Abstract
Noised-induced hearing loss (NIHL) is an acquired, progressive neurological damage caused by exposure to intense noise in various environments including industrial, military and entertaining settings. The prevalence of NIHL is much higher than other occupational injuries in industrialized countries. Recent studies have revealed that genetic factors, together with environmental conditions, also contribute to NIHL. A group of genes which are linked to the susceptibility of NIHL had been uncovered, involving the progression of oxidative stress, potassium ion cycling, cilia structure, heat shock protein 70 (HSP70), DNA damage repair, apoptosis, and some other genes. In this review, we briefly summarized the studies primary in population and some animal researches concerning the susceptible genes of NIHL, intending to give insights into the further exploration of NIHL prevention and individual treatment.
Keywords: genes, noised-induced hearing loss, noise prevention, susceptibility, genetic variants
Introduction
The auditory system helps people to hear sound, understand language, and even distinguish people or objects by recognizing different sounds. Any organic or functional impairment of the auditory pathway can lead to hearing impairment. According to a WHO report (Castaneda et al., 2019), more than 100 million people in East Asia are at risk of disabling hearing loss, leading to lifelong disability, and deafness has become one of the major problems affecting their life quality. Sensorineural hearing loss may be caused by pathological changes in the Corti’s organ of the inner ear, the auditory nerve, or the auditory cortex. It is characterized by the impairment of sound perceptive and analytic ability, and classified as drug-induced hearing loss, presbycusis, hereditary hearing loss, noise-induced hearing loss (NIHL) and others. Although cochlear implant technology has been increasingly advanced in the treatment of hearing loss, its therapeutic effect varies with different lesion sites, therefore, sensorineural hearing loss remains one of the most challenging medical problems.
Noise pollution is one of the seven public hazards in modern society. NIHL, one of the hot spots of social concern, is the second cause of hearing loss in adults, and more than 6% of the global population is affected by NIHL according to WHO data (Sliwinska-Kowalska and Zaborowski, 2017). NIHL is an acquired hearing loss caused by long-term exposure of the auditory system to noise generated by construction, entertainment, industrial production, military equipment or others, and its incidence is only behind presbycusis among all the types of sensorineural hearing loss (Miao et al., 2019). The A-frequency weighting network (dBA) is normally utilized to measure the levels of noise in decibels (dB) of sound pressure, indicating the risk of NIHL (Varela-Nieto et al., 2020). Moreover, the principal requirements for the diagnosis of NIHL are high-frequency hearing impairment, jeopardous amount of noise exposure and recognizable high-frequency audiometric notch or bulge (Coles et al., 2000). Addition to the auditory symptoms such as hearing descending, hearing allergy, tinnitus, the noise damage may also present as mental disorder, digestive disorder or some other organic dysfunction (Skogstad et al., 2016; Hahad et al., 2019).
Long-term noise exposure can lead to damage of peripheral auditory system, including the structure of cochlea hair cells, cilia, supporting cells, and tectorial membrane (Wang et al., 2002), hitting the external layer of hair cells the most, and the Corti’s organ and spiral ganglion may also undergo degenerative changes (Henderson et al., 2006). The main manifestations of which are increased hearing threshold, decreased auditory sensitivity and speech resolution, tinnitus, and auditory hypersensitivity. A “V”-shaped depression appears at 4k Hz on the audiogram, which is called “V”-shaped notch hearing loss (Carroll et al., 2017). Low intensity or short time noise exposure can cause temporary changes of the auditory nerve synaptic transmitter, resulting in temporary hearing loss which could return to normal after the noise ceased, in terms of temporary threshold shift (TTS) (Kurabi et al., 2017). High intensity or long-time noise exposure causes damages on both hair cells and auditory nerve, resulting in hearing loss that could not be restored, which is called permanent threshold shift (PTS), and eventually leads to sensorineural hearing loss (Liberman, 2016).
Long-term noise exposure may also cause damage to the central auditory system, which mainly occurred in the cochlear nucleus, olivary nucleus, medial geniculate body, inferior colliculus, hippocampus and auditory cortex (Kujawa and Liberman, 2009; Eggermont, 2017). Most previous studies believed that the auditory cortex was the most vulnerable part under noise exposure, but Cheng et al. (2016) showed that the hippocampus may be more sensitive than the auditory cortex, mainly manifested as headache, dizziness, irritability, insomnia, memory loss and even serious mental problems (Eraslan et al., 2015). Long-term noise exposure can increase the expression of corticotropin releasing-hormone (CRH) in the hippocampus and decrease the inhibition of the hypothalamic-pituitary-adrenal (HPA) axis, which may worsen depression and anxiety (Valentino et al., 2010).
In this study, we searched papers published in English and Chinese via PubMed, Embase, Scopus, and Web of Science database, intending to provide an overview of current knowledge relevant to the pathogenesis and susceptibility genes to NIHL.
Pathogenesis of Noised-Induced Hearing Loss
Environmental and genetic factors can both contribute to NIHL. Environmental factors include noise intensity, noise spectrum characteristics, noise exposure time, etc. Genetic factors mainly refer to NIHL susceptibility genes. Presently, there are four main theories about the pathogenesis of NIHL, including mechanical theory, vascular theory, metabolic theory, and immunoinflammatory theory.
Mechanical Theory
According to the mechanical theory, the internal tissue structure damage of cochlea caused by noise with over 130 dB intensity is mainly attributed to the mechanical damage (Patterson and Hamernik, 1997). High intensity noise impacts the Corti’s organ and forms a strong liquid eddy current in the cochlear duct, which can cause the rupture of the vestibular membrane and lead to the fusion of endolymph and perilymph. The cytotoxic K+ in endolymph could reach the tympanic scala through the orifice of the cupula cochleae and then reach the lymphatic space of the Corti’s organ, where the contact of K+ with hair cells leads to the destruction of cochlear sensory epithelial cells, atrophy of stria vascularis and degeneration of auditory nerve fibers. The other ways of mechanical injury were the rupture of the reticular laminae of the basilar membrane or the separation of the stereocilium of the outer hair cells from the cuticular plate, which can cause the K+-rich endolymph to come into contact with the hair cells. In more serious cases, noise-induced mechanical force can also cause the Corti’s organ to peel off from the basilar membrane (Spoendlin and Brun, 1973; Rajguru, 2013).
Vascular Theory
Vascular theory believes that long-term strong noise exposure may lead to vasoconstriction around the cochlear sensory epithelium, swelling vascular endothelial cell, narrowing vascular lumen, slowed blood flow velocity, decrease in local blood perfusion, increase in blood viscosity, accumulation of platelets and red blood cells in capillaries, and obvious thickening of capillary walls. All of the aforementioned factors may ultimately lead to cochlear ischemia and hypoxia, resulting in decreased activity of otoprotective enzymes, accumulation of cellular metabolites in cells, and damage to cochlear hair cells and the Corti’s organ (Kim et al., 2018). Significant inner ear injury occurs when the perilymph oxygen partial pressure decreases by about 20% (Wu et al., 2014).
Metabolic Theory
According to the metabolic theory, noise exposure could lead to extensive metabolic changes in the auditory system. Overexpression of free radicals including reactive oxygen species (ROS) and reactive nitrogen species (RNS) in cochlea leads to the formation of lipid peroxides and accelerates hair cells apoptosis (Zhang et al., 2017). The isoconstrictive vascular substances such as isoprostaglandin and 8-iso-prostaglandin F2α could also be released from cochlear vascular system and the Corti’s organ (Honkura et al., 2016). Strong noise exposure results in abnormal influx of K+ ions, leading to depolarization of membrane potential and abnormal influx of Ca2+, which is termed as calcium overload (Vicente-Torres and Schacht, 2006; Wang et al., 2007a). Excitotoxicity caused by large amount of glutamate release leads to edema and vacuolation of inner hair cells, neurotrophic factor deficiency, and mitochondrial dysfunction, inducing acute hair cell damage (Fridberger et al., 1998). Besides, cytokines and chemokines such as tumor necrosis factor (TNF-α), IL-6 and IL-1β are upregulated, which make contributions to the cascading amplification of exogenous and endogenous apoptotic signaling pathways, promoting the release of pro-apoptotic proteins, leading to the activation of Caspase-3, chromatin concentration, and DNA damage. Le Prell et al. (2007) believed that metabolic injury played a key role in the pathogenesis of NIHL.
Immunoinflammatory Theory
Macrophages are the main natural immune cells in the cochlea and are important drivers of inflammation and tissue repair after noise exposure. In normal condition, cochlear macrophages inhabit spiral ligaments, spiral ganglion, basilar membrane and stria vascularis (Shi, 2010). The distribution, phenotype, number, morphology and functional state of cochlear macrophages were significantly changed after noise exposure (He W. et al., 2020). Due to the existence of tight junctions, the infiltration of macrophages and monocytes into the scala media is mainly confined to the scala tympani cavity beneath the basilar membrane, avoiding the damage and apoptosis of hair cells (Frye et al., 2018). After noise exposure, signaling pathway such as Toll-like receptor 4 (TLR-4)/nuclear factor kappa-B (NF-κB) and mitogen-activated protein kinase (MAPK)/c-Jun N-terminal kinase (JNK) were activated in cochleae, leading to upregulation of downstream inflammatory factors and chemokines including TNF-α, IL-6, IL-12, IL-1β, intercellular cell adhesion molecule-1 (ICAM-1), and monocyte chemoattractant protein-1 (MCP-1) (Wang et al., 2007b; Zhang G. et al., 2019). The release of theses cytokines and chemokines set off chain inflammatory reactions (Frye et al., 2019).
Differences in Individual Susceptibility to Noised-Induced Hearing Loss
Noised-induced hearing loss (NIHL) is ranked as the highest incidence of industrial injury in the United States (Ralli et al., 2017), where 16% adult hearing loss are caused by exposure to industrial noise. While NIHL is a typical type of hearing loss, the causes of it are attributable to both environmental and genetic factors. Long-term exposure to noise is a prominent environmental factor for NIHL, but some studies found that not every worker who exposed to the same level of noise would develop NIHL, and the severity of NIHL also varies greatly (Irion, 1981; Hood, 1987). Taylor et al. (1965) detected the hearing threshold of workers in a textile factory and found that the workers with similar length of service had different hearing threshold which ranged from 10 to 70 dB.
In recent years, with studies of large-scale samples, it is known that even for the subjects exposed to the noise environment with similar density and duration, their hearing threshold shifts has significant individual differences (Lu et al., 2005). It reveals that there is a great difference in the susceptibility to NIHL among the population.
Research Methods of Noised-Induced Hearing Loss Susceptible Gene
After a comprehensive analysis of some experimental studies, we summarized the methods of population research for NIHL susceptible genes as follows: sufficient number of subjects with history of noise exposure were selected as the research object, strict inclusion criteria were established, and the population whose hearing threshold locates higher than 25dB was recruited into case group, whose hearing thresholds was less than or equal to 25dB was selected into control group. The candidate genes of the two groups were detected by implementing case-control study.
There are mainly three methods for the selection of candidate NIHL genes: (1) selection of genes that have been preliminarily confirmed in animal models (2) selection of susceptibility genes that have been reported in other types of deafness; and (3) according to the pathogenic mechanism of NIHL, detect relevant genes in the corresponding pathways.
At present, the techniques for detecting susceptible genes include microarray chip, polymerase chain reaction (PCR) -restriction enzyme digestion, quantitative reverse transcription PCR, amplification refractory mutation system (ARMS)-PCR, high-throughput sequencing, and whole-exome sequencing (WES) etc. Genetic screening was carried out and compared between the two population to determine the susceptible genes which might have important influence on the pathogenesis and development of NIHL.
