Abstract
Bacterial blotch of Agaricus bisporus has typically been identified as being caused by either Pseudomonas tolaasii (brown blotch) or Pseudomonas gingeri (ginger blotch). To address the relatedness of pseudomonads able to induce blotch, a pilot study was initiated in which pseudomonads were selectively isolated from mushroom farms throughout New Zealand. Thirty-three pseudomonad isolates were identified as being capable of causing different degrees of discoloration (separable into nine categories) of A. bisporus tissue in a bioassay. These isolates were also identified as unique using repetitive extragenic palindromic PCR and biochemical analysis. Relationships between these 33 blotch-causing organisms (BCO) and a further 22 selected pseudomonad species were inferred by phylogenetic analyses of near-full-length 16S rRNA gene nucleotide sequences. The 33 BCO isolates were observed to be distributed throughout the Pseudomonas fluorescens intrageneric cluster. These results show that in addition to known BCO (P. tolaasii, P. gingeri, and Pseudomonas reactans), a number of diverse pseudomonad species also have the ability to cause blotch diseases with various discolorations. Furthermore, observation of ginger blotch discoloration of A. bisporus being independently caused by many different pseudomonad species impacts on the homogeneity and classification of the previously described P. gingeri.
The genus Pseudomonas (sensu stricto) comprises a taxon of metabolically versatile organisms that are ubiquitous in soil and water and play an important role as plant, animal, and human pathogens (37). Microbial diversity in mushroom farms has previously been reported, with pseudomonads accounting for 10% of bacteria in compost and sometimes more than 50% of bacteria in casing soils (48).
Discoloration of Agaricus bisporus caused by pathogenic pseudomonads, the so-called blotch diseases, are well documented. Pseudomonas tolaasii contamination results in sunken, dark brown lesions (35, 55); Pseudomonas reactans causes mild dark purple to light brown discoloration and a slight surface depression that becomes deeper and darker with age (59); while the pale yellowish red discoloration that develops into a reddish ginger-colored discoloration (ginger blotch disease) is characteristic of Pseudomonas gingeri (60).
Of the blotch-causing pseudomonads, the best characterized is P. tolaasii. P. tolaasii enters the mushroom farm in peat and limestone used in the casing process (63), and, once present, P. tolaasii is able to attach to mycelial surfaces of developing A. bisporus (40, 42). Temperature and relative humidity have been suggested as important environmental conditions that influence the pathogenicity of P. tolaasii within the mushroom farm (50). A minimal application of 2.7 × 106 to 108 CFU · ml−1 of P. tolaasii was reported as the threshold for inducing disease (34), although other thresholds have been proposed (31, 44, 62). Pathogenic P. tolaasii isolates synthesize a low-molecular-weight extracellular toxin, tolaasin, that is the primary bacterial agent responsible for eliciting disease symptoms (3, 32, 43). Tolaasin, a lipodepsipeptide (LDP), causes disruption of cell membranes from a range of cell types and has both ion channel-forming and biosurfactant properties (3, 17, 43). When P. tolaasii is cultured in close proximity to a second pseudomonad, P. reactans, a white precipitate forms between the colonies, defining the white-line in agar (WLA) assay (61). Tolaasin forms a dense white precipitate with the white line inducing principle (WLIP), an LDP produced by P. reactans (30).
Relatedness of pathogenic pseudomonads in the mushroom industry has been addressed in many previous studies. Using three experimental parameters, pathogenicity, physiological properties, and cellular fatty acid composition, P. gingeri, pathogenic P. reactans, and P. tolaasii were found to be related to but distinguishable from Pseudomonas fluorescens biovars III and V (59). Substrate utilization tests, electrophoresis of soluble proteins, and DNA:DNA hybridization experiments showed P. gingeri to form a unique grouping, as did P. tolaasii, saprophytic P. fluorescens biovar II, the so-called white line reacting organisms (including P. reactans), and Pseudomonas agarici (13). In this same study, Goor et al. showed P. tolaasii to be a homogeneous grouping separate from both P. fluorescens and P. reactans. Homogeneity of P. tolaasii as a species was further demonstrated with nucleotide sequence analysis of the small subunit rRNA (16S rRNA gene) (29).
P. gingeri was first described as a new member of the P. fluorescens complex causing ginger blotch disease of A. bisporus (60). Although P. gingeri demonstrates phenotypic similarity to P. tolaasii (e.g., during colony transition from pathogenic to nonpathogenic states [6]), P. gingeri can be distinguished from P. tolaasii by the WLA and pitting assays (61) and the 2-ketogluconate and lipase tests (60). However, since its first description, P. gingeri has received limited attention as to its epidemiology and characterization as a species.
Two continuing observations initiated the pilot study presented in this paper: (i) New Zealand pseudomonads isolated from A. bisporus exhibiting ginger blotch were variable in colony morphology and growth patterns; and (ii) from an agronomist's point of view, ginger blotch lesions exhibited variable discoloration, suggesting that more than one organism was responsible for the discoloration (or that the organism had variable virulence). Based on these observations, this study sought to address the molecular and phenotypic diversity among pseudomonads in the mushroom farm environment that are capable of causing ginger blotch and other blotch-related diseases of A. bisporus.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial isolates used in this study are listed in Table 1. Reference strains were obtained from the National Collection of Plant Pathogenic Bacteria (Harpenden, United Kingdom), and pseudomonad isolates from a milk factory environment were obtained from a previous study (45). All strains used in this study were maintained at −80°C in Kings B liquid medium (KB) (23) with a final concentration of 20% (vol/vol) glycerol. Strains were transferred onto KB medium supplemented with 1.5% agar, incubated at 28°C for 16 h, and maintained at 4°C for short-term use.
TABLE 1.
