Abstract
Background
Prescription medications such as selective serotonin reuptake inhibitors (SSRIs), commonly used to treat depression, are associated with weight gain. The role of pharmacogenomics in predicting SSRI-induced weight gain is unclear.
Methods
In this retrospective cohort study from participants in the Mayo Clinic RIGHT study who were prescribed citalopram, paroxetine, sertraline, or fluoxetine, our aim was to evaluate the association of metabolizer phenotype and total body weight after 6 months of SSRIs initiation. We evaluated the metabolizer phenotypes (poor/intermediate, normal, and rapid/ultra-rapid) of the cytochromes P450 enzymes genes: CYP2C9, CYP2C19, and CYP2D6 known to influence the metabolism of SSRI medications: CYP2C19 for citalopram, CYP2D6 for paroxetine, CYP2D6 and CYP2C19 for sertraline, and CYP2D6 and CYP2C9 fluoxetine. In addition, we assessed the association of metabolizer phenotype and total body weight change at six months following SSRI prescription using parametric analysis of covariance adjusted for baseline body weight and multivariate regression models.
Results
CYP2C19 poor/intermediate metabolizers prescribed citalopram gained significantly more weight than normal or rapid/ultra-rapid metabolizers at 6 months (TBWG %: 2.6 [95% CI 1.3—4.1] vs. 0.4 [95% CI -0.5 – 1.3] vs. -0.1 [-95% CI -1.5—1.1]; p = 0.001). No significant differences in weight outcomes at six months of treatment with paroxetine, sertraline, or fluoxetine were observed by metabolizer status.
Conclusions
Weight gain observed with citalopram may be mediated by CYP2C19 metabolizer status.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-022-02433-x.
Keywords: Pharmacogenomics, Weight gain, CYP metabolizer phenotypes
Background
Obesity is a chronic and complex multifactorial disease associated with multiple metabolic comorbidities, such as type 2 diabetes mellitus, and psychiatric diagnoses such as major depressive and anxiety disorders. There is a bidirectional relation between depression and obesity; thus, patients with obesity have a 55% increased risk of being diagnosed with depression over time, and patients with depression have a 58% increased risk of developing obesity [1].
Selective serotonin reuptake inhibitors (SSRIs) are the first-line treatment for depression. One side effect of their use is body weight gain during short- and long-term management. In fact, it is reported that patients prescribed an SSRI gain 4.2 kg more than non-users after a three-month treatment period [2], and that after 2.5 years of SSRI treatment, there is an increment of 2.5% of the initial body weight [3]. The risk factors associated with weight gain while receiving antidepressant medications are lower BMI at baseline, age under 65, and female gender [4]. However, the mechanisms of weight gain related to antidepressant use are not well known. Possible mechanisms include remission of major depression and increased neurotransmitters such as serotonin, which regulates feeding behaviors, energy expenditure [5, 6], and decreased brown adipose tissue thermogenesis [7].
Genetic variation is one of the factors that can alter a medication's efficacy by influencing its metabolism (i.e., pharmacokinetics), mechanism of action (i.e., pharmacodynamics), and even adverse side effects by gene-drug interactions. Because cytochrome P450 (CYP) enzymes contribute to phase I drug metabolism, CYP enzyme variation significantly impact treatment outcomes [8]. Pharmacogenomics offers the opportunity to optimize treatment considering these polymorphisms to develop a more personalized approach to antidepressant selection while reducing adverse drug events [9]. In 2013, the Clinical Pharmacogenetics Implementation Consortium (CPIC) developed dosing guidelines for paroxetine, citalopram, and sertraline based on their main metabolizer enzymes' phenotype status CYP2C19 and/or CYP2D6. Different CYP enzymes are involved in SSRI metabolism; however, each drug has a dominant metabolizer enzyme. Thus, citalopram is mainly metabolized by CYP2C19, paroxetine by CYP2D6, fluoxetine by CYP2D6 and CYP2C9, and sertraline by CYP2D6 and CYP2C19 [10]. The guidelines recommend a 50% reduction in the starting dose of citalopram, paroxetine, and sertraline in individuals with CYP2D6 or CYP2C19 poor metabolizer phenotype. In addition, the Food and Drug Administration has made recommendations for a maximum dosage of SSRIs in patients with specific metabolizer phenotypes [11]. There are currently little data detailing how CYP2D6 phenotypic status impacts the total amount of fluoxetine; hence, no gene-based dosage recommendations for fluoxetine have been provided.
Pharmacogenomics is a tool to personalize management in multiple areas, such as psychiatry and weight management [9, 12]. Multiple mood disorder studies have evaluated SSRI responsiveness for depression and lithium therapy in bipolar illness in GWAS studies and polygenic risk scores analysis [13–15]. These studies have found the link between genetic variants of obesity and SSRIs treatment response in depression [16]. However, a study investigating pharmacogenomics and weight gain in mood disorders is required. We hypothesized that patients with decreased metabolism of SSRIs by these cytochrome enzymes would be more likely to experience weight gain as a side effect. The study assesses the association between metabolizer phenotype and weight gain six months following SSRI prescription.
Methods
Study design
This retrospective cohort study approved by the Mayo Clinic Institutional Review Board (IRB 19–001,222) included participants from the Right Drug, Right Dose, Right Time (RIGHT) Study who underwent genetic sequencing of pharmacogenomic genes [17]. The RIGHT study included 11,090 participants, of which 60% were female and 97% were White. For the analyses, we considered participants from the RIGHT Study who had been prescribed citalopram, paroxetine, sertraline, or fluoxetine between 2004–2018. From those, we only included patients with a stable weight in the 6 months before starting the SSRI (n = 1,780). Of these, we exclude those who were < 18 years old, patients who did not have at least 6 months of treatment, did not have weight assessed during follow-up, had a history of bariatric surgery, were pregnant, or had a history of anti-obesity therapy (n = 1,117). The final analytic sample included 663 participants (Fig. 1). This study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline (Additional file 2: Table S1).
Fig. 1.
