription activator protein, which use nitrate, a cheap, stable, and abundant inorganic ion, to induce high‐level controlled RPP. We show that target proteins, such as green fluorescent protein, human growth hormone, and single‐chain variable region antibody fragments can be expressed to high levels using our promoter systems. As nitrate levels are high in many commercial fertilizers, we demonstrate that controlled RPP can be achieved using readily available and inexpensive garden products.
Keywords: biopharmaceuticals, Escherichia coli, recombinant protein production, transcription regulation
Most Escherichia coli overexpression vectors used for recombinant protein production (RPP) depend on organic inducers, such as IPTG, which can be expensive and chemically unstable. To cut the cost of RPP, Hothersall and co‐workers have developed new expression systems, which use nitrate, a cheap, stable and abundant inorganic ion, to induce high‐level RPP. As nitrate levels are high in many commercial fertilizers, they demonstrate that controlled RPP can even be achieved using inexpensive garden products purchased from any local store.
1. INTRODUCTION
Many bacterial promoters are highly regulated, and, since the 1970s, biotechnologists have exploited this to control overexpression of foreign proteins in hosts such as Escherichia coli (Nora et al., 2019; Tungekar et al., 2021). Most currently used overexpression vectors depend on organic inducers, for example, sugars or simple conjugates, such as IPTG (isopropyl β‐d‐1‐thiogalactopyranoside). However, IPTG is costly, and is unstable, requiring a refrigerated supply chain. Our aim was to construct a new expression system that permits high‐level expression of biopharmaceuticals in E. coli, induced by a stable inorganic inducer. We reasoned that this would be useful in developing countries, where organic inducers might be unavailable or unduly expensive. Thus, we have developed a suite of vectors carrying promoters induced by the NarL transcription activator, which is triggered by nitrate ions.
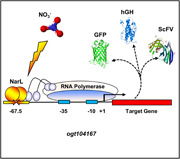
The presence of nitrate in the growth environment triggers phosphorylation and activation of the E. coli NarL protein, resulting in its binding to at least 26 promoter regions, including 11 where it activates transcription (Figure 1a; Constantinidou et al., 2006; Darwin & Stewart, 1996; Santos‐Zavaleta et al., 2019; Stewart, 2003). Most bacterial transcription activators work by binding at their target and then making a direct contact with RNA polymerase (RNAP), which recruits and positions RNAP to the promoter region (Browning & Busby, 2016; Lee et al., 2012). Previous studies with NarL‐dependent promoters showed that activated NarL recognizes a 7‐base sequence element, and most targets consist of two copies of this element organized as an inverted repeat, separated by two base pairs (known as the “7‐2‐7” sequence; Darwin et al., 1997). Some NarL‐dependent promoters are particularly complicated, involving interactions with other transcription factors, for example, the E. coli narG promoter is coregulated by NarL and the anaerobically triggered transcription factor, FNR (Browning et al., 2010; Darwin & Stewart, 1996). However, we identified two promoter regions (yeaR and ogt) where NarL alone is able to activate transcript initiation (Ruanto et al., 2020; Squire et al., 2009). Here, we describe new derivatives of both the ogt and narG promoters, and use them to drive high‐level recombinant protein production (RPP), engineering them to optimize their activity and dependence on both NarL and nitrate.
Figure 1.
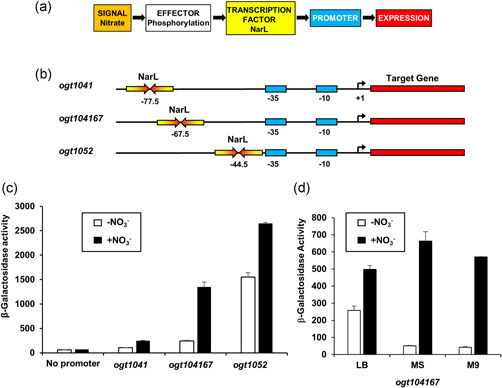
Expression analysis of the ogt promoter fragments used in this study. (a) Control of gene expression by nitrate and the NarL transcription activator protein. The presence of nitrate in the growth medium leads to the phosphorylation of the NarL transcription factor, enabling it to bind to target promoters and control transcript initiation by RNAP (Constantinidou et al., 2006; Darwin & Stewart, 1996; Santos‐Zavaleta et al., 2019; Stewart, 2003). (b) The panel shows schematic representations of the ogt1041, ogt104167 and ogt1052 promoter fragments. The NarL‐binding sites are shown as inverted arrows, −35 and −10 promoter elements are shown as rectangles, and transcript start sites (+1) are indicated by bent arrows. The location of each DNA site for NarL is labeled, according to convention, by the position of the center of the 7–2–7 sequence. Hence, position −77.5 is located between base pairs 77 and 78, upstream from the transcript start. At the ogt1041, ogt104167, and ogt1052 promoters, the single DNA sites for NarL are located at positions −77.5, −66.5, and −44.5, respectively, and thus are 65, 55, and 32 bp, respectively, upstream from the corresponding promoter −10 element. (c) The panel shows measured β‐galactosidase activities in wild‐type JCB387 cells, carrying ogt1041, ogt104167, and ogt1052 promoter fragments cloned into the pRW50 lacZ expression vector. Cells were grown in minimal salts media supplemented with 20 mM sodium nitrate, where indicated. (d) The panel shows measured β‐galactosidase activities in wild‐type JCB387 cells, carrying the ogt104167 promoter fragment cloned into pRW50. Cells were grown in LB medium (LB), minimal salts media (MS), and M9 minimal medium (M9) supplemented with 20 mM sodium nitrate, where indicated. β‐Galactosidase activities are expressed as nmol ONPG hydrolyzed/min/mg dry cell mass and represent the average of three independent experiments. Error bars represent SD
2. MATERIALS AND METHODS
2.1. Bacterial strains, plasmids, and materials
E. coli K‐12 strains, plasmids, and promoter fragments used in this study are listed in Table S1 and oligonucleotide primers are in Table S2. Strains were grown in lysogeny broth (LB) (Sigma), Lennox broth (2% (w/v) peptone (Oxoid), 1% (w/v) yeast extract (Oxoid) and 170 mM NaCl), minimal salts medium (Squire et al., 2009) (minimal salts with 0.4% glycerol, 10% Lennox broth, 20 mM fumarate), or M9 minimal salts media (Sigma), supplemented with 0.36% glucose, 2 mM MgSO4, 0.1 mM CaCl2, all with appropriate antibiotic supplements (ampicillin 100 µg/ml and tetracycline 15 µg/ml).
