Abstract
Clostridium spp. is a large genus of obligate anaerobes and is an extremely heterogeneous group of bacteria that can be classified into 19 clusters. Genetic analyses based on the next-generation sequencing of 16S rRNA genes and metagenome analyses conducted on human feces, mucosal biopsies, and luminal content have shown that the three main groups of strict extremophile anaerobes present in the intestines are Clostridium cluster IV (also known as the Clostridium leptum group), Clostridium cluster XIVa (also known as the Clostridium coccoides group) and Bacteroides. In addition to the mentioned clusters, some C. butyricum strains are also considered beneficial for human health. Moreover, this bacterium has been widely used as a probiotic in Asia (particularly in Japan, Korea, and China). The mentioned commensal Clostridia are involved in the regulation and maintenance of all intestinal functions. In the literature, the development processes of new therapies are described based on commensal Clostridia activity. In addition, some Clostridia are associated with pathogenic processes. Some C. butyricum strains detected in stool samples are involved in botulism cases and have also been implicated in severe diseases such as infant botulism and necrotizing enterocolitis in preterm neonates. The aim of this study is to review reports on the possibility of using Clostridium strains as probiotics, consider their positive impact on human health, and identify the risks associated with the expression of their pathogenic properties.
Keywords: Clostridium, commensal Clostridia, probiotic, Clostridium clusters
1. Introduction
The genus Clostridium is comprised of anaerobic, Gram-positive, rod-shaped, spore-forming bacteria. The first isolation of Clostridium spp. was described in the nineteenth century by Louis Pasteur. He noticed that the new bacterium was able to grow under anaerobic conditions. This microorganism was named Vibrion butyrique because of its ability to produce butyric acid via anaerobic butyric fermentation. At the end of the 19th century, Adam Prażmowski renamed the discovered microorganism Clostridium butyricum [1]. Clostridia are ubiquitous in the environment, where they mainly inhabit soil and water sediment, but also the alimentary tract of humans and animals. The bacteria of the genus Clostridium are often described only as a biological threat and an enemy of mankind. Many of them, however, have positive properties and they can be used in many industries, and also to support different kinds of therapies that are beneficial for human and animal health [1,2]. Clostridia perform a variety of metabolic functions, including the conversion of starch, proteins, and purines into organic acids (i.e., acetic, butyric, and caproic acid), alcohols, CO2, and hydrogen. Clostridium spp., due to their broad and flexible metabolic capacity, are part of many ecosystems of microbial co-culture [3]. These bacteria include groups of very heterogeneous microorganisms. Based on 16S rRNA analyses, a new taxonomic criterion based on phylogenetic analyses has been presented. A total of 19 clusters or groups were identified, leading to the description of five new genera and the proposition of eleven new species combinations [4]. Subsequent studies confirmed and extended this structure, with many organisms formally recognized as species of the genus Clostridium being transferred to new or established genera [5]. The new criterion introduced some asporulate bacteria, such as Roseburia cecicola and Ruminococcus torques (25). Most previous members of Clostridium were assigned to Clostridium cluster I, represented by C. butyricum. The reclassification of organisms from the genus Clostridium still continues, and all changes are updated on a regular basis on the List of Prokaryotic Names with Standing in Nomenclature (http://www.bacterio.net, accessed on 5 May 2022) [5].
The human and animal alimentary tract is inhabited by about 1 kg of commensal microbes. These commensals belong to Bacteria, Archea, Eucarya, and viral particles. Recent advances in next-generation sequencing (NGS) have made it possible to perform phylogenetic 16S rRNA and metagenome analyses of samples from mucosal biopsies, luminal content, and human feces, revealing that 98% of intestinal microbiota are comprised of four major bacterial phyla: Firmicutes, Bacteroides, Protobacteria and Actinobacteria [6,7]. Among these phyla, three main groups of extremophile anaerobes are recognized: Bacteroides, Clostridium cluster XIVa (Clostridium coccoides group), and Clostridium cluster IV (Clostridium leptum group) [8,9,10,11,12,13,14]. The mentioned groups of Clostridium spp. make up a huge share of the total intestinal microbiota (estimated at 10–40%) [6,10,15,16]. In addition to the mentioned clusters, some C. butyricum strains (belonging to cluster I) are also considered to be beneficial for human health, and they represent approximately 10–20% of human fecal samples, as revealed by microbial culture. Moreover, this bacterium has been widely used as a probiotic in Asia (particularly in Japan, Korea, and China). Clostridia are able to produce many beneficial substances for human and animal health, such as organic acids (acetic, butyric, fumaric, and lactic) [1]. Aside from the benefits linked to the positive impact of Clostridium on intestinal homeostasis, this genus still creates doubt because of its possible involvement in the formation of dangerous toxins and its pathogenic activity toward humans and animals. Clostridium spp. are still seen in a positive light as members of the probiotic family.
The aim of this study is to present the potential health benefits of clostridial activity (especially C. leptum, C. coccoides groups, and C. butyricum), and also the risks carried by the expression of their pathogenic properties.