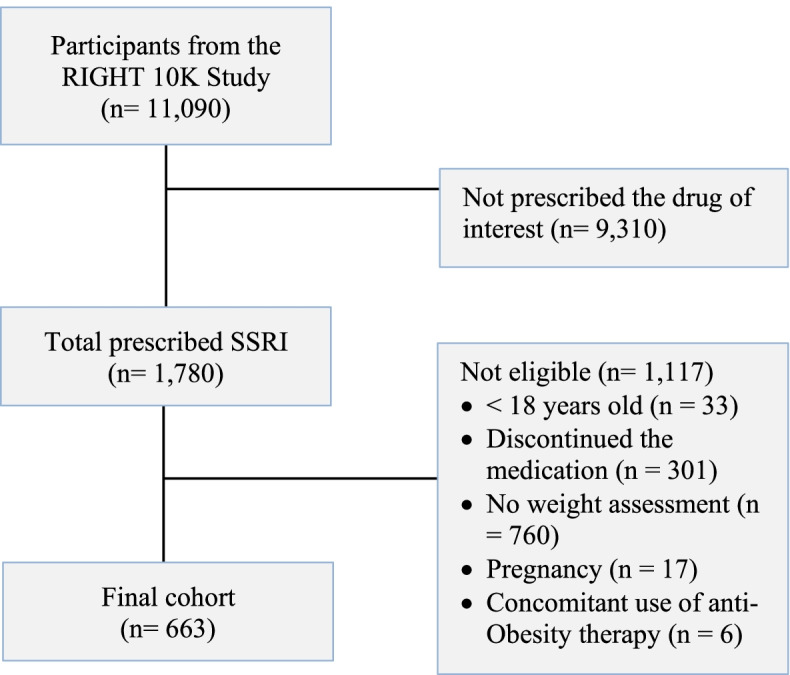
Study cohort
Data collection
Three physicians exhaustively reviewed the electronic medical record (EMR) to confirm inclusion and exclusion criteria. For the 663 participants included in the analysis, medication list, height, and body weight were extracted from the EMR, and comorbidities were extracted with ICD-9 and ICD-10 codes. Race was self-reported by study participants in the EMR, and race categories (American Indian or Alaska Native, Asian, Black or African American, Native Hawaiian or Other Pacific Islander, and White) were defined based on the US Office of Management and Budget’s Revisions to the Standards for the Classification of Federal Data on Race and Ethnicity. Body weight was extracted in kilograms at the time of prescription (± 2 weeks), three (± 2 weeks), and six (± 2 weeks) months of the initial prescription. Body mass index (BMI) was calculated with the formula weight in kilograms divided by height in meters squared.
Exposure
As part of the RIGHT cohort, the Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP)-accredited Baylor College of Medicine's Human Genome Sequencing Center Clinical Laboratory sequenced 77 genes using version 3 (v.3.) of the PGRN-Seq assay (now termed PGx-seq) [18]. CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, DPYD, SLCO1B1, TPMT, UGT1A1, VKORC1, HLA-A, and HLA-B were interpreted and reported by the Personalized Genomics Laboratory[17]. Genes studied are those pertinent to the enzymatic metabolism of the SSRI medications. The cytochrome P450 enzyme genes were CYP2C19 for citalopram; CYP2D6 for paroxetine; CYP2D6 and CYP2C9 for fluoxetine, and CYP2C19 and CYP2D6 sertraline [10]. Genotypes, including both rare and common variants, were translated into diplotypes using star allele nomenclature, when applicable, as described in PharmVar (www.pharmvar.org, last accessed 1/29/2022). Diplotypes were assigned a predicted metabolic phenotype (metabolizer status) using standard clinical laboratory processes, which rely on the assignment of the function of each allele present relative to a normal function (or “wild-type”) allele (Additional file 2: Table S2). Details of the genetic analysis have been previously reported [19]. Metabolizer status was classified as: poor/intermediate, normal (extensive), and rapid/ultra-rapid metabolizer [20]. We evaluate the concomitant use of inducers and inhibitors for P450-mediated metabolism (Additional file 2: Tables S3 and S4)[21].
Outcome measurements
The outcome was calculated using the following formula:
Statistical analysis
Baseline anthropometric and demographics were not normally distributed according to the Shapiro-Wilks test and are summarized as median and interquartile range (IQR). Categorical data are presented as frequency and percentages. Categorical data were analyzed using Pearson's chi-squared test. ANCOVA models were used to assess the difference in total body weight change (%) with metabolizer status and BMI as a covariate. Multiple linear regression was calculated to evaluate the effect of metabolizer status and total body weight change (%) with normal metabolizer as a reference group and in three models, and parameter estimates with standard error (SE) were calculated for poor/intermediate metabolizer and rapid/ultra-rapid metabolizer. Model one included BMI; model two included BMI and age, and model three included BMI, sex, and age. The analysis also excluded patients with concomitant strong and moderate inducers and inhibitors for P450-mediated metabolism. To investigate whether the weight gain varies between BMI groups according to the World Health Organization classification (i.e., underweight and normal weight, overweight, and obesity), we stratified the analysis by normal weight, overweight, and obesity groups. ANCOVA models were used to assess the difference in total body weight change (%) with metabolizer status within these groups with BMI as a covariate, data are presented as mean and 95% confidence intervals (CI). Plots and statistical analyses were performed in SAS 9.2 (SAS Institute Inc., Cary, NC). P-values ≤ 0.05 were considered statistically significant.
Results
The final cohort included 663 participants (age 61 [46 – 72] years, 76% females, and 94% white). The median BMI was 27.8 (24.0 – 32.9) kg/m2, 62% of our cohort had normal weight or overweight, and 38% had obesity (Table 1). The prevalence of the different metabolizer status phenotypes for CYP2C19, CYP2D6, and CYP2C9 by SSRI is described in Table 2 and Additional file 3: Figure S1. The normal metabolizer phenotype was the most frequent in all cytochrome enzymes, except for CYP2D6, where the poor/intermediate metabolizer was the most predominant. The rapid/ultrarapid metabolizer phenotype was the least frequent. There were no significant differences in comorbidities among metabolizer phenotype (Additional file 2: Table S5).
Table 1.
Participant characteristics in all participants and by drug. Data are shown as mean ± standard deviation or percentage
| Total | Citalopram | Fluoxetine | Paroxetine | Sertraline | p-value* | |
|---|---|---|---|---|---|---|
| N = 663 | N = 202 | N = 191 | N = 107 | N = 163 | ||
| Demographics | ||||||
| Age, years | 61 (46 – 72) | 51 (40 – 66) | 65 (53 – 76) | 59 (48 – 69) | 64 (46 – 77) | < 0.001 |
| Gender, females | 507 (76%) | 158 (78%) | 151 (79%) | 80 (75%) | 118 (72%) | 0.44 |
| Race, White | 621 (94%) | 190 (94%) | 181 (95%) | 100 (93%) | 150 (92%) | 0.79 |
| Anthropometrics | ||||||
| Weight, kg | 78.0 (65.3 – 94) | 76.2 (64.3 – 91.1) | 82.0 (69.7 – 97) | 74.2 (62 – 92.7) | 76.7 (65 – 93.7) | 0.03 |
| BMI, kg/m2 | 27.8 (24.0 – 32.9) | 27.2 (23.6 – 32.1) | 30.0 (25.1 – 34.4) | 26.3 (23.8 – 32.5) | 27.7 (23.7 – 32.5) | 0.009 |
| BMI Class | ||||||
| Class, underweight or normal weight | 211 (32%) | 61(30%) | 46 (24%) | 45 (35%) | 59 (36%) | |
| Class, overweight | 202 (30%) | 79 (39%) | 48 (25%) | 25 (23%) | 50 (31%) | |
| Class, obesity | 250 (38%) | 62 (31%) | 97 (51%) | 37 (42%) | 52 (33%) | |
Continuous data are summarized as median (IQR). Categorical data are presented as frequencies and percentages
Abbreviations used: BMI Body mass index
*p-value: calculated with ANOVA
Table 2.
