Abstract
Purpose
The purpose of this study was to identify genetic risk loci for retinal traits, including drusen, in an Amish study population and compare these risk loci to known risk loci of age-related macular degeneration (AMD).
Methods
Participants were recruited from Amish communities in Ohio, Indiana, and Pennsylvania. Each participant underwent a basic health history, ophthalmologic examination, and genotyping. A genomewide association analysis (GWAS) was conducted for the presence and quantity of each of three retinal traits: geographic atrophy, drusen area, and drusen volume. The findings were compared to results from a prior large GWAS of predominantly European-ancestry individuals. Further, a genetic risk score for AMD was used to predict the presence and quantity of the retinal traits.
Results
After quality control, 1074 participants were included in analyses. Six single nucleotide polymorphisms (SNPs) met criteria for genomewide significance and 48 were suggestively associated across three retinal traits. The significant SNPs were not highly correlated with known risk SNPs for AMD. A genetic risk score for AMD provided significant predictive value of the retinal traits.
Conclusions
We identified potential novel genetic risk loci for AMD in a midwestern Amish study population. Additionally, we determined that there is a clear link between the genetic risk of AMD and drusen. Further study, including longitudinal data collection, may improve our ability to define this connection and improve understanding of the biological risk factors underlying drusen development.
Keywords: drusen, age-related macular degeneration (AMD), genetics
Age-related macular degeneration (AMD) is the leading cause of vision loss nationwide and accounts for 8.7% of blindness.1 Although the prevalence of AMD differs based on ancestry, it is estimated to affect approximately 196 million individuals globally and is projected to increase to 288 million in 2040.2 Early stages of AMD may be relatively asymptomatic, but as the disease progresses there can be blurring and distortion of central vision that can eventually result in irreversible central vision loss.3 Late stages of AMD are generally categorized into two subtypes: neovascular AMD (or macular neovascularization4 [MNV]) and geographic atrophy (GA).5 In people with MNV, neovascularization arising from either the choroidal circulation (type 1 or 2 MNV) or the deep retinal circulation (type 3 MNV) is associated with exudation which can result in scarring and loss of the retinal photoreceptors.1 MNV accounts for over half of late-stage AMD cases and approximately 90% of blindness due to AMD.6 GA, characterized by progressive degeneration of the macula, can also cause vision loss but this generally occurs more gradually compared with MNV.1
Although visual impairment in early stages of AMD usually does not have a large impact on quality of life, late-stage AMD can lead to significant functional loss, such as the inability to read, drive, or work. Additionally, vision loss has been linked to many comorbidities, including depression, stroke and heart disease, and coronary artery disease.7,8 Because of this, it is imperative to identify individuals with AMD in early stages of the disease. Current treatments cannot completely reverse an individual's progression of AMD, but treatments, including antivascular endothelial growth factor, can slow or halt the progression of neovascular AMD.9–11 Similarly, there are preventive measures, including modifications to diet, nutritional supplements, and increased physical activity, that help in reducing the risk and rate of progression.12,13
Risk factors for AMD have been characterized extensively.12,14,15 The primary risk factors for AMD include age, cigarette smoking, previous cataract surgery, and genetics. Twin studies have estimated the heritability of AMD to be between 46% and 71%, which is high for a complex disease.16,17 Early genetic studies of AMD identified two primary risk altering loci: CFH18–21 and ARMS2/HTRA1.22,23 Variation in these two genes accounts for over half of the genetic heritability of AMD.24,25 More recent genomewide association studies (GWASs) have revealed as many as 34 independent loci associated with AMD status across both common and rare variants.26
Although small cellular waste deposits between the retinal pigment epithelium (RPE) and Bruch's membrane (BM) known as drusen serve as biomarkers for AMD, recent work confirms their importance in understanding the physiology of AMD.27–29 Drusen are indicative of AMD progression and show potential in better understanding conversion to wet AMD.6,28,30 Larger drusen and a larger drusen area is associated with a higher risk for developing GA, with GA often appearing in regions where these large drusen collapse.31 Although the severity or burden of drusen in the eye were historically assessed using categorical scales on color photographs, advances in imaging, notably optical coherence tomography (OCT), has allowed the volume of drusen to be precisely quantified.
Despite extensive characterization of genetic risk of AMD, risk variants specific to drusen development have not been as intensively explored. Much of the work surrounding the genetic risk of drusen has investigated either certain genes or variants, as opposed to evaluation of risk across the entire human genome.32–34 Here, we performed a GWAS of drusen and GA traits to identify risk variants for both the presence and quantity of drusen. These data were collected as part of the Amish Eye Study, which aims to identify early predictors of progression to AMD by studying individuals at high risk of developing AMD.35,36 The Amish are a founder population that is isolated both genetically and culturally. The population descended from German and Swiss Anabaptists who emigrated from Western Europe to North America throughout the 18th and 19th centuries.37,38 Because of their cultural beliefs, the Amish generally abstain from both smoking and alcohol use after joining the church as adults, reducing the impact of these environmental risk factors. However, some Amish do engage in limited smoking as adolescents. Due to their relative isolation, the Amish provide a valuable population for genetic studies that may be enriched for some rare variation.39 We examined the link between AMD and drusen by investigating the utility of genetic risk scores (GRSs) for AMD to predict retinal and GA traits. Findings from the present study highlight the genetic basis of drusen and GA development, which can help to better understand the role of drusen in AMD development and progression.