2. Clostridium leptum and Clostridium coccoides Groups—Beneficial Activity
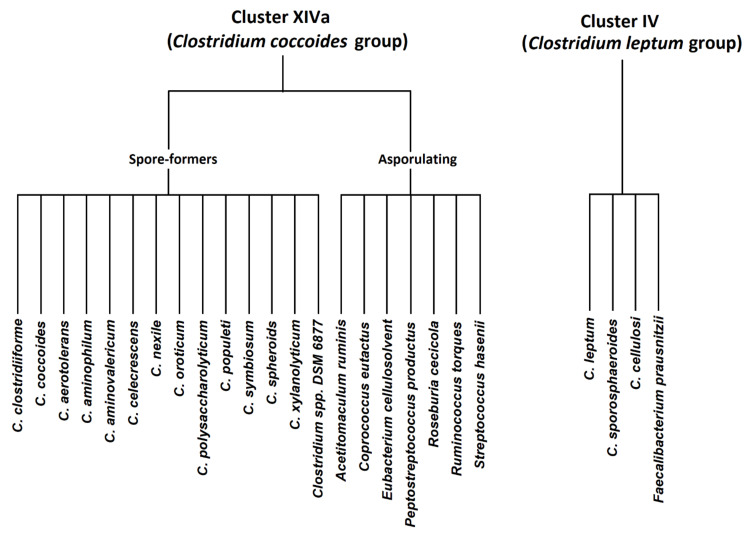
Members in cluster IV (C. leptum group) include C. leptum, C. sporosphaeroides, C. cellulosi, and Faecalibacterium prausnitzii. Clostridium cluster XIVa (Clostridium coccoides group) consists of 21 species, including spore-forming Clostridia (C. coccoides, C. aerotolerans, C. aminophilum, C. aminovalericum, C. celecrescens, C. nexile, C. oroticum, C. polysaccharolyticum, C. populeti, C. symbiosum, C. spheroids, C. xylanolyticum, and Clostridium spp. DSM 6877) and other asporulate species (Acetitomaculum ruminis, Coprococcus eutactus, Eubacterium cellulosolvens, Peptostreptococcus productus, Roseburia cecicola, Ruminococcus torques and Streptococcus hansenii) [2]. The members of both groups are presented graphically in Figure 1.
Figure 1.
Clostridium coccoides and Clostridium leptum groups—graphical presentation.
Clostridium coccoides is considered to be one of the most predominant groups of bacteria in the human intestines. This group consists of different species such as Clostridium, Butyrivibrio, Dorea, Coprococcus, Eubacterium, Ruminococcus and Roseburia, which are classified as high oxygen-sensitive anaerobes. Some of these bacteria are known to be butyrate-producing. The Clostridium coccoides group (cluster XIVa) constitutes almost 60% of mucin-adhered microbiota. The Cluster XIVa group is shown to prevent vancomycin-resistant Enterococcus (VRE) colonization, as demonstrated in an antibiotic-treated mouse model [17,18]. The mentioned groups of Clostridia and their beneficial effects are listed in Table 1.
Table 1.
Cluster IV (Clostridium leptum group) and Cluster XIVa (Clostridium coccoides group) probiotic properties.
| Cluster | Species | Beneficial Properties | Literature | |
|---|---|---|---|---|
| Cluster IV (Clostridium leptum group) |
C. leptum
C. sporosphaeroides C. cellulosi Faecalibacterium prausnitzii |
|
[2,7,8,9,10,11,14,15,19,20,21,22,23,26,27,30,31,32] | |
| Cluster XIVa (Clostridium coccoides group) |
Spore-formers |
C. clostridiiforme C. coccoides C. aerotolerans C. aminophilum C. aminovalericum C. celecrescens C. nexile C. oroticum C. polysaccharolyticum C. populeti C. symbiosum C. spheroids C. xylanolyticum Clostridium spp. DSM 6877 |
|
[2,8,9,10,11,13,14,17,18] |
| Asporulating |
Acetitomaculum ruminis
Coprococcus eutactus Eubacterium cellulosolvent Peptostreptococcus productus Roseburia cecicola Ruminococcus torques Streptococcus hasenii |
|||
Cluster IV (Clostridium leptum group) is considered to be dominant in adult human fecal microflora—its abundance approaches about 16–25%. Characteristic for this group is a synergy with different intestinal microbes in unabsorbed dietary carbohydrate fermentation. The production of short-chain fatty acids (SCFAs) by these bacteria is the major source of energy for the colonic epithelium, consequently regulating intestinal epithelial function. Faecalibacterium prausnitzii is considered to be the most frequently encountered and abundant member of the C. leptum group. A significant production of butyrate through carbohydrate fermentation is attributed to this species. This bacterium is also considered to play an anti-inflammatory function in the gastrointestinal tract. An imbalance in the composition of intestinal microbiota results in the potential for the development of serious gastrointestinal diseases, such as inflammatory bowel diseases (IBD). Ulcerative colitis and Crohn’s disease (CD), in particular, are suspected to result from an imbalance in the natural immune reaction to the luminal microbiota in some individuals, along with genetic disorders [19,20,21,22,23,24]. The mentioned alterations are frequently associated with a significant increase in Bacteroides-Prevotella, an increase or decrease in bifidobacterial, and a decrease in the C. leptum group, especially F. prausnitzii. The mentioned changes were noticed in patients with active Crohn’s disease [25]. Faecalibacterium prausnitzii was also proven to prevent inflammation by blocking the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and the release of interleukin 8 (IL8). This bacterium was also recognized for its anti-inflammatory effect caused by chronic colitis [26].
Umesaki et al. [27] and Atarashi et al. [28] observed that a mixture of commensal Clostridia was able to change the intraepithelial lymphocyte profile in the large intestine. Atarashi et al. [29] noticed that C. leptum and C. coccoides groups were able to induce the accumulation of mucosal Treg cells in the colon and enrich transformation growth factor-β in the colon. They also noticed that the chosen strains of IV, XIVa and XVIII clusters could induce the expansion and differentiation of regulatory Treg cells. The authors reported that the oral administration of a mixture of the mentioned clusters could attenuate allergic diarrhea and colitis in mice [29]. A study by Godefroy et al. [30] suggested that DP8α cells (among them, DP8α T cells, including F. prausnitzii-specific T cells have been recognized) co-expressing CCR6 and CXCR6 cause a decrease in inflammation in inflammatory bowel disease (IBD) patients. Faecalibacterium prausnitzii is able to release an extracellular polymeric matrix (EPM) and can form a biofilm. The EPM is able to trigger the secretion of IL-10 and IL-12, which attenuate inflammation [31]. Moreover, salicylic acid could be produced from salicin fermentation by 40% of F. prausnitzii strains and block the production of IL-8 [32].