Distribution of phenotypes of cytochromes enzymes involved in the metabolism of citalopram, paroxetine, fluoxetine, and sertraline among the participants
| Total | Citalopram | Fluoxetine | Paroxetine | Sertraline | |
|---|---|---|---|---|---|
| N = 663 | N = 202 | N = 191 | N = 107 | N = 163 | |
| CYP2C19 | |||||
| Poor/intermediate metabolizer, n | 196 (30%) | 58 (29%) | 46 (28%) | ||
| Normal metabolizer, n | 268 (40%) | 83 (41%) | 66 (41%) | ||
| Rapid/ultrarapid metabolizer, n | 199 (30%) | 61 (30%) | 51 (31%) | ||
| CYP2D6 | |||||
| Poor/intermediate metabolizer, n | 462 (70%) | 136 (71%) | 70 (65%) | 127 (78%) | |
| Normal metabolizer, n | 191 (29%) | 53 (28.9%) | 34 (32%) | 31 (19%) | |
| Rapid/ultrarapid metabolizer, n | 10 (1%) | 2 (0.1%) | 3 (3%) | 5 (3%) | |
| CYP2C9 | |||||
| Poor/intermediate metabolizer, n | 234 (35%) | 70 (37%) | |||
| Normal metabolizer, n | 429 (65%) | 121 (63%) | |||
Categorical data are presented as frequencies and percentages
The detail medications used which inhibit or induce the CYP 450 enzymes by SSRI can be found in Additional file 2: Tables S3 and S4. From patients taking citalopram, 1 (0.5%) was concomitantly prescribed rifampin, a strong inducer for CYP2C19, and 3 (1.5%) were prescribed fluconazole or fluvoxamine, strong inhibitors for CYP2C19. From patients taking fluoxetine, 1 (0.05%) was concomitantly prescribed terbinafine, a strong inhibitor for CYP2D6, and 11 (5.7%) were prescribed a moderate inhibitor for CYP2C9 or CYP2D6 (i.e., fluconazole [n = 8], and duloxetine [n = 3]). From the patients taking paroxetine, 1 (1%) was prescribed duloxetine, a moderate inhibitor of CYP2D6. From patients taking sertraline, 1 (1%) was concomitantly prescribed phenytoin, a moderate inducer for CYP2C19, and 2 (1%) were prescribed fluconazole a strong inhibitor for CYP2C19.
Total body weight gain % by metabolizer status
The total body weight gain percentage (TBWG %) at six months for the patients prescribed any SSRIs was 0.7% (-1.4 – 2.9). When analyzed by medication prescribed, TBWG % at six months for citalopram was 1.1% (-1.3 – 3.1), paroxetine 0.9% (-1.2 – 3.3), sertraline 1.1% (-1.0 – 2.9), and fluoxetine 0.1% (-1.9 – 2.6). For patients on citalopram, patients who were poor/intermediate CYP2C19 metabolizers gained significantly more weight than normal and rapid/ultrarapid metabolizers (TBWG %: 2.6 [95% CI 1.3—4.1] vs. 0.4 [95% CI -0.5 – 1.3] vs. -0.1 [-95% CI -1.5—1.1], respectively; p = 0.001) (Fig. 2). After excluding 3 patients with an inducer or inhibitor of the CYP 450, patients who were poor/intermediate CYP2C19 metabolizers gained significantly more weight than normal and rapid metabolizers (TBWG %: 2.2 [95% CI 1.1—4.0] vs. 0.4 [95% CI -0.5 – 1.5] vs. -0.2 [95% CI -1.3—1.4], respectively; p = 0.003). No significant difference was found in TBWG percentage at three or six months according to the CYP2D6 phenotype for paroxetine, fluoxetine, and sertraline, CYP2C9 for fluoxetine CYP2C19 for fluoxetine and sertraline (Table 3).
Fig. 2.
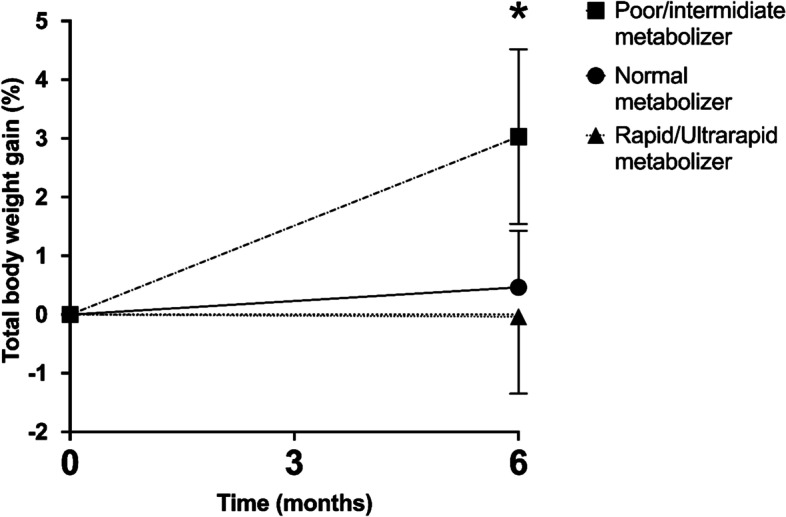
Effect of citalopram on total body weight by CYP2C19 phenotype. *p = 0.001
Table 3.
Total body weight gain percentage by CYP phenotype in participants prescribed with citalopram, paroxetine, fluoxetine, and sertraline
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p- value | |
|---|---|---|---|---|
| CYP2C19 | ||||
| Citalopram | ||||
| TBWG 6 months, % | 2.6 (95% CI 1.3 – 4.1) | 0.4 (95% CI -0.5 – 1.3) | -0.1 (95% CI -1.5 – 1.1) | 0.001 |
| Sertraline | ||||
| TBWG 6 months, % | 0.9 (95% CI -0.09 – 2.1) | 0.4 (95% CI -0.6 – 1.5) | 1.7 (95% CI 0.7 – 2.9) | 0.13 |
| CYP2D6 | ||||
| Paroxetine | ||||
| TBWG 6 months, % | 0.7 (95% CI -0.1 – 1.5) | 1.6 (95% CI 0.2 – 2.9) | 0.7 (95% CI -4.8 – 6.1) | 0.50 |
| Sertraline | ||||
| TBWG 6 months, % | 1.1 (95% CI 0.4 – 1.9) | 0.8 (95% CI -0.3 – 1.8) | 0.9 (95% CI -9.0 – 10.9) | 0.98 |
| Fluoxetine | ||||
| TBWG 6 months, % | 0 (95% CI -0.7 – 0.7) | 0.2 (95% CI -1.2 – 1.5) | 1.3 (95% CI -3 – 3) | 0.84 |
| CYP2C9 | ||||
| Fluoxetine | ||||
| TBWG 6 months, % | 0.5 (95% CI -0.5 – 1.6) | -0.1 (95% CI -1.1 – 0.6) | 0.33 | |
Continuous data are summarized as mean and 95% confidence interval (CI)
Abbreviations used: TBWG Total Body Weight Gain
p-value: calculated with ANCOVA with metabolizer status and BMI as covariates
Total body weight gain % by metabolizer status among BMI groups
Table 4 details the effect of metabolizer status and TBWG % after six months for each medication by BMI group. Patients in the overweight group that were prescribed citalopram who were poor/intermediate CYP2C19 metabolizers gained significantly more weight than normal and rapid metabolizers (TBWG %: 3.0 [95% CI -0.3 – 6.4] vs. -0.3 [95% CI -2.1 – 1.5] vs. 1.3 [95% CI -3.6 – 1.1], respectively; p = 0.02). We did not observe any significant difference in TBWG % between metabolizer status among patients in the underweight, normal weight, or obesity group taking citalopram, fluoxetine, sertraline, or paroxetine (Table 5).
