Abstract
Simple Summary
Immune checkpoint inhibitors (ICIs) markedly improve the survival benefits of advanced melanoma and non-small cell lung cancer (NSCLC). Nevertheless, only a subset of patients could benefit from such a therapy. Novel and effective clinical biomarkers are needed to assess ICI treatment efficacy. Heparan sulfate proteoglycan 2 (HSPG2) is frequently mutated in melanoma and NSCLC. In this study, we comprehensively integrated the pretreatment somatic mutational profiles and clinical information of both tumors and observed that HSPG2 mutations were associated with favorable tumor immunogenicity and immunotherapeutic efficacy. Our study provides a potential clinical molecular biomarker for evaluating ICI therapy responses.
Abstract
Immune checkpoint inhibitors (ICIs) markedly promote the survival outcome of advanced melanoma and non-small cell lung cancer (NSCLC). Clinically, favorable ICI treatment efficacy is noticed only in a smaller proportion of patients. Heparan sulfate proteoglycan 2 (HSPG2) frequently mutates in both tumors. Herein, we aim to investigate the immunotherapeutic and immunological roles of HSPG2 mutations in melanoma and NSCLC. A total of 631 melanoma samples and 109 NSCLC samples with both somatic mutational profiles and clinical immunotherapy data were curated. In addition, by using The Cancer Genome Atlas data, genomic and immunological traits behind HSPG2 mutations were elucidated. Melanoma patients with HSPG2 mutations had a markedly extended ICI outcome than other patients. An association between HSPG2 mutations and the improved outcome was further confirmed in NSCLC. In addition, an elevated ICI response rate was presented in HSPG2-mutated NSCLC patients (81.8% vs. 29.7%, p = 0.002). Subsequent analyses revealed that HSPG2-mutated patients had a favorable abundance of response immunocytes, an inferior abundance of suppression immunocytes, enhanced mutational burden, and interferon response-relevant signaling pathways. We uncovered that HSPG2 mutations were predictive of a better ICI response and associated with preferable immunogenicity, which may be considered as a genomic determinant to customize biotherapy strategies.
Keywords: HSPG2 mutations, immunotherapy, melanoma, NSCLC, clinical biomarker, cancer genomics
1. Introduction
Surgery, radiotherapy, chemotherapy, and targeted therapy are commonly used clinical treatment methods for cancer patients. However, for patients at advanced or metastatic stages, the above treatment modalities may be unsatisfactory [1]. In recent years, the advent of immune checkpoint inhibitors (ICIs) has greatly prolonged the prognosis of cancer patients [2]. The main theory of ICI treatments is to battle tumor cells by activating the immune system [2]. ICI agents have become the clinical first-line treatment strategy for several cancers; nevertheless, their biggest drawback is that only a small percentage of patients can benefit from them [3]. Therefore, selecting a suitable population to receive such ICI treatments is necessary.
At present, multiple biomarkers are determined to evaluate cancer immunotherapeutic efficacy. Programmed cell death 1 ligand 1 (PD-L1) is the first approved molecular biomarker for predicting anti-PD-1/L1 treatment response [4]. Its elevated expression was demonstrated to connect with favorable ICI efficacy [5]. However, in several clinical trials, PD-L1-negative tumors could also benefit from immunotherapy [6]. Tumor mutation burden (TMB) was recently reported to be involved in a better clinical immune therapy outcome [7]. Inconsistent results derived from several studies [8] showed that tumors with high TMB did not exhibit the treatment benefits. The above evidence demonstrated that PD-L1 expression and TMB sometimes are imperfect in predicting immunotherapeutic effects. Recently, multiple novel ICI biomarkers were reported, including gene mutations (e.g., POLE [9], JAK1/2 [10], B2M [10], and MUC16 mutations [11]), specific mutational signatures (e.g., signatures 1, 4, 7, and 11 [12]), and molecular subtypes [13].
Heparan sulfate proteoglycan 2 (HSPG2) encodes the perlecan protein, which comprises a central protein to which three long chains of glycosaminoglycans (heparan sulfate or chondroitin sulfate) are attached. It has been revealed that the perlecan protein exhibits vital roles in multiple biological behaviors via the interaction with prolargin, laminin, collagen type IV, transthyretin, etc. Several recent studies have demonstrated that HSPG2 overexpression was associated with invasion, metastasis, and an inferior survival outcome in triple-negative breast cancer [14], acute myeloid leukemia [15], glioblastoma [16,17], oligoastrocytoma [18], and oligodendroglioma [18]. HSPG2 was also reported to regulate immune and stromal infiltration in glioma [19] and prostate cancer [20]. Lima et al. performed a proteogenomics analysis and found that HSPG2-specific mutations played a protective role in prostate cancer [21]. So far, no studies have revealed the correlation of HSPG2 mutations with immunological features and ICI treatment efficacy in cancers.
Taking into account that ICI treatments are most commonly used for melanoma and NSCLC patients, in this work, we comprehensively integrated the pretreatment somatic mutational profiles from melanoma and NSCLC; furthermore, clinical information after immunotherapy of both tumors was also obtained. Finally, based on 631 melanoma and 109 NSCLC samples, we investigated the immunological and clinical immunotherapeutic implications of HSPG2 alterations. This immunogenomic research might provide useful clues for customizing cancer immunotherapy strategies.
2. Materials and Methods
2.1. Samples Used in This Study
From previous publications, we integrated a total of 631 melanoma [22,23,24,25,26,27,28,29] and 109 NSCLC samples [30,31] with both somatic mutational profiles and ICI treatment information. All included samples in this study were treated with blockade treatment of immune checkpoints (i.e., PD-1/L1, CTLA-4, or combination). Since genomic mutation data were acquired from distinct sequencing and annotating platforms, we re-annotated them with the Oncotator software (developed by Ramos et al., Boston, MA, USA) against the h19 reference genome [32]. In this research, nonsynonymous mutations (i.e., missense mutations, nonsense mutations, frameshift del/ins, in frame del/ins, and splice site mutations) were employed for subsequent analyses. The detailed clinical baseline characteristics and ICI therapy information for melanoma and NSCLC samples are shown in Table S1 and Table S2, respectively.