Susceptibility Genes of Noise-Induced Hearing Loss
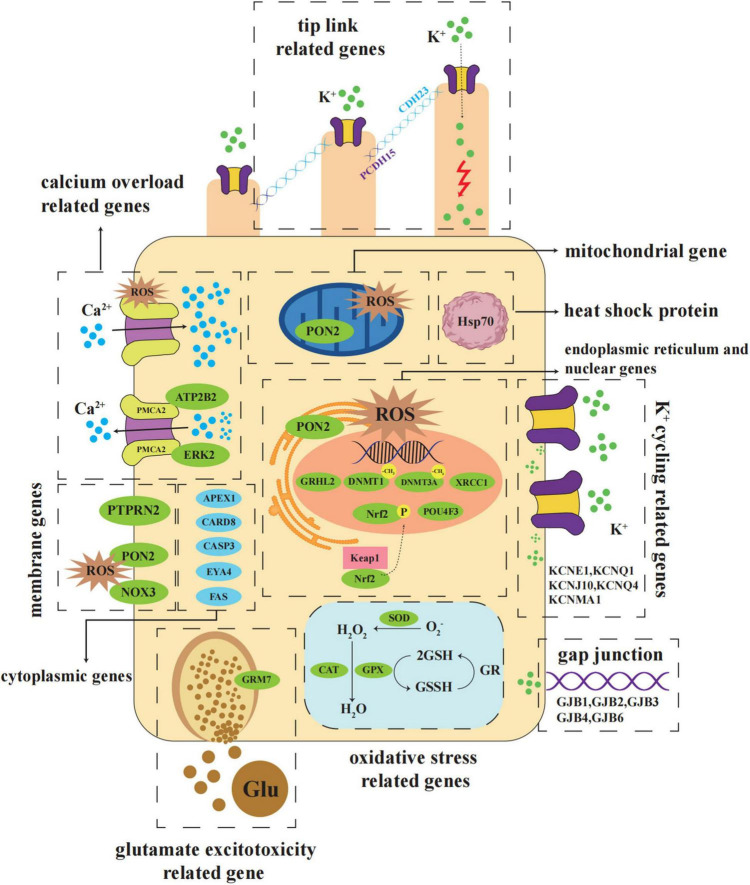
According to the pathogenesis of NIHL, recent studies have revealed a large group of genes that are linked to NIHL involving oxidative stress, potassium ion cycling, cilia structure, heat shock protein genes 70, DNA damage repair, apoptosis, monogenic NIHL genes and others. The distribution of major susceptibility genes and the functions they are involved in is shown in Figure 1. The summary of NIHL susceptible genes and their locus is concluded in Table 1.
FIGURE 1.
Schematic diagram of major NIHL susceptible genes distributed in the hair cells. NIHL susceptible genes are involved in the progression of oxidative stress, potassium ion cycling, calcium overload, glutamate excitotoxicity, DNA damage repair, apoptosis, and other biochemical processes. They are distributed in various locations in cells, including membrane, cytoplasm, nucleus, mitochondria, and endoplasmic reticulum. Abbreviations: Glu, Glutamate; GR, Glutathione reductase; GSH, Glutathione; GSSH, Glutathione oxidized; Keap1, Kelch-like ECH associated protein 1; PMCA2, Plasma membrane calcium-transporting ATPase 2; ROS, Reactive oxygen species.
TABLE 1.
Summary of NIHL susceptible genes and their locus.
| Groups of genes | Gene | Full name | Genetic locus | References |
| Antioxidant genes | APEX1 | Apurinic/Apyrimidinic endodeoxyribonuclease 1 | rs1130409, rs1760944 | Shen et al., 2016; Ding et al., 2019 |
|
ATP2B2 (PMCA2) |
ATPase plasma membrane Ca2+ transporting 2 | rs1719571, rs3209637, rs14154 | Kozel et al., 2002; Li X. et al., 2016; Yan et al., 2013; Zhang S. et al., 2019 | |
| CAT | Catalase | rs769217, rs208679, rs7943316, rs769214, rs475043, rs12273124, rs494024, rs564250 | Konings et al., 2007; Xia et al., 2011; Yang et al., 2015; Li T. et al., 2020 | |
| GPX1 | Glutathione peroxidase 1 | rs1987628 | Wen et al., 2014; Li J. Y. et al., 2020 | |
| GST | Glutathione S-transferase | rs1695, rs1049055, rs10712361 | Lin et al., 2009; Shen et al., 2012; Loukzadeh et al., 2019; Zong et al., 2019; Li Y. H. et al., 2020 | |
| NFE2L2 (NRF2) | Nuclear factor erythroid 2-related factor 2 | rs77684420, rs6726395, rs1962142, rs6721961 | Honkura et al., 2016; Wang et al., 2019 | |
| NOX3 | NADPH Oxidase 3 | rs12195525, rs33652818 | Lavinsky et al., 2015; Xin et al., 2021 | |
| PON2 | Paraoxonase 2 | rs12026, rs7785846, rs12704796, rs987539, rs7493, rs7786401 | Cao et al., 2013; Li X. et al., 2016; Bhatt et al., 2020; Wu et al., 2020; Zhou H. et al., 2020 | |
| SOD1 | Superoxide dismutase 1 | rs2070424, rs10432782 | Liu et al., 2010b | |
| SOD2 | Superoxide dismutase 2 | rs4880, rs2855116 | Ohlemiller et al., 1999; Fortunato et al., 2004; Liu et al., 2010a; Wang et al., 2014; Wang J. et al., 2017 | |
| Potassium ion cycling related genes | KCNQ1 | Potassium voltage-gated channel subfamily Q member 1 | rs800336, rs2056892, rs2011750, rs2283158, rs2283179, rs2283205, rs231899, rs760419, rs163171, rs8234, rs7945327, rs11022922, rs718579, rs463924 | Van Laer et al., 2006; Pawelczyk et al., 2009; Ding et al., 2020 |
| KCNQ4 | Potassium voltage-gated channel subfamily Q member 4 | rs34287852, rs2769256, rs727146, rs4660468, rs12143503, rs4660470 | Van Laer et al., 2006; Pawelczyk et al., 2009; Guo et al., 2018b; Zhou W. H. et al., 2020 | |
| KCNE1 | Potassium voltage-gated channel subfamily E regulatory subunit 1 | rs915539, rs2070358, rs1805127, rs1805128 | Van Laer et al., 2006; Ding et al., 2020 | |
| KCNJ10 | Potassium voltage-gated channel subfamily J member 10 | rs1130183, rs1186675 | Van Laer et al., 2006; Pawelczyk et al., 2009; Bhatt et al., 2020 | |
| KCNMA1 | Potassium calcium-activated channel subfamily M alpha 1 | rs696211, rs1436089 | Konings et al., 2009b; Zhang X. et al., 2019 | |
| GJB1 (Cx32) | Gap Junction Protein Beta 1 | rs747181, rs1997625 | Van Laer et al., 2006; Pawelczyk et al., 2009 | |
| GJB2 (Cx26) | Gap Junction Protein Beta 2 | rs3751385, rs5030700, rs137852540 | Van Laer et al., 2006; Pawelczyk et al., 2009; Zhou et al., 2016 | |
| GJB3 (Cx31) | Gap Junction Protein Beta 3 | rs476220 | Van Laer et al., 2006 | |
| GJB4 (Cx30.3) | Gap Junction Protein Beta 4 | rs1998177, rs755931 | Van Laer et al., 2006; Pawelczyk et al., 2009 | |
| GJB6 (Cx30) | Gap Junction Protein Beta 6 | rs945370, rs2065796, rs2065797 | Van Laer et al., 2006 | |
| SLC12A2 | Solute carrier family 12 member 2 | rs1962291, rs1560637, rs790153, rs790156, rs10089 | Van Laer et al., 2006 | |
| Cilia structure related genes | CDH23 | Cadherin related 23 | rs1227049, rs1227051, rs3802711, rs3752752, rs41281334 | Yang et al., 2006; Kowalski et al., 2014; Yu et al., 2016; Bhatt et al., 2020; Jiao et al., 2020; Jiang et al., 2021 |
| PCDH15 | Protocadherin related 15 | rs11004085, rs7095441, rs1100085, rs10825122, rs1930146, rs2384437, rs4540756, rs2384375 | Konings et al., 2009b; Xu et al., 2017a,b | |
| MYH14 | Myosin heavy chain 14 | rs667907, rs588035 | Konings et al., 2009b; Fu et al., 2016 | |
| Heat shock protein genes 70 | HSPA1A | Heat shock protein family A member 1A | rs1043618, rs1061581 | Li et al., 2017 |
| HSPA1B | Heat shock protein family A member 1B | rs2763979 | Konings et al., 2009a; Chang et al., 2011 | |
| HSPA1L | Heat shock protein family A member 1L | rs2075800, rs2227956 | Chang et al., 2011; Li Y. H. et al., 2016; Li et al., 2017 | |
| DNA damage repair related genes | DNMT1 | DNA methyltransferase 1 | rs2228611 | Guo et al., 2018a |
| DNMT3A | DNA methyltransferase 3 alpha | rs749131, rs1550117 | Guo et al., 2018a | |
| EYA4 | EYA transcriptional coactivator and phosphatase 4 | rs3777781, rs212769, rs3813346, rs9321402, rs9493627 | Zhang et al., 2015; Yang Q. et al., 2016; Yang et al., 2017 | |
| OGG1 | 8-Oxoguanine DNA glycosylase | rs1052133 | Shen et al., 2014 | |
| Apoptosis related genes | CASP3 | Caspase 3 | rs1049216, rs6948 | Wu et al., 2017 |
|
ERK2 (MAPK1) |
Extracellular signal-regulated kinase 2 | Null (animal experiment) | Kurioka et al., 2015 | |
|
JNK1 (MAPK8) |
C-Jun N-terminal kinases 1 | rs11598320, rs8424 | Sun et al., 2021 | |
| Other NIHL susceptible genes | AUTS2 | Activator of transcription and developmental regulator | rs35075890 | Niu et al., 2021 |
| CARD8 | Caspase recruitment domain family member 8 | rs2043211 | Miao et al., 2021 | |
|
DFNA5 (GSDME) |
Gasdermin E | rs2521758 | Zhang et al., 2015 | |
| FAS | Fas cell surface death receptor | rs1468063, rs2862833 | Xu et al., 2021 | |
| FOXO3 | Forkhead box O3 | rs2802292, rs10457180, rs12206094 | Guo et al., 2017, Guo et al., 2018c; Jiao et al., 2017 | |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | rs6489721 | Wan et al., 2020 | |
| GRHL2 | Grainyhead like transcription factor 2 | rs3735715, rs1981361, rs666026, rs611419 | Li X. et al., 2013; Zhang et al., 2015; Xu et al., 2016; Yang Q. Y. et al., 2016; Li X. et al., 2020 | |
| GRM7 | Glutamate metabotropic receptor 7 | rs1485175, rs1920109, rs9826579 | Yu et al., 2018a,b | |
| HDAC2 | Histone deacetylase 2 | rs10499080, rs6568819 | Wang et al., 2021 | |
| HOTAIR | HOX transcript antisense RNA | rs4759314 | Wang B. et al., 2017 | |
| IL-6 | Interleukin 6 | rs1800795 | Braga et al., 2014 | |
| ITGA8 | Integrin subunit alpha 8 | rs10508489 | Xia et al., 2011 | |
| NCL | Nucleolin | rs7598759 | Grondin et al., 2015 | |
| NOTCH1 | Notch receptor 1 | rs3124594, rs3124603 | Ding et al., 2018 | |
| NRN1 | Neuritin 1 | rs3805789 | Liu et al., 2021 | |
| PER1 | Period circadian regulator 1 | rs2585405 | Chen et al., 2021 | |
| POU4F3 | POU class 4 homeobox 3 | rs1368402, rs891969 | Xu et al., 2016 | |
| PTPRN2 | Protein tyrosine phosphatase receptor type N2 | rs10081191 | Niu et al., 2021 | |
| SIK3 | Salt-inducible kinase 3 | rs493134, rs6589574, rs7121898 | Yin et al., 2020 | |
| STAT3 | Signal transducer and activator of transcription 3 | rs1053005 | Gao et al., 2021 | |
| TSP | Thrombospondin | Null (animal experiment) | Smeriglio et al., 2019 | |
| UBAC2 | UBA domain containing 2 | rs3825427 | Wan et al., 2022 | |
| WHRN | Whirlin | rs12339210 | Jiang et al., 2021 | |
| XPO5 | Exportin 5 | rs11077 | Wang et al., 2020 | |
| XRCC1 | X-Ray repair cross complementing 1 | rs1799782 | Ding et al., 2019 |
Antioxidant Genes
According to the metabolic theory, oxidative stress plays a major role in the pathomechanisms of NIHL (Spoendlin and Brun, 1973; Rajguru, 2013; Chen et al., 2020). Mutations of oxidative stress related genes would disturb the balance of the oxidative and antioxidative system in the cochlea, thus fail to eliminate the oxidative damage of ROS, leading to the structural and functional disorders of the cochlea which ultimately result in hearing loss.
ATPase Plasma Membrane Ca2+ Transporting 2 (ATP2B2, PMCA2)
ATP2B2, encoding plasma membrane calcium-transporting ATPase isoform2 (PMCA2), is located on human chromosome region 3p25, and played an important role on intracellular calcium homeostasis (Yang et al., 2013). In an animal experiment, Kozel et al. (2002) hypothesized that Atp2b2+/– mice may be more susceptible to NIHL. Recently, Li X. et al. (2016) designed a study to investigate whether genetic variability in ATP2B2 was associated with high susceptibility to NIHL in Chinese Han nationality population. However, no significant main effect was observed for ATP2B2 gene single-nucleotide polymorphisms (SNPs) (rs1719571, rs3209637 and rs4327369) in their study because of the small sample size. In another case-control study of 760 Chinese textile workers, the results indicated that the rs3209637 C genotype of ATP2B2 may lead to a greatly increased incidence of NIHL. Meanwhile, the analysis also demonstrates that ATP2B2 SNPs (rs1719571, rs14154, and rs3209637) have a great effect on NIHL (Zhang S. et al., 2019).
