Abstract
Soil structure depends on the association between mineral soil particles (sand, silt, and clay) and organic matter, in which aggregates of different size and stability are formed. Although the chemistry of organic materials, total microbial biomass, and different enzyme activities in different soil particle size fractions have been well studied, little information is available on the structure of microbial populations in microhabitats. In this study, topsoil samples of different fertilizer treatments of a long-term field experiment were analyzed. Size fractions of 200 to 63 μm (fine sand fraction), 63 to 2 μm (silt fraction), and 2 to 0.1 μm (clay fraction) were obtained by a combination of low-energy sonication, wet sieving, and repeated centrifugation. Terminal restriction fragment length polymorphism analysis and cloning and sequencing of 16S rRNA genes were used to compare bacterial community structures in different particle size fractions. The microbial community structure was significantly affected by particle size, yielding higher diversity of microbes in small size fractions than in coarse size fractions. The higher biomass previously found in silt and clay fractions could be attributed to higher diversity rather than to better colonization of particular species. Low nutrient availability, protozoan grazing, and competition with fungal organisms may have been responsible for reduced diversities in larger size fractions. Furthermore, larger particle sizes were dominated by α-Proteobacteria, whereas high abundance and diversity of bacteria belonging to the Holophaga/Acidobacterium division were found in smaller size fractions. Although very contrasting organic amendments (green manure, animal manure, sewage sludge, and peat) were examined, our results demonstrated that the bacterial community structure was affected to a greater extent by the particle size fraction than by the kind of fertilizer applied. Therefore, our results demonstrate specific microbe-particle associations that are affected to only a small extent by external factors.
Soil structure depends on the association between mineral soil particles (sand, silt, and clay) and organic matter, in which aggregates of different size and stability are formed (54). Mechanically resistant microaggregates (<250 μm) evolve through interactions of primary particles with microorganisms, plant roots, fungal hyphae, polysaccharides, and humic materials. They build distinctly less stable macroaggregates (>250 μm), which can be destroyed by changing the soil management (54). This kind of aggregation is held together by a network of roots and hyphae (46). The structural organization of soil particles provides a spatially heterogeneous habitat for microorganisms characterized by different substrate, nutrient, and oxygen concentrations and water contents as well as variable pH values (37).
Different methods have been applied to study the distribution of organic matter and microbes in the soil matrix, such as electron microscopy (15), repeated washing of soil aggregates (23), and techniques based on physical soil fractionation (6, 27, 58). Several studies have demonstrated that cell numbers and microbial biomass were most concentrated in the smaller size silt and clay fractions (27, 30, 32, 58). Because the sizes of soil aggregates and soil pores correlate, microbial biomass is also mainly present in micropores (5 to 30 μm) (3, 22, 36). Furthermore, the activities of such soil enzymes as urease, invertase, and alkaline phosphatase were reported to be highest in the silt and clay fractions, whereas the activity of xylanase was found to be highest in the sand fraction (21, 31, 32, 51, 52). The latter enzyme has been suggested to be an indicator of fungal activity (21, 32). Fungal biomass has been demonstrated to be low in comparison with the bacterial biomass in grasslands as well as in an arable field (5, 23), but fungi may play an important role in the initial breakdown of organic matter.
Soil microbial communities show extremely high phenotypic and genotypic diversities, DNA renaturation experiments suggested that there are 4 × 103 to 7 × 103 different genome equivalents per g of soil (55). Microbial diversity has been addressed in several studies (4, 10, 14, 44, 45); less information, however, is available on the structure of microbial populations in microhabitats. For example, Gelsomino et al. (17) revealed in a preliminary experiment that similar bacterial types were distributed over soil aggregates of different sizes obtained by a wet sieving procedure. In addition, Kandeler et al. (32) recently demonstrated by phospholipid fatty acid analysis and by denaturating gradient gel electrophoresis of 16S rRNA genes that the microbial biomass within the clay fraction was mainly due to bacterial colonization. In contrast, a high percentage of fungally derived fatty acids were found in the coarse sand fraction, which was associated with the particulate organic matter. Although these publications gave initial insight into the structure of the microbial community of these specific microhabitats, it is not well understood whether organic matter quality and/or particle size regulates the distribution of specific microbial populations in different particle size fractions.
The aim of the present study was to test whether the microbial population structure responds to changes in the quality of the organic substrates in particle size fractions obtained from soils of a long-ferm fertilizer experiment in Ultuna, Sweden. A physical fractionation procedure following low-energy sonication (51) was combined with analysis of microbial communities in different particle size fractions. DNA-based methods were used because most bacteria found in natural environments are not accessible to cultivation (2). The 16S rRNA gene has become a frequently employed phylogenetic marker to describe microbial diversity in soils without the need of cultivation (10, 14, 49). Terminal restriction length polymorphism (T-RFLP) analysis (7, 11, 38, 47, 50) and cloning and sequencing of 16S rRNA genes were applied to analyze bacterial diversity. Data on the nutrient turnover and soil organic matter fractions and changes in physical soil properties and enzyme activities of this long-term field experiment have been published previously (18–21, 35, 36).
MATERIALS AND METHODS
Site and samples.
Topsoil samples (to a depth of 0 to 20 cm) were taken from the Ultuna long-term field experiment in autumn 1998. A complete compilation of the experiment and the data can be found in the report by Kirchmann et al. (34). The experiment started in 1956, and at that time, the soil (to a depth of 0 to 20 cm) had 15 g of organic carbon kg−1, 1.7 g of nitrogen kg−1, and a pH of 6.6. The experiment tests 14 treatments, each of which was replicated four times in a randomized block design. In this study, samples from seven treatments were analyzed: continuous bare fallow; NoN, plots did not receive nitrogen fertilizers; Ca(NO3)2 (with 80 kg of N ha−1 year−1); GM, green manure; AM, well-decomposed animal manure; peat; and SS, sewage sludge. Organic amendments were analyzed before application, and equal amounts of organic matter (2,000 kg of Ca(NO3)2 ha−1 year−1) were added in 1956, 1960, and 1963 and thereafter were added every second year by hand. In addition, plots received 20 kg of P ha−1 as superphosphate and 35 to 38 kg of K ha−1 as potassium chloride annually in the spring. Cereals, rape crops, and fodder beet were grown alternately.
Fractionation procedure.
The size fractionation procedure is based on the method of Jocteur Monrozier et al. (27) and was performed with sieved samples (≤2 mm) as described in detail by Stemmer et al. (51). Essentially, the soil-water suspension was dispersed by low-energy sonication (output energy of 0.2 kJ g−1) and subsequently fractionated by a combination of wet sieving and repeated centrifugation to avoid disruption of microaggregates. Particle size fractions of 200 to 63 μm (fine sand fraction), 63 to 2 μm (silt fraction), and 2 to 0.1 μm (clay fraction) were obtained without the addition of a flocculant. The fraction samples were freeze-dried.
DNA isolation.
