Abstract
Genetic recombination plays a critical role in the emergence of pathogens with phenotypes such as drug resistance, virulence, and host adaptation. Here, we tested the hypothesis that recombination between sympatric ancestral populations leads to the emergence of divergent variants of the zoonotic parasite Cryptosporidium parvum with modified host ranges. Comparative genomic analyses of 101 isolates have identified seven subpopulations isolated by distance. They appear to be descendants of two ancestral populations, IIa in northwestern Europe and IId from southwestern Asia. Sympatric recombination in areas with both ancestral subtypes and subsequent selective sweeps have led to the emergence of new subpopulations with mosaic genomes and modified host preference. Subtelomeric genes could be involved in the adaptive selection of subpopulations, while copy number variations of genes encoding invasion-associated proteins are potentially associated with modified host ranges. These observations reveal ancestral origins of zoonotic C. parvum and suggest that pathogen import through modern animal farming might promote the emergence of divergent subpopulations of C. parvum with modified host preference.
Keywords: Cryptosporidium parvum, genome evolution, population genetics, recombination, adaptive selection, emerging infection
Introduction
Cryptosporidium spp. are important protozoan parasites responsible for enteric diseases in humans and farm animals (Checkley et al. 2015). In addition to being a primary cause of moderate-to-severe diarrhea in young children in low- and middle-income countries, cryptosporidiosis is one of the most important waterborne diseases in industrialized nations and has been associated with numerous outbreaks of illness each year (Kotloff et al. 2013; Gharpure et al. 2019). Human cryptosporidiosis is mainly caused by Cryptosporidium hominis and Cryptosporidium parvum. The former is mostly a human pathogen, while the latter has been found in a broad range of mammals, responsible for outbreaks of neonatal diarrhea in farm animals (Feng et al. 2018). Therefore, C. parvum is the most important zoonotic Cryptosporidium species.
Sequence analyses of the gene encoding the major invasion-associated 60 kDa mucin glycoprotein (GP60) have identified many subtype families of C. parvum with different geographic distribution and host ranges (Feng et al. 2018). Among them, IIa, IIc, and IId are three common subtype families preferentially found in dairy calves, humans, and small ruminants, respectively. In humans in the Eurasian continent, IIa subtypes are the dominant C. parvum in European countries, IId subtypes are the dominant C. parvum in Middle Eastern countries, while IIc subtypes are mainly found in low- and middle-income countries (Yang et al. 2021). Among them, IId subtypes in different areas appear to have different host preference. Those in China are found mostly in dairy calves, while those in Europe are mainly found in sheep and goats. In the Middle East, however, cattle are commonly infected with both IIa and IId subtypes of C. parvum (Hijjawi et al. 2022).
The evolutionary history of zoonotic C. parvum subtype families remains unclear. This is largely due to the lack of systematic characterizations of C. parvum at other genetic loci. Population genetic studies involving limited genetic loci have mostly identified a panmictic population structure in the IIa subtype family, suggesting that genetic recombination could play an important role in shaping the evolution of C. parvum in industrialized nations (Feng et al. 2018). An epidemic population structure, however, has been observed in the IId subtype family, indicating that its evolution could be different (Zhang, Hu, et al. 2020). Results of limited comparative genomics analysis of several isolates have shown differences in copy numbers and sequences in several invasion-associated proteins between IIa and IId subtype families (Feng et al. 2017).
Advanced characterizations of the evolution of C. parvum have thus far only been done on the human-adapted IIc subtype family. Results of comparative analysis of whole-genome sequences (WGS) from 21 isolates have shown significant population differentiation of the anthroponotic IIc from zoonotic subtypes and the occurrence of adaptive introgression of anthroponotic alleles into one IIc isolate (Nader et al. 2019). Therefore, it is possible that genetic recombination could be important in the evolution of C. parvum. Recently, a comparative genomics study of 114 C. hominis isolates from diverse areas has identified two population lineages with divergent gene flow rates and outbreak potential, indicating that genetic recombination could modify the phenotypic traits of the pathogen in addition to affecting its evolution (Tichkule et al. 2022).
To test the hypothesis that genetic recombination between sympatric zoonotic IIa and IId subtype can lead to the emergence of divergent C. parvum subpopulations with mosaic genomes and modified host range, we have conducted comparative genomics and population genetic analyses of WGS data from 101 isolates of the IIa, IId, and IIc subtypes, mostly newly acquired from the present study. The data generated have provided not only support for the hypothesis but also clues on the contribution of modern animal farming to the emergence of these novel subpopulations of C. parvum.
Results
Genome Variations in C. parvum
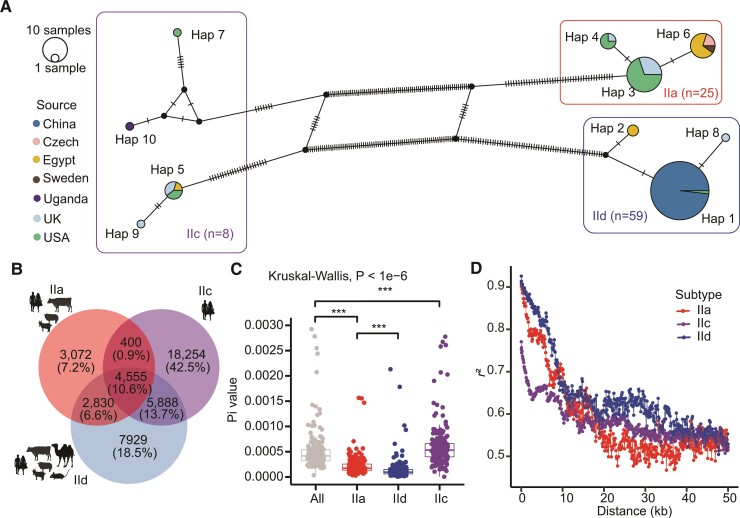
We performed WGS on 91 C. parvum isolates. In addition, we collected 16 published genomes from NCBI and EBI. After mapping to the IOWA II reference genome, 101 sets of high-quality WGS data, including 87 newly acquired and 14 publicly available genomes, were selected (supplementary table S1, Supplementary Material online). The 101 C. parvum isolates included the three major gp60 subtype families IIa (34 genomes), IIc (8 genomes), and IId (59 genomes). Network analysis of the gp60 sequences from the isolates sequenced expectedly revealed the presence of three distinct groups corresponding to the three subtype families IIa, IIc, and IId (fig. 1A). Among them, IIc subtype haplotypes were more divergent from each other, with three of the four haplotypes consisting of only one isolate. IIa samples from the United Kingdom and United States were grouped into two haplotypes, while samples from Egypt, Czech Republic, and Sweden were clustered into another haplotype. Within the IId subtype family, samples from China and one sample from the United States clustered together, while two samples from Egypt and one sample from the United Kingdom formed two different haplotypes.
Fig. 1.
Genomic differences among three major gp60 subtype families of Cryptosporidium parvum. (A) Haplotype network of gp60 sequences extracted from 101 genomes used in the study, including those of IIa, IIc, and IId subtype families from China, Egypt, the Czech Republic, Sweden, Uganda, the United Kingdom, and the United States. Each circle indicates a unique haplotype of C. parvum with the color corresponding to the geographic origin. The size of the circle is proportional to the number of samples with the haplotype. (B) Distribution of shared and subtype family-specific SNPs among three gp60 subtype families with different host ranges, including IIa (mainly found in humans, cattle, sheep, and goats), IIc (mainly found in humans), and IId (found in humans, farm animals, and small rodents). (C) Nucleotide divergence (Pi) within each gp60 subtype family. The Pi values were determined with a 50 kb sliding window along the genome for isolates of four groups, including IIa, IIc, IId, and all samples (***P < 0.001). (D) Decay of genome-wide LD by subtype family. LD between SNPs within a 50 kb distance was calculated for IIa, IIc, and IId subtype families. An equal number of samples were randomly selected in each subtype family for LD calculation to reduce the potential influence of the sample size. Resampling was performed 50 times and the mean values obtained were used in plotting.
Among these genomes, we identified a total of 42,928 variations (37,301 SNPs and 5,627 INDELs, SNPs/INDELs = 6.6:1), including 10,857 among IIa genomes, 29,097 among IIc genomes, and 21,202 among IId genomes (fig. 1B). However, only 4,555 variations (10.6%, 3,644 SNPs and 911 INDELs) were shared by them. Surprisingly, IIc and IId subtype families shared more variations (10,443 or 24.3%) with each other than with IIa subtype family (IIa vs IIc: 4,955 or 11.5%; IIa vs IId: 7,385 or 17.2%), but 16,634 variations (42.5%) were unique to IIc subtype family. Noteworthy, if isolate UKP16 was excluded from the analysis of IIc genomes, variations shared among three subtype families were reduced to 3,293 (8.0%) with the same number of variations (7,385 or 17.9%) shared between IIa and IId. UKP16 (human isolate) was from the zoonotic subtype IIcA5G3j within the otherwise anthroponotic subtype family IIc. This subtype has been found in humans as well as in hedgehogs in Europe and was shown to be a possible recombinant (Sangster et al. 2016; Nader et al. 2019). Similarly, when the human isolate US44513 (IIdA15G1) from the United States was excluded from IId, sequence variations shared between IIc and IId were reduced from 10,443 (24.3%) to 1,167 (3.2%), suggesting the isolate US44513 was likely resulted from recombination between IIc and zoonotic C. parvum (supplementary fig. S1A, Supplementary Material online).
Along the genomes, INDELs were evenly distributed except for some regions of several chromosomes, while SNPs were mainly in the subtelomeric regions of chromosomes (supplementary fig. S1B, Supplementary Material online). INDEL density was especially high in four regions in chromosomes 3 (240k–245k), 5 (305k–310k), and 7 (10k–15k and 950k–955k). These regions included genes encoding three mucin-like proteins, cgd3_720, cgd5_1210, and cgd7_4020.
