Abstract
Background
The incidence of allergic reaction is increasing year by year, but the specific mechanism is still unclear. Paeonia lactiflora Pall.(PLP) is a traditional Chinese medicine with various pharmacological effects such as anti-tumor, anti-inflammatory, and immune regulation. Previous studies have shown that PLP has potential anti-allergic activity. However, there is still no comprehensive analysis of the targeted effects and exact molecular mechanisms of the anti-allergic components of PLP. This study aimed to reveal the mechanism of PLP. in the treatment of type I allergy by combining network pharmacological methods and experimental verification.
Methods
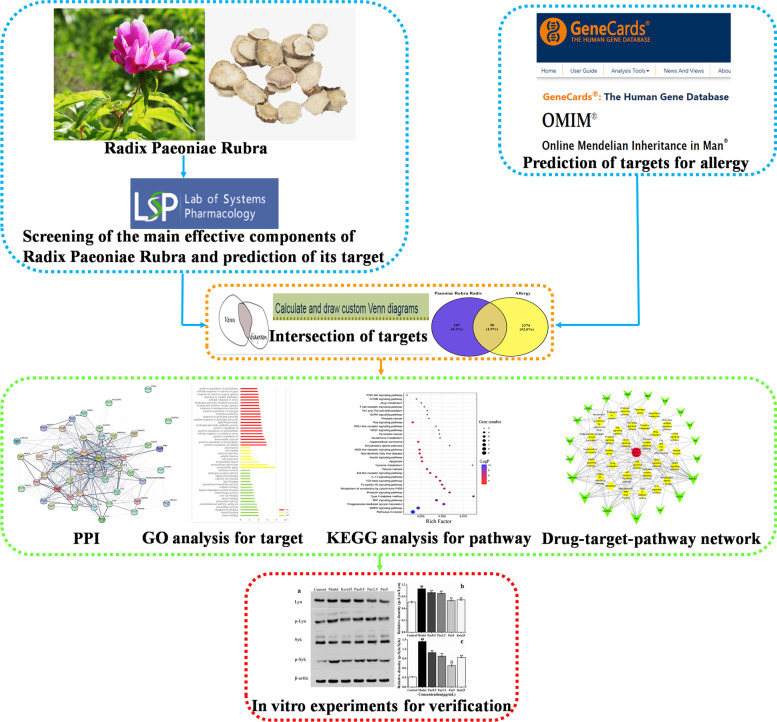
First, we used the traditional Chinese medicine systems pharmacology (TCMSP) database and analysis platform to screen the main components and targets of PLP, and then used databases such as GeneCards to retrieve target information related to ‘allergy’. Protein–protein interaction (PPI) analysis obtained the core target genes in the intersection target, and then imported the intersection target into the David database for gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analysis. Furthermore, the therapeutic effect of paeoniflorin, the main component of PLP, on IgE-induced type I allergy was evaluated in vitro.
Results
GO analysis obtained the main biological processes, cell components and molecular functions involved in the target genes. KEGG analysis screened out MAPK1, MAPK10, MAPK14 and TNF that have a strong correlation with PLP anti-type I allergy, and showed that PLP may pass through signal pathways such as IgE/FcεR I, PI3K/Akt and MAPK to regulate type I allergy. RT-qPCR and Western Blot results confirmed that paeoniflorin can inhibit the expression of key genes and down-regulate the phosphorylation level of proteins in these signal pathways. It further proved the reliability of the results of network pharmacology research.
Conclusion
The results of this study will provide a basis for revealing the multi-dimensional regulatory mechanism of PLP for the treatment of type I allergy and the development of new drugs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12906-022-03677-z.
Keywords: Traditional Chinese medicine, Allergic reaction, IgE/FcεR I
Background
The incidence and mortality of allergic diseases is increasing, and has become a common disease, which greatly affects people's life and physical health. But so far, people have not revealed its exact pathogenesis, and there is no ideal treatment method. At present, glucocorticoid and antihistamine are commonly used clinically to treat allergy, but the curative effect is short and there are many adverse reactions after long-term use [1]. Therefore, it is necessary to continue to explore effective and safe new methods to treat allergic diseases. Last several years, the advantages of traditional Chinese medicine (TCM) with multiple targets and curative effects, and less adverse reactions have attracted the attention of many researchers. TCM is becoming a hot spot in the research and development of drugs to treat allergic diseases [2].
TCM has been used for the treatment of allergic diseases with long history. But due to the complex chemical components and pharmacological effects of TCM, its specific effective substance basis and mechanism are still unclear, which brings huge challenges to the study of the mechanism of TCM to treat allergy. For the past few years, with the in-depth research of TCM and the development of related technologies, the use of TCM in treatment of allergic diseases has gained great recognition and breakthroughs. It has been found that many TCMs and their components have therapeutic effects on allergy, such as Polydatin, Glycyrrhizic acid and Quercetin [3–5]. Treasury of TCM has huge potential for new drug research, and shows excellent application prospects to treat allergy. However, there is still a great deal of potential TCMs with anti-allergic activity waiting to be explored, such as PLP.
The medicinal part of PLP is its dried root, and it has many pharmacological effects such as protecting liver, nerve and heart, anti-tumor, anti-inflammatory and immune regulation. The main active ingredient of PLP is Paeoniflorin (Pae) [6, 7]. Studies have confirmed that PLP and Pae have potential anti-allergic activity [8, 9]. In view of the complexity of the cell signal network involved in allergy, these conclusions should be part of the mechanism for its effectiveness. So the molecular mechanism and specific biological process of PLP anti-allergy still need to be further elucidated. The purpose of this study was to explore the regulation mechanism of multiple genes and multiple pathways in the treatment of type I allergy with PLP.
Network pharmacology is a research method based on multi-directional pharmacology and systems biology, which can analyze the relationship between drugs and diseases at the overall level. Network pharmacology is based on the drug-target-disease network, so as to systematically explore the specific mechanisms of drug to treat diseases. Its greatest advantage is the integration of holistic, dynamic and analysis, which is consistent with the holistic and dialectical treatment principles of TCM [10].
Consequently, our research was based on the network pharmacology to systematically analyze the active ingredients of PLP, allergy-related targets and their pathways to identify potential drug targets and mechanisms. Type I allergy is the most common type of allergy in clinical practice. We used cell models and in vitro experiments to explore the effects and related mechanisms of Pae, the main active ingredient of PLP, in treating type I allergy. Most reports on the relationship between Pae and allergy only focused on showing the inhibitory effects of this compound and lacked in-depth exploration of the underlying mechanism [11, 12].Therefore, in this study, the combined approaches offer deep understanding of the pharmacological mechanisms of PLP, and may provide a novel and efficient way to discover the pharmacological basis and medicinal value of PLP.
Materials and methods
Materials
RBL-2H3 cells were obtained from the ATCC. PrimeScript™ RT reagent Kit, TB Green Kit were purchased from Takara (Beijing, China). The finished product of Paeoniflorin (HPLC ≥ 98%, and is usually extracted from the root of PLP) were purchased from Solarbio (Beijing, China).
Network pharmacology analysis
Screening of the main active ingredients of PLP and acquisition of its targets
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) is a database established based on the framework of TCM system pharmacology, providing 12 important pharmacokinetic properties, such as oral bioavailability (OB) and drug-likeness (DL), which is mainly used to screen and evaluation of pharmaceutical compounds. OB is an important indicator for evaluating whether a drug can be developed, and OB ≥ 30% is considered to have better oral bioavailability. DL can evaluate the possibility of a compound becoming a drug, and DL ≥ 0.18 is considered to have high drug-likeness and may become a new drug [13]. Our method and operation were carried out with reference to relevant literature [14, 15], and the specific steps were as follows: The PLP was imported into the TCMSP database (https://tcmspw.com/tcmsp.php), and all known chemical components contained in the PLP have been retrieved and screened for potential activities, that is, OB ≥ 30%, DL ≥ 0.18. According to the active ingredients obtained after screening, the TCMSP database is used again to retrieve its target.
Acquisition of targets for allergy
The GeneCards (https://www.genecards.org/) database is not only a database that can provide concise genome, proteome, transcription, inheritance and function of all known and predicted human genes, but also an analytical database that combines retrieval, integration, search and display of the information of the human genome [16]. The OMIM database (http://omim.org/) catalogs the genetic components of all known diseases and links them with related genes in the human genome when possible. It provides a reference for further research and genomic analysis tools of cataloging genes [17]. In these two databases, searched with ‘allergy’ as a keyword to find the target of allergy.
Establishment and analysis of protein–protein interaction (PPI) network
Used the Draw Venn database (http://bioinformatics.psb.ugent.be/webtools/Venn/) to take the intersection of the targets obtained in 2.2.1 and 2.2.2, and imported it into the String database (https://string-db.org/). Then used ‘Multiple proteins’ function to establish the PPI network, selected the species as ‘Homo sapiens’, and clicked ‘SEARCH’ and ‘CONTINUE’ options to get the PPI network.
