Abstract
The volumes of intracranial tissues of 40 healthy volunteers acquired from 0.3- and 3-T scanners were compared using intraclass correlation coefficients, correlation analyses, and Bland-Altman analyses. We found high intraclass correlation coefficients, high Pearson’s correlation coefficients, and low percentage biases in all tissues and most of the brain regions, although small differences were observed in some areas. These findings may support the validity of brain volumetry with low-field magnetic resonance imaging.
Keywords: brain volumetry, field strength comparison, low magnetic field magnetic resonance imaging, morphology, neuroimaging
Introduction
MR-based brain volumetry has been widely used in research and clinical settings mainly using 1.5-T or higher magnetic field scanners.1 However, a large number of low-field MR scanners (0.25–1 T) are currently used worldwide because of their cost-effectiveness, safety profile, and reduced metal artifacts.2 Notably, around 2500 out of 5200 facilities with MR scanners in Japan have those with a magnetic field strength less than 1.5 T.3 Low-field MR scanners have also some additional advantages, such as short longitudinal relaxation time, high magnetic field homogeneity, and availability of open MR scanners that are beneficial for subjects with claustrophobia. If MR-based brain volumetry proves to be reliable on low-field MR scanners, we can then exploit the potential of existing scanners and their clinical applications may be further expanded.
Some previous studies have assessed the influence of magneticfield strength on brain volumetry using 1.5- and 3-T MR scanners.4,5 Huppertz et al. reported that the inter-scanner coefficients of variation (CV) of volumes of the various brain structures ranged from 0.66% to 14.7%.4 Briellmann et al. reported that the absolute inter-scanner bias for hippocampal volume measurements obtained from 1.5- and 3-T MR scanners was 6% ± 3.9%.5 These studies have indicated that the compatibility of the volumetry results is reliable among high magnetic fields (≧ 1.5 T). Goto et al. reported that the repeatability of the atlas-based method for brain volumetry was tested at a 0.4-T MR scanner and it was adequate for the estimation of changes in brain volume.6 However, the studies of compatibility of the brain volumes in volumetry between low and high fields are still sparse. The objective of the present study was to investigate the compatibility of the calculated brain volumes in the volumetry conducted between low- and high-field MR scanners. The volumes measured at both 0.3 and 3 T were compared in the 62 regions identified by atlas-based volumetry (ABV) in the same subjects.
Materials and Methods
Subjects
This study was part of the Bunkyo Health Study, including 1629 elderly people aimed at the prevention of disease requiring long-term care.7 Forty healthy volunteers (9 males and 31 females; mean age, 72.1 ± 5.3 years) who did not have major intracranial lesions, such as bleeding, aneurysms, and cerebral infarction, as confirmed by radiologists were included in this study. This study was approved by the local institutional review board of Juntendo University, and written informed consent was obtained from all participants before entering the study.
Devices and scan parameters
All subjects underwent whole-brain 3D T1-weighted imaging with both 0.3-T (AIRIS Vento; Hitachi, Tokyo, Japan) and 3-T (MAGNETOM Prisma; Siemens Healthcare, Erlangen, Germany) clinical MRI systems. All subjects were first scanned on a 0.3-T MR scanner followed by a 3-T MR scanner with a maximum duration of 2 weeks between each acquisition. The T1-weighted images on the 0.3-T MR scanner were obtained using a 3D-gradient echo with inversion recovery sequence with the following parameters: TR = 25 ms; TE = 5.8 ms; inversion time (TI) = 600 ms; flip angle (FA) = 12 degree; number of excitations (NEX) = 1; FOV = 200 × 250 × 250 mm3; resolution = 0.98 × 0.98 × 2 mm3; slice orientation = sagittal; total scan time = 601 s. T1-weighted imaging on the 3-T MR scanner was performed using a magnetization-prepared rapid gradient echo sequence with the following parameters: TR = 2300 ms; TE = 2.32 ms; TI = 900 ms; FA = 8 degree; NEX = 1; FOV = 200 × 250 × 250 mm3; resolution = 0.9 × 0.9 × 0.9 mm3; slice orientation = sagittal; total scan time = 321 s.