For the isolation of DNA, the protocol described by van Elsas and Smalla (57) was slightly modified. Freeze-dried soil (0.3 to 1.0 g) and 0.38 g of lysozyme were resuspended in 0.75 ml of 0.12 M sodium phosphate buffer (pH 8.0), and this mixture was incubated at 37°C for 15 min. After the samples had been cooled on ice, 750 mg of acid-washed glass beads (Sigma; 0.09 to 0.13 mm) was added, and bead beating was performed three times for 90 s at full speed in a mixer mill (type MM2000; 220 V, 50 Hz; Retsch GmbH & Co. KG, Haam, Germany) with intervals of 30 s. Sodium dodecyl sulfate (45 μl of a 20% solution) was added, and the mixture was incubated at room temperature for 15 min. Subsequently, 1 volume of phenol was added, and the mixture was mixed and centrifuged for 5 min at 10,000 × g. The organic phase was extracted again with 0.12 M sodium phosphate solution, and aqueous phases were pooled and extracted with 1 volume of chloroform. After centrifugation for 5 min at 10,000 × g, 0.1 volume of 5 M potassium acetate was added to the aqueous phase, and humic acids were precipitated for 15 min at room temperature. Samples were then centrifuged for 5 min at 10,000 × g, and the supernatant was amended with 0.1 volume of 5 M NaCl and 0.7 volume of isopropanol and incubated for 30 min at −80°C in order to precipitate DNA. DNA was obtained by centrifugation for 10 min at 10,000 × g, and the resulting pellet was washed with 70% ethanol, dried, and resuspended in 80 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). For further purification, spin columns that contained Sepharose CL-6B (Pharmacia) and polyvinylpyrrolidone (20 mg of Sepharose CL-6B ml−1) were prepared. In most cases, passage through two columns was needed to remove all PCR-inhibiting substances.
T-RFLP analysis.
The eubacterial primers 8f (59), labeled at the 5′ end with 6-carboxyfluorescein (6-Fam; MWG), and 518r (38) were used to amplify approximately 530 bp of the 16S rRNA gene. The reactions were carried out with a PTC-100 thermocycler (MJ Research, Inc.), applying an initial denaturation step of 5 min at 95°C followed by 35 cycles of 30 s at 95°C, 1 min of annealing at 54°C, and 2 min of extension at 72°C. The PCR mixtures (50 μl) contained 1× reaction buffer (Gibco, BRL); 200 μM (each) dATP, dCTP, dGTP, and dTTP; 0.15 μM (each) primer; 3 mM MgCl2; 2.5 U of Taq DNA polymerase (Gibco, BRL); and 20 ng of template DNA. PCR products (10 μl [approximately 250 ng of DNA]) were digested for 2 h in 20 μl with 10 U of a combination of the restriction enzymes HhaI and HaeIII (Gibco, BRL). Preliminary experiments with several restriction enzymes with 4-bp recognition sites (AluI, MspI, RsaI, HhaI, and HaeIII; Gibco, BRL) demonstrated that a combination of HaeIII and HhaI yielded a higher number of terminal restriction fragments (T-RFs) than other enzymes. Aliquots (1 μl) were mixed with 0.16 μl of deionized formamide, 0.83 μl of loading buffer (Perkin-Elmer), and 0.3 μl of DNA fragment length standard (Genescan 500 Rox; Perkin-Elmer). The reaction mixtures were denatured at 92°C for 2 min and chilled on ice prior to electrophoresis. Samples (1.5 μl) were run on 5% denaturing polyacrylamide gels, and the fluorescently labeled terminal restriction sizes were analyzed with an ABI 373A automated DNA sequencer (PE Applied Biosystems, Inc., Foster City, Calif.). The lengths of the labeled fragments were determined by comparison with the internal standard.
Analysis of T-RF profiles.
T-RF peaks between 35 and 500 bp and peak heights of ≤50 fluorescence units were included in the analysis according to the range of the size marker. Generally, the error for determining fragment sizes with our automated DNA sequencer was less than 1 bp; however, in some cases, a higher variation was found. Therefore, T-RFs that differed by less than 1.5 bp were clustered, unless individual peaks were detected in a reproducible manner. Three replicate samples of all treatments and particle sizes either were analyzed individually, or a representative sample profile was determined in a similar way as suggested by Dunbar et al. (12). Essentially, the sum of peak heights in each replicate profile was calculated, indicating the total DNA quantity. Total fluorescence was adjusted to the medium DNA quantity by calculating a correction factor. For example, three replicate profiles had total fluorescence values of 4,500, 4,700, and 4,900, and then each peak in the latter profile was multiplied with a factor of 0.96 (i.e., the quotient of 4,700/4,900), and peaks in the first profile were multiplied with a factor of 1.04 (i.e., a quotient of 4,700/4,500). After adjustment, only peaks of ≥50 fluorescence units were considered. In addition, T-RFs were scored as positive only when they were present in at least two of three replicates.
In order to determine similarities between T-RFLP profiles, a binary matrix that recorded the absence and presence of aligned fragments was generated. Estimation of genetic distances as suggested by Nei and Li (43) in combination with the UPGMA (unweighted pair group with mathematical averages) method was used to compare T-RFLP fingerprints. For analysis and tree generation (based on 100 bootstrap replications), the TREECON software package (56) was applied.
Small-subunit rDNA clone libraries.
DNA isolated from the clay fraction of the AM treatment was used to create a 16S ribosomal DNA (rDNA) clone library. 16S rRNA genes were amplified by PCR with the primer 8f (59) under the PCR conditions (59) described above. PCR amplicons approximately 1,500 bp long were ligated into the pGEM-T plasmid vector (Promega) and transformed into Escherichia coli DH5α competent cells. After overnight transformation, single-cell colonies were transferred into 1.5-ml Eppendorf tubes containing 80 μl of TE buffer. Tubes were heated for 10 min at 95°C to lyse cells, chilled on ice, and centrifuged for 2 min at 13,000 rpm. Supernatants were used in subsequent PCRs.
Sequencing of PCR products from cloned inserts.
Insert sequences were amplified by PCR, applying a denaturation step of 5 min at 95°C followed by 30 cycles of 30 s at 95°C, 1 min of annealing at 50°C, and 2 min of extension at 72°C. The PCR mixtures (50 μl) contained 1× reaction buffer (Gibco, BRL); 200 μM (each) dATP, dCTP, dGTP, and dTTP; 0.15 μM (each) primers M13for and M13rev; 2.5 U of Taq DNA polymerase (Gibco, BRL); and 1 μl of supernatant obtained after lysis of transformants. PCR products were purified with the NucleoTraPCR kit (Macheroy-Nagel) according to the manufacturer's instructions and used as a template in sequencing reactions. DNA sequencing was performed with an ABI 373A automated DNA sequencer (PE Applied Biosystems, Inc., Foster City, Calif.) and the ABI PRISM Big Dye terminator cycle sequencing kit (Perkin-Elmer).
Phylogenetic analysis.