Analysis of biological processes and pathway enrichment
The David database (https://david.ncifcrf.gov/) can be used for enrichment analysis of a great quantity of sample genes and proteins, also can simultaneously provide systematic and comprehensive biological information. Through the integration and analysis of information, we can intuitively show the pathway enrichment of target genes, which has become one of the indispensable tools of bioinformatics research. Imported the target obtained in 2.2.3 into the David database for Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. GO analysis is a description of genes in different dimensions and levels, which includes three aspects: biological process (BP), cell component (CC) and molecular function (MF). KEGG is a database that links gene catalogs obtained from fully sequenced genomes with system functions of higher-level cell, species, and ecosystem. KEGG analysis discovers the pathways of drug targets by enriching target genes, thereby obtaining the mechanism of drug treatment of diseases [18]. Selected the species as ‘Homo sapiens’, and conducted target analysis through MF, BP, and CC in GO. Simultaneously selected KEGG in Pathway for pathway analysis, and screened the results with the -LogP ≥ 2 for analysis.
Network establishment
Cytoscape is a mapping software that can be used to establish, analyze, and visualize complex networks. It is often used to analyze the results of network pharmacology. Used Excel to establish data sets of PLP-signal pathway and signal pathway-target, and imported them into Cytoscape to establish the network of PLP-target-signal pathway.
In vitro experiments
Western Blot analysis
Our experimental method was performed with reference to relevant literature [19, 20], and the specific steps were as follows: After culturing RBL-2H3 cells (5 × 105 cells/mL) for 24 h, each group was sensitized with 1 mL of DNP-IgE (0.2 μg/mL). After 12 h, drug groups were replaced with 2 mL of the corresponding drug respectively (Pae 0.5, 2.5, 5 μg/mL, Keto 25 μg/mL). After 1 h, in addition to the normal group, 400 μL of DNP-BSA (0.4 μg/mL) was added for stimulation. After 30 min, extracted total protein and measured its concentration.
The experiment used 8% separating gel, 4% stacking gel, and loaded 30 μg protein sample. After electrophoresis, the cut gel was transferred to the PVDF membrane. The PVDF membrane was blocked with shaking at room temperature for 1 h. After incubation with primary antibodies of Lyn, p-Lyn, Syk, p-Syk and β-actin at 4℃ overnight, the secondary antibodies were incubated at room temperature for 1 h. Visualization was performed by using the ChemiScope Mini 3300 and density analysis was performed with Image J software.
qPCR
The steps were the same as 2.3.1. Then extracted total RNA, removed gDNA from RNA and performed reverse transcription by using PrimeScript™ RT reagent Kit. Used TB Green kit for qPCR reaction. The key genes tested include: Lyn, Syk, Fyn, PLCγ, PI3K, Akt, p38, ERK, JNK, p65 and GAPDH.
Statistical analysis
Results were expressed as the mean ± SD. ANOVA in SPSS 17.0 software was used to assess significant differences between groups (p < 0.05).
Results
Main active ingredients of PLP and its targets
As shown in Table 1, there are 29 main active ingredients of PLP, including Pae, and 157 targets obtained from the TCMSP database.
Table 1.
The main active ingredients of PLP
| Mol ID | Molecule Name | OB (%) | DL | |
|---|---|---|---|---|
| 1 | MOL001002 | ellagic acid | 43.06 | 0.43 |
| 2 | MOL001918 | paeoniflorgenone | 87.59 | 0.37 |
| 3 | MOL001921 | Lactiflorin | 49.12 | 0.8 |
| 4 | MOL001924 | paeoniflorin | 53.87 | 0.79 |
| 5 | MOL001925 | paeoniflorin_qt | 68.18 | 0.4 |
| 6 | MOL002714 | baicalein | 33.52 | 0.21 |
| 7 | MOL002776 | Baicalin | 40.12 | 0.75 |
| 8 | MOL000358 | beta-sitosterol | 36.91 | 0.75 |
| 9 | MOL000359 | sitosterol | 36.91 | 0.75 |
| 10 | MOL004355 | Spinasterol | 42.98 | 0.76 |
| 11 | MOL000449 | Stigmasterol | 43.83 | 0.76 |
| 12 | MOL000492 | ( +)-catechin | 54.83 | 0.24 |
| 13 | MOL006990 | (1S,2S,4R)-trans-2-hydroxy-1,8-cineole-B-D-glucopyranoside | 30.25 | 0.27 |
| 14 | MOL006992 | (2R,3R)-4-methoxyl-distylin | 59.98 | 0.3 |
| 15 | MOL006994 | 1-o-beta-d-glucopyranosyl-8-o-benzoylpaeonisuffrone_qt | 36.01 | 0.3 |
| 16 | MOL006996 | 1-o-beta-d-glucopyranosylpaeonisuffrone_qt | 65.08 | 0.35 |
| 17 | MOL006999 | stigmast-7-en-3-ol | 37.42 | 0.75 |
| 18 | MOL007003 | benzoyl paeoniflorin | 31.14 | 0.54 |
| 19 | MOL007004 | Albiflorin | 30.25 | 0.77 |
| 20 | MOL007005 | Albiflorin_qt | 48.7 | 0.33 |
| 21 | MOL007008 | 4-ethyl-paeoniflorin_qt | 56.87 | 0.44 |
| 22 | MOL007012 | 4-o-methyl-paeoniflorin_qt | 56.7 | 0.43 |
| 23 | MOL007014 | 8-debenzoylpaeonidanin | 31.74 | 0.45 |
| 24 | MOL007016 | Paeoniflorigenone | 65.33 | 0.37 |
| 25 | MOL007018 | 9-ethyl-neo-paeoniaflorin A_qt | 64.42 | 0.3 |
| 26 | MOL007022 | evofolinB | 64.74 | 0.22 |
| 27 | MOL007025 | isobenzoylpaeoniflorin | 31.14 | 0.54 |
| 28 | MOL002883 | Ethyl oleate (NF) | 32.4 | 0.19 |
| 29 | MOL005043 | campest-5-en-3beta-ol | 37.58 | 0.71 |
Target of allergy
Through GeneCards and OMIM database searched, 2424 targets related to ‘allergy’ were obtained (Too much data to show).
Analysis of PPI network
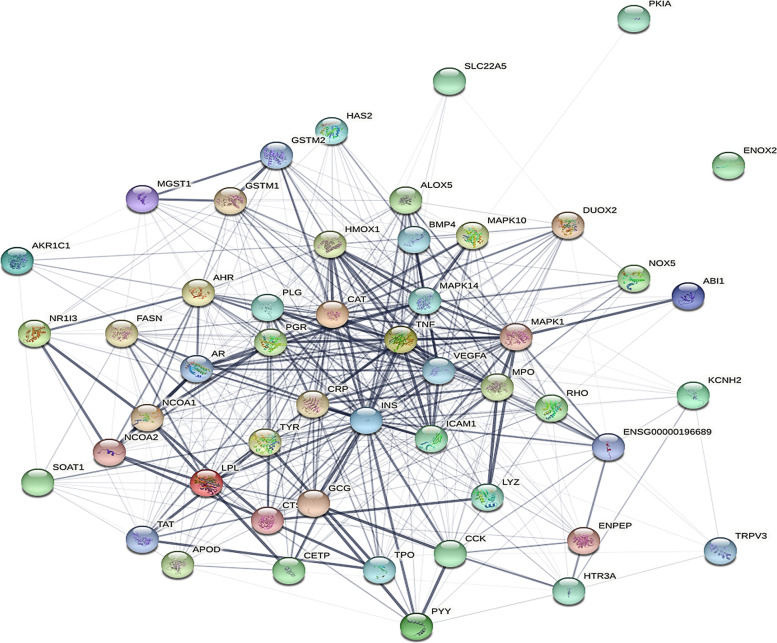
Imported the two target sets obtained in 2.2.1 and 2.2.2 into the Draw Venn database to obtain the intersection (Fig. 1). It is found that there are 50 potential targets of PLP in allergy (as shown in Table 2), which were imported into the String database to establish PPI (as shown in Fig. 2), among which the top 5 interaction relationships according to the number are: INS, TNF, CAT, MAPK1 and VEGFA (Fig. 3).
Fig. 1.
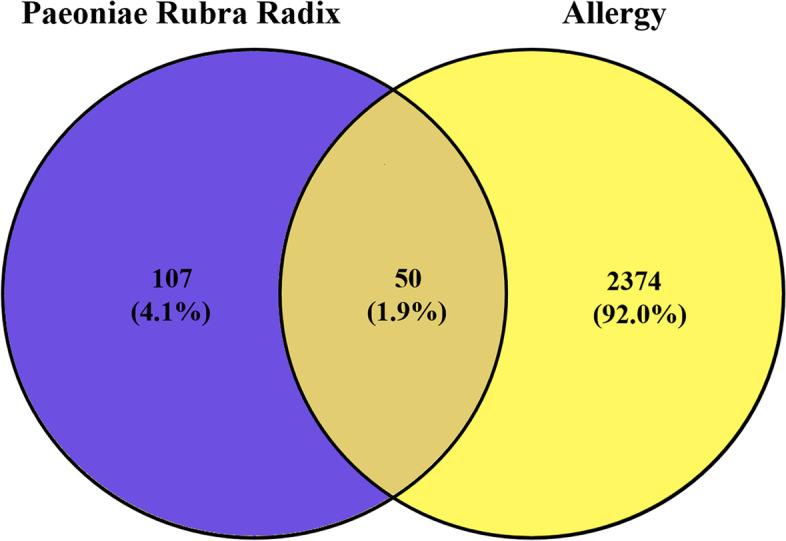
Intersection of the targets of PLP and allergy
Table 2.