Methods
Study Population
Potential subjects included Amish adults living in and around Holmes County, Ohio, Lancaster County, Pennsylvania, and both Elkhart and LaGrange Counties, Indiana. Individuals were recruited as part of the Amish Eye Study, which has been described elsewhere.35,39 Briefly, eligible individuals included those from a family with at least 2 individuals with early or intermediate AMD who were over the age of 50 years. Informed consent was obtained for all individuals. Research complied with the Health Insurance Portability and Accountability Act and the Declaration of Helsinki.
Phenotype Data Collection
Subjects participated in a health history survey and ophthalmologic examination that included spectral domain (SD) OCT volume scans for both eyes. This has been described in detail elsewhere.35,39 Briefly, imaging was performed with the Cirrus OCT device (Carl Zeiss Meditec, Dublin, CA, USA) using a volume scanning protocol (512 A-scans × 128 B-scans, 6 × 6 mm pattern, centered on the fovea). A minimum signal strength was required during the acquisition, and scans were repeated as needed to achieve high-quality scans. Images were then sent to the Doheny Image Reading Center and three specific traits were quantified (GA area within a central 5 mm circle, RPE elevation area within the central 3 and 5 mm circles, and RPE elevation volume within the central 3 and 5 mm circles) automatically by the US Food and Drug Administration (FDA)-cleared Advanced RPE analysis software available in the device. Each AMD case was manually inspected for drusen secondary to AMD. Any cases with vitelliform lesions or serious pigment epithelial detachment (PED) were flagged and findings that were not secondary to AMD were not graded for RPE. As drusen accumulate in the sub-RPE space, the RPE elevation parameters are a reflection of the drusen burden, and thus from here on forward these parameters are referred to as drusen area and drusen volume. A detectable amount of drusen area and volume were considered to be measurements equal to or greater than 0.1 mm2 or mm3, respectively. Individuals not meeting these criteria were considered to have no drusen area or no drusen volume. It is possible that an individual had a detectable amount of drusen area or drusen volume, but not both, if one of the two measures was detectable but the other was not.
Genotyping and Quality Control
Genotyping was performed using a customized Illumina Multi-Ethnic Genotyping Array (MEGAex) to capture over 2 million single nucleotide polymorphisms (SNPs) on 1149 individuals. Before performing an association analysis, we evaluated the quality of SNPs and subjects. The first step of the quality control procedures was applied using KING40 and PLINK.41 Individuals were excluded if they showed < 95% genotypic call rates or the inferred gender did not match the reported gender. Known twins and duplicates were identified through a relatedness check. One randomly chosen twin/duplicate was excluded. Variants with < 95% call rates, non-autosomal, or those with minor allele frequency (MAF) = 0 (i.e., monomorphic) were excluded.
Mendelian inconsistencies were assessed using MARKERINFO in the S.A.G.E. package.42 SNPs with more than five Mendelian errors observed were excluded. Mendelian errors for SNPs with less than five errors were marked as missing within the nuclear family. Hardy-Weinberg equilibrium proportions were not used as a quality filter, as they are difficult to assess accurately in our highly related founder population data. Maximum likelihood estimates of allele frequencies and marker-specific inbreeding coefficients were estimated using FREQ in the S.A.G.E. package42 and SNPs with a minor allele frequency <0.01 were excluded.
The final dataset, after all exclusions were applied, consisted of 1074 individuals with 831,286 genotyped SNPs. Principal components analysis was performed to check for any cryptic population structure using PCAiR in GENESIS.43 Only variants with low linkage disequilibrium (LD) among each other (R2 < 0.2) were used and the first three principal components were investigated. The overall flowchart of this process is shown in Supplementary Figure S1.
Association Analysis
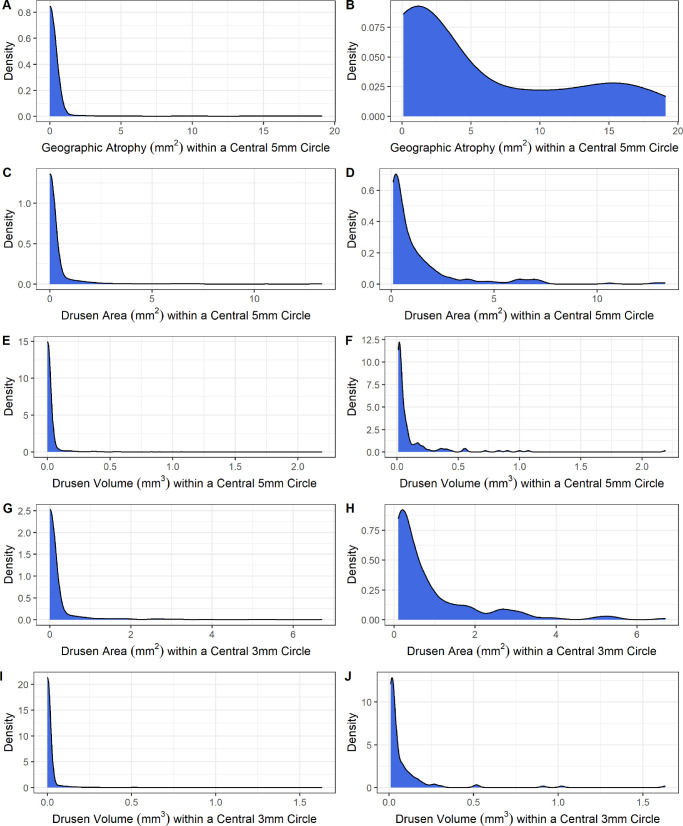
Kinship-adjusted principal components were created using the GENESIS Bioconductor package.43 After checking for potential differences in principal components across demographic variables, 2 models were constructed on each of the 5 traits: GA area within a central 5 mm circle, drusen area within a central 5 mm circle, drusen volume within a central 5 mm circle, drusen area within a central 3 mm circle, and drusen volume within a central 3 mm. The first model (model 1) included a logistic regression approach to predict zero versus non-zero status, referred to as presence of each of the traits. The second model (model 2) was performed on a subset of individuals who had presence of each of the respective traits to remove the effect of the high proportion of zeros in the data (see the Fig.). We utilized a log-transformed Gaussian approach to approximate a normal distribution for model fitting (Supplementary Fig. S2). Only the eye with greater levels of atrophy or drusen for each individual was considered for analysis. We first performed basic logistic (model 1) or linear (model 2) regression to investigate the effect of the age, sex, and ever smoked covariates on each of the outcomes.