The average genetic divergence (Pi) of genomes was 0.00058 for C. parvum. Genetic diversity within the IIc subtype family (Pi = 0.00077), however, was higher than within IId (Pi = 0.00015) and IIa (Pi = 0.00025) subtype families (P < 1e−6 by the Kruskal–Wallis test) (fig. 1C). Genes located in subtelomeric regions of chromosomes were more likely polymorphic (supplementary fig. S1C, Supplementary Material online). Among the 15 highly polymorphic genes identified, three (cgd1_470, cgd6_40 and cgd6_1080) encoded mucin-like glycoproteins proteins. Annotation of the SNPs revealed that 25,946 (69.6%) SNPs were in the coding region, 13,539 (52.2%) of which were nonsynonymous.
Among the three gp60 subtype families, IIa had more rapid linkage disequilibrium (LD) decay than IIc and IId; the shortest physical distance in base pairs halfway between the minimum and maximum LD (LD50) was 6.8, 8.8, and 9.9 kb for IIa, IId, and IIc, respectively (fig. 1D). Among countries with significant numbers of isolates of zoonotic C. parvum, LD in isolates from the United States (LD50, 2.1 kb) and the United Kingdom (2.7 kb) decayed more rapidly than LD in isolates from China (9.3 kb) and Egypt (10.2 kb) (supplementary fig. S2, Supplementary Material online).
Presence of Divergent C. parvum Subpopulations
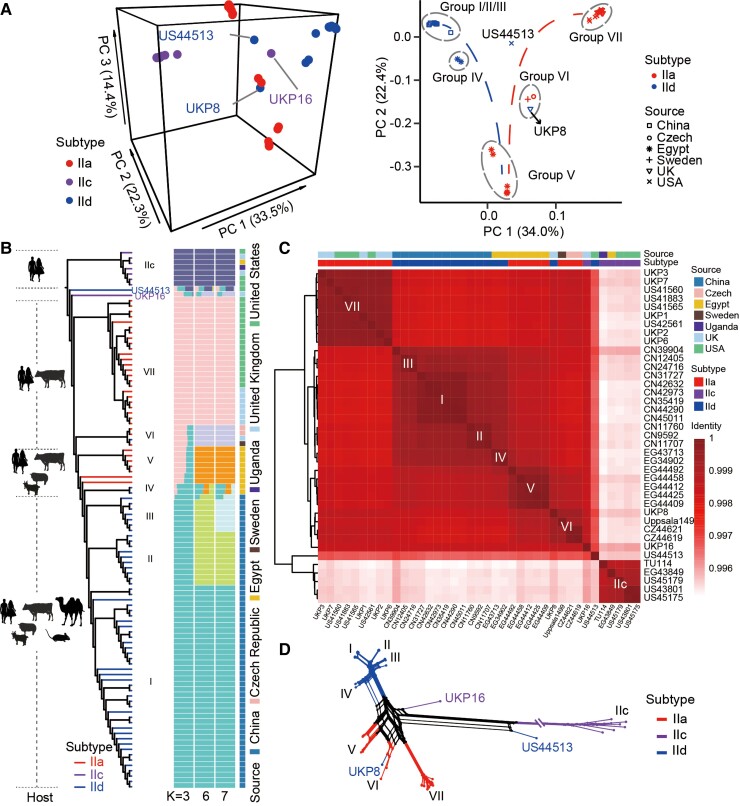
At the whole-genome level, principal component analysis (PCA) analyses showed anthroponotic IIc isolates mostly formed a divergent cluster (the first principal component = 33.5%) (fig. 2A). In contrast, zoonotic isolates (IIa and IId) showed substantial genetic diversity with no complete segregation of genomes by gp60 subtype family (fig. 2A). In an SNP-based maximum-likelihood (ML) tree, eight subpopulations formed among the C. parvum isolates analyzed (fig. 2B). These subpopulations represent mostly geographically associated IIa and IId subtypes, except for the IIc subpopulation, which contains divergent genomes from different countries. The rooted ML tree, with C. hominis as the outgroup, showed a similar phylogenetic relationship, with IIc forming the most divergent cluster (supplementary fig. S3, Supplementary Material online). Additionally, two isolates, UKP16 (IIcA5G3j) and US44513 (IIdA15G1), were divergent from others, being placed between the zoonotic (IIa and IId) and anthroponotic (IIc) subtype families.
Fig. 2.
Population structure of C. parvum. (A) Results of PCA of genomes from all (left) and zoonotic (right) subtype families. The colors and symbols correspond to subtype families and sample sources. Groups are circled with gray dashed lines and marked with I–VII according to the results in (B). (B) Phylogeny and population structure of representative C. parvum isolates with K = 3, 6, and 7. The ML tree was built with 37,301 SNPs and rooted with the IIc clade. Major hosts of the clades are marked at the left panel and the clades are colored by subtype family. Subpopulations are marked with I–VII plus IIc. Each isolate is represented by a single bar in 3, 6, and 7 colored ancestral components depending on the K values used. All results with K values from 3 to 9 can be viewed in supplementary figure S4, Supplementary Material online. The mosaic color bars at the right represent the geographic origins of isolates. (C) Pairwise comparisons of ANI of selected C. parvum genomes. The identity blocks in dark red correspond to phylogenetic clusters in (B). Bars above the heatmap represent the geographic origin and subtype identity of selected genomes. (D) Phylogenetic network of C. parvum genomes based on 20,126 SNPs (with singletons removed). Branches are colored by gp60 subtype as in (B).
STRUCTURE analysis showed a high population homogeneity in most clades, with IIa, IIc, and IId isolates largely differentiated (fig. 2B and supplementary fig. S4, Supplementary Material online). Therefore, most IIa genomes from Europe and the United States, IId genomes from China, and IIa and IId genomes from Egypt formed their own clusters. In contrast, group VI was formed by two subtype families (IIa and IId). Pairwise Fst comparison of genomes among major IIa (group VII), IId (groups I–III), and IIc subtype families revealed the presence of ongoing differentiation events (supplementary fig. S5, Supplementary Material online). Compared with IIa and IId, IIc was the most divergent subtype family with Fst values of over 0.5 across most of the genome. In contrast, Fst values between IIa and IId were uneven with mosaic sequences across each of the eight chromosomes.
Pairwise comparisons of the average nucleotide identity (ANI) of representative genomes revealed a similar relationship among the C. parvum isolates analyzed (fig. 2C). All IId isolates in China clustered into one large clade along with IId isolates from Egypt (group IV, EG34902 and EG43713). The IIa isolates from Egypt (group V), however, were genetically related to both IId isolates from Egypt and China and IIa isolates from the Czech Republic and Sweden (group VI). In addition, group VI contained one IId isolate (UKP8) from the United Kingdom. As reported previously, the IIc isolate UKP16 was genetically more similar to IIa and IId isolates of group VI than to other IIc isolates at the whole-genome level (Nader et al. 2019). In comparison, IIa genomes from the United Kingdom and United States formed another group (VII). As mentioned above, genome US44513 (IId from the United States) was placed between the anthroponotic (IIc) and zoonotic (IIa and IId) subtype families, while the remaining IIc isolates formed the most divergent clade.
Nonetheless, evident admixture was observed in several genomes such as CN39904 (IId in China), EG43713 and EG34902 (IId in Egypt), US44513 (IId in the United States), as well as UKP16 (IIc in the United Kingdom) (fig. 2B and supplementary fig. S4, Supplementary Material online). Results of phylogenetic network analysis of the SNP data suggest that gene flow among some subtype families could be responsible for the formation of divergent C. parvum subpopulations (fig. 2D). Splits were seen between IIa and IId isolates from Egypt, suggesting that although IId isolates from Egypt (group IV) were genetically related to IId from China, they were admixed with sequences of sympatric IIa isolates (group V). There was also gene flow between groups V (IIa in Egypt) and VI (IIa in Czech Republic and Sweden). The zoonotic IIc isolate UKP16 was placed outside of these clusters with splits between it and other IIc isolates. Similar results were obtained from the IId isolate US44513 from the United States, with possible sequence admixture with IIc.
Homogenous Subpopulation Structure of IId Subtype Family in China
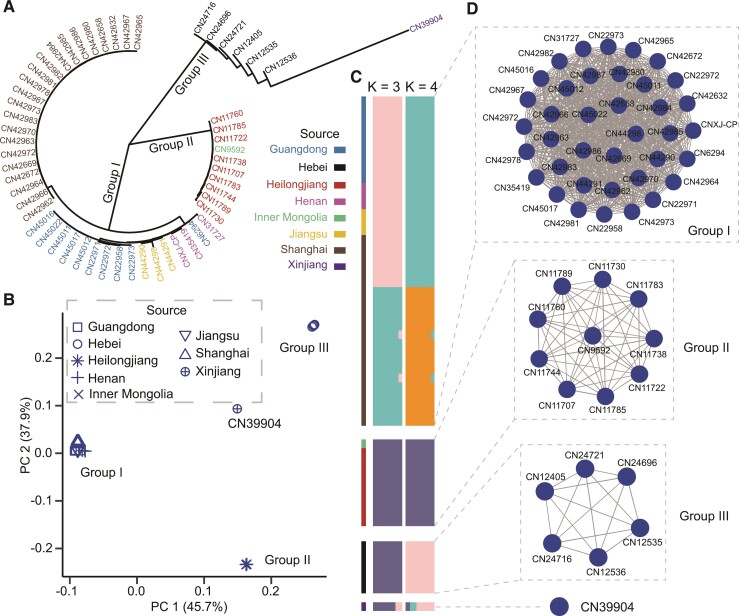
We sequenced 59 isolates from 8 provinces in China covering the east, central, northeast, and northwest areas (supplementary table S1, Supplementary Material online). A total of 2,953 SNPs and 911 INDELs (SNPs/INDELs = 3.2:1) were present among 55 well-assembled genomes, which were far less than the overall sequence diversity in the IId subtype family. The density of INDELs was significantly higher in two areas of chromosomes 5 (cgd5_1210) and 7 (cgd7_4020) encoding mucin glycoproteins. SNPs were mainly found at the 5′ end of chromosomes 4 (cgd4_10 and cgd4_20) and 6 (cgd6_5, cgd6_10 and cgd6_20), and two genes in chromosomes 6 (cgd6_4750) and 7 (cgd7_4500) (supplementary fig. S6A, Supplementary Material online). Neighbor-joining (NJ) tree of concatenated SNPs classified the Chinese isolates into four clades, with isolates from the northeastern province Heilongjiang forming one clade, isolates from the central province Hebei forming another clade, isolates from several eastern provinces (Guangdong, Henan, Jiangsu, and Shanghai) being clustered together, and isolate CN39904 from northwestern Xinjiang being the most divergent (fig. 3A). This was supported by results of PCA (fig. 3B), STRUCTURE (fig. 3C), and phylogenetic network analyses (supplementary fig. S6B, Supplementary Material online). Therefore, the genome differentiation of IId subtypes in China appears to be geographically associated, with few gene flow events from isolates of other origins or little chance from the IIa subtype family. This was supported by the result of LD analysis, which showed slower LD decay among Chinese isolates than among isolates from other countries examined (supplementary fig. S2, Supplementary Material online).