Catalase
Catalase (CAT) is a ubiquitous enzyme in all organisms, functioning as a key antioxidant enzyme in the defense against oxidative stress. Catalase encoded by CAT gene can decompose hydrogen peroxide (H2O2), maintain the balance of redox in the body, and reduce the oxidative damage of cochlea caused by oxidative stress. Yang et al. (2015) screened 719 unrelated Chinese Han adults, including 225 healthy volunteers and 494 noise-exposed workers, and found that rs208679 and rs769217 SNPs were significantly associated with the susceptibility to NIHL. For rs208679 recessive effect, GG genotype showed significantly augmented risk when exposing to noise less than 85dB, while for rs769217 dominant effect, TT/TC combined genotypes significantly increased the risk of NIHL when noise intensity was between 85dB-92dB.
Glutathione Peroxidase 1
The GPX protein belongs to the glutathione peroxidase (GPx) family, which reduces H2O2 and organic hydroperoxides originated from Fenton and Haber Weiss reactions coupling with other glutathione (GSH) and GSH reductase redox systems (Evans and Halliwell, 1999). GPx oxidizes GSH into glutathione oxidized (GSSH), while glutathione reductase (GR) reduces GSSH into GSH. Moreover, H2O2 is catalyzed and broke down into H2O by GPx and CAT to achieve antioxidant effects (Figure 1). Ohlemiller et al. (2000) performed research to investigate the association between cellular Gpx1 gene and the susceptibility to NIHL in mice. The significant results revealed that Gpx-deficient mice showed increased susceptibility to NIHL. Wen et al. (2014) scrutinized the relationship between SNPs of GPX1 gene rs3448, rs1050450, rs1800668, and rs1987628, and the risk of developing NIHL among Chinese Han population. They clarified that GPX1 SNP rs1987628 may be a risk factor of NIHL. Another study of a limited sample set using genotyping kit to analyze the SNPs discovered that the individuals carrying rs1987628 GA genotype of GPX1 had a higher NIHL risk than those carrying the GG genotype (Li J. Y. et al., 2020).
Glutathione S-Transferase
Glutathione S-transferase (GST) can catalyze the binding of a variety of endogenous or exogenous compounds to reduced glutathione, which serves as an important protective antioxidant factor in the cochlea. Shen et al. (2012) analyzed the polymorphism of GST gene in 444 workers with NIHL and 445 workers with normal hearing to find out the relationship between the polymorphism and the susceptibility to NIHL. The results showed that null genotype of GSTM1 rs10712361 had a higher risk of NIHL comparing with wild-type genotype. Lin et al. (2009) found that individuals carrying all genotypes with GSTM1 null, GSTT1 null, and GSTP1 lle (Guo et al., 2017)/lle (Guo et al., 2017) were more susceptible to NIHL.
Nuclear Factor Erythroid 2-Related Factor 2
NRF2, existing widely in tissues, is a key transcription factor in the regulation of oxidative stress. When affected by oxidative stress, NRF2 dissociates from Kelch like epichlorohydrin associated protein 1 (Keap1), a negative regulator of NRF2, and is transferred to the nucleus to recognize and bind antioxidant response elements (ARE) (Sivandzade et al., 2019). Thus, the transcription of downstream antioxidant enzyme genes is initiated, including heme oxygenase 1 (HO-1), superoxide dismutase (SOD), triphosphopyridine nucleotide (NADPH), GST, GR and GPx (He F. et al., 2020). Honkura et al. (2016) explored the contribution of Nrf2 to cochlear protection via Nrf2–/– mice models. They found that Nrf2 deficiency could exacerbate NIHL as auditory brainstem response (ABR) threshold shifts of the Nrf2–/– mice was significantly larger than the wild-type mice at 7 days post-exposure. Although noise exposure does not obviously change the expression of Nrf2 target genes, the potent NRF2-activating drug, CDDO-Im used before the noise exposure could preserve the integrity of hair cells and improve post-exposure hearing level. Wang et al. (2019) found that persons with a G allele (NRF2 tagSNP rs6726395) in addition to rs77684420 and the rs6726395, rs1962142, rs6721961, and rs77684420 haplotype had associations that may be more susceptible to NIHL.
Triphosphopyridine Nucleotide Oxidase-3
The NOX family of ROS-generating NADPH oxidases consists of 7 members: NOX1 to NOX5, DUOX1 and DUOX2. In particular, NOX3 is almost exclusively expressed in the inner ear, and it has been demonstrated to generate superoxide constitutively which is converted to H2O2 by SOD, which can in turn participate in cell signaling events (Krause, 2004; Forman et al., 2010). In a previous study, a significant reduction in the intensity of NOX3 immunolabeling was observed in the inner sulcus region of the cochlea after noise exposure, and down-regulation of NOX3 may represent an endogenous protective mechanism to reduce oxidative stress in the noise-exposed cochlea (Vlajkovic et al., 2013). Xin et al. (2021) conducted a case-control study in five factories in China, and illustrated the association between rs12195525 and NIHL susceptibility. For further exploration, Lavinsky et al. (2015) verified that Nox3 is involved in NIHL susceptibility in Nox3het/Nox3het and Nox3het/ + mutant mice, which was frequency specific at 8 kHz. Besides, the significant and highly potential association of rs33652818 with ABR at 8 and 4 kHz was observed.
Paraoxonase-2
PON2 gene, localized in endoplasmic reticulum (ER), mitochondria and nuclear envelope, is involved in the process of defending ROS, ER stress, mitochondrial superoxide formation, and apoptosis (Altenhofer et al., 2010; Witte et al., 2011). Li X. et al. (2016) studied the polymorphisms of rs12026, rs7785846, and rs12704796 in PON2 in 221 patients with NIHL and 233 subjects with normal hearing by logistic regression analysis. It was found that rs12026 CG and CG + GG genotypes and rs7785846 CT and CT + TT genotypes were highly susceptible to NIHL. Wu et al. (2020) confirmed these results that PON2 gene affects the NIHL susceptibility of cochlea.
Superoxide Dismutase 1 and 2
Superoxide Dismutase (SOD) is an important antioxidant enzyme in organisms and the primary substance for scavenging ROS in the body, which is involved in the reaction of superoxide anion (O2–) and H+ to produce H2O2. It plays an important role on blocking cell damages caused by ROS and repairing the damaged cells in time. Liu et al. (2010a,b) analyzed the audiometric data of 2400 Chinese Han people exposed to occupational noise, and selected the 10% most susceptible and the 10% most resistant individuals as subjects to collect DNA samples. It has been found that the SOD1 AA genotype at the rs2070424 was protective against NIHL, while the SOD1 GG genotype of rs10432782 and the CT genotype of rs4880 (SOD2 V16A SNP) was associated with higher occurrence of NIHL. However, the above results were not in agreement with a former research based on a Swedish population, which suggests that SOD genetic polymorphism may confer a race-specific contribution (Carlsson et al., 2005).
Potassium Ion (K+) Cycling Related Genes
As an important charge carrier in the process of sound sensory conduction, K+ can be secreted to the endolymph, and then utilized by the sensory hair cells of the inner ear through the mechanically sensitive K+ channel, and this ion circulation ensures the generation of hearing. The related genes which has been proved susceptible to NIHL are illustrated in Figure 2.
FIGURE 2.
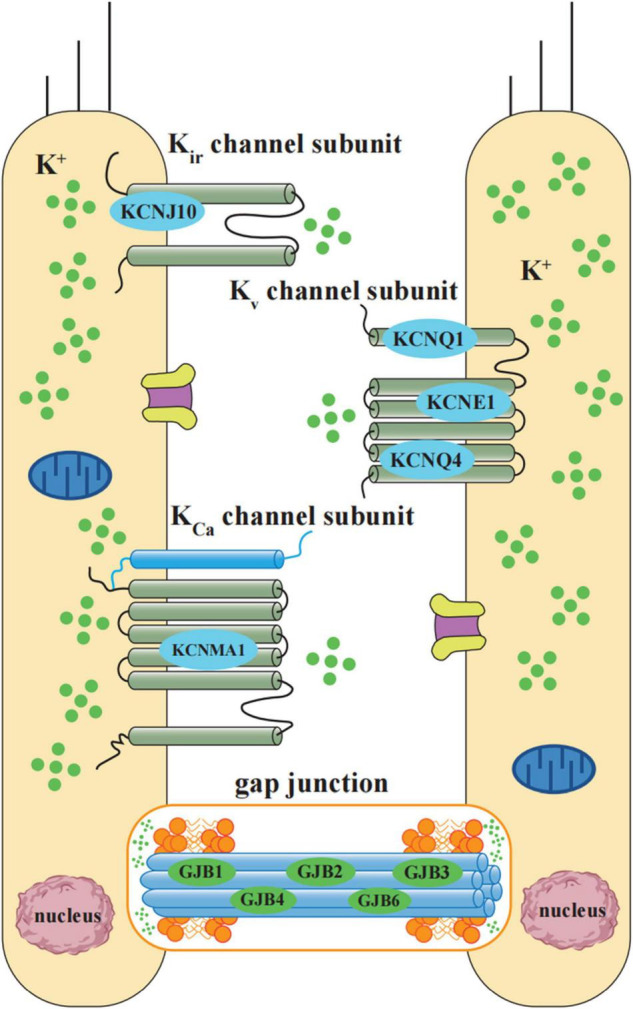
Schematic diagram of potassium ion cycling related NIHL susceptibility genes. K+ cycling related NIHL susceptibility genes include K+ channel proteins and gap junction proteins. According to the operational mechanism and structures, NIHL susceptibility related K+ channels can be classified into 3 groups: inward rectifier (Kir, including KCNJ10), voltage-gated (Kv, including KCNQ1, KCNE1, and KCNQ4), and Ca2+activated (KCa, including KCNMA1). Gap junctions between hair cells and non-sensory cells are primarily formed by a family of connexin proteins, which is encoded by gene GJB1, GJB2, GJB3, GJB4, and GJB6. Gap junction-mediated intercellular communication plays an essential role in K+ exchange.
Potassium Voltage-Gated Channel Subfamily E Regulatory Subunit 1 and Potassium Voltage-Gated Channel Subfamily Q Member 1
KCNE1 encodes a regulatory subunit of the KCNQ1 potassium channel-complex. Both KCNE1 and KCNQ1 are necessary for normal hearing. Pawelczyk et al. (2009) performed a study to clarify the hypothesis that genetic variability in genes of the potassium recycling pathway may be a risk factor for the development of NIHL. The significant results revealed that the AA genotype in rs2070358 appeared more frequently in resistant individuals than in susceptible ones, while genotype GG was more often among susceptible subjects. Recently, another study (Ding et al., 2020) was designed to investigate the association between genetic mutations in the KCNE1 gene and susceptibility to NIHL in the Chinese population. Their results showed that the rs3453 C allele and the rs1805127 G allele were associated with increased susceptibility to NIHL.
Potassium Voltage-Gated Channel Subfamily Q Member 4
Potassium Voltage-Gated Channel Subfamily Q Member 4 (KCNQ4) is a voltage-gated potassium channel that plays essential roles on maintaining ion homeostasis and regulating hair cell membrane potential. Guo et al. (2018b) conducted a genetic association study to scrutinize the association between KCNQ4 polymorphism and susceptibility to NIHL. They detected that rs4660468 CT/TT genotype and T allele may increase the susceptibility. In another study among Chinese population, the SNPs of rs4660468, rs4660470, rs34287852 in KCNQ4 were genotyped by Zhou W. H. et al. (2020). They identified that the risk of developing NIHL in subjects carrying TA genotype of rs4660470 was 2.197 times than the one carrying TT genotypes, suggesting that the mutant allele A of rs4660470 in KCNQ4 may be a risk factor for developing NIHL.
Potassium Inwardly Rectifying Channel Subfamily J Member 10
KCNJ10 encodes the inward-rectifying potassium channel that is expressed in the brain, the inner ear, and kidney. Pawelczyk et al. (2009) conducted a study to explore the putative hypothesis that genetic variations in ten genes associated with the potassium recycling pathway in the inner ear may influence susceptibility to the development of NIHL. Their results discovered that the polymorphism of rs1130183 in KCNJ10 may be a risk factor for the development of NIHL. In addition, Bhatt et al. (2020) performed research to investigate the relationship between candidate genetic variants and NIHL in young musicians, they also identified that KCNJ10 rs1130183 showed significant association with the distortion product otoacoustic emission (DPOAE) signal-to-noise ratio (SNR) in the right ear.
