Abstract
The formation of the leafy heads of Chinese cabbage is an important agricultural factor because it directly affects yield. In this study, we identified two allelic non-heading mutants, nhm4-1 and nhm4-2, from an ethyl methanesulfonate mutagenic population of a heading Chinese cabbage double haploid line “FT.” Using MutMap, Kompetitive Allele-Specific PCR genotyping, and map-based cloning, we found that BraA09g001440.3C was the causal gene for the mutants. BraA09g001440.3C encodes an ent-copalyl diphosphate synthase 1 involved in gibberellin biosynthesis. A single non-synonymous SNP in the seventh and fourth exons of BraA09g001440.3C was responsible for the nhm4-1 and nhm4-2 mutant phenotypes, respectively. Compared with the wild-type “FT,” the gibberellin content in the mutant leaves was significantly reduced. Both mutants showed a tendency to form leafy heads after exogenous GA3 treatment. The two non-heading mutants and the work presented herein demonstrate that gibberellin is related to leafy head formation in Chinese cabbage.
Keywords: Chinese cabbage, leafy head formation, allelic mutations, CPS1, MutMap
Introduction
The leafy head is a unique organ in Chinese cabbage, and the shape, size, uniformity, and density of the leafy head directly affect its commercial value (Zhang et al., 2021). The formation of Chinese cabbage leafy heads is biologically complex, including four developmental stages, the seedling, rosette, folding, and heading stages. Leaves change from flat to upward facing in the rosette stage and start to fold in the folding stage, eventually forming a leafy head in the heading stage (He et al., 2000). The formation of leafy heads is affected by various factors, including the temperature, light intensity, auxin concentration, carbon to nitrogen ratio, and leafy shape (Ito and Kato, 1957; He et al., 2000; Mao et al., 2014).
The mechanism of Chinese cabbage leafy head formation is complex, and there is no current hypothesis that can fully explain the mechanism. However, in recent years, some genes related to leafy head formation in Chinese cabbage have been reported. The target gene BrpSPL9-2 of microRNA brp-miR156 regulates the time of Chinese cabbage heading by shortening the seedling and rosette stages (Wang et al., 2014). BrBRX genes (BrBRX.1, BrBRX.2, and BrBRX.3) control leaf morphological development, and BrBRX.1 and BrBRX.2 genes had similar expression patterns and may be involved in the formation of leafy heads in Brassica rapa (Cheng et al., 2016; Zhang et al., 2021). Yu et al. (2019) explored the expression of BrAN3 before and after the formation of Chinese cabbage leafy heads and found that BrAN3 was significantly expressed in the rosette and heading leaves. According to the different expression patterns of different leaf locations, it was confirmed that BrAN3 could induce the formation of leafy head. Ren et al. (2020) discovered that BcpLH regulates the timing of leafy head formation by integrating important miRNAs.
Plant hormones play important roles in the formation of leafy heads. Cheng et al. (2016) found that four plant hormones (cytokinin, auxin, gibberellin, and jasmonic acid) regulate the formation and development of the leafy head in Chinese cabbage. Yu et al. (2019) reported similar results and proposed that BrAN3 can induce GA, BR, and SA signaling pathways, thereby inhibiting the formation of Chinese cabbage leafy heads. Gao et al. (2017) revealed that auxin transport genes (BrAUX/LAX, BrPIN, and BrPGP) play an important role in leafy head formation in Chinese cabbage by genome-wide annotation and bioinformatics analysis. Li et al. (2019) found that both auxin and abscisic acid signaling pathways play important roles in regulating early leafy head formation. In our previous study (Gao et al., 2020), we found that the non-heading phenotype of Chinese cabbage is caused by mutations in the BraA07g042410.3C gene, which encodes ent-kaurene synthase (KS), a key enzyme involved in gibberellin (GA) biosynthesis. This result indicates that the GA content in Chinese cabbage leaves is related to the formation of leafy heads. The above research results provide a basis for studying the mechanisms of the hormones involved in Chinese cabbage leafy head formation, and further research on genes related to leafy head formation can improve our understanding of the molecular mechanism of leafy head development.