In recent decades, a lot of data have appeared on the 16S rRNA-based profiling of gut microbiota connected to the abundance of the C. leptum group, although the most data are connected to F. prausnitzii and its health benefits in IBD (Crohn’s disease, ulcerative colitis) [20,21,24,33,34,35]. Faenicalibacterium prausnitzii is considered to be an indicative factor (biomarker) of the intestinal condition of the adult human body, and has also been recognized as a prospective predictive factor in Crohn’s disease (CD). An F. prausnitzii deficiency was observed in patients with Crohn’s disease in the ileum [23]. It was noticed that a low level of F. prausnitzii in ileal CD patients undergoing surgery was associated with a higher risk of postoperative recurrence [23,36]. Swidsinski et al. [24] invented a diagnostic test based on the detection of F. prausnitzii and leukocyte count. This test made it possible to distinguish active CD from ulcerative colitis (UC) with sensitivity at the level of 79–80% and a specificity of 98–100%. In the UC patients, a significant increase in leukocytes together with high F. prausnitzii numbers was observed [24]. Bacterial dysbiosis correlated with IBD, with a visible imbalance of the mucosal protective bacteria predominantly in the C. leptum group, including F. prausnitzii [23]. Macafferi et al. [34] reported on the application of rifaximin to CD patients causing remission, and noticed an increase in the level of bifidobacteria and F. prausnitzii. Dorffel et al. [15] showed that other types of treatment such as chemotherapy and interferon a-2b caused the depletion of F. prausnitzii. Additionally, the high-dose cortisol therapy of infliximab is able to restore the number of F. prausnitzii from an undetectable level up to 1.4 × 1010 bacteria/mL within several days [24,37]. The mechanism for modulating the number of F. prausnitzii in patients undergoing treatment for IBD is still unexplained.
3. Clostridium butyricum—Its Probiotic Properties and Ability to Support Treatment
Aside from the groups of Clostridia mentioned above, some strains of C. butyricum (belonging to cluster I) are considered to be probiotic as well. C. butyricum strains are found in various niches, including soil, vegetables and fermented dairy products, and are also natural inhabitants of the intestines of humans and animals. Clostridium butyricum is able to produce SCFAs by fermenting undigested dietary fiber, especially butyrate and acetate. Butyrate is considered to be one of the main fermentation products released by C. butyricum in the butyrate kinase (buk) pathway [38,39]. Short-chain fatty acids released in the colon by microorganisms exert a myriad of effects on host health, including modulating immune homeostasis in the gut, enhancing gastrointestinal barrier function, and alleviating inflammation. Clostridium butyricum has been isolated from 10–20% of the mature human population and is also considered one of the primary colonizers found in the intestines of infants. It has been shown that C. butyricum is able to survive the high acidity of the gastrointestinal environment. Literature data indicate the beneficial effects of the application of C. butyricum, such as promoting faster animal growth and enhancing different immune functions as well as microecological balance [38,39,40]. A preventive effect against Esherichia coli and Clostridioides difficile infections and its influence on the reduction in intestinal damage and permeability were demonstrated [41].
Some non-pathogenic strains of this species have been used as probiotics for decades, e.g., C. butyricum MIYAIRI 588 isolated from human stool samples by Chikaji Miyairi in 1933 and in 1963 from soil samples [42]. This strain is a probiotic commercially available in Japan and Korea, and is used for supporting the treatment of antimicrobial-associated diarrhea [42]. Moreover, the mentioned strain was also authorized as a novel food ingredient by the European Parliament and the Council [43]. Clostridium butyricum seems to be a promising candidate in curing dysbiosis caused by various diseases, such as gastrointestinal, neurological and metabolic disorders, and cancer. The mechanism of its beneficial properties is not fully elucidated; however, some evidence has appeared in the literature linking benefits with the production of SCFAs and their pathways to impact immune homeostasis, as well as the physiology of the gut barrier. It is also possible that C. butyricum has an influence on increasing the number of probiotic bacterial taxa, such as Lactobacillus and Bifidobacterium [39,40,42,44,45].