Gap Junction Protein Beta 2 (Connexin 26, Cx26)
GJB2, encoding a gap junction protein expressed in the inner ear, has been considered to be involved in the potassium recycling pathway in the cochlea. Van Eyken et al. (2007) performed a study to investigate the association between the GJB2 35delG mutation and the development of NIHL. Frustratingly, the results suggested that 35delG carriers had no increased susceptibility to the development of NIHL. However, in an animal study, Zhou et al. (2016) established a Connexin26 knockdown mouse model to investigate the relationship between Connexin26 gene and NIHL. Their results indicated that decreased Connexin26 expression may contribute to the increased susceptibility to NIHL and promote the cell degeneration in the Corti’s organ.
Cilia Structure Related Genes
Tip links of the hair cells play a crucial role in the process of mechano-electrical transduction (MET), transforming the mechanical sound stimuli into electrical signals (Sakaguchi et al., 2009). The main constituent of tip links are cadherin related 23 (CDH23) and procadherin related 15 (PCDH15), atypical members of the cadherin superfamily. Cadherin is a calcium-dependent cellular adhesion glycoprotein, which plays an important role in cell recognition, migration, tissue differentiation, the composition of adult tissues and embryonic development. The polymorphism of those genes is closely related to the susceptibility to NIHL. Besides, the damage of MYH14, located at the tip links between hair cells and hair cells, hair cells and supporting cells, also leads to susceptibility to NIHL.
Cadherin Related 23
Cadherin Related 23 (CDH23) is an important protein which is mainly expressed in the cilia of inner hair cells and vestibular membrane (Wilson et al., 2001). Anchored to ciliated microfilaments by actin, it forms a protein network with myosin VIIA for functional activity (Boeda et al., 2002). Its primary function is to maintain the structure and function of hair cell cilia and the ion composition of endolymph, which ensure the mechanical-electrical conversion of sound waves can be carried out normally during the transduction of sound waves in the inner ear (Siemens et al., 2004). It was evidenced in adult mice that Cdh23 mutant mice were susceptible to NIHL. The results showed that the threshold of compound action potential (CAP) was increased by about 50dB at 12 kHz and 30 kHz frequency, which was more than twice that of wild type mice (Holme and Steel, 2004). Kowalski et al. (2014) selected 314 workers with the worst hearing as the experimental group and 313 workers with the best hearing as the control group from 3860 workers database exposed to the same noise environment. Statistical analysis showed that the genotype of the SNP rs3752752 located in exon 21 was closely related to NIHL susceptibility, in which CC genotype was more common in susceptible population, while CT genotype appeared more frequently in the group with better noise tolerance. Another study (Yang et al., 2006) revealed that individuals with rs3802721TT genotype, rs1227049CC genotype and GG genotype at the end of exon 7 were more susceptible to NIHL.
Procadherin Related 15
PCDH15 encodes a membrane protein that mediates calcium-dependent cell adhesion. It is considered that tip-link is composed of proteins encoded by PCDH15 and CDH23 genes (Rowlands et al., 2000). The protein encoded by the PCDH15 forms the lower part of the tip-link, and the CDH23 forms the upper part. In vitro, the extracellular components of PCDH15 and CDH23 form parallel homodimers, and the homodimers are arranged in a Ca2+ dependent antiparallel manner (Ahmed et al., 2006). In recent years, it has been found that there is a correlation between PCDH15 gene polymorphism and NIHL susceptibility. Zhang et al. (2014) selected 476 workers with NIHL and 475 workers with normal hearing from a factory in China for a case-control study. There is no difference in sex ratio, noise exposure years and exposure intensity between the two groups. It was found that the allele frequency and genotypes of rs1104085 were significantly correlated with NIHL susceptibility, that is, the susceptibility of variant allele CT or CC genotype was significantly lower than that of wild type TT homozygotes. Besides, SNPs of rs1100085, rs10825122, rs1930146, rs2384437, rs4540756, and rs2384375 were also found to have correlations with NIHL.
Myosin Heavy Chain 14
The MYH14 is located on chromosome 19 and encodes myosin-binding protein C. It is an ATP-dependent molecular motor involved in cytoskeletal rearrangement and ion gate control. MYH14 was first identified as the causative gene for neurogenic deafness in 2004 (Donaudy et al., 2004). Konings et al. (2009a) conducted an association study of NIHL based on a candidate gene approach. They found two SNPs in MYH14 (rs667907 and rs588035) that resulted in significant associations in the Polish sample set and significant interactions with noise exposure level in the Swedish sample set. Fu et al. (2016) established Myh14 knockout mice using CRISPR/Cas9 technology and clarified the role of MYH14 in the cochlea and NIHL. They found that Myh14–/– mice were more susceptible to high-intensity noise compared to control mice. After acoustic trauma, more pronounced loss of outer hair cells was observed in Myh14–/– mice than in wild-type controls, suggesting that Myh14 may play a beneficial role in protecting the cochlea after acoustic overstimulation in CBA/CaJ mice.
Heat Shock Protein Genes 70
Heat shock protein genes (HSPs) can be overexpressed in the inner ear by stimulation such as physiological stress, ototoxic drugs, high temperature and noise. Among them, HSP70 is a dominant type of heat stress protein which has great protective effect. Gratton et al. (2011) observed the difference of cochlear membrane labyrinth gene expression between noise-susceptible experimental group and noise-tolerant control group. It was found that the protein contents of HSP70 and HSP40 in the control group were significantly higher than those in the experimental group, indicating that the expression of HSP70 gene may play an important role on protecting animals from NIHL. Lei et al. (2017) used Meta analysis to comprehensively analyze the relationship between HSP70 polymorphism and NIHL susceptibility, and concluded that the polymorphism of rs1061581 and rs2227956 may be closely related to the susceptibility to NIHL. Li et al. (2017) screened 286 NIHL patients by measuring the hearing threshold of iron and steel workers, and selected another 286 normal hearing workers in the same noise environment as the control group. It was found that the proportion of TT genotype of rs2763979 in Chinese Han population was higher than that of CC/TC genotype in the NIHL group.
DNA Damage Repair Related Genes
Eyes Absent Homolog 4
Eyes Absent Homolog 4 (EYA4) is a member of the eye absent family of proteins that encode transcriptional activator-related proteins and plays an important role on regulating tissue-specific differentiation during embryonic development (Borsani et al., 1999). It also participates in a variety of biological activities including maintaining the development and maturation of the Corti’s organ (Wayne et al., 2001). Zhang et al. (2015) investigated the relationship between the polymorphisms of EYA4 and the risk of developing NIHL. The results of this study showed that rs3777781 and rs212769 in the EYA4 gene were significantly associated with the risk of NIHL. In rs3777781, carriers of the AT and AA genotypes had a reduced risk of NIHL compared to subjects carrying the TT genotype. In rs212769, carriers of the AG and AA genotypes had an increased risk of NIHL compared to subjects with the GG genotype. In another case-control study (Yang et al., 2017), subjects carrying the rs3813346 TT genotype had a higher risk of NIHL than subjects carrying the GG genotype in the noise intensity > 85 dB group. In contrast, in the cumulative noise exposure (CNE) > 98 dB-year group, haplotype CGT showed a protective role in the development of NIHL compared to haplotype TGC, suggesting that genetic polymorphisms in the EYA4 gene may be a genetic susceptibility factor for NIHL.
8-Oxoguanine DNA Glycosylase
Human 8-hydroxyguanine glycosylase (hOGG1) is a DNA repair enzyme in the base excision repair pathway, whose main function is to recognize and excise 8-oxo G in the DNA double strand and repair damaged DNA. Shen et al. (2014) designed research to investigate the relationship between the gene polymorphism (hOGG1 Ser326Cys) of rs1052133 and susceptibility to high frequency hearing loss. The hOGG1 Cys/Cys genotype was found to be a possible risk factor for high-frequency hearing loss, and stratified analysis revealed it was also associated with risk factors such as years of work in noisy jobs, noise exposure level and smoking. Thus, they concluded that the hOGG1 Cys/Cys genotype may be a risk factor for high frequency hearing loss in the Chinese Han population.
Apoptosis Related Genes
Extracellular Signal-Regulated Kinase 2
Extracellular signal-regulated kinase (ERK) is a member of the MAPK cascades which is a key signaling pathway that control a multitude of cellular processes such as cell survival, protein synthesis, cell proliferation, growth, migration, and apoptosis (Cargnello and Roux, 2011). Recently, accumulative evidences indicate that ERK is involved in response to cellular stress such as noise exposure. When activated by stimulation, ERK2 transfers from the cytoplasm to the nucleus, result in the activation of downstream transcription factors who would further execute kinds of cellular functions (Seger et al., 1991). Kurioka et al. (2015) revealed that conditional Erk2 knockout mice were more susceptible to noise damage and had slower recovery from NIHL compared to control mice. Furthermore, they detected a significant lower survival rate of inner hair cells in Erk2 knockout mice. Their results suggest that Erk2 is essential to the survival of hair cells in NIHL. However, to the best of our knowledge, the research concerning ERK2 polymorphisms in NIHL population is nearly a piece of blank.
C-Jun N-Terminal Kinases 1
C-Jun N-terminal kinase (JNK), also known as stress-activated protein kinase (SAPK), is a member of the MAPK family (Hollville et al., 2019). The JNK stress pathways are involved in many different intracellular signaling pathways that control diverse cellular processes such as cell growth, differentiation, transformation, and most importantly, apoptosis (Zeke et al., 2016). Sun et al. (2021) conducted a study to explore the effect of JNK1 polymorphisms on the sensitivity of NIHL, and the results indicated that the rs11598320 TT genotype and the rs8428 TT genotype may be associated with a higher risk of NIHL. Interestingly, a previous study has also reported that prednisone, a well-known steroid clinically used in the treatment of hearing loss, could inhibit the IL-1β-induced activation of JNK1 (Hong and Jang, 2014).
Other Noised-Induced Hearing Loss Susceptible Genes
Caspase Recruitment Domain Family Member 8
Inflammation is a complex process that is thought to contribute to the development of NIHL. CARD8 is an important component of the inflammasome and has been implicated in inflammation. Miao et al. (2021) conducted a study to investigate the relationship between CARD8 gene polymorphisms and NIHL risk and to infer the underlying mechanisms. They verified three SNPs (rs2043211, rs1062808 and rs12459322) in a Chinese population including 610 NIHL cases and 612 normal hearing controls. The haplotype AGG (rs2043211-rs1062808-rs12459322), the AA genotype and A allele of rs2043211 were found associated with a reduced risk of NIHL.
Fas Cell Surface Death Receptor
Reactive oxygen species (ROS) production in the cochlea and blood caused by noise exposure leads to the processes of oxidative stress, lipid peroxidation, and DNA damage, during which FAS is activated. Xu et al. (2021) conducted case-control research to investigate the relationship between genetic polymorphisms in the FAS gene and NIHL risk. 692 NIHL workers and 650 controls were genotyped for four SNPs, among which two polymorphisms, rs1468063 and rs2862833, were associated with NIHL. Individuals harboring rs1468063-TT or rs2862833-AA genotypes had a decreased risk of NIHL.
Forkhead Box O3
FOXO3 is a gene with a variety of biological functions and is closely related to mammalian longevity. It regulates specific activation of transcription factors to exert effects on cell differentiation, apoptosis, cell cycle, DNA damage repair and oxidative stress (Stefanetti et al., 2018). Through the study of the animal model of NIHL, Gilels et al. (2017) found that the outer hair cells of Foxo3 knockout mice were more seriously damaged than those of normal mice after the same intensity of noise exposure, and the severity of hearing loss increased significantly, indicating that Foxo3 is an important protective gene for mice to maintain hearing after noise exposure. Guo et al. (2017) conducted research to explore the effects of FOXO3 polymorphisms on individual NIHL susceptibility. The results proved that individuals with the G allele of rs2802292, G allele of rs10457180, T allele of rs12206094 and the haplotype GAC and others (TGT/GGT/GGC/GAT) (rs2802292-rs10457180-rs12206094) are associated with an increased risk of NIHL in a Chinese population. In addition, they revealed that GT-GG genotype in FOXO3 may be a risk factor for occupational NIHL (Guo et al., 2018c). They concluded that the genetic polymorphisms rs2802292, rs10457180, rs12206094 and rs12212067 within FOXO3 have the potential to be biomarkers for noise exposed impairment for workers.
Grainyhead-Like 2
Grainyhead-Like 2 (GRHL2) is a transcription factor that expressed in epithelial tissues, it not only plays a central role in embryonic development, but also contributes to epithelial cell maintenance (Peters et al., 2002). Li X. et al. (2013) conducted a study to evaluate the contribution of the GRHL2 polymorphisms to NIHL susceptibility in a Chinese population and found that the subjects carrying rs611419 AT/TT were more resistant to NIHL compared with those carrying the AA genotype. In addition, another study revealed that the CC genotype of rs1981361 in GRHL2 gene was contributed to a higher risk of NIHL (Xu et al., 2016). Additionally, the fact that the rs3735715 GG genotype had a higher NIHL risk compared with the GA genotype was also verified in another study among Chinese population (Yang Q. Y. et al., 2016).
