Abstract
Cells of Gordonia nitida LE31 grown on 3-methylpyridine degraded 3-ethylpyridine without a lag time and vice versa. Cyclic intermediates were not detected, but formic acid was identified as a metabolite. Degradation of levulinic acid was induced in cells grown on 3-methylpyridine and 3-ethylpyridine. Levulinic aldehyde dehydrogenase and formamidase activities were higher in cells grown on 3-methylpyridine and 3-ethylpyridine than in cells grown on acetate. These data indicate that 3-methylpyridine and 3-ethylpyridine were degraded via a new pathway involving C-2–C-3 ring cleavage.
Alkylpyridines are toxic environmental pollutants commonly found in many surface waters and groundwaters near industries for the production of synthetic liquid fuels (10, 13, 14). Despite their occurrence and toxicity, only a few studies have focused on their biodegradation (3, 12). Metabolism of alkylpyridines may follow one of the three initial reactions: reduction of the aromatic ring (9, 11, 15), oxidation of the aromatic ring (1, 2), and oxidation of the alkyl group (4, 6).
Until now, characterization of the pathways of degradation of 3-substituted alkylpyridines was carried out only with Pseudomonas sp. strain KM3, which degraded 3-methylpyridine (3-MP) by oxidation of the methyl group (4). There has been no report on the pathways for degradation of 3-ethylpyridine (3-EP). In a previous study, a 3-MP- and 3-EP-degrading bacterium was isolated and assigned to a new species, Gordonia nitida (strain LE31), because of its taxonomic distinctiveness (17). This paper describes the identification of a metabolite and of enzyme activities during the degradation of 3-MP and 3-EP by G. nitida LE31, which indicate a new degradation pathway.
Strain LE31 was cultured in minimal salts medium, which contained 0.9 g of K2HPO4, 0.54 g of KH2PO4, 0.25 g of MgSO4 · 7H2O, 0.25 g of KCl, 0.01 g of CaCl2 · 2H2O, 1 ml of trace element solution (16), and 1 ml of selenite-tungstate solution (16) per liter of distilled water. Cultures were carried out aerobically at 30°C on a rotary shaker. Degradation of heterocyclic aromatic compounds was monitored by measuring the UV spectra (Beckman DU60 spectrophotometer) and by high-pressure liquid chromatography (HPLC) as described by Rhee et al. (9). Cell growth was determined by measuring absorbance of the culture broth at 600 nm. When various pyridine derivatives and aromatic compounds were tested as substrates for LE31, only 3-MP and 3-EP (1 mM each) supported growth within 48 h. The following compounds were not used as sole carbon or nitrogen sources: pyridine, pyridine-N-oxide, pyrazine, quinoline, 2-MP, 4-MP, 2-EP, 4-EP, 2,3- dimethylpyridine (2,3-DMP), 2,4-DMP, 2,5-DMP, 2,6-DMP, 3,4-DMP, 3,5-DMP, 2-hydroxypyridine, 3-hydroxypyridine, 4-hydroxypyridine, 2-chloropyridine, 3-chloropyridine, 4-chloropyridine, 3-fluoropyridine, 2-carboxypyridine, 3-carboxypyridine, 4-carboxypyridine, phenol, catechol, aniline, benzene, toluene, xylene, benzoic acid, m-cresol, and chlorobenzene (1 mM each). Among the aliphatic compounds tested, acetate, propionate, butyrate, valerate, isobutyrate, 2-methylbutyrate, isovalerate, lactate, and ethanol (100 ppm each) supported growth. The following compounds were not used as a sole carbon source: piperidine, 3-methylpiperidine, formate, pyruvate, malate, succinate, citrate, glyoxylate, oxalate, glycolate, formamide, methylamine, ethanolamine, glycine, glutamate, glutarate, and methanol (100 ppm each). As is the case for many bacteria that degrade N-heterocyclic aromatic compounds, utilization of pyridine derivatives by LE31 was very restricted (8, 11).