Fig. 3.
Homogenous subpopulation structure of C. parvum in China. (A) NJ tree of 2,953 concatenated SNPs. The color of isolates corresponds to their geographical origins. (B) Outcome of PCA of C. parvum in China. The shape of each node corresponds to the geographical origin of the isolate. (C) Population structure of C. parvum isolates in China. Each isolate is represented by a single bar in colored ancestral components at K = 3 and K = 4. The mosaic color bars at the top represent the geographic origins of the isolates. (D) Relatedness network for pairs of isolates in China with high proportion of IBD sharing. All 55 genomes constructed 3 big clusters with 763 edges and 1 orphan node (CN39904). The edge between two nodes is drawn if the IBD sharing fraction is over 90% of the genome, indicating highly genetic homology.
To test whether isolates in these highly homogeneous subpopulations evolved from similar genetic sources, we undertook the identity by descent (IBD) analysis for relatedness and constructed a relatedness network (fig. 3D and supplementary fig. S7, Supplementary Material online). In IBD analysis, highly related genomes (i.e., fraction of shared IBD over 90%) cluster together, while isolated genomes represent orphans that have evolved from distinct genetic backgrounds. As expected, IId genomes from China formed three groups (I, II, and III) with a high shared identity (mean IBD sharing fraction over 95%), corresponding to the three subpopulations (fig. 3D and supplementary tables S2 and S3, Supplementary Material online). In contrast, most IIa samples (group VII) collected from the United States and United Kingdom formed a separate but less compact cluster, isolates from Egypt did not form IIa- and IId-specific clusters, while other isolates with evident admixture (CN39904, US44513, and UKP16) formed orphan nodes (fig. 3D and supplementary fig. S7, Supplementary Material online). These results suggest that C. parvum isolates from other areas have more complicated ancestral origins than those from China.
Sympatric Recombination between IIa and IId Subtypes in Egypt
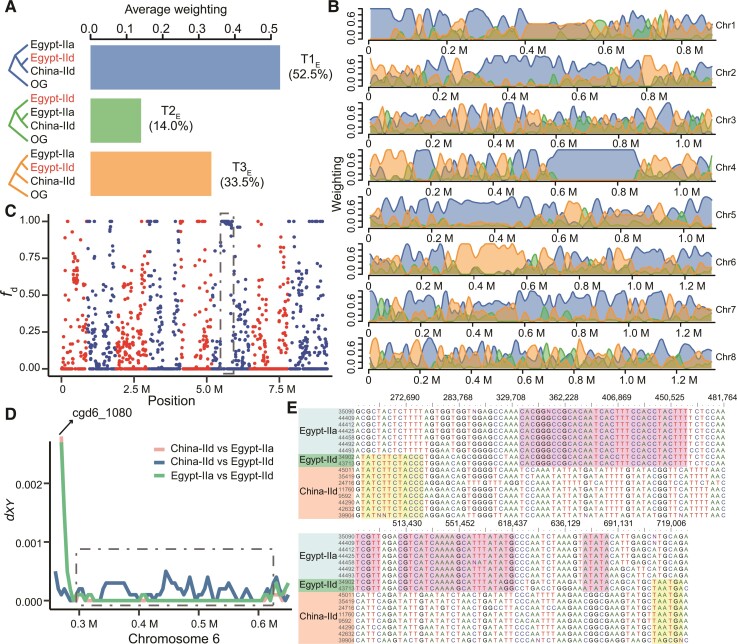
In phylogenetic and PCA analyses and pair-wise genome comparison, IId isolates from Egypt (Egypt-IId) were genetically more like IId isolates in China (China-IId) than other IId isolates in the study (fig. 2 and supplementary fig. S3, Supplementary Material online). Results of the phylogenetic network analysis, however, suggested Egypt-IId might have gene flow from IIa isolates in Egypt (fig. 2D). We hypothesized that Egypt-IId shared ancestors with China-IId but had since gone through some genetic exchanges with the local IIa subpopulation (Egypt-IIa), resulting in unique phylogenetic relationships among them. We assessed phylogenetic topology with a 10 kb sliding window and conducted topology weighting along the China-IId, Egypt-IIa, and Egypt-IId genomes, with C. hominis as the outgroup (OG). The results obtained revealed the existence of three possible phylogenetic relationships among isolates (fig. 4A). T1E, with subpopulations China-IId and Egypt-IId clustered together, was the most prevalent topology (52.5%), which reflected the overall population structure in line with the outcome of other analyses (fig. 4A and B). The second was T3E (33.5%) with the clustering of Egypt-IIa and Egypt-IId together, possibly resulting from both gene flow and incomplete lineage sorting (ILS). Along the genome, we found a large block with a strong signal supporting the topology in chromosome 6 (fig. 4B). To test for whether sequence introgression caused this signal, we further performed fd statistics to study signatures of sequence admixture. Counts of ABBA (Egypt-IId shared the derived allele exclusively with the Egypt-IIa) and BBAA (Egypt-IId shared the derived allele exclusively with the China-IId) patterns were calculated along the genome. Regions with an excess of ABBA pattern (fd close to 1) were indicative of gene flow events between the Egypt-IId and IIa. We identified a large area of chromosome 6 (nucleotides 0.3–0.6 M containing the cgd6_1180 to cgd6_2590 genes) where introgression of IIa sequences occurred in IId isolates from Egypt (fig. 4C). The combined evidence is indicative of the occurrence of gene flow between IId and IIa subtypes as a result of sympatric recombination in Egypt.
Fig. 4.
Sequence introgression in IId isolates of C. parvum from Egypt. (A) Average topology weighting among three subpopulations of C. parvum with Cryptosporidium hominis as the outgroup (OG). T1E accounts for the largest proportion, followed by T3E and T2E. (B) Topology weighting with a 10 kb sliding window along the eight chromosomes in the C. parvum genome. The proportion of each topology along the genome is shaded with colors corresponding to the three topologies in (A). (C) fd statistics of IIa and IId subtype families of C. parvum in Egypt. A 20 kb sliding window with 5 kb steps was used. Values closer to 1 are indicative of introgression between Egypt-IIa and Egypt-IId. The eight chromosomes are colored with alternative red and blue colors. The dashed box marks an introgression region (325k–600k) in which all isolates in Egypt share high ancestral similarity. (D) Sequence divergence among the three C. parvum subpopulations (Egypt-IIa, Egypt-IId, and China-IId) in one region of chromosome 6. In the gp60 (cgd6_1080) region, isolates in Egypt-IId have high sequence identity to China-IId. After it, IIa and IId in Egypt have high sequence identity in the region (0.3–0.6 M) marked with the dashed box corresponding to the box in (C). (E) Alignment of concatenated SNPs in the region marked in (C) and (D).
Pairwise genetic divergence (dxy) analysis was used to further compare sequence divergence among Egypt-IId, Egypt-IIa, and China-IId genomes along the eight chromosomes. It also showed high sequence divergence in the cgd6_1080 gene encoding GP60 and high sequence identity downstream from it between Egypt-IIa and Egypt-IId, confirming the occurrence of IIa introgression in this area within Egypt-IId genomes (fig. 4D). Alignment of concatenated SNPs in a 450 kb region around the area showed a mosaic sequence pattern with nearly 300 kb of high sequence identity between Egypt-IIa and Egypt-IId (fig. 4E). In contrast, upstream from the introgression area, China-IId and Egypt-IId genomes were highly similar, indicating sequence conservation within the IId subtype family in the gp60 region (fig. 4D and E). In fact, IId from China and Egypt had very similar sequences in the entire chromosome 1 and a 270 kb region (cgd4_3160 to cgd4_1940) in chromosome 4 (fig. 4B and supplementary fig. S8, Supplementary Material online).
Comparative genomics analysis of the well-assembled IIa (EG44425) and IId (EG43713) genomes from Egypt identified 3,543 SNPs between them, including seven highly polymorphic genes in chromosomes 1 and 6 (supplementary fig. S9A, Supplementary Material online). They contained three genes encoding mucin glycoproteins, including cgd1_470 (encoding mucin 8) in chromosome 1, and cgd6_40 (encoding mucin MSC6-7) and cgd6_1080 (encoding mucin GP60) in chromosome 6. The remaining genes were cgd1_120 (encoding a secreted protein with a cysteine cluster) and three genes encoding hypothetical proteins (cgd1_460 and cgd1_480 in chromosome 1, and a Chro.60010 homolog in chromosome 6). In contrast, 1,523 SNPs were observed between IId genomes in Egypt (EG43713) and China (CN45011) with no highly polymorphic genes (supplementary fig. S9B, Supplementary Material online). Moreover, IIa subtype family in Egypt was phylogenetically related to IIa subtype in group VI (Czech Republic and Sweden), with 3,117 SNPs and six highly polymorphic genes in chromosomes 1, 4, and 5 between them (supplementary fig. S9C, Supplementary Material online). These genes included one encoding MEDLE protein (cgd6_5490 in chromosome 5), one encoding NFDQ protein (cgd6_5500 in chromosome 5), and four genes encoding putative secretory proteins (cgd1_530 in chromosome1, cgd4_10 and cgd4_20 in chromosome 4 and cgd6_5470 in chromosome 5). These sequence differences in functional subtelomeric genes between genomes could lead to the emergence of unique phenotypic traits in some of the subpopulations. This awaits validations through gene ablation and substitution studies using newly developed genetic tools.