In this study, we found two non-heading mutants (nhm4-1 and nhm4-2) whose phenotypes were consistent with each other. Allelism testing proved that the mutant genes of nhm4-1 and nhm4-2 were allelic. MutMap, Kompetitive Allele-Specific PCR (KASP), and map-based cloning analyses were performed to identify the candidate gene of the mutants. We demonstrated that BraA09g001440.3C (BrCPS1), which encodes an ent-copalyl diphosphate synthase 1 (CPS1) involved in GA biosynthesis, was the candidate gene. The function of BrCPS1 in the leafy head formation of Chinese cabbage was further confirmed by the determination of the GA content in the leaves of two allelic mutants and the spraying with exogenous GA3. These results provide information for understanding the formation mechanism of leafy heads in Chinese cabbage.
Materials and Methods
Plant Materials
In our previous study (Gao et al., 2020), 14 non-heading mutant plants were harvested in an EMS-induced mutagenic population. For this study, we selected two mutants with extremely similar phenotypes, nhm4-1 and nhm4-2.
Genetic Analysis
For genetic analysis, the mutants were crossed with the wild-type “FT” to obtain F1, F2, and BC1 populations. To investigate the genetic characteristics of the mutants, we recorded the phenotypes of each plant in each generation and analyzed the separation rate of the F2 and BC1 populations using the Chi-square (χ2) test.
Allelism Test Between nhm4-1 and nhm4-2
To detect the allelism of the two mutant genes, we conducted an allelism test. Mutants nhm4-1 and nhm4-2 were used as parents for hybridization, and the phenotypes of their hybrid progeny were observed and recorded.
Mutmap Analysis to Determine the Candidate Gene
A modified MutMap method (Abe et al., 2012) was applied for fine mapping and identification of candidate genes for nhm4-1. For MutMap, the DNA of 50 mutant plants in the F2 population was mixed equally as the mutant pool. DNA from the two parental plants and the mutant pool asextracted from fresh leaves at the rosette stage using a DNA secure plant kit (Tiangen, Beijing, China) and resequenced with a NovaSeq 6,000 sequencer (Illumina, San Diego, CA, United States of America).
Low-quality data were filtered from the raw data according to the filtering criteria of our previous study (Gao et al., 2020) to obtain clean reads. Clean reads were mapped to reference genome sequences using BWA software (Li and Durbin, 2010), and SAMtools (Li et al., 2009) was used to sort the alignment file. Insertions and deletions (INDELs) and single-nucleotide polymorphisms (SNPs) were identified using GATK software (McKenna et al., 2010) and ANNOVAR software (Wang et al., 2010). Circos software (Krzywinski et al., 2009) was used to map variation information in the genome. The ΔSNP index across the chromosomes of the B. rapa genome was obtained using sliding-window analysis (with a five-SNP window size and one SNP for each step).
SNP Genotyping by KASP
Kompetitive Allele-Specific PCR was developed for the genotypic assay to detect the co-segregation of each SNP and to confirm the nhm4-1 candidate gene. The allele-specific primers used are shown in Supplementary Table S1. A total of 184 F2 plants were used for KASP genotyping. Of these, 48 plants showed a mutant phenotype and 136 exhibited the wild-type phenotype. The assay was carried out at the Vegetable Research Center of the Agriculture and Forestry Academy in Beijing.
Cloning and Sequence Analysis
The coding sequences of candidate genes were amplified with specific primers (Supplementary Table S2) in the wild-type “FT,” nhm4-1 mutant, and nhm4-2 mutant plants. Primer 5.0 was used to design specific primers and perform gene cloning following the methods of Gao et al. (2020). Sequencing was performed using the Sanger method at GENEWIZ (Suzhou, China). The sequences were aligned using DNAMAN V6 software (Lynnon BioSoft, Montreal, QC, Canada).
Enzyme Activity Assays
Ent-Copalyl diphosphate synthase 1 activity in the leaves from the wild-type and nhm4-1 mutant plants was evaluated using the Plant CPS enzyme-linked immunosorbent assay (ELISA) Kit (Meimian Biotech Co., Ltd., Jiangsu, China) via a double antibody sandwich method following the manufacturer’s instructions. Each material was performed for three biological repeats, and three times the technical repeats were performed in each biological repeat.