When cultured in minimal salts medium with 3-MP or 3-EP as the sole source of carbon, nitrogen, and energy, strain LE31 degraded 3-EP and 3-MP (1 mM each) without producing any aromatic or aliphatic intermediates detectable by scanning of UV spectra (200 to 400 nm) or by HPLC analyses. In the course of 3-EP and 3-MP degradation, ammonium ion was released into the culture liquid. Concentrations of ammonium ion were determined with a test kit (Sigma, St. Louis, Mo.). Since there was no nitrogen source in the culture medium except the alkylpyridines, the presence of ammonium ion in the culture broth indicates that the aromatic ring was cleaved and the nitrogen was released. About 64% of the nitrogen in the pyridine ring was detected as ammonium ion in the culture supernatant after complete degradation of 3-MP and 3-EP. Some of the nitrogen released may have been incorporated into cell mass. Addition of ammonium, nitrite, or nitrate ions (1 mM each) to the medium as an external nitrogen sources did not affect or slightly reduced the rate of degradation of 3-MP or 3-EP (data not shown). Strain LE31 degraded 1 mM 3-MP and 3-EP in 43 and 32 h, respectively. Cultures with greater than 3 mM 3-MP or 4 mM 3-EP were significantly inhibited.
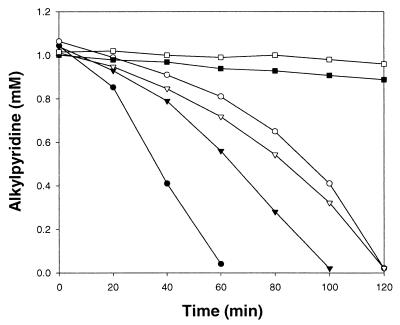
In order to study the induction of the degradation enzymes, cells grown on 3-MP, 3-EP, and sucrose were harvested at the log phase, washed three times with 50 mM potassium phosphate buffer (pH 7.0), suspended in the same buffer, and tested for their ability to degrade 3-MP and 3-EP. Cells grown on 3-MP degraded 3-EP without a lag time and vice versa, indicating that some of the enzymes in the pathways of 3-MP and 3-EP degradation are shared. On the other hand, the culture grown on sucrose did not degrade 3-MP and 3-EP within 2 h (Fig. 1). Shukla (11) reported similar results with 2-MP- or 2-EP-grown cells of an Arthrobacter sp. that degraded both compounds without lag time.
FIG. 1.
Degradation of 3-EP (●, ▾, and ▪) and 3-MP (○, ▿, and □) by washed cells of G. nitida LE31 grown on 3-EP (● and ○), 3-MP (▾ and ▿), and sucrose (▪ and □). Initial cell concentrations were 0.95 g (dry weight) per liter.
Throughout the course of washed-cell experiments, culture supernatants sampled during the degradation of 3-MP and 3-EP were examined to detect metabolic intermediates. Detection of heterocyclic intermediates by the UV scanning and HPLC analysis was unsuccessful. Furthermore, when metabolic inhibitors (5 mM fluoroacetate, 10 mM semicarbazide, 0.3 mM chloramphenicol, and 0.5 mM 2,4-dinitrophenol) were added to the washed-cell cultures, no appreciable amounts of metabolites were detected. Crude cell extracts of LE31 grown on 3-MP and 3-EP were also not able to transform 3-EP and 3-MP (1 mM each). Along with the crude cell extracts, a range of coenzymes were included alone or in combination in some incubations; these were ATP (1 mM), NAD(P)+ (1 mM), NAD(P)H (1 mM), and flavin adenine dinucleotide (0.01 mM). Metal ions tested for support of enzyme activities were Fe2+, Fe3+, K+, Na+, Mn2+, Cu2+, and Zn2+ (0.01 mM each). In all cases, 3-MP and 3-EP were not degraded. Similar results were also reported with 2-MP and pyridine degraders, where researchers were not able to detect any cyclic intermediates and could not detect catalytic activity for the transformation of the pyridine ring in the cell extracts (9, 11, 15).
