Abstract
Two genes encoding β-galactosidase isoenzymes, β-galI and β-galIII, from Bifidobacterium infantis HL96 were revealed on 3.6- and 2.4-kb DNA fragments, respectively, by nucleotide sequence analysis of the two fragments. β-galI (3,069 bp) encodes a 1,022-amino-acid (aa) polypeptide with a predicted molecular mass of 113 kDa. A putative ribosome binding site and a promoter sequence were recognized at the 5′ flanking region of β-galI. Further upstream a partial sequence of an open reading frame revealed a putative lactose permease gene transcribing divergently from β-galI. The β-galIII gene (2,076 bp) encodes a 691-aa polypeptide with a calculated molecular mass of 76 kDa. A rho-independent transcription terminator-like sequence was found 25 bp downstream of the termination codon. The amino acid sequences of β-GalI and β-GalIII are homologous to those found in the LacZ and the LacG families, respectively. The acid-base, nucleophilic, and substrate recognition sites conserved in the LacZ family were found in β-GalI, and a possible acid-base site proposed for the LacG family was located in β-GalIII, which featured a glutamate at residue 160. The coding regions of the β-galI and β-galIII genes were each cloned downstream of a T7 promoter for overexpression in Escherichia coli. The molecular masses of the overexpressed proteins, as estimated by polyacrylamide gel electrophoresis on sodium dodecyl sulfate-polyacrylamide gels, agree with their predicted molecular weights. β-GalI and β-GalIII were specific for β-d-anomer-linked galactoside substrates. Both are more active in response to ONPG (o-nitrophenyl-β-d-galactopyranoside) than in response to lactose, particularly β-GalIII. The galacto-oligosaccharide yield in the reaction catalyzed by β-GalI at 37°C in 20% (wt/vol) lactose solution was 130 mg/ml, which is more than six times higher than the maximum yield obtained with β-GalIII. The structure of the major trisaccharide produced by β-GalI catalysis was characterized as O-β-d-galactopyranosyl-(1-3)-O-β-d-galactopyranosyl-(1-4)-d-glucopyranose (3′-galactosyl-lactose).
Bifidobacterium spp. are immobile gram-positive anaerobic bacteria that were originally isolated by Tissier in the feces of breast-fed infants in 1899 (3). They were once considered bacteria of other genera, such as Bacillus or Lactobacillus, due to their morphological and physiological similarities (3). However, the enzymatic characteristics and molecular genetics of this group are distinct enough to exclude bifidobacteria from other genera (9, 52). Currently, a total of 32 species are included in this genus (25). Bifidobacteria are not only unique in taxonomic terms but also well known for their beneficial probiotic effects, which were initially investigated by Manciaux in 1958. The application of bifidobacteria to food products started in Japan in the 1980s and has become popular worldwide in recent years. Bifidobacterium infantis is one of the species frequently used in bifidobacterium-containing probiotic products. There is an increasing interest in exploiting the enzymatic characteristics and the molecular biology of B. infantis.
β-Galactosidases (EC 3.2.1.23) catalyze both hydrolytic and transgalactosylation reactions (44, 63). Hydrolytic activity has been applied in the food industry for decades for reducing lactose content in milk. However, transgalactosylation activity, which yields galacto-oligosaccharides (GaOS), has been less well studied than the hydrolytic reaction. GaOS synthesized via β-galactosidases have shown beneficial effects on the promotion of the growth of bifidobacteria (38, 56). This has prompted interest in studying other β-galactosidases. The electrophoretic analysis of β-galactosidases from Bifidobacterium species revealed that most of the strains contain more than one β-galactosidase isoenzyme (46). Our own previous studies showed the presence of three isoenzymes (β-GalI, -II, and -III) in B. infantis HL96, which possesses high transgalactosylation activity compared with that of 29 selected strains of bifidobacteria (unpublished data). To identify an enzyme with powerful transgalactosylation activity and to reach a better understanding of the molecular organization of the genes coding for lactose utilization in a useful probiotic strain, β-galactosidase genes of HL96 were cloned in a previous study (22). The molecular properties and enzymatic characteristics, as well as the structure of a main GaOS product, are reported in this study.
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli DH10B (Gibco-BRL) was used as the recipient in all transformation experiments involving the subcloning of pBIG1 and pBIG4. E. coli ER2566 (New England BioLabs Inc.), providing a T7 RNA polymerase, was used as the host for the overexpression of β-galI and β-galIII genes from the T7 promoter. E. coli strains, except for β-galactosidase gene transformants of ER2566, were grown in Luria broth containing the antibiotics required for maintaining the plasmid. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates for the detection of β-galactosidase activity in the transformed E. coli were prepared with Luria broth supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranose (Roche Diagnostics) at 40 μg/ml.
Plasmid preparation.
pBIG1 and pBIG4 (22) contain β-galI and β-galIII genes, respectively, of B. infantis HL96, which is a strain isolated at the Food Research and Development Center, Agriculture and Agri-Food Canada (Ste-Hyacinthe, Québec). Plasmid pBluescript SK(−) (Stratagene Inc.) was used for subcloning the DNA fragments from pBIG1 and pBIG4. pET24(+) (Novagen Inc.) containing a T7 promoter was used for overexpression of β-galI and β-galIII. The double-stranded plasmid DNA used for subcloning and DNA sequencing was prepared by following the Qiagen plasmid purification method.
Subcloning and sequencing.
Restriction maps of pBIG1 and pBIG4 were generated (22), and subclones of pBIG1 and pBIG4 were constructed. Two of the subclones, pG1Bg and pG4B, were constructed by deleting two BglII sites of pBIG1 and by deleting the entire fragment from the right end to the first BamHI site of pBIG4, respectively. Fragments of pG1Bg and pG4B, 3.6 and 2.4 kb, respectively, were subcloned into pBluescript for nucleotide sequence determination; the latter was performed by the dideoxy chain termination method of Sanger et al. (48) using the Sequenase sequencing kit (U.S. Biochemicals). T3 and T7 primers or synthetic oligonucleotides were used as sequencing primers.
Sequence analysis.
Nucleotide sequences were analyzed using the Wisconsin package (version 10.1) from the Genetics Computer Group. Proteins homologous to the deduced amino acid sequences of β-GalI and β-GalIII were retrieved from the nonredundant protein database using the BlastP program, which is available from the National Center for Biotechnology Information. The comparison of β-GalI and β-GalIII with homologous proteins was carried out using the BestFit program (version 1.1) (16), which is available from SeqWeb.
Enzymatic assay and HPLC.