The intestinal barrier plays an important role in maintaining intestinal microbiota tolerance, nutrients, electrolytes, and water absorption, and maintaining the required level of defense against pathogen invasion. It is a specific selective gate that prevents the diffusion of toxins or antigens. The intestinal barrier consists of three layers: the mucus layer, epithelium, and lamina propria [32,46]. The mucus is considered to be the first physical barrier against pathogens. This layer varies in thickness in the different parts of the gastrointestinal tract, and is mainly composed of glycoproteins such as mucins which are secreted by the epithelium [33]. The disintegration of mucin in mice is conductive to the development of colitis and increases the probability of the development of colorectal cancer [47,48]. The protective abilities of C. butyricum have been tested in two mouse model experiments. Long et al. [49] described a significant increase in colonic mucosal thickness after C. butyricum Sx-01 administration. Hagihara et al. [39] also noticed that the application of C. butyricum MYRIAYRI 588 caused an increase in mucin secretion and significantly decreased epithelial damage in colonic tissues during recovery from clindamycin-induced antibiotic-associated diarrhea. The abundant production of butyrate by C. butyricum strains is indicated as the main factor for the increase in mucin secretion, which results in the increased expression of the mucin MUC genes. Clostridium butyricum also has an influence on TJ protein (molecules situated at the tight junctions of the epithelial, endothelial, and myelinated cells) expression across several different disease models [45,50]. The positive effect of C. butyricum on the intestinal epithelium is attributed to butyrate production. The fecal concentration of butyrate increases significantly after C. butyricum supplementation. Additionally, direct supplementation with sodium butyrate showed a beneficial effect on increased intestinal permeability [51,52,53]. Moreover, C. butyricum is also reported to have an influence on the positive effect of immunomodulation of interleukin-17 (IL-17). An enhanced barrier integrity effect on IL-17 production by γδ T cells (intraepithelial T cells that are one of the components of the first line of defense) in the colonic lamina propria was observed after the implementation of C. butyricum [54]. Clostridium butyricum was also shown to promote the production of anti-inflammatory lipid metabolites, e.g., palmitoleic acid, prostaglandin metabolites and also pro-resolving mediators in mouse colonic tissues. The mentioned metabolites, such as protectin D1, participate in the promotion of anti-inflammatory T cells secreting IL-10 in the colon. Moreover, microbial components and metabolites are able to stimulate the release of antimicrobial peptides (AMP) and immunoglobulin A (IgA) which could constitute a chemical barrier against microbial pathogens [17,55].
Another positive aspect of the activity of C. butyricum comes from reports suggesting their preventive action against inflammation through a mechanism involving type 2 immunity, which may support the treatment of type 1 diabetes. Moreover, literature data indicate an inductive effect of native Clostridium spp. such as C. butyricum MIYAIRI 588 on regulatory T cells (Tregs) of the colon [38,42]. Jia et al. [56] have suggested that C. butyricum CGMCC0313.1 (CB0313.1) could induce pancreatic Tregs cells and consequently suppress diabetes in non-obese diabetic (NOD) mice. Those authors noted that protection could include increased Treg counts, the restoration of Th1/Th2/Th17 cell balance, and a change to a less pro-inflammatory immune environment in the gut, pancreatic lymph node, and pancreas. The increase in α4β7 + (intestinal homing receptor) Treg in the pancreatic lymph node (PLN) suggested that the mechanism may include the increased migration of Treg from the gut to the pancreas. In addition, the sequencing of the 16S rRNA genes showed that CB0313.1 increased the Firmicutes/Bacteroidetes ratio and enriched the Clostridial subgroups and the butyrate-producing bacterial subgroups. These results suggest the beneficial preventive effects on type 1 diabetes and the need for further clinical investigation in this area.
Tomita et al. [57] noticed that Clostridium butyricum therapy may affect the therapeutic efficacy of immune checkpoint inhibitors (ICI). They retrospectively evaluated 118 patients with advanced non-small cell lung cancer treated with immune checkpoint blockage. A survival analysis comparing patients given C. butyricum therapy before and after ICI treatment showed the significant prolongation of progression-free survival and overall survival, even in patients who received antibiotic therapy. This study suggests that probiotic therapy with C. butyricum may have a positive impact on the therapeutic efficacy of ICI in patients with cancer [57]. Tian et al. [58] investigated the effect of C. butyricum on adverse events in lung cancer (LC) patients treated with chemotherapy. Systemic therapy is an aggressive treatment that carries a risk of intestinal damage and gastrointestinal reactions. The authors performed an NGS-based analysis of the intestinal microbiome at baseline and after a three-week treatment with C. butyricum, and they found that the incidence of chemotherapy-induced diarrhea was lower in the patient group that was administered C. butyricum compared to the placebo group. In addition, Tian et al. [58] found an increase in the amount of bacteria from SCFA-producing genera in the C. butyricum-supplemented group, with a concomitant tendency toward decreased pathogenic bacteria which was almost the opposite of the observation in the placebo group. The researchers showed that C. butyricum reduced chemotherapy-induced diarrhea in LC patients and decreased systemic inflammatory response [58].
Zhou et al. [59] studied the effect of C. butyricum on colorectal cancer (CRC) progression. In a study on mice, they demonstrated that the supplementation of C. butyricum inhibited the development of CRC in vivo and restrained the proliferation of in vitro CRC cells and the expression of MyD88 (MYD88 innate immune signal transduction adaptor) and NF-κB/p65 (nuclear factor-kappa B subunit/nuclear factor NF-kappa-B P65 subunit). Furthermore, the group of mice that had C. butyricum administered in vivo showed more apoptotic cells in the tumor tissue, lower levels of IL-6, and higher levels of IL-10 compared to the mice in the control group. The administration of the bacteria altered the composition of the intestinal microflora, which was also enriched in the small intestine and tumor tissue [59]. The inhibition of the CRC cell proliferation cell cycle and the promotion of apoptosis by C. butyricum were indicated by Chen et al. [60]. The in vivo inhibition of CRC development was induced by 1,2-dimethylhydrazine dihydrochloride [60]. In addition, the researchers found that the administration of C. butyicum reduced inflammation and improved immune homeostasis in induced CRC. A study by Chen et al. [51] additionally indicated that CRC cells showed decreased proliferation and increased levels of apoptosis in the presence of C. butyricum. In addition, C. butyricum has been shown to inhibit the Wnt/β-catenin signaling pathway (which regulates cellular function such as proliferation, differentiation, migration, genetic stability, apoptosis, and stem cell renewal) and modulate the composition of the gut microbiota [61], and has been associated with a reduction in pathogenic bacteria, as also indicated by Tian et al. [58]. Furthermore, Chen et al. [51] demonstrated that C. butyricum increases the number of bile acid biotransforming bacteria and increases the number of beneficial bacteria, including bacteria that produce short-chain fatty acids. In Table 2, the beneficial properties of non-pathogenic C. butyricum strains are listed.