Figure.
Density plots of five retinal traits. (A) Geographic atrophy (GA) area (mm2) within a central 5 mm circle. (B) GA area (mm2) within a central 5 mm circle, excluding those with zero. (C) Drusen area (mm2) within a central 5 mm circle. (D) Drusen area (mm2) within a central 5 mm circle, excluding those with zero. (E) Drusen volume (mm3) within a central 5 mm circle. (F) Drusen volume (mm3) within a central 5 mm circle, excluding those with zero. (G) Drusen area (mm2) within a central 3 mm circle. (H) Drusen area (mm2) within a central 3 mm circle, excluding those with zero. (I) Drusen volume (mm3) within a central 3 mm circle. (J) Drusen volume (mm3) within a central 3 mm circle, excluding those with zero. Note that the plots of (B), (D), (F), (H), and (J) exclude individuals who do not have any measurable amount of the trait.
Depending on the model, a GWAS was performed with binomial (model 1) or Gaussian family (model 2) with adjustment for age, sex, ever smoked, PC1, and PC2 using GENESIS.43,44 A genetic relationship matrix containing information on the genetic relationship between any two individuals across all genotyped SNPs was constructed and provided as a covariance matrix in each of the models. For model 1, SNPs with a minor allele frequency ≥ 1% were considered. For model 2, only SNPs with minor allele frequency ≥ 5% were considered, given the much smaller sample size of individuals with each of the traits. A Score test was run on the additive effect for each copy of the allele for the SNP (0, 1, or 2 minor alleles) to examine the effect size and significance. Both the Bonferroni correction and the Benjamini-Hochberg (BH) method were considered for correcting for multiple testing in determining genomewide significance. After Bonferroni correction at α = 0.05, the threshold was set at P < 7.28 × 10−8. For the BH method, a false discovery rate (FDR) of 0.2 was used. Additionally, SNPs were considered to have suggestive effects if the Score test gave P values of less than 1.0 × 10−5. After significant and suggestive SNPs were determined, a gene set enrichment analysis was performed to determine if any pathways are enriched at FDR of less than 0.2.45,46
Genetic Risk Score Analysis
A GRS was constructed for each individual using weights derived from the summary statistics of genomewide significant SNPs from a recent large GWAS of AMD.26 Effects of SNPs were included if directly genotyped in our customized Illumina MEGAex47 or if genotyped SNPs were correlated at R2 > 0.8 to a genomewide significant SNP in a European reference population. Three logistic regression models (score only, covariates only, and score and covariates) with the outcome of binary presence (model 3) for each of the five model 1 traits were constructed. Goodness of fit criteria, including area under curve (AUC) and effects of each covariate were calculated. Receiver operating characteristic (ROC) curves were constructed to further evaluate the predictive ability of the risk score. Similarly, three linear regression models (score only, covariates only, and score and covariates) were created for each of the model 2 traits (e.g. using the subsets of individuals with presence of the trait (model 4). Goodness of fit criteria, including R2, and effects of covariates were calculated. Mixed models were also constructed with a genetic relatedness matrix as a covariance matrix to assess the value of the GRS in predicting each of the five traits after adjustment for individual relatedness.
Results
Drusen Characteristics
After quality assurance and control, a total of 1074 Amish individuals were included in the study (see the Fig., Table 1). There were 57.8% of the participants that were women and the mean age at examination was 66 years (range = 50 to 99 years). There were 19.8% of the individuals that reported ever smoking, as defined by smoking more than 100 cigarettes in their lifetime. There were 5.9% of individuals that had any quantity of GA area in their worse eye, whereas 22.6% and 16.5% had drusen area and volume in a 5 mm circle, respectively. We detected an observable drusen area and volume in a 3 mm circle in 18.1% and 13.0% of participants, respectively. We observed only minor differences among the people with observable amounts of each of the retinal traits compared to the overall study population (Supplementary Table S1). The SD OCT results revealed a large proportion of zeros (no GA and no drusen) and a right skew among the non-zeros across all of the five traits (see Supplementary Fig. S2). We characterized the correlation among each of the five retinal traits and observed strong correlation, especially among measures of drusen area and volume (Supplementary Tables S2, S3).
Table 1.
Characteristics of Study Population of Enrolled Adults, Excluding Refused or Inconclusive Responses
| Trait | N (Binary) or Mean (Continuous) | % (Binary) or SD (Continuous) |
|---|---|---|
| Female | 621 | 57.8 |
| Ever smoked (>100 packs lifetime) | 213 | 19.8 |
| Age at time of OCT examination | 66.1 | 10.3 |
| Presence of geographic atrophy within a central 5 mm circle | 59 | 5.9 |
| Presence of drusen area within a central 5 mm circle | 226 | 22.6 |
| Presence of drusen volume within a central 5 mm circle | 166 | 16.5 |
| Presence of drusen area within a central 3 mm circle | 182 | 18.1 |
| Presence of drusen volume within a central 3 mm circle | 131 | 13.0 |
SD, standard deviation; OCT, optical coherence tomography; mm, millimeter.