2.2. Vector construction
To examine expression from each promoter construct, DNA fragments were initially cloned into the low‐copy number lac expression vector, pRW50 (Lodge et al., 1992). The ogt1041, ogt104167, and ogt1052 promoters have been described previously (Ruanto et al., 2020). The NN series of promoters (i.e., NN(−81.5) to NN(−69.5)), which carry a consensus NarL 7–2–7 heptamer sequence upstream of a CRP binding site, centered at position −40.5, were constructed using PCR. Promoter fragments were generated using primers NN(−81.5) to NN(−69.5) with D10527 and pRW50/CC(−40.5) as a template. Purified PCR products were restricted with EcoRI and HindIII and cloned into pRW50. For the narG CC(−40.5) promoter, the full‐length narG promoter (narG223: DNA sequences from −223 to +70) was PCR amplified using primers narGup223 and narGDown, with E. coli K‐12 JCB387 genomic DNA as a template. In the next round of PCR, primers narGup223 and narGCC(−40.5) were used with pRW50/narG223 to amplify the upstream region of the narG promoter. The resultant PCR product was then restricted with EcoRI and BamHI and cloned into pRW50/CC(−40.5) to generate the narG CC(−40.5) promoter. DNA substitutions were introduced into the promoter region of the narG CC(−40.5) promoter by PCR amplifying the downstream of narG CC(−40.5) with primers narGCC(TG) or narGCC(−10) and primer D10527. PCR products were cloned into pRW50/narG CC(−40.5) using HindIII and BamHI restriction sites. All constructs were verified by Sanger DNA sequencing.
For expression of recombinant proteins, the ogt104167 and narG CC(−40.5) promoters were introduced into pET15b/6his‐gfp and pET20b (Novagen) plasmids. Promoters were PCR amplified using primer pairs ogt(BglII) and ogt(XbaI) or narG223(BglII) and CC(XbaI) with the relevant pRW50 construct. Purified PCR products were restricted with BglII and XbaI and cloned into pET15b/6his‐gfp and pET20b, replacing the canonical T7 RNA polymerase promoter. DNA, encoding hGH‐6His and anti‐IL‐1β‐6His scFv, from pHAK1 and pYU49 (Alanen et al., 2015; Matos et al., 2014), respectively, was cloned into pET20b/ogt104167 and pET20b/narG CC(−40.5) using NdeI and SacI (Figures S1 and S2). All constructs were verified by Sanger DNA sequencing. The pET22b vector constructs, which express hGH‐6His, and carry the lac O1O1, lac O3O1, and tac promoters (i.e., PAR1, PAR4L, and PAR8 constructs), were previously described by Hothersall et al. (2021) (Table S1).
2.3. Strain construction
The ΔnarG strain JCB387N11 was constructed using P1 transduction, by transferring the kanamycin resistance gene marker, from E. coli K‐12 strain BW25113 narG::aph into strain JCB387 (Thomason et al., 2007). Kanamycin‐resistant colonies were isolated and the presence of the narG::aph cassette was confirmed, using PCR with primers narGFw and narGRev. The kanamycin resistance cassette was then removed by transforming candidates with plasmid pCP20 (Cherepanov & Wackernagel, 1995).
2.4. β‐Galactosidase assays
pRW50 derivatives, containing lacZ promoter fusions, were transformed into the relevant E. coli K‐12 strains and β‐galactosidase activities were measured using a Miller protocol (Miller, 1972). Single colonies, carrying each construct, were inoculated into Lennox Broth and grown overnight at 37°C with shaking. To assay activities, overnight cultures were inoculated into 5 ml of minimal salts media and grown at 37°C with shaking until an OD650 = 0.5–0.6 (Squire et al., 2009). Where appropriate, the medium was supplemented with 20 mM sodium nitrate. To test the ability of household plant‐growth fertilizers to induce gene expression, solutions of BabyBio (SBM Life Science), Growmore (Westlands), Miracle‐Gro All Purpose and Miracle‐Gro LiquaFeed (Evergreen Garden Care) were added to a final concentration of 1% (v/v or w/v). β‐Galactosidase activities are expressed as nmol ONPG (o‐nitrophenyl‐β‐d‐galactopyranose) hydrolyzed/min/g dry cell mass and represent the average of three independent experiments.
2.5. Recombinant protein overexpression and detection
Cultures of E. coli strain JCB387N11, carrying pET expression plasmids containing the ogt104167 and narG CC(−40.5) promoters and various target genes, were grown with shaking in 10 mL of minimal salts medium, until an OD600 = 0.3–0.5. Protein overexpression was induced by the addition of sodium nitrate and samples were taken after 3 h induction. To test the ability of household fertilizer to induce gene expression, BabyBio (SBM Life Science) was added to a final concentration of 1% (v/v). For pET22b constructs, which carry the lac O1O1, lac O3O1, and tac promoters (Hothersall et al., 2021), RPP was induced by the addition of IPTG to a final concentration of 1 mM. Anaerobic growth conditions were achieved by growing cultures statically in 100 ml minimal salts medium in 100 ml Duran bottles, as in our previous work (Filenko et al., 2007). Cultures were incubated at 37°C without shaking to OD600 = 0.3–0.5, and RPP was then induced by the addition of 20 mM sodium nitrate, with samples taken after 3 h induction.