Post-Processing
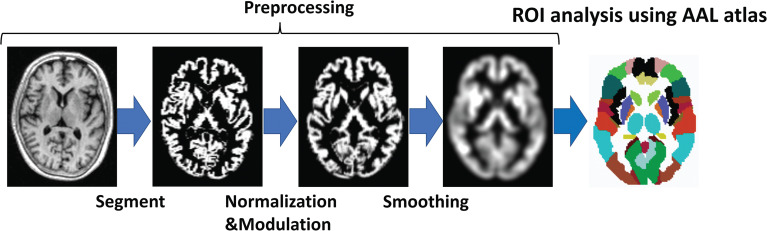
Post-processing was performed using an image analysis script implemented in Linux for the ImPACT program.8 This script utilizes voxel-based morphometry implemented in Statistical Parametric Mapping (SPM) package 12 (http://www.fil.ion.ucl.ac.uk/spm/software/spm12/) in MATLAB R2012b (MathWorks, Natick, MA, USA). In the first step, the image was segmented into six tissues, namely, gray matter (GM), white matter (WM), cerebrospinal fluid (CSF), bone, soft tissue, and air. Then, segmented images obtained at both 0.3 and 3 T were resized to 1.5-mm iso-voxels and aligned between subjects using diffeomorphic anatomical registration through exponentiated lie algebra (DARTEL). After DARTEL, the images were normalized to the Montreal Neurological Institute templates, with the signal intensity modulated to preserve the volume. Lastly, images were smoothened with an 8-mm full width at half-maximum Gaussian kernel (Fig. 1). Spatial smoothing was performed to compensate for anatomical variabilities that were not compensated by spatial normalization and to improve the SNR.
Fig. 1.
Flow diagram of atlas-based analysis. Preprocessing, segmentation to improve the agreement of normalization and evaluate each tissue separately, normalization to evaluate each subject’s data collectively, modulation to convert signal intensity to volume, and smoothening to remove the influence of individual differences among subjects were performed. Afterward, each regional volume was measured using the Automated Anatomical Labeling atlas. AAL, automated anatomical labeling.
Identification of each volume
All statistical analyses were performed with IBM SPSS Statistic Version 21 (IBM, Armonk, NY, USA). The calculated volumes at 0.3 T (V0.3T) and 3 T (V3T) in each tissue region were compared, respectively, to examine how the volumes were affected by the differences between the field strengths. The GM volume (GMV), WM volume (WMV), and CSF volume (CSFV) were calculated by the summation of the volumes in the segmented GM, WM, and CSF images. The total brain volume (TBV) and intracranial volume (ICV) were calculated using Eq 1 and 2, respectively.
| Eq 1 |
| Eq 2 |
Further, ABV was performed to determine the degree of difference between the V0.3T and V3T in the GM subregions of the brain. The GM was parcellated into 116 (ROIs) based on the Automated Anatomical Labeling atlas interpolated to a 1.5-mm iso-voxel, which matches the resolution of the modulated and normalized component images. The atlas-derived regional volumes were measured by calculating the sum of the internal signals for each atlas and recorded. Subsequently, the left and right volumes were integrated, resulting in a total of 62 volumes.
Statistics
To evaluate the degree of difference, intraclass correlation coefficients (ICC; 3,1) in each of V0.3T and V3T were calculated. Pearson’s correlation coefficient (rp) and %Bias were then calculated as supplemental indices for evaluating the difference. The %Bias was calculated using the following equation (Eq 3):
| Eq 3 |
Significances for both ICC and rp were defined as P < 0.01. In this study, based on a prior report,9 the calculated ICCs and rp values were ranked as follows: poor, values < 0.4; fair, values ≥ 0.4 and < 0.6; good, values ≥ 0.6 and < 0.75; excellent, values ≥ 0.75 and ≤ 1.
For each volume, scatter plots of all V0.3T and V3T were created with linear regression lines and Bland-Altman graphs (difference was calculated as the volume obtained from the 0.3-T scanner minus the volume obtained from the 3-T scanner). The inclination of the linear regression line (Ilr), mean difference (dm), standard error of the mean difference (SE), 95% confidence interval (CI), 95% limits of agreement (LOA), and correlation coefficient of Bland-Altman (rBA) (i.e., Pearson’s correlation coefficient between the average and the difference) were also calculated. In addition, the ICCs, rp values, and %Bias of volumes were calculated individually for each of the 116 (original) and 62 (the left and right volumes were integrated) subregions (see supplementary documents for 116 ROI results).