Association Analysis
We found only minor evidence of an association between sex or ever smoked in our Amish data on any of the traits (Supplementary Table S4). Age was strongly associated with each of the outcomes. The model 1 GWAS for presence of drusen area within a central 5 mm circle yielded one genomewide significant SNP (rs8125299, P = 8.22 × 10−8, Supplementary Fig. S3) after correction using the BH method with an FDR of 0.2 (Table 2). No SNPs met genomewide significance for model 1 of drusen volume within a central 3 mm circle. One SNP (rs79746087, P = 6.85 × 10−8, Supplementary Fig. S4) was significant after Bonferroni correction and the BH method for presence of drusen area within a central 3 mm circle (model 1). Three SNPs, including rs7028791 (P = 1.31 × 10−8, Supplementary Fig. S5), rs7850939 (P = 2.27 × 10−8, Supplementary Fig. S6), and rs79746087 (P = 6.87 × 10−8, Supplementary Fig. S7) were significant after Bonferroni correction and use of the BH method in model 1 for drusen volume within a central 3 mm circle. Two SNPs, rs76316680 (P = 1.84 × 10−7, Supplementary Fig. S8) and rs1759824 (P = 3.73 × 10−7, Supplementary Fig. S9) met criteria for genomewide significance for drusen volume in a 3 mm circle using the BH method in model 2 (see Table 2). The P values for each of these SNPs across all measures and models can be found in Supplementary Table S5. There was minimal evidence of inflation based on calculations of the genomic inflation factor for models 1 and 2 of each of the five traits (Supplementary Table S6).
Table 2.
List of Genomewide Significant SNPs After Correction Using the Benjamini-Hochberg Method With False Discovery Rate of 20% for 3 Retinal Traits: Drusen Area Within a Central 5 mm Circle, Drusen Area Within a Central 3 mm Circle, and Drusen Volume Within a Central 3 mm Circle
| Measure | Model | SNP (GrCh37) | Chromosome | Position | Score P Value | Nearest Gene(s) |
|---|---|---|---|---|---|---|
| Drusen area within a central 5 mm circle | 1 | rs8125299 | 20 | 4860658 | 8.22 × 10−8 | SLC23A2 |
| Drusen area within a central 3 mm circle | 1 | rs79746087 | 19 | 2489650 | 6.85 × 10−8 | GADD45B, GNG7 |
| Drusen volume within a central 3 mm circle | 1 | rs7028791 | 9 | 113240481 | 1.31 × 10−8 | SLC31A1 |
| 1 | rs7850939 | 9 | 113242405 | 2.27 × 10−8 | SLC31A1 | |
| 1 | rs79746087 | 19 | 2489650 | 6.87 × 10−8 | GADD45B, GNG7 | |
| 2 | rs76316680 | 4 | 138280638 | 1.84 × 10−7 | DPP10-AS3, DPP10-AS1 | |
| 2 | rs17759824 | 2 | 114942278 | 3.73 × 10−7 | SLC7A11, LINC00499 |
If SNP is exonic, the gene that it is located in is listed. If SNP is intergenic, the nearest upstream and downstream genes are listed.
A total of 35 and 19 SNPs met suggestive criteria (P < 1 × 10−5) for model 1 and model 2, respectively, across the five traits (Supplementary Table S7). Gene set enrichment analysis45,46 performed on the overall list of genes corresponding to suggestive or significant SNPs yielded no enriched gene sets at FDR < 0.20.
Genetic Risk Score Analysis
Twenty-two SNPs met inclusion criteria for the GRS (Supplementary Table S8). Goodness of fit criteria and P values for the score variable on the logistic regression models for presence (non-zero status) of each of the five retinal traits (model 3) as primary outcomes can be found in Table 3. Table 4 includes goodness of fit criteria and risk score P value for the quantity of outcomes in a non-zero subset for each of the five traits (model 4). Inclusion of the risk score generally improved AUC of models and the score term was significant at α = 0.05 among all outcomes except for the GA area.
Table 3.
Area Under the Curve (AUC), Akaike Information Criteria (AIC), and P Values for the Risk Score Variable Across Prediction of Presence (Model 3) of Five Outcomes: Geographic Atrophy (GA), Drusen Area Within a Central 5 mm Circle, and Drusen Volume Within a Central 5 mm Circle
| Measure | Model Type | AUC | AIC | Risk Score P Value | Adjusted Model Risk Score P Value |
|---|---|---|---|---|---|
| Geographic atrophy within a central 5 mm circle | Score only | 0.636 | −76.34 | 4.18 × 10−6 | 2.53 × 10−6 |
| Covariates only | 0.838 | −194.17 | – | – | |
| Score + covariates | 0.865 | −215.22 | 1.43 × 10−6 | 8.64 × 10−6 | |
| Drusen area within a central 5 mm circle | Score only | 0.615 | 1058.6 | 6.79 × 10−11 | 1.23 × 10−9 |
| Covariates only | 0.769 | 911.13 | – | – | |
| Score + covariates | 0.798 | 866.13 | 8.31 × 10−12 | 2.83 × 10−10 | |
| Drusen volume within a central 5 mm circle | Score only | 0.617 | 833.47 | 8.05 × 10−9 | 4.26 × 10−9 |
| Covariates only | 0.808 | 650.64 | – | – | |
| Score + covariates | 0.833 | 615.11 | 1.02 × 10−9 | 6.23 × 10−10 | |
| Drusen area within a central 3 mm circle | Score only | 0.634 | 846.89 | 2.46 × 10−10 | 9.31 × 10−10 |
| Covariates only | 0.789 | 745.79 | – | – | |
| Score + covariates | 0.820 | 647.45 | 4.69 × 10−11 | 1.88 × 10−9 | |
| Drusen volume within a central 3 mm circle | Score only | 0.641 | 582.51 | 3.89 × 10−9 | 2.50 × 10−10 |
| Covariates only | 0.827 | 458.72 | – | – | |
| Score + covariates | 0.862 | 366.09 | 6.66 × 10−10 | 8.44 × 10−10 |
Adjusted risk score P value is provided for a logistic regression model with account for relatedness. When specified, covariates included age at time of examination, sex, and smoking status. All P values are significant at α = 0.05.