The potential targets of PLP in allergy
| Target name | Gene Symbol | |
|---|---|---|
| 1 | androgen receptor | AR |
| 2 | progesterone receptor | PGR |
| 3 | vascular endothelial growth factor a | VEGFA |
| 4 | glutathione s-transferase mu 1 | GSTM1 |
| 5 | transient receptor potential cation channel subfamily v member 1 | TRPV1 |
| 6 | arachidonate 5-lipoxygenase | ALOX5 |
| 7 | catalase | CAT |
| 8 | plasminogen | PLG |
| 9 | thyroid peroxidase | TPO |
| 10 | tumor necrosis factor | TNF |
| 11 | myeloperoxidase | MPO |
| 12 | aryl hydrocarbon receptor | AHR |
| 13 | potassium voltage-gated channel subfamily h member 2 | KCNH2 |
| 14 | 5-hydroxytryptamine receptor 3a | HTR3A |
| 15 | mitogen-activated protein kinase 14 | MAPK14 |
| 16 | cathepsin d | CTSD |
| 17 | solute carrier family 22 member 5 | SLC22A5 |
| 18 | mitogen-activated protein kinase 1 | MAPK1 |
| 19 | intercellular adhesion molecule 1 | ICAM1 |
| 20 | tyrosinase | TYR |
| 21 | c-reactive protein | CRP |
| 22 | insulin | INS |
| 23 | glucagon | GCG |
| 24 | cholecystokinin | CCK |
| 25 | cholesteryl ester transfer protein | CETP |
| 26 | peptide yy | PYY |
| 27 | nuclear receptor subfamily 1 group i member 3 | NR1I3 |
| 28 | hemeoxygenase 1 | HMOX1 |
| 29 | glutathione s-transferase mu 2 | GSTM2 |
| 30 | lysozyme | LYZ |
| 31 | nuclear receptor coactivator 2 | NCOA2 |
| 32 | fatty acid synthase | FASN |
| 33 | aldo-ketoreductase family 1 member c1 | AKR1C1 |
| 34 | tyrosine aminotransferase | TAT |
| 35 | nuclear receptor coactivator 1 | NCOA1 |
| 36 | nadph oxidase 5 | NOX5 |
| 37 | apolipoprotein d | APOD |
| 38 | hyaluronan synthase 2 | HAS2 |
| 39 | microsomal glutathione s-transferase 1 | MGST1 |
| 40 | rhodopsin | RHO |
| 41 | transient receptor potential cation channel subfamily v member 3 | TRPV3 |
| 42 | dual oxidase 2 | DUOX2 |
| 43 | mitogen-activated protein kinase 10 | MAPK10 |
| 44 | ablinteractor 1 | ABI1 |
| 45 | lipoprotein lipase | LPL |
| 46 | sterol o-acyltransferase 1 | SOAT1 |
| 47 | bone morphogenetic protein 4 | BMP4 |
| 48 | camp-dependent protein kinase inhibitor alpha | PKIA |
| 49 | ecto-nox disulfide-thiol exchanger 2 | ENOX2 |
| 50 | glutamylaminopeptidase | ENPEP |
Fig. 2.
PPI network of PLP-allergy target
Fig. 3.
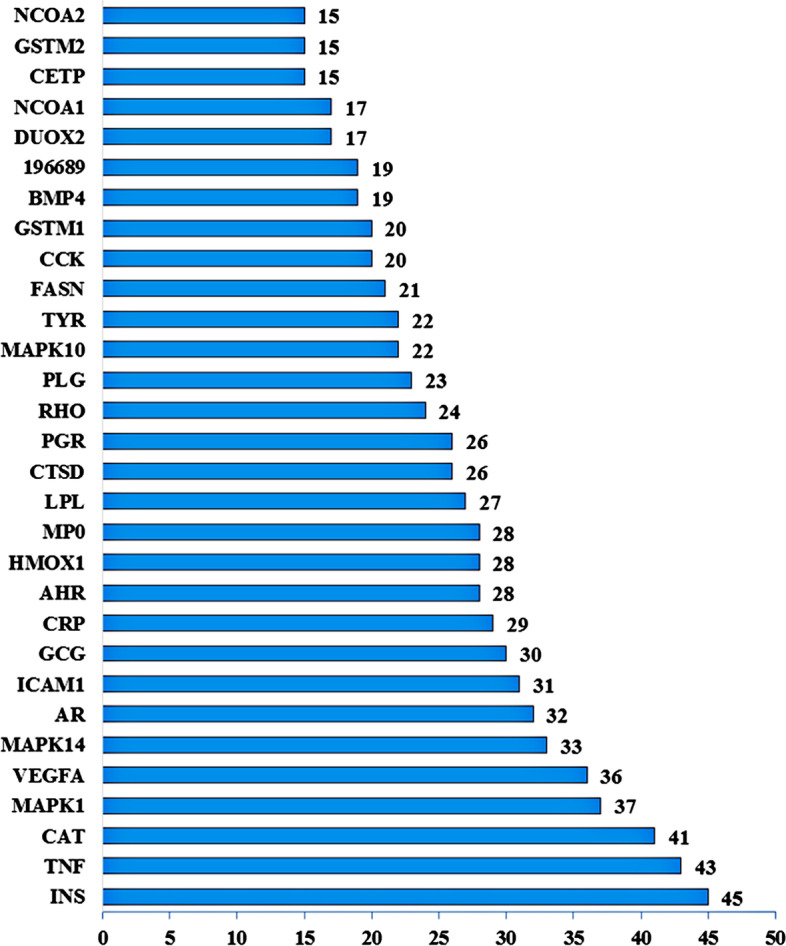
PPI network of top 30 target genes
Analysis of biological process and pathway enrichment
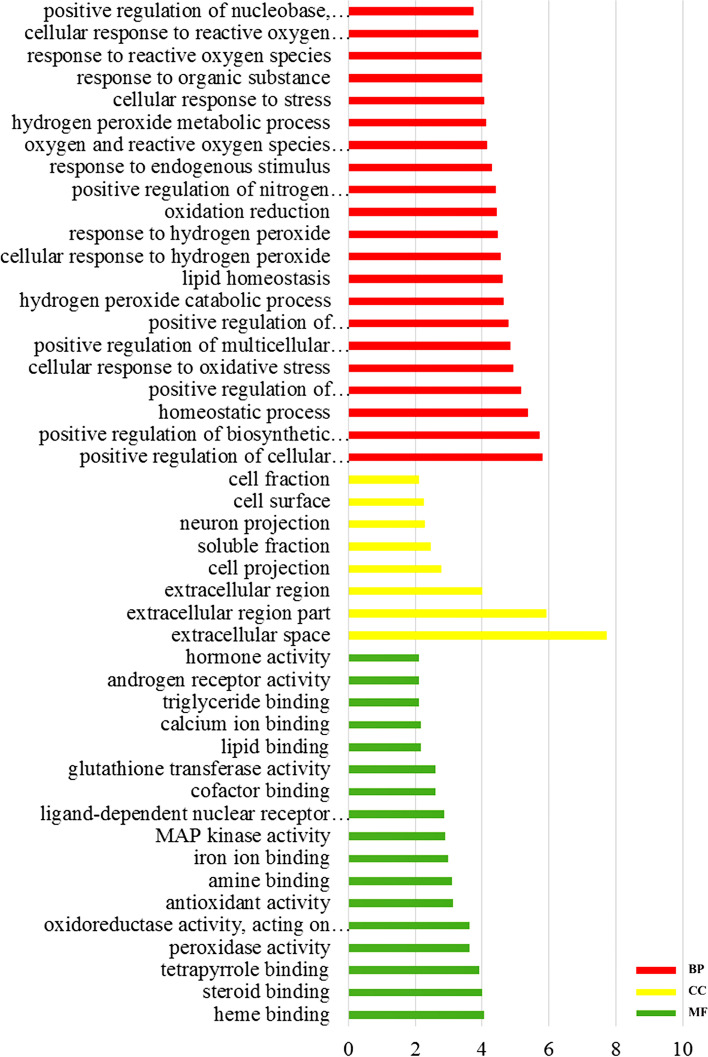
Imported the obtained 50 intersection targets into the David database for GO and KEGG analysis. As shown in Table 3, GO-BP analysis obtained 235 results of PLP anti-allergic effects, 109 of them are -LogP ≥ 2, and the biological processes with the number of genes ≥ 18 are mainly: positive regulation of cell biosynthesis process, positive regulation of polymer biosynthesis and metabolic process, redox, regulation of cell death and apoptosis, transcription regulation, regulation of RNA metabolic process, intracellular signal cascade and so on. GO-CC analysis obtained 27 results, 8 of them are -LogP ≥ 2, and these cell locations with the number of genes ≥ 10 mainly include the extracellular region and the plasma membrane. GO-MF analysis obtained 41 results, and 17 of them are -LogP ≥ 2. The molecular processes involved are antioxidant activity, MAPK activity, binding of Ca2+ and triglycerides and so on. The process with the number of genes ≥ 10 is binding of Ca2+. The visual processing was showed in Fig. 4.