Sequences were subjected to a BLAST analysis (1) with the National Center for Biotechnology Information (NCBI) database and were compared with sequences available in the Ribosomal Database Project (RDP) (41). Alignments with related sequences were done with the Multalin alignment tool available on the web (http://www.toulouse.inra.fr/multalin.html) (9). The TREECON software package (56) was used to calculate distance matrices by the Jukes and Cantor algorthim (28) and to generate phylogenetic trees according to nearest-neighbor criteria.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the NCBI database under accession no. AF388312 to AF388362.
RESULTS
T-RFLP analysis.
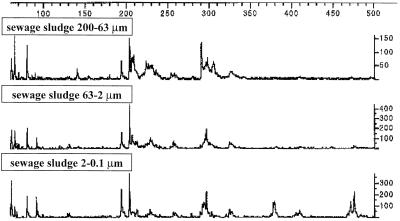
Bacterial population structures in different particle size fractions of a long-term field experiment receiving different fertilizer amendments were determined by T-RFLP analysis (Fig. 1). Replicates showed a certain degree of variation; however, they grouped together when all profiles were compared. A total of 55 T-RFs were detected, and because some fragments were only present in individual replicates, the total number of T-RFs found in the representative sample profiles decreased to 43. The number of T-RFs in individual sample profiles ranged from 7 (fallow and peat, sand fraction) to 26 (AM, clay fraction) (Table 1). In all treatments, the bacterial diversity was lowest in the sand fractions and highest in the clay fractions. The only exception was the treatment with peat as an organic amendment, in which the highest number of T-RFs was found in the silt fraction (Table 1). Treatments involving amendments of GM and AM showed higher diversities, particularly in clay size fractions, than treatments without organic amendments. Additionally, the sand fraction of the GM treatment exhibited an unusually high diversity compared to the sand fractions of other treatments. Some fragments present in several treatments showed far greater fluorescence signals in samples of the GM treatment.
FIG. 1.
16S rDNA-based HaeIII-HhaI T-RFLP fingerprint patterns obtained with three different particle size fractions of soils amended with sewage sludge.
TABLE 1.
Particle size distribution, Corg C/N ratio, and the total number of T-RFs in the Ultuna long-term experiment sampled in 1998a
| Particle size (μm) | Treatmentb | Particle size distribution (g kg of soil−1) | Corg concn (g of Corg kg of bulk soil−1) | C/N ratioc | No. of T-RFs |
|---|---|---|---|---|---|
| 200–63 | Fallow | 170 c | 0.3 a | 15.3 | 7 |
| NoN | 169 c | 0.35 a | 15.8 | 10 | |
| Ca(NO3)2 | 166 abc | 0.48 a | 9.4 | 8 | |
| GM | 165 bc | 0.75 b | 15.2 | 15 | |
| AM | 157 ab | 0.77 b | 5.0 | 9 | |
| SS | 156 a | 1.70 b | 20.1 | 10 | |
| Peat | 165 bc | 2.07 d | 37.5 | 7 | |
| 63–2 | Fallow | 499 a | 5.65 a | 11.3 | 14 |
| NoN | 500 ab | 6.75 ab | 11.1 | 10 | |
| Ca(NO3)2 | 518 ab | 8.44 ab | 11.9 | 19 | |
| GM | 516 ab | 10.49 bc | 13.7 | 18 | |
| AM | 519 b | 12.76 c | 10.0 | 16 | |
| SS | 562 c | 18.86 d | 10.4 | 15 | |
| Peat | 54 c | 21.77 d | 24.0 | 19 | |
| 2–0.1 | Fallow | 251 bc | 3.88 a | 7.8 | 23 |
| NoN | 254 c | 4.19 a | 7.6 | 22 | |
| Ca(NO3)2 | 247 bc | 4.75 b | 7.8 | 22 | |
| GM | 251 bc | 5.40 c | 8.0 | 24 | |
| AM | 248 bc | 6.16 d | 8.2 | 25 | |
| SS | 223 a | 5.90 d | 8.2 | 20 | |
| Peat | 234 b | 6.26 d | 10.0 | 17 |
Means within one column that are not significantly different from each other at P = 0.05 are followed by the same letters.
The pH of bulk soil samples was 6.6 in 1956 and decreased distinctly in the peat- and SS-treated plots. The pH values measured were 6.3, 6.5, 6.9, 6.3, 6.7, 5.8, and 5.8 for fallow, NoN, Ca(NO3)2, GM, AM, SS, and peat, respectively. This effect has already been discussed by Kirchmann et al. (36).
C/N values are from G. Haberhauer, M. H. Gerzabek, and H. Kirchmann, personal communication.
Most T-RFs that were detected in sand particles showed intensive fluorescence signals and comprised fragment sizes of 39, 60, 62, 66, 81, 194, 197, 205, 208, 212, 223, 228, 231, 253, 290, 294, 297, 304, and 379 bp (Table 2 and Fig. 1). Only three T-RFs with sizes of 197 bp (GM), 253 bp (GM and AM), and 290 bp (GM, AM, and SS) were found exclusively in the sand fraction. Most T-RFs detected in the sand fraction colonized silt and clay particles equally well (Table 2). A large number of T-RFs was detected in silt and clay fractions but not in sand particles (Table 2). Bacteria with T-RF sizes of 76 and 311 bp (both fallow) colonized only silt fractions, and various T-RFs were found exclusively in clay particles. These included sizes of 140 bp (GM, AM, and SS), 233 bp (fallow, NoN), 242 bp [fallow, NoN, Ca(NO3)2, GM, and AM], 284 bp (GM and AM), 332 bp (NoN,GM, AM, and peat), 404 bp (peat), 409 bp (SS), and 476 bp (SS). Few treatment-specific T-RFs were found. The treatment with SS as fertilizer showed a strong 205-bp peak in all particle sizes, but lacked fragments of 101, 220, 262, and 329 bp that were found in all other treatments. Furthermore, T-RFs of 95, 236, and 242 bp were detected in neither the peat treatment nor the SS treatment, despite their abundance in other treatments (Table 2). T-RFs with sizes of 228, 231, and 294 bp were found in several treatments, but were enriched when GM was applied as fertilizer.
TABLE 2.