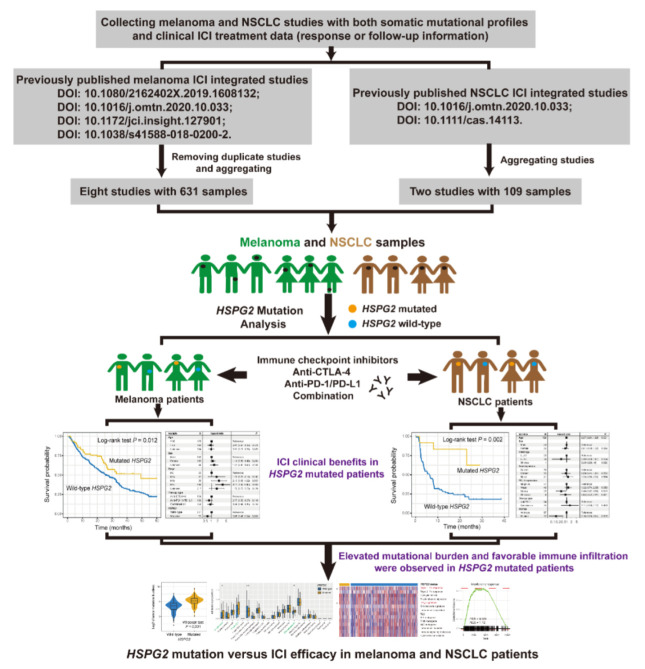
From The Cancer Genome Atlas (TCGA) project (http://xena.ucsc.edu/ accessed on 1 May 2022), we obtained a total of 457 melanoma and 995 NSCLC samples with genomic mutation data, transcriptomic expression profiles, and clinical information. Especially, the log2 transformed and normalized gene expression profiles of both tumors were applied to explore the potential immunological mechanisms behind HSPG2 mutations. The detailed flowchart of this research is shown in Figure 1.
Figure 1.
Workflow of this study. Investigation of the roles of HSPG2 mutations in evaluating ICI treatment efficacy in melanoma and NSCLC.
2.2. Mutational Signatures in the Genome
Mutational signatures were extracted using mutational profiles of melanoma and NSCLC samples based on a method proposed by a recent study [33]. In this method, Bayesian nonnegative matrix factorization (NMF) was used to disassemble mutation feature matrix A with 96 base substitution types into 2 nonnegative matrices W and H (i.e., A ≈ W × H), with W indicating the detected specific mutational signatures and H representing the mutational activities for each signature. The number of columns of matrix W is the number of mutational signatures. The rows of matrix A are the 96 mutational contexts, and its columns are the integrated samples of both cohorts. The 96 mutational contexts are derived from combinations of 6 mutational types (i.e., C > A, C > G, C > T, T > A, T > C, and T > G) and their 5′ and 3′ adjacent bases. The rows and columns of matrix H indicate the individual signatures and their corresponding mutational activities, respectively. The pruning process is performed by introducing the weight parameter λk, which is associated with the kth column of W and the kth row of H. All extracted mutational signatures were then compared with well-annotated signatures stored in the COSMIC database (version 2, Cambridge, UK) using cosine similarity.
2.3. Tumor-Infiltrating Immune Cells
To elucidate the different immune cell infiltrating abundances between HSPG2 mutant and wild-type groups, we employed CIBERSORT and the Angelova et al. algorithm to evaluate infiltrating levels of distinct immunocytes. CIBERSORT uses the LM22 signature, which includes 547 representative genes to assess tumor-infiltrating levels of 22 immunocytes [34]. The Angelova et al. algorithm applies a feature signature with 812 genes to evaluate the infiltration abundance of 31 immunocytes [35]. The detailed characteristic genes for each immune cell subtype are collected in Table S3.
2.4. Immune Infiltration and Immunogenicity-Related Signatures
Recent research presented multiple molecular signatures associated with immune infiltration and tumor immunogenicity. We thus curated the relevant signatures as follows: (1) immune/stromal cell signatures [36]; (2) an immune cell subset of T cells, B cells, and natural killer (NK) cells [37]; (3) T/NK cells, B/plasma cells, and monocyte/dendritic cell enrichment signature [38]; (4) Type 1/2 interferon (IFN) signature [39]; (5) IFNγ signature [40]; (6) T cell-inflamed gene expression profile (GEP) [41]; (7) immune cytolytic activity [39]; (8) immune signaling molecules [37]; (9) cytokines and chemokines [37]; and (10) tertiary lymphoid structures [42]. Detailed characteristic genes for distinct immunogenicity signatures are illustrated in Table S4.
2.5. GSVA and GSEA
Single sample gene set enrichment analysis (ssGSEA) was utilized to evaluate enriched levels of collected immunocyte- and immunogenicity-relevant signatures under the GSVA R package [43]. R DESeq2 package [44] was used to perform whole-genome differential analysis between HSPG2-mutant and wild-type groups. All t values extracted from the differential result were then considered as the input variable to conduct GSEA and acquire specific biological circuits of HSPG2 mutations. Hallmark pathways were applied as the background comparison circuits.
2.6. TMB and NB
Tumor mutation burden (TMB) was defined as the log2 transition of total non-synonymous mutations per megabase in both integrated and TCGA datasets. Neoantigen burden (NB) was calculated according to a recent method [45] for 224 melanoma and 109 NSCLC-integrated samples. From the Cancer Immunome Atlas (TCIA), we acquired the neoantigen data of 340 melanoma and 656 NSCLC samples in the TCGA cohort.