Results
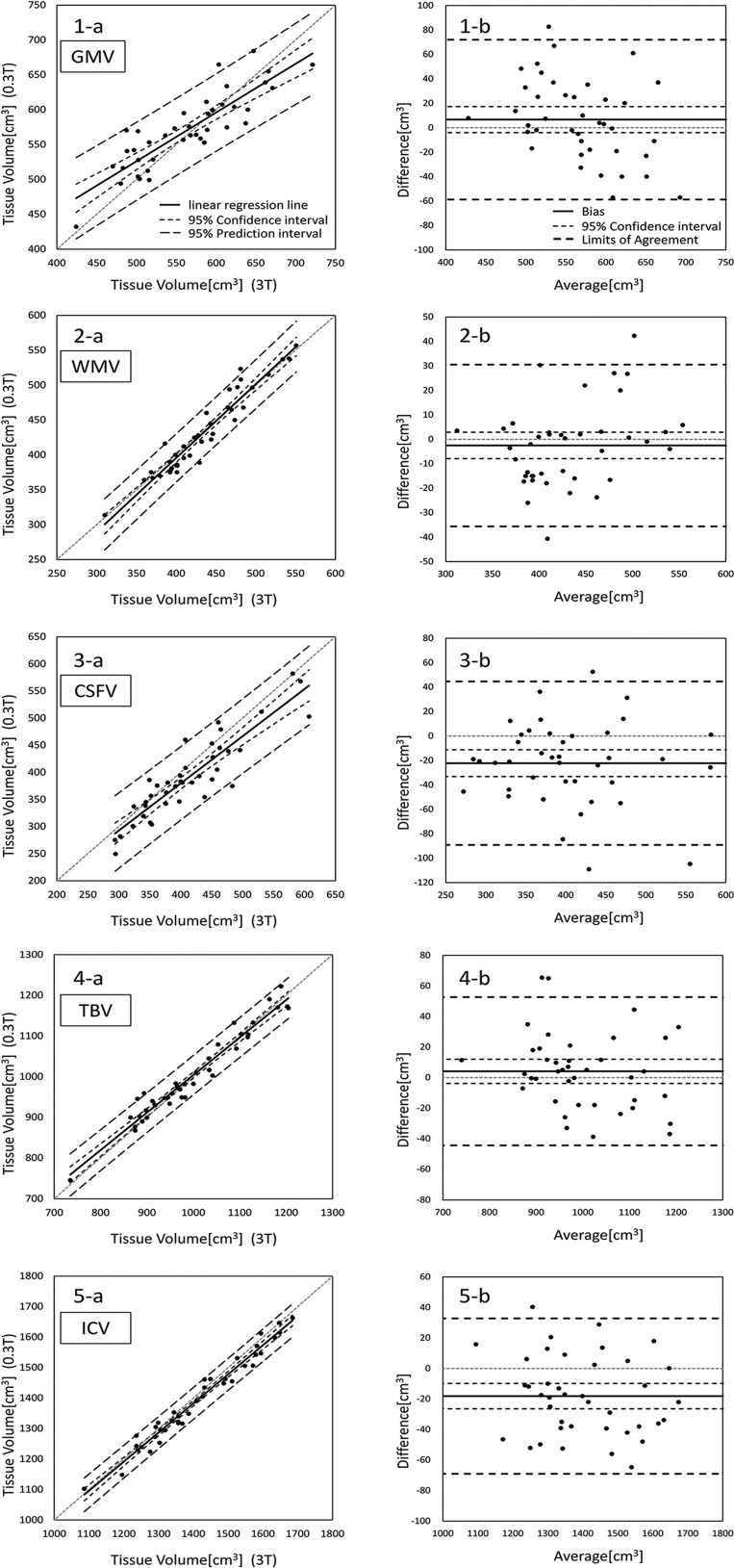
The ICC, rp, %Bias, Ilr, dm, SE, CI, LOA, and rBA values for GMV, WMV, CSFV, TBV, and ICV obtained from the 0.3- and 3-T MR scanners are shown in Table 1. All ICCs and rp values of tissue volumes were significant and assessed as excellent. The %Bias was less than 2% for all tissue volumes, except for CSFV showing a %Bias of 5.37%. The CI ranges of the CSFV (-33.12 to -11.29) and the ICV (-26.39 to -9.78) were all negative (i.e., showed fixed error) (Table 1, Fig. 2). The fixed errors of the CSFV and the ICV indicated that the V0.3T were significantly smaller than the V3T. The rBA of GMV indicated a significant negative proportional bias; in other words, the V0.3T of GM were overestimated for smaller volumes and underestimated for larger volumes compared to the V3T of GM. When compared across different tissues, SEs decreased in the order of CSFV, GMV, and WMV.
Table 1.