β-Galactosidase activity was measured by incubating enzymes with 10 mM o-nitrophenyl-β-d-galactopyranoside (ONPG; Sigma) in 50 mM sodium phosphate buffer (pH 7.5) at 37°C for 10 min and stopping the reaction by adding an equal volume of 1.0 M Na2CO3. The released o-nitrophenol was quantitatively determined by measuring the A420 of the reaction solution. One unit of activity was defined as that amount of enzyme liberating 1 μmol of o-nitrophenol per min, as described previously (22). The hydrolytic activities of the enzymes using various substrates (purchased from Sigma) were studied under the same experimental conditions. Hydrolytic activities in response to lactose, maltose, sucrose, raffinose, and melibiose were determined by analyzing the amount of glucose formed or the amount of substrate utilized in the reaction mixture by high-performance liquid chromatography (HPLC) (Waters system) including a differential refractometer (type R401) and a polymeric column (ION-300). The flow rate was adjusted to 0.5 ml/min, with 0.02 N H2SO4 as the mobile phase, and the elution time was programmed to be 20 min.
Overexpression of β-GalI and β-GalIII.
Overexpression plasmids pEBIG1 and pEBIG4 were constructed by cloning the β-galI and β-galIII genes from PCR-amplified fragments into EcoRI/HindIII and BamHI/NotI sites of pET24, respectively. Upstream and downstream primers pG1UE and pG1DH (5′-CTCTTCCATAATAGAATTCACAACGAGG518 and 5′-AGGCGCCAGCAAGCTTACCTGTGGCGC, respectively) and pG4UB and pG4DN (5′-TGCGTACAAGGATCCTATCGATGCAATG92 and 5′-CGACAGGTGCGGCCGCTGTTGACCCATGAGTG2341, respectively) were used in PCR to amplify β-galI and β-galIII genes from pBIG1 and pBIG4, respectively. These primers annealed with the respective genes at the positions indicated by the numbers showing the 3′ end nucleotides, except pG1DH, which annealed with the BamHI downstream region of pBR322. The numbers are counted from the initial nucleotides of the inserts of pG1Bg and pG4B. Both nucleotide sequences are available in GenBank (see below). The primers created a cloning site (underlined) at each end of the β-galI and β-galIII gene fragments for their cloning into pET24. E. coli ER2566(pEBIG1) and ER2566(pEBIG4) were grown in 2YT medium (1.5% tryptone, 0.75% yeast extract, 0.5% NaCl, 0.5% glycerol) until an optical density at 600 nm of 1.0 was reached. Isopropyl-β-d-thiogalactopyranoside (IPTG;1 mM; Roche Diagnostics) was then added to the culture medium, and incubation at 37°C continued for 2 h. The induced cells were harvested and disrupted by sonication using a microtip set at power level 4 at 30% duty for 2 min with 30-s cooling intervals. The disrupted cells were boiled in sample buffer for 10 min before loading onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. The supernatant after centrifugation at 15,000 × g for 20 min (4°C) was used for enzymatic assays and nondenaturing PAGE.
PAGE and activity staining.
In accordance with the method of Laemmli (28), proteins were analyzed by SDS-PAGE using 10% (wt/vol) nondenaturing polyacrylamide gel and staining by Coomassie blue or activity staining; the latter was performed by incubating the gel in 4-o-methylumbelliferyl-β-d-galactoside (4MeUmG) solution as previously described (22).
GaOS synthesis.
Cell extracts were incubated with 20% (wt/vol) lactose solution in sodium phosphate buffer (pH 7.5) for 30 h at 37°C, with continuous agitation at 100 rpm in a shaker incubator to facilitate hydrolysis. Samples were withdrawn at 5-h intervals and immediately heated at 90°C for 10 min to stop the reaction. The samples were diluted appropriately, filtered, and injected into the HPLC system for GaOS analysis. The rate of production of GaOS was estimated from the total amount of saccharides eluted at retention times between those for standard saccharides lactose and stachyose in the reaction solution. Residual enzyme activities were monitored during the reaction by recovering the enzymes from the reaction mixtures at 5-h intervals and assaying for ONPG hydrolytic activity. Enzymes were recovered by centrifugation (Centricon YM-100; Millipore).
Purification of GaOS.
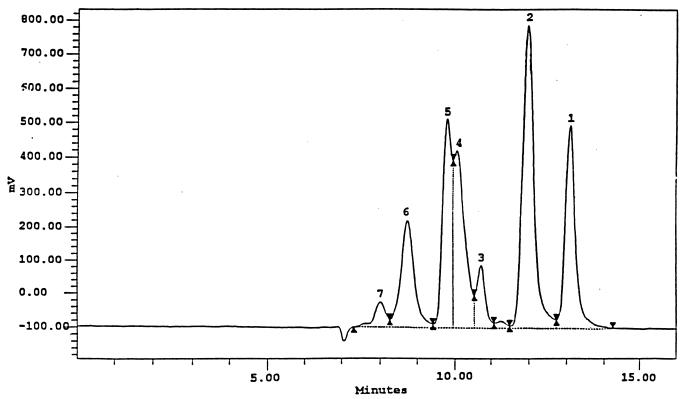
To identify the GaOS produced via the β-GalI reaction, an activated-charcoal column equilibrated with water was used for separating the monosaccharides from the GaOS mixture. This mixture was derived from the lactose hydrolysis reaction using cell extract of ER2566(pEBIG1). The reaction was carried out by incubating the cell extract (∼100-U activity strength) in 20% lactose solution (40 ml in sodium phosphate buffer, pH 7.5) at 37°C for 15 h. The reaction mixture was applied to the column, the column was washed with 300 ml of 3% ethanol, and the GaOS was subsequently eluted using an ethanol gradient (5 to 20%). The collected GaOS fraction was concentrated and further purified by HPLC on an ION-300 column as previously described. GaOS fractions are eluted at a flow rate of 0.5 ml/min, and 200-μl fractions corresponding to the GaOS (see Fig. 5, peak 6) were collected. Fractions from 20 HPLC runs were pooled and freeze-dried for nuclear magnetic resonance (NMR) analysis.
FIG. 5.
HPLC chromatogram of lactose hydrolysis products of β-GalI (peak 1, galactose; peak 2, glucose; peak 3, impurity present in lactose; peak 4, lactose; peaks 5 to 7, GaOS).
NMR spectroscopy.