Total protein samples were routinely prepared by resuspending normalized amounts of cells in 2× Laemmli loading buffer (Sigma), heating at 95°C for 3 min and centrifuging before loading. Normalized protein samples were resolved by reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) and analyzed using Coomassie blue staining and western blot analysis, as in our previous work (Browning et al., 2013). For western blot analysis, 6His‐GFP was detected using anti‐GFP antiserum raised in mouse (Sigma), and an anti‐mouse‐HRP secondary antibody (Sigma), hGH‐6His was detected using anti‐hGH antiserum raised in rabbit (Browning et al., 2019) and an anti‐rabbit‐HRP secondary antibody (Amersham), and anti‐IL‐1β‐6His scFv was detected using anti‐6His (C‐terminal)‐HRP (Invitrogen). Blots were developed using Pierce ECL western blotting analysis substrate and all gels and blots shown are representative experiments. To assess the aggregation of product in inclusion bodies, total, soluble, and insoluble protein samples were also prepared using an Agilent BugBuster, according to the manufacturer's instructions, as in our previous work (Hothersall et al., 2021).
2.6. Flow cytometry analysis
For flow cytometry analysis, 50 ml cultures in minimal salts medium were incubated with shaking at 30 or 37°C until the culture reached OD600 ~0.6 and then RPP was induced by the addition of 20 mM sodium nitrate. Cultures were analyzed using a BD Accuri C6 flow cytometer (Becton Dickinson). Samples were mixed with 0.2‐µm‐filtered PBS, and data were collected at a rate of 1000–4000 events per second, using slow flow and a forward scatter height (FSC‐H) threshold of 10,000 to eliminate noncellular material, until 20,000 events had been recorded per sample. Data were analyzed using CFlow software (BD). GFP fluorescence was detected using a 533/30 BP filter on channel FL1.
Live and dead cells were differentiated using propidium iodide (PI) (Wyre & Overton, 2014). The PI concentration used in the sample (final concentration of 4 μg/ml) and the gating to distinguish between live and dead cells was determined by measuring a mixture of live and dead cells. Dead cells were prepared by taking 2 ml of live cells pelleted by centrifugation at 13,000g for 1 min, washed in phosphate buffered saline (PBS), pelleted at 13,000g for 1 min, and resuspended in 1 ml of 70% ethanol for 5 min at room temperature. Ethanol was removed and the resulting material was suspended in PBS, pelleted at 13,000g and resuspended in 1 ml PBS. PI fluorescence was detected using a 670 LP filter on channel FL3.
3. RESULTS
3.1. Expression of recombinant protein using the ogt104167 promoter
Our previous studies with the ogt promoter region showed that a single 7‐2‐7 DNA site for NarL is sufficient for NarL‐dependent induction of transcription when located 65, 55, or 32 base pairs (bp) upstream from the promoter −10 element, that is, the ogt1041, ogt104167, and ogt1052 promoters, respectively (Figure 1b) (Ruanto et al., 2020). To examine expression from these promoters in more detail, each promoter fragment was cloned into the low copy number lac expression vector, pRW50, to generate lacZ transcriptional fusions that were transformed into the Δlac E. coli K‐12 strain, JCB387. The expression of β‐galactosidase in JCB387, carrying each promoter, was then determined when cells were grown in the presence or absence of 20 mM sodium nitrate. Results in Figure 1c show that although ogt1052 was a highly active promoter, it was poorly coupled to nitrate in the growth media. In contrast, the ogt104167 and ogt1041, promoters were better coupled to nitrate levels, being more tightly regulated.
As the ogt104167 promoter was the more active, we chose this to examine heterologous gene expression. Therefore, the genes encoding His‐tagged green fluorescent protein (6His‐GFP), His‐tagged human growth hormone (hGH‐6His), and a His‐tagged variable fragment of a single‐chain antibody directed against interleukin 1β (anti‐IL‐1β‐6His scFv) (Figures S1 and 2) were cloned into pET vectors carrying ogt104167. As ogt104167 was tightly controlled in minimal media (Figure 1d) expression was examined in minimal salts medium (Squire et al., 2009) and, to reduce the removal of nitrate inducer from the medium, we used an E. coli strain, lacking NarG, the major nitrate reductase. Results in Figure 2 illustrate SDS‐PAGE and western blot analysis of batch‐grown cultures in which RPP was induced for 3 h by the addition of 20 mM sodium nitrate. In each case, significant nitrate‐induced overexpression was seen, with little or no expression detected in the absence of nitrate. Furthermore, expression from ogt104167 increased with increasing nitrate concentration, reaching a maximum at ~5 mM sodium nitrate (Figure 2b). Thus, the ogt104167 promoter is a tightly regulated nitrate‐responsive promoter that can be used to drive expression of heterologous proteins in E. coli.
Figure 2.
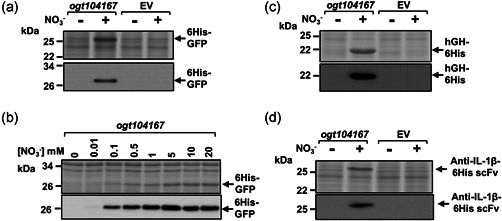
Recombinant protein production driven by the ogt104167 promoter. The figure shows Coomassie blue‐stained sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and western blots (below) that detail the expression of (a, b) recombinant 6His‐GFP, (c) hGH‐6His, and (d) anti‐IL‐1β‐6His scFv in Escherichia coli K‐12 JCB387N11 (ΔnarG) cells grown in minimal salts medium after 3 h induction by the addition of 20 mM sodium nitrate (+) at an OD600 of 0.3–0.5. The DNA encoding each target was cloned into pET expression vectors carrying the ogt104167 promoter fragment. Empty vector controls (EV) are included, where indicated.