New Zealand BCO isolates and reference pseudomonad strains used in this studya
| Designation | Farmb | Assay resultd | Commente | WL(R)f | WL(T)g | Biotypeh | Accession no.i |
|---|---|---|---|---|---|---|---|
| NZ 032 | A | B9 | B | + | − | 0156555 | AF320995 |
| NZ 027 | B | B9 | B | + | − | 0156555 | AF320994 |
| NZ 006 | A | B6 | − | − | 0156555 | AY014800 | |
| P. tolaasii 2192Tj | Nc | B9 | B | + | − | 0156555 | AF320988 |
| NZ 031 | C | B4 | − | − | 0177575 | AY014807 | |
| NZ 009 | A | B3 | − | + | 1357555 | AY014802 | |
| P. reactans 1311 | N | B2 | − | + | 0357555 | AF320987 | |
| NZ 014 | A | B3 | D | − | − | 1147555 | AY014804 |
| NZ 024 | A | B3 | D | − | − | 1147455 | AY014806 |
| NZ 052 | A | B9 | − | + | 0157575 | AY014811 | |
| NZ 062 | A | B9 | Y | − | − | 0157575 | AY014814 |
| NZ 060 | A | B4 | B | − | + | 0157575 | AY014813 |
| NZ 007 | A | B4 | − | + | 0557555 | AY014801 | |
| NZ 081 | B | B1 | − | − | 0757555 | AF388206 | |
| NZ 111 | E | B9 | D | − | − | 0157577 | AY014825 |
| NZ 065 | B | B3 | D | − | + | 0757555 | AY014815 |
| NZ 124 | C | B8 | B | − | − | 0157555 | AY014829 |
| NZ 066 | B | B1 | − | − | 0157555 | AF388209 | |
| NZ 113 | E | B3 | − | − | 0357555 | AY014827 | |
| NZ 064 | A | B3 | − | − | 0357555 | AF388208 | |
| NZ 102 | A | B3 | B | − | − | 0357555 | AY014820 |
| NZ 097 | A | B3 | − | + | 4156577 | AY014818 | |
| NZ 101 | A | B6 | − | + | 0146657 | AY014819 | |
| NZ 096 | A | B2 | − | + | 0157555 | AY014817 | |
| NZ 039 | D | B3 | Y | − | − | 0157555 | AY014808 |
| NZ 104 | A | B3 | − | − | 0157555 | AY014822 | |
| NZ 017 | A | B3 | D | − | − | 1357555 | AY014805 |
| NZ 103 | A | B9 | B | − | − | 0157555 | AY014821 |
| NZ 099 | A | B1 | − | − | 0357555 | AF388207 | |
| NZ 011 | A | B6 | − | − | 0157555 | AY014803 | |
| NZ 112 | E | B3 | − | − | 4140457 | AY014826 | |
| P. gingeri 3147T | N | B5 | Y | − | − | 0356555 | AF320991 |
| NZ 043 | B | B5 | Y | − | − | 0157555 | AY014809 |
| NZ 059 | A | B6 | B | − | − | 0156555 | AY014812 |
| NZ 092 | A | B5 | − | − | 0156555 | AY014816 | |
| NZ 047 | B | B5 | Y | − | − | 0156565 | AY014810 |
For discussion purposes, isolates have been ordered as phylogenetically defined in Fig. 2.
Topographically distinct farm locations designated A to E from which pseudomonads were isolated.
N, reference strains obtained from National Collection of Plant Pathogenic Bacteria, Harpenden, United Kingdom.
Discoloration of mushroom cube bioassay (11) after 48 h where B is blotch and numbers (1 to 9) refer to assigned blotch discolorations depicted in Fig. 1.
D, tissue degradation; Y, yellowing of filter paper; B, browning of filter paper.
White line reaction with P. reactans NCPPB 1311.
White line reaction with P. tolaasii NCPPB 2192T.
API 20 NE numerical profile obtained.
Accession numbers of 16S rRNA nucleotide sequences stored in GenBank.
T indicates the recognized type strain of a species.
Isolation of pseudomonads from mushroom farms.
Sampling from mushroom farms in New Zealand was carried out during the Austral summer of 1999. A section of tissue was excised from mushroom caps exhibiting blotch discolorations and was placed into a McCartney tube containing sterile KB medium (10 ml). One gram of compost or casing material was placed into a McCartney tube containing sterile KB medium (10 ml). Water samples from water reservoirs and frequently used taps on mushroom farms were collected in sterile McCartney bottles. Samples were maintained on ice upon collection and during transport to the laboratory, a period not exceeding 2 h. Samples were incubated for 24 h at 28°C before an aliquot was applied to Gould's agar medium (14). Individual bacterial colonies were purified by passage onto fresh Gould's medium and stored as described above.
Bioassay for pathogenicity.
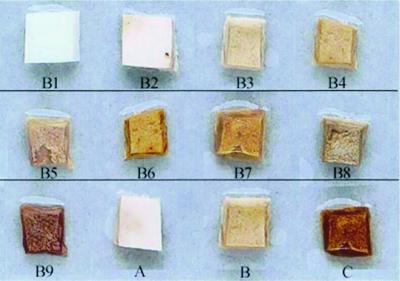
Bioassays were performed as described by Gandy (11) using healthy, 1-day-old A. bisporus. Cubes (1 cm3) of cap tissue were excised with sterile scalpel blades and placed into a sterile petri dish containing a 50-mm-pore-size paper filter dampened with 800 μl of sterile double-distilled water. Four cubes were placed 2 cm apart to eliminate cross-contamination by motile pseudomonads. Bacterial strains were cultured in KB medium to a density of 109 CFU · ml−1, and a 50-μl aliquot of cells was placed onto three cubes. The fourth cube was inoculated with a 50-μl control of uninoculated KB. Petri dishes were sealed with parafilm and incubated under ambient conditions for 24 h. Mushroom caps incubated with bacterial isolates were scored for the degree of blotch discoloration on a scale of B1 to B9 (where B = blotch). To ensure comparable results, a color scale was developed using the revised Munsell standard soil color charts (issued in 1957), where B1 = Hue 2.5Y 8/1; B2 = Hue 2.5Y 7/2; B3 = Hue 2.5Y 6/3; B4 = Hue 2.5Y 5/4; B5 = Hue 2.5Y 4/6; B6 = Hue 2.5Y 4/4; B7 = Hue 2.5Y 3/3; B8 = 2.5Y 3/2; and B9 = Hue 2.5Y 3/1. All bioassays were repeated in triplicate using different sources of A. bisporus tissue. Tissue degradation, coloring of filter paper, and discoloration of the control cube were recorded after 48 h.
WLA assay.
The ability to produce tolaasin was determined using the WLA assay described by Wong and Preece (61). Bacterial colonies were toothpick inoculated at a distance of 7 mm from streaks of the indicator bacteria, either Pseudomonas reactans NCPPB 1311 or Pseudomonas tolaasii NCPPB 2192T. A white precipitation line was observed after 24 to 48 h of incubation at 28°C.