NMR experiments were performed on a Varian Unity 500 NMR spectrometer. Samples were prepared by suspending the GaOS purified from HPLC in D2O (15 mM). Standard two-dimensional techniques including DQF-COSY, TOCSY, ROESY, HSQC, and HMBC were employed, and residual water signals were presaturated with a low-power continuous wave. Proton chemical shifts were referenced with external DSS, and for 13C the relative reference was to external dioxane. In all experiments, spectral windows in the proton dimension were 1,845 Hz and in the carbon dimension were 7,542 Hz. A 1-s relaxation delay was used for all experiments. Quadrature detection was achieved with the States method (54) in indirect dimension, with a spin locking time for ROESY spectra of 500 ms. All the spectra were zero filled to 2,048 by 1,024 points and processed with a squared cosine-bell function in both dimensions for phase-sensitive spectra; for HMBC spectra were processed with an unshifted squared sine-bell function. All the NMR data were acquired with Varian's standard pulse sequence and processed with NMRPipe (8). Spectral assignment was largely facilitated by the use of ANSIG (26, 27).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the β-galI and β-galIII genes are AF192265 and AF192266, respectively.
RESULTS
Nucleotide sequences of β-galI and β-galIII genes.
Subclones of pBIG1 and pBIG4 with deletions were constructed for locating the genes and for analyzing their nucleotide sequences. β-GalI activity was not affected by deletion of the BglII fragment but was abolished by deletion of the XhoI fragment (22), indicating that the β-galI gene is located within the insert of pG1Bg. pBIG4 subclone pG4B, containing a 2.4-kb DNA fragment, was found to possess β-galactosidase activity. The DNA sequences of the fragments cloned in pG1Bg and pG4B revealed two open reading frames (ORFs). The corresponding deduced amino acid sequences suggested that each encodes a β-galactosidase. We designated them β-galI and β-galIII, respectively. Part of the nucleotide sequence of the insert of pG1Bg is shown in Fig. 1. The coding region of β-galI (3,069 bp) was determined to start with ATG at nucleotide 541 and end with TGA at nucleotide 3609. The putative ribosome binding sequence (RBS) AGAAAG (Fig. 1) was found nine bases upstream from the β-galI translation start site. The ORF of β-galIII (2,076 bp) was found to start with ATG at nucleotide 114 and end at TAA at nucleotide 2189. The putative RBS AAGGAA was found 10 bases upstream from the translation start site. Analysis of the nucleotide sequence upstream of β-galI revealed the presence of a coding region, transcribing divergently from β-galI (Fig. 1). The deduced amino acid sequence encoded by this partial ORF showed some similarity with the sequences encoded by the 5′ regions of the lactose permease genes of Leuconostoc lactis (59), Lactococcus lactis (29), Lactobacillus bulgaricus (30), and Streptococcus thermophilus (43). A rho-independent transcription terminator-like region (AACGGCTCCCTTGCTGCTCATCGTAG CAGGGGAGTCGTTTTCGTTT; stem region underlined) was found 25 bp away from the termination codon of β-galIII. The nucleotide sequence between β-galI and the putative lactose permease coding region is AT rich (G+C content: 43%). In contrast, the G+C content of the β-galI coding region is 64.6%. Using NNPP (promoter prediction by neural network) (45), we located two promoter sequences within this AT-rich region (Fig. 1). Further experiments are required to analyze these possible promoters.
FIG. 1.
Partial nucleotide sequence of the insert fragment of pG1Bg. The coding region of β-galI starts from base 541 and ends at base 3609. A putative lactose permease 5′ coding region transcribing divergently from β-galI is located upstream of β-galI; its start codon, ATG, is boxed. The deduced amino acid sequence is shown below the nucleotide sequence. The predicted ribosome binding sites (r.b.) for β-galI and permease are underlined. Possible +1 to −40 sequences for β-galI and permease genes are highlighted. Also indicated is an EcoRI site, upstream of the β-galI gene, created by primer pG1UE in PCR.
Comparison of amino acid sequences.
The alignments of the deduced amino acid sequences of β-GalI and β-GalIII with other homologous β-galactosidases are summarized in Table 1. β-GalI features some sequence homology with β-galactosidases from various microorganisms, including the LacZ and EbgA of E. coli (Table 1). However, β-GalIII appears homologous to other microbial β-galactosidases (Table 1), some of which have been assigned to a new β-galactosidase family, LacG, which includes the Arthrobacter sp. strain B7 β-galactosidase of fragment 12, Bacillus stearothermophilus BgaB, and Bacillus circulans BgaA (14). Bacillus stearothermophilus BgaB was classified as a member of glycosyl hydrolase family 42 based on amino acid sequence, while E. coli LacZ belongs to glycosyl hydrolase family 2 (17). β-GalIII was 52 and 47.6% similar to Bacillus stearothermophilus BgaB and Bacillus circulans BgaA, respectively, whereas its homology with the Arthrobater β-galactosidase was calculated at only 35%. The alignment of β-GalIII with β-GalI or with other β-galactosidases homologous to E. coli LacZ (LacZ family) using the BestFit program failed to provide any significant homology (data not shown). Our study suggests that β-GalIII is a new member of glycosyl hydrolase family 42 and of the LacG family, the members of which have molecular masses ranging between 70 and 80 kDa. Three regions of the E. coli LacZ, two flanking a glutamate molecule at residues 461 and 537 and one flanking a glycine at residue 794, were reported to be the acid-base, nucleophilic, and substrate recognition sites (7, 12, 32). These three regions are well conserved in almost all β-galactosidases of the LacZ family. Similarly, these three conserved regions were revealed in β-GalI, with two glutamates located at residues 469 and 536 and a glycine located at 793 (Fig. 2A). The active sites of LacZ have been well studied. On the other hand, an acid-base site with a glutamate as the key amino acid has been proposed for the LacG family (14, 18). Conservation of this region among β-GalIII and seven other β-galactosidases (Fig. 2B) is not high, as observed by the alignment of the LacZ family. However, a conserved WH-SNEY sequence was revealed in our study. Whether or not Glu160 is involved in enzyme catalysis remains to be determined. The codon usages of β-galI and β-galIII genes are different from that of E. coli, specifically the codons for Gly, Glu, Val, Ala, and Lys. There appears to be a preference for G and C residues in the third bases of codons in B. infantis genes.
TABLE 1.