Table 4.
Total body weight gain by obesity class and CYP phenotype in participants prescribed with citalopram, paroxetine, fluoxetine, and sertraline
| Normal and Underweight Group | ||||||||
| CYP2C19 | ||||||||
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p-value | |||||
| Citalopram | TBWG 6 mths, % | n = 17 | 4.4 (95% CI 1.9 – 6.8) | n = 26 | 1.5 (95% CI -0.3 – 3.4) | n = 18 | 1.5 (95% CI -0.6 – 3.5) | 0.11 |
| Sertraline | TBWG 6 mths, % | n = 12 | 2.6 (95% CI 0.9 – 4.5) | n = 28 | 1.2 (95% CI -0.8 – 3.1) | n = 19 | 2.7 (95% CI 1.1 – 4.3) | 0.32 |
| CYP2D6 | ||||||||
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p-value | |||||
| Paroxetine | TBWG 6 mths, % | n = 33 | 0.6 (95% CI -0.7 – 1.9) | n = 11 | 0.4 (95% CI -2.7 – 3.6) | n = 1 | 3.0 | 0.65 |
| Sertraline | TBWG 6 mths, % | n = 49 | 1.8 (95% CI 0.6 – 3.1) | n = 8 | 1.4 (95% CI -0.5 – 3.4) | n = 2 | 6.7 (95% CI -55.1 – 69.6) | 0.16 |
| Fluoxetine | TBWG 6 mths, % | n = 32 | 1.2 (95% CI -0.3 – 2.7) | n = 14 | 0.9 (95% CI -1.8 – 3.6) | 0.88 | ||
| CYP2C9 | ||||||||
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p-value | |||||
| Fluoxetine | TBWG 6 mths, % | n = 26 | 1.9 (95% CI -0.3 – 4.1) | n = 20 | 0.5 (95% CI -1.1 – 2.1) | 0.60 | ||
| Overweight Group | ||||||||
| CYP2C19 | ||||||||
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p-value | |||||
| Citalopram | TBWG 6 mths, % | n = 21 | 3.0 (95% CI -0.3 – 6.4) | n = 34 | -0.3 (95% CI -2.1 – 1.5) | n = 24 | 1.3 (95% CI -3.6 – 1.1) | 0.02 |
| Sertraline | TBWG 6 mths, % | n = 19 | 1.5 (95% CI -0.2 – 3.1) | n = 17 | 0.7 (95% CI -0.7 – 2.1) | n = 14 | 2.3 (95% CI 0.6 – 4.1) | 0.44 |
| CYP2D6 | ||||||||
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p-value | |||||
| Paroxetine | TBWG 6 mths, % | n = 12 | 1.6 (95% CI -0.8 – 3.9) | n = 12 | 3.5 (95% CI 1.4 – 5.6) | n = 1 | 1.3 | 0.13 |
| Sertraline | TBWG 6 mths, % | n = 40 | 1.3 (95% CI 0.3 – 2.3) | n = 8 | 2.3 (95% CI -0.5 – 3.4) | n = 2 | 0.9 (95% CI -25.0 – 26.8) | 0.69 |
| Fluoxetine | TBWG 6 mths, % | n = 28 | 1.2 (95% CI -2.9 – 0.4) | n = 20 | 0.5 (95% CI -2.2 – 3.2) | 0.21 | ||
| CYP2C9 | ||||||||
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p-value | |||||
| Fluoxetine | TBWG 6 mths, % | n = 16 | 0.6 (95% CI -3.3 – 2.1) | n = 32 | 0.5 (95% CI -2.3 – 1.3) | 0.91 | ||
| Obesity Group | ||||||||
| CYP2C19 | ||||||||
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p-value | |||||
| Citalopram | TBWG 6 mths, % | n = 20 | 0.8 (95% CI -0.5 – 1.9) | n = 23 | 0.4 (95% CI -0.8 – 1.7) | n = 19 | -0.3 (95% CI -2.7 – 2.2) | 0.18 |
| Sertraline | TBWG 6 mths, % | n = 15 | -0.9 (95% CI -3.1 – 1.2) | n = 21 | -0.7 (95% CI -2.6 – 1.2) | n = 18 | 0.6 (95% CI -1.7 – 2.9) | 0.52 |
| CYP2D6 | ||||||||
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p-value | |||||
| Paroxetine | TBWG 6 mths, % | n = 11 | 0.4 (95% CI -0.8 – 1.6) | n = 25 | 0.7 (95% CI -1.3 – 2.6) | n = 1 | 0.4 | 0.98 |
| Sertraline | TBWG 6 mths, % | n = 38 | 0.1 (95% CI -1.6 – 1.3) | n = 15 | 0.3 (95% CI -1.8 – 1.2) | n = 1 | -10.6 | 0.26 |
| Fluoxetine | TBWG 6 mths, % | n = 19 | 0.1 (95% CI -0.9 – 1.0) | n = 76 | -0.9 (95% CI -3.1 – 1.2) | n = 2 | 1.3 (95% CI -33.0 – 35.6) | 0.79 |
| CYP2C9 | ||||||||
| Poor/intermediate metabolizer | Normal metabolizer | Rapid/ultra-rapid metabolizer | p-value | |||||
| Fluoxetine | TBWG 6 mths, % | n = 34 | 0.2 (95% CI -1.1 – 1.5) | n = 63 | -0.3 (95% CI -1.5 – 0.8) | 0.30 | ||
Continuous data are summarized as mean (95% CI)
Abbreviations used: CI confidence interval, TBWG Total Body Weight Gain
p-value: calculated with ANCOVA with metabolizer status and BMI as covariate
Table 5.