3.2. Construction and testing of the narG CC(−40.5) promoter
Although controlled RPP was achieved with nitrate using ogt104167, we wanted to reach higher expression levels, while maintaining regulation by nitrate and NarL. Most bacterial transcription activators work by making a direct contact with RNAP, and this acts as molecular “velcro” to position the RNAP at the promoter (Browning & Busby, 2016; Finkelstein, 2005). At some promoters, different activators work together, with each factor contributing its contact to the “velcro” (Finkelstein, 2005; Lee et al., 2012). This is the case at the E. coli narG promoter where activity is co‐dependent on the binding of FNR and NarL (Walker & DeMoss, 1992, 1994). We decided to focus on developing a similar promoter, dependent on NarL and a second activator, but, as FNR functions only in anaerobic conditions, we used its homolog, CRP (the cyclic AMP receptor protein), that is active in most growth conditions (Li et al., 1998). Previous studies had shown that CRP (like FNR) activates transcription as a dimer, and activates optimally when the spacing between the center of the DNA site for CRP and the promoter −10 element is 29 base pairs (Gaston et al., 1988; Li et al., 1998; Rossiter et al., 2015). However, if the spacing is reduced to 28 base pairs, promoter activity falls to basal levels, but it can be restored by an upstream‐bound activator (Rossiter et al., 2015). Hence, starting with a simple semisynthetic CRP‐dependent promoter, CC(−41.5), carrying a single DNA site for CRP located 29 bp upstream from the promoter −10 element (Gaston et al., 1990; West et al., 1993), we first adjusted the spacing to 28 bp, and then sought to restore activity to the resulting promoter (CC(−40.5)) by inserting a single upstream 7–2–7 DNA site for NarL. However, despite trying a range of locations, we were unable to find a combination that resulted in nitrate‐regulated promoter activity (Figure S3). In contrast, when we inserted an upstream segment from the narG promoter covering the multiple DNA sites for NarL, the activity of the resulting promoter (narG CC(−40.5)) was massively induced by the inclusion of nitrate in the growth media. Figure 3 illustrates the different constructions (Figure 3a), together with assay data for each promoter, using our lacZ‐based expression vector (Figure 3b,c). To confirm that the observed activity of narG CC(−40.5) is due to a single promoter, co‐activated by NarL and CRP, we showed that nitrate‐dependent induction is dependent on NarL (Figure 3b) and that a single base substitution in the promoter −10 element reduces activity to basal levels (Figure S4). Additionally, introduction of CRP carrying the HL159 and KE101 substitutions, which prevent productive interactions between CRP and RNAP (West et al., 1993), also suppresses induction (Figure S4).
Figure 3.
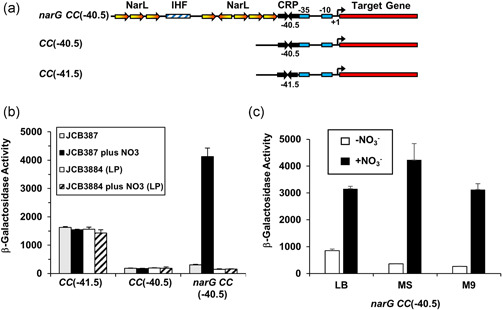
Expression analysis of the narG CC(−40.5) promoter. (a) The panel shows schematic representations of the narG CC(−40.5), CC(−40.5), and CC(−41.5) promoter fragments. The DNA sites for NarL and CRP are shown as arrows, the integration host factor (IHF)‐binding site is depicted as a hatched box and the promoter −35 and −10 elements shown as rectangles. Transcript start sites (+1) are indicated by bent arrows. (b) The panel shows measured β‐galactosidase activities in strain JCB387 and JCB3884 (narL narP) cells, carrying the CC(−41.5), CC(−40.5), and narG CC(−40.5) promoter fragments cloned into the pRW50 lacZ expression vector. Cells were grown in minimal salts media supplemented with 20 mM sodium nitrate (+NO3), where indicated. (c) The panel shows measured β‐galactosidase activities in JCB387 cells, carrying the narG CC(−40.5) promoter fragment cloned into pRW50, with cells grown in LB medium (LB), minimal salts media (MS), or M9 minimal medium (M9) supplemented with 20 mM sodium nitrate, as indicated. β‐galactosidase activities are expressed as nmol ONPG hydrolyzed/min/mg dry cell mass and represent the average of three independent experiments. Error bars represent SD.
3.3. High‐level recombinant protein production using the narG CC(−40.5) promoter
The response of the narG CC(−40.5) promoter to nitrate suggested exploitation in overexpression vectors, and so it was introduced into a plasmid encoding 6His‐GFP. Results in Figure 4 show that, in batch cultures, 6His‐GFP was induced to high levels by nitrate and, as predicted, the narG CC(−40.5) promoter was stronger than ogt104167 (Figure S5). Importantly, cell growth and viability was unaffected by nitrate‐induced 6His‐GFP expression, even at different temperatures (i.e., 30 and 37°C) for extended periods of time (Figure 5a–d). Analysis of individual cells by flow cytometry indicated that 6His‐GFP induction occurred homogeneously within the bacterial cell population (Figure 5e). Note that some low‐level expression of 6His‐GFP occurs in the absence of nitrate (Figure 4) but, because expression depends on CRP, it is subject to catabolite repression and can be suppressed by the inclusion of glucose in the growth medium (Figure 4c) (Hothersall et al., 2021; Kaur et al., 2018).
Figure 4.
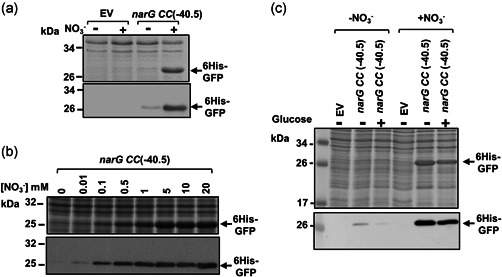
Expression of 6His‐GFP using the narG CC(−40.5) promoter. The figure shows Coomassie blue‐stained sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and western blots (below) of JCB387N11 (ΔnarG) cells expressing 6His‐GFP, using the narG CC(−40.5) promoter. The DNA encoding 6His‐GFP was cloned into pET expression vectors carrying the narG CC(−40.5) promoter fragment. Cells were grown in minimal salts media and RPP was initiated for 3 h (a, c) by the addition of 20 mM sodium nitrate or (b) by a range of sodium nitrate concentrations. In (c), cultures were supplemented with 0.4% glucose, where indicated. Empty vector controls (EV) were included.