Metabolic Glutamate Receptor 7 Gene
Metabolic Glutamate Receptor 7 Gene (GRM7) is mainly responsible for glutamate-mediated postsynaptic excitation of neurons. In order to study the effect of GRM7 polymorphism on NIHL susceptibility, Yu et al. (2018a) selected 292 NIHL patients and 584 workers with normal hearing in a steel factory as subjects. It is found that the C allele genotype of the rs1485175 mutant of GRM7 gene plays an important role in reducing the incidence of NIHL. Permutation test of generalized multiple dimensionality reduction (GMDR) suggested that rs1920109, rs1485175 and rs9826579 might interact with each other in the pathogenesis of NIHL.
HOX Transcript Antisense RNA
LncRNA HOTAIR is a non-coding RNA that plays a crucial role in RNA processing, gene regulation, chromatin modification, gene transcription, post-transcriptional regulation (Kalwa et al., 2016). It is involved in the alterations of oxidative stress levels, cell proliferation, cell cycle progression and apoptosis. As its expression level is always dysregulated in variety of cancers, it is considered to be used as a potential biomarker (Yang et al., 2019). In order to explore the effect of HOTAIR polymorphisms on the NIHL susceptibility, three tag SNPs of the HOTAIR (rs874945, rs4759314 and rs7958904) were genotyped in a Chinese population including 570 NIHL cases and 570 controls (Wang B. et al., 2017). The results showed that individuals with the G allele of HOTAIR tagSNP rs4759314 and the haplotype (rs874945, rs4759314 and rs7958904) were associated with an increased risk of NIHL.
POU Class 4 Homeobox 3
POU Class 4 Homeobox 3 (POU4F3), also known as Bm3.1 or Bm3c, is a transcription factor which is important for the maturation, differentiation and survival of inner ear hair cells. Xu et al. (2016) performed a matched case-control study to explore the relationship between SNPs in the POU4F3 gene and susceptibility to high frequency hearing loss in a Chinese population. They revealed that when CNE > 95 dB, individuals carrying the AA genotype had an increased risk of hearing loss compared to the CC/CA genotype at SNP rs1368402. Compared to the AA/GA genotype at rs891969, the GG genotype revealed to be a risk genotype.
Concluding Remarks
As the death of hair cells in the cochlear is irreversible, and NIHL is a completely preventable disease, it is particularly important to prevent the potential hearing impairment in advance through possible screening and evaluation. The explore of NIHL susceptible genes offers an opportunity to decrease the incidence of hearing loss by risk assessment as early as infant. The incidence of NIHL would be significantly reduced by distributing the susceptible individuals away from intense noise exposure. For example, factories could assign different employees according to their genotype of NIHL susceptible genes to avoid the occupational impairment; NIHL susceptibility screening could also be applied during conscription.
Although dozens of possible susceptibility genes related to NIHL have been screened, there is still a big gap between practical application and researches. Taylor et al. (1965) first established a linear regression model between noise exposure and high frequency hearing threshold in 1965. It was found that the distribution of NIHL susceptibility in the population showed a unimodal left bias, and there was no single peak on the right side of the main peak (susceptible area), suggesting that the susceptibility is related to many factors and is likely to be affected by multiple minor genes, which increases the difficulty of the study on susceptibility genes.
In relation to the screening of NIHL susceptibility genes, there are some limitations whether using animal research or population study. For animal research, although it has the advantages of short test cycle and easy to obtain materials, the results must be verified in the population. For population study, family analysis is the most effective method to study susceptibility genes, but medical ethics cannot expose all subjects to noise environment, so pedigree analysis cannot be used in the study, only NIHL susceptibility genes can be searched in the genome. Besides, due to many factors, such as regional diversity, ethnic differences, study sample size and gene interaction, inconsistent research conclusions is a commonplace, resulting in limited clinical reference value. Most studies have been conducted in a single population, so further analysis of the correlation between different populations is essential.
Currently, only a handful of NIHL susceptibility genes have been uncovered, and existing studies suggest that NIHL may be caused by accumulative abnormal influence of multiple genes. Further in-depth researches are needed to explore gene-gene interaction and find comprehensive and dominant susceptibility genes among numerous NIHL susceptibility genes. Although there are still great difficulties and challenges in the study of NIHL susceptibility genes, with the further research on new genetic research methods, such as next-generation DNA sequencing (NGS) and high-throughput genotyping array, more susceptibility genes related to NIHL will be found. The luminant prospect of designing of molecular probes that can be used for clinical detection of NIHL susceptible individuals is awaiting on the way.
In conclusion, genetic factor plays a vital role on the pathogenesis of NIHL. NIHL susceptible genes can be used for better identification of potential risks and prevent the occurrence of NIHL. Through the continuous screening of genetic variants in the susceptibility of NIHL, new susceptibility genes will come to light, and ideally, get into the stage of clinical application, which lays a solid foundation for the accurate screening of high-risk population and the reduction of NIHL incidence.
Author Contributions
X-MC and X-MX drafted the manuscript. NY, W-WG, and S-LY collected the literature. X-MX summarized the literature. X-MC tabulated the data and drew the figures. Q-QJ revised the manuscript. Q-QJ and S-MY took responsibility for the integrity of the data and the accuracy of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Our deepest gratitude goes to colleagues from our laboratory.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 82000981 and 81970897).
References
- Ahmed Z. M., Goodyear R., Riazuddin S., Lagziel A., Legan P. K., Behra M., et al. (2006). The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J. Neurosci. 26 7022–7034. 10.1523/JNEUROSCI.1163-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhofer S., Witte I., Teiber J. F., Wilgenbus P., Pautz A., Li H., et al. (2010). One enzyme, two functions: PON2 prevents mitochondrial superoxide formation and apoptosis independent from its lactonase activity. J. Biol. Chem. 285 24398–24403. 10.1074/jbc.M110.118604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt I. S., Dias R., Washnik N., Wang J., Guthrie O., Skelton M., et al. (2020). Association analysis of candidate gene polymorphisms and audiometric measures of noise-induced hearing loss in young musicians. Otol. Neurotol. 41 e538–e547. 10.1097/MAO.0000000000002615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeda B., El-Amraoui A., Bahloul A., Goodyear R., Daviet L., Blanchard S., et al. (2002). Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 21 6689–6699. 10.1093/emboj/cdf689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G., DeGrandi A., Ballabio A., Bulfone A., Bernard L., Banfi S., et al. (1999). EYA4, a novel vertebrate gene related to Drosophila eyes absent. Hum. Mol. Genet. 8 11–23. 10.1093/hmg/8.1.11 [DOI] [PubMed] [Google Scholar]
- Braga M. P., Maciel S. M., Marchiori L. L., Poli-Frederico R. C. (2014). Associacao entre interleucina-6 polimorfismo na regiao de -174 G/C e perda auditiva em idosos com historia de exposicao ao ruido ocupacional. Rev. Bras. Otorrinolaringol. 80 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. L., Li X. T., Zhong L., Shen H. X., Ding L., Liu J., et al. (2013). Association between single nucleotide polymorphisms of PON2 gene and susceptibility to occupational noise-induced deafness among Chinese Han population exposed to high noise levels. Chin. J. Ind. Hyg. Occup. Dis. 31 734–739. [PubMed] [Google Scholar]
- Cargnello M., Roux P. P. (2011). Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75 50–83. 10.1128/mmbr.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson P. I., Van Laer L., Borg E., Bondeson M. L., Thys M., Fransen E., et al. (2005). The influence of genetic variation in oxidative stress genes on human noise susceptibility. Hear. Res. 202 87–96. 10.1016/j.heares.2004.09.005 [DOI] [PubMed] [Google Scholar]
- Carroll Y. I., Eichwald J., Scinicariello F., Hoffman H. J., Deitchman S., Radke M. S., et al. (2017). Vital signs: noise-induced hearing loss among adults - United States 2011-2012. MMWR Morb. Mortal. Wkly. Rep. 66 139–144. 10.15585/mmwr.mm6605e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda R., Natarajan S., Jeong S. Y., Hong B. N., Kang T. H. (2019). Traditional oriental medicine for sensorineural hearing loss: can ethnopharmacology contribute to potential drug discovery? J. Ethnopharmacol. 231 409–428. 10.1016/j.jep.2018.11.016 [DOI] [PubMed] [Google Scholar]
- Chang N. C., Ho C. K., Lin H. Y., Yu M. L., Chien C. Y., Ho K. Y. (2011). Association of polymorphisms of heat shock protein 70 with susceptibility to noise-induced hearing loss in the Taiwanese population. Audiol. Neurootol. 16 168–174. 10.1159/000317119 [DOI] [PubMed] [Google Scholar]
- Chen H., Ding X., Ding E., Chen M., Wang H., Yang G., et al. (2021). A missense variant rs2585405 in clock gene PER1 is associated with the increased risk of noise-induced hearing loss in a Chinese occupational population. BMC Med. Genomics 14:221. 10.1186/s12920-021-01075-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. M., Ji S. F., Liu Y. H., Xue X. M., Xu J., Gu Z. H., et al. (2020). Ginsenoside Rd ameliorates auditory cortex injury associated with military aviation noise-induced hearing loss by activating SIRT1/PGC-1alpha signaling pathway. Front. Physiol. 11:788. 10.3389/fphys.2020.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Wang S. H., Huang Y., Liao X. M. (2016). The hippocampus may be more susceptible to environmental noise than the auditory cortex. Hear. Res. 333 93–97. 10.1016/j.heares.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Coles R. R., Lutman M. E., Buffin J. T. (2000). Guidelines on the diagnosis of noise-induced hearing loss for medicolegal purposes. Clin. Otolaryngol. Allied Sci. 25 264–273. 10.1046/j.1365-2273.2000.00368.x [DOI] [PubMed] [Google Scholar]
- Ding E., Guo J., Ge X., Sheng R., Chen J., Zhang H., et al. (2019). Analysis of polymorphisms associated with base excision repair in patients susceptible and resistant to noise-induced hearing loss. Dis. Markers 2019:9327106. 10.1155/2019/9327106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding E., Liu J., Shen H., Gong W., Zhang H., Song H., et al. (2018). Notch polymorphisms associated with sensitivity of noise induced hearing loss among Chinese textile factory workers. BMC Med. Genet. 19:168. 10.1186/s12881-018-0676-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding E., Wang H., Han L., Zhang H., Zhu B. (2020). Variations in the potassium voltage-gated channel subfamily E regulatory subunit 1 gene associated with noise-induced hearing loss in the Chinese population. Environ. Sci. Pollut. Res. Int. 27 18822–18830. 10.1007/s11356-020-08422-y [DOI] [PubMed] [Google Scholar]
- Donaudy F., Snoeckx R., Pfister M., Zenner H. P., Blin N., Di Stazio M., et al. (2004). Non-muscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4). Am. J. Hum. Genet. 74 770–776. 10.1086/383285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermont J. J. (2017). Effects of long-term non-traumatic noise exposure on the adult central auditory system. Hearing problems without hearing loss. Hear. Res. 352 12–22. 10.1016/j.heares.2016.10.015 [DOI] [PubMed] [Google Scholar]