Multiple regression variate analysis. Total body weight gain by obesity class and CYP phenotype in participants prescribed with citalopram, paroxetine, fluoxetine, and sertraline
| CYP2C19 | |||||||||||||
| Model 1 | Model 2 | Model 3 | |||||||||||
| Poor/intermediate metabolizer | Rapid/ultra-rapid metabolizer | Poor/intermediate metabolizer | Rapid/ultra-rapid metabolizer | Poor/intermediate metabolizer | Rapid/ultra-rapid metabolizer | ||||||||
| PE | p-value* | PE | p-value* | PE | p-value+ | PE | p-value+ | PE | p-value++ | PE | p-value++ | ||
| Citalopram | TBWG 6 mths, % | 1.7 (0.5) | 0.001 | -1.2 (0.5) | 0.01 | 1.7 (0.6) | 0.001 | -1.2 (0.5) | 0.02 | 1.7 (0.5) | 0.001 | -1.2 (0.5) | 0.02 |
| Sertraline | TBWG 6 mths, % | -0.03 (0.4) | 0.94 | 0.7 (0.4) | 0.08 | 0.01 (0.4) | 0.99 | 0.7 (0.4) | 0.10 | 0.01 (0.4) | 0.99 | 0.7 (0.4) | 0.11 |
| CYP2D6 | |||||||||||||
| Model 1 | Model 2 | Model 3 | |||||||||||
| Poor/intermediate metabolizer | Rapid/ultra-rapid metabolizer | Poor/intermediate metabolizer | Rapid/ultra-rapid metabolizer | Poor/intermediate metabolizer | Rapid/ultra-rapid metabolizer | ||||||||
| PE | p-value* | PE | p-value* | PE | p-value+ | PE | p-value+ | PE | p-value++ | PE | p-value++ | ||
| Paroxetine | TBWG 6 mths, % | -0.3 (1.4) | 0.82 | -0.3 (0.7) | 0.71 | -0.4 (0.7) | 0.61 | -0.1 (1.4) | 0.96 | 0.4 (0.7) | 0.61 | -0.1 (1.4) | 0.96 |
| Sertraline | TBWG 6 mths, % | 0.1 (0.6) | 0.76 | -0.3 (1.2) | 0.80 | 0.1 (0.6) | 0.89 | -0.3 (1.2) | 0.77 | 0.1 (0.6) | 0.91 | 0.3 (1.2) | 0.77 |
| Fluoxetine | TBWG 6 mths, % | -0.6 (1.1) | 0.58 | 1.2 (2.2) | 0.57 | -0.6 (1.1) | 0.57 | 1.3 (2.2) | 0.50 | -0.6 (1.1) | 0.60 | 1.2 (2.2) | 0.57 |
| CYP2C9 | |||||||||||||
| Model 1 | Model 2 | Model 3 | |||||||||||
| Poor/intermediate metabolizer | Rapid/ultra-rapid metabolizer | Poor/intermediate metabolizer | Rapid/ultra-rapid metabolizer | Poor/intermediate metabolizer | Rapid/ultra-rapid metabolizer | ||||||||
| PE | p-value* | PE | p-value* | PE | p-value+ | PE | p-value+ | PE | p-value++ | PE | p-value++ | ||
| Fluoxetine | TBWG 6 mths, % | 0.3 (0.3) | 0.33 | 0.3 (0.6) | 0.33 | 0.3 (0.3) | 0.39 | ||||||
p-value: multiveriate linear regression with normal metabolizar as reference group
* model including BMI
+ model including BMI and age
++ model including BMI, age and sex
Abbreviations used: BMI body mass index; PE parameter estimates, TBWG Total Body Weight Gain
Effect of metabolizer status on total body weight gain %
Multiple linear regression was calculated to evaluate the effect of metabolizer status and TBWG % after six months for each medication. For citalopram, when adjusting for BMI, poor/intermediate CYP2C19 metabolizer status resulted in a weight gain of 1.7% (Standard Error [SE] 0.5; p = 0.001), while rapid/ultrarapid metabolizer status resulted in a decrease of 1.2% (SE 0.5; p = 0.01). This effect remained significant after adjusting for BMI and age where poor/intermediate CYP2C19 metabolizer status resulted in a weight gain of 1.7% (Standard Error [SE] 0.6; p = 0.001), while rapid/ultrarapid metabolizer status resulted in a decrease of 1.2% (SE 0.5; p = 0.02). This trend was also seen after adjusting for BMI, sex, and age where poor/intermediate CYP2C19 metabolizer status resulted in a weight gain of 1.7% (Standard Error [SE] 0.5; p = 0.001), while rapid/ultrarapid metabolizer status resulted in a decrease of 1.2% (SE 0.5; p = 0.02). No significant effect was found for CYP2D6 phenotype for paroxetine, fluoxetine, and sertraline, CYP2C9 for fluoxetine, CYP2C19 for fluoxetine, and sertraline when adjusting for BMI, BMI and age, or BMI, age, and sex.
Discussion
The current study identified that poor/intermediate metabolizer status for CYP2C19 is associated with a 1.7% more weight gain after 6 months than normal metabolizers in patients taking citalopram. This study shows that this remains significant among patients with overweight, where patients with poor/intermediate metabolizer status for CYP2C19 and taking citalopram had a TBWG of 3.0%. There were no disparities in comorbidities across individuals with various metabolizer statuses, and the difference in weight change remained after excluding patients who were using a CYP 450 inducer or inhibitor concurrently, highlighting the importance of the metabolizer status. Our results are generally consistent with previous studies showing the effect of citalopram on body weight [4, 22]. Aldrich et al. conducted a retrospective study using an electronic medical record of 263 youth with anxiety and depression prescribed citalopram. They showed a significant association between poor CYP2C19 metabolizer phenotype and earlier weight gain after 45 days of treatment [23]. The weight changes related to other antidepressants were not connected with the other pharmacogenomic genes of interest.
Our findings are also consistent with previous studies in which fluoxetine and sertraline have shown minimal effects on weight gain [24]. Conversely, paroxetine has demonstrated a greater risk of weight gain. Serretti et al. reported that the mean weight difference during 8 months of treatment was 2.73 kg for paroxetine [25]. These findings were not replicated in our cohort treated with paroxetine; this discrepancy might be explained by the different intervals of the observations in the two studies.
A number of reasons complicate weight changes in individuals receiving depression medication; they may indicate an improvement in those who have lost weight due to their depression, but they can also be a side effect of the treatment. In our study, weight gain was seen in overweight patients on citalopram with poor/intermediate CYP2C19 metabolizer status. Previous research has found that participants who considered their weight status as overweight were more likely to gain weight in the future [26]. We found that weight increase in patients treated with citalopram was significant even after controlling for BMI, indicating the importance of metabolizer status. As a result, it is critical to identify individuals who are prone to weight gain and risk factors that may contribute to it. It is crucial to underline that a decision tool such as pharmacogenomics may be more effective in these individuals as an ad hoc instrument.
Multiple drugs with indications for chronic weight management have been authorized with improved safety profiles [27]. However, in a patient-centered care model, it is important to recognize barriers that may decrease to less effective and efficient weight management and that negatively impact weight loss outcomes. One of these barriers is weight gain as a medications’ side effect, i.e. obesogenic drugs [28]. According to the findings of a patient survey, the most common reason for discontinuing antidepressant medication is a lack of effectiveness. However, up to 27% of patients who reported noncompliance discontinued the drug due to weight increase [29]. Previous research has looked at the link between metabolizer status and medicine discontinuation; however, no convincing relationship has been identified due to study design and sample collection [30]. More research is needed to determine the true impact of metabolizer status on drug discontinuation, particularly in individuals who are overweight and using SSRIs.
Despite their widespread usage of antidepressant medicine, initial drug selection success might be lower. According to the findings of the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) trial, only one-third of the patients achieved remission within the first treatment level [31]. Previous studies have found the clinical benefits of using pharmacogenomics to tailor the therapeutic approach in patients with major depressive disorder and anxiety [32–37]. Poor metabolizers taking escitalopram resulted in a greater rate of therapeutic failure, indicating the potential clinical value of CYP2C19 genotyping for individualization of escitalopram [38]. However, there has not been any difference in adverse drug events reported between pharmacogenomic tailored treatment compared with controls [8, 9, 32].