Comparison of β-GalI and β-GalIII of B. infantis with other β-galactosidases
| Enzyme source | No. of aab | Similarity (%) | Identity (%) | Referencea |
|---|---|---|---|---|
| B. infantis β-GalI | 1,022 | 100 | 100 | |
| Streptococcus thermophilus | 1,026 | 55.7 | 46.3 | 51 |
| Lactobacillus bulgaricus | 1,007 | 54.8 | 45.8 | 50 |
| Clostridium acetobutylicum | 897 | 53 | 44.6 | 15 |
| Bacillus megaterium | 1,034 | 49 | 43.3 | |
| Thermotoga maritima | 1,087 | 44 | 35.2 | 37 |
| E. coli EbgA | 1,031 | 43 | 32.7 | 55 |
| Arthrobacter sp. strain B7, fragment 15 | 1,015 | 43 | 34.8 | 58 |
| Lactococcus lactis | 998 | 42.9 | 34.3 | 29 |
| Klebsiella pneumoniae | 1,034 | 42.6 | 34.6 | 6 |
| Enterobacter cloacae | 1,028 | 42.2 | 34.5 | 36 |
| E. coli LacZ | 1,024 | 41.7 | 34 | 24 |
| Kluyveromyces lactis | 1,025 | 41.7 | 32 | 42 |
| B. infantis β-GalIII | 691 | 100 | 100 | |
| Bacillus stearothermophilus | 672 | 52 | 41.3 | 19 |
| Caldicellulosiruptor sp. strain 14B | 678 | 49.4 | 39 | |
| Thermotoga maritima | 672 | 48.2 | 36.8 | 37 |
| Bacillus circulans BgaA | 675 | 47.6 | 35.5 | |
| Bacillus subtilis | 687 | 46 | 33.1 | |
| Clostridium perfringens | 676 | 46 | 36.3 | |
| Thermus sp. strain T2 BgaA | 645 | 43.9 | 35.7 | 60 |
FIG. 2.
(A) Multiple alignment of the possible acid-base, nucleophilic, and substrate recognition sites of β-GalI and of 12 listed β-galactosidases. Abbreviations: ec, Enterobacter cloacae; e, E. coli LacZ; k, Klebsiella pneumoniae; la, Lactococcus lactis; s, Streptococcus thermophilus; clo, Clostridium acetobutylicum; l, Lactobacillus bulgaricus; bif, B. infantis β-GalI; bm, Bacillus megaterium; t, Thermotoga maritima; ee, E. coli EbgA; kl, Kluyveromyces lactis; art, Arthrobacter sp. strain B7 fragment 15. (B) Multiple alignment of the possible acid-base site of β-GalIII and seven other β-galactosidases. Abbreviations: bsu, Bacillus subtilis; cp, Clostridium perfringens; cal, Caldicellulosiruptor sp. strain 14B; bc, Bacillus circulans BgaA; tm, Thermotoga maritima; bs, Bacillus stearothermophilus; bif, B. infantis β-GalIII; t, Thermus sp. strain T2 BgaA. Conserved amino acids are highlighted. ↓, amino acids proposed to be the key residues in the active sites. Multiple alignment was done using the PrettyBox program of the Genetics Computer Group.
Overexpression of β-galI and β-galIII genes in E. coli
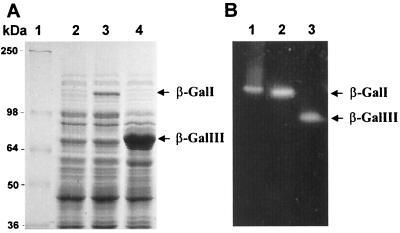
To further investigate the two β-galactosidase gene products, we made use of the T7 RNA polymerase expression system for overexpressing β-galI and β-galIII genes in E. coli. The coding regions of β-galI and β-galIII were cloned into pET24, under the control of a T7 promoter. Gene expression in E. coli was induced by IPTG and analyzed by both SDS-PAGE and nondenaturing PAGE. A protein with an apparent molecular mass of approximately 115 kDa was overproduced from pEBIG1 (Fig. 3A), in close agreement with the predicted molecular weight of β-GalI. The apparent molecular mass of the protein overproduced from pBIG4 is estimated at 76 kDa (Fig. 3A), also in agreement with the predicted molecular weight of β-GalIII. Activity staining of the nondenaturing PAGE with 4MeUmG (Fig. 3B) confirmed the presence of β-GalI and β-GalIII.
FIG. 3.
(A) SDS-PAGE analysis of β-galI and β-galIII genes overexpressed in E. coli ER2566 under 1 mM IPTG induction. Lane 1, marker proteins (molecular masses are indicated); lanes 2, 3, and 4, 1 mM IPTG-treated whole-cell lysates of E. coli ER2566 containing pET24 (control), pEBIG1, and pEBIG4, respectively. Arrows, overexpressed proteins of β-GalI and β-GalIII. (B) Activity staining on nondenaturing polyacrylamide gel. Lane 1, commercial E. coli β-galactosidase (positive control); lanes 2 and 3, soluble fractions of IPTG-induced E. coli ER2566 containing pEBIG1 (lane 2) or pEBIG4 (lane 3) from sonication treatment.
Substrate specificity.
The hydrolytic activities of β-GalI and β-GalIII in response to various glycosides were analyzed and compared with the hydrolysis of ONPG (Table 2). β-GalI and β-GalIII were highly active in response to ONPG, which is a β-d-anomer-linked galactoside. Other substrates having an α-d linkage or not having a galactose in the glycon moiety were not hydrolyzed or were slightly hydrolyzed by β-GalIII in the case of p-nitrophenyl-β-d-galacturonide. Under identical reaction conditions, β-GalI hydrolyzed lactose at a rate approximately 60% that of ONPG. In contrast, hydrolysis of lactose by β-GalIII proceeded at a rate less than 10% that of ONPG hydrolysis. Maltose, sucrose, raffinose, and melibiose were apparently not hydrolyzed by β-GalI or by β-GalIII.
TABLE 2.
Relative hydrolytic rates of various substrates by β-GalI and β-GalIII
| Substratea | Relative
hydrolytic rate (%)b for:
|
|
|---|---|---|
| β-GalI | β-GalIII | |
| ONPG | 100 | 100 |
| Lactose | 62 | 9.3 |
| PNP-β-d-galacturonide | 0c | 2.4 |
| PNP-β-d-glucopyranoside | 0 | 0 |
| PNP-β-d-glucuronide | 0 | 0 |
| ONP-α-d-galactopyranoside | 0 | 0 |
| PNP-α-d-glucopyranoside | 0 | 0 |
PNP, p-nitrophenyl.
The ONPG hydrolytic activities of the crude β-GalI and β-GalIII were 102 and 236 U/mg, respectively. They were defined as 100% and were used for comparison with those of other substrates.
Zero activity means no activity was detected or the activity was lower than 1% of that of ONPG.
GaOS synthesis.