Representative sample T-RFLP profiles of three particle size fractions obtained from soils that have received different fertilizer applicationsa
| bp | Fallow
|
NoN
|
Ca-nitrate
|
GM
|
AM
|
SS
|
Peat
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| s | si | c | s | si | c | s | si | c | s | si | c | s | si | c | s | si | c | s | si | c | |
| 39 | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ |
| 60 | ■ | □ | □ | □ | □ | □ | ■ | □ | ■ | □ | ■ | □ | □ | ▴ | □ | ■ | ■ | □ | |||
| 62 | ■ | ▴ | ■ | □ | ▴ | ■ | ▴ | ■ | ▴ | □ | ▴ | ▴ | ▴ | ▴ | |||||||
| 66 | ▴ | □ | □ | ■ | □ | ▴ | □ | ▴ | ▴ | ▴ | □ | ■ | ■ | ■ | ■ | □ | ■ | ■ | |||
| 69 | □ | □ | □ | □ | ■ | ||||||||||||||||
| 76 | □ | ||||||||||||||||||||
| 81 | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ■ | ▴ | ■ | ■ | ▴ | ▴ | ■ | ▴ |
| 92 | □ | □ | ■ | □ | □ | ||||||||||||||||
| 95 | ▴ | ▴ | □ | ■ | ▴ | ||||||||||||||||
| 101 | □ | □ | □ | ▴ | □ | □ | ■ | □ | □ | ■ | □ | ||||||||||
| 140 | □ | □ | □ | ||||||||||||||||||
| 194 | ▴ | ▴ | ▴ | ▴ | ▴ | ■ | ▴ | ▴ | ▴ | ▴ | ▴ | ▴ | ■ | ■ | ▴ | □ | ■ | ▴ | ▴ | ▴ | □ |
| 197 | □ | ||||||||||||||||||||
| 205 | □ | ▴ | ▴ | ▴ | |||||||||||||||||
| 208 | ■ | □ | ▴ | ■ | ■ | ▴ | ▴ | ■ | □ | ■ | ▴ | □ | ■ | ■ | ■ | ■ | ▴ | □ | ▴ | ||
| 212 | □ | □ | □ | □ | □ | □ | ■ | ||||||||||||||
| 216 | ▴ | □ | ▴ | ||||||||||||||||||
| 220 | □ | ■ | □ | □ | □ | ■ | ▴ | ■ | ▴ | □ | □ | ||||||||||
| 223 | □ | □ | □ | □ | □ | □ | |||||||||||||||
| 228 | ■ | ▴ | ■ | □ | ■ | □ | □ | ▴ | □ | ▴ | ■ | ▴ | ■ | ■ | □ | □ | ■ | □ | |||
| 231 | □ | ▴ | ▴ | □ | ▴ | □ | ▴ | ▴ | ▴ | ▴ | ▴ | ■ | ▴ | ■ | □ | □ | |||||
| 233 | ■ | ■ | |||||||||||||||||||
| 236 | ■ | ■ | ■ | ■ | ■ | ■ | ▴ | ■ | ▴ | ||||||||||||
| 242 | □ | □ | □ | ▴ | □ | ||||||||||||||||
| 253 | □ | □ | |||||||||||||||||||
| 257 | □ | □ | □ | □ | □ | □ | |||||||||||||||
| 262 | □ | □ | □ | □ | □ | ■ | □ | □ | |||||||||||||
| 284 | □ | ||||||||||||||||||||
| 290 | □ | □ | ■ | ||||||||||||||||||
| 294 | □ | □ | ■ | □ | □ | □ | ■ | □ | ■ | ■ | ■ | □ | □ | ■ | ■ | ■ | ■ | □ | |||
| 297 | □ | □ | □ | □ | □ | ■ | □ | ■ | ■ | ▴ | □ | ||||||||||
| 301 | ■ | □ | □ | ■ | □ | ||||||||||||||||
| 304 | □ | ■ | |||||||||||||||||||
| 311 | □ | ||||||||||||||||||||
| 326 | □ | □ | □ | □ | □ | □ | □ | ||||||||||||||
| 329 | □ | □ | □ | □ | □ | ▴ | □ | ■ | □ | ||||||||||||
| 332 | □ | ■ | ■ | □ | |||||||||||||||||
| 379 | ■ | ■ | |||||||||||||||||||
| 404 | □ | ||||||||||||||||||||
| 409 | □ | ||||||||||||||||||||
| 413 | ▴ | □ | □ | ||||||||||||||||||
| 471 | □ | ■ | |||||||||||||||||||
| 476 | ■ | ||||||||||||||||||||
Particle size fractions: s, sand; si, silt; c, clay. □, 50 to 99 fluorescence units; ■, 100 to 149 fluorescence units; ▴, >149 fluorescence units.
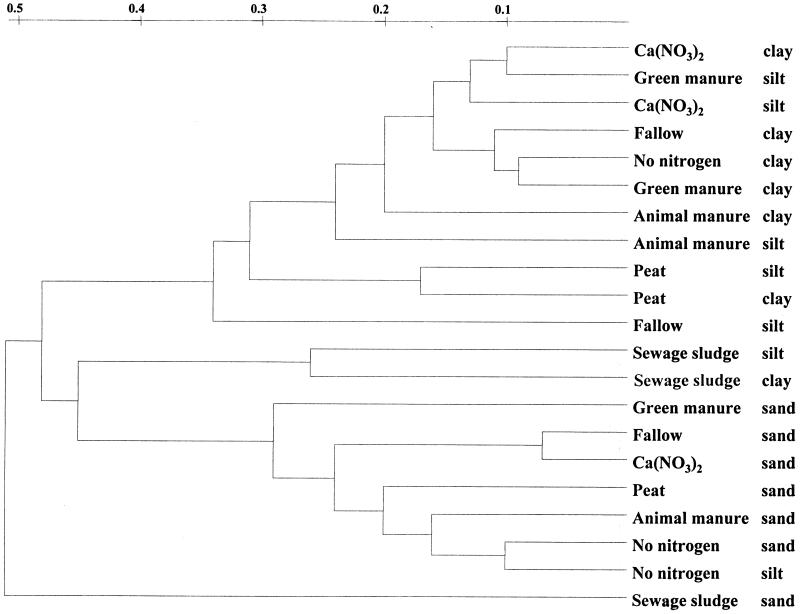
Cluster analysis revealed high similarity of bacterial populations located in the sand fractions of all treatments; only the community structure of the treatment that received SS was not represented in that cluster (Fig. 2). The cluster that comprised sand microbial communities also included the bacteria found in the silt fraction of the treatment without nitrogen application. The second cluster comprised bacterial communities inhabiting the silt and clay fractions. Bacterial communities of the SS treatment were quite distinct compared to other treatments and showed high similarities in the silt and clay fractions.
FIG. 2.
UPGMA dendrogram generated from all representative T-RFLP sample profiles. The scale indicates the distance level.
Sequence analysis and phylogenetic assignment.