RNA Isolation and Quantitative Real-Time PCR
For analysis of the relative expression levels of the candidate gene, total RNA was extracted from the cotyledons, first true leaves, third true leaves, sixth true leaves, rosette leaves, and heading leaves of wild-type “FT,” mutant nhm4-1, and mutant nhm4-2 plants using an RNA extraction kit (Aidlab, Beijing, China). First-strand cDNA was synthesized using the FastQuant RT kit (Tiangen, Beijing, China) and quantitative real-time PCR (qRT-PCR) was performed using an Ultras SYBR Mixture (CWBIO, Beijing, China) and the Quant Studio 6 Flex Real-Time PCR System (ABI, Los Angeles, CA, United States of America). The Actin gene (F: 5′-ATCTACGAGGGTTATGCT-3′; R: 5′-CCACTGAGGACGATGTTT-3′) was used as the reference gene. Each experiment was independently performed with three technical replicates and three biological replicates. The relative gene expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The primer sequences used for qRT-PCR amplification are shown in Supplementary Table S3.
Measurement of Endogenous GA Content and Treatment of Exogenous GA3
According to our previous methods (Gao et al., 2020), the content of endogenous GA in the leaves of wild-type “FT,” mutant nhm4-1, and mutant nhm4-2 plants was determined by liquid chromatography-tandem mass chromatography (Chen et al., 2012). The response of the mutants nhm4-1 and nhm4-2 to GA was determined by spraying exogenous GA3 solution (500 mg/L). The plants were sprayed with GA3 at 3-day intervals once the cotyledons were fully expanded, and the treatments ended before the rosette stage. Seedlings sprayed with an equal volume of ddH2O without GA3 were used as controls.
Results
Phenotypic Characterization and Inheritance Analysis of nhm4-1 and nhm4-2
The phenotype of the mutant nhm4-1 was highly consistent with that of the mutant nhm4-2. The leaves of the mutant plants showed geotropic growth throughout the developmental stages and could not form leafy heads at the heading stage, unlike those of the wild-type “FT” plants (Figure 1).
Figure 1.
Phenotype observation of the wild-type “FT” and mutant plants. (A) Phenotypes of wild-type “FT,” mutant nhm4-1, and mutant nhm4-2 plants during the rosette stage. (B) Phenotypes of wild-type “FT,” mutant nhm4-1, and mutant nhm4-2 plants at the heading stage. Bar = 5 cm.
As shown in Table 1, the phenotype of all plants in the F1 generation was consistent with that of the wild-type “FT.” The segregation ratio of the F2 generation was 3:1, while that of the BC1 generation was approximately 1:1. These results indicate that the mutation phenotype of the nhm4-1 and nhm4-2 mutants was controlled by a single recessive nuclear gene.
Table 1.
Genetic analysis of mutants nhm4-1 and nhm4-2.
| Generations | Total plants | Mutant plants | Wild type plants | Segregation ratio | χ 2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| nhm4-1 | nhm4-2 | nhm4-1 | nhm4-2 | nhm4-1 | nhm4-2 | nhm4-1 | nhm4-2 | nhm4-1 | nhm4-2 | |
| P1(WT) | 50 | 50 | 0 | 0 | 50 | 50 | ||||
| P2(nhm4-1 or nhm4-2) | 50 | 50 | 50 | 50 | 0 | 0 | ||||
| F1(P1 × P2) | 30 | 30 | 0 | 0 | 30 | 30 | ||||
| F1(P2 × P1) | 30 | 30 | 0 | 0 | 30 | 30 | ||||
| BC1(F1 × P1) | 60 | 70 | 0 | 0 | 60 | 70 | ||||
| BC1(F1× P2) | 85 | 70 | 45 | 32 | 40 | 38 | 1.12:1 | 1.18:1 | 0.18 | 0.35 |
| F2 | 350 | 250 | 82 | 59 | 268 | 191 | 3.26:1 | 3.23:1 | 0.38 | 0.19 |
Allelism Testing
We crossed mutants nhm4-1 and nhm4-2 to detect the allelism of the two. Both F1 populations exhibited a mutant phenotype after hybridization, indicating that the mutant genes of nhm4-1 and nhm4-2 are allelic (Figure 2).
Figure 2.
Allelism test of mutant nhm4-1 and mutant nhm4-2. (A) Wild-type “FT.” (B) Mutant nhm4-1 × mutant nhm4-2. (C) Mutant nhm4-2 × mutant nhm4-1. The plants are shown at 35 days after sowing (DAS). Bar = 5 cm.