Presence of Other Divergent IId Subpopulations
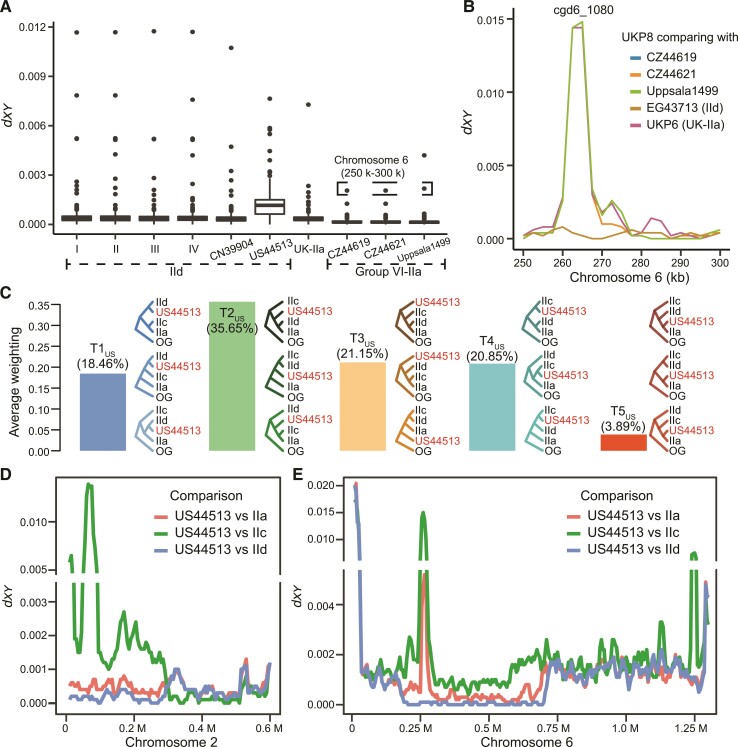
Among the IId isolates analyzed, the UK IId isolate (UKP8) and USA IId isolate (US44513) were genetically divergent from other IId genomes in China and Egypt (fig. 2). In phylogenetic and PCA analyses and pairwise comparison of genomic sequences, UKP8 clustered with IIa isolates from the Czech Republic (CZ44619 and CZ44621) and Sweden (Uppsala1499), forming group VI (figs. 2 and 5A). In direct genome comparisons, UKP8 was mostly identical to IIa isolates within the group, except for the gp60 area (fig. 5B). In contrast, there were 3,879 SNPs between UKP8 and the local IIa isolates (UKP6 as the reference) from group VII, with two mucin genes in chromosomes 6 (cgd6_40 and cgd6_1080) being highly polymorphic (supplementary fig. S9D, Supplementary Material online).
Fig. 5.
Evolution of C. parvum IId subtype family in the United Kingdom (UKP8) and the United States (US44513). (A) Sequence divergence (dxy) between UKP8 and the other genomes from different groups, including IId genomes from US44513 and groups I to IV (left), and between UKP8 and IIa genomes from UK within group VII and IIa genomes within group VI (right). Dxy values were calculated with a 10 kb window. Outliers marked with dashed box represent divergent genes in a 50 kb region in chromosome 6 between the UKP8 and IIa genomes in group VI. (B) Dxy between UKP8 and five genomes in the gp60 (cgd6_1080) region. Three genomes from group VI, EG43713 (IId from Egypt) in group IV, and UKP6 (IIa from UK) in group VII were selected for comparison using a 5 kb sliding window with 2.5 kb steps. The maximum sequence similarity or difference is seen in the gp60 (cgd6_1080) region. (C) Average topology weighting among three gp60 subtype families and US44513, including local IIa genomes in the United States (IIa), IId genomes in China (IId), and IIc genomes in the United States (IIc), with C. hominis as the outgroup (OG). The 15 possible topologies were classified into five types according to the position of US44513 in phylogenetic trees. (D) Genetic similarity of US44513 to the three gp60 subtype families in the first half of chromosome 2. The mosaic pattern of dxy reflects complex sequence introgressions in US44513. The alignment of concatenated SNPs in this region is shown in supplementary figure S11B, Supplementary Material online. (E) High genetic similarity and divergence between US44513 and other subtypes within chromosome 6. In a region including the gp60 locus (0.18–0.72 M), US44513 is highly similar to IId genomes. The alignment of concatenated SNPs in the region is shown in supplementary figure S11C, Supplementary Material online. Downstream from this region to the end of chromosome 6, US44513 has sequence very different from all other genomes. The dxy statistics were calculated using a 20 kb sliding window with 5 kb steps.
Similarly, the sole IId isolate from the United States, US44513, was most divergent from other IIa and IId isolates in phylogenetic and PCA analyses and pairwise genome comparisons (fig. 2A–C). STRUCTURE analysis indicated the likely presence of multiple ancestral components, with IIa (group VII), IId (group I), and IIc accounting for more than 90% when K = 7 (fig. 2B and supplementary fig. S4, Supplementary Material online). We therefore hypothesized that US44513 evolved from a complex evolutionary process among IIa, IIc, and IId subtype families, and performed topology weighting on IIa (group VII), IId (group I), IIc and US44513 genomes (fig. 5C). Among five basic topologies generated with the isolate, T2US, in which US44513 was located between the anthroponotic IIc and zoonotic IIa and IId, was most common (35.7%). The second was T3US (21.2%), in which US44513 was located outside other populations, representing unsampled C. parvum. Other common topologies included T4US (20.9%) with US44513 forming a cluster with IIc, and T1US (18.5%) with US44513 forming a cluster with IId. Along the mosaic genome, three large continuous sequence blocks were seen in chromosomes 1, 2, and 6 (supplementary fig. S10, Supplementary Material online). In chromosome 1, a region (99k–120k) showed high identity between US44513 and IIa genomes, especially in the otherwise highly polymorphic gene cgd1_470, which encodes mucin 8 (supplementary fig. S11A, Supplementary Material online). In chromosome 2, the sequence in the first 300 kb in the US44513 genome was similar to the IId genome. Afterwards (300k–530k), it was more similar to the IIc genome (fig. 5D, supplementary figs. S10 and S11B, Supplementary Material online). In chromosome 6, a large region (180k–715k) showed high identity between US44513 and IId subtype genomes (fig. 5E, supplementary figs. S10 and S11C, Supplementary Material online). After this region and until the end of chromosome 6, the genome sequence of US44513 was significantly different from all other genomes analyzed, indicating an unknown source had contributed to its emergence (fig. 5E). Therefore, US44513 had a complex history, involving multiple recombination events among IIa, IIc, and IId subtype families.
Geospatial Diversification of Zoonotic C. parvum
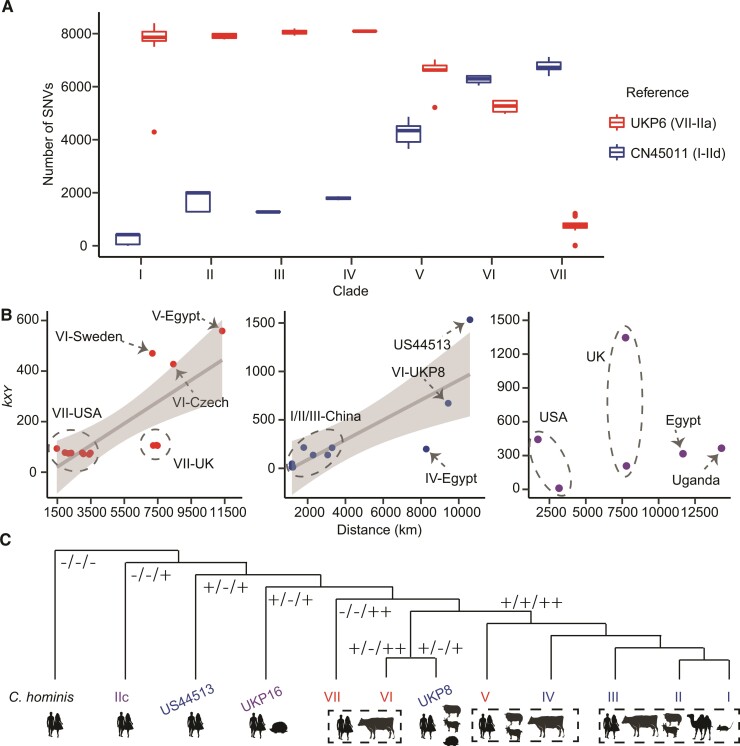
The evidence presented above suggests parasite populations in China (group I) and the United Kingdom (group VII) might represent the ancestral IId and IIa subtype families of zoonotic C. parvum, respectively. There were sequence introgressions between them as IId moved westward and IIa eastward in the Eurasian continent, creating recombinant extant subpopulations in areas where they coexisted (fig. 2A). Using one well-assembled IIa genome in group VII (UKP6) as the reference genome, the number of SNPs in IIa isolates increased from group VI to V (fig. 6A). The reverse was seen when a well-assembled IId genome in group I (CN45011) was used as the reference IId genome in the SNP analysis. Therefore, there was a positive correlation between geographic distances and nucleotide differences (kxy) in IIa (r = 0.76, P = 0.0014) an IId (r = 0.81, P = 0.0041) genomes (fig. 6B). This genetic isolation by geographic distance, however, was not observed in the anthroponotic IIc subtype family (r = 0.08).
Fig. 6.