2.7. Statistical Analysis
R software was utilized to conduct relevant analyses. Mutational features of specified genes were exhibited with a waterfall plot, which is embedded in the maftools package [46]. The pheatmap package was employed to achieve heatmap exhibition of different immunogenicity signatures in two HSPG2 subgroups. Survival plots were obtained using the Kaplan–Meier method, and the Logrank test was applied to evaluate significant differences. Multivariate logistic and Cox regression models with multiple confounders taken into consideration were performed using the forest model package. The relationships of continuous and categorical factors with HSPG2 mutations were estimated with the Wilcoxon rank sum test and Fisher’s exact test, respectively. The detailed sample size and cohorts used for specific HSPG2-related analyses are shown in Table S5.
3. Results
3.1. HSPG2 Mutations of Melanoma
Among the 631 pooled melanoma samples, 193 (30.6%) exhibited the ICI status of complete response (CR) or partial response (PR); 430 (68.1%) had the response of stable disease (SD) or progressive disease (PD), and immunotherapy response data of the remanent samples (1.3%) were not available. The mutational landscape of these melanoma samples was characterized by C > T substitutions (Figure S1). Mutational features of significantly mutated melanoma genes concerning HSPG2 mutations are illustrated in Figure S1. HSPG2 is frequently mutated, accounting for 80 of 631 patients (12.7%). Amino acid changes produced by HSPG2 alterations are illustrated with a lollipop plot (Figure S2).
3.2. HSPG2 Mutations Associated with Melanoma ICI Outcome
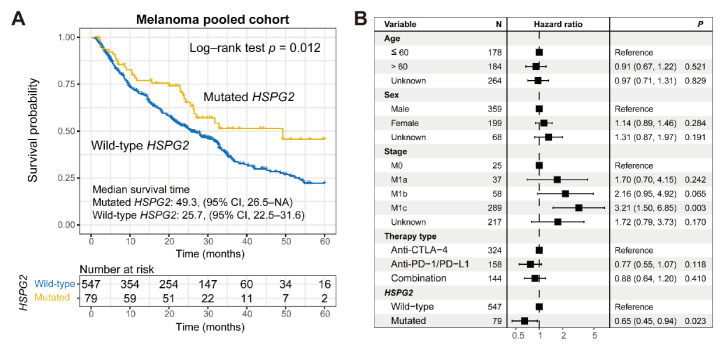
A melanoma univariate prognosis analysis revealed that patients with HSPG2 mutations presented a significantly more prolonged outcome than HSPG2 wild-type patients (median survival time: 49.3 vs. 25.7 months, Log–rank test p = 0.012; Figure 2A). Multivariate Cox regression analysis adjusted for age, sex, stage, and treatment type further corroborated this connection (HR: 0.65, 95% CI: 0.45–0.94, p = 0.023; Figure 2B). ICI predictive implications of HSPG2 mutations in the included individual cohorts and distinct therapy types are exhibited in Figure S3 and Figure S4, respectively.
Figure 2.
Association of HSPG2 mutations with ICI treatment outcome in melanoma. (A) Kaplan-Meier survival curves of melanoma patients with and without HSPG2 mutations. (B) Representation of multivariate Cox regression model of HSPG2 mutations with multiple confounding factors adjusted.
3.3. Connection of HSPG2 Mutations with Mutational Burden in Melanoma
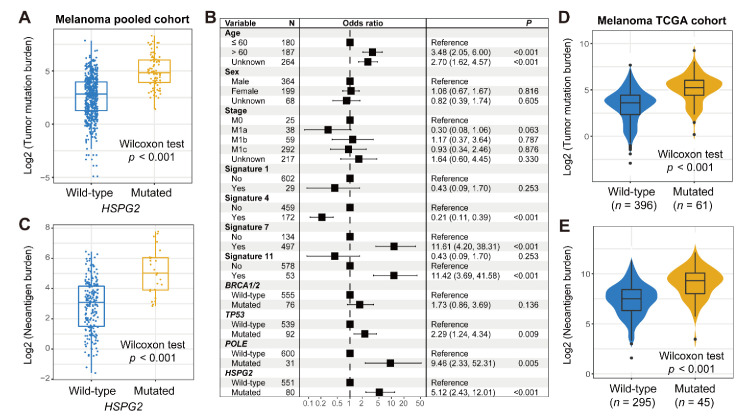
TMB was recently identified as a promising marker to evaluate immune treatment efficacy in advanced cancers. We thus investigated the correlation between HSPG2 mutations and melanoma TMB. The results show that HSPG2-mutated patients harbored a markedly more elevated TMB than wild-type patients (Wilcoxon rank sum test p < 0.001; Figure 3A). Genomic mutational signatures play vital roles in regulating genome stability. Based on melanoma mutational profiles, we determined a total of four mutational signatures by employing the NMF method; they are signatures 1, 4, 7, and 11. Specific mutational activities of the above four signatures for each patient are curated in Table S6. We subsequently performed a multivariate logistic model by incorporating clinical features and determined mutational signatures and DNA repair gene mutations to acquire an accurate connection between HSPG2 mutations and TMB. The results demonstrate that the connection of HSPG2 mutations with TMB was still existent in the multivariate analysis (OR: 5.12, 95% CI: 2.43–12.01, p < 0.001; Figure 3B). In addition, a higher NB was also enriched in HSPG2-mutated patients (Wilcoxon rank sum test p < 0.001; Figure 3C). We also used the mutational profiles of melanoma samples from the TCGA cohort and observed a significantly elevated TMB and NB in HSPG2-mutated patients (Wilcoxon rank sum test both p < 0.001; Figure 3D,E).