Identification of the Candidate Gene Responsible for nhm4-1
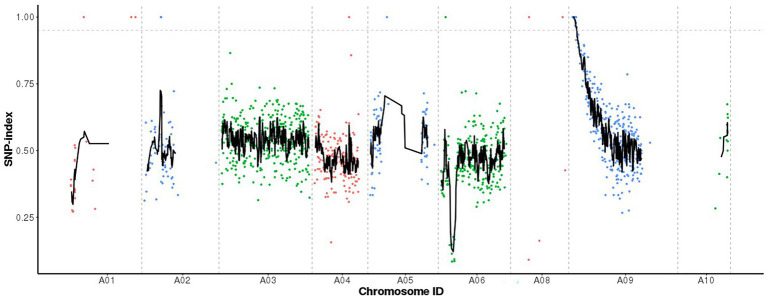
The wild-type “FT,” the mutant nhm4-1, and a mutant pool containing 50 homozygous recessive F2 mutant plants were resequenced, resulting in 94,719,968, 118,123,996, and 270,862,892 clean reads, respectively. A total of 98.00, 98.43, and 98.91% of the clean reads in the wild-type “FT,” mutant nhm4-1, and mutant pools, respectively, were mapped to the Chinese cabbage v. 3.0 reference genome (BRAD1). Based on the alignment to the reference genome sequence, the mutation analysis software GATK (McKenna et al., 2010) was used to extract all potential polymorphic SNP sites in the genome. Circos software was used to draw the variation information on the genome, and it was found that SNP was mainly distributed on chromosome A03 and A09 (Supplementary Figure S1). This was followed by further filtering and screening, after which 1,587 high-quality SNPs were obtained.
When the SNP index was 0.95 as the threshold, we located a 1.59 Mb (867,020-2,457,084) candidate region on chromosome A09 (Figure 3). Five SNP mutations occurred in the exon, of which only two SNPs (SNP A09, 900,112 and SNP A09, 1,723,490) caused non-synonymous amino acid changes (Table 2). SNP A09, 900,112 (C–T) was located in the BraA09g001440.3C and SNP A09, 1,723,490 (G–A) was located in the BraA09g002790.3C.
Figure 3.
Identification of candidate gene by MutMap. The X-axis represents ten chromosomes, the Y-axis represents the SNP index value, and the dotted pink line is the index threshold (0.95).
Table 2.
Candidate SNP information from the candidate region.
| Chromosome | Position | Gene ID | Variation base | SNP index | Exon ID | Mutation type | Annotation information |
|---|---|---|---|---|---|---|---|
| A09 | 900,112 | BraA09g001440.3C | C–T | 1 | Exon 7 | Non-synonymous SNV | ent-copalyl diphosphate synthase |
| A09 | 1,639,305 | BraA09g002590.3C | C–T | 1 | Exon 1 | synonymous SNV | E3 ubiquitin-protein ligase RMA1 |
| A09 | 1,723,490 | BraA09g002790.3C | G–A | 1 | Exon 1 | Non-synonymous SNV | Cell division cycle protein 123 homolog |
| A09 | 2,003,067 | BraA09g003180.3C | G–A | 0.98 | Exon 1 | synonymous SNV | Succinate dehydrogenase |
| A09 | 2,193,985 | BraA09g003550.3C | G–A | 0.96 | Exon 9 | synonymous SNV | – |
KASP Analysis
To confirm the candidate SNP, primers were designed based on the mutation information of these two SNPs and then applied to F2 populations. Genotyping analysis was performed using KASP, and the association between these two SNPs and the mutant phenotypes was verified. The genotypic assay showed that SNP A09, 900,112 of BraA09g001440.3C was the T:T genotype in the 48 mutant phenotypic plants and the C:T or C:C genotypes in the 136 wild-type phenotypic plants (Supplementary Table S4), implying that SNP A09, 900,112 co-segregated with the mutant phenotype. However, a recombinant was found at SNP A09, 1,723,490 of BraA09g002790.3C. The A:A genotype and the A:G genotype were detected in the mutant phenotypic plants. Thus, this SNP did not co-segregate with the mutant phenotype. These results confirm that BraA09g001440.3C harbors SNP A09, 900,112, and is the most likely candidate gene of the nhm4-1 mutant. Gene annotation indicates that BraA09g001440.3C is a homologous gene of Arabidopsis CPS1 (At4g02780) and encodes the CPS1 enzyme, which catalyzes the conversion of geranylgeranyl pyrophosphate (GGPP) to copalyl pyrophosphate (CPP) in GA biosynthesis. In this study, the candidate gene of the nhm4-1 mutant is referred to as BrCPS1.