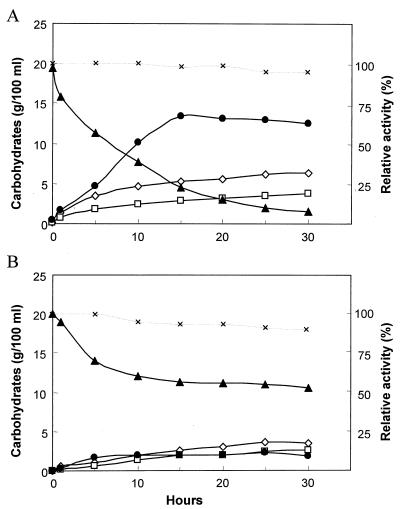
We also monitored the time course of GaOS production during lactose hydrolysis by β-GalI and β-GalIII (Fig. 4). Results showed that the maximal concentration of GaOS produced by β-GalI was 130 mg/ml, this being obtained after a 15-h incubation period. On the other hand, the maximum concentration of GaOS produced by β-GalIII was 21 mg/ml. Three different types of GaOS with retention times of 8.08, 8.82, and 9.69 min (Fig. 5) were eluted between lactose and stachyose (retention time was 7.95 min for the latter). Oligosaccharides larger than stachyose were not observed under the HPLC conditions described in Materials and Methods. The ONPG hydrolytic activities of β-GalI and β- GalIII were constant throughout the reactions (Fig. 4), which suggested that both enzymes remained active during the reaction in 20% lactose at 37°C for 30 h.
FIG. 4.
Time course of GaOS production during lactose hydrolysis by β-GalI (A) and β-GalIII (B). Symbols: ⋄, glucose; □, galactose; ▴, lactose; ●, GaOS. ×, residual ONPG hydrolytic activities of the recovered enzymes compared with the original activities (defined as 100%) at 0 h.
NMR analysis of GaOS.
The GaOS of peak 6 (Fig. 5) was further purified and determined by a two-dimensional NMR technique to be O-β-d-galactopyranosyl-(1-3)-O-β-d-galactopyranosyl-(1-4)-d-glucopyranose. Since the sample contains minor di- and tetrasaccharide impurities, which substantially obscured the spectral assignment, reference trisaccharide O-β-d-galactopyranosyl-(1-4)-O-β-d-galactopyranosyl-(1-4)-d-glucopyranose (4′-galactosyl-lactose) was used in the analysis. Proton assignments were verified from standard homonuclear experiments DQF-COSY and TOCSY, and 13C assignments were then verified from the HSQC experiment, which detects the C-H correlation. Due to the weak J couplings between H4 and H5 of galactose (J < 1 Hz) (20), the assignments of H5 and H6 on the galactose units were deduced from the HMBC spectrum. NOE experiments were primarily used for sequencing the sugars. The largest NOE is usually observed on the linkage protons, although other NOEs can also occur for protons near the linkage site. Since the NOE sizes at 500 MHz for trisaccharides are usually negative and small, ROESY spectra were collected instead. All the chemical shifts are tabulated in Table 3.
TABLE 3.
Proton and carbon-13 chemical shifts for GaOS and the reference compound
| Unit | Reference compound
|
GaOS
|
||
|---|---|---|---|---|
| Proton shift | Carbon shift | Proton shift | Carbon shift | |
| 3-Glu-α-(1) | 5.179 | 92.87 | 5.207 | 92.92 |
| 3-Glu-α-(2) | 3.533 | 72.44 | 3.567 | 72.11 |
| 3-Glu-α-(3) | 3.791 | 72.45 | 3.825 | 72.43 |
| 3-Glu-α-(4) | 3.605 | 72.42 | ||
| 3-Glu-α-(5) | 3.903 | 71.18 | 3.945 | 71.13 |
| 3-Glu-α-(6) | 3.827 | 60.99 | 3.856 | 61 |
| 3-Glu-β-(1) | 4.618 | 96.81 | 4.647 | 96.83 |
| 3-Glu-β-(2) | 3.236 | 74.86 | 3.266 | 74.85 |
| 3-Glu-β-(3) | 3.594 | 75.44 | 3.630 | 75.38 |
| 3-Glu-β-(4) | 3.709 | 75.53 | 3.731 | 76.04 |
| 3-Glu-β-(5) | 3.551 | 75.84 | 3.605 | 75.85 |
| 3-Glu-β-(6) | 3.760, 3.911 | 61.1 | 3.792, 3.945 | 61.11 |
| 2-Gal-(1) | 4.438 | 103.93 | 4.490 | 103.61 |
| 2-Gal-(2) | 3.581 | 72.43 | 3.692 | 71.22 |
| 2-Gal-(3) | 3.734 | 74.02 | 3.825 | 82.96 |
| 2-Gal-(4) | 4.150 | 78.22 | 4.185 | 69.5 |
| 2-Gal-(5) | 3.611 | 79.43 | 3.669 | 79.21 |
| 2-Gal-(6) | 3.788 | 61.8 | 3.826–3.704 | 62.05 |
| 1-Gal-(1) | 4.556 | 105.25 | 4.594 | 105.34 |
| 1-Gal-(2) | 3.529 | 72.43 | 3.589 | 72.11 |
| 1-Gal-(3) | 3.616 | 73.85 | 3.655 | 73.63 |
| 1-Gal-(4) | 3.862 | 69.67 | 3.913 | 69.63 |
| 1-Gal-(5) | 3.633 | 76.21 | 3.677 | 76.14 |
| 1-Gal-(6) | 3.720 | 62.03 | 3.826–3.704 | 62.05 |
The linkage site between two galactoses (Gal1-Gal2-Glu) was determined by comparing the 13C chemical shifts of the GaOS and of the reference compound, since the linkage site often exhibits a dramatic chemical shift (47); this was also confirmed by the NOE experiments. The significant drift of the 13C chemical shift at C3 of 2-Gal together with the large NOE size between the H1 of 1-Gal and the H3 of 2-Gal unambiguously established the presence of a 1,3 link. On the other hand, no significant 13C chemical shift variations were observed for the Glu unit of GaOS when comparison with the reference compound was made. Clear NOE was detected between H1 of 2-Gal and H4 of Glu. We therefore concluded that the linking between 2-Gal and Glu occurs at the 1,4 sites. Figure 6 summarizes all the important NOEs for the determination of linking sites in the GaOS.
FIG. 6.