Figure 3.
Association of HSPG2 mutations with mutational burden in melanoma. (A) Distinct TMB distribution in HSPG2-mutated versus wild-type subgroups. (B) Multivariate logistic regression model of HSPG2 mutations was performed with clinical and genomic confounders taken into consideration. (C) Distinct NB distribution in HSPG2-mutated versus wild-type subgroups. Connection of HSPG2 mutations with (D) TMB and (E) NB based on the data from the TCGA melanoma cohort.
3.4. Validation in NSCLC
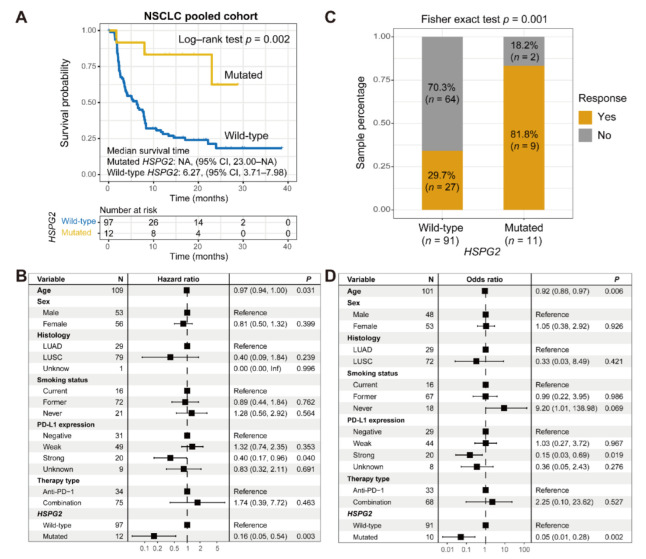
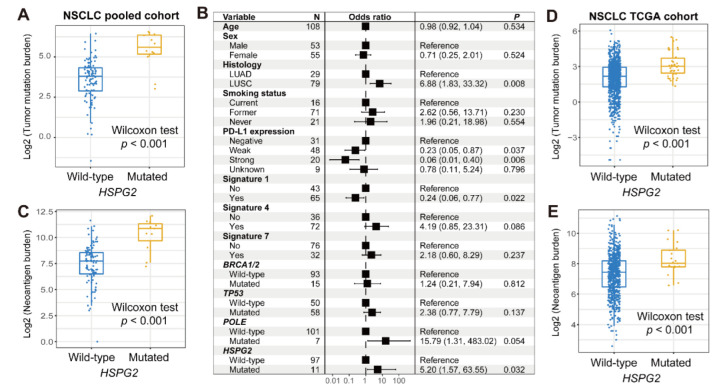
Among the 109 collected NSCLC patients, 36 (33.0%) harbored the CR/PR immunotherapeutic responses. HSPG2 was also frequently mutant in NSCLC, accounting for 12 of 109 (11.0%) patients. A prognosis analysis showed that HSPG2 mutations predicted a significantly improved ICI outcome in NSCLC (Log–rank test p = 0.002; Figure 4A). We further conducted a multivariate-adjusted Cox regression analysis with multiple confounding factors (e.g., age, sex, smoking status, histology, and PD-L1 expression) taken into consideration, and the consistent association between HSPG2 mutations and the favorable outcome was also observed (HR: 0.16, 95% CI: 0.05–0.54, p = 0.003; Figure 4B). ICI prognostic implications of HSPG2 mutations in distinct treatment types are illustrated in Figure S5. An immunotherapeutic response analysis revealed that an enhanced ICI response rate was enriched in patients with HSPG2 mutations (81.8 vs. 29.7%, Fisher’s exact test p = 0.001; Figure 4C). Consistently, in a multivariate logistic analysis, this link was still significant after adjusting for other variables (OR: 0.05, 95% CI: 0.01–0.28, p = 0.002; Figure 4D).
Figure 4.
Association of HSPG2 mutations with ICI treatment efficacy in NSCLC. (A) Kaplan–Meier survival curves of NSCLC patients with and without HSPG2 mutations. (B) Representation of multivariate Cox regression model of HSPG2 mutations with multiple confounding factors adjusted. (C) ICI response rate exhibition of HSPG2-mutated and wild-type groups. (D) Multivariate logistic regression model of HSPG2 mutations was performed with clinical and genomic variables taken into consideration.
We further explored the association of HSPG2 mutations with the mutational burden in NSCLC. The results show that a markedly higher TMB was observed in patients with HSPG2 mutations (Wilcoxon rank sum test p < 0.001; Figure 5A). Mutational signatures 1, 4, and 7 in NSCLC mutational profiles were extracted, and their mutational activities are exhibited in Table S7. A multivariate-adjusted logistic model with multiple confounding factors taken into account was conducted, and the results indicate that the higher TMB was still enriched in HSPG2-mutated patients (OR: 5.20, 95% CI: 1.57–63.55, p = 0.032; Figure 5B). In addition, HSPG2 mutations were also identified to be linked with an elevated NB (Wilcoxon rank sum test p < 0.001; Figure 5C). In the TCGA NSCLC cohort, consistently, the associations of HSPG2 mutations with higher TMB and NB were also noticed (Wilcoxon rank sum test both p < 0.001; Figure 5D,E).
Figure 5.
Association of HSPG2 mutations with mutational burden in NSCLC. (A) Distinct TMB distribution in HSPG2-mutated versus wild-type subgroups. (B) Multivariate logistic regression model of HSPG2 mutations was performed with clinical and genomic confounders taken into consideration. (C) Distinct NB distribution in HSPG2-mutated versus wild-type subgroups. Connection of HSPG2 mutations with (D) TMB and (E) NB based on the data from the TCGA NSCLC cohort.