Statistical values for comparisons of tissue volumes between the 0.3- and 3-T scanners
| Correlation analysis | Bland-Altman results | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ICC | rp | %Bias | Ilr | dm | SE | CI | LOA | rBA | |
| GMV | 0.848* | 0.868* | 1.18 | 0.698 | 6.65 | 33.36 | -4.02 to 17.3 | -58.74 to 72.04 | -0.403* |
| WMV | 0.954* | 0.959* | 0.58 | 1.058 | -2.52 | 16.87 | -7.92 to 2.87 | -35.58 to 30.54 | 0.328 |
| CSFV | 0.901* | 0.902* | 5.37 | 0.87 | -22.21 | 34.13 | -33.12 to -11.29 | -89.11 to 44.69 | -0.081 |
| TBV | 0.974* | 0.975* | 0.41 | 0.924 | 4.13 | 24.78 | -3.80 to 12.05 | -44.45 to 52.70 | -0.239 |
| ICV | 0.984* | 0.984* | 1.28 | 0.955 | -18.08 | 25.97 | -26.39 to -9.78 | -68.99 to 32.82 | -0.167 |
CI, confidence interval at 95%; CSFV, cerebrospinal fluid volume; dm, mean difference; GMV, gray matter volume; ICC, interclass correlation coefficient; Ilr, inclination of the linear regression line; ICV, intracranial volume. P < .01 was defined as a significant correlation and was represented with an asterisk; LOA, limits of agreement at 95%; rBA, correlation coefficient of Bland-Altman; rp, Pearson’s correlation coefficient; SE, standard error of the mean difference; TBV: total brain volume; WMV, white matter volume.
Fig. 2.
The left row (a) shows the scatter plots with lines of equality, and the right row (b) shows the Bland-Altman graph (difference was calculated as the volume obtained from the 0.3-T scanner minus the volume obtained from the 3-T scanner) of each volume (1, GMV; 2, WMV; 3, CSFV; 4, TBV; 5, ICV) compared between 0.3- and 3-T scanners. CSFV, cerebrospinal fluid volume; GMV, gray matter volume; ICV, intracranial volume; TBV, total brain volume; WMV, white matter volume.
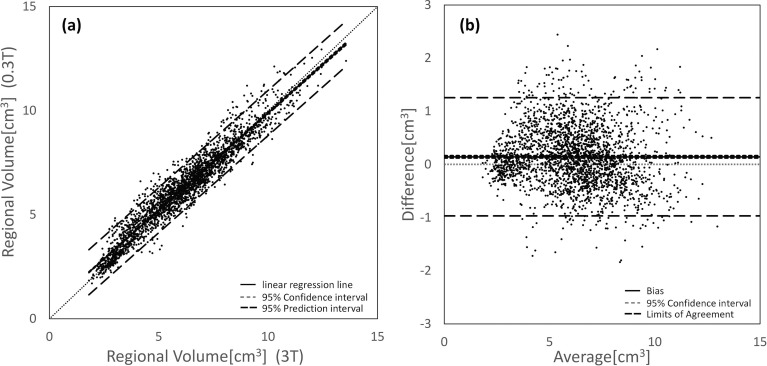
In the examination of the ABV, the ICCs and rp values acquired with the V0.3T and V3T were significant and assessed as excellent for all subregional volumes (ICC = 0.980; rp = 0.960; Fig. 3). A comparison of all subregional V0.3T and V3T yielded the following results: Ilr = 0.934; dm = 0.142; SE = 0.567; CI = 0.120 to 0.165; LOA = -0.970 to 1.254; and rBA = -0.099 (P > 0.01). The CI range indicated that the average subregional V0.3T were significantly larger compared to the V3T (i.e., fixed error).
Fig. 3.
(a) Scatter plot and (b) Bland-Altman graph (difference was calculated as the volume obtained from the 0.3-T scanner minus the volume obtained from the 3-T scanner) of all subregional volumes compared between the 0.3- and 3-T scanners. The subregional volumes (62 ROIs for 40 subjects) acquired with the 0.3- and 3-T scanners showed only small differences and high linearity indicated by the ICC and Pearson’s correlation coefficients (ICC = 0.980; rp = 0.960). ICC, intraclass correlation coefficients; rp, Pearson’s correlation coefficient.
In the comparison between the V0.3T and V3T in individual subregions, the ICCs, rp values, and %Biases are summarized in Tables 2, 3, and 4, respectively. Both the ICCs and rp values of the subregional V0.3T and V3T were significant for 61 of the 62 ROIs (Tables 2 and 3), indicating reliable compatibility. The Vermis 1_2 showed the lowest ICC and rp value, and the highest %Bias and the V0.3T were larger than the V3T (Tables 2–4). The ICCs were evaluated as excellent for 55 ROIs, good for 5 ROIs, and fair for 1 ROI (Table 2). The rp values were evaluated as excellent for 57 ROIs and good for 4 ROIs (Table 3).
Table 2.