AUC, area under the curve; AIC, Akaike Information Criteria; GA, geographic atrophy.
Table 4.
Multiple R2, Adjusted R2, and P Values for Risk Score Variable Across Quantities of Non-Zero Subset (Model 4) of 5 Outcomes: Geographic Atrophy Within a Central 5 mm Circle, Drusen Area Within a Central 5 mm Circle, Drusen Volume Within a Central 5 mm Circle, Drusen Area Within a Central 3 mm Circle, and Drusen Volume Within a Central 3 mm Circle
| Measure | Model Type | Multiple R2 | Adjusted R2 | Risk Score P Value | Adjusted Model Risk Score P Value |
|---|---|---|---|---|---|
| Geographic atrophy | Score only | 0.020 | 0.003 | 0.285 | 0.221 |
| Covariates only | 0.115 | 0.050 | – | ||
| Score + covariates | 0.169 | 0.090 | 0.071 | 0.027* | |
| Drusen area within a central 5 mm circle | Score only | 0.017 | 0.012 | 0.053 | 0.049* |
| Covariates only | 0.158 | 0.142 | – | ||
| Score + covariates | 0.185 | 0.166 | 0.007* | 0.008* | |
| Drusen volume within a central 5 mm circle | Score only | 0.019 | 0.013 | 0.074 | 0.161 |
| Covariates only | 0.076 | 0.053 | – | ||
| Score + covariates | 0.105 | 0.077 | 0.022* | 0.076 | |
| Drusen area within a central 3 mm circle | Score only | 0.030 | 0.024 | 0.023* | 0.040* |
| Covariates only | 0.058 | 0.037 | – | ||
| Score + covariates | 0.092 | 0.064 | 0.014* | 0.013* | |
| Drusen volume within a central 3 mm circle | Score only | 0.044 | 0.036 | 0.021* | 0.050* |
| Covariates only | 0.034 | 0.003 | – | ||
| Score + covariates | 0.074 | 0.034 | 0.034* | 0.048* |
Adjusted risk score P value is provided for a Gaussian regression model with account for relatedness. When specified, covariates included age at time of exam, sex, and smoking status. An asterisk (*) represents that the P value is significant at α = 0.05.
Discussion
This study evaluated the genetic risk for presence and quantity of three AMD traits: GA area, drusen area, and drusen volume. The results indicated that these traits are suggestively associated with a large quantity of SNPs across the genome in Amish adults, after adjusting for age, sex, ever smoked, and two principal components, although the significance of the effects were varied potentially due to a rather small sample size of individuals with a non-zero amount of each of the traits. Seven SNPs, including one SNP repeated between model 1 of drusen area and volume (mm3) in a 3 mm circle centered, were significant after correction with FDR < 0.20 (BH). Few studies have previously shown a genetic influence on drusen, particularly at the whole genome level.27,33 Within those, the LIPC and ABCA1 genes have been shown to be associated with large drusen development.33 Additionally, a GRS for AMD has been linked to baseline drusen measures but not progression of drusen.34
The most suggestive signals and the signals with the smallest P values were found when considering the presence/absence (model 1) of each of the retinal traits, rather than quantity, although this could at least in part be explained by the greater sample sizes, and hence, the power to detect smaller effects, when including all individuals as opposed to solely those with non-zero values for the traits.
One SNP, rs79746087, was significant for both the drusen area and the drusen volume within a central 3 mm circle, indicating a significant role across measures. It was also suggestive for the drusen area and the volume within a central 5 mm circle. Although these are not surprising based on the correlated outcomes (see Supplementary Table S2), it is promising to find a region that is implicated in each of the drusen area and volume measurements. It is located in an intergenic region between GADD45B and GNG7. There is evidence that it may be an expression quantitative trait locus eQTL but with no direct links to the eye or macular degeneration.48 The first of these, GADD45B, has been implicated in growth arrest49 and DNA damage response including protection from retinal ganglion cell injuries.50–52 GNG7 is associated with ischemic injuries of the retina.53 Other genes with known function located close to our suggestive GWAS loci include SLC23A2, SLC31A1, SLC7A11, DPP10-AS3, and DPP10-AS1. The solute carrier (SLC) family of genes are generally implicated in bodily transport of various metabolites, including amino acids, vitamin C, and copper. They have been previously implicated in various health outcomes relating to the eye, including drusen size and glaucoma.54–59 DPP10-AS3 and DPP10-AS1 are long non-coding RNA genes that play a role in the development of diabetic retinopathy and various cancers.60–64 Although none of the genomewide significant SNPs have been directly implicated in AMD,26 3 of them are located within 6 megabases of a known AMD SNP but only in weak linkage disequilibrium in a European reference population65 (Supplementary Table S9). Namely, rs79746087 is between CNN2 and C3, whereas rs7850939 and rs7028791 are downstream of ABCA1. Previous data suggests that the genetic profile of early AMD may be different than from late AMD, making it possible that the pathways implicated in drusen might be more closely aligned with early AMD.66,67 It is also possible that these regions may be implicated in the development of drusen but not closely associated with AMD diagnosis.