3.5. Immunologic Features behind HSPG2 Mutations
In melanoma, we performed analyses of immune infiltration, immunogenicity signatures, and pathway enrichment to elucidate the potential immunological mechanisms of HSPG2 mutations. The CIBERSORT method revealed that the significantly increased infiltration levels of CD8 T cells, M1 macrophages, and B naive cells were enriched in the HSPG2- mutated subgroup (all p < 0.05; Figure 6A). According to the results from the Angelova et al. algorithm, activated CD4/8 T cells, central/effector memory CD8 T cells, and dendritic cells exhibited an enhanced infiltration in HSPG2-mutated patients (all p < 0.05; Figure 6B); however, a decreased infiltration of mast cells, which were recently reported to associate with immune suppression, was observed in these mutated patients (p < 0.05; Figure 6B).
Figure 6.
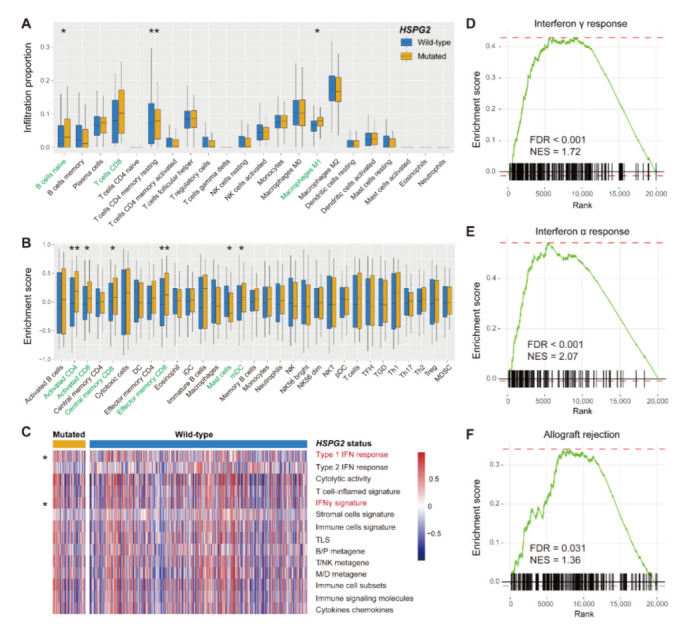
Immunocyte infiltration and signaling circuits behind HSPG2 mutations in melanoma. (A) CIBERSORT method inferred the distinct infiltrating levels of 22 immune cells based on HSPG2 mutational status. Immune cells highlighted with green are significantly differentially infiltrated. (B) Angelova et al. algorithm inferred the distinct infiltrating levels of 31 immune cells based on HSPG2 mutational status. (C) Distinct enrichment distribution of 14 curated immune signatures in HSPG2-mutated versus wild-type subgroups. Signatures highlighted with red are significantly differentially enriched. (D–F) Significantly enriched signaling pathways connected with HSPG2 mutations. * p < 0.05, ** p < 0.01.
We then composed a heatmap with distinct immune signature enrichment in two HSPG2 groups (Figure 6C). We found that the type I interferon response and interferon gamma signatures were significantly presented in HSPG2-mutated patients (both p < 0.05). Further GSEA analysis verified the results that interferon gamma and alpha responses were enriched in patients with HSPG2 mutations (both FDR < 0.001; Figure 6D,E and Figure S6). In addition, the immune response-related circuit of allograft rejection also appeared in the HSPG2-mutated subgroup (FDR = 0.031; Figure 6F and Figure S6).
Subsequently, we investigated the immune infiltration status of HSPG2 mutations in NSCLC. Consistent with the findings for melanoma, the markedly increased infiltration levels of immune response cells (e.g., central memory CD4/8 T cells, effector memory CD8 T cells, and cytotoxic cells) and decreased infiltration levels of immune suppressive cells (e.g., regulatory T cells) were observed in NSCLC patients harboring HSPG2 mutations (all p < 0.05; Figure S7A,B).
4. Discussion
Immune checkpoint blockade agents promote survival for cancer patients; however, an obvious disadvantage of such a treatment is the lower response rate. Therefore, identifying patients who are suitable to receive ICI treatments is clinically necessary. Immunotherapies are commonly used for melanoma and NSCLC; in this study, by integrating somatic mutational profiles and clinical therapy information for the above two cancers, we determined that HSPG2 mutations were predictive of favorable immunogenicity and ICI efficacy. The findings obtained from this work would provide a potential molecular biomarker for evaluating immunotherapeutic efficacy.
Activated, central/effector memory CD4/8 T cells were previously demonstrated to play a positive role in promoting tumor immune responses [47,48]. Two macrophage subtypes (i.e., M1 and M2) exhibit inverse immune regulation functions, with M1 macrophages associating with immune response and M2 associating with immune suppression [49,50]. Regulatory T cells are classical immune suppressive cells and mediate the tumor immune escape [51]. Mast cells, an immunocyte subtype, played distinct roles (i.e., immune promotion and inhibition) under distinct signaling regulation [52]. Recently reported immunocyte infiltration evaluation methods (e.g., CIBERSORT and Angelova et al.) give us a chance to depict the immune infiltration landscape across all included samples and to investigate the association between HSPG2 mutations and specific immune cell infiltration. In this study, we observed that an elevated infiltration of immune response cells (e.g., CD8 T cells and M1 macrophages) and a decreased infiltration of immune suppressive cells (e.g., regulatory T cells and mast cells) were enriched in HSPG2- mutated patients, which suggests that HSPG2 mutations mediate a favorable immune infiltration and tumor microenvironment.