Figure 5.
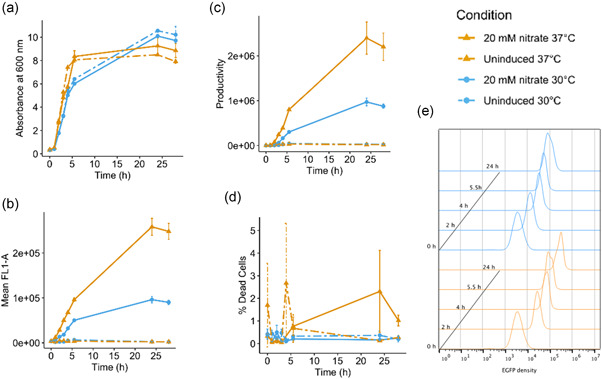
Analysis of 6His‐GFP expression from the narG CC(−40.5) promoter in E. coli JCB387N11 cells at 30 or 37°C. The figure shows the (a) growth, (b) mean GFP fluorescence measured by flow cytometry, (c) productivity (green fluorescence multiplied by OD600), and (d) percentage dead cells of 50 ml JCB387N11 (ΔnarG) cultures expressing 6His‐GFP, using the narG CC(−40.5) promoter. Cells were grown in minimal salts media at either 30°C (blue) or 37°C (orange) for 25 h. Recombinant protein production was initiated by the addition of sodium nitrate to a final concentration of 20 mM, as cells reached an OD600 of ~0.6. Values represent the average values from duplicate flasks and error bars represent ±SD. (e) Flow cytometry analysis of green fluorescence from cells expressing 6His‐GFP grown and induced at either 30°C (blue) or 37°C (orange). Data are plotted as histograms showing the number of cells with different green fluorescence (FL1‐A) values.
3.4. Expression of biopharmaceuticals using the narG CC(−40.5) promoter
To examine expression from the narG CC(−40.5) promoter further, vectors were constructed to express hGH‐6His and the anti‐IL‐1β‐6His scFv. Like 6His‐GFP, expression of both hGH‐6His and the anti‐IL‐1β‐6His scFv was induced by nitrate (Figure 6a,b). Since the correct folding of hGH and scFv requires the formation of disulfide bonds, which is not favored in the reducing environment of E. coli cytoplasm, the majority of hGH‐6His and anti‐IL‐1β‐6His scFv was insoluble (Figure 6c). Hence, these targets were expressed at a lower temperature in E. coli SHuffle Express cells, a genetically modified E. coli strain that enables cytoplasmic disulfide bond formation. Under these conditions, hGH‐6His and the anti‐IL‐1β‐6His scFv were soluble (Figure 6d), highlighting the functionality of this expression system in different strains with different growth regimes. Furthermore, in the conditions that we have tried, RPP driven by the narG CC(−40.5) and ogt104167 promoters is comparable to that of other promoters (e.g., the tac promoter and other lac‐based promoters; Hothersall et al., 2021), and has minimal effect on bacterial growth (Figure 7a,b).
Figure 6.
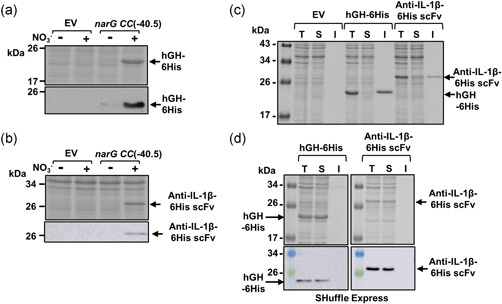
Solubility of recombinant hGH‐6His and anti‐IL‐1β‐6His scFv expressed in Escherichia coli JCB387N11 and SHuffle Express cells. (a, b) Coomassie blue‐stained sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) gels and western blots (below), which detail the expression of recombinant hGH‐6His and anti‐IL‐1β‐6His scFv, respectively, in E. coli K‐12 JCB387N11 (ΔnarG) cells grown in minimal salts medium after 3 h induction by the addition of 20 mM sodium nitrate (+). (c, d) Coomassie blue‐stained SDS‐PAGE gels investigating the solubility of hGH‐6His and anti‐IL‐1β‐6His scFv expressed in JCB387N11 (ΔnarG) and SHuffle Express cells, respectively, using the narG CC(−40.5) promoter. Cultures were grown in minimal salts medium and protein production was induced by the addition of 20 mM sodium nitrate for 3 h. Harvested cells were lysed to prepare total (T), soluble (S), and insoluble (I) protein samples. In (d), a western blot (below) is included, showing the detection of hGH‐6His and anti‐IL‐1β‐6His scFv. Empty vector controls (EV) are indicated.
Figure 7.