ICC in the 62 regional volumes measured using 0.3- and 3-T scanners, shown in the descending order of ICC
| Rank | Brain region | ICC | Rank | Brain region | ICC | Rank | Brain region | ICC |
|---|---|---|---|---|---|---|---|---|
| 1 | Vermis_9 | 0.995* | 22 | Cingulum_Post | 0.894* | 43 | Precuneus | 0.841* |
| 2 | Cerebelum_9 | 0.981* | 23 | Cerebelum_10 | 0.893* | 44 | Angular | 0.839* |
| 3 | Cerebelum_6 | 0.954* | 24 | Frontal_Inf_Orb | 0.886* | 45 | Frontal_Sup | 0.837* |
| 4 | Fusiform | 0.95* | 25 | Frontal_Inf_Tri | 0.885* | 46 | Calcarine | 0.832* |
| 5 | Vermis_8 | 0.949* | 26 | Temporal_Sup | 0.884* | 47 | SupraMarginal | 0.812* |
| 6 | Heschl | 0.942* | 27 | Lingual | 0.879* | 48 | Caudate | 0.808* |
| 7 | Cerebelum_8 | 0.941* | 28 | Temporal_Inf | 0.878* | 49 | Frontal_Mid | 0.804* |
| 8 | Amygdala | 0.934* | 29 | Temporal_Pole_Mid | 0.878* | 50 | Cerebelum_3 | 0.796* |
| 9 | Cerebelum_Crus2 | 0.933* | 30 | Insula | 0.875* | 51 | Cuneus | 0.795* |
| 10 | Vermis_6 | 0.932* | 31 | Vermis_7 | 0.875* | 52 | Occipital_Mid | 0.795* |
| 11 | Cerebelum_7b | 0.932* | 32 | ParaHippocampal | 0.874* | 53 | Temporal_Mid | 0.784* |
| 12 | Olfactory | 0.93* | 33 | Frontal_Mid_Orb | 0.874* | 54 | Thalamus | 0.779* |
| 13 | Putamen | 0.927* | 34 | Cerebelum_Crus1 | 0.872* | 55 | Parietal_Inf | 0.753* |
| 14 | Rolandic_Oper | 0.926* | 35 | Temporal_Pole_Sup | 0.869* | 56 | Postcentral | 0.741* |
| 15 | Cerebelum_4_5 | 0.921* | 36 | Cingulum_Ant | 0.866* | 57 | Vermis_10 | 0.696* |
| 16 | Rectus | 0.92* | 37 | Cingulum_Mid | 0.864* | 58 | Pallidum | 0.683* |
| 17 | Frontal_Inf_Oper | 0.914* | 38 | Hippocampus | 0.863* | 59 | Parietal_Sup | 0.667* |
| 18 | Vermis_4_5 | 0.909* | 39 | Occipital_Sup | 0.862* | 60 | Paracentralobule | 0.605* |
| 19 | Frontal_Sup_Orb | 0.904* | 40 | Frontal_Sup_Medial | 0.846* | 61 | Precentral | 0.572* |
| 20 | Frontal_Med_Orb | 0.904* | 41 | Occipital_Inf | 0.843* | 62 | Vermis_1_2 | 0.069 |
| 21 | Vermis_3 | 0.901* | 42 | Supp_Motor_Area | 0.841* |
P < .01 was defined as significant correlation and was represented with an asterisk. ICC, Intraclass correlation coefficient.
Table 3.
Pearson’s correlation coefficients for the 62 regional volumes measured using 0.3- and 3-T scanners, shown in descending order of correlation coefficients
| Rank | Brain region | Pearson | Rank | Brain region | Pearson | Rank | Brain region | Pearson |
|---|---|---|---|---|---|---|---|---|
| 1 | Vermis_9 | 0.995* | 22 | Frontal_Med_Orb | 0.904* | 43 | Precuneus | 0.848* |
| 2 | Cerebelum_9 | 0.984* | 23 | Vermis_3 | 0.904* | 44 | Frontal_Sup_Medial | 0.847* |
| 3 | Fusiform | 0.962* | 24 | Temporal_Pole_Mid | 0.899* | 45 | Angular | 0.847* |
| 4 | Cerebelum_6 | 0.955* | 25 | Cingulum_Post | 0.899* | 46 | Supp_Motor_Area | 0.844* |
| 5 | Vermis_8 | 0.955* | 26 | Frontal_Inf_Orb | 0.896* | 47 | Frontal_Sup | 0.843* |
| 6 | Heschl | 0.95* | 27 | Temporal_Sup | 0.888* | 48 | SupraMarginal | 0.84* |
| 7 | Cerebelum_8 | 0.944* | 28 | Frontal_Inf_Tri | 0.886* | 49 | Calcarine | 0.837* |
| 8 | Amygdala | 0.944* | 29 | Temporal_Inf | 0.885* | 50 | Cuneus | 0.821* |
| 9 | Vermis_6 | 0.939* | 30 | Insula | 0.879* | 51 | Thalamus | 0.819* |
| 10 | Olfactory | 0.936* | 31 | ParaHippocampal | 0.878* | 52 | Frontal_Mid | 0.811* |