Table 2.
Probiotic properties of non-pathogenic Clostridium butyricum strains.
| Probiotic Properties | Literature | |
|---|---|---|
| C. butyricum |
|
[15,17,29,38,39,41,42,44,45,47,48,49,50,51,53,54,56,57,58,59,60] |
1 SCFA = short-chain fatty acids; 2 CI = immune checkpoint inhibitors; 3 CRC = colorectal cancer.
4. Pathogenic Character of Clostridia and Safety Aspects
Despite the fact that most Clostridia are commensal bacteria, some of them can cause serious health disturbances in humans and animals, such as botulism and necrotic enterocolitis.
Botulism is a paralyzing disease caused by botulinum neurotoxins (BoNTs). The production of BoNT is generally attributed to C. botulinum species [62]. The BoNT molecule is a component of a protoxin complex with several associated protein molecules, all of which are encoded by the botulinum gene cluster. Some of these genes are also found in other Clostridium species and some have moved between different plasmids within the same physiological group of Clostridia. This indicates that the horizontal transfer of genes encoding the botulinum cluster occurs between species of Clostridium. The abundance of mobile elements is likely connected to accelerated genome plasticity and gene transfer events [63,64,65].
Despite the fact that C. butyricum is considered a commensal, non-pathogenic bacterium, cases of botulism in infants have been reported and associated with this species. The first case of botulism in infants caused by C. butyricum capable of producing BoNT type E was reported in Italy [62]. The isolated toxigenic organisms differed significantly from C. botulinum type E. The visible microorganism in each case resembled C. butyricum, but produced a neurotoxin that could not be distinguished from botulinum toxin type E by its effect on mice and its neutralization by botulinum type E antitoxin [15]. Since these cases, C. butyricum has been associated with botulism in many other countries, including China, Japan, India, USA, and Ireland [66]. The operon encoding BoNT type E in the toxigenic C. butyricum is very similar to the operon encoding BoNT type E in C. botulinum group II strains [64].
Aside from botulism, C. butyricum strains have been associated with necrotizing enterocolitis (NEC). This was first described by Howard et al. [67] in 1977, who noted C. butyricum isolates in stool and blood cultures of preterm neonates. The symptoms of NEC manifest with gastrointestinal bleeding, abdominal distension, mucosal ulcerations, necrosis, portal venous gas, and pneumatosis intestinalis. Generally, NEC causes high morbidity and mortality. The genome sequencing of the pathogenic C. butyricum strains identified different toxin genes [38,68]. Cassir et al. [38] identified four genes encoding polypeptides in NEC-associated C. butyricum strains that were very similar to hemolysins shared by Brachyspira hyodysenteriae, the etiological agent of swine dysentery. Among those hemolysins, the pore-forming β-hemolysin is considered to be the main virulence factor capable of inducing enterocyte necrotic lesions via the culture supernatant. However, the participation of β-hemolysin in the etiology of NEC is not fully elucidated. Studies conducted on an axenic chicken model showed the reproducibility of NEC with C. butyricum pathogenic human neonatal-derived strains [69]. The experimental findings suggested that C. butyricum could play a primary role in NEC pathogenesis. The bacterium is suspected to ferment carbohydrates depending on the lactase deficiency of preterm neonates [70,71]. The occurrence of NEC caused by C. butyricum has been associated mainly with preterm neonates; however, recently, Sato et al. [35] described this in an 84-year-old Japanese man during hospitalization for the treatment of stab wounds.
5. Conclusions
Clostridium species, especially clusters IV, XIVa, and C. butyricum, are commensal bacteria and are common in human and animal intestines. The mentioned groups and C. butyricum can exert anti-inflammatory effects and maintain intestinal health by releasing their components and metabolites, especially SCFAs. These microorganisms could find supporting application in IBD, diabetes, and even cancer therapy. Therefore, these strains have broad prospects as probiotics in the future. The next-generation sequencing of 16S RNA genes and metagenomic analyses provide increasing insight into the content of particular clostridial groups in the intestinal biomes of individuals with various diseases. In-depth analyses of the impact of the presence and content of specific groups of Clostridia will certainly contribute to our knowledge of how these bacteria affect disease processes, treatment, and recovery. The properties that Clostridia possess seem to be valuable in the probiotic field. However, despite this, there are still some doubts about the safety of Clostridia application (especially C. butyricum) to improve health. Thorough research is still needed on the molecular aspects of physiology and genetic diversity, and on horizontal gene transfer between pathogenic and non-pathogenic strains of Clostridia and related species.