Although our GWAS of retinal traits revealed signals near known AMD risk SNPs, we further investigated the utility of using the known AMD-associated loci26 to predict the presence/absence and quantity of each of the three traits. We were ultimately able to capture at least one risk variant from each of the major regions or a correlated SNP (see Supplementary Fig. S8) and create a GRS for each individual based on these AMD risk variants. Our finding that the GRS improved each model's ability to predict their respective outcome suggests that the underlying genetic architecture of AMD is implicated in drusen development, and further suggests that there is a genetic influence on development of drusen. Further, we provide evidence that some of these genetic risk factors may be similar to those of AMD. There is great potential for using drusen as a biomarker for AMD due to the noninvasive nature of the OCT examination.
Use of the Amish as a study population has the inherent strength in that can detect effects that are small in a larger, general population but are amplified within this founder population. By combining OCT data with SNP-level data, we are the first to provide an extensive analysis of how risk variants may affect drusen development in a founder population. However, we are limited by a relatively large proportion of zeros among each of the measures. The power to detect smaller effects and rare variants will be increased by obtaining longitudinal data to measure progression of the retinal traits over time. Further, use of whole genome sequencing could allow for better detection of rare variants within the genomewide significant loci, as the Amish may demonstrate variation that differs from a general European ancestry population.
We conclude that there is an underlying genetic component to drusen development that may be somewhat separated from the biological pathways considered in AMD development. With the relatively high heritability of AMD for a complex disease, it is not surprising that drusen may also be inherited in a similar manner.16,25 Work on the genetic risk of drusen could aid in improving understanding of drusen as a biomarker for genetic risk of AMD. Building this knowledge could serve to improve screening and treatment to prevent AMD by more thorough incorporation of drusen burden into such practices.
Supplementary Material
Acknowledgments
The authors thank the Amish families for their willing participation in our study. The authors used the Anabaptist Genealogy Database68 and information from the Swiss Anabaptist Genealogy Association to help determine relationships.
Supported by National Institutes of Health/National Eye Institute (NEI), grants EY023164, EY022310, and EY012118. The authors acknowledge the resources provided by the Department of Population and Quantitative Health Sciences, School of Medicine at Case Western Reserve University, the Doheny Imaging Reading Center at the Doheny Eye Institute, the Departments of Ophthalmology and Genetics at the University of Pennsylvania, and the John P. Hussman Institute for Human Genomics at University of Miami, Miller School of Medicine.
Disclosure: M.D. Osterman, None; Y.E. Song, None; M. Nittala, None; S.R. Sadda, Amgen (C), Allergan (C), Novartis (C), Regeneron (C), Centervue (C), Heidelberg (C), Heidelberg Engineering (F), Optos (F), Nidek (F), Topcon (F), Genentech/Roche (C), Oxurion (C), Bayer (C), 4DMT (C), Optos (C), Carl Zeiss Meditec (F), Centervue (F); W.K. Scott, None; D. Stambolian, None; M.A. Pericak-Vance, None; J.L. Haines, None
References
- 1. Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY.. Age-related macular degeneration. Lancet. 2012; 379(9827): 1728–1738. [DOI] [PubMed] [Google Scholar]
- 2. Wong WL, Su X, Li X, et al.. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob Health. 2014; 2(2): e106–e116. [DOI] [PubMed] [Google Scholar]
- 3. Marmor DJ, Marmor MF.. Simulating vision with and without macular disease. Arch Ophthalmol. 2010; 128(1): 117–125. [DOI] [PubMed] [Google Scholar]
- 4. Spaide RF, Jaffe GJ, Sarraf D, et al.. Consensus Nomenclature for Reporting Neovascular Age-Related Macular Degeneration Data: Consensus on Neovascular Age-Related Macular Degeneration Nomenclature Study Group. Ophthalmology. 2020; 127(5): 616–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klein R, Davis MD, Magli YL, Segal P, Klein BEK, Hubbard L.. The Wisconsin Age-related Maculopathy Grading System. Ophthalmology. 1991; 98(7): 1128–1134. [DOI] [PubMed] [Google Scholar]
- 6. Kassoff A, Kassoff J, Buehler J, et al.. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001; 119(10): 1417–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamoureux EL, Mitchell P, Rees G, et al.. Impact of early and late age-related macular degeneration on vision-specific functioning. Br J Ophthalmol. 2011; 95(5): 666–670. [DOI] [PubMed] [Google Scholar]
- 8. Cheng Q, Saaddine JB, Klein R, Rothenberg R, Chou CF, Il'yasova D. Early Age-related Macular Degeneration with Cardiovascular and Renal Comorbidities: An Analysis of the National Health and Nutrition Examination Survey, 2005–2008. Ophthalmic Epidemiol. 2017; 24(6): 413–419. [DOI] [PubMed] [Google Scholar]
- 9. Comparison of Age-related Macular Degeneration Treatments Trials (CATT) Research Group; Maguire MG, Martin DF, et al.. Five-Year Outcomes with Anti–Vascular Endothelial Growth Factor Treatment of Neovascular Age-Related Macular Degeneration: The Comparison of Age-Related Macular Degeneration Treatments Trials. In Ophthalmology. 2016; 123(8): 1751–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Villegas VM, Aranguren LA, Kovach JL, Schwartz SG, Flynn HW Jr. Current advances in the treatment of neovascular age-related macular degeneration. Expert Opin Drug Deliv. 2017; 14(2): 273–282. [DOI] [PubMed] [Google Scholar]