Previous studies have retrospectively assessed side effects and metabolizer status with antidepressants such as tricyclic antidepressants (TCAs), serotonin-noradrenaline reuptake inhibitors (SNRIs), and SSRIs [39, 40]. In terms of side effects and CYP2C19 and citalopram/escitalopram, a meta-analysis of 2037 patients found that, compared to normal metabolizers, CYP2C19 poor metabolizers had a greater risk of gastrointestinal, neurological, and sexual adverse effects [41]. The Patient Rated Inventory of Side Effects (PRISE), which includes weight gain among other items and covers 9 categories and 32 items, is a typical measure used to evaluate side effects in most pharmacogenomics studies. Most research, however, focuses on side effects associated with a specific organ or system, which may restrict the relationship between pharmacogenomic studies with weight gain. Another study has looked at variations in the genes CYP2D6, CYP2C19, and CYP2C9 in patients taking SSRIs and tolerability and did not find a clear pharmacogenetic explanation for side effects [30]. In a recent study of 9500 participants, poor metabolizers were at higher risk of side effects adding to the evidence for a link between CYP2C19 metabolism and SSRI tolerability [42]. However, tolerability and side effects were evaluated by a survey using a qualitative assessment considering weight gain. The relationship between poor CYP2C19 metabolizer status and early weight gain documented in the medical record in children using escitalopram or citalopram has been described, adding to the data connecting metabolizer status and weight increase [43]. This is the first study to objectively evaluate one of the common side effects of SSRIs, regardless of the response to treatment in adults.
Although studies on the association between citalopram blood concentrations and pharmaceutical effectiveness and tolerability are lacking, it is usually assumed that a 50% difference in blood concentration will have a clinical impact [10, 44]. Because of the increase in concentration for CYP2C19 poor metabolizers compared to normal metabolizers, there is an implied risk of adverse outcomes, and the recommendation is to reduce the citalopram dose by 50% [10]. Further studies are needed to evaluate drug blood concentrations and weight gain according to metabolizer status.
Our study has some limitations. First, the low prevalence of some CYP enzyme phenotypes (CYP2D6 rapid/ultrarapid metabolizer) and the small sample size might cause a type II error in assessing our primary outcome. This limits the ability to detect a difference in TBWG % with other enzyme phenotypes. Importantly, given the nature of the study, we could not include all patients prescribed an SSRI because a few had no follow up at our institution. Second, the generalizability of the data is limited by the retrospective nature of our study and does not establish a causal link between CYP enzyme phenotypes and weight gain. Third, it is difficult to investigate characteristics such as drug compliance since medical record data varies so much between health care providers, and the influence of polypharmacy on patient outcomes was not evaluated. This is an important aspect to consider in future research. Previously, tricyclic antidepressants, which have a more profound effect on bodyweight, weight gain, have been a clear limitation of compliance to the medication. Weight gain in our cohort may have resulted in noncompliance, concealing the differences in weight in our cohort among drugs that have previously been associated with higher weight gain. Fourth, there are ascertainment biases inherent to a study conducted in tertiary care centers with a study population that is predominately White. Fifth, the mood response to the SSRI was not formally recorded with validated questionnaires, and this outcome was unclear from retrospective chart review during data gathering. Thus, response to treatment of depression could confound weight gain or weight loss.
In addition to the CYP genes, other genes related to the serotonin and norepinephrine signaling have been implicated with the therapeutic responses to SSRI. Previous studies have evaluated the effect of genetic variants on change in depressive symptoms and found significant associations with several variants in the serotonin receptor gene (HTR2A) and the response to escitalopram, the norepinephrine transporter gene (SLC6A2) and the response to nortriptyline, and the glucocorticoid receptor gene (NR3C1) and the response to both nortriptyline and escitalopram [45]. Here, we did not evaluate the effect of genetic variants that may affect the response to drugs with different mechanisms of action. Studies have also examined genes related to weight gain and SSRIs, such as catechol-O-methyltransferase (COMT), tryptophan hydroxylase 1 (TPH1), HTR2C, and serotonin transporter gene (SLC6A4). The evidence shows that GG COMT and AA TPH1 genotypes have more weight gain outcomes than HTR2C and SLC6A4 polymorphism[9, 16, 37]. Our research did not cover the effect of other genes involved in the SSRI metabolisms; therefore, further research focusing on other enzymes involved in the SSRI metabolism is needed to understand the variability in weight gain response to this class of medication. The study’s strengths include a high level of detail regarding CYP genotypes polymorphisms and weight loss outcomes and complications after bariatric surgery. It is, to our knowledge, the largest research evaluating weight change outcomes, combining the administration of CYP inducers and inhibitors at the same time.
Conclusions
In conclusion, we have performed a retrospective pharmacogenomics study to understand the SSRIs’ common weight gain side effects. We showed that the CYP2C19 genotype might explain weight gain in citalopram patients, and it might become a projection tool for preventing weight gain and obesity, particularly in patients who are overweight. Further studies are needed to validate this observation in prospective trials.
Supplementary Information
Additional file 2: Table S1. STROBE reporting check list. Table S2. Variants/star alleles considered in this study. Table S3. Clinical inducers and inhibitors for P450-mediated metabolism considered. [46]. Table S4. Distribution of concomitant use of clinical inducers and inhibitors for P450-mediated metabolism by antidepressant. Table S5. Distribution of comorbidities by metabolizer status. CYP2C9>
Additional file 3: Figure S1. Distribution of phenotypes of cytochromes enzymes involved in the metabolism of citalopram, paroxetine, fluoxetine, and sertraline among the participants.
Abbreviations
- BMI
Body mass index
- CPIC
Clinical Pharmacogenetics Implementation Consortium
- CYP
Cytochrome P450
- EMR
Electronic medical record
- IQR
Interquartile range
- PE
Parameter estimates
- RIGHT
Right Drug, Right Dose, Right Time
- SSRIs
Selective serotonin reuptake inhibitors
- TBWG
Total Body Weight Gain
Authors’ contributions
SS, MLR, LC, AA, and SB co-conceptualized and co-designed the study, drafted the initial data, and critically reviewed the manuscript. AA and SB coordinated and supervised data collection, and critically reviwed the manuscript. PD performed the statistical analysis. DGI, MDH, SB, AM, and MC provided critical feedback on study design and critically reviwed the manuscript. All authors read and approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Funding
Dr. Acosta is supported by NIH (NIH K23-DK114460). Dr. Camilleri receives funding related to obesity from the National Institutes of Health (NIH RO1-DK67071).
Availability of data and materials
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Declarations
Ethics approval and consent to participate
All participants provided written informed consent for genotyping analysis and further utilization of their data as part of the Mayo Clinic Biobank and the RIGHT Study. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the Mayo Clinic Institutional Review Board (IRB 19–001222), formal consent was waived due to minimal risk.
Consent for publication
Not applicable.