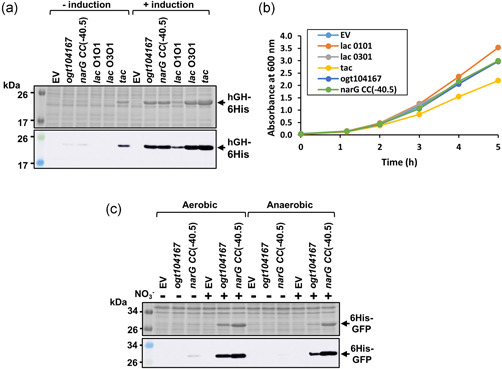
Comparison of protein expression from the ogt104167 and narG CC(−40.5) promoters with lac‐based expression systems and different expression regimes. (a) Coomassie blue‐stained sodium dodecyl sulfate polyacrylamide (SDS‐PAGE) gel electrophoresis gel and western blot (below) that detail hGH‐6His expression and (b) growth profiles of induced cultures expressing hGH‐6His from ogt104167 and narG CC(−40.5) in comparison to the low strength lac O1O1 promoter, the medium strength lac O3O1 promoter, and the strong tac promoter (Hothersall et al., 2021) in E. coli K‐12 JCB387N11 (ΔnarG) cells. Cells were grown in minimal salts medium to an OD600 of 0.3–0.5. and protein production was induced for 3 h (+) by the addition of either 20 mM sodium nitrate to ogt104167, narG CC(−40.5), and the empty vector control (EV), or 1 mM IPTG to the tac and lac‐based vectors. (c) Coomassie blue‐stained SDS‐PAGE gel and western blot (below) that detail 6His‐GFP expression from ogt104167 and narG CC(−40.5) under anaerobic and aerobic conditions. E. coli K‐12 JCB387N11 (ΔnarG) cells were grown in minimal salts medium and protein production was induced for 3 h (+) by the addition of 20 mM sodium nitrate. Representative gels and growth curves are shown.
4. DISCUSSION
Our overarching aim was to develop a robust regulated expression system for foreign proteins in E. coli. While decades of research have led to the discovery of scores of new regulators, very few have been exploited for biotechnology. Here, building on our previous work with NarL, we have developed two synthetic promoters whose activity is triggered by the addition of nitrate ions to the growth media. In previous reports, E. coli narG promoter derivatives have been exploited to drive RPP in anaerobic conditions (Hwang et al., 2017, 2018; Kim et al., 2011). Note that these previous studies focused on induction driven by anaerobiosis (dependent on FNR) rather than induction driven by nitrate (dependent on NarL). Here we have exploited the upstream sequences from the E. coli narG promoter to confer nitrate‐dependence on a core promoter cassette (dependent on CRP), thereby uncoupling nitrate‐dependent induction from anaerobic induction. This results in expression systems that can operate under both aerobic and anaerobic conditions (Figure 7c).
For many commercial expression systems, the inducer represents a significant cost. Since sodium nitrate costs less than one dollar per kilogram, we believe that the new vectors described here will be useful in locations where infrastructure is limiting. Additionally, since nitrate levels are high in many commercial fertilizers, recombinant protein can also be induced using inexpensive garden products available in local stores (Figure 8), and the facility to induce RPP without needing pure chemicals could prove useful in some situations. Thus, as well as cutting the cost of industrial RPP, our new promoters may have applications for protein production outside of the laboratory in the realm of DIY biology (Landrain et al., 2013).
Figure 8.
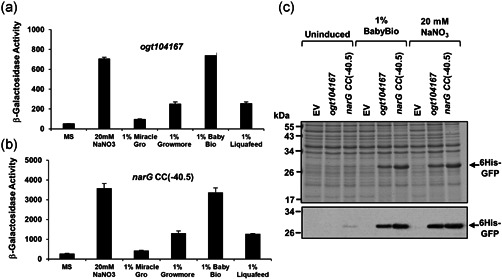
Induction of the ogt104167 and narG CC(−40.5) promoters using garden fertilizers. The figure shows measured β‐galactosidase activities in wild‐type JCB387 cells, carrying either the (a) ogt104167 or (b) the narG CC(−40.5) promoter fragments cloned into pRW50. Cells were grown in minimal salts media supplemented with 20 mM sodium nitrate or household fertilizers (Miracle‐Gro All Purpose, Growmore, BabyBio and Miracle‐Gro LiquaFeed) to a final concentration of 1% (v/v or w/v). β‐Galactosidase activities are expressed as nmol ONPG hydrolyzed/min/mg dry cell mass and represent the average of three independent experiments. Error bars represent SD. Panel (c) shows a Coomassie blue stained sodium dodecyl sulfate polyacrylamide gel electrophoresis gel and western blot (below), which detail the expression of recombinant 6His‐GFP in E. coli K‐12 JCB387N11 (ΔnarG). Cells were grown in minimal salts medium and RPP was induced for 3 h by the addition of 20 mM sodium nitrate or 1% (v/v) BabyBio fertilizer. Empty vector controls (EV) are also included.
CONFLICT OF INTERESTS
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
Joanne Hothersall, Tim W. Overton, Colin Robinson, Stephen J. W. Busby, and Douglas F. Browning devised the research program. Joanne Hothersall, Sandie Lai, Nan Zhang, Rita E. Godfrey, Patcharawarin Ruanto, Sarah Bischoff, and Douglas F. Browning performed the experiments. Joanne Hothersall, Stephen J. W. Busby, and Douglas F. Browning wrote the manuscript with input from all the authors.
Supporting information
Supplementary information.
ACKNOWLEDGMENTS
J. H. was supported by an Industrial Biotechnology Catalyst (Innovate UK, BBSRC, EPSRC) (BB/M018261/1) to promote the translation, development, and commercialization of innovative Industrial Biotechnology processes, and by a BBSRC IAA Follow‐on‐Fund award (BBSRC IAA BB/S506709/1). D. F. B was supported by BBSRC grants BB/M018261/1 and BB/R017689/1. S. L. was supported by a studentship from the BBSRC Midlands Integrative Biosciences Training Programme. This study was supported by Industrial Biotechnology Catalyst Grant (Innovate UK, BBSRC, EPSRC) (BB/M018261/1); BBSRC IAA Follow‐on‐Fund award (BBSRC IAA BB/S506709/1); BBSRC grant BB/R017689/1; BBSRC Midlands Integrative Biosciences Training Programme.
Hothersall, J. , Lai, S. , Zhang, N. , Godfrey, R. E. , Ruanto, P. , Bischoff, S. , Robinson, C. , Overton, T. W. , Busby, S. J. W. , & Browning, D. F. (2022). Inexpensive protein overexpression driven by the NarL transcription activator protein. Biotechnology and Bioengineering, 119, 1614–1623. 10.1002/bit.28071
Contributor Information
Stephen J. W. Busby, Email: s.j.w.busby@bham.ac.uk.