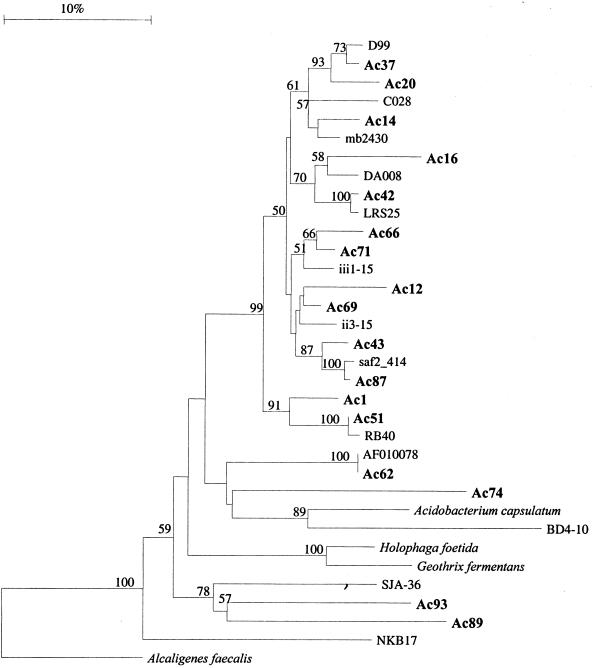
Partial sequence information was obtained for 51 16S rRNA genes covering approximately 500 bp derived from the clay size fraction of the AM treatment that showed highest diversity in the T-RFLP analysis. Twenty-four sequences showed high similarity to RDP database entries (similarity score [Sab] values > 0.8) (Table 3), and 30 sequences showed at least 95% similarity to known sequences deposited in the NCBI database. The remaining sequences were only moderately related (90 to 94% similarity) to known 16S rRNA genes and represented members of yet undescribed bacterial divisions, deeply branching members of described divisions, or chimeric sequences. Phylogenetic analysis revealed that clones were not evenly distributed among different bacterial divisions. Bacteria belonging to the Holophaga/Acidobacterium division accounted for 37% of the clones examined, whereas 27% could be classified as high-G+C or low-G+C gram-positives. The remaining clones belonged to the α-, γ-, and δ-Proteobacteria (18%), Verrucomicrobiales (12%), the Flexibacter/Cytophaga/Bacteroides division (4%), and Planctomycetales (2%). A phylogenetic tree comprising the three cultured members of the Holophaga/Acidobacterium cluster, sequences obtained in this study, and other environmental clones is depicted in Fig. 3. Because this tree is based only on partial sequences, it demonstrates the tremendous diversity of bacteria belonging to the Holophaga/Acidobacterium division rather than definitive phylogenetic relationships.
TABLE 3.
Phylogenetic assignment of clones with Sab values > 0.8 to RDP database entriesa
| Clone | Closest NCBI database matchb | % Similarity | Putative phylum | RDP placementc | T-RF length (bp) |
|---|---|---|---|---|---|
| Ac62 | Uncultured eubacterium (AF010078) | 99 | Holophaga/Acidobacterium | Unclassified | 92 |
| Ac74 | Agricultural soil bacterium SC-I-49 (AJ252638) | 98 | Holophaga/Acidobacterium | Unclassified | 95 |
| Ac20 | Uncultured eubacterium (AF010016) | 99 | Holophaga/Acidobacterium | Unclassified | 214 |
| Ac14 | Unclassified clone (Z95735) | 96 | Holophaga/Acidobacterium | Unclassified | 221 |
| Ac1 | Uncultured bacterium Riz 104 (AJ244314) | 97 | Holophaga/Acidobacterium | Unclassified | 222 |
| Ac69 | Unclassified clone (Z95725) | 94 | Holophaga/Acidobacterium | Unclassified | 222 |
| Ac92 | Unclassified clone (Z95728) | 97 | Holophaga/Acidobacterium | Unclassified | 222 |
| Ac42 | Uncultured eubacterium (AJ232842) | 97 | Holophaga/Acidobacterium | Unclassified | 222 |
| Ac51 | Clone RB40 (Z95721) | 98 | Holophaga/Acidobacterium | Environmental clone RB40 group | 222 |
| Ac71 | Agricultural soil bacterium SC-I-19 (AJ252622) | 96 | Holophaga/Acidobacterium | Environmental clone RB40 group | 222 |
| Ac87 | Grassland soil clone saf2 414 (AF078262) | 98 | Holophaga/Acidobacterium | Environmental clone RB40 group | 233 |
| Ac43 | Soil bacterium SC-I-19 (AJ252622) | 96 | Holophaga/Acidobacterium | Environmental clone RB40 group | 235 |
| Ac50 | Arthrobacter globiformis (M23411) | 97 | High-G+C gram-positives | Arthrobacter + relatives | 229 |
| Ac56 | Streptomycetaceae (X87316) | 96 | High-G+C gram-positives | Streptomyces + relatives | 76 |
| Ac52 | Clone Riz 1098 (AJ244354) | 98 | Low-G+C gram-positives | B. megaterium group | 232 |
| Ac11 | Bacillus benzoevorans (D78311) | 97 | Low-G+C gram-positives | B. megaterium group | 231 |
| Ac31 | Clostridium lituseburense (M59107) | 97 | Low-G+C gram-positives | C. lituseburense group | 290 |
| Ac36 | Uncultured copper smeltery bacterium D62 (AF337830) | 96 | α-Proteobacteria | Sphingomonas group | 88 |
| Ac48 | Metal-contaminated soil clone (AF145868) | 99 | α-Proteobacteria | Unclassified | 39 |
| Ac68 | Uncultured eubacterium (AF270944) | 97 | α-Proteobacteria | Rhizobium-Agrobacterium group | 195 |
| Ac38 | Rhizosphere clone RSC-II-42 (AJ252682) | 96 | γ-Proteobacteria | Xanthomonas group | 39 |
| Ac67 | Clone EA25 (U51864) | 99 | Verrucomicrobiales | Prosthecobacter group | 237 |
| Ac57 | Uncultured bacterium DA101 (Y07576) | 95 | Verrucomicrobiales | Prosthecobacter group | 237 |
Tentative phylogenetic placement and percent similarity values were determined by BLAST and are based on approximately 500 bp of the 16S rRNA gene sequence for each clone.
Accession numbers of closest database matches are given in parentheses.
The tentative phylogenetic placement was determined by using the Sequence Match option in the RDP.
FIG. 3.
Neighbor-joining phylogenetic tree based on 382 nucleotides of the 16S rRNA gene of clones showing highest similarity with bacteria belonging to the Holophaga/Acidobacterium division. Sequences obtained in this study are printed in boldface letters. The percentages of 100 bootstrap replicates are shown at the left nodes when at least 50%. The tree demonstrates the tremendous diversity of clones belonging to this group rather than definitive phylogenetic relationships. The accession numbers of the 16S rDNA sequences used are AF337850 (D99), AF013527 (C028), Z95735 (mb2430), Y12597 (DA008), AJ232842 (LRS25), Z95728 (iii1–15), Z95725 (ii3–15), AF078262 (saf2 414), Z95721 (RB40), AB015560 (BD4–10), AJ009461 (SJA-36), AB013269 (NKB17), D26171 (Acidobacterium capsulatum), X77215 (Holophaga foetida), and U41563 (Geothrix fermentans). AF155147 (Alcaligenes faecalis) was used as an outgroup.
All theoretical HaeIII-HhaI T-RFs of sequenced clones were found in community T-RFLP profiles. The only exception was a theoretical T-RF size of 132 bp that was not detected in the T-RFLP analysis. Several sequences showed identical T-RF sizes (Table 4). A relationship between phylogeny and location in soil was found. Some bacterial groups such as the Holophaga/Acidobacterium division and most Prosthecobacter members were mainly present in smaller size fractions (silt and clay), whereas Proteobacteria were evenly distributed in all fractions (Table 4).
TABLE 4.