Table 3.
GO analysis of anti-allergic reactions of PLP
| Name | -LogP | |
|---|---|---|
| BP | positive regulation of cellular biosynthetic process | 5.799403 |
| BP | positive regulation of biosynthetic process | 5.733428 |
| BP | homeostatic process | 5.383481 |
| BP | positive regulation of macromolecule biosynthetic process | 5.155981 |
| BP | positive regulation of macromolecule metabolic process | 4.799018 |
| BP | oxidation reduction | 4.436033 |
| BP | positive regulation of nitrogen compound metabolic process | 4.407209 |
| BP | cellular response to stress | 4.071929 |
| BP | response to organic substance | 3.994682 |
| BP | positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 3.749874 |
| BP | regulation of cell death | 3.558830 |
| BP | regulation of transcription from RNA polymerase II promoter | 3.25929 |
| BP | regulation of apoptosis | 2.946298 |
| BP | regulation of programmed cell death | 2.915996 |
| BP | regulation of transcription, DNA-dependent | 2.394503 |
| BP | regulation of RNA metabolic process | 2.310271 |
| BP | intracellular signaling cascade | 2.143719 |
| BP | regulation of transcription | 2.129554 |
| CC | extracellular space | 7.734715 |
| CC | extracellular region part | 5.92365 |
| CC | extracellular region | 3.996677 |
| CC | cell projection | 2.76938 |
| CC | soluble fraction | 2.461935 |
| CC | neuron projection | 2.298664 |
| CC | cell surface | 2.26695 |
| CC | cell fraction | 2.123144 |
| MF | heme binding | 4.051026 |
| MF | steroid binding | 4.016238 |
| MF | tetrapyrrole binding | 3.919531 |
| MF | peroxidase activity | 3.634473 |
| MF | oxidoreductase activity, acting on peroxide as acceptor | 3.634473 |
| MF | antioxidant activity | 3.137301 |
| MF | amine binding | 3.113086 |
| MF | iron ion binding | 2.997328 |
| MF | MAP kinase activity | 2.908816 |
| MF | ligand-dependent nuclear receptor activity | 2.870518 |
| MF | cofactor binding | 2.621835 |
| MF | glutathione transferase activity | 2.595378 |
| MF | lipid binding | 2.177942 |
| MF | calcium ion binding | 2.170265 |
| MF | triglyceride binding | 2.122953 |
| MF | androgen receptor activity | 2.122953 |
| MF | hormone activity | 2.107126 |
Fig. 4.
Results of GO analysis
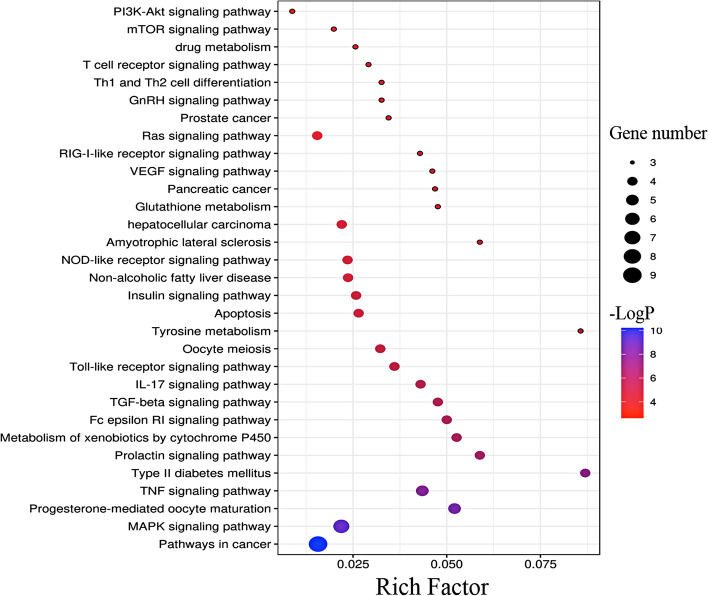
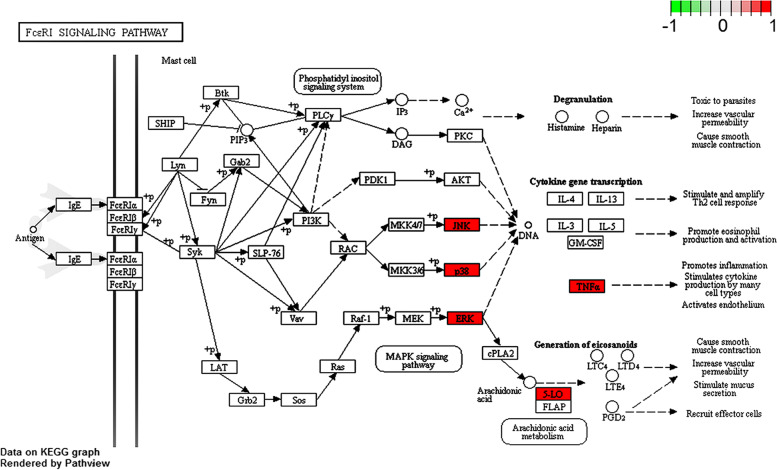
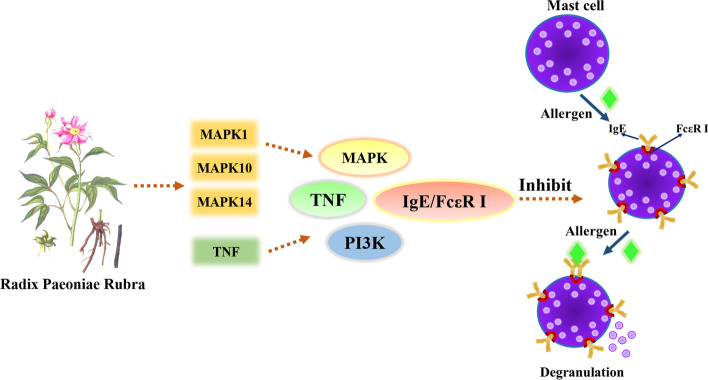
Through KEGG analysis, 31 related pathways were obtained (Table 4). The top 13 signal pathways according to the number of genes mainly include: tumor-related signal pathway, MAPK signal pathway, TNF signal pathway, liver cancer signal pathway, type II diabetes-related signal pathway, lactation signal pathway, FcεR I signal pathway and IL-17 signal pathway. In addition, the anti-allergic effect of PLP may also be related to Th cell differentiation and PI3K/Akt signal pathway. Visualized the above-mentioned signal pathways with the Metascape database (http://metascape.org/gp/index.html), and obtained the bubble chart of related pathways of PLP anti-allergic effect (Fig. 5), in which the values of Rich Factor and -LogP both are positively correlated with the degree of enrichment. Moreover, the important targets of MAPK 1, MAPK 10, MAPK 14 and TNF are mainly distributed in the FcεR I signal pathway that is related to allergic reaction (Fig. 6, and the copyright of this KEGG pathway picture belongs to Kanehisa Laboratory).
Table 4.
KEGG analysis of anti-allergic reactions of PLP
| Pathway name | -LogP | Gene number | |
|---|---|---|---|
| 1 | Pathways in cancer | 10.3275 | 9 |
| 2 | MAPK signaling pathway | 8.970662 | 7 |
| 3 | Progesterone-mediated oocyte maturation | 8.320762 | 5 |
| 4 | TNF signaling pathway | 7.92438 | 5 |
| 5 | Metabolism of xenobiotics by cytochrome P450 | 6.724768 | 4 |
| 6 | hepatocellular carcinoma | 5.207091 | 4 |
| 7 | Type II diabetes mellitus | 7.614715 | 4 |
| 8 | Prolactin signaling pathway | 6.920665 | 4 |
| 9 | Fc epsilon RI signaling pathway | 6.634641 | 4 |
| 10 | IL-17 signaling pathway | 6.370812 | 4 |
| 11 | Toll-like receptor signaling pathway | 6.062156 | 4 |
| 12 | Apoptosis | 5.528667 | 4 |
| 13 | Insulin signaling pathway | 5.483541 | 4 |
| 14 | Non-alcoholic fatty liver disease | 5.33452 | 4 |
| 15 | NOD-like receptor signaling pathway | 5.324366 | 4 |
| 16 | Ras signaling pathway | 4.60425 | 4 |
| 17 | TGF-beta signaling pathway | 6.549032 | 4 |
| 18 | Oocyte meiosis | 5.869675 | 4 |
| 19 | Glutathione metabolism | 4.988936 | 3 |
| 20 | drug metabolism | 4.182712 | 3 |
| 21 | Pancreatic cancer | 4.968272 | 3 |
| 22 | VEGF signaling pathway | 4.947937 | 3 |
| 23 | RIG-I-like receptor signaling pathway | 4.850846 | 3 |
| 24 | Th1 and Th2 cell differentiation | 4.494327 | 3 |
| 25 | GnRH signaling pathway | 4.494327 | 3 |
| 26 | T cell receptor signaling pathway | 4.347688 | 3 |
| 27 | mTOR signaling pathway | 3.854131 | 3 |
| 28 | PI3K-Akt signaling pathway | 2.819689 | 3 |
| 29 | Prostate cancer | 4.567031 | 3 |
| 30 | Tyrosine metabolism | 5.766723 | 3 |
| 31 | Amyotrophic lateral sclerosis | 5.267027 | 3 |
Fig. 5.