Douglas F. Browning, Email: d.browning@aston.ac.uk.
DATA AVAILABILITY STATEMENT
Data available in article supplementary material.
REFERENCES
- Alanen, H. I. , Walker, K. L. , Lourdes Velez Suberbie, M. , Matos, C. F. , Bönisch, S. , Freedman, R. B. , Keshavarz‐Moore, E. , Ruddock, L. W. , & Robinson, C. (2015). Efficient export of human growth hormone, interferon alpha2b and antibody fragments to the periplasm by the Escherichia coli Tat pathway in the absence of prior disulfide bond formation. Biochimica et Biophysica Acta/General Subjects, 1853(3), 756–763. 10.1016/j.bbamcr.2014.12.027 [DOI] [PubMed] [Google Scholar]
- Browning, D. F. , & Busby, S. J. (2016). Local and global regulation of transcription initiation in bacteria. Nature Reviews Microbiology, 14(10), 638–650. 10.1038/nrmicro.2016.103 [DOI] [PubMed] [Google Scholar]
- Browning, D. F. , Godfrey, R. E. , Richards, K. L. , Robinson, C. , & Busby, S. J. W. (2019). Exploitation of the Escherichia coli lac operon promoter for controlled recombinant protein production. Biochemical Society Transactions, 47(2), 755–763. 10.1042/bst20190059 [DOI] [PubMed]
- Browning, D. F. , Grainger, D. C. , & Busby, S. J. (2010). Effects of nucleoid‐associated proteins on bacterial chromosome structure and gene expression. Current Opinion in Microbiology, 13(6), 773–780. 10.1016/j.mib.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Browning, D. F. , Matthews, S. A. , Rossiter, A. E. , Sevastsyanovich, Y. R. , Jeeves, M. , Mason, J. L. , Wells, T. J. , Wardius, C. A. , Knowles, T. J. , Cunningham, A. F. , Bavro, V. N. , Overduin, M. , & Henderson, I. R. (2013). Mutational and topological analysis of the Escherichia coli BamA protein. PLoS One, 8(12), e84512. 10.1371/journal.pone.0084512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov, P. P. , & Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp‐catalyzed excision of the antibiotic‐resistance determinant. Gene, 158(1), 9–14. [DOI] [PubMed] [Google Scholar]
- Constantinidou, C. , Hobman, J. L. , Griffiths, L. , Patel, M. D. , Penn, C. W. , Cole, J. A. , & Overton, T. W. (2006). A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. Journal of Biological Chemistry, 281(8), 4802–4815. 10.1074/jbc.M512312200 [DOI] [PubMed] [Google Scholar]
- Darwin, A. J. , & Stewart, V. (1996). The NAR modulon systems: Nitrate and nitrite regulation of anaerobic gene expression. In Lin E. C. C., & Lynch A. S. (Eds.), Regulation of gene expression in Escherichia coli (pp. 343–359). Springer. [Google Scholar]
- Darwin, A. J. , Tyson, K. L. , Busby, S. J. , & Stewart, V. (1997). Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K‐12 depends on DNA binding site arrangement. Molecular Microbiology, 25(3), 583–595. [DOI] [PubMed] [Google Scholar]
- Filenko, N. , Spiro, S. , Browning, D. F. , Squire, D. , Overton, T. W. , Cole, J. , & Constantinidou, C. (2007). The NCR regulon of Escherichia coli K‐12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. Journal of Bacteriology, 189(12), 4410–4417. 10.1128/jb.00080-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, J. M. (2005). Velcro rules. Nature Reviews Molecular Cell Biology, 6(1), S14. 10.1038/nrm1803 [DOI] [Google Scholar]
- Gaston, K. , Bell, A. , Kolb, A. , Buc, H. , & Busby, S. (1990). Stringent spacing requirements for transcription activation by CRP. Cell, 62(4), 733–743. [DOI] [PubMed] [Google Scholar]
- Gaston, K. , Chan, B. , Kolb, A. , Fox, J. , & Busby, S. (1988). Alterations in the binding site of the cyclic AMP receptor protein at the Escherichia coli galactose operon regulatory region. Biochemical Journal, 253(3), 809–818. 10.1042/bj2530809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothersall, J. , Godfrey, R. E. , Fanitsios, C. , Overton, T. W. , Busby, S. J. W. , & Browning, D. F. (2021). The PAR promoter expression system: Modified lac promoters for controlled recombinant protein production in Escherichia coli . New Biotechnology, 64, 1–8. 10.1016/j.nbt.2021.05.001 [DOI] [PubMed] [Google Scholar]
- Hwang, H. J. , Kim, J. W. , Ju, S. Y. , Park, J. H. , & Lee, P. C. (2017). Application of an oxygen‐inducible nar promoter system in metabolic engineering for production of biochemicals in Escherichia coli . Biotechnology and Bioengineering, 114(2), 468–473. 10.1002/bit.26082 [DOI] [PubMed] [Google Scholar]
- Hwang, H. J. , Lee, S. Y. , & Lee, P. C. (2018). Engineering and application of synthetic nar promoter for fine‐tuning the expression of metabolic pathway genes in Escherichia coli . Biotechnology for Biofuels, 11, 103. 10.1186/s13068-018-1104-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, J. , Kumar, A. , & Kaur, J. (2018). Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. International Journal of Biological Macromolecules, 106, 803–822. 10.1016/j.ijbiomac.2017.08.080 [DOI] [PubMed] [Google Scholar]