Multiple studies have revealed the potential implications of TMB for evaluating cancer immunotherapeutic efficacy [8,12,13,53]. However, in clinical practice, the determination of TMB needs to conduct whole-exome mutational detection [54]. Another disadvantage of the TMB application is the uncertain cut-off values in distinct cancer types [54]. Therefore, easier surrogates are necessary for such clinical settings. Recent research has shown that mutations in single genes, such as POLE [9], NLRP3 [55], COL3A1 [12], and PTPRT [54] could predict tumor TMB and immunotherapeutic response. In this study, HSPG2 mutations were also determined to link with an elevated mutational burden and a preferable ICI efficacy in melanoma and NSCLC, which indicates that HSPG2 mutations may be a possible indicator for TMB and cancer immune treatment response.
The findings derived from this work show that HSPG2 mutations were connected with an improved outcome in melanoma and NSCLC patients under an immunotherapy setting. We then investigated whether HSPG2 mutations play roles in the above two cancers treated with conventional chemotherapies in the TCGA cohort. The results demonstrate that no significant associations between HSPG2 mutations and patient prognosis were observed in both two tumors (multivariate-adjusted both p > 0.05; Figure S8). These findings suggest that HSPG2 mutations play a predictive role regarding cancer immunotherapeutic efficacy rather than a prognostic role.
In this study, based on the integrated ICI-treated 631 melanoma and 109 NSCLC samples, we observed that HSPG2 mutations were predictive of a favorable ICI treatment outcome, which provides evidence for customizing immunotherapeutic strategies. By using the same pooled melanoma and NSCLC cohorts [12,54,56], we also discovered other mutations of the genes FAT1, COL3A1, NRAS, NARS2, DCC, and PTPRT were associated with better ICI response and outcome. In addition, the ICI efficacy-related mutational signatures and molecular subtypes were also determined. The above findings emphasize the importance of clinically expanded cohorts to uncover novel molecular determinants of response to immunotherapies.
There are several limitations to this study. First, the integration of melanoma and NSCLC data was based on multiple distinct datasets, which might produce some biases during data analyses. Second, only two cancer types were included in this study, and no more available cancers were used for validation. Three, the connection between HSPG2 mutations and ICI efficacy lacked functional experiments and in-house verification.
5. Conclusions
Collectively, based on the aggregated melanoma and NSCLC immunotherapeutic cohorts, we discovered that HSPG2 mutations were associated with better tumor immunogenicity and ICI treatment efficacy. The results from this genomic association study suggest that HSPG2 mutations may be considered as a possible molecular biomarker for assessing immune treatment responses.
Acknowledgments
Qinghua Wang thanks Wenjing Zhang for her company over the past 10 years and for giving birth to a lovely baby.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14143495/s1, Figure S1: Mutational patterns of HSPG2 and common melanoma driver genes illustrated with waterfall plot; Figure S2: Detailed amino acid changes induced by HSPG2 mutations in the integrated melanoma cohort; Figure S3: Kaplan-Meier survival analyses of HSPG2 mutations in individual ICI-treated melanoma cohorts; Figure S4: Kaplan-Meier survival analyses of HSPG2 mutations in distinct ICI treatment types in melanoma; Figure S5: Kaplan-Meier survival analyses of HSPG2 mutations in individual ICI-treated NSCLC cohorts; Figure S6: Significantly enriched signaling pathways in HSPG2-mutated subgroups in melanoma. Immune response pathways are highlighted with green; Figure S7: Immune infiltration associated with HSPG2 mutations in NSCLC. (A) Distinct infiltration of 22 immunocytes of HSPG2-mutated and wild-type groups evaluated with CIBERSORT algorithm. Immunocytes highlighted with green are significantly differentially infiltrated. (B) Distinct infiltration of 31 immunocytes of two HSPG2 groups evaluated with Angelova et al. method; Figure S8: Prognostic capacities of HSPG2 mutations in (A) melanoma and (B) NSCLC patients derived from the TCGA project; Table S1: Clinical characteristics of 631 pooled melanoma patients treated with ICI agents; Table S2: Clinical characteristics of 109 pooled NSCLC patients treated with ICI agents; Table S3: Specific genes used for enrichment analysis in each infiltrating immune cell subtype; Table S4: Specific genes used for enrichment analysis in each curated immune-related signature; Table S5: Sample size and cohorts for specific HSPG2-related analyses in this study; Table S6: The extracted 4 mutational signatures and their mutational activities in the pooled melanoma cohort; Table S7: The extracted 3 mutational signatures and their mutational activities in the pooled NSCLC cohort.