API 20 NE strip analysis.
The Analytical Profile Index (API) 20 NE micro-method for the identification of nonfastidious gram-negative rods, using 8 conventional biochemical tests and 12 carbohydrate assimilation tests, was performed as described by the manufacturer (Bio Merieux). The numerical profiles obtained from pseudomonad strains were compared to the profiles stored in the 1999 Analytical Profile Index Software database (Bio Merieux).
Genomic DNA isolation and standard PCR conditions.
DNA was isolated from pure cultures of bacteria using the Wizard Genomic DNA Isolation Kit (Promega) and stored at 4°C until required. All PCR amplifications were carried out in a Perkin-Elmer 9700 thermocycler. Unless stated otherwise, a standard PCR reaction mixture (25 μl total) consisted of 1× buffer (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 2.5 mM MgCl2, 0.01% gelatin, and 0.1% Triton X-100), deoxyribonucleotide triphosphates (dATP, dCTP, dGTP, dTTP) at a final concentration of 200 μM, 0.625 U of Taq DNA polymerase (Roche Molecular Biochemicals), oligonucleotide primers at a final concentration of 2 mM, and 100 ng of template DNA. Thermocycling consisted of 30 cycles (1 min, 94°C; 1 min, 55°C; 1 min, 72°C). Prior to cycling, samples were heated at 94°C for 5 min and the extension step was increased to 5 min, 72°C as part of the terminal cycle. Primers and deoxyribonucleotide triphosphates were removed from PCR products using the High Pure PCR product purification kit (Roche Molecular Biochemicals).
REP-PCR.
The primers (REP1R-I and REP2-1) and protocols used for repetitive extragenic palindromic PCR (REP-PCR) were those described by de Bruijn (7).
PCR amplification and DNA nucleotide sequencing of the 16S rRNA gene.
Primers U16A and U16B (57) were used to amplify the nearly complete 16S rRNA gene (approximately 1,480 bp). Direct nucleotide sequencing of this gene was achieved using primers U16A, U16B, and F357, F945, R1087, and R518 (25) in combination with the Big Dye Terminator Kit and an ABI Prism 3TIXLCPE (PE Biosystems). All 16S rRNA gene sequences analyzed in this study were confirmed by determining contiguous overlapping sequences of PCR DNA.
Phylogenetic analysis of 16S rRNA gene nucleotide sequences.
The 16S rRNA gene nucleotide sequences obtained in this study were aligned with 22 further pseudomonad 16S rRNA sequences (Table 2) obtained from GenBank using the nucleotide alignment software Clustal W (54). Phylogenetic trees were constructed with neighbor joining (47) and evolutionary distances calculated according to the method of Jukes and Cantor (20) using the software package Treecon for Windows version 1.3b (56). Bootstrap analysis (9) was carried out using 500 replicates. Acinetobacter calcoaceticus ATCC 23055 was included for single-sequence (forced) outgroup rooting of the tree.
TABLE 2.
Twenty-two validly described species of the genus Pseudomonas (sensu stricto) (29) used in 16S rRNA gene phylogenetic analysis
| Bacterial isolate | Strain designationa | GenBank accession no. |
|---|---|---|
| P. aeruginosa | LMG 1242Tb | Z76651 |
| P. agarici | LMG 2112T | Z76652 |
| P. alcaligenes | LMG 1224T | Z76653 |
| P. amygdali | LMG 2123T | Z76654 |
| P. asplenii | LMG 2137T | Z76655 |
| P. aureofaciens | DSM 6698T | Z76656 |
| P. balearica | DSM 6083T | U26418 |
| P. chlororaphis | LMG 5004T | Z76657 |
| P. cichorii | LMG 2162T | Z76658 |
| P. citronellolis | DSM 50332T | Z76659 |
| P. coronafaciens | LMG 13190T | Z76660 |
| P. ficuserectae | LMG 5694T | Z76661 |
| P. flavescens | NCPPB 3063T | U01916 |
| P. fluorescens biotype A | DSM 50090T | Z76662 |
| P. marginalis pv. marginalis | LMG 2210T | Z76663 |
| P. mendocina | LMG 1223T | Z76664 |
| P. oleovorans | DSM 1045T | Z76665 |
| P. putida biotype A | DSM 291T | Z76667 |
| P.s. stutzeri | CCUG 11256T | U26262 |
| P. syringae pv. syringae | LMG 1247t1T | Z76669 |
| P. tolaasii | LMG 2342T | Z76670 |
| P. viridiflava | LMG 2352T | Z76671 |
| Acinetobacter calcoaceticus | ATCC 23055 | Z93434 |
ATCC, American Type Culture Collection, Rockville, Maryland; DSM, Deutsche Sammlung von Mikroorganismen, Gottingen, Germany; LMG, Laboratorium voor Microbiologie en Genetica, Rijksuniversiteit, Gent, Belgium; IAM, Institute of Applied Microbiology, Tokyo, Japan; NCPPB, National Collection of Plant Pathogenic Bacteria, Harpenden, United Kingdom.
T, type strain.
Nucleotide sequence accession number.
The 16S rRNA gene sequences determined in this study have been deposited with GenBank under the accession numbers listed in Table 1.
RESULTS
Selective isolation of pseudomonads capable of causing blotch of A. bisporus.
Pseudomonads were selectively isolated from blotched mushrooms and various substrates (compost, casing, and water samples) from a major mushroom farm (50 isolates) and three smaller mushroom farms (15 isolates from each) within New Zealand. Ninety-five isolates were assessed for the ability to discolor and/or damage A. bisporus tissue in a bioassay with comparison to reference strains (Fig. 1). Bioassays showed that 76 of the 95 pseudomonad isolates caused discolorations of A. bisporus tissue to varying degrees (B2, 3.1% [n = 3]; B3, 36.8% [n = 35]; B4, 10.5% [n = 10]; B5, 11.6% [n = 11]; B6, 11.6% [n = 11]; B8, 2.1% [n = 2]; B9, 4.2% [n = 4]). Pseudomonads capable of causing discoloration in bioassays were termed blotch-causing organisms (BCO). The remaining 19 of the 95 isolates exhibited discolorations consistent with negative controls (B1) and were therefore considered nonpathogenic isolates.
FIG. 1.