Cloning and Sequence Analysis of BrCPS1
Since MutMap and KASP analyses supported BrCPS1 as the most likely candidate gene of Brnhm4-1, we cloned the coding sequence of BrCPS1 from the wild-type “FT,” mutant nhm4-1, and mutant nhm4-2. Gene annotation showed that BrCPS1 was 7,928 bp in length and consisted of 15 exons (Figure 4). Sequence alignment of the cDNAs and deduced amino acids between the wild-type and mutants is shown in Figure 5. Sequence comparison showed that a single base substitution occurred at position A09, 900,112 (C to T) in nhm4-1, resulting in an amino acid to change from leucine (L) to phenylalanine (F). However, the BrCPS1 clone in mutant nhm4-2 plants showed a single-nucleotide mutation in the fourth exon (A09, 898,783; G–A), resulting in an amino acid to change from glycine (G) to aspartic acid (D). This differs from the mutation site of the nhm4-1 mutant, suggesting that nhm4-1 and nhm4-2 are allelic mutations of the same BrCPS1 gene.
Figure 4.
Gene structure of BraA09g001440.3C. The blue boxes represent exons and the black lines represent introns.
Figure 5.
Alignment of coding and amino acid sequences of BrCPS1. The red frames show the sites where mutant nhm4-1 and mutant nhm4-2 occurred non-synonymous mutations.
In this study, we also cloned the candidate gene BraA09g002790.3C (SNP A09, 1,723,490) in mutants nhm4-1 and nhm4-2. There was a single base mutation (G–A) in nhm4-1 at position A09, 1,723,490, which is consistent with the MutMap results. There was no change in the mutant nhm4-2. These results further confirm that BrCPS1 is the gene responsible for the non-heading phenotype.
Enzyme Activity Assays
To investigate whether the activity of CPS1 was changed in mutant nhm4-1, we measured the activity of CPS1 in wild-type and mutant nhm4-1. The activity of CPS1 was significantly decreased in the mutant nhm4-1 compared with the wild-type (Supplementary Figure S2).
Analysis of BrCPS1 Expression Patterns by qRT-PCR
To investigate whether the mutation site affects gene expression, we used qRT-PCR to analyze the expression level of BrCPS1 in the cotyledons, true leaves, rosette leaves, and heading leaves of wild-type “FT,” mutant nhm4-1, and mutant nhm4-2 plants. The expression levels of BrCPS1 were reduced at all stages of leaf development in both mutants, especially in the rosette leaves, which differ from wild-type “FT” results (Figure 6).
Figure 6.
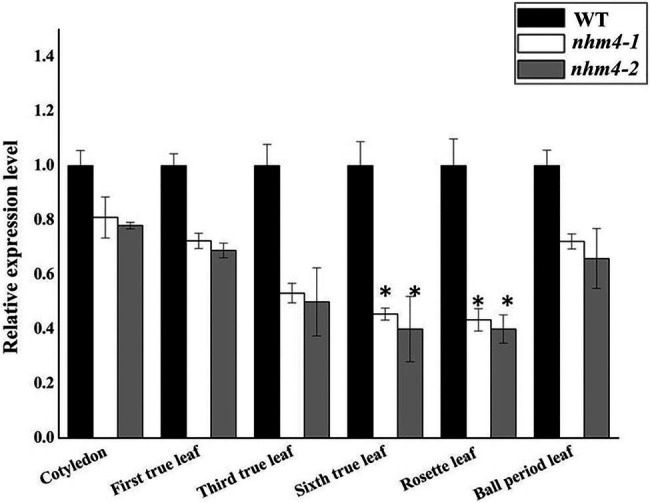
The expression levels of BrCPS1. Expression levels of genes in wild-type “FT” were used as reference for relative expression. * refers to significant differences in expression levels at p < 0.05 (Student’s t-test).