- 11. Reid CA, Nettesheim ER, Connor TB, Lipinski DM.. Development of an inducible anti-VEGF rAAV gene therapy strategy for the treatment of wet AMD. Sci Rep. 2018; 8(1): 11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Al-Zamil WM, Yassin SA.. Recent developments in age-related macular degeneration: A review. Clin Interv Aging. 2017; 12: 1313–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Wang VM, Chan CC.. The role of anti-inflammatory agents in age-related macular degeneration (AMD) treatment. Eye. 2011; 25(2): 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chakravarthy U, Wong TY, Fletcher A, et al.. Clinical risk factors for age-related macular degeneration: A systematic review and meta-analysis. BMC Ophthalmol. 2010; 10: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tomany SC, Wang JJ, Van Leeuwen R, et al.. Risk factors for incident age-related macular degeneration: Pooled findings from 3 continents. Ophthalmology. 2004; 111(7): 1280–1287. [DOI] [PubMed] [Google Scholar]
- 16. Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: Relative roles of genetic and environmental influences. Arch Ophthalmol. 2005; 123(3): 321–327. [DOI] [PubMed] [Google Scholar]
- 17. Meyers SM, Greene T, Gutman FA.. A twin study of age-related macular degeneration. Am J Ophthalmol. 1995; 120(6): 757–766. [DOI] [PubMed] [Google Scholar]
- 18. Klein RJ, Zeiss C, Chew EY, et al.. Complement factor H polymorphism in age-related macular degeneration. Science. 2005; 308(5720): 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haines JL, Hauser MA, Schmidt S, et al.. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005; 308(5720): 419–421. [DOI] [PubMed] [Google Scholar]
- 20. Edwards AO, Ritter R, Abel KJ, Manning A, Panhuysen C, Farrer LA.. Complement factor H polymorphism and age-related macular degeneration. Science. 2005; 308(5720): 421–424. [DOI] [PubMed] [Google Scholar]
- 21. Hageman GS, Anderson DH, Johnson LV., et al.. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci. 2005; 102(20): 7227–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rivera A, Fisher SA, Fritsche LG, et al.. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005; 14(21): 3227–3236. [DOI] [PubMed] [Google Scholar]
- 23. Fritsche LG, Loenhardt T, Janssen A, et al.. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008; 40(7): 892–896. [DOI] [PubMed] [Google Scholar]
- 24. De Jong EK, Geerlings MJ, Den Hollander AI. Age-related macular degeneration, Genetics and Genomics of Eye Disease. Published online 2020: 155–180, doi: 10.1016/B978-0-12-816222-4.00010-1. [DOI]
- 25. DeAngelis MM, Owen LA, Morrison MA, et al.. Genetics of age-related macular degeneration (AMD). Hum Mol Genet. 2017; 26(R1): R45–R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fritsche LG, Igl W, Bailey JNC, et al.. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016; 48(2): 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hageman GS, Luthert PJ, Victor Chong NH, Johnson L V., Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch's membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001; 20(6): 705–732. [DOI] [PubMed] [Google Scholar]
- 28. Stanton CM, Wright AF.. Inflammatory biomarkers for AMD. Adv Exp Med Biol. 2014; 801: 251–257. [DOI] [PubMed] [Google Scholar]
- 29. De Jong PTVM. Elusive drusen and changing terminology of AMD review-article. Eye. 2018; 32(5): 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Or C, Heier JS, Boyer D, et al.. Vascularized drusen: a cross-sectional study. Int J Retin Vitr. 2019; 5(1): 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bowes Rickman C, Farsiu S, Toth CA, Klingeborn M.. Dry age-related macular degeneration: Mechanisms, therapeutic targets, and imaging. Investig Ophthalmol Vis Sci. 2013; 54(14): ORSF68–ORSF80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagner EK, Raychaudhuri S, Villalonga MB, et al.. Mapping rare, deleterious mutations in Factor H: Association with early onset, drusen burden, and lower antigenic levels in familial AMD. Sci Rep. 2016; 6(January): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Y, Reynolds R, Fagerness J, Rosner B, Daly MJ, Seddon JM.. Association of variants in the LIPC and ABCA1 genes with intermediate and large drusen and advanced age-related macular degeneration. Investig Ophthalmol Vis Sci. 2011; 52(7): 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffman JD, Van Grinsven MJJP, Li C, et al.. Genetic association analysis of drusen progression. Investig Ophthalmol Vis Sci. 2016; 57(4): 2225–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nittala MG, Velaga SB, Hariri A, et al.. Retinal sensitivity using microperimetry in age-related macular degeneration in an Amish population. Ophthalmic Surg Lasers Imaging Retin. 2019; 50(9): E236–E241. [DOI] [PubMed] [Google Scholar]
- 36. Corvi F, Srinivas S, Nittala MG, et al.. Reproducibility of qualitative assessment of drusen volume in eyes with age related macular degeneration. Eye. 2021; 35(9): 2594–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mook MA, Hostetler JA.. Amish Society. J Am Folk. JHU Press; 1993. Published online 1969, doi: 10.2307/539066. [DOI] [Google Scholar]
- 38. Hoffman JD, Cooke Bailey JN, D'Aoust L, et al.. Rare complement factor H variant associated with age-related macular degeneration in the Amish. Investig Ophthalmol Vis Sci. 2014; 55(7): 4455–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sardell RJ, Nittala MG, Adams LD, et al.. Heritability of Choroidal Thickness in the Amish. Ophthalmology. 2016; 123(12): 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM.. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010; 26(22): 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Purcell S, Neale B, Todd-Brown K, et al.. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81(3): 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. S.A.G.E. Statistical Analysis for Genetic Epidemiology, Release 6.4. Published online 2016. Available at: http://darwin.cwru.edu.