Phylogenetic assignment and theoretical T-RF sizes of 51 16S rRNA gene sequences obtained from the clay fraction of the AM treatment as well as the location in soil of corresponding T-RFs in T-RFLP profiles
| Phylogenetic assignment | Theoretical T-RF size (bp) | Size of corresponding T-RF in T-RFLP (bp) | Location in soil |
|---|---|---|---|
| Holophaga/Acidobacterium | 37 | 39 | All fractions (may also derive from gram-positives and Proteobacteria) |
| 76a | 76 | Silt and clay (may also derive from gram-positives) | |
| 92 | 92 | Silt and clay | |
| 96a | 96 | Silt and clay | |
| 206 | 205 | All fractions (may also derive from Proteobacteria) | |
| 214 | 216 | Silt and clay | |
| 220 | 220 | Silt and clay | |
| 222 | 223 | Mainly silt and clay (may also derive from Prosthecobacter) | |
| 233a | 233 | Clay | |
| 234 | 236 | Silt and clay | |
| 235 | 236 | Silt and clay (may also derive from Prosthecobacter or Flexibacter/Cytophaga/Bacteroides) | |
| 242 | 242 | Clay | |
| Verrucomicrobiales (Prosthecobacter) | 100 | 101 | Silt and clay |
| 208 | 208 | All fractions | |
| 222 | 228 | Mainly silt and clay (may also derive from Holophaga/ Acidobacterium) | |
| 225b | |||
| 236 | 236 | Silt and clay (may also derive from Holophaga/Acidobacterium) | |
| 237 | 236 | Silt and clay | |
| Gram-positives | |||
| High G+C | 37 | 39 | All fractions (may also derive from Holophaga/Acidobacterium or Proteobacteria) |
| 38 | 39 | All fractions (may also derive from Holophaga/Acidobacterium or Proteobacteria) | |
| 63 | 62 | Mainly silt and clay | |
| 76a | 76 | Silt and clay (may also derive from Holophaga/Acidobacterium) | |
| 132c | |||
| 210a | |||
| 229 | 228 | All fractions | |
| Low G+C | 88a | ||
| 231 | 231 | All fractions | |
| 232 | 231 | All fractions | |
| 240 | 242 | Clay | |
| 284 | 284 | Clay | |
| 290d | 290 | Mainly sand | |
| Proteobacteria | |||
| α | 39 | 39 | All fractions (may also derive from Holophaga/Acidobacterium or gram-positives) |
| 64 | 66 | All fractions | |
| 83 | 81 | All fractions | |
| 195 | 194 | All fractions | |
| γ | 39 | 39 | All fractions (may also derive from Holophaga/Acidobacterium or gram-positives) |
| 69b | 69 | Silt and clay | |
| δ | 211a | 212 | All fractions |
| Unclassified | 206 | 205 | All fractions (may also derive from Holophaga/Acidobacterium) |
| Planctomycetes | 230 | 231 | All fractions |
| Flexibacter/Cytophaga/ Bacteroides | 213a | 212 | All fractions |
| 235 | 236 | Silt and clay (may also derive from Holophaga/Acidobacterium and Prosthecobacter) |
Was detected in the T-RFLP profile of the AM clay fraction, but was lost during the normalization procedure.
Was not found in the T-RFLP profile of the AM clay fraction.
Was not detected in any T-RFLP profile.
DISCUSSION
Association of bacteria with soil particles.
Several studies reported a higher microbial biomass in smaller size fractions (27, 30, 32, 58). Our results clearly demonstrated that not only biomass, but also bacterial community structure, was significantly affected by particle size and that smaller size fractions host higher diversities of microbes than larger size particles. The higher biomass in silt and clay fractions could be attributed to a higher diversity rather than to better colonization by particular species. This was demonstrated by comparable fluorescence intensities of several T-RF peaks in all fractions.
It has been suggested that finer size particles provide a protective habitat for microorganisms through pore size exclusion of predators (protozoa) (13, 48). Recently, it was demonstrated that the predation regimen could act as a major structuring force for the bacterial community composition in an aquatic system (29). In that study, bacteria belonging to the α and γ subdivisions of the Proteobacteria proved to be grazing resistant, mainly due to the formation of 3- to 6-μm rods, which probably exceeded the size of bacteria edible by protozoa. In addition, filamentous bacteria belonging to the β-Proteobacteria and to the Cytophaga-Flavobacterium cluster proved to resist grazing. Furthermore, partial protection of Rhizobium leguminosarum from protozoan grazing in soil due to the addition of bentonite clay was observed (26). In our experiment, microfaunal predation may have represented a selective pressure on the community structure in sand fractions. Because the most abundant T-RFs (39, 81, and 194 bp) were derived mainly from α-Proteobacteria, we assume that resistance to grazing may explain the great abundance of some bacterial groups in sand particles.
Alternatively, higher nutrient availability in smaller size particles may have caused higher bacterial diversities. According to van Gestel et al. (58), the vicinity between microbes, organic matter, and clay is required for the survival of microbes, in which organic matter and clay particles provide substrates and nutrients. The enrichment of organic matter and microbial biomass in finer fractions seems to be a consequence of aggregate formation along with the decomposition of particulate organic matter (36). Therefore, T-RFs found in the sand fraction may represent bacterial species being better adapted to limited nutrient conditions or possessing the capacity to utilize a wider range of substrates. They may be able to degrade high-molecular-weight organic molecules derived from the initial breakdown of plant material. Phylogenetic assignment of sequenced 16S rRNA genes indicated a great abundance of α-Proteobacteria, such as Sphingomonas (T-RF of 81 bp), and bacteria belonging to the Rhizobium-Agrobacterium group (T-RF of 195 bp). Some members of the α-Proteobacteria utilize a wide range of substrates, and the genus Sphingomonas is particularly known for its ability to degrade aromatic compounds (16, 33). Because sand size particles seem to be preferentially colonized by fungi (32), many bacteria were probably outcompeted by eukaryotic organisms in this fraction. It is further assumed that community composition was affected by the oxygen concentration. Sequence analysis indicated the presence of aerobic bacteria, as well as strict anaerobes such as clostridia, in clay particles, suggesting that particles with sizes smaller than 2 μm provided a niche for aerobes as well as for anaerobes, whereas larger particle sizes were dominated by aerobic microbes. Recently, the development of biofilms in soils consisting of a dense lawn of clay aggregates containing one or more bacteria, phyllosilicates, and grains of iron oxides was observed (40). Clay particles were held together by an extracellular polysaccharide matrix and were arranged as hutches that served as housing for microbes. These “clay hutches” have been proposed to represent a minimal nutritional sphere for autochthonous bacteria and may at least partly explain the higher microbial diversity in clay size particles.
The Holophaga/Acidobacterium division was the most abundant and diverse group among analyzed clones obtained from the clay fraction of the AM treatment. This bacterial group was also dominant in other soils (4, 10, 39). In this study, members of the Holophaga/Acidobacterium division mainly colonized silt and clay fractions. Because this phylum includes Holophaga foetida, Geothrix fermetans, and Acidobacterium capsulatum as the only cultivated species so far, very little information on the role of these microbes in natural environments is available. In addition, members of the Verrucomicrobiales, showing highest similarity to the genus Prosthecobacter, were found mainly in silt and clay fractions. One characteristic of this genus is the formation of prosthecae, which consist of narrow, cytoplasm-containing extensions of the cell wall. They confer several advantages to aerobic heterotrophic bacteria, such as enhanced respiration and nutrient uptake, as well as improved attachment to solid substrates (25). Furthermore, the known members of the genus Prosthecobacter utilize only a narrow range of carbohydrates as their sole carbon source (25), which may explain their location in smaller size fractions.