Analysis of GA Content in Mutants nhm4-1 and nhm4-2
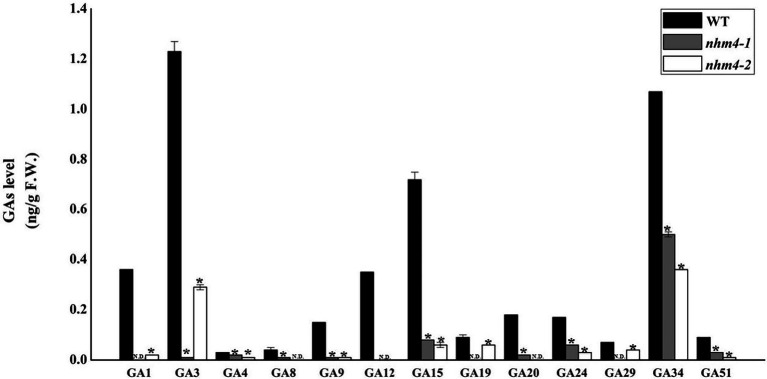
We determined the GA content of 18 endogenous GAs (GA1, GA3, GA4, GA5, GA6, GA7, GA8, GA9, GA12, GA15, GA19, GA20, GA24, GA29, GA34, GA44, GA51, and GA53) in the leaves of wild-type and mutant plants. GA5, GA6, GA7, GA44, and GA53 were not detected in either the wild-type or mutant strains (Figure 7). GA12 was detected only in the wild-type and not in either of the mutants. The levels of GAs (GA9, GA15, GA20, and GA24) in the bioactive GA biosynthesis pathway were significantly decreased in the mutants than in the wild-type.
Figure 7.
Determination of endogenous GA content in the leaves of wild-type “FT” and mutant plants. The content of GA in wild-type “FT” was used as control. * represents significant differences at p < 0.01 (Student’s t-test). N.D., not detectable.
Effects of Exogenous Application of GA3 on the Non-heading Phenotype
Based on the determination results of endogenous GA content, we investigated the responses of the mutants to exogenous GA3 application. After exogenous spraying of GA3, we observed that the leaves of the mutants had grown upward, similar to the wild-type leaves in the rosette stage, which demonstrates a tendency to form a leafy head (Figure 8). Consequently, leafy head formation is related to GA biosynthesis or the deactivation pathway.
Figure 8.
Phenotypic responses of nhm4-1 and nhm4-2 to exogenous GA3 application. Bar = 5 cm.
Discussion
Chinese cabbage is an important economic vegetable, and the formation of Chinese cabbage leafy heads significantly affects yield and quality. In this study, we identified a pair of allele non-heading mutants: nhm4-1 and nhm4-2. Based on the MutMap, KASP, and map-based cloning results, we indicated that the mutation of CPS1, a key enzyme of GA biosynthesis, causes a non-heading phenotype. These results revealed that the formation of leafy heads is related to GA biosynthesis in Chinese cabbage.
Non-heading mutants are important resources for studying the mechanism of leafy head formation in Chinese cabbage. Selecting a suitable mutagenic material is an important prerequisite for the creation of mutants. Inbred lines are commonly used as mutagenic materials in Brassica crops (Barro et al., 2001; Ferrie et al., 2008; Liu et al., 2010). However, in this study, a heading Chinese cabbage double haploid (DH) line was employed as the mutagenic material to create non-heading mutants. The genetic background of the DH line was homozygous, which is beneficial for screening mutants. Moreover, the genetic background was highly consistent between the wild-type “FT” and the mutants, and genetic differences only occurred at the mutation sites, which is helpful for functional genomics investigation in plants.
The application of allelic mutants to identify gene functions has been proven to be effective in lettuce (Huo et al., 2016), sorghum (Jiao et al., 2018), maize (Zhao et al., 2018), and rice (Lee et al., 2019). Zhang et al. (2018) identified two round-leaf mutants, rl-1 and rl-2, from an EMS mutagenic population in cucumber. The candidate for rl-1 was identified as CsPID, which encodes a Ser/Thr protein kinase. The results of map-based cloning showed that the rl-2 gene was also located on the same candidate gene CsPID, but the mutation site was different from that of rl-1. Allelism tests also confirmed that rl-1 and rl-2 were alleles. The use of allelic mutants rl-1 and rl-2 strongly proved that the CsPID gene is a candidate gene for controlling the formation of round leaves. As a reference, the plant materials in this study were two allelic mutants, and we used the same method to verify the function of the mutated gene.