Cube pathogenicity bioassays (11) to determine which BCO isolates are capable of inducing discoloration of A. bisporus tissue. Pictured are cubes within the assigned color scale, B1 through B9 (B1, cube inoculated with KB alone). The following reference strains are included for comparison: A, P. reactans NCPPB 1311 (B2); B, P. gingeri NCPPB 3147T (B5); C, P. tolaasii NCPPB 2192T (B9).
Refined selection of 33 isolates.
Since the focus of this pilot study was to determine the diversity of pseudomonads capable of causing blotch, 33 isolates were selected based on the following criteria: (i) they caused variable discolorations of A. bisporus tissue in bioassay, and (ii) they exhibited variance in colony morphology and growth patterns. Analysis of REP-PCR results ensured that each of the 33 selected BCO isolates was unique.
LDP production.
LDP production was assessed using the WLA (Table 1). Eight of the thirty-three BCO isolates were observed to produce WLA+ reactions with P. tolaasii, consistent with the type strain of P. reactans. Two isolates (NZ 027 and NZ 032) produced a WLA+ result with P. reactans, as would be expected of P. tolaasii isolates.
Biochemical analysis of BCO isolates.
Tentative species identification of 33 BCO isolates was initially based on API 20 NE biochemical analysis. Comparison of API 20 NE biotypes to the corresponding API 20 NE database identified NZ 112 and NZ 101 as P. putida, NZ 097 as Burkholderia cepacia, and the remaining 30 BCO isolates as P. fluorescens (Table 1). However, further discrimination of the BCO isolates was necessary due to the lack of discriminatory capability of the API 20 NE test. For instance, the type strain of P. tolaasii (NCPPB 2192T) could not be distinguished from P. fluorescens.
Phylogenetic characterization of BCO isolates.
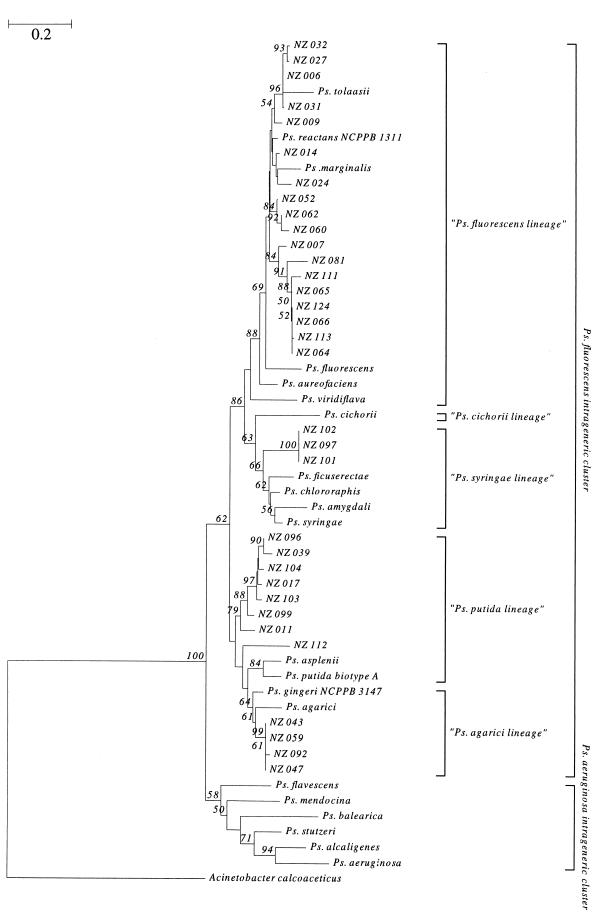
Figure 2 shows the inferred phylogenetic relationships derived from a neighbor-joining analysis of the pairwise comparisons among the 16S rRNA gene sequences of the selected 33 BCO isolates from this survey with 22 validly described species of the genus Pseudomonas (sensu stricto) (Table 2). All 33 BCO isolates from this study clustered within the P. fluorescens intrageneric cluster, not within the P. aeruginosa intrageneric cluster (as defined by Moore et al. [29]). Only four BCO isolates (NZ 043, NZ 059, NZ 092, and NZ 047) were observed to group closely with the previously identified typed strain of P. gingeri, NCPPB 3147, within the P. agarici lineage (Fig. 2). The remaining 29 isolates were observed to distribute throughout the P. fluorescens intrageneric cluster, with 18 (the majority) isolates falling in the P. fluorescens lineage, 8 within the P. putida lineage (5 forming a tight cluster), and 3 within the Pseudomonas syringae lineage. Two nonpathogenic (B1) isolates (NZ 081 and NZ 099) were observed to show a high level of relatedness to BCO clusters, and two B1 isolates (NZ 064 and NZ 066) grouped tightly within a single BCO cluster. Consistent with the WLA+ results, NZ 027 and NZ 032 grouped closely with P. tolaasii, strongly suggesting identity as P. tolaasii. Isolates that reacted with P. tolaasii (therefore classed as WLIP organisms, including P. reactans [30]) were located mainly in the P. fluorescens lineage; however, there was also intrageneric clustering within the P. syringae and Pseudomonas putida lineages as well. Isolates grouping with P. gingeri NCPPB 3147T (NZ 043, NZ 059, NZ 092, and NZ 047) exhibited discolorations consistent with ginger blotch (defined in this study as B4 to B6 based on a single color scale factor: either side of the B5 discoloration caused by P. gingeri NCPPB 3147T). However, B4 to B6 discolorations were also observed with BCO isolates in the P. fluorescens lineage (NZ 006, B6; NZ 031, B4; NZ 007, B4; and NZ 060, B4), the P. syringae lineage (NZ 101, B6), and the P. putida lineage (NZ 011, B6) (Fig. 2).
FIG. 2.
The inferred phylogenetic relationships between the BCO isolates from this study and 22 validly described members of the genus Pseudomonas (sensu stricto). Evolutionary distances were determined with pairwise dissimilarities of the 16S rRNA gene sequences, and the dendrogram was generated using the neighbor-joining algorithm. Two major intrageneric clusters and five evolutionary lineages are defined as described by Moore et al. (29). Bootstrap proportions of confidence are represented as percentages for those branchings with values greater than 50%.
DISCUSSION
Previous studies addressing the relatedness of pathogenic pseudomonads to cultivated A. bisporus are well documented. As a result of these studies, P. tolaasii has been well characterized and its homogeneity as a species has been established. However, P. gingeri has received limited attention since it was first described by Wong et al. (60).