| 11 | Cerebelum_Crus2 | 0.936* | 32 | Frontal_Mid_Orb | 0.876* | 53 | Occipital_Mid | 0.807* |
| 12 | Putamen | 0.934* | 33 | Vermis_7 | 0.875* | 54 | Temporal_Mid | 0.794* |
| 13 | Cerebelum_7b | 0.932* | 34 | Cerebelum_Crus1 | 0.874* | 55 | Parietal_Inf | 0.785* |
| 14 | Cerebelum_10 | 0.931* | 35 | Occipital_Sup | 0.873* | 56 | Postcentral | 0.76* |
| 15 | Rolandic_Oper | 0.929* | 36 | Temporal_Pole_Sup | 0.872* | 57 | Vermis_10 | 0.752* |
| 16 | Cerebelum_4_5 | 0.922* | 37 | Hippocampus | 0.871* | 58 | Pallidum | 0.694* |
| 17 | Rectus | 0.921* | 38 | Cingulum_Ant | 0.866* | 59 | Parietal_Sup | 0.683* |
| 18 | Frontal_Inf_Oper | 0.916* | 39 | Cerebelum_3 | 0.865* | 60 | Paracentralobule | 0.619* |
| 19 | Lingual | 0.913* | 40 | Cingulum_Mid | 0.864* | 61 | Precentral | 0.605* |
| 20 | Vermis_4_5 | 0.909* | 41 | Occipital_Inf | 0.855* | 62 | Vermis_1_2 | 0.070 |
| 21 | Frontal_Sup_Orb | 0.907* | 42 | Caudate | 0.851* |
P < .01 was defined as significant correlation and was represented with an asterisk.
Table 4.
%Bias for the 62 regional volumes measured using 0.3- and 3-T scanners, shown in ascending order of %Bias
| Rank | Brain region | %Bias | Rank | Brain region | %Bias | Rank | Brain region | %Bias |
|---|---|---|---|---|---|---|---|---|
| 1 | Frontal_Inf_Oper | 0.10 | 22 | Cerebelum_9 | 3.32 | 43 | Amygdala | 7.14 |
| 2 | Calcarine | 0.77 | 23 | Cerebelum_8 | 3.46 | 44 | Lingual | 7.17 |
| 3 | Frontal_Inf_Tri | 1.08 | 24 | Supp_Motor_Area | 3.49 | 45 | Vermis_8 | 8.15 |
| 4 | Temporal_Inf | 1.13 | 25 | Olfactory | 3.54 | 46 | Fusiform | 8.20 |
| 5 | Parietal_Inf | 1.52 | 26 | Cerebelum_4_5 | 3.93 | 47 | Putamen | 8.26 |
| 6 | Angular | 1.66 | 27 | Temporal_Pole_Mid | 3.94 | 48 | Rectus | 8.95 |
| 7 | Parietal_Sup | 1.69 | 28 | Vermis_4_5 | 4.24 | 49 | Cingulum_Ant | 9.21 |
| 8 | Cerebelum | 1.72 | 29 | Cerebelum_7b | 4.36 | 50 | ParaHippocampal | 9.38 |
| 9 | Temporal_Sup | 2.12 | 30 | Cingulum_Post | 4.37 | 51 | Pallidum | 10.42 |
| 10 | Cuneus | 2.15 | 31 | Occipital_Inf | 4.53 | 52 | Frontal_Med_Orb | 10.42 |
| 11 | Frontal_Mid | 2.17 | 32 | Frontal_Inf_Orb | 4.56 | 53 | Heschl | 10.88 |
| 12 | Postcentral | 2.23 | 33 | Frontal_Sup | 4.67 | 54 | Frontal_Mid_Orb | 11.59 |
| 13 | Temporal_Mid | 2.26 | 34 | SupraMarginal | 5.33 | 55 | Occipital_Sup | 12.82 |
| 14 | Temporal_Pole_Sup | 2.36 | 35 | Rolandic_Oper | 5.81 | 56 | Precentral | 13.33 |
| 15 | Precuneus | 2.56 | 36 | Cerebelum_Crus1 | 5.97 | 57 | Paracentralobule | 13.38 |
| 16 | Hippocampus | 3.07 | 37 | Insula | 5.99 | 58 | Frontal_Sup_Orb | 15.00 |
| 17 | Caudate | 3.09 | 38 | Cerebelum_6 | 6.38 | 59 | Cerebelum_3 | 17.79 |
| 18 | Cingulum_Mid | 3.16 | 39 | Vermis_9 | 6.45 | 60 | Vermis_10 | 23.73 |
| 19 | Cerebelum_Crus | 3.24 | 40 | Occipital_Mid | 6.78 | 61 | Vermis_3 | 26.80 |
| 20 | Vermis_6 | 3.29 | 41 | Thalamus | 6.82 | 62 | Vermis_1_2 | 44.77 |
| 21 | Vermis_7 | 3.29 | 42 | Frontal_Sup_Medial | 7.08 |
Discussion
In this study, we compared the V0.3T and V3T of the brain tissues (GMV, WMV, CSFV, TBV, and ICV) and the subregional V0.3T and V3T and investigated how the magnetic field strengths affected the measured volumes. Overall, our results demonstrated that the volumes were comparable and warrant the future use of a low-field MR scanner for brain volumetry.