Enrichment analysis of pathways
Fig. 6.
Important target genes are mainly distributed in the FcεR I signal pathway
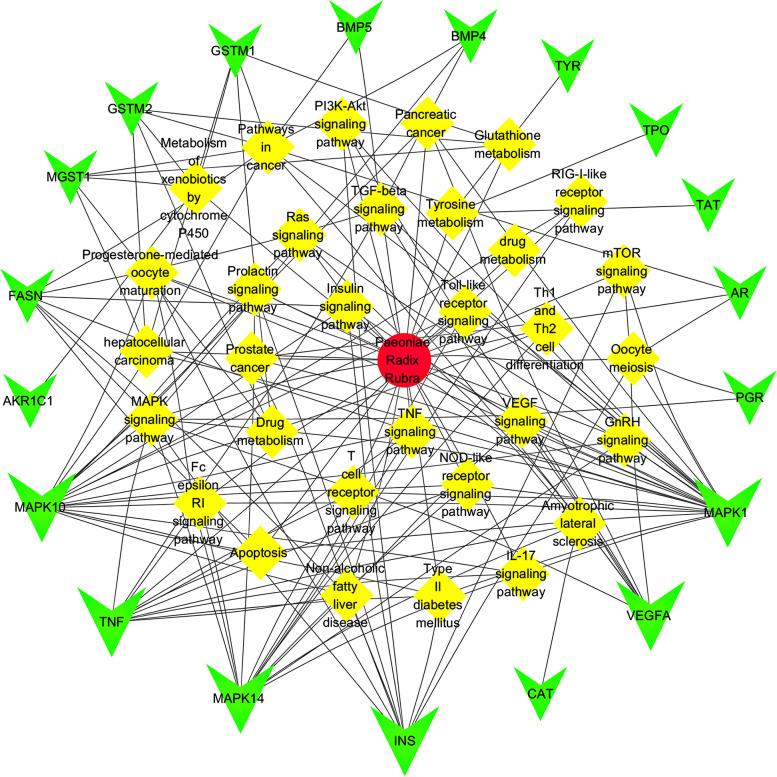
Network of PLP-target-signal pathway
Cytoscape was used for establish the network of PLP-target-signal pathway (Fig. 7). Red represents PLP, yellow represents signal pathway, and green represents intersection target. There are 52 nodes and 153 edges in this figure. In topological metrics analysis, node centrality is a widely used measurement with three main metrics: degree, closeness, and betweeness. These three topological metrics were selected as candidate targets. After comprehensively analyzing the values of the three metrics for each target in this network, it was found that the top four targets were MAPK 1, MAPK 10, MAPK 14 and TNF (Table 5). Therefore, they are considered as important candidate targets of PLP for the treatment of allergy.
Fig. 7.
Network of PLP-target-pathway
Table 5.
Topological metrics analysis of network
| Nude | Degree | Closeness | Betweeness | |
|---|---|---|---|---|
| 1 | MAPK 1 | 24 | 0.53125 | 0.15264486 |
| 2 | MAPK 10 | 18 | 0.47222222 | 0.06938654 |
| 3 | MAPK 14 | 14 | 0.4047619 | 0.03426261 |
| 4 | TNF | 12 | 0.3984375 | 0.02919505 |
| 5 | Pathways in cancer | 10 | 0.43965517 | 0.09264678 |
| 6 | MAPK signaling pathway | 10 | 0.49038462 | 0.06359126 |
| 7 | INS | 10 | 0.38059701 | 0.01816986 |
| 8 | VEGFA | 8 | 0.3984375 | 0.0178217 |
| 9 | TGF-beta signaling pathway | 7 | 0.46363636 | 0.04071533 |
| 10 | FASN | 7 | 0.39230769 | 0.01303181 |
| 11 | TNF signaling pathway | 6 | 0.45535714 | 0.01066349 |
| 12 | Metabolism of xenobiotics by cytochrome P450 | 5 | 0.45535714 | 0.06255236 |
| 13 | hepatocellular carcinoma | 5 | 0.45535714 | 0.03820949 |
| 14 | Oocyte meiosis | 5 | 0.44736842 | 0.02674978 |
| 15 | GSTM 1 | 5 | 0.36428571 | 0.01750897 |
| 16 | GSTM 2 | 5 | 0.36428571 | 0.01750897 |
| 17 | MGST 1 | 5 | 0.36428571 | 0.01750897 |
| 18 | Non-alcoholic fatty liver disease | 5 | 0.44736842 | 0.00885468 |
| 19 | Insulin signaling pathway | 5 | 0.44736842 | 0.00812833 |
| 20 | Apoptosis | 5 | 0.44736842 | 0.00743339 |
| 21 | Type II diabetes mellitus | 5 | 0.44736842 | 0.00686433 |
| 22 | Progesterone-mediated oocyte maturation | 5 | 0.44736842 | 0.00659734 |
| 23 | Prolactin signaling pathway | 5 | 0.44736842 | 0.00659734 |
| 24 | Fc epsilon R I signaling pathway | 5 | 0.44736842 | 0.00553748 |
| 25 | IL-17 signaling pathway | 5 | 0.44736842 | 0.00553748 |
| 26 | Toll-like receptor signaling pathway | 5 | 0.44736842 | 0.00553748 |
| 27 | NOD-like receptor signaling pathway | 5 | 0.44736842 | 0.00553748 |
| 28 | Tyrosine metabolism | 4 | 0.43220339 | 0.11529412 |
| 29 | Amyotrophic lateral sclerosis | 4 | 0.43220339 | 0.04152907 |
| 30 | Glutathione metabolism | 4 | 0.44736842 | 0.02333667 |
| 31 | Prostate cancer | 4 | 0.43965517 | 0.01329679 |
| 32 | mTOR signaling pathway | 4 | 0.43965517 | 0.00569641 |
| 33 | PI3K-Akt signaling pathway | 4 | 0.43965517 | 0.00569641 |
| 34 | VEGF signaling pathway | 4 | 0.43965517 | 0.00551107 |
| 35 | Pancreatic cancer | 4 | 0.43965517 | 0.00455056 |
| 36 | Ras signaling pathway | 4 | 0.43965517 | 0.00455056 |
| 37 | T cell receptor signaling pathway | 4 | 0.43965517 | 0.00387705 |
| 38 | Th1 and Th2 cell differentiation | 4 | 0.43965517 | 0.00311393 |
| 39 | GnRH signaling pathway | 4 | 0.43965517 | 0.00311393 |
| 40 | AR | 3 | 0.35915493 | 0.00368863 |
| 41 | BMP 4 | 3 | 0.36956522 | 0.00260806 |
| 42 | RIG-I-like receptor signaling pathway | 3 | 0.43220339 | 0.00193453 |
| 43 | Drug metabolism | 3 | 0.27419355 | 4.71E-04 |
| 44 | BMP 5 | 2 | 0.34931507 | 7.81E-04 |
| 45 | PGR | 2 | 0.34 | 5.39E-04 |
| 46 | drug metabolism | 1 | 0.41129032 | 0 |
| 47 | AKR1C1 | 1 | 0.31481481 | 0 |
| 48 | TYR | 1 | 0.30357143 | 0 |
| 49 | TPO | 1 | 0.30357143 | 0 |
| 50 | TAT | 1 | 0.30357143 | 0 |
| 51 | CAT | 1 | 0.30357143 | 0 |
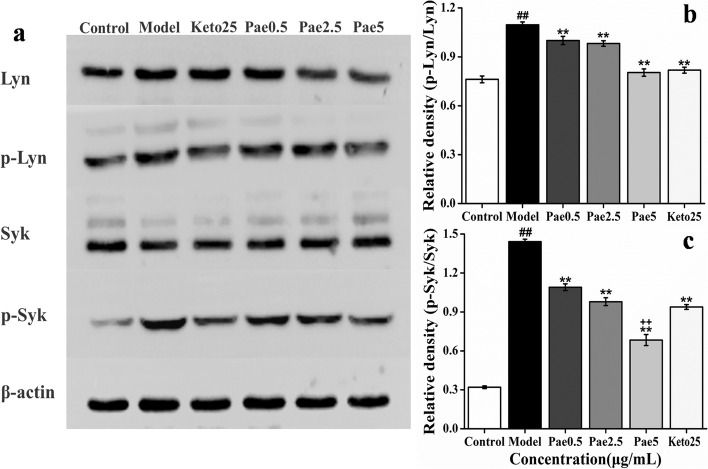
Pae can inhibit the phosphorylation of Lyn and Syk proteins when RBL-2H3 cells degranulation
Pae can inhibit the phosphorylation levels of Lyn and Syk proteins during the degranulation of RBL-2H3 cells in a dose-dependent manner (Fig. 8 and Additional file 1, 2, 3, 4, 5: Fig.S1-5). The inhibitory effect of 5 μg/mL Pae on phosphorylation of Syk protein was significantly stronger than positive control group (Keto group).