Gibberellin is a diterpenoid plant hormone that has various regulatory effects on plant growth, such as germination, stem elongation, and flowering (Yamaguchi, 2008; Hedden and Thomas, 2012). CPS is the first gene that enters the GA biosynthesis pathway. Because of its key role in GA biosynthesis, many different CPS genes have been identified in species including Arabidopsis thaliana (Sun and Kamiya, 1994), Zea mays (Bensen et al., 1995), and Oryza sativa (Otomo et al., 2004; Prisic et al., 2004; Xu et al., 2004). However, in this study, based on the results of endogenous GA content determination, we speculated that the mutation of the BrCPS1 gene caused the inability to produce and accumulate bioactive GAs (GA1, GA3, GA4, and GA7) in the leaves, which made the leaves appear to grow geotropically and thus could not form leafy heads. We conducted exogenous GA3 spraying experiments on the mutants, and found that after exogenous supplementation of GA3, the leaves of mutant grew upward similar to the wild-type at the rosette stage, which proved it has a tendency to form the leafy head and GA3 played a role in the process of leafy head formation, and the upward leaves are the necessary process of leafy head formation. Consistent with our previous research, the mutation of KS, an important enzyme, catalyzes the second step in the cyclization of GGPP into ent-kaurene in the GA synthesis pathway, also led to a non-heading phenotype of Chinese cabbage (Gao et al., 2020). Both mutated genes are key enzymes in the starting site of GA biosynthesis. Mutations in both genes resulted in decreased GA content in the mutants, and inhibited the leafy head formation, further demonstrating that the GA content in the leaves is an important factor affecting leafy head formation.
In general, combined with the results of previous studies, we believe that the important enzymes involved in the synthesis of GA have mutated, hindering the biosynthesis of GA and resulting in the inability to produce and accumulate biologically active GAs. The lack of GA in Chinese cabbage leaves prevents the formation of leafy heads. These results can contribute to further our understanding of the molecular mechanisms of Chinese cabbage leafy head formation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, SAMN26241760, https://www.ncbi.nlm.nih.gov/, SAMN26241761, https://www.ncbi.nlm.nih.gov/, SAMN26241762.
Author Contributions
YG and GQ have equally contributed to this study. HF and YG designed the experiments. YG conducted the experiments, performed the data analysis, and wrote the manuscript. GQ, MZ, and ZL helped create the mutants. WF and SH assisted in the date analysis. HF revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by the National Natural Science Foundation of China (Grant No. 31730082).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank editage (www.editage.cn) for English language editing.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.889798/full#supplementary-material
References
- Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., Kanzaki H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178. doi: 10.1038/nbt.2095, PMID: [DOI] [PubMed] [Google Scholar]
- Barro F., Fernandez-Escobar J., De La Vega M., Martin A. (2001). Doubled haploid lines of Brassica carinata with modified erucic acid content through mutagenesis by EMS treatment of isolated microspores. Plant Breed. 120, 262–264. doi: 10.1046/j.1439-0523.2001.00602.x [DOI] [Google Scholar]
- Bensen R. J., Johal G. S., Crane V. C., Tossberg J. T., Schnable P. S., Meeley R. B., et al. (1995). Cloning and characterization of the maize An1 gene. Plant Cell 7, 75–84. doi: 10.1105/tpc.7.1.75, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. L., Fu X. M., Liu J. Q., Ye T. T., Hou S. Y., Huang Y. Q., et al. (2012). Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 905, 67–74. doi: 10.1016/j.jchromb.2012.08.005, PMID: [DOI] [PubMed] [Google Scholar]
- Cheng F., Sun R., Hou X., Zheng H., Zhang F., Zhang Y., et al. (2016). Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 48, 1218–1224. doi: 10.1038/ng.3634, PMID: [DOI] [PubMed] [Google Scholar]
- Ferrie A. M. R., Taylor D. C., MacKenzie S. L., Rakow G., Raney J. P., Keller W. A. (2008). Microspore mutagenesis of Brassica species for fatty acid modifications: a preliminary evaluation. Plant Breed. 127, 501–506. doi: 10.1111/j.1439-0523.2008.01502.x [DOI] [Google Scholar]
- Gao Y., Huang S., Qu G., Fu W., Zhang M., Liu Z., et al. (2020). The mutation of ent-kaurene synthase, a key enzyme involved in gibberellin biosynthesis, confers a non-heading phenotype to Chinese cabbage (Brassica rapa L. ssp. pekinensis). Hortic. Res. 7:178. doi: 10.1038/s41438-020-00399-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L. W., Lyu S. W., Tang J., Zhou D. Y., Bonnema G., Xiao D., et al. (2017). Genome-wide analysis of auxin transport genes identifies the hormone responsive patterns associated with leafy head formation in Chinese cabbage. Sci. Rep. 7:42229. doi: 10.1038/srep42229, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. K., Xue W. X., Sun Y. D., Yu X. H., Liu P. L. (2000). Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Res. 10, 151–160. doi: 10.1038/sj.cr.7290044, PMID: [DOI] [PubMed] [Google Scholar]