Initiated by costly New Zealand outbreaks of ginger discolorations of A. bisporus, both on beds and postharvest, characterization of the causal organism(s) of ginger blotch in New Zealand mushroom farms was sought. Observation of pseudomonads with variable colony morphology and growth patterns isolated from mushrooms exhibiting ginger blotch symptoms raised the question of whether ginger blotch of A. bisporus is caused by a homogeneous species previously described as P. gingeri. Initially, 95 pseudomonads were isolated from mushroom farms throughout New Zealand that were associated with discolorations of A. bisporus. In order to determine pathogenic potential, the ability of each isolate to discolor and/or damage A. bisporus tissue was assessed in a bioassay with comparison to reference strains (Table 1). Bioassay results of these 95 isolates showed many to cause ginger blotch discolorations (B4 to B6). However, many isolates were also observed to cause different degrees of A. bisporus discoloration (B1 to B3 and B7 to B9). For this reason, the focus of this study was diverted from ginger blotch to blotch of A. bisporus in general to ascertain the prevalence of pseudomonad species capable of causing blotch diseases. The 33 pseudomonad isolates selected for phylogenetic analysis were chosen because they all exhibited variable colony morphology and growth patterns in culture and/or caused various discolorations of A. bisporus tissue in bioassay. No data were collected to determine the distribution of BCO isolates throughout New Zealand or to identify the predominant causal BCO strain.
Using chromametric measurements, variations of discoloration of A. bisporus tissue have been previously established for different pseudomonads. P. tolaasii was shown to evoke a specific color change (independent of the concentration of bacteria applied), while P. gingeri and P. reactans induced discolorations that were different from each other's and from P. tolaasii's (49). Bioassays in this study were carried out in triplicate using different sources of A. bisporus tissue in each replicate. This was to ensure accurate results of A. bisporus tissue discoloration, since amounts of tyrosinase (a major enzyme associated with the brown discoloration of A. bisporus [19]) have been shown to vary depending on spatial location and developmental stages of the mushroom (26). Known concentrations (107 CFU · ml−1) of pseudomonad isolates were inoculated in each bioassay to facilitate comparisons between results, since differences in bacterial loadings on caps are thought to influence the development of the discolorations (49). Discolorations observed in the bioassay could, therefore, be assumed to be the result of individual strain virulence factors alone.
Species similarities of the 33 BCO isolates were initially determined by biochemical analysis. While the carbon assimilation tests and production of enzyme intermediates included in the API 20 NE strip do not effectively discriminate between isolates of P. gingeri, P. tolaasii, and P. reactans or between biovars of P. fluorescens isolates, clear biotype differences between many of the 33 BCO isolates were observed (Table 1). For this reason, API 20 NE strips were useful in determining differences in biochemical phenotypes of the BCO isolates but did not facilitate species identification. Therefore, further discrimination was required.
To determine the phylogenetic relationship of the 33 selected BCO isolates, comparison of nearly complete nucleotide sequences of the 16S rRNA gene was used in this study, since it is considered an effective method for defining prokaryotic genotypic relatedness and resolving taxonomic identities (10, 16, 29). The topology of the dendrogram derived from analysis of the 16S rRNA sequences obtained in this study (Fig. 2) is in agreement with the intrageneric structure described by Moore et al. (29). The 33 BCO isolates were observed to distribute throughout the P. fluorescens intrageneric cluster, supporting studies of Soler-Rivas et al. (49) in which distantly related pseudomonads (P. tolaasii, P. gingeri, and P. reactans) caused different degrees of discoloration. However, our study further demonstrates the extent of the diversity of pseudomonads capable of inducing discoloration of A. bisporus. Our findings also show that the same degree of discoloration may be caused by dissimilar species of pseudomonads, suggesting that the factor(s) causative of inducing a particular discoloration of A. bisporus are not exclusive to a particular pseudomonad species.
Since pseudomonads are arguably the most diverse and ecologically significant group of bacteria (38), the observation of distantly related pseudomonad species having similar disease phenotypes is not unexpected. Numerous species within the genus Pseudomonas have been classified (36, 39, 53), and many new species continue to be identified by methods that have been revised in response to advances in DNA technology (including DNA-DNA hybridization [58] and gene sequence analysis of 16S rRNA [24, 29] and gyrB and rpoD [64]). Such methods have aided taxonomic resolution, but as this study has shown, they may also introduce discrepancies between phenotypic and genotypic analyses. For example, Moore et al. (29) demonstrated that seven phenotypically indistinguishable genomovars of Pseudomonas stutzeri contain up to six nucleotide differences within the 16S rRNA gene, and Yamamoto et al. (64) also observed that many phenotypic traits of pseudomonad species did not reflect their phylogenetic relationships (e.g., Pseudomonas corrugata was observed in the P. fluorescens complex and Pseudomonas amygdali in the P. syringae complex).
As has been discussed by Maynard Smith (28), there is much debate on the naming of bacterial species given the difficulty of defining a “bacterial species concept” due to recognition of the significant contribution of recombination to bacterial population genetics. Therefore, 16S rRNA gene analyses carried out in this study were intended to provide an indication of BCO species similarity only and cannot account for the acquisition of genes and accessory genetic elements (plasmids, transposons, integrens, and phages) by lateral gene transfer (33, 52), classical “spontaneous” mutation (27), and recombination (15, 22)—all of which are important sources of bacterial evolution and species diversity.
Most members of the pseudomonas genus produce active extracellular enzymes that have been associated with plant disease, including proteinases and lipases. P. tolaasii was found to produce a proteinase very similar to those secreted by other pseudomonad species (8), which, although the effect of this proteinase in mushroom infection is unknown, may facilitate the damage caused in the mushroom (2). Lipases have also been shown to facilitate bacterial infections by disrupting host membranes, and P. tolaasii, like many other pathogenic pseudomonads, produces an extracellular heat-stable monomeric metallo-lipase with a mass of 670 kDa (1). However, its involvement in mushroom infection is unresolved. Like P. tolaasii, it may be assumed that the BCO isolates in this study produce extracellular enzymes, including proteinases and lipases, which are likely to be involved in the discoloration of A. bisporus. The degrees of discoloration observed in this study may be a result of different combinations of extracellular enzymes produced by the BCO isolates facilitating different enzymatic activation of A. bisporus tissue.