Fig. 8.
Effect of Pae on the phosphorylation of Lyn and Syk (n = 3). a Western Blot detected the phosphorylation of Lyn and Syk in RBL-2H3 cells. b Density analysis of Lyn. c Density analysis of Syk. ##p < 0.01 vs control; **p < 0.01 vs model; ++p < 0.01 vs Keto
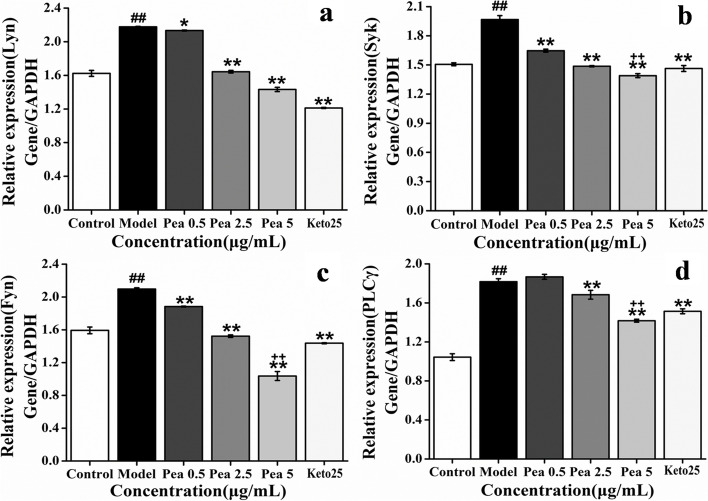
Effect of Pae on the expression of genes when RBL-2H3 cells degranulation
Pae can inhibit the expression of Lyn, Syk, Fyn and PLCγ genes when the degranulation of RBL-2H3 cells in a dose-dependent manner (Fig. 9). The inhibitory effect of 5 μg/mL Pae on Syk, Fyn and PLCγ was stronger than Keto group.
Fig. 9.
Effect of Pae on the expression of Lyn, Syk, Fyn and PLCγ in the IgE signal pathway (n = 3). a Lyn; b Syk; c Fyn; d PLCγ. ##p < 0.01 vs control; *p < 0.05, **p < 0.01 vs model; ++p < 0.01 vs Keto
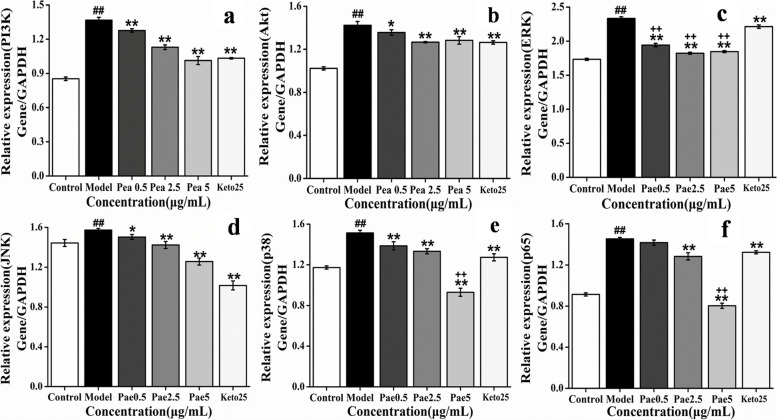
Pae can inhibit the expression of PI3K, Akt, ERK, JNK, p38 and p65 genes when the degranulation of RBL-2H3 cells in a dose-dependent manner (except Akt and ERK). The inhibitory effect of 5 μg/mL Pae on ERK, p38 and p65 was stronger than Keto group (Fig. 10).
Fig. 10.
Effect of Pae on the expression of PI3K, Akt, ERK, JNK, p38 and p65 (n = 3). (a) PI3K; (b) Akt; (c) ERK; (d) JNK; (e) p38; (f) p65.##p < 0.01 vs control; *p < 0.05, **p < 0.01 vs model; ++p < 0.01 vs Keto
Discussion
The characteristics of multi-component, multi-target and the interaction of each component of TCM make it a complex system, and network pharmacology is a more comprehensive and systematic research technology that aims to reveal the complexity of biological systems, drugs and diseases, which has certain similarities with TCM, and is becoming a hot spot in TCM research [21]. Zhang Z Y [14] used the method of network pharmacology to obtain the key targets and possible mechanisms of Siwu Decoction to treat breast cancer, which provided a basis for the development of anti-breast cancer drugs. Changying J [15] successfully predicted the active ingredients and main targets of Qinghuo Rougan Decoction to treat uveit is through network pharmacology. Because network pharmacology is particularly suitable for reflecting and explaining the interaction of multi-component and multi-targets of TCM, it points out a novel direction for the modernization research of TCM, and is expected to bring novel opportunities for promoting the exploration of the multi-component mechanism of TCM and the development of modern TCM.
As one of the TCMs that can be used in dietary supplement, PLP has been found to have anti-inflammatory, anti-tumor and immune regulation effects. So it has been widely used to treat many diseases. PLP is often combined with other TCMs in the treatment of allergy. Shaoyao Gancao Decoction and Xiaoqinglong Decoction are classic prescriptions with anti-allergic effects and have good therapeutic effects, and both contain PLP. Therefore, it is speculated that PLP may have anti-allergic activity, but the mechanism of its treatment of allergy has not been fully understood. However, considering that PLP has the characteristics of multiple components and multiple targets based on the theory of TCM, experimental research alone cannot systematically reveal the biological mechanism of PLP anti-type I allergy, and the holistic characteristics of network pharmacology are suitable for this research. Different from previous studies, this research used network pharmacology to predict the efficacious ingredients and key mechanisms of PLP anti-type I allergy, and then conducted in vitro experiments for verification.
The TCMSP database contains 499 TCMs included in the Chinese Pharmacopoeia and their 29,384 components, 3311 targets and 837 related diseases. Each component provides pharmacokinetic data, as well as potential targets and related disease information, so that the relationship network of drug-target-disease can be obtained, which provides a new platform for the in-depth study of the pharmacological mechanism of TCM [22]. In order to obtain more accurate compounds for more in-depth research, we selected compounds with OB ≥ 30% and DL ≥ 0.18 as potential active ingredients, and obtained 29 main active ingredients and 157 targets of PLP, among which Pae is one of the main effective ingredients, which has high OB and DL values. Moreover, the existing research on PLP mainly focused on Pae, indicating that the data analysis has high reliability. GeneCards and OMIM databases are often used to screen disease-related targets. Using these two databases to search will help to obtain more comprehensive and detailed disease targets and improve accuracy. Through searching, we found 2424 targets related to ‘allergy’. GO and KEGG analysis are often used to analyze the function of target genes and related enrichment pathways. They are the most important data analysis in the network pharmacology system, and it is also a key step for network pharmacology to reveal the mechanism of drug to treat diseases [23]. By sorting out the intersection of targets, there are 50 possible targets for PLP anti-allergy. Through GO-BP analysis, the biological processes involved in the anti-allergic effect of PLP mainly include: positive regulation of cell biosynthesis, regulation of cell death and apoptosis, and intracellular signal cascades. GO-CC analysis showed that the cellular location of the anti-allergic effect of PLP mainly included the extracellular area and plasma membrane. GO-MF analysis showed that the molecular processes involved in the anti-allergic effect of PLP are antioxidant activity, MAPK activity, binding of Ca2+ and triglycerides and so on, among which the Ca2+ concentration is closely related to the occurrence of type I allergy. KEGG analysis obtained 31 related pathways of PLP anti-allergy, including the FcεR I signal pathway that is closely related to type I allergy, which researchers are familiar with, indicated that PLP has the potential to treat allergy, and also verified the reliability of network pharmacological analysis. The results concurrently showed that PLP may regulate allergy through signal pathways such as MAPK, TNF, PI3K/Akt, apoptosis and Th cell differentiation.
The obtained network of drug-target-pathway contains 52 nodes and 153 edges, among which MAPK 1, MAPK 10, MAPK 14 and TNF have high topological metrics and may be key targets. Combined with the results of KEGG analysis, it is found that these four important targets are distributed in the FcεR I signal pathway. MAPK 1, MAPK 10, and MAPK 14 belong to the MAPK family and are the integration points of many biochemical signals. They regulate cell proliferation, differentiation, and transcriptional regulation, and are closely related to multiple signal pathways involved in the regulation of allergy. TNF is related to various diseases such as allergy, autoimmune diseases, and tumors. Therefore, it is speculated that PLP may exert its inhibitory effect on allergy mainly through these targets and FcεR I signal pathway, and Pae, as the main component of PLP, may also inhibit the degranulation of mast cells (MC) by acting on these targets and pathways, and then play a therapeutic effect on type I allergy. Furthermore, the research on the chemical components and mechanism of PLP used for immune regulation and anti-inflammation is mainly focused on Pae [24, 25], so Pae was selected as the representative of PLP as the research object of subsequent in vitro experiments.