Author Contributions
Conceptualization, T.G., A.G. and P.D.; writing—original draft preparation, T.G. and P.D.; writing—review and editing, A.G., P.K. and K.K.; supervision, P.K. and K.K.; project administration, T.G. and A.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Samul D., Worsztynowicz P., Leja K., Grajek W. Beneficial and harmful roles of bacteria from the Clostridium genus. Acta Biochim. Pol. 2013;60:515–521. doi: 10.18388/abp.2013_2015. [DOI] [PubMed] [Google Scholar]
- 2.Guo P., Zhang K., Ma X., He P. Clostridium species as probiotics: Potentials and challenges. J. Anim. Sci. Biotechnol. 2020;11:24. doi: 10.1186/s40104-019-0402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du Y., Zou W., Zhang K., Ye G., Yang J. Advances and Applications of Clostridium Co-culture Systems in Biotechnology. Front. Microbiol. 2020;11:560223. doi: 10.3389/fmicb.2020.560223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins M.D., Lawson P.A., Willems A., Cordoba J.J., Fernandez-Garayzabal J., Garcia P., Cai J., Hippe H., Farrow J.A.E. The phylogeny of the genus Clostridium: Proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 5.Lawson P.A., Rainey F.A. Proposal to restrict the genus Clostridium Prazmowski to Clostridium butyricum and related species. Int. J. Syst. Evol. Microbiol. 2016;66:1009–1016. doi: 10.1099/ijsem. [DOI] [PubMed] [Google Scholar]
- 6.Lopetuso L.R., Scaldaferri F., Petito V., Gasbarrini A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tap J., Mondot S., Levenez F., Pelletier E., Caron C., Furet J.P., Ugarte E., Munoz-Tamayo R., Paslier D.L., Nalin R., et al. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 8.Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 9.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank D.N., St. Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., Gordon J.I., Relman D.A., Fraser-Liggett C.M., Nelson K.E. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hold G.L., Pryde S.E., Russell V.J., Furrie E., Flint H.J. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 2002;39:33–39. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- 13.Ley R.E., Backhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajilic-Stojanovic M., Smidt H., de Vos W.M. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 15.Dorffel Y., Swidsinski A., Loening-Baucke V., Wiedenmann B., Pavel M. Common biostructure of the colonic microbiota in neuroendocrine tumors and Crohn’s disease and the effect of therapy. Inflamm. Bowel Dis. 2011;18:1663–1671. doi: 10.1002/ibd.21923. [DOI] [PubMed] [Google Scholar]
- 16.Manson J.M., Rauch M., Gilmore M.S. The commensal microbiology of the gastrointestinal tract. Adv. Exp. Med. Biol. 2008;635:15–28. doi: 10.1007/978-0-387-09550-9_2. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi H., Sakamoto M., Kitahara M., Benno Y. Diversity of the Clostridium coccoides group in human fecal microbiota as determined by 16S rRNA gene library. FEMS Microbiol. Lett. 2006;257:202–207. doi: 10.1111/j.1574-6968.2006.00171.x. [DOI] [PubMed] [Google Scholar]
- 18.Livanos A.E., Snider E.J., Whittier S., Chong D.H., Wang T.C., Abrams J.A., Freedberg D.E. Rapid gastrointestinal loss of Clostridial Clusters IV and XIVa in the ICU associates with an expansion of gut pathogens. PLoS ONE. 2018;13:e0200322. doi: 10.1371/journal.pone.0200322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cucchiara S., Iebba V., Conte M.P., Schippa S. The microbiota in inflammatory bowel disease in different age groups. Dig. Dis. 2009;27:252–258. doi: 10.1159/000228558. [DOI] [PubMed] [Google Scholar]
- 20.Kang S., Denman S.E., Morrison M., Yu Z., Dore J., Leclerc M., McSweeney C.S. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm. Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- 21.Mondot S., Kang S., Furet J.P., Aguirre de Carcer D., McSweeney C., Morrison M., Marteau P., Doré J., Leclerc M. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm. Bowel Dis. 2011;17:185–192. doi: 10.1002/ibd.21436. [DOI] [PubMed] [Google Scholar]
- 22.Schwiertz A., Jacobi M., Frick J.S., Richter M., Rusch K., Köhler H. Microbiota in pediatric inflammatory bowel disease. J. Pediatr. 2010;157:240–244. doi: 10.1016/j.jpeds.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 23.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L.G., Gratadoux J.J., Blugeon S., Bridonneau C., Furet J.P., Corthier G., et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swidsinski A., Loening-Baucke V., Vaneechoutte M., Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 25.Kabeerdoss J., Sankaran V., Pugazhendhi S., Ramakrishna B.S. Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: A case-control study in India. BMC Gastroenterol. 2013;13:20. doi: 10.1186/1471-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breyner N.M., Michon C., de Sousa C., Vilas Boas P.B., Chain F., Azevedo V.A., Langella P., Chatel J.M. Microbial anti-Inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-Induced Colitis model in mice through inhibition of NF-κB pathway. Front. Microbiol. 2017;8:114. doi: 10.3389/fmicb.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umesaki Y., Setoyama H., Matsumoto S., Imaoka A., Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 1999;67:3504–3511. doi: 10.1128/IAI.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., Fukuda S., Saito T., Narushima S., Hase K., et al. Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 30.Godefroy E., Alameddine J., Montassier E., Mathé J., Desfrançois-Noël J., Marec N., Bossard C., Jarry A., Bridonneau C., Le Roy A., et al. Expression of CCR6 and CXCR6 by gut-derived CD4/CD8α Tregulatory cells, which are decreased in blood samples from patients with inflammatory bowel diseases. Gastroenterology. 2018;155:1205–1217. doi: 10.1053/j.gastro.2018.06.078. [DOI] [PubMed] [Google Scholar]