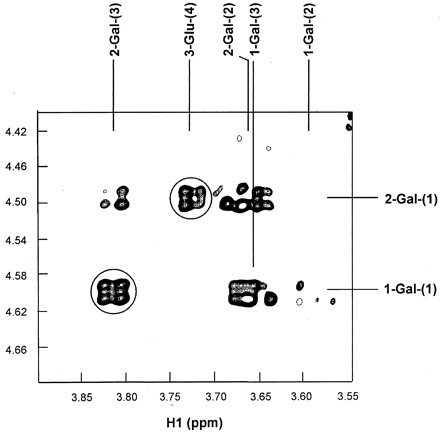
Identification of the glycosidation site from the ROESY spectrum of GaOS. The interresidue cross peaks are circled. The spectrum was collected with the mixing time of 500 ms at 500 MHz.
DISCUSSION
Two β-galactosidase isoenzymes of B. infantis HL96, termed β-GalI and β-GalIII, each of which possesses unique genetic and biochemical properties, were characterized. Although the physiological significance of isoenzymes is not generally understood, isoenzymes with different properties have been identified in several bacteria, such as Arthrobacter spp. (31), Thermotoga maritima (33), and Bacillus circulans (34). The first two were reported to have β-galactosidase isoenzymes homologous to those from different families of β-galactosidases, and two β-galactosidases from Bacillus circulans differed considerably in terms of their GaOS-producing activity. This is but one of a limited number of comparisons of transgalactosylation activities among β-galactosidase isoenzymes. The advantages of having isoenzymes with different properties in terms of gene expression and of hydrolytic and synthetic activities may provide the microorganisms with improved adaptability in changing growth conditions. Alternatively, each isoenzyme may be responsible for a particular hydrolytic or synthetic reaction, such as those involved in the metabolism of lactose or other carbohydrates. Another possibility is that β-galactosidase isoenzyme genes may have arisen through transfer or evolution of genetic material among microorganisms. This may explain the high homology among β-galactosidases from different sources. The cloning and sequencing of β-galI and β-galIII genes of B. infantis HL96 is a logical starting point for exploring the molecular biology of B. infantis. Gene isolation allows reverse genetic analysis of the physiological significance of each isoenzyme. It becomes possible to knock out β-galI or β-galIII in B. infantis to observe how the mutation would affect cell metabolism. This may provide clues to the importance of β-GalI and β-GalIII in B. infantis.
Comparison between β-GalI and the β-galactosidase from Bifidobacterium longum (GenBank accession no. AJ242596) revealed a homology of 98%. A difference of only 35 amino acids was found. The high similarity between β-galactosidases from B. infantis and from B. longum is not surprising. Immunological and DNA-DNA hybridization approaches have demonstrated that B. infantis and B. longum have a close phylogenetic relationship (49, 53).
β-Galactosidase genes are commonly in the same operon with genes encoding permeases, such as the lac genes of E. coli (4), Klebsiella pneumoniae (6), and Lactobacillus bulgaricus (30). β-galI is unlikely to be in the same operon with a permease gene; instead a permease-gene-like gene transcribing divergently from β-galI was found upstream of β-galI. Although the 3′ flanking region of β-galI was not analyzed in this study, according to the nucleotide sequence of β-gal from B. longum, a tRNA-Pro is located at the downstream region. There is a possibility that the permease gene may be linked with the β-galIII gene and transcribed in an operon using a promoter different from that for the β-galI gene. So far, not enough nucleotide sequence data are available to exclude this possibility.
The formation of GaOS during lactose hydrolysis by β-galactosidases from yeast, bacteria, and fungi has been studied by several investigators (1, 5, 13, 21, 35, 41, 57, 61). Those studies showed that the yields of GaOS were mainly affected by enzyme sources and substrate concentrations. We found that β-GalI produced much higher GaOS than β-GalIII. The yield of GaOS could not be significantly improved by increasing the amount of β-GalIII in the reaction or by varying conditions such as reaction temperature, pH, and lactose concentration. On the other hand, GaOS production using β-GalI was increased 40-fold when the lactose concentration was increased from 1 to 20% (data not shown). The yield of GaOS by β-GalI was higher than those reported using the β-galactosidase of Thermus aquaticus (2), B. bifidum (10), Bacillus circulans (34), Aspergillus oryzae (44), and Kluyveromyces lactis (23), which were less than 42 mg/ml. The total concentration of GaOS (mainly tri- and tetrasaccharides) produced by the yeast Rhodotorula minuta β-galactosidase was about 78 mg/ml (40), which was lower than the total GaOS but higher than the concentration (66 mg/ml) of tri- and tetrasaccharides (Fig. 5, peaks 6 and 7) produced by β-GalI.
The major trisaccharide produced with β-GalI was 3′-galactosyl-lactose. The trisaccharide produced by β-galactosidase from B. bifidum was also identified as 3′-galactosyl-lactose (11). However, 4′-galactosyl-lactose and 6′-galactosyl-lactose were reported to be preferentially formed during the transgalactosylation reactions (1, 39, 40, 62), and several β-galactosidases were found to produce more than one type of GaOS (1, 57, 62). Comparing the prebiotic effects of 3′-galactosyl-lactose, 4′-galactosyl-lactose, and 6′-galactosyl-lactose is essential to determine their relative values as nutraceutical additives for humans or animals. If saccharides with β(1-3) linkages are the only transgalactosylation reaction products obtained with β-GalI, such high enzyme stereoselectivity would be beneficial for synthesizing certain specific valuable compounds. The characterization of other GaOS produced with β-GalI and the optimization of reaction conditions to increase GaOS yield are under way.
β-GalIII exhibits lower hydrolytic activity in response to lactose than in response to ONPG. It is unclear whether the β-galactosidases of the LacZ family are more active in hydrolyzing lactose and whether they possess higher transgalactosylation activity than those of the LacG family. More β-galactosidases belonging to the LacZ and LacG families are being characterized, and this should provide more knowledge of the catalytic mechanism of β-galactosidases. A full understanding of the catalytic mechanisms of LacZ and LacG is especially valuable to clarify the correlation of structure and function of such an industrially useful enzyme.
ACKNOWLEDGMENTS
This work was partly supported by a NSERC strategic grant for research and NSERC doctorate fellowship awarded to M. N. Hung.