As this pilot study was initiated to determine the source of ginger blotch disease, there was interest in BCO isolates capable of causing discolorations that are comparable to those caused by P. gingeri NCPPB 3147T (B4 to B6). Four BCO isolates were observed to group closely with P. gingeri reference strains based on the 16S rRNA phylogenetic analysis. These isolates were also observed to cause B4 to B6 discolorations, suggesting that isolates closely related to P. gingeri have the ability to induce A. bisporus discoloration consistent with ginger blotch. However, a further six BCO isolates causing B4 to B6 discolorations were also observed to distribute within the P. fluorescens lineage, the P. syringae lineage, and the P. putida lineage. This result suggests that the organism previously described as P. gingeri is not solely responsible for ginger blotch of A. bisporus and that the disease can be caused by a number of different pseudomonads.
As production of LDP is also a pathogenicity factor associated with A. bisporus discoloration, each BCO isolate was assayed for its ability to secrete an LDP capable of forming a white line precipitate with either P. reactans or P. tolaasii. Observation of eight BCO isolates producing WLA+ reactions with P. tolaasii suggested that they produced LDP similar to the WLIP described by Mortishire-Smith et al. (30). Analysis of these eight BCO isolates within Fig. 2 revealed no commonality of LDP production or discoloration among closely related isolates. For example, although NZ 052 and NZ 062 grouped closely together, only NZ 052 produced a WLA+ reaction with P. tolaasii. Isolates NZ 096, NZ 103, and NZ 104 also clustered tightly, yet they exhibited different bioassay discolorations, and of the three, only NZ 096 was WLA+ with P. tolaasii. Although these two groupings exhibit high phylogenetic similarity, they display quite different pathogenicities (including the presence of the potential virulence factor WLIP), which may suggest that these isolates have acquired traits responsible for differing degrees of A. bisporus discolorations. This is further demonstrated by observation of nonpathogenic (B1) isolates showing close phylogenetic relatedness to BCO isolates able to cause disease. Studies have revealed that horizontal transfer and recombination of virulence genes play a major role in generating genetic diversity among bacterial species (21), and horizontal gene acquisition could also be an explanation of why highly related (based on 16S rRNA) BCO isolates in this study have different virulence potentials.
P. tolaasii has routinely been distinguished from other pseudomonads by its ability to cause dark brown discolorations on mushrooms and by a positive WLA with P. reactans (18, 36, 46, 61, 65). However, an earlier study (12) identified a mushroom farm isolate, Pseudomonas NZI7, as being WLA+ (with P. reactans) and causing discoloration of A. bisporus tissue comparable to that caused by P. tolaasii. Although based on these criteria NZI7 would normally be identified as P. tolaasii, NZI7 was shown to be genetically more similar to P. syringae than to P. tolaasii (12). For this reason, NZ 027 and NZ 032 were included in this study, and although they were not assumed to be P. tolaasii isolates based solely on WLA+ results with P. reactans and B9 discolorations, they did group with P. tolaasii. However, with the exception of NZ 006, other B8 to B9 BCO isolates showed little phylogenetic relatedness to P. tolaasii and were observed to distribute widely throughout the P. fluorescens intrageneric cluster. As B8-B9 BCO isolates were WLA− with P. reactans (except for NZ 027 and NZ 032), it was assumed that browning was due not to tolaasin production but most likely to an extracellularly produced factor(s) that (i) causes tyrosinase activation and production of brown melanins similarly to tolaasin (4, 19, 51) and/or (ii) reduces enzymatic activity in A. bisporus tissue, which has been suggested to be due to protease activity (49). Proteases have been known to degrade tyrosinases (5), and a protease from P. tolaasii has been isolated and is speculated to facilitate damage to the mushroom (2).
Also included in this study were three pseudomonad isolates from a New Zealand milk factory environment (NZ 111, NZ 112, and NZ 113) that caused A. bisporus discoloration as efficiently as the BCO isolates from mushroom farms. This observation (i) demonstrates that the ability to cause discoloration of A. bisporus is not a trait acquired for evolutionary survival within pseudomonads present in the mushroom environment and (ii) further supports the previous discussions asserting that the factor(s) causing blotch discolorations are likely to be a combination of extracellular enzymes common to many different pseudomonads. This finding raises the question of whether other bacterial species may also induce discolorations of A. bisporus and points to the need for adequate biosecurity of mushroom farms.
The purpose of this study was to address the relatedness of pseudomonads capable of inducing blotch diseases of A. bisporus. The results have confirmed previous reports that blotch disease may be caused by different species of pseudomonads. Furthermore, these results have identified three major findings: (i) the diversity of pseudomonads capable of causing blotch discolorations of A. bisporus is considerably more extensive than previously thought; (ii) the organism previously described as P. gingeri is not solely responsible for ginger discolorations of A. bisporus (ginger blotch); and (iii) a particular blotch discoloration may be caused by more than a single pseudomonad species. These findings affect the future classification of P. gingeri, because a major phenotypic characteristic of this species is its ability to induce a ginger discoloration of A. bisporus. These results also have implications for the control of general blotch diseases of A. bisporus since a single causal organism cannot now be targeted. Since certain pseudomonads are considered beneficial in the commercial cultivation of A. bisporus (41), the elimination of all pseudomonads from a farm environment is neither desirable nor practical. Therefore, blotch disease of A. bisporus may prove difficult to manage, and continued research of BCO organisms may better resolve the commonality of virulence factors and the environmental conditions that promote disease.
ACKNOWLEDGMENTS
This work was supported by the PGSF, and samples were provided by the Commercial Mushroom Growers Federation (New Zealand) Ltd.
We also thank Paul Rainey for discussion on the paper and Simon Bulman and David Bean for critical comments.