Evolution of genomes of zoonotic C. parvum by geographic distance. (A) Average number of SNPs identified through mapping of sequencing reads to the representative IIa genome (UKP6 from UK) in group VII and IId genome (CN45011 from China) in group II. (B) Distribution of nucleotide differences (kxy) by geographical distance in pairwise comparison of C. parvum genomes among isolates within IIa (left), IId (middle), and IIc (right) subtype families examined in this study. Positive correlations were detected in IIa (r = 0.76, R2 = 0.55, P = 0.0014) and IId (r = 0.81, R2 = 0.62, P = 0.0041) but not in IIc (r = 0.08). (C) Topology of each population or isolates with different host ranges. The tree represents phylogenetic relationships shown in figure 2B. Symbols at branches indicate the presence (+) and absence (−) of three subtelomeric genes (cgd3_10 paralog encoding a SKSR secreted protein, and cgd3_4260 paralog and cgd6_5520-5510 encoding insulinase-like proteases). Symbols under the tree correspond to the major hosts of the C. parvum subpopulations as well as C. hominis.
The sequence introgressions between IIa and IId subtype families have resulted in the appearance of sympatric IIa and IId genomes in Egypt and European countries with significant sequence identity between each other but divergence from their ancestral populations. This likely has changed the host preference of the parasite populations, such as from the broad host range of IId in China to the small ruminant-adapted IId in European countries (fig. 6C). Direct genome comparisons indicated gains and losses of two subtelomeric genes encoding an SKSR protein (cgd3_10 paralog) and an insulinase-like protease (with high sequence identity to the cgd3_4260 gene) among subpopulations. Both are present in IId isolates from China. The former was gained by IIa isolates in Egypt and Czech Republic as the consequence of sympatric recombination with IId, while the latter was lost in IId isolates in the United Kingdom and United States due to sympatric recombination with IIa (supplementary table S4, Supplementary Material online). As these two protein families have been identified as secreted pathogenesis determinants in Cryptosporidium spp. (Xu et al. 2020), they could be responsible for the modified host ranges of the local C. parvum subpopulations.
On rare occasions, genetic recombination between zoonotic IIa/IId and anthroponotic IIc subtypes has led to the appearance of special lineages such as US44513 in the United States or UKP16 in the United Kingdom, which have genetic features, and perhaps biological traits, very unlike other members of their gp60 subtype families. These two IId isolates maintain the subtelomeric gene encoding the SKSR protein (cgd3_10 paralog) described above, but have lost two subtelomeric genes encoding insulinase-like proteases: a cgd3_4260 paralog and one of the two copies of cgd6_5520-5510 (fig. 6C and supplementary table S4, Supplementary Material online). As a result, they might have a narrower host range than their ancestors in China.
Selective Sweep and Evolution of Zoonotic C. parvum
Genetic selection could have played a role in the adaptive evolution of C. parvum identified above. Results of the dN/dS analysis of 11 representative genomes (supplementary table S1, Supplementary Material online) identified 75 genes under positive selection, including mostly genes with low Pi values, suggesting that selective sweeps might have occurred during the evolution of C. parvum (supplementary fig. S12 and table S5, Supplementary Material online). The highly polymorphic genes with high Pi values, in contrast, were largely under purifying or balance selection (supplementary table S6, Supplementary Material online). In Tajima’s D analysis of the genomes, we found significantly different Tajima’s D values among subtype families (P < 1e−6 in the Kruskal–Wallis test) (supplementary fig. S13, Supplementary Material online). A strong skew toward low-frequency variants was observed in the IIc subtype family, indicating the occurrence of population expansion, possibly after recovery from a recent selective sweep. Values of Tajima’s D were normally distributed in the IIa subtype family (P = 0.58 in the Kolmogorov–Smirnov test). In comparison to IIa, the right-skewed distribution in the IId subtype family suggested that there were more regions under selective sweep in IId than in IIa.
In the analysis of selective sweeps within the IIa and IId subtype families (excluding the recombinants UKP8 and US44513), IId (groups I–IV) had more selective regions than IIa (groups V–VII), in agreement with the results of Tajima’s D analysis (fig. 7A and supplementary fig. S14, Supplementary Material online). Between the two subtype families, IId has almost twice more regions with strong selective sweep signals (876 kb covering 67 genes) than IIa (482 kb covering 31 genes) (supplementary tables S7 and S8, Supplementary Material online). As expected, sequence variations were smaller in the group under selective sweep than in the group without selective sweep, and such regions in the IId family were highly conservative compared with the IIa subtype family (supplementary tables S7 and S8, Supplementary Material online).
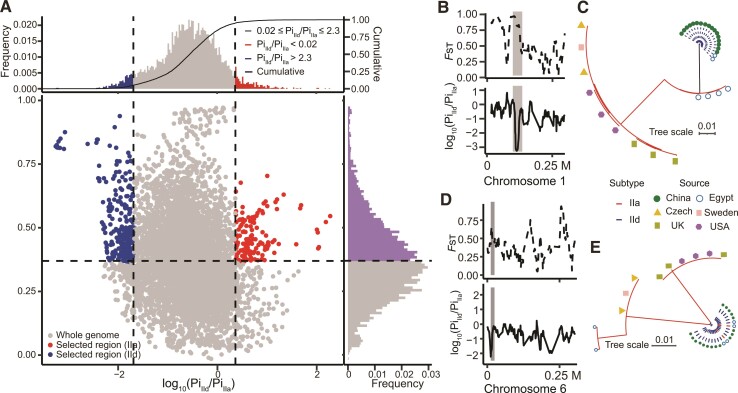
Fig. 7.
Selective sweep in zoonotic C. parvum. (A) Distribution of Pi ratios (IId/IIa) and Fst values calculated in a 10 kb sliding window with 1 kb steps, with the Pi ratio values being logarithmically transformed. Points located to the left (IId genomes) and right tails (IIa genomes) of the empirical Pi ratio distribution (left and right dash lines, respectively) and above the mean Fst value (horizontal dashed line) are considered selected regions. Two examples of regions with strong selective sweep signals in chromosomes 1 (B and C) and 6 (D and E) of the IId genomes are shown. The ML trees were built with SNPs located in selected regions (shaded area) using representative isolates in each group, with the source of the isolates being indicated.
Among the regions under selective sweep in the IId subtype family, two regions included genes encoding mucin-like proteins. In chromosome 1, a 24 kb block (108–132 kb) was identified, including the mucin gene cgd1_470 (mean Fst = 0.84, mean PiIId/PiIIa = 0.0029) (fig. 7B). Among the 386 overall SNPs in the region, 197 were identified in cgd1_470. Between the two subtype families, only 4 SNPs were present in the IId genomes compared with 222 SNPs in the IIa genomes. Phylogenetic analysis of these SNPs revealed clear differentiation between IIa and IId (fig. 7C). With the removal of IIa genomes from Egypt, the number of SNPs in the remaining IIa isolates was reduced to 14, indicating the genomic region went through recombination between IIa from Europe and IId from Asia. The second region with selective sweep (mean Fst = 0.57, mean PiIId/PiIIa = 0.013) was in a block (∼nucleotides 12,000–25,000) in chromosome 6 around the cgd6_40 gene, which also encodes a mucin glycoprotein (fig. 7D). Among the 112 overall SNPs identified among zoonotic C. parvum genomes, only 1 SNP were detected in the IId genomes compared with 111 SNPs in the IIa genomes. Phylogenetic analysis of the SNPs placed two IIa isolates from Egypt into the IId clade, and the other two IIa isolates from Egypt into the clade formed by IIa isolates from the Czech Republic and Sweden, indicating the occurrence of genetic recombination between IIa from Europe and local IId (fig. 7E). Selective sweep in these two regions covering mucin proteins could play an important role in parasites adaptation to the local environment.
Discussion
The comparative genomics and population genetic characterizations of divergent C. parvum subtypes in the study have been facilitated by the generation of high-quality whole-genome sequence data from 87 isolates. Analyses of these and other available data indicate that genetic recombination between sympatric populations shapes the evolution of this protozoan pathogen. In addition to the well-established host-adapted IIa, IIc, and IId subtype families, comparative genomics and population genetic analyses of 101 genomes (including 14 from public databases) have identified several other subpopulations of zoonotic C. parvum in diverse areas. Some of the subpopulations have mosaic genomes, leading to discordance in phylogenetic relationships between gp60 and whole-genome SNPs. This is largely due to sympatric recombination among the three major ancestral populations in areas where they coexist. As expected, sequence introgression is more common between the zoonotic IIa and IId genomes. Occasional recombination, however, occurs between the two zoonotic subtype families and the anthroponotic IIc subtypes, resulting in the formation of recombinants with modified host preference. Selective sweeps might have played a further role in the adaptive evolution of the generated subpopulations.
The data generated suggest that IId in China (groups I–III) could represent the ancestral population of the IId subtype family while the prevalent IIa in western Europe (group VII) could be the origin of IIa subtype family. They could have evolved from a common ancestry and formed the two largest clades in zoonotic C. parvum. In phylogenetic and PCA analyses, they formed two monophyletic clusters that were placed in the opposite ends of the trees and graphs, with the recombinant subpopulations dispersed between them. The IIa subtypes in group VII and IId subtypes in groups I–III have largely shown genome homogeneity in STRUCTURE and IBD analyses and the absence of sequence introgressions from other subtype families. In agreement with their ancestral nature, IIa subtypes are widely present in western Europe and IId subtypes in China, with low occurrence or absence of the other zoonotic subtype family in each area (Guo et al. 2022b). In contrast, other subpopulations of zoonotic C. parvum have mosaic genomes due to sequence introgression, and are detected in areas where both IIa and IId subtypes are highly prevalent (such as Sweden, the Czech Republic, and Egypt).
Sympatric recombination between ancestral IIa and IId subtype families of C. parvum has apparently led to the formation of these new subpopulations. This is especially obvious in Egypt, where both IIa and IId are common in farm animals (Hijjawi et al. 2022). Data generated from the present study indicate that while the genomes of the IId subtype family in Egypt are similar to IId genomes in China (with only ∼1,500 SNPs), they also shared a high genetic identity to genomes of the IIa isolates in Egypt (with ∼3,500 SNPs). Sequence introgression between IIa (group V) and IId (group IV) subtype families in Egypt is especially obvious in a 300 kb block downstream from the gp60 gene in chromosome 6. Concurrence of divergent subpopulations of C. parvum in the same geographic area provides the opportunity for sequence introgression via recombination during meiosis, as indicated recently in vitro (Wilke et al. 2019). The high prevalence of C. parvum in farm animals and waterborne and foodborne transmission of the pathogens facilitate the occurrence of coinfection with mixed C. parvum populations and subsequent genetic recombination (Divis et al. 2018).