- Kim, N. J. , Choi, J. H. , Kim, Y. C. , Lee, J. , Lee, S. Y. , Chang, H. N. , & Lee, P. C. (2011). Development of anaerobically inducible nar promoter expression vectors for the expression of recombinant proteins in Escherichia coli . Journal of Biotechnology, 151(1), 102–107. 10.1016/j.jbiotec.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Landrain, T. , Meyer, M. , Perez, A. M. , & Sussan, R. (2013). Do‐it‐yourself biology: challenges and promises for an open science and technology movement. Systems and Synthetic Biology, 7(3), 115–126. 10.1007/s11693-013-9116-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. J. , Minchin, S. D. , & Busby, S. J. (2012). Activating transcription in bacteria. Annual Review of Microbiology, 66, 125–152. 10.1146/annurev-micro-092611-150012 [DOI] [PubMed] [Google Scholar]
- Li, B. , Wing, H. , Lee, D. , Wu, H. C. , & Busby, S. (1998). Transcription activation by Escherichia coli FNR protein: Similarities to, and differences from, the CRP paradigm. Nucleic Acids Research, 26(9), 2075–2081. 10.1093/nar/26.9.2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge, J. , Fear, J. , Busby, S. , Gunasekaran, P. , & Kamini, N. R. (1992). Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiology Letters, 74(2–3), 271–276. [DOI] [PubMed] [Google Scholar]
- Matos, C. F. , Robinson, C. , Alanen, H. I. , Prus, P. , Uchida, Y. , Ruddock, L. W. , Freedman, R. B. , & Keshavarz‐Moore, E. (2014). Efficient export of prefolded, disulfide‐bonded recombinant proteins to the periplasm by the Tat pathway in Escherichia coli CyDisCo strains. Biotechnology Progress, 30(2), 281–290. 10.1002/btpr.1858 [DOI] [PubMed] [Google Scholar]
- Miller, J. (1972). Experiments in molecular genetics. Cold Spring Harbor Laboratory. [Google Scholar]
- Nora, L. C. , Westmann, C. A. , Martins‐Santana, L. , Alves, L. F. , Monteiro, L. M. O. , Guazzaroni, M. E. , & Silva‐Rocha, R. (2019). The art of vector engineering: towards the construction of next‐generation genetic tools. Microbial Biotechnology, 12(1), 125–147. 10.1111/1751-7915.13318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossiter, A. E. , Godfrey, R. E. , Connolly, J. A. , Busby, S. J. , Henderson, I. R. , & Browning, D. F. (2015). Expression of different bacterial cytotoxins is controlled by two global transcription factors, CRP and Fis, that co‐operate in a shared‐recruitment mechanism. Biochemical Journal, 466(2), 323–335. 10.1042/bj20141315 [DOI] [PubMed] [Google Scholar]
- Ruanto, P. , Chismon, D. L. , Hothersall, J. , Godfrey, R. E. , Lee, D. J. , Busby, S. J. W. , & Browning, D. F. (2020). Activation by NarL at the Escherichia coli ogt promoter. Biochemical Journal, 477(15), 2807–2820. 10.1042/bcj20200408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos‐Zavaleta, A. , Salgado, H. , Gama‐Castro, S. , Sánchez‐Pérez, M. , Gómez‐Romero, L. , Ledezma‐Tejeida, D. , García‐Sotelo, J. S. , Alquicira‐Hernández, K. , Muñiz‐Rascado, L. J. , Peña‐Loredo, P. , Ishida‐Gutiérrez, C. , Velázquez‐Ramírez, D. A. , Del Moral‐Chávez, V. , Bonavides‐Martínez, C. , Méndez‐Cruz, C. F. , Galagan, J. , & Collado‐Vides, J. (2019). RegulonDB v 10.5: Tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K‐12. Nucleic Acids Research, 47(D1), D212–d220. 10.1093/nar/gky1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire, D. J. , Xu, M. , Cole, J. A. , Busby, S. J. , & Browning, D. F. (2009). Competition between NarL‐dependent activation and Fis‐dependent repression controls expression from the Escherichia coli yeaR and ogt promoters. Biochemical Journal, 420(2), 249–257. 10.1042/BJ20090183 [DOI] [PubMed] [Google Scholar]
- Stewart, V. (2003). Biochemical Society Special Lecture. Nitrate‐ and nitrite‐responsive sensors NarX and NarQ of proteobacteria. Biochemical Society Transactions, 31(Pt 1), 1–10. 10.1042/bst0310001 [DOI] [PubMed] [Google Scholar]
- Thomason, L. C. , Costantino, N. , & Court, D. L. (2007). E. coli genome manipulation by P1 transduction. Current Protocols in Molecular Biology. Chapter 1, Unit 1.17. 10.1002/0471142727.mb0117s79 [DOI] [PubMed] [Google Scholar]
- Tungekar, A. A. , Castillo‐Corujo, A. , & Ruddock, L. W. (2021). So you want to express your protein in Escherichia coli? Essays in Biochemistry, 65, 247–260. 10.1042/ebc20200170 [DOI] [PubMed] [Google Scholar]
- Walker, M. S. , & DeMoss, J. A. (1992). Role of alternative promoter elements in transcription from the nar promoter of Escherichia coli. Journal of Bacteriology, 174(4), 1119–1123. 10.1128/jb.174.4.1119-1123.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, M. S. , & DeMoss, J. A. (1994). NarL‐phosphate must bind to multiple upstream sites to activate transcription from the narG promoter of Escherichia coli . Molecular Microbiology, 14(4), 633–641. 10.1111/j.1365-2958.1994.tb01302.x [DOI] [PubMed] [Google Scholar]
- West, D. , Williams, R. , Rhodius, V. , Bell, A. , Sharma, N. , Zou, C. , Fujita, N. , Ishihama, A. , & Busby, S. (1993). Interactions between the Escherichia coli cyclic AMP receptor protein and RNA polymerase at class II promoters. Molecular Microbiology, 10(4), 789–797. [DOI] [PubMed] [Google Scholar]
- Wyre, C. , & Overton, T. W. (2014). Use of a stress‐minimisation paradigm in high cell density fed‐batch Escherichia coli fermentations to optimise recombinant protein production. Journal of Industrial Microbiology and Biotechnology, 41(9), 1391–1404. 10.1007/s10295-014-1489-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Data Availability Statement
Data available in article supplementary material.