REFERENCES
- 1.Asp N G, Burvall A, Dahlqvist A, Hallgren P, Lundblad A. Oligosaccharide formation during hydrolysis of lactose with Saccharomyces lactis lactase (MaxilactR): part 2—oligosaccharide structures. Food Chem. 1980;5:147–153. [Google Scholar]
- 2.Berger J L, Lee B H, Lacroix C. Purification, properties and characterization of a high-molecular-mass β-galactosidase isoenzyme from Thermus aquaticusYT-1. Biotechnol Appl Biochem. 1997;25:29–41. [Google Scholar]
- 3.Bezkorovainy A. Classification of bifidobacteria. In: Bezkorovainy A, Miller-Catchpole R, editors. Biochemistry and physiology of bifidobacteria. Boca Raton, Fla: CRC Press Inc.; 1989. pp. 2–28. [Google Scholar]
- 4.Buchel D E, Groneborn B, Muller-Hill B. Sequence of the lactose permease gene. Nature. 1980;283:541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- 5.Burvall A, Asp N-G, Dahlqvist A. Oligosaccharide formation during hydrolysis of lactose with Saccharomyces lactis lactase (MaxilactR): part 1—quantitative aspects. Food Chem. 1979;4:243–250. [Google Scholar]
- 6.Buvinger W E, Riley M. Nucleotide sequence of Klebsiella pneumoniae lacgenes. J Bacteriol. 1985;163:850–857. doi: 10.1128/jb.163.3.850-857.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cupples C G, Miller J H, Huber R E. Determination of the roles of Glu-461 in β-galactosidase (Escherichia coli) using site-specific mutagenesis. J Biol Chem. 1990;265:5512–5518. [PubMed] [Google Scholar]
- 8.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 9.de Vries W, Stouthamer A H. Pathway of glucose fermentation in relation to the taxonomy of bifidobacteria. J Bacteriol. 1967;93:574–576. doi: 10.1128/jb.93.2.574-576.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumortier V, Brassart C, Bouquelet S. Purification and properties of a β-D-galactosidase from Bifidobacterium bifidumexhibiting a transgalactosylation reaction. Biotechnol Appl Biochem. 1994;19:341–354. [Google Scholar]
- 11.Dumortier V, Montreuil J, Bouquelet S. Primary structure of ten galactosides formed by transgalactosylation during lactose hydrolysis by Bifidobacterium bifidum. Carbohydr Res. 1990;201:115–123. doi: 10.1016/0008-6215(90)84228-m. [DOI] [PubMed] [Google Scholar]
- 12.Gebler J C, Aebersold R, Withers S G. Glu-537, not Glu-461, is the nucleophile in the active site of (lacZ) β-galactosidase from Escherichia coli. J Biol Chem. 1992;267:11126–11130. [PubMed] [Google Scholar]
- 13.Greenberg N A, Mahoney R R. Formation of oligosaccharides by β-galactosidase from Streptococcus thermophilus. Food Chem. 1983;10:195–204. [Google Scholar]
- 14.Gutshall K R, Trimbur D E, Kasmir J J, Brenchley J E. Analysis of a novel gene and β-galactosidase isoenzyme from a psychrotrophic Arthrobacterisolate. J Bacteriol. 1995;177:1981–1988. doi: 10.1128/jb.177.8.1981-1988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock K R, Rockman E, Young C A, Pearce L, Maddox I S, Scott D B. Expression and nucleotide sequence of the Clostridium acetobutylicum β-galactosidase gene cloned in Escherichia coli. J Bacteriol. 1991;173:3084–3095. doi: 10.1128/jb.173.10.3084-3095.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon J P. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA. 1995;92:7090–7094. doi: 10.1073/pnas.92.15.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirata H, Fukazawa T, Negoro S, Okada H. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J Bacteriol. 1986;166:722–727. doi: 10.1128/jb.166.3.722-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homans S W. 1H NMR studies of oligosaccharides in NMR of macromolecules: a practical approach. Oxford, United Kingdom: Oxford University Press; 1993. [Google Scholar]
- 21.Huber R H, Wellenfels K. A quantitation of the factors which affect the hydrolase and transgalactosylase activities of β-galactosidase (E. coli) on lactose. Biochemistry. 1976;15:1994–2001. doi: 10.1021/bi00654a029. [DOI] [PubMed] [Google Scholar]
- 22.Hung M N, Lee B H. Cloning and expression of β-galactosidase genes from Bifidobacterium infantis into Escherichia coli. Biotechnol Lett. 1998;20:659–662. [Google Scholar]
- 23.Jeon I J, Mantha V R. High performance liquid chromatography analysis of oligosaccharides formed during β-galactosidase action on lactose. J Dairy Sci. 1985;68:581–588. [Google Scholar]
- 24.Kalnins A, Otto K, Ruther U, Muller-Hall B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2:593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufmann P, Pfefferkorn A, Teuber M, Meile L. Identification and quantification of Bifidobacteriumspecies isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl Environ Microbiol. 1997;63:1268–1273. doi: 10.1128/aem.63.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraulis P J. ANSIG: a program for the assignment of protein 1H 2D NMR spectra by interactive graphics. J Magn Reson. 1989;24:627–633. [Google Scholar]
- 27.Kraulis P J, Domaille P J, Campbell-Burk S L, van Aken T, Laue E D. Solution structure and dynamics of Ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lee J M, Chung D K, Park J H, Lee W K, Chang H C, Kim J H, Lee H J. Cloning and nucleotide sequence of the β-galactosidase gene from Lactococcus lactisssp. lactis ATCC7962. Biotechnol Lett. 1997;19:179–183. [Google Scholar]
- 30.Leong-Morgenthaler P, Zwahlen M C, Hottinger H. Lactose metabolism in Lactobacillus bulgaricus: analysis of the primary structure and expression of the gene involved. J Bacteriol. 1991;173:1951–1957. doi: 10.1128/jb.173.6.1951-1957.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loveland J, Gutshall K, Kasmir J, Prema P, Brenchley J E. Characterization of psychrotrophic microorganisms with β-galactosidase activities. Appl Environ Microbiol. 1994;60:12–18. doi: 10.1128/aem.60.1.12-18.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Bilbao M, Holdsworth R E, Edwards L A, Huber R E. A highly reactive beta-galactosidase (Escherichia coli) resulting from a substitution of an aspartic acid for Gly-794. J Biol Chem. 1991;266:4979–4986. [PubMed] [Google Scholar]
- 33.Moore J B, Markiewicz P, Miller J H. Identification and sequencing of the Thermotoga maritima lacZgene, part of a divergently transcribed operon. Gene. 1994;147:101–106. doi: 10.1016/0378-1119(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 34.Mozaffar Z, Nakanishi K, Matsuno R, Kamikubo T. Purification and properties of β-galactosidases from Bacillus circulans. Agric Biol Chem. 1984;48:3053–3061. [Google Scholar]