Genetic recombination between IIa and IId subtype families might have led to the formation of other divergent IId subpopulations in Europe. In the present study, the UKP8 (of the IIdA22G1 subtype at the gp60 locus) genome unexpectedly clustered together with three IIa genomes from the Czech Republic and Sweden rather than other IId genomes. Sequence comparison revealed a high sequence identity among these four genomes within group VI except for the gp60 region. The IIa isolates in the group have substantial differences (with ∼3,700 SNPs) from the ancestral IIa subtypes in group VII, indicating that like in Egypt, the genetic exchange could occur between the IIa isolates in Sweden and the Czech Republic and sympatric IId isolates in these countries. Within Europe, Sweden is known to have a high occurrence of both IIa and IId subtypes in humans and cattle (Lebbad et al. 2021). Interestingly, several recently published C. parvum genomes from Italy and Slovenia, including IT-C395 (IIaA18R1), IT-C366 (IIdA17G2R1), and Slo4 (IIaA20G1R1), clustered with UKP8, supporting the conclusion from the present study (Corsi et al. 2022). In these European countries, IId subtypes are mainly seen in small ruminants (Guo et al. 2021; Lebbad et al. 2021). The latter, however, can also be infected with IIa subtypes, allowing genetic admixture between the two zoonotic subtype families.
Sequence introgression can apparently occur between zoonotic and anthroponotic subtype families of C. parvum despite the recent designation of the latter as a separate subspecies (Nader et al. 2019). In the present study, comparative genomics analyses indicate a very divergent IId isolate from the United States, US44513 (IIdA15G1), is phylogenetically more related to the IIc genomes than to IId genomes. The ancestral IId sequences are mostly seen in two large blocks in chromosomes 2 and 6. In addition, some IIa sequences are present in chromosome 1, especially regions around the gene cgd1_470 encoding a mucin-like protein. As shown previously and again in the present study, the genome of the IIc isolate UKP16 (IIcA5G3j subtype) has sequence introgression from zoonotic C. parvum (Nader et al. 2019). These sequence introgression and substitutions could have gone through adaptive selection, as selective sweeps were identified in several genomics areas, especially those containing genes encoding mucin glycoproteins. Such a selection frequently leads to reduced genetic diversity in the vicinity of the selected locus, which is the hallmark of selective sweep (Weetman et al. 2015).
Genetic recombination has been identified to play a major role in the evolution and transmission of other apicomplexans such as Plasmodium and Toxoplasma. In Plasmodium falciparum, genetic recombination is common due to the common concurrence of multiple lineages, especially in areas with high transmission intensity therefore high complexity of infection (Nkhoma et al. 2020). Recombination between lineages enables the introgression of haplotypes associated with antimalarial drug resistance, leading to the rapid dispersal of resistant progenies under selection pressure from drug usage (Amato et al. 2018). This is likely to be the case with other phenotypic traits, as experimental genetic crosses of P. falciparum isolates have led to the generations of progenies with modified growth, virulence, and mosquito infectivity in addition to drug resistance (Ranford-Cartwright and Mwangi 2012). In other Plasmodium species, sympatric recombination has been identified in two host-adapted lineages of zoonotic P. knowlesi in Malaysia (Divis et al. 2018). Similarly, the genetic cross of type II and type III strains of Toxoplasma gondii can lead to the generation of progenies that are more virulent than both parental strains (Grigg et al. 2001). This appears to be the case in a study of field isolates from southern sea otters with fatal toxoplasmosis, which showed the sexual recombination can generate virulent stains (Kennard et al. 2021).
The sequence introgressions and host-adapted subtype families could have potentially modified the host ranges of C. parvum. Between the two major zoonotic subtype families of C. parvum, IIa is mainly found in cattle and IId in sheep and goats (Feng et al. 2018). Geographic variations, however, exist in the host preferences of these two subtype families. For example, within the IId subtype family, subtypes in China have a broad host range, including ruminants, camels, equine, primates, and rodents. Similarly, IId subtypes in northern African and Mideast countries (including Egypt), some East European countries and Sweden are found in cattle in addition to sheep and goats. In contrast, in western and southern Europe, IId subtypes are mainly found in lambs and goat kids (Feng et al. 2018). This agrees with the identification of several divergent IIa and IId subpopulations in the present study. More molecular epidemiological studies with a sampling of a broad range of animals in diverse areas and advanced characterization of isolates are needed to confirm this suggestion.
Gains and losses of subtelomeric genes encoding some protein families could potentially contribute to differences in host preference among the C. parvum subpopulations. These copy number variations mainly involve genes encoding two secretary protein families (insulinase-like proteases and SKSR proteins). As subpopulations of zoonotic C. parvum with more copies of the two gene families have broader host ranges, it was suggested that these secretory proteins might be involved in host adaptation in Cryptosporidium spp. (Guo, Tang, et al. 2015; Xu et al. 2019). The gain of a copy of the SKSR gene (a paralog of cgd3_10 in chromosome 3) was observed in the IIc isolate UKP16 (IIcA5G3j subtype), which has been found in hedgehogs in Europe. This indicates that the gene gain could have increased the infectivity of the IIc isolate to animals. Confirmation of this theory can be achieved using the genetic editing tools newly developed for Cryptosporidium spp. (Sangster et al. 2016).
Animal domestication and modern animal farming could have promoted the dispersal of zoonotic C. parvum (Guo et al. 2022a) (supplementary fig. S15, Supplementary Material online). Although the subpopulation of C. parvum in China might represent the ancestral IId subtypes due to the absence of introgression of IIa sequences, they could have initially originated from the Middle East and Mediterranean basin (Fertile Crescent) where the domestication of taurine cattle occurred in the Neolithic age (Zhang, Lenstra, et al. 2020). This is supported by the distribution of IId subtypes in cattle, which is mostly in North African and Middle Eastern countries including Egypt. These subtypes have apparently dispersed eastwards all the way to China and westwards through Turkey and Eastern European countries to Sweden. The lower IId subtype diversity in China compared with North African and Middle Eastern countries, probably a result of recent selective sweeps, supports this theory on the origin of IId subtypes (Guo et al. 2022b; Hijjawi et al. 2022). The occurrence of IId subtypes in cattle in these areas coincides with less intensive cattle farming. In contrast, IIa subtypes are widely distributed in industrialized nations around the world where dairy farming is most intensive. As the most successful Holstein–Friesian dairy cattle were introduced to North America, Australia, New Zealand, and other areas from northwestern Europe in the late 20th century, the animal import could have played a role in the dispersal of IIa subtypes around the world (Guo et al. 2022a). This is supported by the genomic similarity of the dominant IIa subpopulation (group VII) between the United Kingdom and United States in phylogenetic, PCA, and IBD analyses in the present study. Recent phylogeographic analysis of Escherichia coli O157:H7 has suggested that the introduction of Holstein–Friesian cattle might be the driving force for the global spread of the novel pathogen from the Netherlands (Franz et al. 2019). Therefore, the dissemination of IIa subtypes in industrialized nations might have changed not only the population structure of C. parvum through sympatric recombination with concurrent IId subtypes but also the importance of zoonotic transmission in cryptosporidiosis epidemiology. Additional evolutionary genomics studies with more systematic sampling are needed to validate the origins of zoonotic C. parvum subtype families in farm animals and the role of animal trade in their dispersal around the world. Efforts should be made to prevent the introduction of IIa subtypes into areas where animal farming is less intensive, such as China and other Asian and African countries.
In conclusion, data from the study shed light on the effects of genetic recombination and the subsequent selective adaptation on the emergence of divergent C. parvum subpopulations with modified host range. Prior to this, the limited availability of genomes had prevented the characterization of the population structure and evolutionary history of C. parvum across diverse areas and the impact of agricultural activities. More data should be collected around the world to track the global dispersal of zoonotic C. parvum. In addition, advanced studies on genetic determinants of host range in C. parvum and related species using forward and reverse genetic tools should be conducted to improve our understanding of their cross-species and zoonotic transmission.
Materials and Methods
Whole Genome Sequencing of C. parvum
Cryptosporidium parvum-positive samples from humans and animals were used in the study. They were diagnosed as C. parvum by PCR and sequence analysis of the small subunit ribosomal RNA gene and further subtyped by sequence analysis of the gp60 gene (Xiao et al. 2009). Oocysts were isolated from positive fecal materials using immunomagnetic beads (Dynabeads anti-Cryptosporidium kit, Invitrogen, Oslo, Norway). DNA was extracted from the purified oocysts using QIAamp DNA minikit (Qiagen Sciences, Hilden, Germany) and amplified using a REPLI-g Midi kit (Qiagen Sciences) as described (Guo, Li, et al. 2015). Altogether, 91 C. parvum genomes were sequenced using the 100 or 250 bp paired-end technology and the standard Illumina library preparation procedures on an Illumina HiSeq 2500 (Illumina, San Diego, CA, USA), with a minimum 100-fold coverage of the expected genome.
WGS Data from Public Databases
Public WGS data of Cryptosporidium spp. used in this study were downloaded from the National Center for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov) and the European Bioinformatics Institute (EBI) website (www.ebi.ac.uk/). Twenty-one sets of WGS data, including 5 C. hominis samples and 16 C. parvum samples, were selected. The downloaded Sequence Read Archive (SRA) data were converted into the fastq format using fastq-dump of SRA Toolkit (https://github.com/ncbi/sra-tools).