- Hedden P., Thomas S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. doi: 10.1042/BJ20120245 [DOI] [PubMed] [Google Scholar]
- Huo H., Henry I. M., Coppoolse E. R., Verhoef-Post M., Schut J. W., de Rooij H., et al. (2016). Rapid identification of lettuce seed germination mutants by bulked segregant analysis and whole genome sequencing. Plant J. 88, 345–360. doi: 10.1111/tpj.13267, PMID: [DOI] [PubMed] [Google Scholar]
- Ito H., Kato T. (1957). Studies on the head formation of Chinese cabbage: histological and physiological studies of head formation. J. Jpn. Soc. Hortic. Sci. 26, 154–162. doi: 10.2503/jjshs.26.154 [DOI] [Google Scholar]
- Jiao Y., Burow G., Gladman N., Acosta-Martinez V., Chen J., Burke J., et al. (2018). Efficient identification of causal mutations through sequencing of bulked F2 from two allelic bloomless mutants of Sorghum bicolor. Front. Plant Sci. 8:2267. doi: 10.3389/fpls.2017.02267, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Piao R., Lee Y., Kim B., Seo J., Lee D., et al. (2019). Identification and characterization of LARGE EMBRYO, a new gene controlling Embryo size in Rice (Oryza sativa L.). Rice. 12:22. doi: 10.1186/s12284-019-0277-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010). Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang X., Lu Y., Feng D., Gu A., Wang S., et al. (2019). Characterization of non-heading mutation in heading Chinese cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 10:112. doi: 10.3389/fpls.2019.00112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wang J., Huang T., Wang F., Yuan F., Cheng X., et al. (2010). A missense mutation in the VHYNP motif of a DELLA protein causes a semi-dwarf mutant phenotype in Brassica napus. Theor. Appl. Genet. 121, 249–258. doi: 10.1007/s00122-010-1306-9, PMID: [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mao Y., Wu F., Yu X., Bai J., Zhong W., He Y. (2014). MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 164, 710–720. doi: 10.1104/pp.113.228007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo K., Kenmoku H., Oikawa H., König W. A., Toshima H., Mitsuhashi W., et al. (2004). Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J. 39, 886–893. doi: 10.1111/j.1365-313X.2004.02175.x, PMID: [DOI] [PubMed] [Google Scholar]
- Prisic S., Xu M., Wilderman P. R., Peters R. J. (2004). Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 136, 4228–4236. doi: 10.1104/pp.104.050567, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Wu F., Bai J., Li X., Yang X., Xue W., et al. (2020). BcpLH organizes a specific subset of microRNAs to form a leafy head in Chinese cabbage (Brassica rapa ssp. pekinensis). Hortic Res. 7:1. doi: 10.1038/s41438-019-0222-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T. P., Kamiya Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6, 1509–1518. doi: 10.1105/tpc.6.10.1509, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. doi: 10.1093/nar/gkq603, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu F., Bai J., He Y. (2014). BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 12, 312–321. doi: 10.1111/pbi.12138, PMID: [DOI] [PubMed] [Google Scholar]
- Xu M., Hillwig M. L., Prisic S., Coates R. M., Peters R. J. (2004). Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 39, 309–318. doi: 10.1111/j.1365-313X.2004.02137.x, PMID: [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804 [DOI] [PubMed] [Google Scholar]
- Yu J., Gao L., Liu W., Song L., Xiao D., Liu T., et al. (2019). Transcription Coactivator ANGUSTIFOLIA3 (AN3) regulates leafy head formation in Chinese cabbage. Front. Plant Sci. 10:520. doi: 10.3389/fpls.2019.00520, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Chen F., Zhao Z., Hu L., Liu H., Cheng Z., et al. (2018). Mutations in CsPID encoding a Ser/Thr protein kinase are responsible for round leaf shape in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 131, 1379–1389. doi: 10.1007/s00122-018-3084-8, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liang J., Cai X., Chen H., Wu J., Lin R., et al. (2021). Divergence of three BRX homoeologs in Brassica rapa and its effect on leaf morphology. Hortic Res. 8:68. doi: 10.1038/s41438-021-00504-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhang Y., Wang L., Wang X., Xu W., Gao X., et al. (2018). Mapping and functional analysis of a maize Silkless mutant sk-A7110. Front. Plant Sci. 9:1227. doi: 10.3389/fpls.2018.01227, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, SAMN26241760, https://www.ncbi.nlm.nih.gov/, SAMN26241761, https://www.ncbi.nlm.nih.gov/, SAMN26241762.