REFERENCES
- 1.Baral A, Fox P F. Isolation and characterization of an extracellular lipase from Pseudomonas tolaasii. Food Chem. 1997;58:33–38. [Google Scholar]
- 2.Baral A, Fox P F, O'Connor T P. Isolation and characterization of an extracellular proteinase from Pseudomonas tolaasii. Phytochemistry. 1995;39:757–762. [Google Scholar]
- 3.Brodey C L, Rainey P B, Tester M, Johnstone K. Bacterial blotch disease of the cultivated mushroom is caused by an ion-channel forming lipodepsipeptide toxin. Mol Plant-Microbe Interact. 1991;4:407–411. [Google Scholar]
- 4.Burton K S. The effects of pre- and post-harvest development on mushroom tyrosinase. J Hortic Sci. 1988;63:255–260. [Google Scholar]
- 5.Burton K S, Love M E, Smith J F. Biochemical changes associated with mushroom quality in Agaricus spp. Enzyme Microb Technol. 1993;15:736–741. [Google Scholar]
- 6.Cutri S S, MaCauley B J, Roberts W P. Characteristics of pathogenic non-fluorescent (smooth) and non-pathogenic fluorescent (rough) forms of Pseudomonas tolaasii and Pseudomonas gingeri. J Appl Bacteriol. 1984;57:291–298. [Google Scholar]
- 7.De Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairbairn D J, Law B A. Proteinases of psychrotropic bacteria: their production, properties, effects and control. J Dairy Res. 1986;53:139. doi: 10.1017/s0022029900024742. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 10.Fox G E, Stackebrandt E, Hespell R B, Gibson J, Maniloff J, Dyer T A, Wolfe R S, Balch W E, Tanner R S, Magrum L J, Zablen L B, Blakemore R, Gupta R, Bonen L, Lewis B J, Stahl D A, Luehrsen K R, Chen K N, Woese C R. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 11.Gandy D G. A technique for screening bacteria causing brown blotch of cultivated mushrooms. Rep Glasshouse Crops Res Inst. 1968;1968:150–154. [Google Scholar]
- 12.Godfrey S A C, Marshall J W, Klena J D. Genetic characterisation of Pseudomonas “NZ17”—a novel pathogen that results in a brown blotch disease of Agaricus bisporus. J Appl Microbiol. 2001;91:412–420. doi: 10.1046/j.1365-2672.2001.01398.x. [DOI] [PubMed] [Google Scholar]
- 13.Goor M, Vantomme R, Swings J, Gillis M, Kersters K, De Ley J. Phenotypic and genotypic diversity of Pseudomonas tolaasii and white line reacting organisms isolated from cultivated mushrooms. J Gen Microbiol. 1986;132:2249–2264. [Google Scholar]
- 14.Gould W D, Hagedorn C, Bardinelli T R, Zablotowicz R M. New selective media for enumeration and recovery of fluorescent pseudomonads from various habitats. Appl Environ Microbiol. 1985;49:28–32. doi: 10.1128/aem.49.1.28-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haubold B, Rainey P B. Genetic and ecotypic structure of a fluorescent Pseudomonas population. Mol Ecol. 1996;5:747–761. [Google Scholar]
- 16.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 17.Hutchison M L, Johnstone K. Evidence for the involvement of the surface active properties of the extracellular toxin tolaasin in the manifestation of brown blotch disease symptoms by Pseudomonas tolaasii on Agaricus bisporus. Physiol Mol Plant Pathol. 1993;42:373–384. [Google Scholar]
- 18.Janse J D, Derks J H J, Spit B E, van der Tuin W R. Classification of fluorescent soft rot Pseudomonas bacteria, including P. marginalis strains, using whole cell fatty acid analysis. Syst Appl Microbiol. 1992;15:538–553. [Google Scholar]
- 19.Jolivet S, Voiland A, Pellon G, Arpin N. Main factors involved in the browning of Agaricus bisporus. Mushroom Sci. 1995;14:695–702. [Google Scholar]
- 20.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 21.Kehoe M A, Kapur V, Whatmore A M, Musser J M. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 1996;4:436–443. doi: 10.1016/0966-842x(96)10058-5. [DOI] [PubMed] [Google Scholar]
- 22.Kiewitz C, Tummler B. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J Bacteriol. 2000;182:3125–3135. doi: 10.1128/jb.182.11.3125-3135.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescens. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 24.Laguerre G, Rigottier-Gois L, Lemanceau P. Fluorescent Pseudomonas species categorized by using polymerase chain reaction (PCR)/restriction fragment analysis of 16S rDNA. Mol Ecol. 1994;3:479–487. doi: 10.1111/j.1365-294x.1994.tb00126.x. [DOI] [PubMed] [Google Scholar]
- 25.Lane D J. 16S/23S sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley; 1991. pp. 115–175. [Google Scholar]
- 26.Leeuwen J V, Wichers H J. Tyrosinase activity and isoform composition in separate tissues during development of Agaricus bisporus fruit bodies. Mycol Res. 1999;103:413–418. [Google Scholar]
- 27.Levin B R, Bergstrom C T. Bacteria are different: observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc Natl Acad Sci USA. 2000;97:6981–6985. doi: 10.1073/pnas.97.13.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maynard Smith J. Do bacteria have population genetics? In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. 52. Symposium 52. Cambridge, United Kingdom: The Society for General Microbiology; 1995. pp. 1–12. [Google Scholar]
- 29.Moore E R B, Mau M, Arnscheidt A, Bottger E C, Hutson R A, Collins M D, Van de Peer Y, De Wachter R, Timmis K N. The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst Appl Microbiol. 1996;19:478–492. [Google Scholar]
- 30.Mortishire-Smith R J, Nutkins J C, Packman L C, Brodey C L, Rainey P B, Johnstone K, Williams D H. Determination of the structure of an extracellular peptide produced by the mushroom saprotroph Pseudomonas reactans. Tetrahedron. 1991;47:3645–3654. [Google Scholar]
- 31.Nair N G, Bradley J K. Mushroom blotch bacterium during cultivation. Mushroom J. 1980;90:201–203. [Google Scholar]
- 32.Nutkins J C, Mortishire-Smith R J, Packman L C, Brodey C L, Rainey P B, Johnstone K, Williams D H. Structure determination of tolaasin, an extracellular lipodepsipeptide produced by the mushroom pathogen Pseudomonas tolaasii Paine. J Am Chem Soc. 1991;113:2621–2627. [Google Scholar]