Variant Analysis
The sequence reads were trimmed for adapters and Phred scores below 20 using bbduk from bbtools v37.66 (sourceforge.net/projects/bbmap/), with reads shorter than 65 bp being discarded. The cleaned reads were mapped to the reference IOWA II genome using bwa v0.7.17 (https://github.com/lh3/bwa) with the bwa-mem algorithm and sorted with SAMtools v1.11 (Danecek et al. 2021). Duplicated reads were removed using Sambamba v0.6.7 (Tarasov et al. 2015) and the mapping depth was calculated using bedtools v2.28 (bedtools.readthedocs.io/en/latest/). Only isolates with mapping depth over 12× and genome coverage over 96% were selected for further analyses. Eventually, 106 WGS genomes were included in the final comparative genomics analyses, including 5 C. hominis genomes as the outgroup (supplementary table S1, Supplementary Material online).
Sequence variations were identified using mpileup of BCFtools v1.11 (Danecek et al. 2021) from the sorted bam files with INDELs being realigned with GATK v3.8 (DePristo et al. 2011) for each bam file. Low-quality variants were filtered with a similar stringency threshold with BCFtools filter (including %QUAL≥30, AVG(FMT/DP) > 25 and MQ≥30) and only biallelic SNPs were retained (Nader et al. 2019). In addition, SNPs within 3 bp of an INDEL were filtered. Finally, variants with <5% missing sites were used for subsequent analyses. Variants were annotated with SnpEff v4.3 (Cingolani et al. 2012). Reads of each C. parvum genome were further mapped to a well-assembled IIa genome from group VII (US42561) and a IId genome from group I (CN45011). The SNPs among isolates were identified in a similar way.
Phylogenetic and Population Structure Analyses
The gp60 sequences obtained were aligned with MUSCLE program implemented in MEGA v7 (Kumar et al. 2016). Haplotypes in the alignments were identified using DnaSP v6 (Rozas et al. 2017). Integer NJ haplotype network was built with PopArt (http://popart.otago.ac.nz/index.html). An unrooted maximum likelihood (ML) tree was built with concatenated whole-genome SNPs from 101 C. parvum isolates using IQ-TREE v2.1.2 (Minh et al. 2020) with 1,000 bootstrap. Substitution models were auto-selected with ModelFinder Plus (MFP) and were optimized with option -bnni. A rooted ML tree was built with C. hominis as the outgroup. An NJ tree of SNPs from the IId subtype in China was also built with MEGA. In addition, SNPs located in specific regions were extracted with VCFtools v0.1.17 (Danecek et al. 2011) and concatenated to build ML trees. The trees generated were visualized with iTol (https://itol.embl.de/). We also built 10 kb sliding window trees along the genomes of selected groups, with topology weighting being calculated for each genealogy with Twisst (Martin and Van Belleghem 2017). A phylogenetic network tree was built from the SNP matrix using SplitsTree v4 (Huson and Bryant 2006) after singletons were removed.
PCA was done with SNPs under LD threshold 0.4 in 10 kb windows using SNPrelate (Zheng et al. 2012). The same SNP set was also used in the analysis of population structure with fastSTRUCTURE (Raj et al. 2014). Population sizes (K) of 3–9 were tested, and the appropriate number of model complexity was chosen by built-in script, leading to the identification of the best model at K = 6 and the maximized marginal likelihood at K = 7. IBD analysis was performed to assess the relatedness of isolates using hmmIBD v2.0.4 (Schaffner et al. 2018). Samples with IBD sharing >90% of their genomes were considered highly related. Networks of the outcome were generated based on the relatedness of isolates and visualized in Cytoscape v3.5.1 (https://cytoscape.org/).
Population Genetic Analyses
LD among different subtype groups was calculated using PopLDdecay v3.3 (Zhang et al. 2019) with rarefaction. The calculation was performed on a random sampling of the equal number of isolates 50 times. The average r2 values were used to evaluate the overall LD decay. Nucleotide divergence (Pi), INDEL density, and Fst were calculated using VCFtools. A sliding window analysis of genetic divergence (dxy) was conducted using script popgenWindows.py (https://github.com/simonhmartin/genomics_general). The average genetic divergence (kxy) was calculated using DnaSP. SNPs between local IIa and IId subtype isolates were identified by read mapping to the reference IOWA II genome and comparison of the mapping outcome using in-house scripts.
Genomic regions with a selective sweep in zoonotic C. parvum were identified using the combined analyses of the distribution of Pi ratio (IId vs IIa) and Fst values (Li et al. 2013). Regions with significantly low and high Pi ratios (the 5% left and right tails) of the empirical distribution and higher than the average Fst value were considered signals of selective sweeps.
Genome Assembly and Comparative Genomic Analyses
The cleaned reads of C. parvum were de novo assembled using SPAdes v3.11.1 (Bankevich et al. 2012) with the careful mode to reduce potential mismatches and short indels with k-mers of 21, 33, and 55. Contigs of the draft genomes were mapped to the reference genome using MUMmer v3.23 (http://mummer.sourceforge.net/). The minimum alignment identity was set to 80% and alignment length shorter than 500 bp was discarded. To improve the quality of assemblies, Pilon v1.23 (Walker et al. 2014) was used to correct mistakes, including base errors, small indels, gaps, and local misassembles. The draft and final genomes were evaluated using QUAST v4.6 (Gurevich et al. 2013). The subtype identity of the genomes was verified by Blast search of the gp60 sequence. Genomes were annotated based on sequence homology using exonerate v2.2 (Slater and Birney 2005) with the model protein2genome selected and the initial incomplete hits being further refined. The gene-based dN/dS (ω) among 11 well-assembled representative genomes was calculated using HyPhy v2.2.4 and MG94xREV model (https://www.hyphy.org/).
ANI was calculated using pyani v0.2.10 (http://widdowquinn.github.io/pyani/). To mitigate the sample size imbalance, we selected representative assemblies from each subpopulation. A 1 kb sliding window was employed to identify highly polymorphic genes with sequence differences over 2% (20 SNPs/1,000 bp). To evaluate gene gains and losses in the subtelomeric regions, eight representative assemblies were aligned with the reference genome using Mauve v2.4.0 (Darling et al. 2004). The alignments were checked manually for gene insertions and deletions, with the unmapped contigs being analyzed using blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify additional gene insertions.
Introgression Event Detection
The fd statistic was used to identify potential introgression genomic segments of IId in Egypt from local IIa isolates (Martin et al. 2015). A four-taxon tree topology ([{China-IId, Egypt-IId}, Egypt-IIa], OG), with C. hominis as the outgroup (OG), was used to identify introgression events. Considering the ancestral allele of one group as A while the derived alleles as B, an excess of ABBA pattern relative to BABA indicated gene flow between Egypt-IId and Egypt-IIa. The fd statistic was calculated in a 20 kb sliding window with 5 kb step using script ABBABABAwindow.py (https://github.com/simonhmartin/genomics_general). Negative values of D statistic and missing fd values were converted to 0. The standard error for the ABBA-BABA estimates was calculated using the weighted block jack-knife method.
Statistical Analysis
The geographic distance among regions was calculated from data in Wikipedia (https://www.wikipedia.org/). The correlation between kxy and distance within each subtype family was calculated and assessed using the F-test. The Kruskal–Wallis test was used to compare differences in nucleotide divergence and Tajima’s D distribution among groups, while the Kolmogorov–Smirnov test was used for the normality test. All statistical analyses were conducted in R v4.0.2 (https://www.r-project.org/).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Johanna L. Nader and Thomas C. Mathers for consultations on SNP filtration. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. This work was supported in part by the Guangdong Major Project of Basic and Applied Basic Research (2020B0301030007), National Natural Science Foundation of China (31820103014, 32150710530, U1901208), 111 Project (D20008), and Innovation Team Project of Guangdong Universities (2019KCXTD001). The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Contributor Information
Tianpeng Wang, State Key Laboratory of Agrobiotechnology, College of Biological Sciences, China Agricultural University, Beijing, China.
Yaqiong Guo, Guangdong Laboratory for Lingnan Modern Agriculture, Center for Emerging and Zoonotic Diseases, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China.
Dawn M Roellig, Division of Foodborne, Waterborne, and Environmental Diseases, Centers for Disease Control and Prevention, Atlanta, GA, USA.
Na Li, Guangdong Laboratory for Lingnan Modern Agriculture, Center for Emerging and Zoonotic Diseases, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China.
Mónica Santín, Environmental Microbial and Food Safety Laboratory, Beltsville Agricultural Research Center, Agricultural Research Service, US Department of Agriculture, Beltsville, MD, USA.
Jason Lombard, Center for Epidemiology and Animal Health, Veterinary Services, Animal and Plant Health Inspection Service, US Department of Agriculture, Fort Collins, CO, USA.
Martin Kváč, Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, Ceske Budejovice, Czech Republic.
Doaa Naguib, Department of Hygiene and Zoonoses, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt.
Ziding Zhang, State Key Laboratory of Agrobiotechnology, College of Biological Sciences, China Agricultural University, Beijing, China.
Yaoyu Feng, Guangdong Laboratory for Lingnan Modern Agriculture, Center for Emerging and Zoonotic Diseases, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China.
Lihua Xiao, Guangdong Laboratory for Lingnan Modern Agriculture, Center for Emerging and Zoonotic Diseases, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China.
Author Contributions
L.X., Y.F., and Z.Z. conceived this project and secured funds; Y.G., D.M.R., M.S., J.L., M.K., D.N., N.L., Y.F., and L.X. collected the samples; Y.G. and N.L. isolated oocysts and performed the whole-genome sequencing; D.M.R. provided technical assistance; T.W., Z.Z., and L.X. performed bioinformatics analyses; T.W. and L.X. prepared the draft manuscript; all authors participated in the revision of the manuscript and approved the final submission.
Data Availability
Raw reads of newly sequenced genomes in this study are available at NCBI under the BioProject PRJNA759721. The accession numbers are listed in supplementary table S1, Supplementary Material online. Additional data related to this paper may be requested from the authors.