In addition to the OB values mentioned above. Studies have reported that the absorption permeability and absorption rate of Pae are approximately the same between various sites in the small intestine. And the absorption mechanism is passive diffusion. After oral administration of Pae, it is mainly absorbed in the form of metabolites of paeonimetabolin-I (PM-I) and paeoniflorgenin (PG). Shaoyao Gancao Decoction (a dose equivalent to Pae 25 mg/kg) was administered to rats, and the peak plasma concentrations (Cmax) of Pae and PM-I were 0.21 and 2.05 mg/L, respectively. In addition, the study also found that Baishao decoction (a dose equivalent to Pae 110 mg/kg) was administered to rats, and the Cmax of PG was as high as 8 mg/L. The peak time (Tmax) of PM-I and PG were 3.0 h and 10 min, respectively. Pae has strong hydrophilicity, weak lipophilicity, and weak transmembrane absorption ability, but it can quickly reach the brain tissue through the blood–brain barrier. The mean AUC of Pae was 615.7 mg/min·L. Pae is less affected by liver metabolism, but can be degraded by glycosidases and anaerobic bacteria in intestine [26]. At present, drug research mostly focuses on the effect on the absorption of Pae, and there are few reports on the effect on the tissue distribution characteristics, metabolic pathways and metabolites of Pae.
RBL-2H3 cells possess the biological characteristics of MCs. And RBL-2H3 cells are used as the classic model for studying degranulation reaction in vitro. Therefore, after considering various factors, we finally chose RBL-2H3 cells as the cell model. To improve the reliability of the results, we chose Keto as the positive control drug. It has a strong anti-allergic effect, and can inhibit the release of allergic mediators from MCs and stabilize their membranes. Keto can also block Ca2+ channels and inhibit IgE synthesis. Thus, it is often used as a positive control drug in anti-allergy experiments.
According to different pathogenesis, allergy can be divided into 4 types, among which type I allergy is the most common in life [27]. The pathogenesis of type I allergy is complicated, and the specific and comprehensive regulation mechanism is still unclear. IgE/FcεR I is a classic signal pathway that directly regulates type I allergy. There are many studies on it, but the signal network that it participates in the development of type I allergy still needs to be perfected and supplemented. This study focused on the IgE/FcεR I signal pathway, and selected the other more important signal pathways in the results of network pharmacology for analysis, so as to prove the possible mechanism of PLP to treat type I allergy.
The classic IgE/FcεR I signal pathway includes Syk, Lyn and Fyn, among which Lyn and Syk as initial signals to participate in the activation of MC, and they have become key therapeutic targets for allergic diseases. Activated Syk can finally activate PLCγ and PI3K, which can cause the degranulation of MC [28, 29]. Fyn is the upstream of IgE/FcεR I signal pathway. The cross-linking of FcεR I can activate Fyn-dependent Gab2, and Gab2 can bind to PI3K, which will eventually activate Akt [30, 31]. In this study, the results of Western Blot and RT-qPCR showed that Pae can inhibit the phosphorylation of Lyn and Syk proteins and the expression of Lyn, Syk, Fyn, PLCγ, PI3K and Akt genes when the degranulation of MC. This result is consistent with the predicted results of network pharmacology, indicating that the network pharmacology method established in this study has good credibility, demonstrating that Pae can inhibit IgE/FcεR I and PI3K/Akt signal pathways.
When the IgE/FcεR I signal pathway is activated, it will directly or indirectly activate the MAPK and NF-κB signal pathways [32, 33]. MAPK includes JNK, ERK and p38 [34]. They mediate extracellular and nuclear signal transduction pathways, which can promote the activation of cytoplasmic phospholipase A2 and transfer to the cell membrane, thereby prompting MC to secrete biologically active mediators [35]. NF-κB is formed by p50 and p65, and is also closely related to MC degranulation [36]. Li L [37] found that allergy can be treated by inhibiting MAPK and NF-κB signal pathways. In this experiment, RT-qPCR was used to detect the effect of Pae on the expression of ERK, JNK, p38 and p65 genes when MC degranulation, showing that Pae can inhibit the expression of JNK, p38 and p65, but its inhibitory effect on ERK is weak, suggesting that Pae's inhibitory effect may be selective. These convincing evidences show that the mechanism of Pae on type I allergy is multi-target and multi-pathway, which is consistent with the experimental results of others we mentioned above. Our study revealed Pae has inhibitory effects on the key genes of in the downstream signal pathway of IgE/FcεR I, further confirming the multi-dimensional regulatory mechanism of Pae to treat allergy, which provides new support and reference for the study of the mechanism of PLP in the treatment of type I allergy.
Conclusions
In summary, it was speculated that MAPK 1, MAPK 10, MAPK 14 and TNF may be the key targets of PLP to treat allergy. By interacting with these targets, PLP regulates FcεR I, MAPK, TNF, PI3K/Akt and Th cell differentiation and other signal pathways to participate in the occurrence and development of type I allergy (Fig. 11). Moreover, according to the results of Western Blot and RT-qPCR, Pae has been proven to have a therapeutic effect on type I allergy, which is achieved by regulating IgE/FcεR I and downstream signal pathways. These results of this study will offer a great opportunity for the deep understanding of the pharmacological mechanisms of PLP (Fig. 12). But there is no doubt that in order to fully reveal the mechanism of PLP and Pae, further in-depth research is needed. Further studies were planned where other cell and animal models related to type I allergy will be established to verify its inhibitory effect on type I allergy, which can provide a theoretical basis for the development of related fields and new drugs research.
Fig. 11.
The provable mechanism of PLP anti-Type I allergy derived from this study
Fig. 12.
Graphical abstract of this paper
Supplementary Information
Additional file 1: Fig.S1. Original image of the expression of Lyn in RBL-2H3 cells detected by Western Blot.
Additional file 2: Fig.S2. Original image of the expression of p-Lyn in RBL-2H3 cells detected by Western Blot.
Additional file 3: Fig.S3. Original image of the expression of Syk in RBL-2H3 cells detected by Western Blot.
Additional file 4: Fig.S4. Original image of the expression of p-Syk in RBL-2H3 cells detected by Western Blot.
Additional file 5: Fig.S5. Original image of the expression of β-actin in RBL-2H3 cells detected by Western Blot.
Acknowledgements
Not applicable
Abbreviations
- TCM
Traditional Chinese medicine
- BP
Biological process
- CC
Cell component
- DL
Drug-likeness
- GO
Gene Ontology
- IgE
Immunoglobulin E
- Keto
Ketotifen fumarate
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MC
Mast cells
- MF
Molecular function
- OB
Oral bioavailability
- Pae
Paeoniflorin
- PLCγ
Phospholipase C γ
- PLP
Paeonia lactiflora Pall.
- PPI
Protein–protein interaction
- TCMSP
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform
- PM-I
Paeonimetabolin-I
- PG
Paeoniflorgenin
Authors’ contributions
Y Z and Z L designed the research project; Y Z, H L, X L, Y S and Y S performed the experiments; Y Z, H L, Y Z and Z L analyzed the data and wrote the manuscript; and all authors contributed to the preparation of the manuscript. Y Z, H L, Y Z and Z L revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by key research and development projects in Hebei Province [grant number 20372702D]; Natural Science Foundation of Hebei Province [grant number H2019201455, H2020201018].
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yang Zhao and Hui Li are contributed equally to this work.
Contributor Information
Hui Li, Email: lihui@pkuih.edu.cn.
Yanfen Zhang, Email: zhangjing@hbu.edu.cn.
Zhongcheng Liu, Email: liuzc@hbu.edu.cn.