Although cyclic intermediates were not detected, appreciable amounts of an organic acid were produced during the degradation of 3-MP and 3-EP (5 mM each) during high-density washed-cell culture. The acid had a retention time similar to that of formic acid when analyzed by HPLC using an Aminex HPX 87H column in an isocratic condition (0.6 ml/min) with 5 mM sulfuric acid in water as a mobile phase and using a UV detector (210 nm). The metabolite was recovered by HPLC fractionation and subjected to mass spectrometry. Mass spectra were obtained with an Autospec-UltimaE mass spectrometer (Micromass, Manchester, United Kingdom) by the direct-introduction probe method. Considering that the compound had a molecular ion and a mass spectrum similar to those of formic acid, we identified it as formic acid (Table 1). Quantification of formic acid in the culture liquids by the HPLC and a biochemical method (7) showed similar results. Up to 3.3 and 1.9 mM formic acid was transiently accumulated during the degradation of 5.4 mM 3-MP and 4.7 mM 3-EP, respectively. Accumulation of formic acid may be due to the relative weakness of the activities of the enzyme for degradation of formic acid (probably formate dehydrogenase) compared to other enzymes involved in the degradation pathway.
TABLE 1.
Mass spectra of the metabolite produced during degradation of 3-EP by G. nitida LE31 and of formic acid
| Compound | m/z (% relative intensity) |
|---|---|
| Metabolite | 46 (1.6) [M+], 45 (6.3), 44 (15.1), 43 (18.1), 41 (9.1), 40 (3.8), 32 (33.8), 31 (8.6), 29 (6.7), 28 (100), 27 (3.1) |
| Formic acid | 46 (3.1) [M+], 45 (11.3), 44 (28.7), 43 (1.8), 41 (5.1), 40 (3.5), 32 (31.6), 31 (15.3), 29 (6.6), 28 (100), 27 (3.7) |
According to Korosteleva et al. (4), Pseudomonas sp. strain KM-3, which metabolized 3-MP via oxidation of the methyl side chain, readily degraded 3-carboxypyridine. However, LE31 was not able to use 3-carboxypyridine in the growth substrate test and washed-cell experiments (data not shown). Also, since no aromatic intermediate was detected and none of the oxygenated pyridine derivatives were attacked by LE31, it is not likely that degradation of 3-EP and 3-MP was initiated by oxidation of the pyridine ring. Degradation of 3-MP and 3-EP may also not proceed by complete reduction of the aromatic ring because 3-methylpiperidine, the fully reduced derivative of 3-MP, was also not degraded. Instead, ring cleavage preceded or accompanied by reduction of the pyridine ring is a more likely mechanism. During the degradation of pyridine by Bacillus strain 4, formic acid was also identified as a metabolite (15). In this case, although cyclic intermediates were not detected, pyridine was thought to be initially reduced to 1,4-dihydropyridine before ring cleavage. Shukla (11) also proposed a similar reaction for the degradation of 2-MP by the Arthrobacter sp. that they studied. Considering these findings, LE31 may degrade 3-MP and 3-EP via a pathway similar to that of Bacillus strain 4.
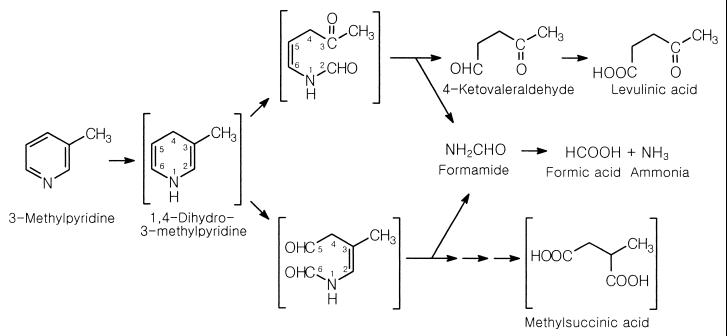
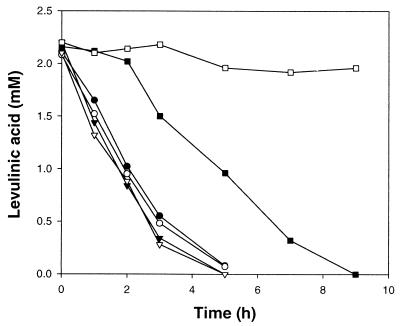
Theoretically, the detection of formic acid during the degradation of 3-MP and 3-EP implies C-2–C3 or C-5–C6 ring cleavage (Fig. 2), which is similar to the pathway suggested for pyridine degradation (15). Therefore, studies on the activities for the degradation of tentative ring cleavage products could be helpful for the identification of the precise pathway. Putative intermediates of 3-MP degradation, i.e., methylsuccinic acid (C-5–C6 cleavage), levulinic acid (C-2–C3 cleavage), and 2-methylglutaric acid (putative product of N–C-1 or N–C-6 cleavage), were tested for utilization by high-density washed cells grown on 3-MP and 3-EP. Among them, only levulinic acid was degraded by the cells grown on 3-MP and 3-EP. The potential involvement of levulinic acid in the pathway was confirmed by a washed-cell experiment. Cells grown on 3-MP or 3-EP readily degraded levulinic acid regardless of the presence of chloramphenicol (Fig. 3). However, in the absence of chloramphenicol, cells grown on sucrose degraded levulinic acid after a 2-h lag time, and when chloramphenicol was present, levulinic acid was not degraded within 9 h, indicating that levulinic acid-degrading enzymes were induced in cells grown on 3-MP and 3-EP (Fig. 3).