Response of bacteria in particle size fractions to organic amendments.
In previous studies, the effects of the different treatments in the long-term field experiment in Ultuna were shown to vary significantly with respect to organic matter turnover (19, 21). The specific layout of the experiment, relating manure applications to equal amounts of organic carbon, allows a direct comparison of treatments with respect to breakdown of organic matter by soil organisms. Referring to carbon turnover, treatments could be ranked by decreasing digestibility: GM > AM > SS > peat. Particularly the latter two treatments showed a considerable accumulation of organic carbon (Corg). For peat, it was demonstrated that turnover is slow, and therefore Corg derived from peat has accumulated in soil. This finding was supported by the large amounts of Corg derived from peat in the silt fraction compared to those in clay size particles (21). Our results of the microbial population analysis reflect what has been reported for Corg turnover. In general, diversities were highest in soils treated with GM and AM and lowest in soils that received no or inorganic fertilizers and thus corresponded to the digestibility of the fertilizer applied. Amendments with SS and peat as fertilizer resulted in quite unique bacterial population structures that were due to different compositions rather than to increased or decreased diversities. Both treatments exhibited very low pH values (pH 5.8), which were probably responsible for some community changes. Soils amended with peat were also characterized by a high C/N ratio. Some T-RFs were not detected in soils amended with SS and peat despite their presence in other treatments. They included members of the Holophaga/Acidobacterium division (95, 236, and 242 bp), Prosthecobacter (236 bp), or low G+C gram-positives (242 bp). These bacteria may not have been able to withstand the acidic environment. Other bacteria belonging to the Holophaga/Acidobacterium division and the genus Prosthecobacter were able to colonize most treatments, except soils containing SS. The sensitivity of some microbial groups to the generally high heavy metal content of SS may explain this finding. The high abundance of a bacterial group with a T-RF of 205 bp in SS-treated soils was striking. Two clones with a matching T-RF were found: one fell into the Holophaga/Acidobacterium division, whereas the other showed the highest similarity to Proteobacteria. The high diversity of bacteria in sand size particles of soils amended with GM may have been responsible for the fast turnover of organic matter. In addition, a particularly high abundance of some gram-positive bacterial species, including Arthrobacter globiformis and Bacillus species, was found in the GM treatment.
Conclusions.
Community analysis by T-RFLP of 16S rRNA genes proved to be a highly suitable and sensitive tool with which to investigate the microbial community structures in different particle size fractions and treatments. Three normalized replicate samples of each fraction and treatment showed comparable population profiles and grouped well in a cluster analysis. Quantitative T-RFLP analysis has to be treated with caution due to biases inherent to PCR amplification (53) and variations in the copy number of the 16S rRNA gene in different bacterial species (8). However, with awareness of these limitations, T-RFLP analysis can be used for a semiquantitative analysis of bacterial community structures (12, 47). In this experiment, the microbial composition was mainly affected by the particle size fraction and did respond to a lesser extent to organic amendments. Therefore, our results demonstrate specific microbe-particle associations that are affected to a smaller extent by external factors such as fertilization or heavy metal pollution (32). Knowledge of the microbial community structure represents a first step toward understanding soil function in response to the environment. In addition to community structure, the analysis of functional genes within a given population will greatly increase our comprehension of the role of bacteria in soil processes important for geochemical dynamics of elements, specifically carbon, nitrogen, and sulfur.
ACKNOWLEDGMENTS
This work represents a complementary study to a project that was funded by the Austrian Science Foundation (Fonds zur Förderung der wissenschaftlichen Forschung). A. Sessitsch received an APART fellowship funded by the Austrian Academy of Sciences.
We thank Brigitta Temmel for preparing the particle size fractions. We are grateful to Levente Bodrossy for critically reading the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amato M, Ladd J N. Decomposition of 14C-labelled glucose and legume material in soils: properties influencing the accumulation of organic residue C and microbial biomass C. Soil Biol Biochem. 1992;24:455–466. [Google Scholar]
- 4.Borneman J, Skroch P W, O'Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brussaard L, Bouwman L A, Geurs M, Hassink J, Zwart K B. Biomass composition and temporal dynamics of soil organisms of a silt loam soil under conventional and integrated management. Neth J Agric Sci. 1990;38:283–302. [Google Scholar]
- 6.Christensen B T. Physical fractionation of soil and organic matter in primary particle size and density separates. Adv Soil Sci. 1992;20:1–90. [Google Scholar]
- 7.Clement B G, Kehl L E, DeBord K L, Kitts C L. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods. 1998;31:135–142. [Google Scholar]
- 8.Cole S T, Girons I S. Bacterial genomics. FEMS Microbiol Rev. 1994;14:139–160. doi: 10.1111/j.1574-6976.1994.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 9.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunbar J, Takala S, Barns S M, Davis J A, Kuske C R. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol. 1999;65:1662–1669. doi: 10.1128/aem.65.4.1662-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunbar J, Ticknor L O, Kuske C R. Assessment of microbial diversity in four Southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl Environ Microbiol. 2000;66:2943–2950. doi: 10.1128/aem.66.7.2943-2950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar J, Ticknor L O, Kuske C R. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol. 2001;67:190–197. doi: 10.1128/AEM.67.1.190-197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliot E T, Anderson R V, Coleman D C, Cole C V. Habitable pore space and microbial trophic interactions. Oikos. 1980;35:327–335. [Google Scholar]
- 14.Felske A, Wolterink A, van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster R C. Microenvironments of soil microorganisms. Biol Fertil Soils. 1988;6:189–203. [Google Scholar]
- 16.Fredrickson J K, Balkwill D L, Drake G R, Romine M F, Ringelberg D B, White D C. Aromatic-degrading Sphingomonas isolates from the deep subsurface. Appl Environ Microbiol. 1995;61:1917–1922. doi: 10.1128/aem.61.5.1917-1922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelsomino A, Keijzer-Wolters A C, Cacco G, van Elsas J D. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J Microbiol Methods. 1999;38:1–15. doi: 10.1016/s0167-7012(99)00054-8. [DOI] [PubMed] [Google Scholar]
- 18.Gerzabek M H, Kirchmann H, Pichlmayer F. Response of soil aggregate stability to manure amendments in a long-term experiment in Ultuna, Sweden. Z Pflanzenernsaehr Bodenkd. 1995;158:257–260. [Google Scholar]
- 19.Gerzabek M H, Pichlmayer F, Kirchmann H, Haberhauer G. The response of soil organic matter to manure amendments in a long-term experiment in Ultuna, Sweden. Eur J Soil Sci. 1997;48:273–282. [Google Scholar]
- 20.Gerzabek M H, Kirchmann H, Haberhauer G, Pichelmayer F. The response of soil nitrogen and 15N natural abundance to different amendments in a long-term experiment at Ultuna, Sweden. Agronomie Agric Environ. 1999;19:457–466. [Google Scholar]
- 21.Gerzabek, M. H., G. Haberhauer, E. Kandeler, A. Sessitsch, and H. Kirchmann. Response of organic matter pools and enzyme activities in particle size fractions to organic amendments in a long-term field experiment. In Proceedings of the Third Symposium ISMOM, Naples-Capri, Italy, in press.