- 31.Miquel S., Leclerc M., Martin R., Chain F., Lenoir M., Raguideau S.S., Hudault S., Bridonneau C., Northen T., Bowen B., et al. Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. MBio. 2015;6:e00300-15. doi: 10.1128/mBio.00300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunyady B., Mezey E., Palkovits M. Gastrointestinal immunology: Cell types in the lamina propria—A morphological review. Acta Physiol. Hung. 2000;87:305–328. [PubMed] [Google Scholar]
- 33.Corfield A.P. The Interaction of the Gut Microbiota with the Mucus Barrier in Health and Disease in Human. Microorganisms. 2018;6:78. doi: 10.3390/microorganisms6030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maccaferri S., Vitali B., Klinder A., Kolida S., Ndagijimana M., Laghi L., Calanni F., Brigidi P., Gibson G.R., Costabile A. Rifaximin modulates the colonic microbiota of patients with Crohn’s disease: An in vitro approach using a continuous culture colonic model system. J. Antimicrob. Chemother. 2010;65:2556–2565. doi: 10.1093/jac/dkr080. [DOI] [PubMed] [Google Scholar]
- 35.Sato Y., Kujirai D., Emoto K., Yagami T., Yamada T., Izumi M., Ano M., Kase K., Kobayashi K. Necrotizing enterocolitis associated with Clostridium butyricum in a Japanese man. Acute. Med. Surg. 2018;5:194–198. doi: 10.1002/ams2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokol H., Seksik P., Furet J.P., Firmesse O., Nion-Larmurier I., Beaugerie L., Cosnes J., Corthier G., Marteau P., Doré J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel. Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 37.Miquel S., Martín R., Rossi O., Bermúdez-Humarán L.G., Chatel J.M., Sokol H., Thomas M., Wells J.M., Langella P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013;16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Cassir N., Benamar S., Khalil J.B., Croce O., Saint-Faust M., Jacquot A., Million M., Azza S., Armstrong N., Henry M., et al. Clostridium butyricum strains and dysbiosis linked to necrotizing enterocolitis in preterm neonates. Clin. Infect. Dis. 2015;61:1107–1115. doi: 10.1093/cid/civ468. [DOI] [PubMed] [Google Scholar]
- 39.Hagihara M., Kuroki Y., Ariyoshi T., Higashi S., Fukuda K., Yamashita R., Matsumoto A., Mori T., Mimura K., Yamaguchi N., et al. Clostridium butyricum Modulates the Microbiome to Protect Intestinal Barrier Function in Mice with Antibiotic-Induced Dysbiosis. iScience. 2020;23:100772. doi: 10.1016/j.isci.2019.100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao R.X., Zhu X.X., Wan C.M., Wang Z.L., Wen Y., Li Y.Y. Effect of Clostridium butyricum supplementation on the development of intestinal flora and the immune system of neonatal mice. Exp. Ther. Med. 2018;15:1081–1086. doi: 10.3892/etm.2017.5461.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassir N., Benamar S., La Scola B. Clostridium butyricum: From beneficial to a new emerging pathogen. Clin. Microbiol. Infect. 2016;22:37–45. doi: 10.1016/j.cmi.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 42.Stoeva M.K., Garcia-So J., Justice N., Myers J., Tyagi S., Nemchek M., McMurdie P.J., Kolterman O., Eid J. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes. 2021;13:1907272. doi: 10.1080/19490976.2021.1907272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The European Commission Commission Implementing Decision of 11 December 2014 authorising the placing on the market of Clostridium butyricum (CBM 588) as a novel food ingredient under Regulation (EC) No 258/97 of the European Parliament and of the Council (notified under document C(2014) 9345) OJEU. 2014;57:153. [Google Scholar]
- 44.Hagihara M., Yamashita R., Matsumoto A., Mori T., Kuroki Y., Kudo H., Oka K., Takahashi M., Nonogaki T., Yamagishi Y., et al. The impact of Clostridium butyricum MIYAIRI 588 on the murine gut microbiome and colonic tissue. Anaerobe. 2018;54:8–18. doi: 10.1016/j.anaerobe.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 45.Pan L.L., Niu W., Fang X., Liang W., Li H., Chen W., Zhang H., Bhatia M., Sun J. Clostridium butyricum strains suppress experimental acute pancreatitis by maintaining intestinal homeostasis. Mol. Nutr. Food Res. 2019;63:e1801419. doi: 10.1002/mnfr.201801419. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad R., Sorrell M.F., Batra S.K., Dhawan P., Singh A.B. Gut permeability and mucosal inflammation: Bad, good or context dependent. Mucosal Immunol. 2017;10:307–317. doi: 10.1038/mi.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Der Sluis M., De Koning B.A.E., De Bruijn A.C.J.M., Velcich A., Meijerink J.P.P., Van Goudoever J.B., Büller H.A., Dekker J., Van Seuningen I., Renes I.B., et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 48.Velcich A., Yang W., Heyer J., Fragale A., Nicholas C., Viani S., Kucherlapati R., Lipkin M., Yang K., Augenlicht L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 49.Long M., Yang S., Li P., Song X., Pan J., He J., Zhang Y., Wu R. Combined use of C. butyricum Sx-01 and L. salivarius C-1-3 improves intestinal health and reeduces the amount of lipids in serum via modulation of gut microbiota in mice. Nutrients. 2018;10:810. doi: 10.3390/nu10070810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Sun J., Du J., Wang F., Fang R., Yu C., Xiong J., Chen W., Lu Z., Liu J. Clostridium butyricum exerts a neuroprotective effect in a mouse model of traumatic brain injury via the gut-brain axis. Neurogastroenterol. Motil. 2018;30:e13260. doi: 10.1111/nmo.13260. [DOI] [PubMed] [Google Scholar]
- 51.Chen D., Jin D., Huang S., Wu J., Xu M., Liu T., Dong W., Liu X., Wang S., Zhong W., et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020;469:456–467. doi: 10.1016/j.canlet.2019.11.019. [DOI] [PubMed] [Google Scholar]