References
- Amato R, Pearson RD, Almagro-Garcia J, Amaratunga C, Lim P, Suon S, Sreng S, Drury E, Stalker J, Miotto O, et al. . 2018. Origins of the current outbreak of multidrug-resistant malaria in southeast Asia: a retrospective genetic study. Lancet Infect Dis. 18:337–345. doi: 10.1016/S1473-3099(18)30068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. . 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, et al. . 2015. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis. 15:85–94. doi: 10.1016/S1473-3099(14)70772-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi GI, Tichkule S, Sannella AR, Vatta P, Asnicar F, Segata N, Jex AR, van Oosterhout C, Caccio SM.. 2022. Recent genetic exchanges and admixture shape the genome and population structure of the zoonotic pathogen Cryptosporidium parvum. Mol Ecol. 34:997–1011. doi: 10.1111/mec.16556 [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. . 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. . 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:1–4. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. doi: 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. . 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–498. doi: 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divis PCS, Duffy CW, Kadir KA, Singh B, Conway DJ. 2018. Genome-wide mosaicism in divergence between zoonotic malaria parasite subpopulations with separate sympatric transmission cycles. Mol Ecol. 27:860–870. doi: 10.1111/mec.14477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Li N, Roellig DM, Kelley A, Liu G, Amer S, Tang K, Zhang L, Xiao L. 2017. Comparative genomic analysis of the IId subtype family of Cryptosporidium parvum. Int J Parasitol. 47:281–290. doi: 10.1016/j.ijpara.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Ryan UM, Xiao L. 2018. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 34:997–1011. doi: 10.1016/j.pt.2018.07.009 [DOI] [PubMed] [Google Scholar]
- Franz E, Rotariu O, Lopes BS, MacRae M, Bono JL, Laing C, Gannon V, Soderlund R, van Hoek A, Friesema I, et al. . 2019. Phylogeographic analysis reveals multiple international transmission events have driven the global emergence of Escherichia coli O157:H7. Clin Infect Dis. 69:428–437. doi: 10.1093/cid/ciy919 [DOI] [PubMed] [Google Scholar]
- Gharpure R, Perez A, Miller AD, Wikswo ME, Silver R, Hlavsa MC. 2019. Cryptosporidiosis outbreaks—United States, 2009–2017. Morb Mortal Weekly Rep. 19:2650–2654. doi: 10.15585/mmwr.mm6825a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC. 2001. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294:161–165. doi: 10.1126/science.1061888 [DOI] [PubMed] [Google Scholar]
- Guo Y, Li N, Lysen C, Frace M, Tang K, Sammons S, Roellig DM, Feng Y, Xiao L. 2015. Isolation and enrichment of Cryptosporidium DNA and verification of DNA purity for whole-genome sequencing. J Clin Microbiol. 53:641–647. doi: 10.1128/JCM.02962-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li N, Ryan U, Feng Y, Xiao L. 2021. Small ruminants and zoonotic cryptosporidiosis. Parasitol Res. 120:4189–4198. doi: 10.1007/s00436-021-07116-9 [DOI] [PubMed] [Google Scholar]
- Guo Y, Ryan U, Feng Y, Xiao L. 2022a. Association of common zoonotic pathogens with concentrated animal feeding operations. Front Microbiol. 12:810142. doi: 10.3389/fmicb.2021.810142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Ryan U, Feng Y, Xiao L. 2022b. Emergence of zoonotic Cryptosporidium parvum in China. Trends Parasitol. 38:335–343. doi: 10.1016/j.pt.2021.12.002 [DOI] [PubMed] [Google Scholar]
- Guo Y, Tang K, Rowe LA, Li N, Roellig DM, Knipe K, Frace M, Yang C, Feng Y, Xiao L. 2015. Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. BMC Genomics 16:320. doi: 10.1186/s12864-015-1517-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijjawi N, Zahedi A, Al-Falah M, Ryan U. 2022. A review of the molecular epidemiology of Cryptosporidium spp. and Giardia duodenalis in the Middle East and North Africa (MENA) region. Infect Genet Evol. 98:105212. doi: 10.1016/j.meegid.2022.105212 [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23:254–267. doi: 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Kennard A, Miller M, Khan A, Quinones M, Miller N, Sundar N, James E, Roos D, Conrad P, Grigg M. 2021. Virulence shift in a sexual clade of Type X Toxoplasma infecting Southern Sea Otters. bioRxiv:2021.03.31.437793; doi: 10.1101/2021.03.31.437793 [DOI] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, et al. . 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbad M, Winiecka-Krusnell J, Stensvold CR, Beser J. 2021. High diversity of Cryptosporidium species and subtypes identified in cryptosporidiosis acquired in Sweden and abroad. Pathogens 10:523. doi: 10.3390/pathogens10050523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y, Wang T, Yeung CK, Chen L, Ma J, et al. . 2013. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 45:1431–1438. doi: 10.1038/ng.2811 [DOI] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD. 2015. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 32:244–257. doi: 10.1093/molbev/msu269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Van Belleghem SM. 2017. Exploring evolutionary relationships across the genome using topology weighting. Genetics 206:429–438. doi: 10.1534/genetics.116.194720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R.. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. doi: 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader JL, Mathers TC, Ward BJ, Pachebat JA, Swain MT, Robinson G, Chalmers RM, Hunter PR, van Oosterhout C, Tyler KM. 2019. Evolutionary genomics of anthroponosis in Cryptosporidium. Nat Microbiol. 4:826–836. doi: 10.1038/s41564-019-0377-x [DOI] [PubMed] [Google Scholar]
- Nkhoma SC, Trevino SG, Gorena KM, Nair S, Khoswe S, Jett C, Garcia R, Daniel B, Dia A, Terlouw DJ, et al. . 2020. Co-transmission of related malaria parasite lineages shapes within-host parasite diversity. Cell Host Microbe 27:93–103.e4. doi: 10.1016/j.chom.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Stephens M, Pritchard JK. 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197:573–589. doi: 10.1534/genetics.114.164350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranford-Cartwright LC, Mwangi JM. 2012. Analysis of malaria parasite phenotypes using experimental genetic crosses of Plasmodium falciparum. Int J Parasitol. 42:529–534. doi: 10.1016/j.ijpara.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34:3299–3302. doi: 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- Sangster L, Blake DP, Robinson G, Hopkins TC, Sa RC, Cunningham AA, Chalmers RM, Lawson B. 2016. Detection and molecular characterisation of Cryptosporidium parvum in British European hedgehogs (Erinaceus europaeus). Vet Parasitol. 217:39–44. doi: 10.1016/j.vetpar.2015.12.006 [DOI] [PubMed] [Google Scholar]
- Schaffner SF, Taylor AR, Wong W, Wirth DF, Neafsey DE. 2018. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar J. 17:196. doi: 10.1186/s12936-018-2349-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31. doi: 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. 2015. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31:2032–2034. doi: 10.1093/bioinformatics/btv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichkule S, Caccio SM, Robinson G, Chalmers RM, Mueller I, Emery-Corbin SJ, Eibach D, Tyler KM, van Oosterhout C, Jex AR. 2022. Global population genomics of two subspecies of Cryptosporidium hominis during 500 years of evolution. Mol Biol Evol. 39:msac056. doi: 10.1093/molbev/msac056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. . 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, Mitchell SN, Wilding CS, Birks DP, Yawson AE, Essandoh J, Mawejje HD, Djogbenou LS, Steen K, Rippon EJ, et al. . 2015. Contemporary evolution of resistance at the major insecticide target site gene Ace-1 by mutation and copy number variation in the malaria mosquito Anopheles gambiae. Mol Ecol. 24:2656–2672. doi: 10.1111/mec.13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke G, Funkhouser-Jones LJ, Wang Y, Ravindran S, Wang Q, Beatty WL, Baldridge MT, VanDussen KL, Shen B, Kuhlenschmidt MS, et al. . 2019. A stem-cell-derived platform enables complete Cryptosporidium development in vitro and genetic tractability. Cell Host Microbe 26:123–134.e8. doi: 10.1016/j.chom.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Hlavsa MC, Yoder J, Ewers C, Dearen T, Yang W, Nett R, Harris S, Brend SM, Harris M, et al. . 2009. Subtype analysis of Cryptosporidium specimens from sporadic cases in Colorado, Idaho, New Mexico, and Iowa in 2007: widespread occurrence of one Cryptosporidium hominis subtype and case history of an infection with the Cryptosporidium horse genotype. J Clin Microbiol. 47:3017–3020. doi: 10.1128/JCM.00226-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Guo Y, Roellig DM, Feng Y, Xiao L. 2019. Comparative analysis reveals conservation in genome organization among intestinal Cryptosporidium species and sequence divergence in potential secreted pathogenesis determinants among major human-infecting species. BMC Genomics 20:406. doi: 10.1186/s12864-019-5788-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Li N, Guo Y, Feng Y, Xiao L. 2020. Comparative genomic analysis of three intestinal species reveals reductions in secreted pathogenesis determinants in bovine-specific and non-pathogenic Cryptosporidium species. Microb Genomics 6:e000379. doi: 10.1099/mgen.0.000379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Guo Y, Xiao L, Feng Y. 2021. Molecular epidemiology of human cryptosporidiosis in low- and middle-income countries. Clin Microbiol Rev. 34:e00087-19. doi: 10.1128/CMR.00087-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Dong S-S, Xu J-Y, He W-M, Yang T-L. 2019. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35:1786–1788. doi: 10.1093/bioinformatics/bty875 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hu S, Zhao W, Guo Y, Li N, Zheng Z, Zhang L, Kvac M, Xiao L, Feng Y. 2020. Population structure and geographical segregation of Cryptosporidium parvum IId subtypes in cattle in China. Parasites Vectors 13:425. doi: 10.1186/s13071-020-04303-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Lenstra JA, Zhang S, Liu W, Liu J. 2020. Evolution and domestication of the Bovini species. Anim Genet. 51:637–657. doi: 10.1111/age.12974 [DOI] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328. doi: 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads of newly sequenced genomes in this study are available at NCBI under the BioProject PRJNA759721. The accession numbers are listed in supplementary table S1, Supplementary Material online. Additional data related to this paper may be requested from the authors.