FIG. 2.
Proposed pathways for degradation of 3-MP by G. nitida LE31. Compounds in brackets are hypothetical.
FIG. 3.
Degradation of levulinic acid by washed cells of G. nitida LE31 grown on 3-EP (● and ○), 3-MP (▾ and ▿), and sucrose (▪ and □). Initial cell concentrations were 0.76 g (dry weight) per liter. Closed symbols, no chloramphenicol; open symbols, 200 ppm chloramphenicol.
In order to confirm the possibility of C-2–C3 cleavage, activities of enzymes involved in the putative degradation pathway were compared in crude cell extracts prepared from cells grown on 3-MP, 3-EP, and acetate. The enzyme activities tested were levulinic aldehyde dehydrogenase (C-2–C3 cleavage), methylsuccinic aldehyde dehydrogenase (C-5–C6 cleavage), 2-methylglutaric aldehyde dehydrogenase (N–C-1 or N–C-6 cleavage), and formamidase (C-2–C-3 or C-5–C-6 cleavage). Enzyme activities were measured in crude extracts of cells harvested in the log phase of growth. Preparation of crude cell extracts and assay of isocitrate dehydrogenase (a tricarboxylic acid cycle enzyme used as a control) and formamidase were carried out as described by Rhee et al. (9). Protein contents were determined by the method of Lowry et al. (5). Activities of the aldehyde dehydrogenases were measured by the oxidation of NAD(P)H at 340 nm with the reduction of the corresponding carboxylic acids. When NADPH was used, activities of the aldehyde dehydrogenases were less than 1 mU in all cases (data not shown). However, when NADH was used, levulinic aldehyde dehydrogenase activity was apparently higher among the aldehyde dehydrogenases, and it was especially higher in 3-MP- and 3-EP-grown cells than that in acetate-grown cells (Table 2). Accordingly, levulinic aldehyde dehydrogenase may be involved in the pathway of 3-MP and 3-EP degradation. The induction of levulinic acid degradation (Fig. 3) and expression of levulinic aldehyde dehydrogenase activity (Table 2) indicate that 3-MP and 3-EP may be degraded via C-2–C-3 ring cleavage which produced levulinic acid and formic acid as intermediates of 3-MP degradation (Fig. 2). Along with the detection of formic acid as an intermediate, induction of formamidase activity (Table 2) also supports the pathways proposed in the Fig. 2. Interestingly, many of the results in this study, i.e., (i) production of formic acid from both 3-MP and 3-EP, (ii) induction of levulinic acid degradation in washed-cell experiments, and (iii) activities of the enzymes tested, support the idea that 3-MP and 3-EP were degraded by the same enzyme system.
TABLE 2.
Enzyme activities in extracts of G. nitida LE31 grown on 3-EP, 3-MP, and acetate as sole sources of carbon
| Enzyme | Product measured | Sp act (mUa/mg of protein) for the following growth substrate:
|
||
|---|---|---|---|---|
| 3-EP | 3-MP | Acetate | ||
| Isocitrate dehydrogenase | NADH | 7 | 3 | 49 |
| Levulinic aldehyde dehydrogenase | NAD | 28 | 24 | 11 |
| Methylsuccinic aldehyde dehydrogenase | NAD | <1 | <1 | 1 |
| 2-Methylglutaric aldehyde dehydrogenase | NAD | 2 | <1 | <1 |
| Formamidase | NH3 | 24 | 19 | <1 |
One unit is defined as the amount of enzyme that produces 1 μmol of product per min.