Author Contributions
Conceptualization, Q.W. (Qinghua Wang); methodology, Q.W. (Qinghua Wang), W.Z., Z.L., S.W. and F.S.; software, W.Z. and Y.K.; validation, W.Z. and Y.R.; data curation, Q.W. (Qiang Wang), J.L., H.Q. and W.Z.; writing—original draft preparation, W.Z.; writing—review and editing, Q.W. (Qinghua Wang) and S.W.; visualization, C.S. and Y.L.; supervision, Q.W. (Qinghua Wang); project administration, Q.W. (Qinghua Wang); funding acquisition, Q.W. (Qinghua Wang) and S.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genomic and clinical data used in this study were obtained from previously published studies and can be obtained by contacting the corresponding author under reasonable requests. The codes used for reproducing the results of this study can be acquired by contacting the first authors.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Medicine and Health Science and Technology Development Plan Project of Shandong Province (grant number 202112050480), National Natural Science Foundation of China (grant number 32000495), and Natural Science Foundation of Shandong Province (grant number ZR2020MH202).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tsimberidou A.M., Fountzilas E., Nikanjam M., Kurzrock R. Review of precision cancer medicine: Evolution of the treatment paradigm. Cancer Treat. Rev. 2020;86:102019. doi: 10.1016/j.ctrv.2020.102019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y. Cancer immunotherapy: Harnessing the immune system to battle cancer. J. Clin. Investig. 2015;125:3335–3337. doi: 10.1172/JCI83871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castro A., Pyke R.M., Zhang X., Thompson W.K., Day C.P., Alexandrov L.B., Zanetti M., Carter H. Strength of immune selection in tumors varies with sex and age. Nat. Commun. 2020;11:4128. doi: 10.1038/s41467-020-17981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doroshow D.B., Bhalla S., Beasley M.B., Sholl L.M., Kerr K.M., Gnjatic S., Wistuba I.I., Rimm D.L., Tsao M.S., Hirsch F.R. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2021;18:345–362. doi: 10.1038/s41571-021-00473-5. [DOI] [PubMed] [Google Scholar]
- 5.Patel S.P., Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 6.Sharma P., Callahan M.K., Bono P., Kim J., Spiliopoulou P., Calvo E., Pillai R.N., Ott P.A., de Braud F., Morse M., et al. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): A multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol. 2016;17:1590–1598. doi: 10.1016/S1470-2045(16)30496-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarchoan M., Hopkins A., Jaffee E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samstein R.M., Lee C.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y., Barron D.A., Zehir A., Jordan E.J., Omuro A., et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F., Zhao Q., Wang Y.N., Jin Y., He M.M., Liu Z.X., Xu R.H. Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. JAMA Oncol. 2019;5:1504–1506. doi: 10.1001/jamaoncol.2019.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torrejon D.Y., Abril-Rodriguez G., Champhekar A.S., Tsoi J., Campbell K.M., Kalbasi A., Parisi G., Zaretsky J.M., Garcia-Diaz A., Puig-Saus C., et al. Overcoming Genetically Based Resistance Mechanisms to PD-1 Blockade. Cancer Discov. 2020;10:1140–1157. doi: 10.1158/2159-8290.CD-19-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q., Yang Y., Yang M., Li X., Chen K. High mutation load, immune-activated microenvironment, favorable outcome, and better immunotherapeutic efficacy in melanoma patients harboring MUC16/CA125 mutations. Aging. 2020;12:10827–10843. doi: 10.18632/aging.103296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W., Kong Y., Li Y., Shi F., Lyu J., Sheng C., Wang S., Wang Q. Novel Molecular Determinants of Response or Resistance to Immune Checkpoint Inhibitor Therapies in Melanoma. Front. Immunol. 2021;12:798474. doi: 10.3389/fimmu.2021.798474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi F., Zhang W., Yang Y., Yang Y., Zhao J., Xie M., Sheng C., Wang S., Wang Q. Sex Disparities of Genomic Determinants in Response to Immune Checkpoint Inhibitors in Melanoma. Front. Immunol. 2021;12:721409. doi: 10.3389/fimmu.2021.721409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalscheuer S., Khanna V., Kim H., Li S., Sachdev D., DeCarlo A., Yang D., Panyam J. Discovery of HSPG2 (Perlecan) as a Therapeutic Target in Triple Negative Breast Cancer. Sci. Rep. 2019;9:12492. doi: 10.1038/s41598-019-48993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X., Liang S., Zhan Q., Yang L., Chi J., Wang L. HSPG2 overexpression independently predicts poor survival in patients with acute myeloid leukemia. Cell Death Dis. 2020;11:492. doi: 10.1038/s41419-020-2694-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazanskaya G.M., Tsidulko A.Y., Volkov A.M., Kiselev R.S., Suhovskih A.V., Kobozev V.V., Gaytan A.S., Aidagulova S.V., Krivoshapkin A.L., Grigorieva E.V. Heparan sulfate accumulation and perlecan/HSPG2 up-regulation in tumour tissue predict low relapse-free survival for patients with glioblastoma. Histochem. Cell Biol. 2018;149:235–244. doi: 10.1007/s00418-018-1631-7. [DOI] [PubMed] [Google Scholar]
- 17.Dzikowski L., Mirzaei R., Sarkar S., Kumar M., Bose P., Bellail A., Hao C., Yong V.W. Fibrinogen in the glioblastoma microenvironment contributes to the invasiveness of brain tumor-initiating cells. Brain Pathol. 2021;31:e12947. doi: 10.1111/bpa.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma X.L., Shang F., Ni W., Zhu J., Luo B., Zhang Y.Q. Increased HSPG2 expression independently predicts poor survival in patients with oligoastrocytoma and oligodendroglioma. Eur. Rev. Med. Pharmacol. Sci. 2018;22:6853–6863. doi: 10.26355/eurrev_201810_16154. [DOI] [PubMed] [Google Scholar]