References
- 1.Modgill V, Badyal DK, Verghese A. Effificacy and safety of montelukast add-on therapy in allergic rhinitis. Methods Find Exp Clin Pharmacol. 2010;32:669–674. doi: 10.1358/mf.2010.32.9.1533686. [DOI] [PubMed] [Google Scholar]
- 2.Liu CX, Liu R, Fan HR, Xiao XF, Chen XP, Xu HY, et al. Network pharmacology bridges traditional application and modern development of traditional Chinese medicine. Chinese Herb Med. 2015;7:3–17. doi: 10.1016/S1674-6384(15)60014-4. [DOI] [Google Scholar]
- 3.Ye J, Piao H, Jiang J, Jin G, Zheng M, Yang J, et al. Polydatin inhibits mast cell-mediated allergic inflammation by targeting PI3K/Akt, MAPK, NF-κB and Nrf2/HO-1 pathways. Sci Rep. 2017;7:11895. doi: 10.1038/s41598-017-12252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han S, Sun L, He F, Che H. Anti-Allergic activity of glycyrrhizic acid on IgE-mediated allergic reaction by regulation of allergy-related immune cells. Sci Rep. 2017;7:7222. doi: 10.1038/s41598-017-07833-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding YY, Che DL, Li CM, Cao J, Wang JM, Peng Y, et al. Quercetin inhibits Mrgprx2-induced pseudo-allergic reaction via PLCγ-IP3R related Ca2+ fluctuations. Int Immunopharmacol. 2019;66:185–197. doi: 10.1016/j.intimp.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Niu K, Liu Y, Zhou Z, Wu X, Wang H, Yan J. Antitumor Effects of Paeoniflorin on Hippo Signaling Pathway in Gastric Cancer Cells. J Oncol. 2021;2021:1–12. doi: 10.1155/2021/4724938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu SH, Wu DG, Chen YW. Chemical constituents and bioactivities of plants from the genus Paeonia. Chem Bio divers. 2010;7:90–104. doi: 10.1002/cbdv.200800148. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Zhang Y, Wang J, Liu R, Zhang J, Dong K, et al. Paeoniflorin inhibits MRGPRX2-mediated pseudo-allergic reaction via calcium signaling pathway. Phytother Res. 2020;34:401–408. doi: 10.1002/ptr.6531. [DOI] [PubMed] [Google Scholar]
- 9.Lee B, Shin YW, Bae EA, Han SJ, Kim JS, Kang SS, et al. Antiallergic effect of the root of Paeonia lactiflora and its constituents paeoniflorin and paeonol. Arch Pharm Res. 2008;31:445–450. doi: 10.1007/s12272-001-1177-6. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Guo W, Cheung F, Tan HY, Wang N, Feng Y. Integrating network pharmacology and experimental models to investigate the efficacy of Coptidis and Scutellaria containing HuanglianJiedu decoction on hepatocellular carcinoma. Am J Chin Med. 2020;48:161–182. doi: 10.1142/S0192415X20500093. [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Nan C. Paeoniflorin inhibits mast cell-mediated allergic inflammation in allergic rhinitis. J Cell Biochem. 2018;119:8636–8642. doi: 10.1002/jcb.27135. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Hu S, Ge S, Wang J, He L. Paeoniflorin inhibits IgE-mediated allergic reactions by suppressing the degranulation of mast cells though binding with FcϵRI alpha subunits. Eur J Pharmacol. 2020;886:173415. doi: 10.1016/j.ejphar.2020.173415. [DOI] [PubMed] [Google Scholar]
- 13.Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang ZY, Liu J, Liu YF, Shi DN, He YS, Zhao PW. Virtual screening of the multi-gene regulatory molecular mechanism of Si-Wu-tang against non-triple-negative breast cancer based on network pharmacology combined with experimental validation. J Ethnopharmacol. 2021;269:113696. doi: 10.1016/j.jep.2020.113696. [DOI] [PubMed] [Google Scholar]
- 15.Jing C, Sun Z, Xie X, Zhang X, Bi H. Network pharmacology-based identification of the key mechanism of Qinghuo Rougan Formula acting on uveiti. Biomed Pharmacother. 2019;120:109381. doi: 10.1016/j.biopha.2019.109381. [DOI] [PubMed] [Google Scholar]
- 16.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The Genecards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 17.Ada H, Scott AF, Joanna A, Carol B, David V, Mckusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;2005:514–517. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M. Molecular network analysis of diseases and drugs in KEGG. Methods Mol Biol. 2013;939:263–275. doi: 10.1007/978-1-62703-107-3_17. [DOI] [PubMed] [Google Scholar]
- 19.Han EJ, Fernando I, Kim EA, Kim J, Jung K, Kim SY, et al. 5-Bromo-3,4-dihydroxybenzaldehyde from Polysiphonia morrowii attenuate IgE/BSA-stimulated mast cell activation and passive cutaneous anaphylaxis in mice. Biochem Pharmacol. 2020;178:114087. doi: 10.1016/j.bcp.2020.114087. [DOI] [PubMed] [Google Scholar]
- 20.Do HJ, Hwang YJ, Yang HJ, Park KI. Effect of rhus verniciflua extract on IgE-antigen-mediated allergic reaction in rat basophilic leukemic RBL-2H3 mast cells and passive cutaneous anaphylaxis in mice. Evid Based Complement Alternat Med. 2019;2019:6497691. doi: 10.1155/2019/6497691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Huang C, Shi P, Xu H, Xu F. Mechanism of Chinese yam for the treatment of aging-related diseases based on network pharmacology. Euro J Integr Med. 2021;41:101254. doi: 10.1016/j.eujim.2020.101254. [DOI] [Google Scholar]
- 22.Dai W, Sun Y, Zhong G. A network pharmacology approach to estimate the active ingredients and potential targets of cuscutae semen in the treatment of Osteoporosis. Med Sci Monit. 2020;26:e920485. doi: 10.12659/MSM.920485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou ZD, Xie SP, Saw WT, Ho PGH, Wang HY, Zhou L, et al. The therapeutic implications of tea polyphenols against dopamine (DA) neuron degeneration in parkinson'sdisease (PD) Cells. 2019;8:911. doi: 10.3390/cells8080911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji L, Hou X, Liu W, Xian D, Jiang Z, Huang K, et al. Paeoniflorin inhibits activation of the IRAK1-NF-κB signaling pathway in peritoneal macrophages from lupus-prone MRL/lpr mice. Microb Pathog. 2018;124:223–229. doi: 10.1016/j.micpath.2018.08.051. [DOI] [PubMed] [Google Scholar]
- 25.Kong X, Leng D, Liang G, Zheng H, Wang Q, Shen Y, et al. Paeoniflorin augments systemic Candida albicans infection through inhibiting Th1 and Th17 cell expression in a mouse model. Int Immunopharmacol. 2018;60:76–83. doi: 10.1016/j.intimp.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Liu ZQ, Jiang ZH, Liu L, Hu M. Mechanisms responsible for poor oral bioavailability of paeoniflorin: role of intestinal disposition and interactions with sinomenine. Pharm Res. 2006;23:2768–2780. doi: 10.1007/s11095-006-9100-8. [DOI] [PubMed] [Google Scholar]
- 27.Coombs R, Gell P. The classification of allergic reactions underlying disease: In Gell PGH, Coombs RRA (eds). Clin Aspects Immunol. 1963;317–37.
- 28.Shim SY. Suppressive effects of vaccinium angustifolium root extract via down-regulation of activation of Syk, Lyn, and NF-κB in FcεR I-mediated allergic reactions. Prev Nutr Food Sci. 2018;23:30–34. doi: 10.3746/pnf.2018.23.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woolhiser M, Okayama Y, Gilfillan A, Metcalfe D. IgG-dependent activation of human mast cells following up-regulation of FcγRI by IFN-γ. Eur J Immunol. 2015;31:3298–3307. doi: 10.1002/1521-4141(200111)31:11<3298::AID-IMMU3298>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Gomez G, Gonzalez-Espinosa C, Odom S, Baez G, Cid ME, Ryan JJ, et al. Impaired FcεR I-dependent gene expression and defective eicosanoid and cytokine production as a consequence of fyn deficiency in mast cells. J Immunol. 2005;175:7602–7610. doi: 10.4049/jimmunol.175.11.7602. [DOI] [PubMed] [Google Scholar]
- 31.Choi WS, Kim YM, Combs C, Frohman MA, Beaven MA. Phospholipases D1 and D2 regulate different phases of exocytosis in mast cells. J Immunol. 2002;168:5682–5689. doi: 10.4049/jimmunol.168.11.5682. [DOI] [PubMed] [Google Scholar]
- 32.Gilfillan AM, Rivera J. The tyrosine kinase network regulating mast cell activation. Immunol Rev. 2009;228:149–169. doi: 10.1111/j.1600-065X.2008.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Liu Y. Regulation of innate immune response by MAP kinase phosphatase-1. Cell Signal. 2007;19:1372–1382. doi: 10.1016/j.cellsig.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trinh TA, Seo YH, Choi S, Lee J, Kang KS. Protective effect of osmundacetone against neurological cell death caused by oxidative glutamate toxicity. Biomol. 2021;11:328. doi: 10.3390/biom11020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faaaai HK, Naranjo A, Smelkinson M, Yun BM, Tobio A, Boyden S, et al. Mechanical activation of ADGRE2 causes calcium-dependent activation of PI3K and MAPK pathways driving mast cell degranulation and PGD2 production. J Allergy Clin Immunol. 2020;145:AB186. [Google Scholar]
- 36.Leitner PD, Vietor I, Huber LA, Valovka T. Fluorescent thermal shift-based method for detection of NF-κB binding to double-stranded DNA. Sci Rep. 2021;11:2331. doi: 10.1038/s41598-021-81743-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Jin G, Jiang J, Zheng M, Jin Y, Lin Z, et al. Cornuside inhibits mast cell-mediated allergic response by down-regulating MAPK and NF-κB signaling pathways. Biochem Biophys Res Commun. 2016;473:408–414. doi: 10.1016/j.bbrc.2016.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig.S1. Original image of the expression of Lyn in RBL-2H3 cells detected by Western Blot.
Additional file 2: Fig.S2. Original image of the expression of p-Lyn in RBL-2H3 cells detected by Western Blot.
Additional file 3: Fig.S3. Original image of the expression of Syk in RBL-2H3 cells detected by Western Blot.
Additional file 4: Fig.S4. Original image of the expression of p-Syk in RBL-2H3 cells detected by Western Blot.
Additional file 5: Fig.S5. Original image of the expression of β-actin in RBL-2H3 cells detected by Western Blot.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.