- 43. Gogarten SM, Sofer T, Chen H, et al.. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019; 35(24): 5346–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. R Development Core Team R. R: A Language and Environment for Statistical Computing. 2011. Available at: https://www.semanticscholar.org/paper/R%3A-A-language-and-environment-for-statistical-Team/659408b243cec55de8d0a3bc51b81173007aa89b. [Google Scholar]
- 45. Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B.. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003; 34(3): 267–273. [DOI] [PubMed] [Google Scholar]
- 46. Subramanian A, Tamayo P, Mootha VK, et al.. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005; 102(43): 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Multi-Ethnic Genotyping Array Consortium. Available at: https://www.illumina.com/science/consortia/human-consortia/multi-ethnic-genotyping-consortium.html. Accessed May 25, 2020.
- 48. Aguet F, Barbeira AN, Bonazzola R, et al.. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020; 369(6509): 1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A.. Growth Arrest and DNA-Damage-Inducible, Beta (GADD45b)-Mediated DNA Demethylation in Major Psychosis. Neuropsychopharmacol. 2011; 37(2): 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu B, Sun X, Suyeoka G, Garcia JGN, Leiderman YI.. TGFβ Signaling Induces Expression of Gadd45b in Retinal Ganglion Cells. Invest Ophthalmol Vis Sci. 2013; 54(2): 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu B, Chen H, Neufeld AH.. Upregulation of Gadd45b in the Aging Retina. Invest Ophthalmol Vis Sci. 2007; 48(13): 46. [Google Scholar]
- 52. Liu B, Suyeoka G, Papa S, Franzoso G, Neufeld AH.. Growth arrest and DNA damage protein 45b (Gadd45b) protects retinal ganglion cells from injuries. Neurobiol Dis. 2009; 33(1): 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Andreeva K, Zhang M, Fan W, et al.. Time-dependent Gene Profiling Indicates the Presence of Different Phases for Ischemia/Reperfusion Injury in Retina. Ophthalmol Eye Dis. 2014; 6: 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Merle BMJ, Rosner B, Seddon JM.. Genetic Susceptibility, Diet Quality, and Two-Step Progression in Drusen Size. Invest Ophthalmol Vis Sci. 2020; 61(17): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Erichsen HC, Engel SAM, Eck PK, et al.. Genetic variation in the sodium-dependent vitamin C transporters, SLC23A1, and SLC23A2 and risk for preterm delivery. Am J Epidemiol. 2006; 163(3): 245–254. [DOI] [PubMed] [Google Scholar]
- 56. Chen AA, Marsit CJ, Christensen BC, et al.. Genetic variation in the vitamin C transporter, SLC23A2, modifies the risk of HPV16-associated head and neck cancer. Carcinogenesis. 2009; 30(6): 977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zanon-Moreno V, Ciancotti-Olivares L, Asencio J, et al.. Association between a SLC23A2 gene variation, plasma vitamin C levels, and risk of glaucoma in a Mediterranean population. Mol Vis. 2011; 17(July): 2997–3004. [PMC free article] [PubMed] [Google Scholar]
- 58. Chintala S, Li W, Lamoreux ML, et al.. Slc7a11 gene controls production of pheomelanin pigment and proliferation of cultured cells. Proc Natl Acad Sci USA. 2005; 102(31): 10964–10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang Y, Dai Z, Barbacioru C, Sadée W.. Cystine-glutamate transporter SLC7A11 in cancer chemosensitivity and chemoresistance. Cancer Res. 2005; 65(16): 7446–7454. [DOI] [PubMed] [Google Scholar]
- 60. Pollack S, Igo RP, Jensen RA, et al.. Multiethnic genome-wide association study of diabetic retinopathy using liability threshold modeling of duration of diabetes and glycemic control. Diabetes. 2019; 68(2): 441–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ong MS, Cai W, Tan TZ, Huang RY, Hooi SC, Yap CT, Kumar AP. Long non-coding RNA landscape in colorectal cancer. RNA & disease. 2019; 9: 6. [Google Scholar]
- 62. Tian H, Pan J, Fang S, et al.. Epigenetic Regulation Contributes to the Oncogenic Role of DPP10-AS1 in Lung Cancer. SSRN Electron J. Available at: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3421596. [Google Scholar]
- 63. Zhou Y, Zhou Z, Ji Z, Yan W, Li H, Yu X. Tetramethylpyrazine reduces prostate cancer malignancy through inactivation of the DPP10‑AS1/CBP/FOXM1 signaling pathway. Int J Oncol. 2020; 57(1): 314–324. [DOI] [PubMed] [Google Scholar]
- 64. Liu G, Zhao H, Song Q, Li G, Lin S, Xiong S.. Long non-coding RNA DPP10-AS1 exerts anti-tumor effects on colon cancer via the upregulation of ADCY1 by regulating microRNA-127-3p. Aging (Albany NY). 2021; 13(7): 9748–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Machiela MJ, Chanock SJ.. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015; 31(21): 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Holliday EG, Smith AV, Cornes BK, et al.. Insights into the Genetic Architecture of Early Stage Age-Related Macular Degeneration: A Genome-Wide Association Study Meta-Analysis. PLoS One. 2013; 8(1): e53830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thee EF, Meester-Smoor MA, Luttikhuizen DT, et al.. Performance of classification systems for age-related macular degeneration in the Rotterdam study. Transl Vis Sci Technol. 2020; 9(2): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Agarwala R, Biesecker LG, Schäffer AA.. Anabaptist genealogy database. Am J Med Genet - Semin Med Genet. 2003; 121C(1): 32–37. [DOI] [PubMed] [Google Scholar]
- 69. Pruim RJ, Welch RP, Sanna S, et al.. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2011; 27(13): 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.