The data indicate that the degradation of 3-MP and 3-EP by G. nitida LE31 seems to proceed via a new pathway involving ring cleavage between carbons 2 and 3.
REFERENCES
- 1.Feng Y, Kaiser J-P, Minard R D, Bollag J-M. Microbial transformation of ethylpyridines. Biodegradation. 1994;5:121–128. [Google Scholar]
- 2.Kaiser J-P, Minard R D, Bollag J-M. Transformation of 3- and 4-picoline under sulfate-reducing conditions. Appl Environ Microbiol. 1993;59:701–705. doi: 10.1128/aem.59.3.701-705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser J-P, Feng Y, Bollag J-M. Microbial metabolism of pyridine quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol Rev. 1996;60:483–498. doi: 10.1128/mr.60.3.483-498.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korosteleva L A, Kost A N, Vorob'eva L I, Modyanova L V, Terent'ev P B, Kulikov N S. Microbiological degradation of pyridine and 3-methylpyridine. Appl Biochem Microbiol. 1981;17:276–283. [Google Scholar]
- 5.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 6.Modyanova L V, Vorob'eva L I, Shibilkina O K, Dovgilevich E V, Terent'ev P B, Kost A N. Microbiological transformations of nitrogen-containing heterocyclic compounds. I. Hydroxylation of isomeric methyl- and dimethylpyridines by certain microscopic fungi. Sov Biotechnol. 1990;3:24–27. [Google Scholar]
- 7.Ogata M, Iwamoto T, Kawai T. Enzymatic assay of urinary formic acid as an index of methanol exposure. Ind Health. 1989;27:125–129. doi: 10.2486/indhealth.27.125. [DOI] [PubMed] [Google Scholar]
- 8.O'Loughlin E J, Sims G K, Traina S J. Biodegradation of 2-methyl, 2-ethyl, and 2-hydroxypyridine by an Arthrobacter sp. isolated from subsurface sediment. Biodegradation. 1999;10:93–104. doi: 10.1023/a:1008309026751. [DOI] [PubMed] [Google Scholar]
- 9.Rhee S-K, Lee G M, Yoon J-H, Park Y-H, Bae H-S, Lee S-T. Anaerobic and aerobic degradation of pyridine by a newly isolated denitrifying bacterium. Appl Environ Microbiol. 1997;63:2578–2585. doi: 10.1128/aem.63.7.2578-2585.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riley R G, Garland T R, Shiosaki K, Mann D C, Wildung R E. Alkylpyridines in surface waters, ground waters, and subsoils of a drainage located adjacent to an oil shale facility. Environ Sci Technol. 1981;15:697–701. doi: 10.1021/es00088a009. [DOI] [PubMed] [Google Scholar]
- 11.Shukla O P. Microbial decomposition of α-picoline. Indian J Biochem Biophys. 1974;11:192–200. [PubMed] [Google Scholar]
- 12.Sims G K, O'Loughlin E J. Degradation of pyridines in the environment. Crit Rev Environ Control. 1989;19:309–340. [Google Scholar]
- 13.Stuermer D H, Ng D J, Morris C J. Organic contaminants in groundwater near an underground coal gasification site in northeastern Wyoming. Environ Sci Technol. 1982;16:582–587. doi: 10.1021/es00103a009. [DOI] [PubMed] [Google Scholar]
- 14.Turney G L, Goerlitz O F. Organic contamination in groundwater at gas works park, Seattle, Washington. Ground Water Monit Rev. 1990;10:187–198. [Google Scholar]
- 15.Watson G K, Cain R B. Microbial metabolism of the pyridine ring. Metabolic pathways of pyridine biodegradation by soil bacteria. Biochem J. 1975;146:157–172. doi: 10.1042/bj1460157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harber W, Schleifer K H, editors. The prokaryotes. Berlin, Germany: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]
- 17.Yoon J-H, Lee J J, Kang S-S, Takeuchi M, Shin Y K, Lee S T, Kang K H, Park Y-H. Gordonia nitida sp. nov., a bacterium that degrades 3-ethylpyridine and 3-methylpyridine. Int J Syst Evol Microbiol. 2000;50:1203–1210. doi: 10.1099/00207713-50-3-1203. [DOI] [PubMed] [Google Scholar]