- Eraslan E., Akyazi I., Erg L. E. E., Matur E. (2015). Noise stress changes mRNA expressions of corticotropin-releasing hormone, its receptors in amygdala, and anxiety-related behaviors. Noise Health 17 141–147. 10.4103/1463-1741.155838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P., Halliwell B. (1999). Free radicals and hearing. Cause, consequence, and criteria. Ann. N. Y. Acad. Sci. 884 19–40. 10.1111/j.1749-6632.1999.tb08633.x [DOI] [PubMed] [Google Scholar]
- Forman H. J., Maiorino M., Ursini F. (2010). Signaling functions of reactive oxygen species. Biochemistry 49 835–842. 10.1021/bi9020378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato G., Marciano E., Zarrilli F., Mazzaccara C., Intrieri M., Calcagno G., et al. (2004). Paraoxonase and superoxide dismutase gene polymorphisms and noise-induced hearing loss. Clin. Chem. 50 2012–2018. 10.1373/clinchem.2004.037788 [DOI] [PubMed] [Google Scholar]
- Fridberger A., Flock A., Ulfendahl M., Flock B. (1998). Acoustic overstimulation increases outer hair cell Ca2+ concentrations and causes dynamic contractions of the hearing organ. Proc. Natl. Acad. Sci. U.S.A. 95 7127–7132. 10.1073/pnas.95.12.7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M. D., Ryan A. F., Kurabi A. (2019). Inflammation associated with noise-induced hearing loss. J. Acoust. Soc. Am. 146:4020. 10.1121/1.5132545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M. D., Zhang C., Hu B. H. (2018). Lower level noise exposure that produces only TTS modulates the immune homeostasis of cochlear macrophages. J. Neuroimmunol. 323 152–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Zhang L., Jin Y., Sun X., Zhang A., Wen Z., et al. (2016). Loss of Myh14 increases susceptibility to noise-induced hearing loss in CBA/CaJ mice. Neural Plast. 2016:6720420. 10.1155/2016/6720420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D. F., Wang B. S., Sun D. W., Wang N., Guo J. D., Zhu B. L. (2021). Targeted binding of rs1053005 locus of STAT3 with miR-452-3p and the association between STAT3 gene polymorphism and noise-induced hearing loss. Chin. J. Ind. Hyg. Occup. Dis. 39 412–417. 10.3760/cma.j.cn121094-20210203-00077 [DOI] [PubMed] [Google Scholar]
- Gilels F., Paquette S. T., Beaulac H. J., Bullen A., White P. M. (2017). Severe hearing loss and outer hair cell death in homozygous Foxo3 knockout mice after moderate noise exposure. Sci. Rep. 7:1054. 10.1038/s41598-017-01142-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton M. A., Eleftheriadou A., Garcia J., Verduzco E., Martin G. K., Lonsbury-Martin B. L., et al. (2011). Noise-induced changes in gene expression in the cochleae of mice differing in their susceptibility to noise damage. Hear. Res. 277 211–226. 10.1016/j.heares.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin Y., Bortoni M. E., Sepulveda R., Ghelfi E., Bartos A., Cotanche D., et al. (2015). Genetic polymorphisms associated with hearing threshold shift in subjects during first encounter with occupational impulse noise. PLoS One 10:e0130827. 10.1371/journal.pone.0130827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ding E., Bai Y., Zhang H., Shen H., Wang J., et al. (2017). Association of genetic variations in FOXO3 gene with susceptibility to noise induced hearing loss in a Chinese population. PLoS One 12:e0189186. 10.1371/journal.pone.0189186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Cheng J., Sheng R., Cai W., Xin Y., Ding E., et al. (2018a). Association of DNMT3a’s single nucleotide polymorphisms with susceptibility to noise-induced hearing loss in Chinese workers. J. Hyg. Res. 47 638–643. [PubMed] [Google Scholar]
- Guo H., Ding E., Sheng R., Cheng J., Cai W., Guo J., et al. (2018b). Genetic variation in KCNQ4 gene is associated with susceptibility to noise-induced hearing loss in a Chinese population. Environ. Toxicol. Pharmacol. 63 55–59. 10.1016/j.etap.2018.08.009 [DOI] [PubMed] [Google Scholar]
- Guo H. R., Ding E. M., Cai W. Y., Guo J. D., Wang N., Zhang H. D., et al. (2018c). Correlation between FOXO3 gene polymorphisms and susceptibility to occupational noise-induced deafness. Chin. J. Ind. Hyg. Occup. Dis. 36 492–496. 10.3760/cma.j.issn.1001-9391.2018.07.003 [DOI] [PubMed] [Google Scholar]
- Hahad O., Prochaska J. H., Daiber A., Muenzel T. (2019). Environmental noise-induced effects on stress hormones, oxidative stress, and vascular dysfunction: key factors in the relationship between cerebrocardiovascular and psychological disorders. Oxid. Med. Cell. Longev. 2019:4623109. 10.1155/2019/4623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Ru X., Wen T. (2020). NRF2, a transcription factor for stress response and beyond. Int. J. Mol. Sci. 21:4777. 10.3390/ijms21134777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Yu J., Sun Y., Kong W. (2020). Macrophages in noise-exposed cochlea: changes, regulation and the potential role. Aging Dis. 11 191–199. 10.14336/AD.2019.0723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D., Bielefeld E. C., Harris K. C., Hu B. H. (2006). The role of oxidative stress in noise-induced hearing loss. Ear Hear. 27 1–19. 10.1097/01.aud.0000191942.36672.f3 [DOI] [PubMed] [Google Scholar]
- Hollville E., Romero S. E., Deshmukh M. (2019). Apoptotic cell death regulation in neurons. FEBS J. 286 3276–3298. 10.1111/febs.14970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holme R. H., Steel K. P. (2004). Progressive hearing loss and increased susceptibility to noise-induced hearing loss in mice carrying a Cdh23 but not a Myo7a mutation. J. Assoc. Res. Otolaryngol. 5 66–79. 10.1007/s10162-003-4021-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Jang B. C. (2014). Prednisone inhibits the IL-1β-induced expression of COX-2 in HEI-OC1 murine auditory cells through the inhibition of ERK-1/2, JNK-1 and AP-1 activity. Int. J. Mol. Med. 34 1640–1646. 10.3892/ijmm.2014.1967 [DOI] [PubMed] [Google Scholar]
- Honkura Y., Matsuo H., Murakami S., Sakiyama M., Mizutari K., Shiotani A., et al. (2016). NRF2 is a key target for prevention of noise-induced hearing loss by reducing oxidative damage of cochlea. Sci. Rep. 6:19329. 10.1038/srep19329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J. D. (1987). Hearing acuity and susceptibility to noise-induced hearing loss. Br. J. Audiol. 21 175–181. 10.3109/03005368709076403 [DOI] [PubMed] [Google Scholar]
- Irion H. (1981). Acute bilateral hearing loss during a pop concert: consideration for differential diagnosis. Adv. Otorhinolaryngol. 27 121–129. 10.1159/000400334 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Fa B., Zhang X., Wang J., Feng Y., Shi H., et al. (2021). Identifying genetic risk variants associated with noise-induced hearing loss based on a novel strategy for evaluating individual susceptibility. Hear. Res. 407:108281. 10.1016/j.heares.2021.108281 [DOI] [PubMed] [Google Scholar]
- Jiao J., Gu G. Z., Chen G. S., Li Y. H., Zhang H. L., Yang Q. Y., et al. (2017). Investigation into the relationship between mitochondrial 12 S rRNA gene, tRNA gene and cytochrome oxidase gene variations and the risk of noise-induced hearing loss. Chin. J. Prev. Med. 51 34–40. 10.3760/cma.j.issn.0253-9624.2017.01.008 [DOI] [PubMed] [Google Scholar]
- Jiao J., Gu G. Z., Chen G. S., Zhang H. L., Wu H., Li Y. H., et al. (2020). Relationship research among CDH23 gene and the risk of noise-induced hearing loss. Chin. J. Ind. Hyg. Occup. Dis. 38 84–90. 10.3760/cma.j.issn.1001-9391.2020.02.002 [DOI] [PubMed] [Google Scholar]
- Kalwa M., Hanzelmann S., Otto S., Kuo C. C., Franzen J., Joussen S., et al. (2016). The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 44 10631–10643. 10.1093/nar/gkw802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Xia A., Grillet N., Applegate B. E., Oghalai J. S. (2018). Osmotic stabilization prevents cochlear synaptopathy after blast trauma. Proc. Natl. Acad. Sci. U.S.A. 115 E4853–E4860. 10.1073/pnas.1720121115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings A., Van Laer L., Pawelczyk M., Carlsson P. I., Bondeson M. L., Rajkowska E., et al. (2007). Association between variations in CAT and noise-induced hearing loss in two independent noise-exposed populations. Hum. Mol. Genet. 16 1872–1883. 10.1093/hmg/ddm135 [DOI] [PubMed] [Google Scholar]
- Konings A., Van Laer L., Michel S., Pawelczyk M., Carlsson P. I., Bondeson M. L., et al. (2009a). Variations in HSP70 genes associated with noise-induced hearing loss in two independent populations. Eur. J. Hum. Genet. 17 329–335. 10.1038/ejhg.2008.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings A., Van Laer L., Wiktorek-Smagur A., Rajkowska E., Pawelczyk M., Carlsson P. I., et al. (2009b). Candidate gene association study for noise-induced hearing loss in two independent noise-exposed populations. Ann. Hum. Genet. 73 215–224. 10.1111/j.1469-1809.2008.00499.x [DOI] [PubMed] [Google Scholar]
- Kowalski T. J., Pawelczyk M., Rajkowska E., Dudarewicz A., Sliwinska-Kowalska M. (2014). Genetic variants of CDH23 associated with noise-induced hearing loss. Otol. Neurotol. 35 358–365. 10.1097/MAO.0b013e3182a00332 [DOI] [PubMed] [Google Scholar]
- Kozel P. J., Davis R. R., Krieg E. F., Shull G. E., Erway L. C. (2002). Deficiency in plasma membrane calcium ATPase isoform 2 increases susceptibility to noise-induced hearing loss in mice. Hear. Res. 164 231–239. 10.1016/s0378-5955(01)00420-8 [DOI] [PubMed] [Google Scholar]
- Krause K. H. (2004). Tissue distribution and putative physiological function of NOX family NADPH oxidases. Jpn. J. Infect. Dis. 57 S28–S29. [PubMed] [Google Scholar]
- Kujawa S. G., Liberman M. C. (2009). Adding insult to injury: cochlear nerve degeneration after “temporary” noise-induced hearing loss. J. Neurosci. 29 14077–14085. 10.1523/JNEUROSCI.2845-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabi A., Keithley E. M., Housley G. D., Ryan A. F., Wong A. C. (2017). Cellular mechanisms of noise-induced hearing loss. Hear. Res. 349 129–137. 10.1016/j.heares.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurioka T., Matsunobu T., Satoh Y., Niwa K., Endo S., Fujioka M., et al. (2015). ERK2 mediates inner hair cell survival and decreases susceptibility to noise-induced hearing loss. Sci. Rep. 5:16839. 10.1038/srep16839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinsky J., Crow A. L., Pan C., Wang J., Aaron K. A., Ho M. K., et al. (2015). Genome-wide association study identifies nox3 as a critical gene for susceptibility to noise-induced hearing loss. PLoS Genet. 11:e1005094. 10.1371/journal.pgen.1005094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Prell C. G., Yamashita D., Minami S. B., Yamasoba T., Miller J. M. (2007). Mechanisms of noise-induced hearing loss indicate multiple methods of prevention. Hear. Res. 226 22–43. 10.1016/j.heares.2006.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S., Huang L., Liu Y., Xu L., Wang D., Yang L. (2017). Association between polymorphisms of heat-shock protein 70 genes and noise-induced hearing loss: a meta-analysis. PLoS One 12:e0188539. 10.1371/journal.pone.0188539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Y., Jiao J., Chen G. S., Gu G. Z., Zhang H. L., Yu S. F. (2020). Association between GPX1 gene polymorphisms and noise-induced hearing loss. Chin. J. Ind. Hyg. Occup. Dis. 38 116–120. 10.3760/cma.j.issn.1001-9391.2020.02.008 [DOI] [PubMed] [Google Scholar]
- Li T., Chen G., Jiao J., Zhou W., Wu H., Gu G., et al. (2020). Association between single nucleotides polymorphism of catalase gene and susceptibility to noise-induced hearing loss in occupational population. J. Hyg. Res. 49 716–723. 10.19813/j.cnki.weishengyanjiu.2020.05.004 [DOI] [PubMed] [Google Scholar]