Competing interests
Dr. Andres Acosta is a stockholder in Gila Therapeutics and Phenomix Sciences; he serves as a consultant for Rhythm Pharmaceuticals, General Mills. Dr. Camilleri is a stockholder in Phenomix Sciences and Enterin and serves as a consultant to Takeda, Allergan, Kallyope, and Arena with compensation to his employer, Mayo Clinic. Drs. Sneha Singh, Maria L Ricardo-Silgado, Lizeth Cifuentes, Daniel Gonzalez-Izundegui, Maria Daniela Hurtado, Ann Moyer, and Suzette Bielinski have no disclosures.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Maria L. Ricardo-Silgado and Sneha Singh contributed equally.
References
- 1.Luppino FS, de Wit LM, Bouvy PF, Stijnen T, Cuijpers P, Penninx BWJH, et al. Overweight, Obesity, and Depression: A Systematic Review and Meta-analysis of Longitudinal Studies. Arch Gen Psychiatry. 2010;67(3):220–229. doi: 10.1001/archgenpsychiatry.2010.2. [DOI] [PubMed] [Google Scholar]
- 2.Noordam R, Aarts N, Tiemeier H, Hofman A, Stricker BH, Visser LE. Sex-specific association between antidepressant use and body weight in a population-based study in older adults. J Clin Psychiatry. 2015;76(6):e745–51. Epub 2015/07/02. doi: 10.4088/JCP.13m08896. PubMed PMID: 26132681. [DOI] [PubMed]
- 3.Demyttenaere K, Jaspers L. Review: Bupropion and SSRI-induced side effects. J Psychopharmacol. 2008;22(7):792–804. Epub 2008/03/01. doi: 10.1177/0269881107083798. PubMed PMID: 18308785. [DOI] [PubMed]
- 4.Uguz F, Sahingoz M, Gungor B, Aksoy F, Askin R. Weight gain and associated factors in patients using newer antidepressant drugs. Gen Hosp Psychiatry. 2015;37(1):46–48. doi: 10.1016/j.genhosppsych.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 5.De Long NE, Stepita RA, Taylor VH, Holloway AC. Major depressive disorder and diabetes: does serotonin bridge the gap? Curr Diabetes Rev. 2015;11(2):71–8. doi: 10.2174/1573399811666150223123053. [DOI] [PubMed] [Google Scholar]
- 6.Gill H, Gill B, El-Halabi S, Chen-Li D, Lipsitz O, Rosenblat JD, et al. Antidepressant Medications and Weight Change: A Narrative Review. Obesity (Silver Spring). 2020. Epub 2020/10/07. doi: 10.1002/oby.22969. PubMed PMID: 33022115. [DOI] [PubMed]
- 7.Young RL, Lumsden AL, Keating DJ. Gut serotonin is a regulator of obesity and metabolism. Gastroenterology. 2015;149(1):253–255. doi: 10.1053/j.gastro.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Brethauer SA, Kim J, el Chaar M, Papasavas P, Eisenberg D, Rogers A, et al. Standardized outcomes reporting in metabolic and bariatric surgery. Surg Obes Relat Dis. 2015;11(3):489–506. Epub 2015/06/22. doi: 10.1016/j.soard.2015.02.003. PubMed PMID: 26093765. [DOI] [PubMed]
- 9.Singh S, Ricardo-Silgado ML, Bielinski SJ, Acosta A. Pharmacogenomics of Medication Induced Weight Gain and Antiobesity Medications. Obesity. 2021;29(2):265–273. doi: 10.1002/oby.23068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks JK, Bishop JR, Sangkuhl K, Müller DJ, Ji Y, Leckband SG, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conrado DJ, Rogers HL, Zineh I, Pacanowski MA. Consistency of drug-drug and gene-drug interaction information in US FDA-approved drug labels. Pharmacogenomics. 2013;14(2):215–23. Epub 2013/01/19. doi: 10.2217/pgs.12.203. PubMed PMID: 23327581. [DOI] [PubMed]
- 12.Amare AT, Schubert KO, Baune BT. Pharmacogenomics in the treatment of mood disorders: strategies and opportunities for personalized psychiatry. EPMA Journal. 2017;8(3):211–227. doi: 10.1007/s13167-017-0112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahon FJ, Buervenich S, Charney D, Lipsky R, Rush AJ, Wilson AF, et al. Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. Am J Hum Genet. 2006;78(5):804–14. Epub 2006/04/28. doi: 10.1086/503820. PubMed PMID: 16642436; PubMed Central PMCID: PMCPMC1474035. [DOI] [PMC free article] [PubMed]
- 14.Ising M, Lucae S, Binder EB, Bettecken T, Uhr M, Ripke S, et al. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry. 2009;66(9):966–75. Epub 2009/09/09. doi: 10.1001/archgenpsychiatry.2009.95. PubMed PMID: 19736353; PubMed Central PMCID: PMCPMC4465570. [DOI] [PMC free article] [PubMed]
- 15.Amare AT, Schubert KO, Tekola-Ayele F, Hsu Y-H, Sangkuhl K, Jenkins G, et al. Association of the polygenic scores for personality traits and response to selective serotonin reuptake inhibitors in patients with major depressive disorder. Front Psych. 2018;9:65. doi: 10.3389/fpsyt.2018.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amare AT, Schubert KO, Tekola-Ayele F, Hsu Y-H, Sangkuhl K, Jenkins G, et al. The association of obesity and coronary artery disease genes with response to SSRIs treatment in major depression. J Neural Transm. 2019;126(1):35–45. doi: 10.1007/s00702-018-01966-x. [DOI] [PubMed] [Google Scholar]
- 17.Bielinski SJ, St Sauver JL, Olson JE, Larson NB, Black JL, Scherer SE, et al. Cohort Profile: The Right Drug, Right Dose, Right Time: Using Genomic Data to Individualize Treatment Protocol (RIGHT Protocol). Int J Epidemiol. 2020;49(1):23–4k. Epub 2019/08/06. doi: 10.1093/ije/dyz123. PubMed PMID: 31378813; PubMed Central PMCID: PMCPMC7124480. [DOI] [PMC free article] [PubMed]
- 18.Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S. PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet Genomics. 2016;26(4):161–8. Epub 2016/01/07. doi: 10.1097/fpc.0000000000000202. PubMed PMID: 26736087; PubMed Central PMCID: PMCPMC4935646. [DOI] [PMC free article] [PubMed]
- 19.Lopes JL, Harris K, Karow MB, Peterson SE, Kluge ML, Kotzer KE, et al. Targeted Genotyping in Clinical Pharmacogenomics: What Is Missing? J Mol Diagn. 2022. Epub 2022/01/19. doi: 10.1016/j.jmoldx.2021.11.008. PubMed PMID: 35041929. [DOI] [PMC free article] [PubMed]
- 20.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Verbeurgt P, Mamiya T, Oesterheld J. How common are drug and gene interactions? Prevalence in a sample of 1143 patients with CYP2C9, CYP2C19 and CYP2D6 genotyping. Pharmacogenomics. 2014;15(5):655–665. doi: 10.2217/pgs.14.6. [DOI] [PubMed] [Google Scholar]
- 22.Blumenthal SR, Castro VM, Clements CC, Rosenfield HR, Murphy SN, Fava M, et al. An electronic health records study of long-term weight gain following antidepressant use. JAMA Psychiat. 2014;71(8):889–896. doi: 10.1001/jamapsychiatry.2014.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aldrich SL, Poweleit EA, Prows CA, Martin LJ, Strawn JR, Ramsey LB. Influence of CYP2C19 metabolizer status on escitalopram/citalopram tolerability and response in youth with anxiety and depressive disorders. Front Pharmacol. 2019;10:99. doi: 10.3389/fphar.2019.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dent R, Blackmore A, Peterson J, Habib R, Kay GP, Gervais A, et al. Changes in body weight and psychotropic drugs: a systematic synthesis of the literature. PLoS ONE. 2012;7(6):e36889. doi: 10.1371/journal.pone.0036889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serretti A, Mandelli L, Laura M. Antidepressants and body weight: a comprehensive review and meta-analysis. The Journal of clinical psychiatry. 2010;71(10):0-. [DOI] [PubMed]