The V0.3T and V3T of brain tissues showed small differences and high linearities as indicated by the excellent ICCs and rp values. Among all the tissues, only the CSFV showed a %Bias of over 2%. In addition, the CI ranges of the CSFV and the ICV were all negative, which showed a fixed error. The results revealed that the V0.3T was significantly smaller than the V3T. This discrepancy may be due to the influence of the central brightening effect in CSF; the central brightening effect is larger with a higher field strength. Bernsteine et al. reported that central brightening of ~30% was observed on head imaging at 3 T, while only ~5% was observed at 1.5 T.10 Thus, a reduced influence from the central brightening effect can be advantageous for brain volumetry at a low magnetic field compared with that at a high magnetic field. ABV is often corrected by dividing each volume by ICV to eliminate the influence of various brain sizes across subjects. However, considering that the ICV was significantly larger on the 3-T MR scanner than on the 0.3-T MR scanner, the suppression of the influence of the central brightening effect should be considered before the ABV correction using ICV.11
At 0.3 T, the GMV was overestimated for smaller volumes and underestimated for larger volumes (i.e., proportional error) compared to that at 3 T. This may be due to the difference in magnetic field homogeneity, resolution, and distortion. Furthermore, the SEs were larger in the GMV than in the WMV (Table 1), which might have been due to the difference in the size of the GM and WM segmentation errors.
Subsequently, ABV was performed to investigate the degree of difference in the subregional V0.3T and V3T. There was a significant fixed error between the subregional V0.3T and V3T, where the V0.3T were larger than the V3T. The confirmed fixed error might be partly induced due to the difference in the SNR and the partial volume effect (PVE). On the other hand, the GM volumes in the 62 subregions showed that the degree of difference was small with high linearity as assessed by excellent ICCs and rp values. We believe that the fixed error can be neglected because it was much smaller than the calculated volumes of the subregions and size of the fixed error was smaller than SE.
In the examination of individual subregional V0.3T and V3T, both the ICCs and rp values were significant in the 61 out of 62 subregions. Only the Vermis 1_2 showed no significant correlation, the lowest ICC and rp value, and the highest %Bias; the V0.3T of Vermis 1_2 were larger than the V3T. This was probably due to its small size, leading to a large spatial normalization error12 and larger PVE. Another reason may be the difference in magnetic field homogeneity and distortion between the fields.
The rankings were excellent in the 55 subregions for ICC and 57 subregions for rp, respectively. Three regions were ranked differently between the ICC and rp, where the postcentral and Vermis_10 showed ICC = good; rp = excellent, and the precentral showed ICC = fair; rp = good. Lower ICCs are caused by lower linearity or higher bias or both. We can identify that the cause of the lower ICCs in the postcentral and Vermis_10 was higher bias rather than lower linearity because the rp was excellent. Regarding the precentral, the lower ICC seemed to be caused by both lower linearity and higher bias. Higher bias might be let by systematic biases that may be due to spatial normalization errors and differences in the SNR and PVE.
In clinical settings, evaluation of the hippocampal volume with ABV is important for evaluation of cognitive diseases or other neurological disorders. For example, patients with epilepsy and hippocampal sclerosis can be expected to demonstrate ≤ 30% hippocampal volume loss.5 Decreases in the hippocampal volumes in the range of 5%–10% have been observed in patients with temporal lobe epilepsy and with psychiatric disorders.5 In the study by Briellmann et al., no difference was detected between 1.5- and 3-T MR scanners in the measure of hippocampal volumes.5 The present study also showed that there was only a small difference (ICC = 0.863, %Bias ≤ 5%) between the hippocampal V0.3T and V3T. Therefore, the evaluation of hippocampal volume by ABV using a 0.3-T MR scanner may be sufficient for clinical applications.