- Li X., Cao J., Wang J., Song H., Ji G., Dong Q., et al. (2016). PON2 and ATP2B2 gene polymorphisms with noise-induced hearing loss. J. Thorac. Dis. 8 430–438. 10.21037/jtd.2016.02.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Huo X., Liu K., Li X., Wang M., Chu H., et al. (2013). Association between genetic variations in GRHL2 and noise-induced hearing loss in Chinese high intensity noise exposed workers: a case-control analysis. Ind. Health 51 612–621. 10.2486/indhealth.2012-0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhu Z., Li W., Wei L., Zhao B., Hao Z. (2020). Polymorphism in GRHL2 gene may contribute to noise-induced hearing loss susceptibility: a meta-analysis. Braz. J. Otorhinolaryngol. 86 370–375. 10.1016/j.bjorl.2019.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yu S., Gu G., Chen G., Zheng Y., Jiao J., et al. (2017). Polymorphisms of heat shock protein 70 genes (HSPA1A, HSPA1B and HSPA1L) and susceptibility of noise-induced hearing loss in a Chinese population: a case-control study. PLoS One 12:e0171722. 10.1371/journal.pone.0171722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. H., Chen G. S., Jiao J., Zhou W. H., Wu H., Gu G. Z., et al. (2016). Association between single nucleotide polymorphismsin human heat shock protein 70 gene and susceptibility to noise-induced hearing loss. Chin. J. Ind. Hyg. Occup. Dis. 34 889–894. 10.3760/cma.j.issn.1001-9391.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Li Y. H., Gu G. Z., Zhou W. H., Wu H., Chen G. S., Zhang H. L., et al. (2020). Association between GSTP1 gave polymorphisms and susceptibility to noise-induced hearing loss. Chin. J. Ind. Hyg. Occup. Dis. 38 120–124. 10.3760/cma.j.issn.1001-9391.2020.02.009 [DOI] [PubMed] [Google Scholar]
- Liberman M. C. (2016). Noise-induced hearing loss: permanent versus temporary threshold shifts and the effects of hair cell versus neuronal degeneration. Adv. Exp. Med. Biol. 875 1–7. 10.1007/978-1-4939-2981-8_1 [DOI] [PubMed] [Google Scholar]
- Lin C. Y., Wu J. L., Shih T. S., Tsai P. J., Sun Y. M., Guo Y. L. (2009). Glutathione S-transferase M1, T1, and P1 polymorphisms as susceptibility factors for noise-induced temporary threshold shift. Hear. Res. 257 8–15. 10.1016/j.heares.2009.07.008 [DOI] [PubMed] [Google Scholar]
- Liu Y. M., Li X. D., Guo X., Liu B., Lin A. H., Ding Y. L., et al. (2010a). SOD2 V16A SNP in the mitochondrial targeting sequence is associated with noise induced hearing loss in Chinese workers. Dis. Markers 28 137–147. 10.3233/DMA-2010-0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. M., Li X. D., Guo X., Liu B., Lin A. H., Rao S. Q. (2010b). Association between polymorphisms in SOD1 and noise-induced hearing loss in Chinese workers. Acta Otolaryngol. 130 477–486. 10.3109/00016480903253587 [DOI] [PubMed] [Google Scholar]
- Liu S. Y., Song W. Q., Xin J. R., Li Z., Lei S., Chen Y. Q., et al. (2021). NRN1 and CAT gene polymorphisms, complex noise, and lifestyles interactively affect the risk of noise-induced hearing loss. Biomed. Environ. Sci. 34 705–718. 10.3967/bes2021.098 [DOI] [PubMed] [Google Scholar]
- Loukzadeh Z., Sani H. E., Sheikhha M. H., Ratki F. M. (2019). Association of GST gene polymorphism and noise-induced hearing loss: GST gene polymorphism and NIHL. AIMS Public Health 6 546–553. 10.3934/publichealth.2019.4.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Cheng X., Li Y., Zeng L., Zhao Y. (2005). Evaluation of individual susceptibility to noise-induced hearing loss in textile workers in China. Arch. Environ. Occup. Health 60 287–294. 10.3200/AEOH.60.6.287-294 [DOI] [PubMed] [Google Scholar]
- Miao L., Ji J., Wan L., Zhang J., Yin L., Pu Y. (2019). An overview of research trends and genetic polymorphisms for noise-induced hearing loss from 2009 to 2018. Environ. Sci. Pollut. Res. Int. 26 34754–34774. 10.1007/s11356-019-06470-7 [DOI] [PubMed] [Google Scholar]
- Miao L., Wang B., Ji J., Wan L., Yin L., Zhu B., et al. (2021). CARD8 polymorphism rs2043211 protects against noise-induced hearing loss by causing the dysfunction of CARD8 protein. Environ. Sci. Pollut. Res. Int. 28 8626–8636. 10.1007/s11356-020-11193-1 [DOI] [PubMed] [Google Scholar]
- Niu Y., Xie C., Du Z., Zeng J., Chen H., Jin L., et al. (2021). Genome-wide association study identifies 7q11.22 and 7q36.3 associated with noise-induced hearing loss among Chinese population. J. Cell. Mol. Med. 25 411–420. 10.1111/jcmm.16094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller K. K., McFadden S. L., Ding D. L., Flood D. G., Reaume A. G., Hoffman E. K., et al. (1999). Targeted deletion of the cytosolic Cu/Zn-superoxide dismutase gene (Sod1) increases susceptibility to noise-induced hearing loss. Audiol. Neurootol. 4 237–246. 10.1159/000013847 [DOI] [PubMed] [Google Scholar]
- Ohlemiller K. K., McFadden S. L., Ding D. L., Lear P. M., Ho Y. S. (2000). Targeted mutation of the gene for cellular glutathione peroxidase (Gpx1) increases noise-induced hearing loss in mice. J. Assoc. Res. Otolaryngol. 1 243–254. 10.1007/s101620010043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson J. H., Jr., Hamernik R. P. (1997). Blast overpressure induced structural and functional changes in the auditory system. Toxicology 121 29–40. [DOI] [PubMed] [Google Scholar]
- Pawelczyk M., Van Laer L., Fransen E., Rajkowska E., Konings A., Carlsson P. I., et al. (2009). Analysis of gene polymorphisms associated with K ion circulation in the inner ear of patients susceptible and resistant to noise-induced hearing loss. Ann. Hum. Genet. 73(Pt 4), 411–421. 10.1111/j.1469-1809.2009.00521.x [DOI] [PubMed] [Google Scholar]
- Peters L. M., Anderson D. W., Griffith A. J., Grundfast K. M., San Agustin T. B., Madeo A. C., et al. (2002). Mutation of a transcription factor, TFCP2L3, causes progressive autosomal dominant hearing loss, DFNA28. Hum. Mol. Genet. 11 2877–2885. 10.1093/hmg/11.23.2877 [DOI] [PubMed] [Google Scholar]
- Rajguru R. (2013). Military aircrew and noise-induced hearing loss: prevention and management. Aviat. Space Environ. Med. 84 1268–1276. 10.3357/asem.3503.2013 [DOI] [PubMed] [Google Scholar]
- Ralli M., Balla M. P., Greco A., Altissimi G., Ricci P., Turchetta R., et al. (2017). Work-related noise exposure in a cohort of patients with chronic tinnitus: analysis of demographic and audiological characteristics. Int. J. Environ. Res. Public Health 14:1035. 10.3390/ijerph14091035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands T. M., Symonds J. M., Farookhi R., Blaschuk O. W. (2000). Cadherins: crucial regulators of structure and function in reproductive tissues. Rev. Reprod. 5 53–61. 10.1530/ror.0.0050053 [DOI] [PubMed] [Google Scholar]
- Sakaguchi H., Tokita J., Muller U., Kachar B. (2009). Tip links in hair cells: molecular composition and role in hearing loss. Curr. Opin. Otolaryngol. Head Neck Surg. 17 388–393. 10.1097/MOO.0b013e3283303472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger R., Ahn N. G., Boulton T. G., Yancopoulos G. D., Panayotatos N., Radziejewska E., et al. (1991). Microtubule-associated protein 2 kinases, ERK1 and ERK2, undergo autophosphorylation on both tyrosine and threonine residues: implications for their mechanism of activation. Proc. Natl. Acad. Sci. U.S.A. 88 6142–6146. 10.1073/pnas.88.14.6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Cao J., Hong Z., Liu K., Shi J., Ding L., et al. (2014). A functional Ser326Cys polymorphism in hOGG1 is associated with noise-induced hearing loss in a Chinese population. PLoS One 9:e89662. 10.1371/journal.pone.0089662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Dou J., Han L., Bai Y., Li Q., Hong Z., et al. (2016). Genetic variation in APE1 gene promoter is associated with noise-induced hearing loss in a Chinese population. Int. Arch. Occup. Environ. Health 89 621–628. 10.1007/s00420-015-1100-8 [DOI] [PubMed] [Google Scholar]
- Shen H., Huo X., Liu K., Li X., Gong W., Zhang H., et al. (2012). Genetic variation in GSTM1 is associated with susceptibility to noise-induced hearing loss in a Chinese population. J. Occup. Environ. Med. 54 1157–1162. 10.1097/JOM.0b013e31825902ce [DOI] [PubMed] [Google Scholar]
- Shi X. (2010). Resident macrophages in the cochlear blood-labyrinth barrier and their renewal via migration of bone-marrow-derived cells. Cell Tissue Res. 342 21–30. 10.1007/s00441-010-1040-2 [DOI] [PubMed] [Google Scholar]
- Siemens J., Lillo C., Dumont R. A., Reynolds A., Williams D. S., Gillespie P. G., et al. (2004). Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature 428 950–955. 10.1038/nature02483 [DOI] [PubMed] [Google Scholar]
- Sivandzade F., Prasad S., Bhalerao A., Cucullo L. (2019). NRF2 and NF-B interplay in cerebrovascular and neurodegenerative disorders: molecular mechanisms and possible therapeutic approaches. Redox Biol. 21:101059. 10.1016/j.redox.2018.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogstad M., Johannessen H. A., Tynes T., I, Mehlum S., Nordby K. C., Lie A. (2016). Systematic review of the cardiovascular effects of occupational noise. Occup. Med. 66:500. 10.1093/occmed/kqw113 [DOI] [PubMed] [Google Scholar]
- Sliwinska-Kowalska M., Zaborowski K. (2017). WHO environmental noise guidelines for the European region: a systematic review on environmental noise and permanent hearing loss and tinnitus. Int. J. Environ. Res. Public Health 14:1139. 10.3390/ijerph14101139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeriglio P., Wangsawihardja F. V., Leu R., Mustapha M. (2019). TSP1 and TSP2 have unique and overlapping roles in protecting against noise-induced auditory synaptopathy. Neuroscience 408 68–80. 10.1016/j.neuroscience.2019.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoendlin H., Brun J. P. (1973). Relation of structural damage to exposure time and intensity in acoustic trauma. Acta Otolaryngol. 75 220–226. 10.3109/00016487309139699 [DOI] [PubMed] [Google Scholar]
- Stefanetti R. J., Voisin S., Russell A., Lamon S. (2018). Recent advances in understanding the role of FOXO3. F1000Res. 7:F1000 Faculty Rev-1372. 10.12688/f1000research.15258.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Wang B., Guo H., Wang N., Gao D., Zhu B. (2021). Single nucleotide polymorphisms in JNK1 are associated with susceptibility to noise-induced hearing loss in a Chinese population. Int. Arch. Occup. Environ. Health 94 833–842. 10.1007/s00420-020-01644-0 [DOI] [PubMed] [Google Scholar]
- Taylor W., Pearson J., Mair A., Burns W. (1965). Study of noise and hearing in jute weaving. J. Acoust. Soc. Am. 38 113–120. 10.1121/1.1909580 [DOI] [PubMed] [Google Scholar]
- Valentino R. J., Lucki I., Van Bockstaele E. (2010). Corticotropin-releasing factor in the dorsal raphe nucleus: linking stress coping and addiction. Brain Res. 1314 29–37. 10.1016/j.brainres.2009.09.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eyken E., Van Laer L., Fransen E., Topsakal V., Hendrickx J. J., Demeester K., et al. (2007). The contribution of GJB2 (Connexin 26) 35delG to age-related hearing impairment and noise-induced hearing loss. Otol. Neurotol. 28 970–975. 10.197/MAO.0b013e3180dca1b9 [DOI] [PubMed] [Google Scholar]
- Van Laer L., Carlsson P. I., Ottschytsch N., Bondeson M. L., Konings A., Vandevelde A., et al. (2006). The contribution of genes involved in potassium-recycling in the inner ear to noise-induced hearing loss. Hum. Mutat. 27 786–795. 10.1002/humu.20360 [DOI] [PubMed] [Google Scholar]
- Varela-Nieto I., Murillo-Cuesta S., Calvino M., Cediel R., Lassaletta L. (2020). Drug development for noise-induced hearing loss. Expert Opin. Drug Discov. 15 1457–1471. 10.1080/17460441.2020.1806232 [DOI] [PubMed] [Google Scholar]
- Vicente-Torres M. A., Schacht J. (2006). A BAD link to mitochondrial cell death in the cochlea of mice with noise-induced hearing loss. J. Neurosci. Res. 83 1564–1572. 10.1002/jnr.20832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlajkovic S. M., Lin S. C., Wong A. C., Wackrow B., Thorne P. R. (2013). Noise-induced changes in expression levels of NADPH oxidases in the cochlea. Hear. Res. 304 145–152. 10.1016/j.heares.2013.07.012 [DOI] [PubMed] [Google Scholar]
- Wan L., Wang B., Zhang J., Zhu B., Pu Y. (2020). Associations of genetic variation in glyceraldehyde 3-phosphate dehydrogenase gene with noise-induced hearing loss in a Chinese population: a case-control study. Int. J. Environ. Res. Public Health 17:2899. 10.3390/ijerph17082899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Zhang L., Sun P., Han L., Zhang H., Zhu B., et al. (2022). Association between UBAC2 gene polymorphism and the risk of noise-induced hearing loss: a cross-sectional study. Environ. Sci. Pollut. Res. Int. 29 32947–32958. 10.1007/s11356-021-18360-y [DOI] [PubMed] [Google Scholar]