- 52.Chen G., Ran X., Li B., Li Y., He D., Huang B., Fu S., Liu J., Wang W. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model. EBioMedicine. 2018;30:317–325. doi: 10.1016/j.ebiom.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng W., Wu Y., Chen G., Fu S., Li B., Huang B., Wang D., Wang W., Liu J. Sodium butyrate attenuates diarrhea in weaned piglets and promotes tight junction protein expression in colon in a GPR109A-dependent manner. Cell. Physiol. Biochem. 2018;47:1617–1629. doi: 10.1159/000490981. [DOI] [PubMed] [Google Scholar]
- 54.Lee J.S., Tato C.M., Joyce-Shaikh B., Gulen M.F., Cayatte C., Chen Y., Blumenschein W.M., Judo M., Ayanoglu G., McClanahan T.K., et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity. 2015;43:727–738. doi: 10.1016/j.immuni.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gobbetti T., Dalli J., Colas R.A., Federici Canova D., Aursnes M., Bonnet D., Alric L., Vergnolle N., Deraison C., Hansen T.V., et al. Protectin D1n-3 DPA and resolvin D5n-3 DPA are effectors of intestinal protection. Proc. Natl. Acad. Sci. USA. 2017;114:3963–3968. doi: 10.1073/pnas.1617290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia L., Shan K., Pan L.L., Feng N., Lv Z., Sun Y., Li J., Wu C., Zhang H., Chen W., et al. Clostridium butyricum CGMCC0313.1 protects against autoimmune diabetes by modulating intestinal immune homeostasis and inducing pancreatic regulatory T cells. Front. Immunol. 2017;8:1345. doi: 10.3389/fimmu.2017.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomita Y., Ikeda T., Sakata S., Saruwatari K., Sato R., Iyama S., Jodai T., Akaike K., Ishizuka S., Saeki S., et al. Association of probiotic Clostridium butyricum therapy with survival and response to immune checkpoint blockade in patients with lung cancer. Cancer Immunol. Res. 2020;8:1236–1242. doi: 10.1158/2326-6066.CIR-20-0051. [DOI] [PubMed] [Google Scholar]
- 58.Tian Y., Li M., Song W., Jiang R., Li Y.Q. Effects of probiotics on chemotherapy in patients with lung cancer. Oncol. Lett. 2019;17:2836–2848. doi: 10.3892/ol.2019.9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou M., Yuan W., Yang B., Pei W., Ma J., Feng Q. Clostridium butyricum inhibits the progression of colorectal cancer and alleviates intestinal inflammation via the myeloid differentiation factor 88 (MyD88)-nuclear factor-kappa B (NF-κB) signaling pathway. Ann. Transl. Med. 2022;10:478. doi: 10.21037/atm-22-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Z.F., Ai L.Y., Wang J.L., Ren L.L., Yu Y.N., Xu J., Chen H.Y., Yu J., Li M., Qin W.X., et al. Probiotics Clostridium butyricum and Bacillus subtilis ameliorate intestinal tumorigenesis. Future Microbiol. 2015;10:1433–1445. doi: 10.2217/fmb.15.66. [DOI] [PubMed] [Google Scholar]
- 61.Pai S.G., Carneiro B.A., Mota J.M., Costa R., Leite C.A., Barroso-Sousa R., Kaplan J.B., Chae Y.K., Giles F.J. Wnt/beta-catenin pathway: Modulating anticancer immune response. J. Hematol. Oncol. 2017;10:101. doi: 10.1186/s13045-017-0471-6doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fenicia L., Anniballi F., Aureli P. Intestinal toxemia botulism in Italy, 1984–2005. Eur. J. Clin. Microbiol. Infect. Dis. 2007;26:385–394. doi: 10.1007/s10096-007-0301-9. [DOI] [PubMed] [Google Scholar]
- 63.Hill K.K., Xie G., Foley B.T., Smith T.J., Munk A.C., Bruce D., Smith L.A., Brettin T.S., Detter J.C. Recombination and insertion events involving the botulinum neurotoxin complex genes in Clostridium botulinum types A, B, E and F and Clostridium butyricum type E strains. BMC Biol. 2009;7:66. doi: 10.1186/1741-7007-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skarin H., Segerman B. Horizontal gene transfer of toxin genes in Clostridium botulinum: Involvement of mobile elements and plasmids. Mob. Genet. Elem. 2011;1:213–215. doi: 10.4161/mge.1.3.17617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Y., Sugiyama H., Johnson E.A. Transfer of neurotoxigenicity from Clostridium butyricum to a nontoxigenic Clostridium botulinum type E-like strain. Appl. Environ. Microbiol. 1993;59:3825–3831. doi: 10.1128/aem.59.11.3825-3831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dykes J.K., Luquez C., Raphael B.H., McCroskey L., Maslanka S.E. Laboratory investigation of the first case of C. butyricum type E botulism in the United States. J. Clin. Microbiol. 2015;53:3363–3365. doi: 10.1128/JCM.01351-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howard F., Bradley J., Flynn D., Noone P., Szawatkowski M. Outbreak of necrotising enterocolitis caused by Clostridium butyricum. Lancet. 1977;310:1099–1102. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- 68.Kwok J.S., Ip M., Chan T.F., Lam W.Y., Tsui S.K. Draft genome sequence of Clostridium butyricum strain NOR 33234, isolated from an elderly patient with diarrhea. Genome Announc. 2014;2:e01356-14. doi: 10.1128/genomeA.01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Popoff M.R., Szylit O., Ravisse P., Dabard J., Ohayon H. Experimental cecitis in gnotoxenic chickens monoassociated with Clostridium butyricum strains isolated from patients with neonatal necrotizing enterocolitis. Infect. Immun. 1985;47:697–703. doi: 10.1128/iai.47.3.697-703.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szylit O., Butel M.J., Rimbault A. An experimental model of necrotising enterocolitis. Lancet. 1997;350:33–34. doi: 10.1016/S0140-6736(05)66243-5. [DOI] [PubMed] [Google Scholar]
- 71.Thymann T., Møller H.K., Stoll B., Støy A.C., Buddington R.K., Bering S.B., Jensen B.B., Olutoye O.O., Siggers R.H., Mølbak L., et al. Carbohydrate maldigestion induces necrotizing enterocolitis in preterm pigs. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;297:G1115–G1125. doi: 10.1152/ajpgi.00261.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.