- 22.Hassink J, Bouman L A, Zwart K B, Bloem J, Brussaard L. Relationships between soil texture, physical protection of organic matter, soil biota, and C and N mineralization in grassland soils. Geoderma. 1993;57:105–128. [Google Scholar]
- 23.Hassink J, Bouwman L A, Zwart K B, Brussaard L. Relationships between habitable pore space, soil biota and mineralization rates in grassland soils. Soil Biol Biochem. 1993;25:47–55. [Google Scholar]
- 24.Hattori T. Soil aggregates as habitats of microorganisms. Rep Inst Agric Res Tohuku Univ. 1988;37:23–36. [Google Scholar]
- 25.Hedlund B P, Gosink J J, Staley J T. Phylogeny of Prosthecobacter, the fusiform caulobacters: members of a recently discovered division of the Bacteria. Int J Syst Bacteriol. 1996;46:960–966. doi: 10.1099/00207713-46-4-960. [DOI] [PubMed] [Google Scholar]
- 26.Heynen C E, van Elsas J D, Kuikman P J, van Veen J A. Dynamics of Rhizobium leguminosarum biovar trifolii introduced into soil: the effect of bentonite clay on predation by protozoa. Soil Biol Biochem. 1988;20:483–488. [Google Scholar]
- 27.Jocteur Monrozier L, Ladd J N, Fitzpatrick A W, Foster R C, Raupach M. Components and microbial biomass content of size fractions in soils of contrasting aggregation. Geoderma. 1991;49:37–62. [Google Scholar]
- 28.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H H, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 29.Jürgens K, Pernthaler J, Schalla S, Amann R. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl Environ Microbiol. 1999;65:1241–1250. doi: 10.1128/aem.65.3.1241-1250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanazawa S, Filip Z. Distribution of microorganisms, total biomass, and enzyme activities in different particles of brown soil. Microb Ecol. 1986;12:205–215. doi: 10.1007/BF02011205. [DOI] [PubMed] [Google Scholar]
- 31.Kandeler E, Stemmer M, Klimanek E-M. Response of soil microbial biomass, urease and xylanase within particle-size fractions to long-term soil management. Soil Biol Biochem. 1999;31:261–273. [Google Scholar]
- 32.Kandeler E, Tscherko D, Bruce K D, Stemmer M, Hobbs P J, Bardgett R D, Amelung W. Structure and function of the soil microbial community in microhabitats of a heavy metal polluted soil. Soil Biol Biochem. 2000;32:390–400. [Google Scholar]
- 33.Kim E, Aversano P J, Romine M F, Schneider R P, Zylstra G J. Homology between genes for aromatic hydrocarbon degradation in surface and deep-subsurface Sphingomonas strains. Appl Environ Microbiol. 1996;62:1467–1470. doi: 10.1128/aem.62.4.1467-1470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirchmann H, Persson J, Carlgren K. The Ultuna long-term soil organic matter experiment, 1956–1991. Reports and dissertation 17. Uppsala, Sweden: Department of Soil Sciences, Swedish University of Agricultural Sciences; 1994. [Google Scholar]
- 35.Kirchmann H, Pichlmayer F, Gerzabek M H. Sulfur balances and sulfur-34 abundance in a long-term fertilizer experiment. Soil Sci Soc Am J. 1996;60:174–178. [Google Scholar]
- 36.Kirchmann H, Gerzabek M H. Relationship between soil organic matter and micropores in a long-term experiment at Ultuna, Sweden. J Plant Nutr Soil Sci. 1999;162:493–498. [Google Scholar]
- 37.Ladd J N, Foster R C, Nannipieri P, Oades J M. Soil structure and biological activity. Soil Biochem. 1996;9:23–78. [Google Scholar]
- 38.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction length polymorphisms of genes encoding 16S rDNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 40.Lünsdorf H, Erb R W, Abraham W-R, Timmis K N. “Clay hutches”: a novel interaction between bacteria and clay minerals. Environ Microbiol. 2000;2:161–168. doi: 10.1046/j.1462-2920.2000.00086.x. [DOI] [PubMed] [Google Scholar]
- 41.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marsh T L, Saxman P, Cole J, Tiedje J. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl Environ Microbiol. 2000;66:3616–3620. doi: 10.1128/aem.66.8.3616-3620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nüsslein K, Tiedje J M. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl Environ Microbiol. 1998;64:1283–1289. doi: 10.1128/aem.64.4.1283-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nüsslein K, Tiedje J M. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl Environ Microbiol. 1999;65:3622–3626. doi: 10.1128/aem.65.8.3622-3626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oades J M. Soil organic matter and structural stability: mechanisms and implications for management. Plant Soil. 1984;76:319–337. [Google Scholar]
- 47.Osborn A M, Moore E R B, Timmis K N. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 48.Postma J, van Veen J A. Habitable pore space and survival of Rhizobium leguminosarum biovar trifolii introduced into soil. Microb Ecol. 1990;19:149–161. doi: 10.1007/BF02012096. [DOI] [PubMed] [Google Scholar]
- 49.Ramírez-Saad H C, Sessitsch A, de Vos W M, Akkermans A D L. Bacterial community changes and enrichment of Burkholderia-like bacteria induced by chlorinated benzoates in a peat-forest soil-microcosm. Syst Appl Microbiol. 2000;23:591–598. doi: 10.1016/S0723-2020(00)80035-1. [DOI] [PubMed] [Google Scholar]
- 50.Scala D J, Kerkhof L J. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl Environ Microbiol. 2000;66:1980–1986. doi: 10.1128/aem.66.5.1980-1986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stemmer M, Gerzabek M H, Kandeler E. Organic matter and enzyme activity in particle size fractions of soils obtained after low-energy sonication. Soil Biol Biochem. 1998;30:9–17. [Google Scholar]
- 52.Stemmer M, Gerzabek M H, Kandeler E. Invertase and xylanase activity of bulk soil and particle-size fractions during maize straw decomposition. Soil Biol Biochem. 1999;31:9–18. [Google Scholar]
- 53.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tisdall J M, Oades J M. Organic matter and water stable aggregates in soils. J Soil Sci. 1982;33:141–163. [Google Scholar]
- 55.Torsvik V, Goksøyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van de Peer Y, de Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 57.van Elsas J D, Smalla K. Extraction of microbial community DNA from soils. In: de Bruijn F J, van Elsas J D, Akkermans A D L, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–11. [Google Scholar]
- 58.van Gestel M, Merckx R, Vlassek K. Spatial distribution of microbial biomass in microaggregates of a silty-loam soil and the relation with the resistance of microorganisms to soil drying. Soil Biol Biochem. 1996;28:503–510. [Google Scholar]
- 59.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]