- 33.Ochman H, Lawrence J G, Groisman E A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 34.Olivier J M, Mamoun M, Munsh P. Standardization of a method to assess mushroom blotch resistance in cultivated and wild Agaricus bisporus. Can J Plant Pathol. 1997;19:36–42. [Google Scholar]
- 35.Paine S G. Studies in bacteriosis. II. A brown blotch disease of cultivated mushrooms. Ann Appl Biol. 1919;5:206–219. [Google Scholar]
- 36.Palleroni N J. Genus I. Pseudomonas Migula 1894. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams and Wilkins; 1984. [Google Scholar]
- 37.Palleroni N J. Human and animal-pathogenic pseudomonads. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes, a handbook on the biology of bacteria, ecophysiology, isolation, identification and applications. 2nd ed. Vol. 3. New York, N.Y: Springer; 1992. [Google Scholar]
- 38.Palleroni N J. Pseudomonas classification: a new case history in the taxonomy of gram-negative bacteria. Antonie Leeuwenhoek. 1993;64:231–251. doi: 10.1007/BF00873084. [DOI] [PubMed] [Google Scholar]
- 39.Palleroni N J. Taxonomy of the pseudomonads. In: Sokatch J R, editor. The bacteria, vol. 10: The biology of pseudomonas. London, United Kingdom: Academic Press, Inc.; 1986. pp. 3–20. [Google Scholar]
- 40.Preece T F, Wong W C. Quantitative and scanning electron microscope observations on the attachment of Pseudomonas tolaasii and other bacteria to the surface of Agaricus bisporus. Physiol Plant Pathol. 1982;21:251–257. [Google Scholar]
- 41.Rainey P B. Ph.D. thesis. Christchurch, New Zealand: University of Canterbury; 1989. [Google Scholar]
- 42.Rainey P B. Phenotypic variation of Pseudomonas putida and P. tolaasii affects attachment to Agaricus bisporus mycelium. J Gen Microbiol. 1991;137:2769–2779. doi: 10.1099/00221287-137-12-2769. [DOI] [PubMed] [Google Scholar]
- 43.Rainey P B, Brodey C L, Johnstone K. Biological properties and spectrum of activity of tolaasin, a lipodepsipeptide toxin produced by the mushroom pathogen Pseudomonas tolaasii. Physiol Mol Plant Pathol. 1991;39:57–70. [Google Scholar]
- 44.Rainey P B, Brodey C L, Johnstone K. Biology of Pseudomonas tolaasii, cause of brown blotch disease of the cultivated mushroom. Adv Plant Pathol. 1992;8:95–117. [Google Scholar]
- 45.Reid H D. M.S. thesis. Christchurch, New Zealand: University of Canterbury; 1997. [Google Scholar]
- 46.Rhodes M E. The characterization of Pseudomonas fluorescens. J Gen Microbiol. 1959;21:221–263. doi: 10.1099/00221287-25-3-331. [DOI] [PubMed] [Google Scholar]
- 47.Saitou N, Nei M. The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Samson R, Houdeau G, Khanna P, Guillaumes J, Olivier J M. Proceedings of the International Symposium on Scientific and Technical Aspects of Cultivating Edible Fungi, 1986. Amsterdam, The Netherlands: Pennsylvania State University, University Park, Pa. Elsevier Science Publishers B.V.; 1987. Variability of fluorescent pseudomonas populations in composts and casing soils used from mushroom cultures; pp. 19–25. [Google Scholar]
- 49.Soler-Rivas C, Arpin N, Olivier J M, Wichers H J. Discoloration and tyrosinase activity in Agaricus bisporus fruit bodies infected with various pathogens. Mycol Res. 2000;104:351–356. [Google Scholar]
- 50.Soler-Rivas C, Jolivet S, Arpin N, Olivier J M, Wichers H J. Biochemical and physiological aspects of brown blotch disease of Agaricus bisporus. FEMS Microbiol Rev. 1999;23:591–614. doi: 10.1111/j.1574-6976.1999.tb00415.x. [DOI] [PubMed] [Google Scholar]
- 51.Soulier L, Foret V, Arpin N. Occurrence of agaritine and gamma-glutaminyl-4-hydroxybenzene (GHB) in the fructifying mycelium of Agaricus bisporus. Mycol Res. 1993;97:529–532. [Google Scholar]
- 52.Spiers A J, Buckling A, Rainey P B. The causes of Pseudomonas diversity. Microbiology. 2000;146:2345–2350. doi: 10.1099/00221287-146-10-2345. [DOI] [PubMed] [Google Scholar]
- 53.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 54.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolaas A G. A bacterial disease of cultivated mushroom. Phytopathology. 1915;5:51–54. [Google Scholar]
- 56.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 57.Wang G C-Y, Wang Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology. 1996;142:1107–1114. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]
- 58.Wayne L G, Brenner D J, Clowell R R, et al. International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 59.Wells J M, Sapers G M, Fett W F, Butterfield J E, Jones J B, Bouzar H, Miller F C. Postharvest discoloration of the cultivated mushroom Agaricus bisporus caused by Pseudomonas tolaasii, P. ‘reactans,’ and P. ‘gingeri.’. Phytopathology. 1996;86:1098–1104. [Google Scholar]
- 60.Wong W C, Fletcher J T, Unsworth B A, Preece T F. A note on ginger blotch, a new bacterial disease of the cultivated mushroom, Agaricus bisporus. J Appl Bacteriol. 1982;52:43–48. [Google Scholar]
- 61.Wong W C, Preece T F. Identification of Pseudomonas tolaasii: the white line test in agar and mushroom block rapid pitting tests. J Appl Bacteriol. 1979;47:401–407. [Google Scholar]
- 62.Wong W C, Preece T F. Pseudomonas tolaasii in cultivated mushroom (Agaricus bisporus) crops: numbers of bacterium and symptom development on mushrooms grown in various environments after artificial inoculation. J Appl Bacteriol. 1982;53:87–96. [Google Scholar]
- 63.Wong W C, Preece T F. Pseudomonas tolaasii in mushroom crops: a note on primary and secondary sources of the bacterium on a commercial farm in England. J Appl Bacteriol. 1980;49:305–314. [Google Scholar]
- 64.Yamamoto S, Kasai H, Arnold D L, Jackson R W, Vivian A, Harayama S. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology. 2000;146:2385–2394. doi: 10.1099/00221287-146-10-2385. [DOI] [PubMed] [Google Scholar]
- 65.Zarkower P A, Wuest P J, Royse D J, Myers B. Phenotypic traits of fluorescent pseudomonads causing bacterial blotch of Agaricus bisporus mushrooms and other mushroom-derived fluorescent pseudomonads. Can J Microbiol. 1983;30:360–367. [Google Scholar]