- 26.Robinson E, Hunger J, Daly M. Perceived weight status and risk of weight gain across life in US and UK adults. Int J Obes. 2015;39(12):1721–1726. doi: 10.1038/ijo.2015.143. [DOI] [PubMed] [Google Scholar]
- 27.Fujioka K. Safety and tolerability of medications approved for chronic weight management. Obesity. 2015;23:S7. doi: 10.1002/oby.21094. [DOI] [PubMed] [Google Scholar]
- 28.Fastenau J, Kolotkin RL, Fujioka K, Alba M, Canovatchel W, Traina S. A call to action to inform patient-centred approaches to obesity management: Development of a disease-illness model. Clinical obesity. 2019;9(3):e12309. doi: 10.1111/cob.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashton AK, Jamerson BD, Weinstein WL, Wagoner C. Antidepressant-related adverse effects impacting treatment compliance: results of a patient survey. Curr Ther Res. 2005;66(2):96–106. doi: 10.1016/j.curtheres.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maggo S, Kennedy MA, Barczyk ZA, Miller AL, Rucklidge JJ, Mulder RT, et al. Common CYP2D6, CYP2C9, and CYP2C19 gene variants, health anxiety, and neuroticism are not associated with self-reported antidepressant side effects. Frontiers in Genetics. 2019:1199. [DOI] [PMC free article] [PubMed]
- 31.Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR* D: implications for clinical practice. Am J Psychiatry. 2006;163(1):28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 32.Bradley P, Shiekh M, Mehra V, Vrbicky K, Layle S, Olson MC, et al. Improved efficacy with targeted pharmacogenetic-guided treatment of patients with depression and anxiety: A randomized clinical trial demonstrating clinical utility. J Psychiatr Res. 2018;96:100–107. doi: 10.1016/j.jpsychires.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 33.Winner JG, Carhart JM, Altar CA, Goldfarb S, Allen JD, Lavezzari G, et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr Med Res Opin. 2015;31(9):1633–1643. doi: 10.1185/03007995.2015.1063483. [DOI] [PubMed] [Google Scholar]
- 34.Winner J, Allen J, Altar CA, Spahic-Mihajlovic A. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl Psychiatry. 2013;3(3):e242-e. doi: 10.1038/tp.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winner JG, Carhart JM, Altar A, Allen JD, Dechairo BM. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219–227. [PubMed] [Google Scholar]
- 36.Hall-Flavin D, Winner J, Allen J, Jordan J, Nesheim R, Snyder K, et al. Using a pharmacogenomic algorithm to guide the treatment of depression. Transl Psychiatry. 2012;2(10):e172-e. doi: 10.1038/tp.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh AB. Improved antidepressant remission in major depression via a pharmacokinetic pathway polygene pharmacogenetic report. Clinical Psychopharmacology and Neuroscience. 2015;13(2):150. doi: 10.9758/cpn.2015.13.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jukić MM, Haslemo T, Molden E, Ingelman-Sundberg M. Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: a retrospective study based on 2,087 patients. Am J Psychiatry. 2018;175(5):463–470. doi: 10.1176/appi.ajp.2017.17050550. [DOI] [PubMed] [Google Scholar]
- 39.Steimer W, Zopf K, von Amelunxen S, Pfeiffer H, Bachofer J, Popp J, et al. Amitriptyline or not, that is the question: pharmacogenetic testing of CYP2D6 and CYP2C19 identifies patients with low or high risk for side effects in amitriptyline therapy. Clin Chem. 2005;51(2):376–385. doi: 10.1373/clinchem.2004.041327. [DOI] [PubMed] [Google Scholar]
- 40.Müller DJ, Kekin I, Kao AC, Brandl EJ. Towards the implementation of CYP2D6 and CYP2C19 genotypes in clinical practice: update and report from a pharmacogenetic service clinic. Int Rev Psychiatry. 2013;25(5):554–571. doi: 10.3109/09540261.2013.838944. [DOI] [PubMed] [Google Scholar]
- 41.Fabbri C, Tansey KE, Perlis RH, Hauser J, Henigsberg N, Maier W, et al. Effect of cytochrome CYP2C19 metabolizing activity on antidepressant response and side effects: Meta-analysis of data from genome-wide association studies. Eur Neuropsychopharmacol. 2018;28(8):945–954. doi: 10.1016/j.euroneuro.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Campos AI, Byrne EM, Mitchell BL, Wray NR, Lind PA, Licinio J, et al. Impact of CYP2C19 metaboliser status on SSRI response: a retrospective study of 9500 participants of the Australian Genetics of Depression Study. The Pharmacogenomics Journal. 2022:1–6. [DOI] [PMC free article] [PubMed]
- 43.Aldrich SL, Poweleit EA, Prows CA, Martin LJ, Strawn JR, Ramsey LB. Influence of CYP2C19 metabolizer status on escitalopram/citalopram tolerability and response in youth with anxiety and depressive disorders. Frontiers in Pharmacology. 2019:99. [DOI] [PMC free article] [PubMed]
- 44.Funk KA, Bostwick JR. A comparison of the risk of QT prolongation among SSRIs. Ann Pharmacother. 2013;47(10):1330–1341. doi: 10.1177/1060028013501994. [DOI] [PubMed] [Google Scholar]
- 45.Uher R, Huezo-Diaz P, Perroud N, Smith R, Rietschel M, Mors O, et al. Genetic predictors of response to antidepressants in the GENDEP project. Pharmacogenomics J. 2009;9(4):225–233. doi: 10.1038/tpj.2009.12. [DOI] [PubMed] [Google Scholar]
- 46.FDA. Drug Development and Drug Interactions | Table of Substrates, Inhibitors and Inducers FDA03/10/2020. Available from: https://www.fda.gov/drugs/drug-interactions-labeling/drug-development-and-drug-interactions-table-substrates-inhibitors-and-inducers#table3-3.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 2: Table S1. STROBE reporting check list. Table S2. Variants/star alleles considered in this study. Table S3. Clinical inducers and inhibitors for P450-mediated metabolism considered. [46]. Table S4. Distribution of concomitant use of clinical inducers and inhibitors for P450-mediated metabolism by antidepressant. Table S5. Distribution of comorbidities by metabolizer status. CYP2C9>
Additional file 3: Figure S1. Distribution of phenotypes of cytochromes enzymes involved in the metabolism of citalopram, paroxetine, fluoxetine, and sertraline among the participants.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.