- Wang Y., Hirose K., Liberman M. C. (2002). Dynamics of noise-induced cellular injury and repair in the mouse cochlea. J. Assoc. Res. Otolaryngol. 3 248–268. 10.1007/s101620020028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Pignol B., Chabrier P. E., Saido T., Lloyd R., Tang Y., et al. (2007a). A novel dual inhibitor of calpains and lipid peroxidation (BN82270) rescues the cochlea from sound trauma. Neuropharmacology 52 1426–1437. 10.1016/j.neuropharm.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Wang J., Ruel J., Ladrech S., Bonny C., van de Water T. R., Puel J. L. (2007b). Inhibition of the c-Jun N-terminal kinase-mediated mitochondrial cell death pathway restores auditory function in sound-exposed animals. Mol. Pharmacol. 71 654–666. 10.1124/mol.106.028936 [DOI] [PubMed] [Google Scholar]
- Wang S. L., Yu L. G., Liu R. P., Zhu W. Z., Gao W. M., Xue L. P., et al. (2014). Gene-gene interaction of GJB2, SOD2, and CAT on occupational noise-induced hearing loss in Chinese Han population. Biomed. Environ. Sci. 27 965–968. 10.3967/bes2014.131 [DOI] [PubMed] [Google Scholar]
- Wang B., Ding E., Shen H., Wang J., Sun K., Chen S., et al. (2017). Association of TagSNP in lncRNA HOTAIR with susceptibility to noise-induced hearing loss in a Chinese population. Hear. Res. 347 41–46. 10.1016/j.heares.2017.02.007 [DOI] [PubMed] [Google Scholar]
- Wang J., Li J., Peng K., Fu Z. Y., Tang J., Yang M. J., et al. (2017). Association of the C47T polymorphism in superoxide dismutase gene 2 with noise-induced hearing loss: a meta-analysis. Braz. J. Otorhinolaryngol. 83 80–87. 10.1016/j.bjorl.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. S., Xu K., Zhang H., Pu Y. P., Yin L. H., Han L., et al. (2019). Association between NFE2L2 gene polymorphisms and noise-induced hearing loss in a Chinese population. Biomed. Environ. Sci. 32 465–470. 10.3967/bes2019.063 [DOI] [PubMed] [Google Scholar]
- Wang N., Wang B., Guo J., Zhang S., Han L., Zhang J., et al. (2020). Single-nucleotide polymorphisms in XPO5 are associated with noise-induced hearing loss in a Chinese population. Biochem. Res. Int. 2020:9589310. 10.1155/2020/9589310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang G., Sun D., Wang B., Chen H., Chen M., et al. (2021). Histone deacetylase 2 polymorphisms associated with noise-induced hearing loss in Chinese workers. Environ. Sci. Pollut. Res. Int. 28 38254–38262. 10.1007/s11356-021-13486-5 [DOI] [PubMed] [Google Scholar]
- Wayne S., Robertson N. G., DeClau F., Chen N., Verhoeven K., Prasad S., et al. (2001). Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet. 10 195–200. 10.1093/hmg/10.3.195 [DOI] [PubMed] [Google Scholar]
- Wen X., Qiu C., Li X., Lin H., Huang Y. (2014). Association between GPX-1 single nucleotide polymorphisms and susceptibility to noise-induced hearing loss among Chinese Han population. Chin. J. Ind. Hyg. Occup. Dis. 32 568–572. [PubMed] [Google Scholar]
- Wilson S. M., Householder D. B., Coppola V., Tessarollo L., Fritzsch B., Lee E. C., et al. (2001). Mutations in Cdh23 cause non-syndromic hearing loss in waltzer mice. Genomics 74 228–233. 10.1006/geno.2001.6554 [DOI] [PubMed] [Google Scholar]
- Witte I., Altenhofer S., Wilgenbus P., Amort J., Clement A. M., Pautz A., et al. (2011). Beyond reduction of atherosclerosis: PON2 provides apoptosis resistance and stabilizes tumor cells. Cell Death Dis. 2:e112. 10.1038/cddis.2010.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. S., Yu J. N., Jiao J., Chen G. S., Zhang C. Y., Yu S. F. (2020). Association between PON2 gene polymorphisms and susceptibility to noise-induced hearing loss. Chin. J. Ind. Hyg. Occup. Dis. 38 128–132. 10.3760/cma.j.issn.1001-9391.2020.02.011 [DOI] [PubMed] [Google Scholar]
- Wu Y., Ni J., Qi M., Cao C., Shao Y., Xu L., et al. (2017). Associations of genetic variation in CASP3 gene with noise-induced hearing loss in a Chinese population: a case-control study. Environ. Health 16:78. 10.1186/s12940-017-0280-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. X., Zhu G. X., Liu X. Q., Sun F., Zhou K., Wang S., et al. (2014). Noise alters guinea pig’s blood-labyrinth barrier ultrastructure and permeability along with a decrease of cochlear Claudin-5 and Occludin. BMC Neurosci. 15:136. 10.1186/s12868-014-0136-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G., Gao W., Ji K., Liu S., Wan B., Luo J., et al. (2011). Single nucleotide polymorphisms analysis of noise-induced hearing loss using three-dimensional polyacrylamide gel-based microarray method. J. Biomed. Nanotechnol. 7 807–812. [DOI] [PubMed] [Google Scholar]
- Xin J. R., Chen Y. Q., Liu S. Y., Qian P. Y., Zhao T. Y., Wang H. Y., et al. (2021). Association between polymorphism of CASP and NOX3 with risk of noise-induced hearing loss. Chin. J. Ind. Hyg. Occup. Dis. 39 819–824. 10.3760/cma.j.cn121094-20200710-00390 [DOI] [PubMed] [Google Scholar]
- Xu S., Wang B., Han L., Pu Y., Zhu B., Zhang J. (2021). Polymorphisms in the FAS gene are associated with susceptibility to noise-induced hearing loss. Environ. Sci. Pollut. Res. Int. 28 21754–21765. 10.1007/s11356-020-12028-9 [DOI] [PubMed] [Google Scholar]
- Xu X., Yang Q., Jiao J., He L., Yu S., Wang J., et al. (2016). Genetic variation in POU4F3 and GRHL2 associated with noise-induced hearing loss in Chinese population: a case-control study. Int. J. Environ. Res. Public Health 13:561. 10.3390/ijerph13060561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. R., Wang J. J., Yang Q. Y., Jiao J., He L. H., Yu S. F., et al. (2017a). The Effect of PCDH15 gene variations on the risk of noise-induced hearing loss in a Chinese population. Biomed. Environ. Sci. 30 143–146. 10.3967/bes2017.019 [DOI] [PubMed] [Google Scholar]
- Xu X. R., Yang Q. Y., Jiao J., Zheng Y. X., He L. H., Yu S. F., et al. (2017b). Association between variations in protocadherin 15 gene and occupational noise-induced hearing loss. Chin. J. Prev. Med. 51 20–26. 10.3760/cma.j.issn.0253-9624.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Yan D., Zhu Y., Walsh T., Xie D., Yuan H., Sirmaci A., et al. (2013). Mutation of the ATP-gated P2X(2) receptor leads to progressive hearing loss and increased susceptibility to noise. Proc. Natl. Acad. Sci. U.S.A. 110 2228–2233. 10.1073/pnas.1222285110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang J., Wang X., Wang C., Chen J., Qian Y., et al. (2015). Identification of functional tag single nucleotide polymorphisms within the entire CAT gene and their clinical relevance in patients with noise-induced hearing loss. Int. J. Clin. Exp. Pathol. 8 2852–2863. [PMC free article] [PubMed] [Google Scholar]
- Yang L., Peng X., Li Y., Zhang X., Ma Y., Wu C., et al. (2019). Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol. Cancer 18:78. 10.1186/s12943-019-0990-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Tan H., Zheng J. R., Wang F., Jiang C., He M., et al. (2006). Association of cadherin CDH23 gene polymorphisms with noise induced hearing loss in Chinese workers. J. Hyg. Res. 35 19–22. [PubMed] [Google Scholar]
- Yang Q., Xu X., Jiao J., Zheng Y., He L., Yu S., et al. (2016). Genetic variation in EYA4 on the risk of noise-induced hearing loss in Chinese steelworks firm sample. Occup. Environ. Med. 73 823–828. 10.1136/oemed-2016-103613 [DOI] [PubMed] [Google Scholar]
- Yang Q. Y., Xu X. R., Jiao J., He L. H., Yu S. F., Gu G. Z., et al. (2016). Association between grainyhead-like 2 gene polymorphisms and noise-induced hearing loss. J. Peking Univ. 48 409–413. [PubMed] [Google Scholar]
- Yang Q. Y., Xu X. R., Jiao J., Zheng Y. X., He L. H., Yu S. F., et al. (2017). Association between eye absent homolog 4 gene polymorphisms and occupational noise-induced hearing loss. Chin. J. Prev. Med. 51 27–33. 10.3760/cma.j.issn.0253-9624.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Yang W., Liu J., Zheng F., Jia M., Zhao L., Lu T., et al. (2013). The evidence for association of ATP2B2 polymorphisms with autism in Chinese Han population. PLoS One 8:e61021. 10.1371/journal.pone.0061021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Guo J., Ding E., Zhang H., Han L., Zhu B. (2020). Salt-inducible kinase 3 haplotypes associated with noise-induced hearing loss in Chinese workers. Audiol. Neurootol. 25 200–208. 10.1159/000506066 [DOI] [PubMed] [Google Scholar]
- Yu J. N., Wu S. S., He C. H., Zhang C. Y., Mu H. X., Ma W. S., et al. (2016). Association between CDH23 gene polymorphisms and susceptibility to noise-induced hearing loss in the Chinese population: a meta-analysis. Chin. J. Ind. Hyg. Occup. Dis. 34 920–923. 10.3760/cma.j.issn.1001-9391.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Yu P., Jiao J., Chen G., Zhou W., Zhang H., Wu H., et al. (2018a). Effect of GRM7 polymorphisms on the development of noise-induced hearing loss in Chinese Han workers: a nested case-control study. BMC Med. Genet. 19:4. 10.1186/s12881-017-0515-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Jiao J., Gu G., Chen G., Zhang H., Zhou W., et al. (2018b). Relationship between GRM7 gene polymorphisms and the susceptibility to noise-induced hearing loss. J. Hyg. Res. 47 218–227. [PubMed] [Google Scholar]
- Zeke A., Misheva M., Reményi A., Bogoyevitch M. A. (2016). JNK signaling: regulation and functions based on complex protein-protein partnerships. Microbiol. Mol. Biol. Rev. 80 793–835. 10.1128/mmbr.00043-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Zheng H., Pyykko I., Zou J. (2019). The TLR-4/NF-kappaB signaling pathway activation in cochlear inflammation of rats with noise-induced hearing loss. Hear. Res. 379 59–68. [DOI] [PubMed] [Google Scholar]
- Zhang J., Song Y. L., Tian K. Y., Qiu J. H. (2017). Minocycline attenuates noise-induced hearing loss in rats. Neurosci. Lett. 639 31–35. 10.1016/j.neulet.2016.12.039 [DOI] [PubMed] [Google Scholar]
- Zhang S., Ding E., Yin H., Zhang H., Zhu B. (2019). Research and discussion on the relationships between noise-induced hearing loss and ATP2B2 gene polymorphism. Int. J. Genomics 2019:5048943. 10.1155/2019/5048943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Liu Y., Zhang L., Yang Z., Shao Y., Jiang C., et al. (2014). Genetic variations in protocadherin 15 and their interactions with noise exposure associated with noise-induced hearing loss in Chinese population. Environ. Res. 135 247–252. 10.1016/j.envres.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu Y., Zhang L., Yang Z., Yang L., Wang X., et al. (2015). Associations of genetic variations in EYA4, GRHL2 and DFNA5 with noise-induced hearing loss in Chinese population: a case-control study. Environ. Health 14:77. 10.1186/s12940-015-0063-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ni Y., Liu Y., Zhang L., Zhang M., Fang X., et al. (2019). Screening of noise-induced hearing loss (NIHL)-associated SNPs and the assessment of its genetic susceptibility. Environ. Health 18:30. 10.1186/s12940-019-0471-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Zhou J., Li H., Hui C., Bi J. (2020). Paraoxonase 3 gene polymorphisms are associated with occupational noise-induced deafness: a matched case-control study from China. PLoS One 15:e0240615. 10.1371/journal.pone.0240615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. H., Gu G. Z., Wu H., Li Y. H., Chen G. S., Zhang H. L., et al. (2020). Prediction of KCNQ4 gene polymorphism varies with CNE or noise exposure duration on the Risk of NIHL-Cox model analysis based on cohort study. Chin. J. Ind. Hyg. Occup. Dis. 38 111–116. 10.3760/cma.j.issn.1001-9391.2020.02.007 [DOI] [PubMed] [Google Scholar]
- Zhou X. X., Chen S., Xie L., Ji Y. Z., Wu X., Wang W. W., et al. (2016). Reduced Connexin26 in the mature cochlea increases susceptibility to noise-induced hearing lossin mice. Int. J. Mol. Sci. 17:301. 10.3390/ijms17030301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong S., Zeng X., Guan Y., Liu T., Luo P., Wan F., et al. (2019). Association of glutathione s-transferase M1 and T1 gene polymorphisms with the susceptibility to acquired sensorineural hearing loss: a systematic review and meta-analysis. Sci. Rep. 9:833. 10.1038/s41598-018-37386-w [DOI] [PMC free article] [PubMed] [Google Scholar]