- 19.Tian Y., Ke Y., Ma Y. High expression of stromal signatures correlated with macrophage infiltration, angiogenesis and poor prognosis in glioma microenvironment. PeerJ. 2020;8:e9038. doi: 10.7717/peerj.9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grindel B.J., Martinez J.R., Tellman T.V., Harrington D.A., Zafar H., Nakhleh L., Chung L.W., Farach-Carson M.C. Matrilysin/MMP-7 Cleavage of Perlecan/HSPG2 Complexed with Semaphorin 3A Supports FAK-Mediated Stromal Invasion by Prostate Cancer Cells. Sci. Rep. 2018;8:7262. doi: 10.1038/s41598-018-25435-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lima T., Barros A.S., Trindade F., Ferreira R., Leite-Moreira A., Barros-Silva D., Jeronimo C., Araujo L., Henrique R., Vitorino R., et al. Application of Proteogenomics to Urine Analysis towards the Identification of Novel Biomarkers of Prostate Cancer: An Exploratory Study. Cancers. 2022;14:2001. doi: 10.3390/cancers14082001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., Walsh L.A., Postow M.A., Wong P., Ho T.S., et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L., Sucker A., Hillen U., Foppen M.H.G., Goldinger S.M., et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao D., Margolis C.A., Vokes N.I., Liu D., Taylor-Weiner A., Wankowicz S.M., Adeegbe D., Keliher D., Schilling B., Tracy A., et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 2018;50:1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roh W., Chen P.L., Reuben A., Spencer C.N., Prieto P.A., Miller J.P., Gopalakrishnan V., Wang F., Cooper Z.A., Reddy S.M., et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 2017;9:eaah3560. doi: 10.1126/scitranslmed.aah3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D., Schilling B., Liu D., Sucker A., Livingstone E., Jerby-Arnon L., Zimmer L., Gutzmer R., Satzger I., Loquai C., et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019;25:1916–1927. doi: 10.1038/s41591-019-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaretsky J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., Torrejon D.Y., Abril-Rodriguez G., Sandoval S., Barthly L., et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riaz N., Havel J.J., Makarov V., Desrichard A., Urba W.J., Sims J.S., Hodi F.S., Martin-Algarra S., Mandal R., Sharfman W.H., et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017;171:934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S., et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hellmann M.D., Nathanson T., Rizvi H., Creelan B.C., Sanchez-Vega F., Ahuja A., Ni A., Novik J.B., Mangarin L.M.B., Abu-Akeel M., et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33:843–852.e844. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos A.H., Lichtenstein L., Gupta M., Lawrence M.S., Pugh T.J., Saksena G., Meyerson M., Getz G. Oncotator: Cancer variant annotation tool. Hum. Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J., Mouw K.W., Polak P., Braunstein L.Z., Kamburov A., Kwiatkowski D.J., Rosenberg J.E., Van Allen E.M., D’Andrea A., Getz G. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet. 2016;48:600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angelova M., Charoentong P., Hackl H., Fischer M.L., Snajder R., Krogsdam A.M., Waldner M.J., Bindea G., Mlecnik B., Galon J., et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshihara K., Shahmoradgoli M., Martinez E., Vegesna R., Kim H., Torres-Garcia W., Trevino V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Network Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagalla S., Chou J.W., Willingham M.C., Ruiz J., Vaughn J.P., Dubey P., Lash T.L., Hamilton-Dutoit S.J., Bergh J., Sotiriou C., et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 2013;14:R34. doi: 10.1186/gb-2013-14-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Z.Y., Zhong W.Z., Zhang X.C., Su J., Xie Z., Liu S.Y., Tu H.Y., Chen H.J., Sun Y.L., Zhou Q., et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 41.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., Albright A., Cheng J.D., Kang S.P., Shankaran V., et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finkin S., Yuan D., Stein I., Taniguchi K., Weber A., Unger K., Browning J.L., Goossens N., Nakagawa S., Gunasekaran G., et al. Ectopic lymphoid structures function as microniches for tumor progenitor cells in hepatocellular carcinoma. Nat. Immunol. 2015;16:1235–1244. doi: 10.1038/ni.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanzelmann S., Castelo R., Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balachandran V.P., Luksza M., Zhao J.N., Makarov V., Moral J.A., Remark R., Herbst B., Askan G., Bhanot U., Senbabaoglu Y., et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caserta S., Borger J.G., Zamoyska R. Central and effector memory CD4 and CD8 T-cell responses to tumor-associated antigens. Crit. Rev. Immunol. 2012;32:97–126. doi: 10.1615/CritRevImmunol.v32.i2.10. [DOI] [PubMed] [Google Scholar]
- 48.Mueller S.N., Gebhardt T., Carbone F.R., Heath W.R. Memory T cell subsets, migration patterns, and tissue residence. Ann. Rev. Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- 49.Genin M., Clement F., Fattaccioli A., Raes M., Michiels C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer. 2015;15:577. doi: 10.1186/s12885-015-1546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Locati M., Curtale G., Mantovani A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Ann. Rev. Pathol. 2020;15:123–147. doi: 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka A., Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27:109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Derakhshani A., Vahidian F., Alihasanzadeh M., Mokhtarzadeh A., Lotfi Nezhad P., Baradaran B. Mast cells: A double-edged sword in cancer. Immunol. Lett. 2019;209:28–35. doi: 10.1016/j.imlet.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 53.Klempner S.J., Fabrizio D., Bane S., Reinhart M., Peoples T., Ali S.M., Sokol E.S., Frampton G., Schrock A.B., Anhorn R., et al. Tumor Mutational Burden as a Predictive Biomarker for Response to Immune Checkpoint Inhibitors: A Review of Current Evidence. Oncologist. 2020;25:e147–e159. doi: 10.1634/theoncologist.2019-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang W., Shi F., Kong Y., Li Y., Sheng C., Wang S., Wang Q. Association of PTPRT mutations with immune checkpoint inhibitors response and outcome in melanoma and non-small cell lung cancer. Cancer Med. 2022;11:676–691. doi: 10.1002/cam4.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q., Lyu J., Zhang W., Shi F., Ren Y., Mao Q., Liu Y., Li Y., Wang S. Immunological and clinical immunotherapy implications of NLRP3 mutations in melanoma. Aging. 2021;13:24271–24289. doi: 10.18632/aging.203678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W., Tang Y., Guo Y., Kong Y., Shi F., Sheng C., Wang S., Wang Q. Favorable immune checkpoint inhibitor outcome of patients with melanoma and NSCLC harboring FAT1 mutations. NPJ Precis. Oncol. 2022;6:46. doi: 10.1038/s41698-022-00292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic and clinical data used in this study were obtained from previously published studies and can be obtained by contacting the corresponding author under reasonable requests. The codes used for reproducing the results of this study can be acquired by contacting the first authors.