- 35.Mozaffar Z, Nakanishi K, Matsuno R. Formation of oligosaccharides during hydrolysis of lactose in milk using β-galactosidase from Bacillus circulans. J Food Sci. 1985;50:1602–1606. [Google Scholar]
- 36.Nagano H, Kawaguchi T, Omori M, Shoji Z, Arai M. Molecular cloning and nucleotide sequence of the β-galactosidase gene from Enterobacter cloacaeGAO. Biosci Biotechnol Biochem. 1994;58:1866–1869. doi: 10.1271/bbb.58.1866. [DOI] [PubMed] [Google Scholar]
- 37.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, White O, Salzberg S L, Smith H O, Venter J C, Fraser C M. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 38.Ohtsuka K, Benno Y, Endo K, Ozawa O, Ueda H, Uchida T, Mitsuoka T. Effects of 4′galactosyl-lactose intake on human fecal microflora. Bifidus. 1989;2:143–149. [Google Scholar]
- 39.Onishi N, Kira I, Yokozeki K. Galacto-oligosaccharide production from lactose by Sirobasidium magnumCBS6803. Lett Appl Microbiol. 1996;23:253–256. doi: 10.1111/j.1472-765x.1996.tb00077.x. [DOI] [PubMed] [Google Scholar]
- 40.Onishi N, Tanaka T. Purification and properties of a galacto- and gluco-oligosaccharide-producing β-glycosidase from Rhodotorula minutaIFO879. J Ferment Bioeng. 1996;82:439–443. [Google Scholar]
- 41.Onishi N, Yamashiro A, Yokozeki K. Production of galacto-oligosaccharide from lactose by Sterigmatomyces elviaeCBS8119. Appl Environ Microbiol. 1995;61:4022–4025. doi: 10.1128/aem.61.11.4022-4025.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poch O, L'Hote H, Dallery V, Debeaux F, Fleer R, Sodoyer R. Sequence of the Kluyveromyces lactisβ-galactosidase: comparison with prokaryotic enzymes and secondary structure analysis. Gene. 1992;118:55–63. doi: 10.1016/0378-1119(92)90248-n. [DOI] [PubMed] [Google Scholar]
- 43.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989;171:244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prenosil J E, Stuker E, Bourne J R. Formation of oligosaccharides during enzymatic lactose hydrolysis. Part 1: state of art. Biotechnol Bioeng. 1987;30:1019–1025. doi: 10.1002/bit.260300904. [DOI] [PubMed] [Google Scholar]
- 45.Reese M G. Diploma thesis. Heidelberg, Germany: German Cancer Research Center; 1994. [Google Scholar]
- 46.Roy D, Berger J L, Reuter G. Characterization of dairy-related Bifidobacteriumspp. based on their β-galactosidase electrophoretic patterns. Int J Food Microbiol. 1994;23:55–70. doi: 10.1016/0168-1605(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 47.Sabesan S, Paulson J C. Combined chemical and enzymatic synthesis of sialyloligosaccharides and characterization by 500-MHz and proton and carbon-13 NMR spectroscopy. J Am Chem Soc. 1986;108:2068–2080. [Google Scholar]
- 48.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scardovi V, Trovatelli L D, Zani G, Crociani F, Matteuzzi D. Deoxyribonucleic acid homology relationships among species of the genus Bifidobacterium. Int J Syst Bacteriol. 1971;21:276–294. [Google Scholar]
- 50.Schmidt B F, Adams R M, Requadt C, Power S, Mainzer S E. Expression and nucleotide sequence of the Lactobacillus bulgaricus β-galactosidase gene cloned in Escherichia coli. J Bacteriol. 1989;171:625–635. doi: 10.1128/jb.171.2.625-635.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroeder C J, Robert C, Lenzen G, McKay L L, Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricusβ-galactosidase sequences. J Gen Microbiol. 1991;137:369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- 52.Sebald M, Gasser F, Werner H. DNA base composition and classification. Application to group of bifidobacteria and related genera. Ann Inst Pasteur. 1965;109:251–269. [PubMed] [Google Scholar]
- 53.Sgorbati B, London J. Demonstration of phylogenetic relatedness among members of the genus Bifidobacteriumusing the enzyme transaldolase as an evolutionary marker. Int J Syst Bacteriol. 1982;32:37–42. [Google Scholar]
- 54.States D J, Haberkom R A, Ruben D J. A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J Magn Reson. 1982;48:286–292. [Google Scholar]
- 55.Stokes H W, Betts P W, Hall B G. Sequences of ebgA gene of Escherichia coli: comparison with the lacZgene. Mol Biol Evol. 1985;2:469–477. doi: 10.1093/oxfordjournals.molbev.a040372. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka R, Takayama H, Morotomi M, Kuroshima T, Ueyama S, Matsumoto K, Kuroda A, Mutai M. Effects of administration of TOS and Bifidobacterium breve4006 on the human fecal flora. Bifidobact Microflora. 1985;2:17–24. [Google Scholar]
- 57.Toba T, Yokota A, Adachi S. Oligosaccharide structures formed during the hydrolysis of lactose by Aspergillus oryzaeβ-galactosidase. Food Chem. 1985;16:147–162. [Google Scholar]
- 58.Trimbur D E, Gutshall K R, Prema P, Brenchley J E. Characterization of a psychrotrophic Arthrobactergene and its cold-active β-galactosidase. Appl Environ Microbiol. 1994;60:4544–4552. doi: 10.1128/aem.60.12.4544-4552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaughan E E, David S, de Vos W M. The lactose transporter in Leuconostoc lactisis a new member of the LacS subfamily of galactoside-pentose-hexuronide translocators. Appl Environ Microbiol. 1996;62:1574–1582. doi: 10.1128/aem.62.5.1574-1582.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vian A, Carrascosa A V, Garcia J L, Cortes E. Structure of the β-galactosidase gene from Thermus sp. strain T2: expression in Escherichia coliand purification in a single step of an active fusion protein. Appl Environ Microbiol. 1998;64:2187–2191. doi: 10.1128/aem.64.6.2187-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wierzbicki L E, Kosikowski F V. Formation of oligosaccharides during β-galactosidase action on lactose. J Dairy Sci. 1973;56:1400–1404. doi: 10.3168/jds.S0022-0302(73)85372-X. [DOI] [PubMed] [Google Scholar]
- 62.Yanahira S, Kobayashi T, Suguri T, Nakakoshi M, Miura S, Ishikawa H, Nakajima I. Formation of oligosaccharides from lactose by Bacillus circulansβ-galactosidase. Biosci Biotechnol Biochem. 1995;59:1021–1026. doi: 10.1271/bbb.59.1021. [DOI] [PubMed] [Google Scholar]
- 63.Zarate S, Lopez-Leiva M H. Oligosaccharides formation during lactose enzymatic hydrolysis: a review of literature. J Food Prot. 1990;53:262–268. doi: 10.4315/0362-028X-53.3.262. [DOI] [PubMed] [Google Scholar]