This study has some limitations. First, only healthy subjects were enrolled in this study. The acceptable range of error cannot be unambiguously defined because the quality required in each situation is different. Thus, a future study should evaluate the difference in measured volumes at low- and high-field strengths in clinical settings. Second, the images obtained from both scanners were different not only with respect to magnetic field strength but also with respect to various conditions, such as SNR, resolution, sequence, magnetic field homogeneity, and distortion, all of which may affect volumetry. Even though it is difficult to completely separate the effects of each factor on volumetry, the results of past studies may aid in the interpretation of the results of our study.4,5,6,10,11,13 Finally, voxel-based morphometry using 3-T MR scanners has been reported to be useful, and in this study, the volumes obtained with 3-T were used as reference standards; however, their accuracy for brain volumetry has not been fully established yet.
Conclusion
In almost all the regions that we tested, except for small structures in the cerebellum, the V0.3T and V3T were comparable in the present study. Our findings would indicate that the volumetry at low-field MRI is appropriate compared with that at high-field MRI.
Acknowledgments
This work was funded by ImPACT Program of Council for Science, Technology and Innovation (Cabinet Office, Government of Japan) and was supported in part by a High Technology Research Center Grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) and MEXT-Supported Program for Strategic Research Foundation at Private Universities, 2014–2018. I would like to thank Arata Obuchi of Hitachi Medical Corporation for assistance in setting the MRI scan parameters.
Footnotes
Conflicts of Interest
Yujiro Otsuka is an employee of Milliman Inc., Tokyo office, Tokyo, Japan. The remaining authors declare no conflicts of interest associated with this manuscript.
References
- 1.Price S, Paviour D, Scahill R, et al. Voxel-based morphometry detects patterns of atrophy that help differentiate progressive supranuclear palsy and Parkinson’s disease. Neuroimage 2004; 23:663–669. [DOI] [PubMed] [Google Scholar]
- 2.Marques JP, Simonis FFJ, Webb AG. Low-field MRI: an MR physics perspective. J Magn Reson Imaging 2019; 49:1528–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ministry of Health, Labor and Welfare Heisei 23 [2011] Medical Facility [Static/Dynamic] Survey Outline of Hospital Report, available from http://www.mhlw.go.jp/toukei/saikin/hw/iryosd/11/, as of August 28, 2020. [Google Scholar]
- 4.Huppertz HJ, Kröll-Seger J, Klöppel S, et al. Intra- and interscanner variability of automated voxel-based volumetry based on a 3D probabilistic atlas of human cerebral structures. Neuroimage 2010; 49:2216–2224. [DOI] [PubMed] [Google Scholar]
- 5.Briellmann RS, Syngeniotis A, Jackson GD. Comparison of hippocampal volumetry at 1.5 tesla and at 3 tesla. Epilepsia 2001; 42:1021–1024. [DOI] [PubMed] [Google Scholar]
- 6.Goto M, Suzuki M, Mizukami S, et al. Repeatability of brain volume measurements made with the Atlas-based method from T1-weighted images acquired using a 0.4 tesla low field MR scanner. Magn Reson Med Sci 2016; 15:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Someya Y, Tamura Y, Kaga H, et al. Skeletal muscle function and need for long-term care of urban elderly people in Japan (the Bunkyo Health Study): a prospective cohort study. BMJ Open 2019; 9:e031584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemoto K, Oka H, Fukuda H, et al. MRI-based brain healthcare quotients: a bridge between neural and behavioral analyses for keeping the brain healthy. PLoS One 2017; 12:e0187137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lévy S, Guertin MC, Khatibi A, et al. ; Test-retest reliability of myelin imaging in the human spinal cord: measurement errors versus region- and aging-induced variations. PLoS One 2018; 13:e0189944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein MA, Huston J, Ward HA. Imaging artifacts at 3.0T. J Magn Reson Imaging 2006; 24:735–746. [DOI] [PubMed] [Google Scholar]
- 11.Keihaninejad S, Heckemann RA, Fagiolo G, et al. ; Alzheimer’s disease neuroimaging initiative. A robust method to estimate the intracranial volume across MRI field strengths (1.5T and 3T). Neuroimage 2010; 50:1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagiwara A, Fujita S, Ohno Y, et al. Variability and standardization of quantitative imaging: monoparametric to multiparametric quantification, radiomics, and artificial intelligence. Invest Radiol 2020; 55:601–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andica C, Hagiwara A, Hori M, et al. Automated brain tissue and myelin volumetry based on quantitative MR imaging with various in-plane resolutions. J Neuroradiol 2018; 45:164–168. [DOI] [PubMed] [Google Scholar]